Abstract
Elucidating resistance mechanisms for therapeutic monoclonal antibodies (MAbs) is challenging, because they are difficult to study in non-human models. We therefore developed a strategy to genetically map in vitro drug sensitivity, identifying genes that alter responsiveness to rituximab, a therapeutic anti-CD20 MAb that provides significant benefit to patients with B-cell malignancies. We discovered novel loci with genome-wide mapping analyses and functionally validated one of these genes, CBLB, which causes rituximab resistance when knocked down in lymphoma cells. This study demonstrates the utility of genome-wide mapping to discover novel biological mechanisms of potential clinical advantage.
INTRODUCTION
Drug resistance, often categorized by intrinsic or acquired mechanisms, remains one of the largest challenges in the curative treatment of cancer. The clinical pattern of acquired resistance is driven by a combination of selective pressures from molecularly targeted drug therapy—for example, monoclonal antibodies (MAbs)1 and kinase inhibitors2—and tumor heterogeneity.3,4 In addition to acquired resistance, variations in clinical outcomes are undoubtedly confounded by heritable genetic factors intrinsic to the patient as well.
Many studies in chemotherapeutic resistance synergize large-scale, high-throughput omics and informatics to identify novel genes and signaling pathways and decipher molecular signatures for tumor responsiveness and patient variability during drug treatment.4 The machinery of resistance spans diverse molecular mechanisms from drug efflux and metabolism to cell survival and death pathways. Although the direct translation of in vitro results to clinical efficacy can be tenuous, key observations are gleaned from experiments involving cancerous and healthy cell lines to model drug susceptibility and resistance mechanisms.5
MAbs are a standard component of cancer therapy, specifically targeting malignant and normal B-cells via complement- or antibody-dependent effector pathways and apoptosis,6 yet resistance mechanisms frequently render them ineffective. There is some evidence associating FCGR7–9 and C1QA10 polymorphisms with MAb-mediated effectiveness in vitro and in vivo. These polymorphisms presumably affect the effector portions of antibody-dependent and complement-dependent cytotoxicity (CDC) pathways, respectively. However, mechanisms within the target cell itself that lead to MAb resistance remain elusive.
We approached the problem of MAb resistance in vitro using a high-throughput CDC assay using immortalized non-malignant as well as cancerous cell lines. First, we investigated the heritability of rituximab and ofatumumab response, to ensure the phenotype is indeed mappable. Next, we used public information on genotype and gene expression data to analyze associations between loci/genes and drug response. Our multitiered analysis strategy indicated CBLB as a gene of interest. Finally, we used RNA silencing in several immortalized non-malignant and cancerous cell lines to functionally validate the role of CBLB expression levels on rituximab response. We are the first to report this role of CBLB as a potential mediator for rituximab sensitivity, utilizing the power of an unbiased genome-wide strategy to find previously unsuspected mediators of rituximab response.
MATERIALS AND METHODS
Cell lines and cell culture
Lymphoma cell lines were obtained from the Lineberger Comprehensive Cancer Center Tissue Culture Facility or ATCC. Epstein–Barr virus-immortalized human lymphoblastoid cell lines (LCLs) were obtained from two independent sources: a collection of families (100 samples, 13 families) from Centre d’Etude du Polymorphisme Human (CEPH) obtained from the Coriell Institute (Camden, NJ, USA) and an unrelated collection (486 samples) obtained from unrelated Caucasian participants of the cholesterol and Pharmacogenetics clinical trial as described previously11 from the Children’s Hospital Oakland Research Initiative (CHORI). All cell lines were maintained in RPMI media containing 15% fetal bovine serum.
Genotyping quality control
Genotyping for the cholesterol and pharmacogenetics study has been described elsewhere.11,12 Each individual was genotyped for either 314 621 or 620 901 dinle-nucleotide polymorphisms (SNPs), using HumanHap300 (Illumina, Inc., San Diego, CA, USA) bead chip or HumanQuad610 (Illumina, Inc., San Diego, CA, USA) bead chip platforms, respectively. In addition, imputation was previously performed11 on the data for 2.5 million SNPs from HapMap Release 22, using the Caucasian CEPH reference population and the software program MACH.13
Quality control for the genotype data was described elsewhere.14 Briefly, PLINK was used to filter out SNPs whose genotyping rate was below 90%, whose minor allele frequency was below 0.05 or whose P-value from a Hardy–Weinberg test for equilibrium was below 10−5. The data after quality control contained 2 100 684 SNPs for subsequent analysis. Principal component analysis was used to correct for population stratification. First, SNPs in high linkage disequilibrium were removed using PLINK before the principle components analysis.15 Specifically, a window of 50 SNPs, a step size of 5 SNPs and a pairwise r2 threshold of 0.7 was used. From this step, ~ 81% of the remaining SNPs were removed, leaving a total of 395 033 SNPs available for principal component analysis. In addition, several outlier individuals were also removed. Principal component analysis was performed on this set of SNPs using EIGENSTRAT.16 The linkage disequilibrium pruning was used only for the principal component analysis and was not used as a filter for association analysis. The complete set of 2 100 684 SNPs that passed quality control was used for association analysis.
Genotype data for each cell line was downloaded from version 10 of the CEPH database using error checked markers.17 Genetic map information was downloaded from the Marshfield database.18 Error checking for Mendelian incompatibility, misspecified relationships and unlikely recombinations was performed, as previously described.19,20 A combined total of 8269 SNPs and microsatellite markers were used for linkage analysis. Marker selection for linkage analysis was based on genomic location and overall informativeness. Markers were selected using default settings in the MarkerSet tool.21
CDC assay and quality control
LCLs were seeded at 5 × 104 cells in either a 96-well plate (CEPH) or 8.9 × 104 cells in 384-well plate (CHORI) in a total volume of 50 μl or 100 μl media containing 25% pooled human serum with or without 10 μg ml−1 rituximab. Proliferation was then measured using Alamar Blue (ThermoScientific Biosource, Waltham, MA, USA) according to manufacturer’s instructions. Dye was added to samples and incubated overnight. Fluorescence was measured with excitation at 535 nm and emission at 595 nm on Infinite F200 microplate reader with Connect Stacker (Tecan Group Ltd, Morrisville, NC, USA) and I Control software (Morrisville, NC, USA, Version 1.6). Proliferation was expressed relative to samples without antibody.
Of the 534 plates containing MAb data, 18 were complete replicates in order to determine the reproducibility of the data. Correlations between replicates were strong for rituximab (R2 = 0.85), ofatumumab (R2 = 0.89) and complement (R2 = 0.54). This data was consistent with the reproducibility seen in previous similar experiments.14 Some LCLs were noted and rejected as outliers based on pairwise comparison across the three treatment options. A few LCLs were noted as having rituximab and ofatumumab viabilities more than 3 s.d. from the mean. These seven LCLs took place on the experimental same day and were flagged and removed from subsequent analysis.
Raw cell viabilities for each cell line were normalized according to negative and positive controls to reflect the proportion of cells remaining alive after treatment:
where i denotes the LCL sapmle, j ∈ {complement; rituximab; ofatumamab} indicates the treatment, RFUij is the relative fluorescence it from LCL i that was exposed to treatment j, Negij is the negative control for LCL i and treatment j, Posij is the positive control for LCL i and treatment j. For all treatments, the positive control is 10% dimethyl sulfoxide. For complement treatment without drug, the negative control is the RFU for water only. For treatment with 10 μg ml−1 rituximab or ofatumumab, the negative control is the RFU for complement treatment without drug.
After normalization, a number of cell lines were flagged and removed from subsequent analysis. Two cell lines whose raw complement viability was less than two standard deviations from the raw mean 10% dimethyl sulfoxide were eliminated. Three plates for ofatumumab and rituximab were also removed from analysis due to overinflated normalized raw viabilities.
Normalized complement viabilities correlated weakly with both normalized drug viabilities, which demonstrates the value of adding normalized complement viabilities as a covariate in subsequent analyses. Normalized viabilities for both drugs are correlated (R2 = 0.73) which suggested a multivariate analysis of covariance model could be an informative analysis.
Heritability calculation
Heritability was calculated by variance components analysis as implemented in MERLIN 1.1.2 (University of Michigan, Center for Statistical Genetics, Ann Arbor, MI, USA).22 First, the genetic relationship matrix is estimated between pairs of individuals. Second, restricted maximum likelihood analysis is performed to estimate the variance explained by SNPs used to estimate the genetic relationship matrix. Using the Genome-wide Complex Trait Analysis23 heritability was estimated for drug response in the group of unrelated individuals. Model covariates were used to account for experimental batch effects, growth rate estimation and complement.
Genome-wide association studies
Genome-wide association studies (GWAS) was performed using the MAGWAS software package.24 The software employs a multivariate analysis of covariance model design. Three different association analyses were performed: two association analyses representing each drug individually and one association analyses which jointly models both drug responses as a vector. Genotypic and phenotypic data from the 486 unrelated individuals were used in each model. Model covariates were included to account for the potentially confounding effects from combining the two genotyping technologies—the first two PCs and experimental batch effects: the growth rate estimation and adjusted complement. The adaptive permutation technique was applied with b = 10 000 000 000 and r = 121.25
Linkage analysis
Linkage analysis was also done with MERLIN 1.1.2. In order to try to address the mode of inheritance of rituximab sensitivity, segregation analysis was performed for a range of potential genetic models. Quantitative trait models were fitted allowing for no familial effects (sporadic), a single major gene (dominant, recessive, or codominant), two major genes, polygenes only or major genes plus polygenic effects (mixed models). The models allowing for a major gene assume that the phenotype is normally distributed within each genotype, with mean varying according to genotype but equal within-genotype variances. Analyses were carried out using PAPv5.26 Maximizations were determined from several starting values and those at boundary values were scrutinized further to confirm as far as possible that the true maxima were achieved and results were evaluated in terms of differences in twice the log likelihood for each fitted model compared to the baseline sporadic model. Nested models were compared using the likelihood ratio test and non-nested models using the Akaike information criterion.27
A permutation-based approach was used for defining LOD score cutoffs, indicating significant evidence for linkage. To estimate the probability of obtaining false-positive evidence of linkage for each drug and dose combination under the null hypothesis of no linkage to observed phenotypes, we conducted gene-dropping permutations using MERLIN. Marker data were simulated under the null hypothesis of no linkage or association to observed phenotypes while retaining the same pedigree structures, maps, marker allele frequencies and missing data patterns. We simulated 10 000 replicates for each phenotype of interest (discussed above) and conducted linkage analyses as described earlier for each replication. Based on these simulations, null distributions for each phenotype, for each chromosome were constructed and significance cutoffs were calculated for each phenotype corresponding to P-values ≤ 0.05.
Gene expression data
We obtained raw CEL files from Gene Expression Omnibus record GSE12626. Expression profiles from 57 of the original 100 LCLs typed on Affymetrix Human Genome U133A 2.0 arrays (Waltham, MA, USA) were downloaded and normalized using robust multi-array average software implemented by the Bioconductor package in R,22 version 2.12.1. Quantitative Significance Analysis of Microarrays revealed 13 genes whose expression was correlated with viability in rituximab, using a false discovery rate cutoff of <0.001%.
CBLB knockdown
Stable CBLB knockdown cell lines were prepared by transduction with lentivirus encoding CBLB-specific short hairpin RNA (shRNA sequences) (Open Biosystems TRC1 library) obtained through the Lenti-shRNA Core Facility (University of North Carolina). Corresponding control cell lines were prepared using a scrambled shRNA sequence. To obtain virus, low-passage HEK293T cells were transfected with pLKO.1 plasmids encoding five individual CBLB shRNAs, the packaging plasmid pMDG.2 and the envelope plasmid pCMV-VSV-G at a ratio of 1:0.75:0.25 with Fugene-6 transfection reagent (Promega, Madison, WI, USA). The medium was replaced with fresh media 18 h after transfection and viral particles were collected twice at 24 h intervals thereafter. Viral supernatants from the five transfections and two collection times were then pooled and filtered through a 0.45 μm cellulose acetate filter. Cells were then transduced with viral supernatant supplemented with polybrene (4 μg ml−1; Sigma-Aldrich, St Louis, MO, USA). After 48 h, positive selection for transduced cells was then conducted using 1 μg ml−1 puromycin for 10 days.
Reverse-transcriptase PCR for MS4A1 (CD20) and CBLB
Quantitative PCR was performed as previously described28 using 5 × 106 cells as starting material and additionally included primers for CBLB (CblbF: 5′-TTACGGCATGGCAGGAGTCGGA-3′ and CblbR: 5′-CGGGTCTCTGGAAGG CACGC-3′).
Western blotting for CD20 and CBLB
Western blotting was performed as previously described28 using a rabbit anti-CD20 antibody (Thermo Scientific, Waltham, MA, USA) and a mouse anti-Cbl-b (G-1) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Anti-β-actin (Sigma, St Louis, MO, USA) was used as a load control. Anti-horseradish peroxidase-conjugated secondary antibodies were used as needed with ECL or ECL Prime detection reagent (GE Healthcare Bio-Sciences, Corp., Piscataway Township, NJ USA) to visualize immunoreactive bands. Protein levels were quantified from scanned autoradiographs followed by densitometry using NIH Image 1.61 (http://macgui.com/downloads/?file_id=25143).
Immunofluoresence for CD20
Cells (5 × 105) were washed and resuspended in RPMI with 1% fetal bovine serum at 4 °C for 20 min. Ten microliters of Fluor-conjugated antibodies and, for some experiments, FITC-cholera toxin B subunit (Sigma) were then added for an additional 30 min. Cells were then washed and resuspended in 50 μl RPMI. Cells were incubated at 37 °C for the indicated times then fixed with ice-cold 4% paraformaldehyde for 20 min. Cells were washed and placed on poly-d-lysine-coated slides (Fisher Scientific, Pittsburgh, PA, USA). Coverslips were mounted with Vectashield mounting medium with DAPI (Vector Laboratories, Inc. Bulingame, CA, USA). Cells were visualized using a Zeiss fluorescent Axioskop 2 microscope with a × 40 1.3 numerical aperture objective and Axiocam camera (Thornwood, NY, USA).
RESULTS
Assessment of drug sensitivity and heritability
The response of LCLs in the presence or absence of 10 μg ml−1 rituximab or 10 μg ml−1 ofatumumab treatment was quantitated using AlamarBlue to indicate the viability/cellular health of remaining cells (summarized in Supplementary Table 1). Across both populations, CHORI and CEPH, the viability after rituximab treatment ranged from 7.4% to 138.6% with a median of 51.5%. Ofatumumab response showed increased levels of CDC, but the results were comparable with viability ranging from 0.2% to 106.5% and a median of 32.0%. For each population, there were strong correlations between rituximab and ofatumumab responses: R2 = 0.65 and R2 = 0.72 for CEPH and CHORI, respectively.
Publicly available pedigree information identified 62 out of 100 of the CEPH cell lines as members of complete trios. These individuals were used to estimate heritability: H2 = 21.53% and H2 = 21.07% for rituximab and ofatumumab, respectively. An alternative approach, using the Genome-wide Complex Trait Analysis software23 and the 486 unrelated samples, estimated heritability of rituximab and ofatumumab sensitivity at H2 = 34.54% and H2 = 35.05%, respectively.
Satisfied with the heritability of cellular sensitivity to MAbs, we sought to generate candidate genes for functional validation experiments through genome wide association, linkage, and gene expression analysis.
Association analysis
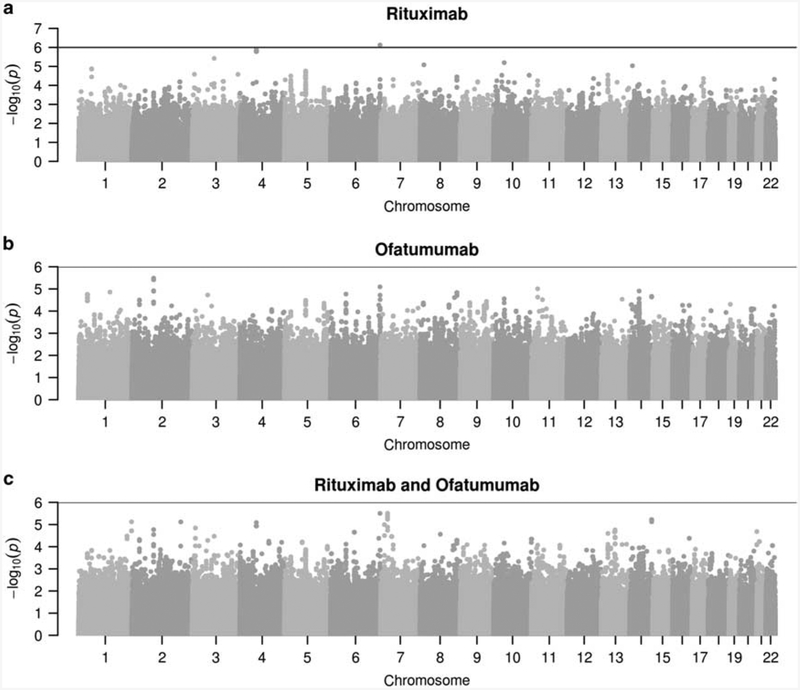
Using genotype data on 2.1 million SNPs for the CHORI samples, three separate GWAS were performed: (a) rituximab response, (b) ofatumumab response and (c) the vector of both responses modeled jointly. With a suggestive threshold cutoff (− log10(P) >6), there were five peak associations detected across all three GWAS (summarized in Supplementary Table 2).
Four out of five of the peak associations occurred on SNPs with rare homozygous major or minor genotypes. Filtering out rare variants—that is, any SNP with few heterozygous or homozygous genotypes (nAA<20, nAa<20 or naa<20)—resulted in one statistically significant result association with rituximab response. The SNP, rs9295079, exists in the gene encoding region for SMOC2 on chromosome 6.
As an alternative to filtering out rare variant associations, we used an adaptive permutation method25 to further test these associations. After applying the adaptive permutation testing, three out of five of peak associations surpassed the suggestive significance threshold cutoff based on the effective number of markers across the genome (− log10(P)>6).29,30 The SNP, rs10070859, had suggestive significance for ofatumumab and the combined analysis. This SNP is within 100 Kbp of the gene encoding region for APC. The results of all three GWAS are illustrated in Figure 1 where all suggestive association results (− log10(p)>6) have the final P-values from the permutation testing results.
Figure 1.
Manhattan plot for (a) rituximab, (b) ofatumumab, and (c) multivariate (rituximab + ofatumumab) GWAS. The negative log transform of P-values for three GWAS are given for 2.1 million SNPs. Nominal P-values are provided for all cases except any significant associations (− log10(p)>6). For all significant associations, the P-values are adjusted for multiple comparisons via permutation testing.
Linkage analysis
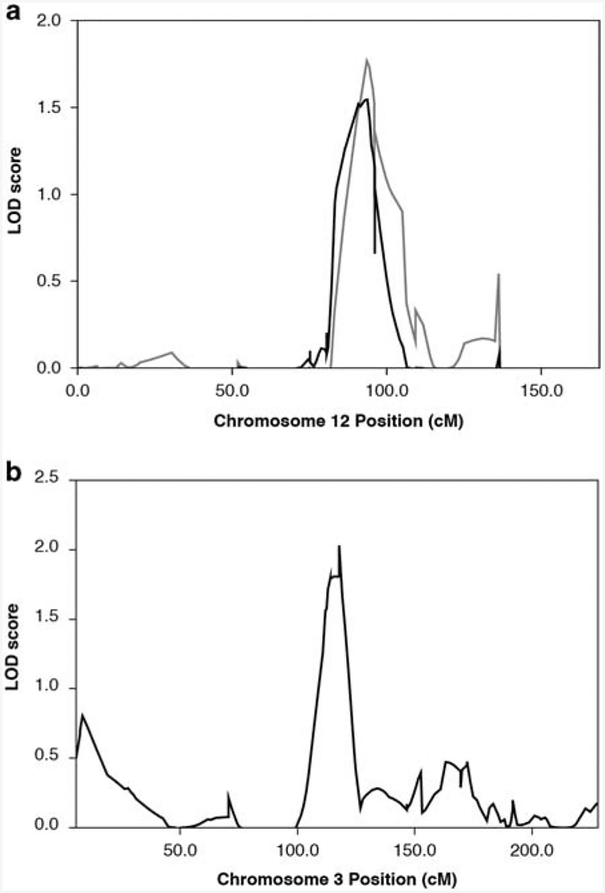
Linkage analysis performed on the CEPH revealed one significant linkage peak on chromosome 12 for rituximab and two significant peaks for ofatumumab, on chromosome 12 and 3 (Figure 2). Significance thresholds were determined by 10 000 simulations and several quantitative trait models were explored to determine which mode of inheritance best fit these data. Although these results reemphasized a genetic component of drug response (with heritability estimates under each model ranging from 0–58%), there was not a clear model that best fit the data. The single locus mixed model and two-locus mixed model estimates were each significant compared to the sporadic model (both P<0.001), but were not significantly different from each other (P<0.344).
Figure 2.
Linkage mapping of rituximab sensitivity. (a) Chromosome 12 LOD scores plotted on the y axis are shown with chromosome coordinates along the x axis in centimorgans (cM). The gray trace is for ofatumumab; black for rituximab. They perfectly overlap at peaks centered at 94cM. (b) Chromosome 3 LOD score peak reached significance only for ofatumumab sensitivity.
Correlations between gene expression and MAb sensitivity
Using publicly available gene expression profiling data, expression profiles from 57 of the original 100 LCLs typed on Affymetrix Human Genome U133A 2.0 arrays were downloaded and normalized. Quantitative Significance Analysis of Microarrays revealed 13 genes whose expression was correlated with viability in rituximab (Supplementary Table 3) and 25 genes whose expression was correlated with viability in ofatumumab (Supplementary Table 4), using a false discovery rate cutoff of <0.001%. All were negatively correlated with viability—that is, as gene expression increased, rituximab/ofatumumab sensitivity increased. CBLB, present in both tables, was the only gene that was also implicated in the union of linkage peaks and genome-wide expression differences. Hence, CBLB was chosen for further functional validation.
CBLB knockdown increases rituximab resistance and alters localization of CD20
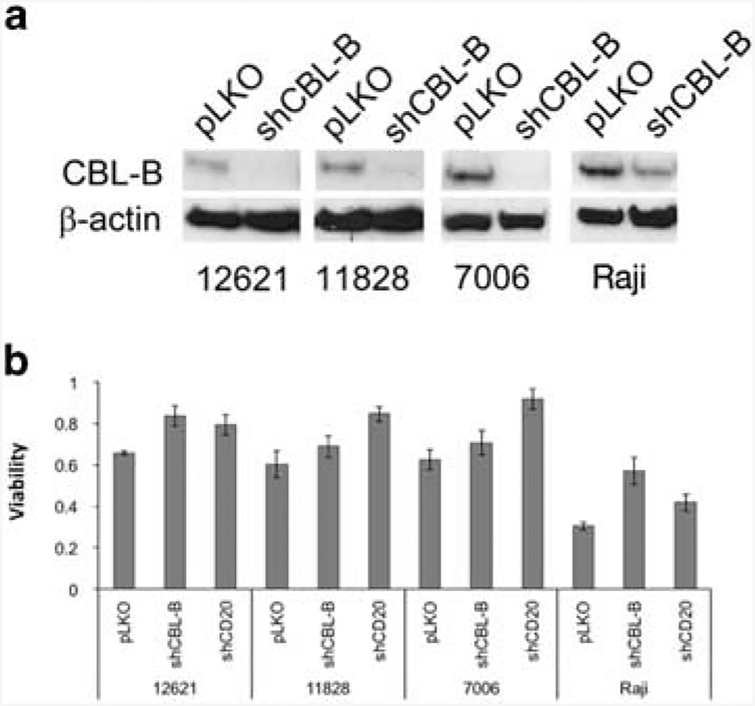
To validate CBLB, identified as both a candidate gene in the chromosome 3 linkage peak and independently as a gene whose expression is correlated with rituximab sensitivity, we reduced protein expression in a number of malignant and non-malignant B-cell lines using an shRNA construct. Knockdown of CBLB resulted in >95% reduction in CBLB protein levels, as evidenced by western blotting analysis (Figure 3a). Resulting cell lines, both LCLs and lymphoma cell lines, were more resistant to both rituximab (Figure 3b) and ofatumumab (Supplementary Figure 1).
Figure 3.
Cbl-b knockdown increases rituximab resistance. (a) Cbl-b protein levels are shown by western blot (four representative cell lines are shown) in cells with empty vector (pLKO) or Cbl-b knockdown (shCBL-B). (b) Viability in rituximab CDC assay, relative to cells with no rituximab added. Three lymphoblastoid and one lymphoma cell line, each with empty vector (pLKO), CBLB shRNA (shCBL) or MS4A1 shRNA (shCD20) are shown.
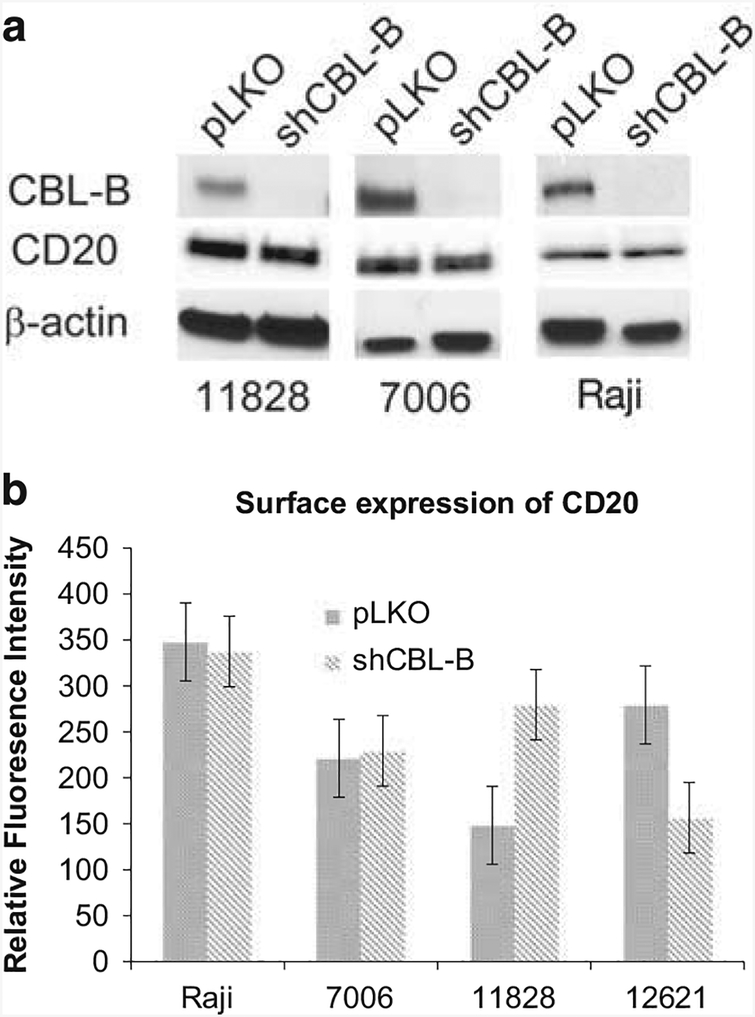
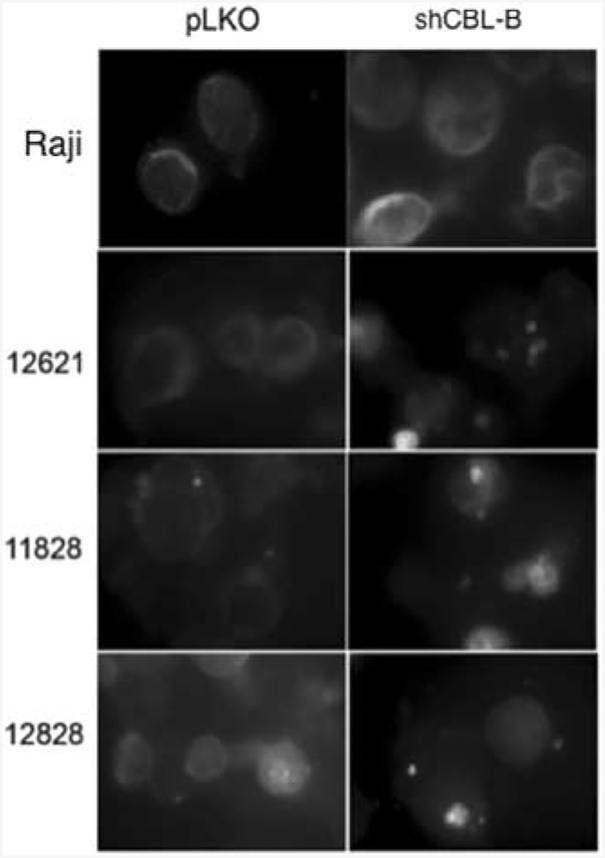
It has been shown previously that CD20 surface protein expression levels are correlated with sensitivity to anti-CD20 antibodies.31–33 To determine whether absence of CBLB alters CD20 expression, we measured total cellular protein levels of CD20, which were not altered by CBLB knockdown (Figure 4a). Furthermore, when we quantified surface expression of CD20, it was not reduced in CBLB knockdown cells (Figure 4b). To further examine CD20 localization as a possible mechanism for the effects of reduced CBLB levels, we also performed immunofluorescence in CBLB knockdown cells. CD20 localization was altered in cells with CBLB knockdown with a greater number of concentrated ‘patches’ in the membrane staining, while in the wild-type cells, the membrane localization of CD20 was more diffuse (Figure 5), This may suggest a potential mechanism for CBLB’s effects that could be explored in future studies.
Figure 4.
CD20 expression is not diminished by Cbl-b knockdown. (a) Western blot showing total Cbl-b and CD20 levels in three representative cell lines with and without Cbl-b knockdown. (b) Flow cytometry to measure cell surface expression of CD20 in four representative cell lines shows no decrease in cells with Cbl-b knockdown (shCBL).
Figure 5.
CD20 localization is altered in cells with Cbl-b knockdown. Left column shows CD20 localization in four control cell lines (empty vector) and right column shows the same cell lines with Cbl-b knockdown.
DISCUSSION
Our results provide some hypotheses and new insight into mechanisms of resistance for the MAbs, rituximab and ofatumumab. We have approached the problem using combined approaches involving heritability, GWAS, linkage analysis and functional validation with knockdown models showing altered localization of CD20.
Deciphering intrinsic and acquired resistance is a primary goal in precision or personalized medicine for cancer therapy: predicting the best treatment strategy based on the genetics and molecular features of individual cases. In vitro studies linking genotype to phenotype via GWAS and linkage analysis are common approaches to identify key loci in small molecule drug resistance and susceptibility. Here we sought to discover some of the genetic factors in MAb sensitivity, specifically rituximab and ofatumumab sensitivity, using multiple approaches—that is, association, linkage and gene expression analysis—and discovered a previously undescribed role for rituximab drug sensitivity mediated by CBLB, which was functionally validated through additional experimental work. Two other genes, SMOC2 and APC, were associated with variability in drug response across individuals and may warrant additional investigation in future work. These results suggest interesting insights into the etiology of response to anti-CD20 MAbs. The linkage results indicate that there are both shared regions of the genome linked to response and at least one region that is unique to ofatumumab. However, a candidate gene from that region, CBLB, appears to mediate both rituximab and ofatumumab, suggesting the specificity to ofatumumab could be merely a statistical one. The gene expression data results also reflect potential differences between the two MAbs in that there are a larger number of genes whose expression is significantly different for ofatumumab (25 in total) than for rituximab.13 This may be due to the higher CDC activity that has been reported for ofatumumab.34
Successful previous studies mapping genes to drug resistance have utilized candidate gene approaches relying on a priori assumptions. These approaches, although undoubtedly useful, cannot capture the unanticipated genetic factors involved in drug resistance. To the best of our knowledge, our study is the first to map a drug resistance gene in humans using an agnostic approach without assumption about mechanism. We have systematically run the gamut—from heritability to gene expression, association and linkage analysis followed by functional validation via genetic knockdown and immunohistochemistry. We have shown that CBLB expression affects localization of CD20 and susceptibility to CDC induced by MAb treatment.
As is the case with any in vitro study, the in vivo significance of the suggested genes of interest must be investigated. The application of these results to the clinical setting is yet unknown. There are notable confounders in extrapolating in vitro results to in vivo actionable items. Generally speaking, there are obvious immunologic and systemic features lacking in in vitro systems which would affect B-cell survival and fate in vivo. For example, natural killer cells and IL2 are powerful mediators of rituximab response.35 Moreover, polymorphisms in FCGR3A seem to affect rituximab response and natural killer cell-mediated lysis.36 Here we investigated one type of cell death by MAbs—i.e., CDC; however, it is well known that MAbs also work through apoptotic and antibody-dependent cell death mechanisms.
Although cell lines do not always lead directly to clinical efficacy prediction, they are often successfully used for hypothesis generation and elaborating on existing hypotheses.37 The LCLs used in our study came from healthy individuals. Although there is strong precedent that interindividual drug resistance and susceptibility can be understood using healthy donors, there are other factors that affect tumor resistance in vivo. For instance, tumor acquired alterations and tumor heterogeneity have an important role in drug resistance that is not reflected in our model system. However, this can also be an advantage, since our system is able to isolate and focus only on the effects of heritable (that is, germline) variability on drug response.
High-throughput in vitro assays, omics and informatics provide a compelling alternative approach to study cancer resistance mechanisms. We have successfully used the LCL model to explore two clinically important MAbs: rituximab and ofatumumab. Our current work has uncovered new mechanistic insight into the relationship between MAb susceptibility, CBLB and CD20. However, the full mechanism through which CBLB expression leads to differential localization of CD20 and confirmation that this is indeed the reason for altered anti-CD20 antibody susceptibility will require additional studies. As CBLB is a component of complement-dependent cell death by anti-CD20 MAbs, it will also be interesting to explore additional newly developed anti-CD20 antibodies, some of which have altered CDC effectiveness, to further test the hypotheses generated in our current work. Understanding the role of CBLB in MAb treatment could pave the way to using patient genotypes to enable decision making in the clinical setting: determining which anti-CD20 antibody would be most effective in which patients, or finding ways to restore rituximab sensitivity in patients who have inherited or acquired resistance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported a Mentored Research Scholar Grant in Applied and Clinical Research (MSRG-12-086-01-TBG) from the American Cancer Society, an R01 Grant (5R01 CA185372) from the National Cancer Institute to KLR and an RO1 (5R01CA161608) from the National Cancer Institute to AMR and HLM.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
REFERENCES
- 1.Shuptrine CW, Surana R, Weiner LM. Monoclonal antibodies for the treatment of cancer. Semin Cancer Biol 2012; 22: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng KP, Hillmer AM, Chuah CTH, Juan WC, Ko TK, Teo ASM et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med 2012; 18: 521–528. [DOI] [PubMed] [Google Scholar]
- 3.Turner NC, Reis-Filho JS. Genetic heterogeneity and cancer drug resistance. Lancet Oncol 2012; 13: e178–e185. [DOI] [PubMed] [Google Scholar]
- 4.Lee AJX, Swanton C. Tumour heterogeneity and drug resistance: personalising cancer medicine through functional genomics. Biochem Pharmacol 2012; 83: 1013–1020. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez de Castro D, Clarke PA, Al-Lazikani B, Workman P. Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance. Clin Pharmacol Ther 2013; 93: 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glennie MJ, French RR, Cragg MS, Taylor RP. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol Immunol 2007; 44: 3823–3837. [DOI] [PubMed] [Google Scholar]
- 7.Weng W-K, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol 2003; 21: 3940–3947. [DOI] [PubMed] [Google Scholar]
- 8.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 2002; 99: 754–758. [DOI] [PubMed] [Google Scholar]
- 9.Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol 2013; 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Racila E, Link BK, Weng W-K, Witzig TE, Ansell S, Maurer MJ et al. A polymorphism in the complement component C1qA correlates with prolonged response following rituximab therapy of follicular lymphoma. Clin Cancer Res 2008; 14: 6697–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation 2008; 118: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS ONE 2010; 5: e9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet 2009; 10: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown CC, Havener TM, Medina MW, Jack JR, Krauss RM, McLeod HL et al. Genome-wide association and pharmacological profiling of 29 anticancer agents using lymphoblastoid cell lines. Pharmacogenomics 2014; 15: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 2010; 34: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 17.HGDP-CEPH Human Genome Diversity Cell Line Panel. 2016. (cited 1 January 2015). Available from http://www.cephb.fr/en/hgdp_panel.php.
- 18.Marshfield Genetic Map Mammalian Genotyping Service. 1995. 2006. Available from http://research.marshfieldclinic.org/genetics/home/index.asp.
- 19.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci USA 2004; 101: 11809–11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters EJ, Motsinger-Reif A, Havener TM, Everitt L, Hardison NE, Watson VG et al. Pharmacogenomic characterization of US FDA-approved cytotoxic drugs. Pharmacogenomics 2011; 12: 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demeure O, Lecerf F. MarkerSet: a marker selection tool based on markers location and informativity in experimental designs. BMC Res Notes 2008; 1: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30: 97–101. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown CC, Havener TM, Medina MW, Krauss RM, McLeod HL, Motsinger-Reif AA. Multivariate methods and software for association mapping in dose-response genome-wide association studies. BioData Min 2012; 5: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Che R, Jack JR, Motsinger-Reif AA, Brown CC. An adaptive permutation approach for genome-wide association study: evaluation and recommendations for use. BioData Min 2014; 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snow GL, Wijsman EM. Pedigree analysis package (PAP) vs. MORGAN: model selection and hypothesis testing on a large pedigree. Genet Epidemiol 1998; 15 (4): 355–369. [DOI] [PubMed] [Google Scholar]
- 27.Akaike H A new look at the statistical model identification. IEEE Trans Automat Contr 1974; 19: 716–723. [Google Scholar]
- 28.Small GW, McLeod HL, Richards KL. Analysis of innate and acquired resistance to anti-CD20 antibodies in malignant and nonmalignant B cells. PeerJ 2013; 1: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 2008; 32: 381–385. [DOI] [PubMed] [Google Scholar]
- 30.Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 2008; 9: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis TA, Czerwinski DK, Levy R. Therapy of B-cell lymphoma with anti-CD20 antibodies can result in the loss of CD20 antigen expression. Clin Cancer Res 1999; 5: 611–615. [PubMed] [Google Scholar]
- 32.Foran JM, Norton andrew J, Micallef INM, Taussig DC, Amess JAL, Rohatiner AZS et al. Loss of CD20 expression following treatment with rituximab (chimaeric monoclonal anti-CD20): a retrospective cohort analysis. Br J Haematol 2001; 114: 881–883. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy GA, Tey S-K, Cobcroft R, Marlton P, Cull G, Grimmett K et al. Incidence and nature of CD20-negative relapses following rituximab therapy in aggressive B-cell non-Hodgkin’s lymphoma: a retrospective review. Br J Haematol 2002; 119: 412–416. [DOI] [PubMed] [Google Scholar]
- 34.Pawluczkowycz AW, Beurskens FJ, Beum PV, Lindorfer MA, van de Winkel JGJ, Parren PWHI et al. Binding of submaximal C1q promotes complement-dependent cytotoxicity (CDC) of B cells opsonized with anti-CD20 mAbs ofatumumab (OFA) or rituximab (RTX): considerably higher levels of CDC are induced by OFA than by RTX. J Immunol 2009; 183: 749–758. [DOI] [PubMed] [Google Scholar]
- 35.Golay J, Manganini M, Facchinetti V, Gramigna R, Broady R, Borleri G et al. Rituximab-mediated antibody-dependent cellular cytotoxicity against neoplastic B cells is stimulated strongly by interleukin-2. Haematologica 2003; 88: 1002–1012. [PubMed] [Google Scholar]
- 36.Dall’Ozzo S Rituximab-dependent cytotoxicity by natural killer cells: influence of FCGR3A polymorphism on the concentration-effect relationship. Cancer Res 2004; 64: 4664–4669. [DOI] [PubMed] [Google Scholar]
- 37.Weinstein JN, Lorenzi PL. Cancer: discrepancies in drug sensitivity. Nature 2013; 504: 381–383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.