Abstract
RNA thermodynamics play an important role in determining the 2D and 3D structures of RNA. Internal loops of the sequence, 5’-GMNU/3’-UNMG, are relatively unstable thermodynamically. Here, five GU flanked 2×2 nucleotide internal loops were structurally investigated in order to reveal determinants for their instability. The internal loops investigated are: 5’-GCAU/3’-UACG, 5’-UUCG/3’-GCUU, 5’-GCUU/3’-UUCG, 5’-GUCU/3’-UCUG, and 5’-GCCU/3’-UCCG. Two-dimensional NMR spectra indicate the absence of GU wobble base pairing in 5’-GCUU/3’-UUCG, 5’-GUCU/3’-UCUG and 5’-GCCU/3’-UCCG. The 5’-GCUU/3’-UUCG loop has an unusual conformation of the GU base pairs, in which U’s O2 carbonyl forms a bifurcated hydrogen bond with G’s amino and imino protons. The internal loop of 5’-GUCU/3’-UCUG displays a shifted configuration in which GC pairs flank a U-U pair and several U’s are in fast exchange between positions inside and outside the helix. In contrast, 5’-GCAU/3-UACG and 5’-UUCG/3’-GCUU both have the expected GU wobble base pairs flanking the internal loop. Evidently, GU base pairs flanking internal loops are more likely to display atypical structures relative to Watson-Crick base pairs flanking internal loops. This appears to be more likely when the G of the GU pair is 5’ to the loop. Such unusual structures could serve as recognition elements for biological function and as benchmarks for structure prediction methods.
Graphical Abstract
INTRODUCTION
Improved prediction of RNA 3D structure depends on appropriate experimentally determined structures to test the validity of predicted structures. In general, de novo prediction of 3D RNA structure lags far behind the accuracy of predicting 3D protein structure.1 In part, this is due to inaccuracies within non-canonical regions.1 In contrast to Watson-Crick interactions, which are handled with remarkable accuracy, the physics of non-canonical interactions is not perfectly understood. The sheer diversity2 of non-canonical regions makes their prediction especially difficult and a more comprehensive understanding of these regions would facilitate better predictions of 3D structure and dynamics.
RNA thermodynamics inform the understanding of both secondary structure and individual base-base interactions. Many non-canonical secondary structure elements have experimentally determined thermodynamics. For common secondary structure elements, such as two by two nucleotide (2×2 nt) internal loops, the thermodynamics are sequence dependent.3–7 The stability of 2×2 nt internal loops is dependent upon both the identity of the non-canonical base pairs and interactions between loop and canonical flanking pairs. When a 2×2 nt internal loops is flanked by GU base pairs, it tends to be less stable than the same internal loop flanked by AU/UA or GC/CG base pairs.4, 7
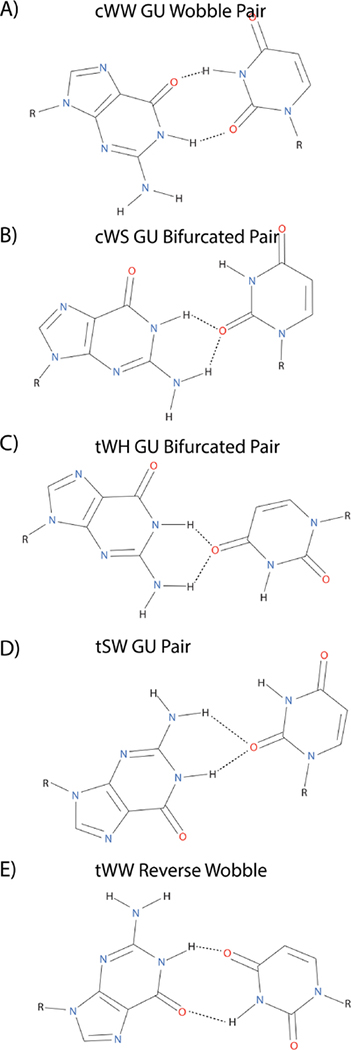
GU base pairs are ubiquitous in RNA structure.8, 9 They are, by far, the most common non-Watson-Crick base pair. The most common orientation of the GU base pair is the cis wobble base pair, in which the Watson-Crick edge of both the G and U interact with one another, forming two inter-base hydrogen bonds (Figure 1A).8 The importance of GU wobble pairs in biology is well documented.9 For example, a GU pair plays an important role in self-cleavage of the HDV ribozyme.10, 11 However, the common wobble is not the only conformation that a GU base pair can adopt (Figure 1). Bifurcated GU pairs have been observed in several instances.12, 13 In these pairs, U’s O2 carbonyl inserts itself between the G amino and imino protons (Figure 1B). Similarly, another type of bifurcated GU pair has also been observed in crystal and NMR structures, in which U’s O4 carbonyl inserts itself between the G amino and G imino protons (Figure 1C). These possible GU conformations illustrate the plasticity of G-U pairing. Due to their rarity in reported structures, not much is known about the thermodynamics or sequence context for these non-wobble GU pairings.
Figure 1.
Structure of several different GU base pairs.2 (A) cWW GU wobble pair, (B) cWS GU bifurcated pair as in the structure of the ribosome (PDB code: 1FFK, residues 966–1002),14 (C) tWH GU bifurcated base pair as in 5S rRNA (PDB code: 364D, residues 74–102),12 (D) tSW GU pair as in the UUCG hairpin (PDB code: 1HLX, residues 9–12),13 (E) tWW Reverse wobble pair (PDB code: 1FFK, residues 1966–1970)14. Note: sugars are excluded from the base pair images despite the involvement of some sugars in hydrogen bonding to a base.
Here, the structures of duplexes containing 2×2 nt internal loops flanked by GU pairs were examined with NMR in order to determine reasons for their instability. Internal loops studied are: 5’-GCAU/3’-UACG, 5’-UUCG/3’-GCUU, 5’-GCUU/3’-UUCG, 5’-GUCU/3’-UCUG, and 5’-GCCU/3’-UCCG. Measured nearest neighbor parameters for the examined internal loops range between 3.58 and 5.37 kcal/mol at 37 °C.4, 7 The 2×2 nt internal loops discussed here are all considered category one internal loops, i.e. loops that are unlikely to have a non-canonical pair with two strong hydrogen bonds between the bases.15 Two-dimensional NMR revealed GU pairs bordering the internal loops that deviated from the expected GU wobble structure for 5’-GCUU/3’-UUCG, 5’-GUCU/3’-UCUG, and 5’-GCCU/3’-UCCG. The 5’-UUCG/3’-GCUU and 5’-GCAU/3’-UACG loops both exhibited the expected wobble GU pairing flanking the internal loop, showing that not all GU pairs flanking category one internal loops fail to form a wobble pairing. The altered interaction patterns of many GU pairs flanking their respective internal loop helps explain previous thermodynamic measurements and may aid in the determination of where non-wobble GU pairs are more likely to occur.
MATERIALS AND METHODS
RNA preparation
Sequences were chosen from previous melting experiments.4, 7 RNA oligonucleotides were purchased from Dharmacon. Sequences were: ((5-CGGGCAUCCG)2, GCAU for short), (5’-CGCAUUCGACGC/3’-GCGUGCUUUGCG, UUCG for short), (5’-CGCAGCUUACGC/3’-GCGUUUCGUGCG, GCUU for short), (5’-CGCAGUCUACGC/3’-GCGUUCUGUGCG, GUCU for short), and (5’-CGCAGCCUACGC/3’-GCGUUCCGUGCG, GCCU for short). RNAs were dissolved in an NMR buffer composed of 80 mM NaCl, 50 mM NaH2PO4, and 0.05 mM Na2EDTA (pH=6.1–6.4). Exchange to D2O was accomplished by lyophilization. Non-self complementary and self complementary duplexes each used a duplex concentration of approximately 1 mM (thus individual strands in a non-self complementary duplex were about 1 mM each and the total strand concentration was roughly 2 mM). Samples for spectra in which Mg2+ was present were created by adding MgCl2 to yield a final Mg2+ concentration of 2mM (see Figures S1–3).
NMR Spectroscopy
NMR spectra were collected on 500 or 600 MHz Varian Inova spectrometers. One-dimensional spectra were obtained with a 1–1-spin-echo water suppression pulse.16 Two-dimensional spectra in H2O were acquired using a WATERGATE pulse with flipback for water suppression.17, 18 A typical set of experiments for each sequence involved NOESY experiments with mixing times of 50, 175, and 400 ms, along with a TOCSY experiment with a mixing time of 32 ms. For (5’-CGGGCAUCCG)2 and 5’-CGCAGCUUACGC/3’-GCGUUUCGUGCG, 13C-1H sensitivity enhanced HSQC (at natural abundance) and 31P-1H HETCOR spectra were collected in order to help assign sugar residues. Two-dimensional data were processed with NMRPipe.19 NMRFAM-SPARKY was used to display and assign protons in the 2D data sets.20 Proton assignment was accomplished by previously described methods.21 Chemical shift/assignment tables are available in supporting information (Table S1–5).
Restraint generation
Restraints were generated from assigned NOE resonances at mixing times of 50 ms or 175 ms. When possible, NOE distances were determined using the 1/r6 NOE volume relationship with cytosine and uracil H5-H6 cross-peak volumes as reference distance (2.45 Å). The sum-over-box integration method in NMRFAM-SPARKY was used in calculating NOE volumes.20 However, spectral overlap prevented measurement of many such peaks, and so a majority of NOE volumes were binned into 4 categories: strong (1.5 Å - 2.7 Å), medium (2.0 Å - 3.5 Å), weak (2 Å - 4.7 Å), and very weak (typically used for spin diffusion peaks, 2.5 Å - 6 Å). Dihedral angles were restrained to typical A-form RNA values where the data indicated canonical pairs, and for angles with direct scalar-coupling evidence.22 Tables S6–8 contain restraints used in simulated annealing.
Simulated Annealing
A-form RNA starting structures were obtained using NUCGEN.23 Structures were generated with NMR restraints in Generalized Born implicit solvent.24 The simulated annealing algorithm used 3 phases over 100 ps. First, the RNA was held at 3,000 K for 5,000 steps (5 ps). Next, the RNA was linearly ramped down to 100 K over 93,000 equal temperature steps (93 ps). Lastly, the RNA was held at 0 K for 2,000 steps (2 ps). Force constants used for distance and dihedral restraints were 30 kcal/(mol*Å2) and 30 kcal/(mol*rad2), respectively. Weights applied to the restraints were 0.1 for the first simulated annealing phase and 1.0 for the two final phases. Once a set of structures was generated, the structures were sorted by restraint energy. From the set of structures with no distance violations > 0.1 Å, the 20 structures with lowest restraint energy were retained in the final ensemble. Pymol was used in analyzing and creating figures based upon the ensembles of structures.25 Structural statistics for the three modeled internal loop structures can be seen in Table S9.
3D Structure Prediction with ROSIE
Three-dimensional structures were predicted with the ROSIE server’s FARFAR (RNA De Novo) protocol.26–29 Structures were predicted for which an NMR based structure was determined: (5’-CGGGCAUCCG)2, 5’-CGCAUUCGACGC/3’-GCGUGCUUUGCG, and 5’-CGCAGCUUACGC/3’-GCGUUUCGUGCG. Runs were performed with and without the chemical shifts for non-exchangeable protons included. In each run, the secondary structure was provided and the GU pairs were notated as being a part of the loop. The two options “Allow Bulge” and “Use Updated (2012) Force Field” were turned on for each individual run. For 5’-CGCAGCUUACGC/3’-GCGUUUCGUGCG, 1000 structures were calculated within each run. For the other two sequences, 200 structures were calculated in each individual run. Lowest energy structures from ROSIE were compared with those from simulated annealing by calculating RMSD values using the lowest energy NMR derived model (the 8 residues of the internal loop only) as the reference.
RESULTS
GCAU Self-Complementary Duplex
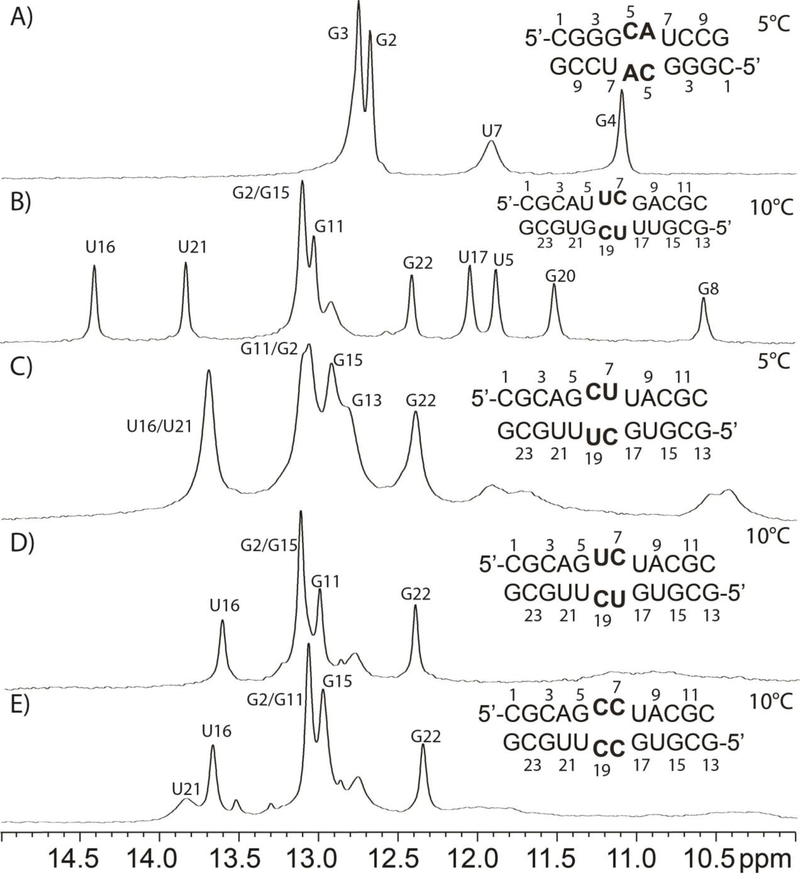
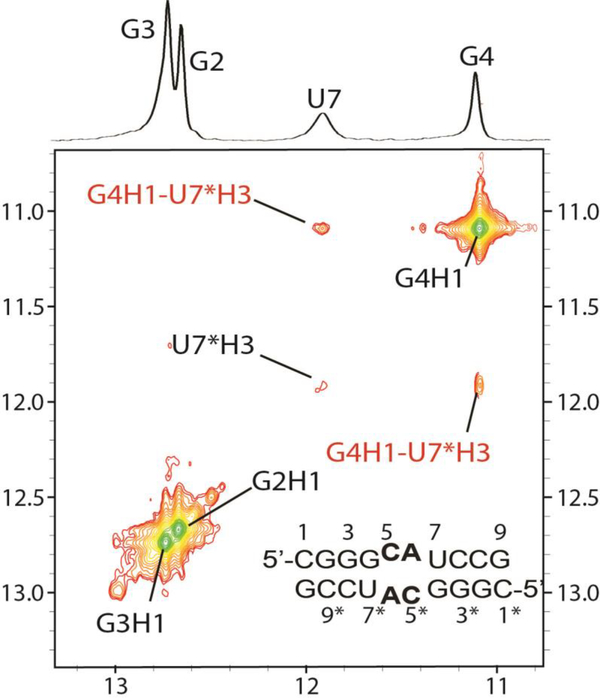
The GCAU duplex, (5’-CGGGCAUCCG)2, is self-complementary with GU base pairs flanking two CA pairs (Figure 2A). The imino proton spectrum for GCAU has four individual resonances (Figure 2A). The resonances at 12.67 and 12.74 ppm are assigned to the G imino protons of stem GC pairs (Figure 3), G2-C9* and G3-C8*, respectively, where * represents the other strand. G4’s imino proton (11.09 ppm) was assigned by a large cross-peak to the G4 amino and cross-peaks to C5H1’ and C8*H1’. The U7 imino proton (Figure 3) is extremely broad and thus likely to be heavily influenced by exchange with water. The diagonal peak for U7 within the 50 ms NOESY spectrum is very weak, but its cross-peak with G4 is stronger (Figure 3). The latter cross-peak is consistent with a G4-U7* wobble pair.30 The proximity of this pair to the internal loop may allow momentary opening of the helix, allowing the U7 imino proton to exchange with water.
Figure 2.
1D NMR spectra of duplexes containing 2×2 internal loops flanked by GU “pairs”.
Figure 3.
Imino 2D NOESY spectrum (50 ms) of GCAU duplex revealing GU wobble pairs (cross-peaks labeled in red).
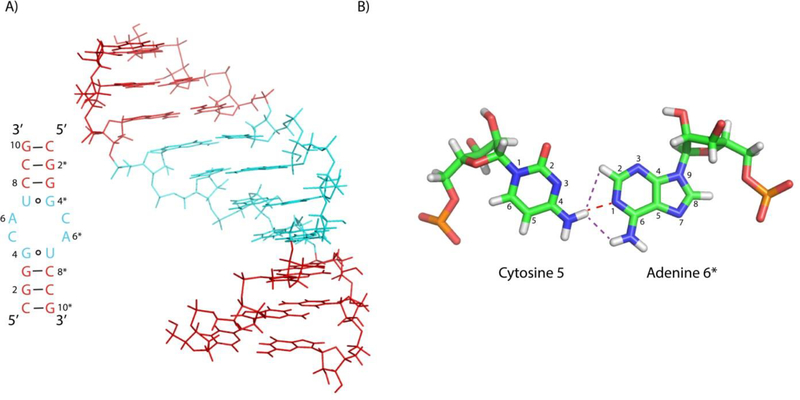
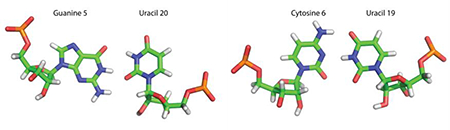
Structures of the CA pairs are established by several NOEs (Figure S4) in a 75 ms NOESY spectrum. The C5 amino protons, C5H41 and C5H42, both show NOEs to A6*H2 and the A6* amino group. The adenine amino resonances are identified by an NOE (likely due to spin diffusion) to A6H2. The modeled duplex (Figure 4A) has a CA pair with a single hydrogen bond between the C5 amino and A6N1 (Figure 4B). If the cytosine aminos were free to rotate rapidly and were not hydrogen bonded, then there would be only one C5 amino resonance. Thus, this hydrogen bond is further confirmed by the two C5 amino resonances. In contrast, the A6 aminos have only one resonance, indicating that they are not hydrogen bonded. This is reflected in the modeled structure of the CA pair (Figure 4B).
Figure 4.
(A) Structure of GCAU duplex. Internal loop is shaded cyan and the adjoining stems are shaded red. (B) Structure of the single H-bond C-A base pair (cWW). The red dashed line represents the single hydrogen bond and purple dashed lines represent two of the observed NOEs.
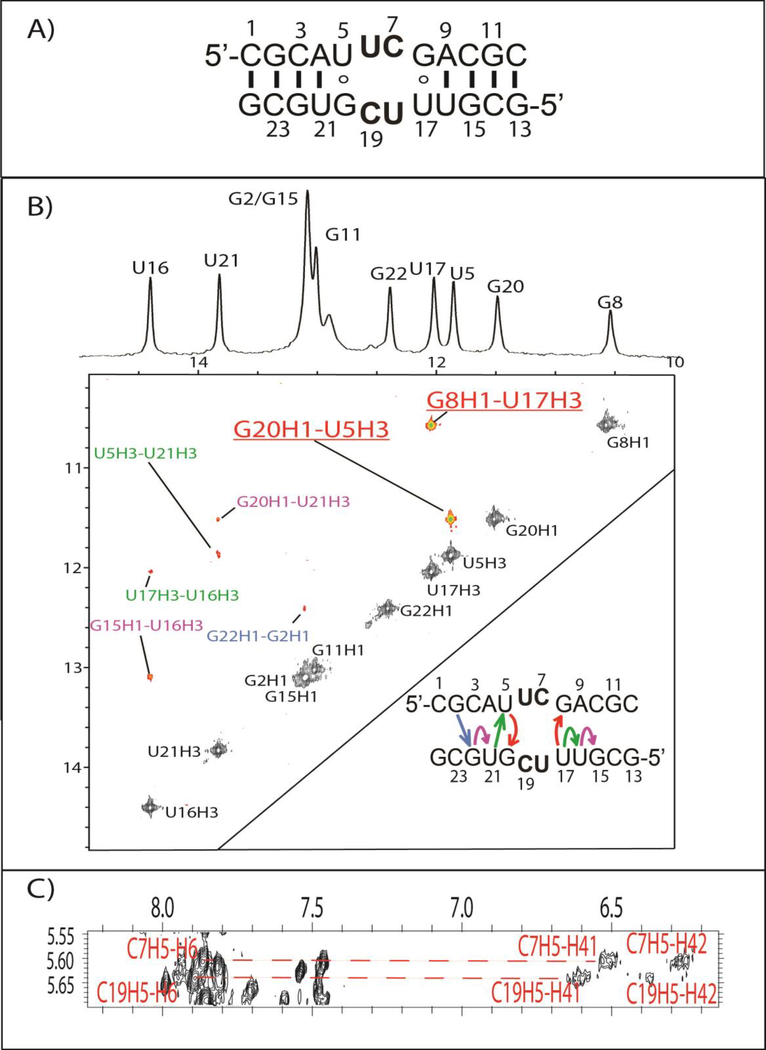
UUCG Non-Self-Complementary Duplex
The UUCG duplex, (5’-CGCAUUCGACGC/3’-GCGUGCUUUGCG), is non-self-complementary with a 5’-UUCG/3’-GCUU internal loop (Figures 2B and 5A). The 1D imino proton spectrum has chemical shifts similar to the other non-self-complementary duplexes containing a similar stem (Figure 2B–E). Evidently, the duplexes in Figure 2B–E have similar stem structures, as expected based upon their sequences. Unlike GCAU (Figure 3), GU wobble pairs adjacent to the internal loop displayed strong imino diagonal peaks as well as strong cross-peaks (U5H3-G20H1 and U17H3-G8H1, Figure 5B). The NMR restrained model accurately recreates the hydrogen bonding of the GU wobble pairs, as can be seen in Figure 6A.
Figure 5.
(A) The predicted secondary structure of the UUCG duplex. (B) The 2D imino NOESY spectrum of UUCG duplex, which features two GU wobble peaks shown in red/underline. Colored NOEs in the NOESY spectrum correspond to imino-imino NOEs shown in the secondary structure in the corner of panel B. (C) CH5-H42/H41 peaks for the internal loop. Two resonances can be seen for each amino, likely, due to the presence of a hydrogen bond in the formation of UC pairs.
Figure 6.
(A) U5-G20 wobble pair in the UUCG duplex. (B) Modeled cWW U6-C19 non-canonical pair structure. The two potential hydrogen bonds are shown in red dashed lines. (C) Ensemble of duplex models for the UUCG duplex.
The UUCG internal loop contains two UC pairs. Both internal loop cytosines, C7 and C19, have two amino resonances, with each having a cross-peak to its own CH5 (Figure 5C). The number of C7 and C19 resonances implies these aminos are hydrogen bonded, as seen in other UC pairings.31, 32 Canonical A-form cross-peaks are seen for all residues within the loop and near it, indicating that the loop residues are within the helix and in an A-form like conformation. However, despite indication that the UC pairs form, no imino resonance is observed for U6 or U18 in the 1D spectrum (Figure 5B). Evidently, the U imino protons within the loop readily exchange with water. Modeling of this pair (Figure 6B) produced a more closed configuration of the UC pairs than has been previously modeled31 or seen.32 Specifically, the hydrogen bond between U6/U18 H3 and C19/C7 N3 in Figure 6B is missing or replaced by a bridging water.31, 32 In the absence of explicit water during modeling, the UC pair cannot be mediated by a bridging water as is seen in the Holbrook et al. crystal structure.32 The crystal structure, however, does not preclude the existence of UC pairing with two hydrogen bonds (Figure 6B). It is possible that the pair is in dynamic equilibrium in solution between the two possible CU pair structures. The modeled UUCG structure has U6/U18 O4 - C19/7 amino hydrogen bonds, consistent with the other structures,31, 32 even though that hydrogen bond was not constrained. It is important to note, however, the lack of interbase NOEs between C7/C19 and U18/U6 when considering the modeled structure.
GCUU Non-Self-Complementary Duplex
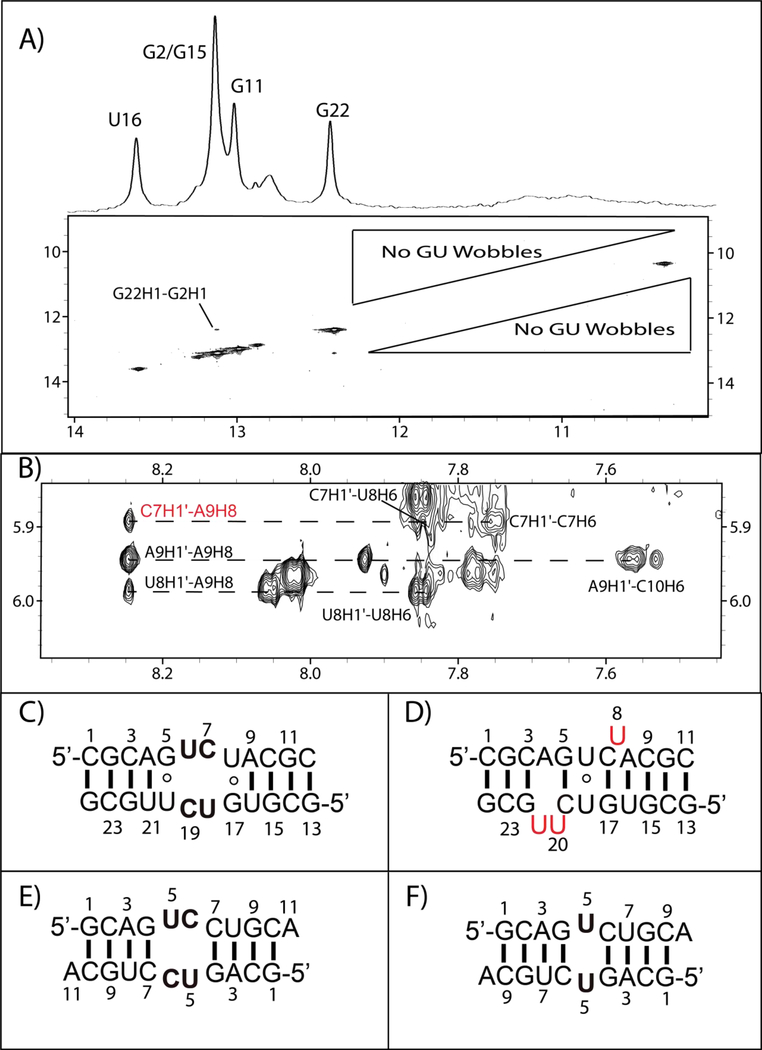
Figure 2C shows the predicted secondary structure and 1D imino proton spectrum for the non-self-complementary duplex, GCUU, containing the 5’-GCUU/3’-UUCG internal loop. A majority of the stem forms as expected, with GC/AU base pairs displaying the expected canonical set of NOEs (Figure 7A and B). Noticeably absent from the 2D spectra, however, are peaks corresponding to GU wobbles for G5/U20 and G17/U8 (Figure 7B). None of the broad peaks below 12 ppm in the 1D spectrum (Figure 2C) yield an imino-imino cross-peak. These broad peaks are not observed in 2D spectra until temperatures below 15 °C, at which point all peaks become extremely broad. Thus no definitive assignment of these broad imino peaks could be made. At higher temperatures (Figure S5), the duplex begins melting and no new peaks emerge.
Figure 7.
(A) Predicted structure for GCUU. Arrows in cyan indicate residues that show cross-strand H1’-H1’ NOESY cross-peaks indicative of bifurcated GU pair formation. (B) 2D imino spectrum that reveals the structure outside of the internal loop formed as expected, despite the lack of GU wobble pairing in the loop region. (C) A NOE between G5’s amino and U20H1’ consistent with bifurcated pair formation. (D) H1’-H1’ walk region for both strands in red and green. The cross strand H1’-H1’ resonances that result from narrowing of the minor groove are shown in cyan.
Despite the lack of a GU wobble cross-peak, G5 amino cross-peaks provide some indication of the pairing scheme for the GU pairs. In a 400 ms NOESY spectrum, the G5 amino shows NOEs to C6H1’, U21H1’, and U20H1’ (Figure 7A and C). In GU wobble pairs and similar structures,14, 33, 34 the G amino and H1’ of the paired U are < 5 Å apart so can yield an NOE. In previous NMR spectra35, 36 and in the UUCG duplex (Figure 2B), this potential NOE is very weak or nonexistent. Thus, the absence of a G5-U20 imino-imino NOE reveals that the G5-U20 pair is likely not in a wobble pair, but the G5 amino to U20H1’ NOE implies a conformation similar to a wobble. Unfortunately, the corresponding G17 amino to G8H1’ NOE could not be located.
Some GU wobble-like structures (Figure 1B) are associated with a narrowed minor groove for residues surrounding the GU pair.14, 33 This narrowing in certain GU pairs was predicted in molecular dynamics simulations of the 5’-GAGU/3’UGAG internal loop.33 Narrowing of the minor groove brings H1’ protons of nearby residues close that would normally be greater than 5 Å apart. More specifically, if the minor groove is narrowed in the GCUU duplex, then H1’ atoms are expected to be within 5 Å of one another for C6/U20, U7/U19, and U8/C18. In D2O, C6H1’-U20H1’ and U8H1’-C18H1’ NOEs can be clearly seen (Figure 7D) and differentiated from other peaks within the H1’ to H1’ walk. A potential U7H1’-U19H1’ cross-peak cannot be seen due to overlap of the H1’ chemical shifts, as expected due to the pseudo-symmetry of the CU pairs. Similar cross-strand H1’ to H1’ NOEs were not observed in the 5’-GCAU/3’-UACG or 5’-UUCG/3’-GCUU duplexes.
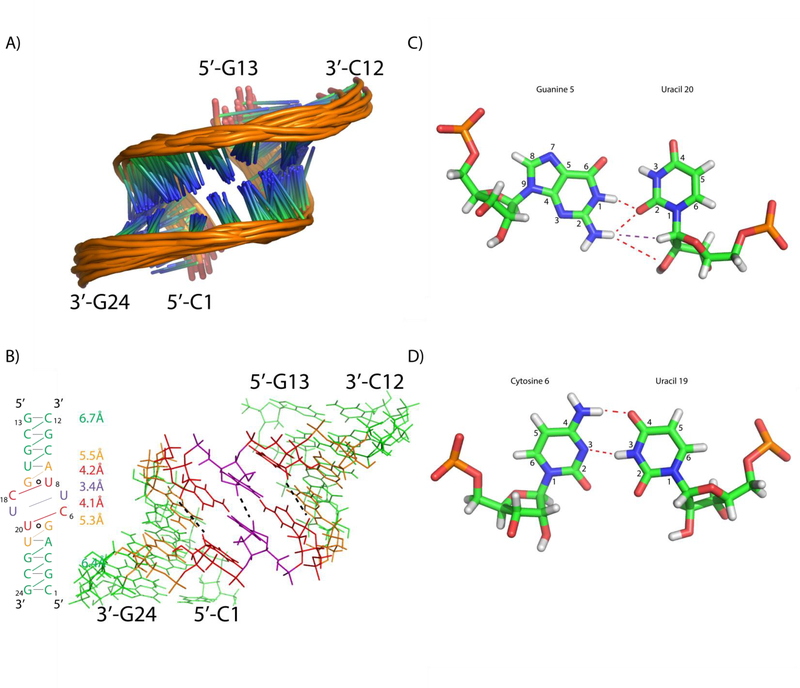
Interestingly, the expected symmetry of the GU pairs is lacking. U8’s sugar has 30–40% C2’ endo character, as indicated by a 3.5 Hz scalar-coupling between H1’ and H2’. This contrasts with U20, where the weaker H1’-H2’ coupling is 2.0 Hz. Further evidence of asymmetry in the loop region is provided by a small U7H1’-A9H8 NOE, indicating that U8 can occasionally flip out from the helix. A corresponding peak indicating U20 can flip out, however, is missing. This information, together with the fact that the G17 amino is not observed while G5 is, indicates that the two GU pairs are not equivalent. One possible reason for this could be the underlying sequence surrounding each GU pair. U20 is in the middle of a long stretch of pyrmidines (C18-U19-U20-U21), whereas U8 has an adjacent purine (C6-U7-U8-A9). Differential stacking interactions may exist for the two GU pairs bordering the internal loop, due to the differences in surrounding sequence.
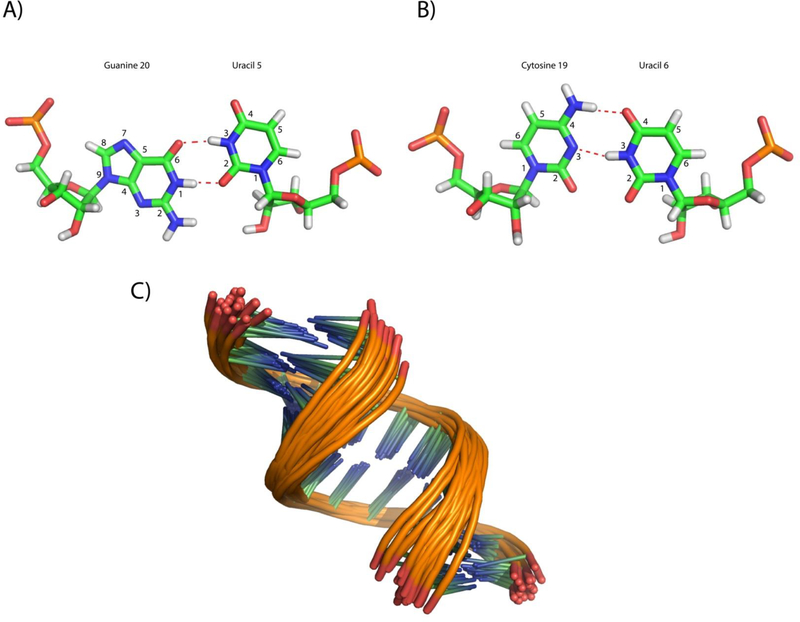
Modeling of the GCUU duplex (Figure 8A) yielded structures with bifurcated GU pairs similar to those observed in the GAGU minor structure and the ribosome (Figure 1B and 8B).14, 33 The minor groove was narrowed in the modeled structures due to distance restraints placed on C6H1’-U20H1’ and U8H1’-C18H1’ (restrained with a lower bound of 3 Å and an upper bound of 5 Å). The average distances for C6H1’-U20H1’ and U8H1’-C18H1’ in the final structural ensemble are 4.1 Å and 4.2 Å, respectively (Figure 8B). These distances contrast with the values in the stem where similar H1’-H1’ distances average 6.4 Å for the lower stem (G2H1’-G24H1’, C3H1’-C23H1’, and A4H1’-G22H1’) and 6.7 Å for the upper stem (C10H1’-U16H1’, G11H1’-G15H1’, C12H1’-C14H1’). G5H1’-U21H1’ and A9H1’-G17H1’ have values intermediate between those seen for the stems and loop (5.3±0.6 Å and 5.5±0.4 Å, respectively), showing that these residues may be where the deviation of the groove begins. Small NOEs are seen that may be due to the narrow groove in this intermediate width (G5H1’-U21H1’ and A9H1’-G17H1’, Figure 7D).
Figure 8.
(A) Ensemble of duplex models for the GCUU duplex. (B) Ensemble averaged H1’-H1’ distances are listed for the corresponding part of the duplex (color coded to the duplex on the right side). In the 3D structure, three H1’-H1’ connections are shown with black dashed lines in the structure on the right. (C) cWS GU Bifurcated pair derived from modeling of GCUU. Red lines indicate the presence of a H-bond and purple lines indicate an observed NOE (D) Modeled cWW CU pair of GCUU duplex.
Modeling revealed CU pairs oriented with the U7 and U19 H3 imino protons hydrogen bonded to the C18 and C6 N3 atoms, respectively (Figure 8D). Much like the UC pairs in UUCG (Figure 5B), it is likely that the actual structure of the CU pairs is more open than that observed in the GCUU model. A lack of identifiable resonances due to U7 or U19 H3 indicate that the imino protons exchange with water. The crystal structure by Holbrook et al. shows UC pairing mediated by a water between the U H3 and C N3, which is consistent with U7 and U19 being able to readily exchange with water.32 Additionally, a hydrogen bond occurs between the C18/C6 amino protons and the U7/U19 carbonyl (O4) (Figure 8D) as seen in other CU pairs.31, 32 Despite a lack of NOE data regarding the U7-C18 and C6-U19 pairs, the force field creates a UC pair that is similar to previously observed UC pairs.
Modeling also shows that there may be a weak H-bond between the G5 amino and U20 sugar’s O2’ (Figure 8C). The average acceptor-donor distance across the 20 models was 3.8 Å, indicating the presence of a weak H-bond.37 This orientation is supported by the existence of the G5 amino to U20H1’ NOE, which brings G5’s amino spatially close to U20’s sugar. The model has U20’s O2’ involved in the potential hydrogen bond, but U20’s hydroxyl proton could not be identified unequivocally.
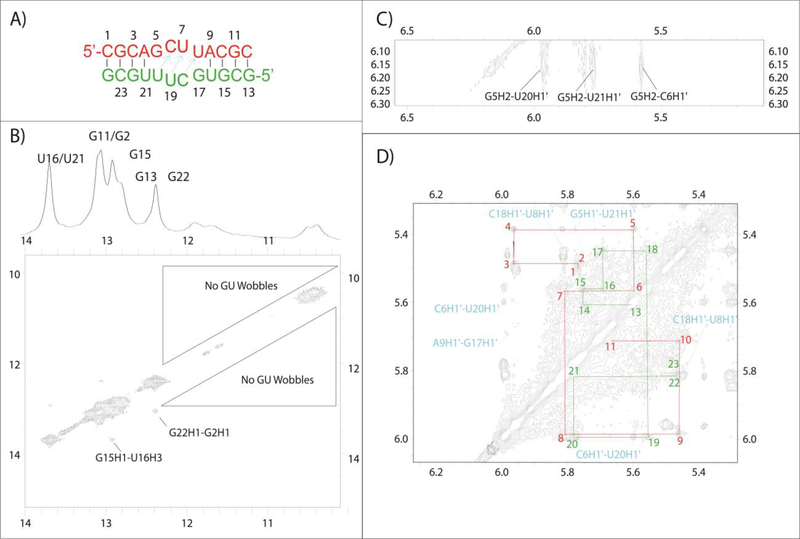
GUCU Non-Self-Complementary Duplex
The non-self-complementary GUCU duplex, (5’-CGCAGUCUACGC/3’-GCGUUCUGUGCG, Figure 2D), was assigned in the usual manner. The 1D spectrum (Figure 2D) implies the duplex has a stem structure similar those with the same stem sequence (Figure 2B–E). Except for the A4-U21 pair, the GC and AU stem components formed. As with the 5’-GCUU/3’-UUCG internal loop, NMR spectra for 5’- GUCU/3’-UCUG did not show an imino-imino cross-peak expected for GU wobble pairs (Figure 9A). The 1D spectrum for the GUCU duplex (Figure 2D) at 10 °C lacked resonances from the loop region (G5, U6, U8, G17, U18, and U20). Evidently, these resonances were too broad to be observed.
Figure 9.
(A) 2D NOESY imino region lacking NOEs that would indicate wobble base pairs. (B) NOEs indicating that U8 is in can flip out from the helix. (C) Predicted structure of GUCU duplex. (D) Secondary structure model for GUCU duplex. All red U’s may not be flipped out at the same time. Middle GC pairs may not always be present. (E) & (F) Model duplexes (GUCC and GUC) created to compare chemical shifts of the internal loop residues with those of the GUCU duplex.
In the GUCU duplex, there is evidence that U8 is sometimes flipped out of the helix. A direct NOE is observed between C7H1’ and A9H8 in a 400 ms NOESY spectrum (Figure 9B). This peak is accompanied by the expected canonical peak between U8H1’ and A9H8 (Figure 9B). The H1’-H2’ scalar-coupling for U8 is 4 Hz, indicating that the sugar is dynamic with about 40% C2’ endo character. Also observed is an NOE between C7H2’ and U8H6, which is a canonical A-form peak that would not be expected if U8 was always flipped out.38 Since U8 has only one set of resonances, there is fast exchange between conformations with U8 in and out of the helix. If the C7H1’-A9H8 NOE is only present when U8 flips out and if C7H1’-A9H8 has a similar distance to most sequential Watson-Crick stem nH1’-(n+1)H6/H8 cross-peaks, then an approximate percentage of the flipped out structure can be obtained by dividing the NOE volume of the C7H1’-A9H8 by an average of the volumes from Watson-Crick stem nH1’-(n+1)H6/H8 NOEs. With these assumptions, U8 is flipped out approximately 40% of the time.
Based on the pseudo-symmetry of the duplex, it would seem likely for U20 to flip out as well. However, a potential NOE between C19H1’ and U21H6 cannot be observed due to overlap. Interestingly, a small NOE cross-peak between U20H1’ and G22H8 is observed, consistent with U21 flipping out of the duplex some of the time (Figure S6). This is unexpected, because the predicted secondary structure has an A4-U21 base pair. The ability of U21 to flip out is also supported by lack of a U21 imino resonance expected for an A4-U21 base pair (Figure 2D).
The chemical shifts of C7 and C19 amino groups are reminiscent of cytosine aminos in Watson-Crick GC pairs (see Tables 1 and S4). Within the 1D imino proton spectrum (Figure 2D), there is no evidence for additional GC base pairs, but this could be due to imino proton exchange with water within a dynamic internal loop. This observation left two possible secondary structures for the duplex (Figure 9C and D): (a) one in which two UC pairs are flanked by GU pairs, and/or (b) a duplex with two unexpected GC pairs flanking a central U6-U18 pair, where U8, U20 and U21 all have the ability to flip out of the duplex.
Table 1.
| Sequence | Residue | H1’ | H2’ | H41 | H42 | H5 | H6 |
|---|---|---|---|---|---|---|---|
| GUCC | C6 | 5.57 | 4.55 | 7.23 | 6.95 | 5.60 | 7.66 |
| GUCU | C7 | 5.88 | 4.44 | 8.20 | 7.14 | 5.75 | 7.75 |
| C19 | 5.77 | 4.32 | 8.06 | 7.24 | 5.80 | 7.87 |
To differentiate between the two possibilities, a self-complementary duplex (GUCC), (5’-GCAGUCCUGCA)2, was designed that contains Watson-Crick GC base pairs flanking a UC tandem mismatch (Figure 9E). Placing GC pairs next to the UC tandem mismatch forces formation of tandem UC pairs while retaining a G 5’ of the U. This allows for chemical shift comparisons between cytosine aminos in a UC pair in 5’-GUCC/3’-CCUG (Figure 9E) and in 5’-GUCU/3’-UCUG. The C of the UC pair in 5’-GUCC/3’-CCUG displays amino H41 resonances that differ by ≥ 0.8 ppm from the resonances of the internal loop C H41 aminos in 5’-GUCU/3’-UCUG (Table 1). The cytosine amino chemical shifts in 5’-GUCC/3’-CCUG are closer to those for 5’-UUCG/3’-GCUU where stable UC pairs form (C7H41 is 6.52 ppm and C19H41 is 6.61 ppm, see Table S2). Additionally, the U H1’, H2’, and H5 chemical shifts of 5’-GUCC/3’-CCUG do not compare favorably with the chemical shifts observed for 5’-GUCU/3’-UCUG (Table 2). An additional duplex (GUC, Figure 9F), (5‘-GCAGUCUGCA)2, was designed to measure the chemical shifts of a UU pair flanked by GC pairs as postulated for the possible structure shown in Figure 9D. Chemical shift comparisons between the U in the GUC duplex and U6/U18 in the GUCU duplex showed astonishing similarity (Table 2). Furthermore, a −1°C imino proton spectrum of GUCU revealed a small peak at 10.36 ppm, equivalent to the shift for U5 in GUC (Figure S7). Together, these model duplexes point towards a high population of an internal loop with GC pairs surrounding a central U-U pair, as in Figure 9D, with dynamic flanking U residues (U8/U20/U21) that are flipped out approximately 40% of the time.
Table 2.
Comparison of uracil chemical shifts between GUCU (Figure 9C/D), GUC (Figure 9F), and GUCC (Figure 9E).
| Sequence | Residue | H1’ | H2’ | H3 | H5 | H6 |
|---|---|---|---|---|---|---|
| GUCC | U5 | 5.608 | 4.415 | NDa | 4.915 | 7.551 |
| GUCU | U6 | 5.454 | 4.027 | (10.36)b | 5.12 | 7.489 |
| U18 | 5.397 | 4.166 | (10.36)b | 5.144 | 7.533 | |
| GUC | U5 | 5.384 | 4.143 | 10.39 | 5.122 | 7.426 |
Resonance is too broad to be measured
The assignment of U6 and U18 imino resonances (shown in parenthesis) is based on a small peak at −1°C (Figure S7).
GCCU Non-Self-Complementary Duplex
Figure 2E shows the predicted secondary structure and the 1D imino proton spectrum of the non-self-complementary-duplex, GCCU, containing the 5’-GCCU/3’-UCCG internal loop. As with GCUU (Figure 2C) and GUCU (Figure 2D), resonances for the expected Watson-Crick pairs are present, but no GU wobble imino NOE peaks were observed in a 2D spectrum (Figure S8).
Two dimensional NMR spectra reveal that the GCCU duplex exists in an equilibrium between minor self-complementary duplexes and the major non-self complementary duplex (Figure S9).7 The minor duplex structures cannot be titrated out of solution by addition of the designed non-self-complementary strand (Figure S10). These minor duplexes consitute < 15% combined of total strand concentration, but complicate analysis of the designed 2×2 nt internal loop because a large number of peaks are present in the 2D NMR spectra. Though the percentage of minor duplexes is small, their resonances can be comparable in NOE volume because they are self-complementary duplexes. Thus, their NOE volumes are doubled relative to the volumes for the major non-self-complementary duplex. These minor conformations and extensive chemical shift overlap prevented assignment of many of the H1’ resonances in the GCCU non-self-complementary duplex (Table S5). The stability of these alternative conformations has been previously noted,7 with both having a melting temperature similar to that of the non-self-complementary duplex.
Addition of 2mM MgCl2 to the GCUU, GUCU, and GCCU duplexes does not change chemical shifts of imino proton resonances
GU pairs can sometimes bind cations.39–41 To see if Mg2+ could affect the GU pairing in the GCUU, GUCU, and GCCU duplexes, 1D NMR spectra were taken in the presence of 2 mM MgCl2 (Figures S1–S3). No significant changes were detected.
3D Structure Prediction with ROSIE and Comparison to NMR Derived Models
For the GCAU, UUCG, and GCUU duplexes, comparisons were made between structures modeled by simulated annealing and the FARFAR protocol in Rosetta26, 29 without and with the aid of chemical shifts.28 For all three duplexes, lowest energy predictions derived from chemical shifts agreed with the non-canonical base-base interactions from simulated annealing (Table 3). For 5’-GCUU/3’-UUCG, even the narrowed minor groove (Figure 8) was reflected in the predictions with and without chemical shifts. For 5’-UUCG/3’-GCUU the predictions displayed a narrow minor groove with all loop cross-strand H1’-H1’ distances between 5.0 and 6.0Å. No H1’-H1’ cross strand NOEs were detected in this region, consistent with the prediction of limited narrowing. For the GCUU duplex (Figure 2C), the lowest energy structure in the absence of chemical shifts showed an unusual G5-U20 pairing in which the imino protons of G5 and U20 pointed directly at one another (Figure S11). Adding chemical shifts changed this pairing to be intermediate between a bifurcated (Figure 1B) and a wobble (Figure 1A) pair.
Table 3.
Comparisons between NMR and FARFAR structures for Internal Loops
| Sequence | Chemical shifts restraints? | Loop RMSDa (Å) |
|---|---|---|
| GCAU | N | 0.940 |
| Y | 0.934 | |
| UUCG | N | 1.554 |
| Y | 1.594 | |
| GCUU | N | 1.731 |
| Y | 1.676 |
RMSD measurements were taken upon the 2×2 internal loop as well as the flanking GU pairs
DISCUSSION
GU pairs (Figure 1) are frequently observed in RNA structure and are the third most common base pair.42–44 GU pairs are so common that they are often considered the third canonical pairing in RNA. More specifically, for 2×2 nt internal loops, at least one GU pair is flanking the loop approximately 25% of the time.5 The thermodynamic stability of nearest neighbor combinations with one GU base pair are similar to nearest neighbor combinations with one AU pair.45 However, despite their apparent thermodynamic stability when adjacent to Watson-Crick pairs, 2×2 nt internal loops with a (5’-GMNU)2 motif are less thermodynamically stable than category one 2×2 nt internal loops with a (5’-AMNU)2 motif.7, 15 Therefore, three dimensional structures of (5’-GMNU)2 internal loops can be informative about causes for destabilization of this class of internal loops. The duplexes investigated here reveal several non-wobble pairing schemes for GU “pairs” flanking 2×2 nt internal loops. Such unusual structures could provide recognition elements for biological function and benchmarks for testing methods for de novo predictions of 3D structure.
The 5’-GCAU/3’-UACG internal loop has wobble GU pairs
The GCAU duplex (Figure 2A) contains two CA pairs flanked by two wobble GU pairs. An NOE between G4H1 and U7H3 is consistent with the GU pairs flanking the internal loop being in a wobble conformation. However, the signal strength/volume of the U7H3 peak (Figure 3) suggests that U7 may be strongly affected by exchange with water. The measured nearest neighbor parameter of 3.7 kcal/mol for (5’-GCAU)2 is similar to that for other category one 2×2 nt internal loops flanked by GU wobble or AU pairs.3–5, 7 The 5’-GCAU/3’-UACG internal loop contains a pyrimidine-purine pair (Figure 4A), which may allow more stability because there is minimal backbone distortion.
The 5’-UUCG/3’-GCUU internal loop has wobble GU pairs
The UUCG duplex also has flanking GU wobble pairs closing the 5’-UUCG/3’-GCUU internal loop (Figures 5B and 6A). The 2D NMR spectra have strong imino-imino NOE cross-peaks for the two GUs flanking the internal loop: U5H3-G20H1 and U17H3-G8H1 (Figure 5B). This was the only internal loop studied here with the U of the GU pair 5’ of the internal loop. At the end of helices, a wobble GU with this orientation has better stacking interactions with Watson-Crick pairs,46 which may favor the wobble configuration’s stacking interactions (Figure S12). A database of tandem mismatches from various RNA secondary structures has the U of GU pairs 5’ of the loop much more often than the G 5’ of the loop.5 With the exception of the 5’-UUCG/3’-GCUU internal loop, which is the least thermodynamically favorable of all 5’-UMNG/3’-GNMU loops, 5’-UMNG/3’-GMNU loops are more stable than 5’-GMNU/3’-UNMG loops.7 The exception when compared to 5’-GUCU/3’-UCUG is due to the formation of GC rather than GU pairs (Figure 9D). Evidently, the orientation of GU pairs flanking internal loops is important for the thermodynamics of the internal loop and might also be important for determining whether flanking GUs adopt a wobble conformation.
The 5’-GCUU/3’-UUCG internal loop has bifurcated GU pairs
The GCUU duplex did not show NOE cross-peaks indicative of GU wobble formation (Figure 7B). Modeling of the 5’-GCUU/3’-UUCG internal loop revealed that the GU pairs flanking the internal loop form a bifurcated GU pair (Figures 1B and 8B), accompanied by local minor groove narrowing (Figure 8C). This narrowing was clearly observed in the form of abnormal cross strand H1’ to H1’ NOEs (Figure 7D). H1’-H1’ NOEs, combined with a G amino to U H1’ NOE from a potential GU pair, and/or lack of an identifiable imino from U and/or G could provide an NMR hallmark for identification of similar bifurcated pairs in other RNAs.
The GU bifurcated pairs are similar to wobble pairs (Figures 1A, 1B and 8C). The major difference between the two types of base pairs is that in the GU bifurcated pair U O2 carbonyl is bifurcated by a G H1 imino and an amino hydrogen, whereas a GU wobble has a single hydrogen bond between U O2 carbonyl and G H1 imino proton. Additionally, in the bifurcated pair, the U H3 imino proton/G O6 carbonyl hydrogen bond is weakened or not present. The ensemble averaged distance between U N3 (donor) and G O6 (acceptor) in these pairs was 4.4 Å, which would not be considered a hydrogen bond.37 Base pair analysis using DSSR47 reveals a large pair opening parameter, relative to that seen in typical GU wobbles. Using this large opening parameter as a structural search criterion in conjunction with RNA FRABASE,48 several other potential bifurcated GU pairs were identified within crystal/NMR structures. Table S10 lists them along with their corresponding base pair parameters. The crystal data suggest that bifurcated GU pairs can form in contexts other than flanking loops.
While similar in structure to wobble GU pairs, bifurcated GU pairs have different properties. The bifurcated pair anchors the G and U bases with H-bonds in three instead of four locations as with GU wobble pairs. This would likely lead to additional dynamics in a GU bifurcated pair, which helps explain the lack of an imino resonance observed for the pair. Furthermore, having one 3-center H-bond instead of two 2-center H-bonds would likely affect the thermodynamics of pairing.49 The pair has different stacking orientations, as well, due to the different orientation of the G and U bases. In addition to narrowing the minor groove, the recognition face for a cWS GU bifurcated pair within the major groove is altered relative to that of a wobble pairing. In the bifurcated pair, the G carbonyl and U imino hydrogen are not hydrogen bonded and are displayed within the major groove, providing a unique recognition interface for interaction with other molecules. For example, the carbonyl and amino groups in the side chains of Asn and Gln can fit in the major groove of such a bifurcated GU pair.
The 5’-GUCU/3’-UCUG internal loop forms GC pairs
The GUCU duplex displayed a stark departure from the expected GU pairs because evidence was found for two GC pairs flanking a central U-U pair (Figure 9D). The altered configuration within the internal loop is reminiscent of the major structure of the 5’-GAGU/3’-UGAG “internal loop”, in which the U of the expected GU pairs flip out of the helix.38, 50 This leaves sheared GG pairs flanking a central A-A stack. In order for the altered configurations in 5’-GAGU/3’-UGAG and 5’-GUCU/3’-UCUG to be present in solution, the expected structure containing wobble GU pairs must be relatively unfavorable. The GUCU internal loop is dramatically different from 5’-UUCG/3’-GCUU, which instead contains two GU wobble pairs in the reverse orientation. These two duplexes are another example of the significance of orientation of GU pairs flanking an internal loop.
The 5’-GCCU/3’-UCCG internal loop does not have GC or wobble GU pairs
NMR spectra (Figures 2E, S8–10) for the 5’-GCCU/3’-UCCG internal loop were especially difficult to interpret due to the existence of small amounts of self-complementary duplexes in addition to the intended non-self-complementary duplex. However, spectra reveal that GU wobble pairs do not form, much like 5’-GUCU/3’-UCUG and 5’-GCUU/3’-UUCG.
The loop sequence, 5’-GCCU/3’-UCCG, has the potential to form a conformation similar to that of 5’-GUCU/3’-UCUG (Figure 9D), with a central pair of C6-C18 flanked by G5-C19 and G17-C7 GC pairs. While no definitive identifications could be made for all four of the C residues within the internal loop, A9H8 was definitively assigned. There is some NOE volume consistent with a flipped out U8, but a vast majority of the volume corresponding to A9H8 was due to the canonical U8H1’-A9H8 peak. Evidently, a majority of the RNA was in the expected secondary structure and not in an alternative pairing configuration like 5’-GUCU/3’UCUG. This is likely due to the fact that single U-U pairs are more stable than single C-C pairs.51, 52
GU pairs with the U 5’ of an internal loop are typically wobble pairs
GU pairs surrounded by Watson-Crick pairs in sequence alignments typically form wobble structures.8, 9, 45 When flanking internal loops, however, they have the potential to form different hydrogen bonds12, 14, 33, 38, 50 (Figure 8B) and even different pairings (Figure 9D). Such unexpected structures could provide recognition elements for RNA function. Thus, it is important to develop rules for predicting proclivity of GU pairs to form wobble or unusual structures. Although the database of such structures is small,53 it allows proposing working hypotheses for predicting when GU pairs in aligned sequences will or will not form wobble pairs, at least for length symmetric internal loops. In general, wobble GU pairs form when the sequence has the U of a GU pair 5’ to the loop.3, 35, 50, 54 This could be partially due to the fact that 5’-wU/3’-cG nearest neighbors are an average of 0.4 kcal/mol more stable at 37 °C than 5’-wG/3’-cU nearest neighbors, where wc represent a Watson-Crick pair.45
GU pairs with the G 5’ of an internal loop can form non-wobble pairs
Here, the three duplexes without GU wobble pairs have the G 5’ to the internal loop, and addition of Mg2+ did not affect the 1D imino proton spectra (Figure S1–3). All three of these duplexes also contained an AU pair preceding the GU pair. Previous 1D NMR spectra provide four additional examples of GU pairs in a 5’-AG-Internal loop/3’-UU-Internal loop or 5’-UG-Internal loop/3’-AU-Internal loop motif that do not yield GU wobbles.7 All these examples have 5’-AG/3’-UU and/or 5’-UG/3’-AU nearest neighbors, which are the least stable of nearest neighbor containing a GU next to a Watson-Crick pair.45 Combined, these results suggest that sequences in a 5’-AG-Internal loop/3’-UU-Internal loop or 5’-UG-Internal loop/3’-AU-Internal loop motif may be unlikely to form GU wobbles. Furthermore, it is worth noting that these internal loops are less thermodynamically stable than expected from prediction.7 Loops of the 5’G orientation are rare in known secondary structures, perhaps due to their unfavorable stability relative to other loops.5
A particularly interesting internal loop was studied in the duplex: 5’-GAGGAAGGCGA/3’-PCUCUAAUUGCU, where P represents purine nucleoside.55 The loop is found in group I self-splicing introns from Candida albicans and Candida dubliniensis. In this loop, the GU with the G 5’ to the (AA)2 loop does not form a wobble pair and its amino group is likely involved in a stabilizing H-bond (such as in the pair in Figure 8C). Evidently, an AU pair preceding a GU with the G 5’ to a loop is not essential to allow formation of a non-wobble pair. The other GU pair adjacent to the (AA)2 loop also does not form a stable wobble pair, but is dynamic. The third GU pair has a wobble conformation. It is not surprising that its adjacent GU pair is not a wobble. When flanked by Watson-Crick pairs, the 5’-GG/3’-UU nearest neighbor free energy increment is favorable by only 0.25 kcal/mol at 37 °C.45 Apparently, the 5’-GAAGG/3’-UAAUU loop can be considered a 3×3 nt loop closed by two GU pairs with different orientations and structures.
Implications for prediction of RNA secondary structure
In the absence of GU wobble pairing adjacent to internal loops, it is possible to consider the non-wobble GU pairs as being a part of the loops. NMR and thermodynamic data shown here (Table 4) give a physical reason for considering non-wobble GU pairs as part of the internal loop. For the duplexes discussed here, many internal loops flanked by non-wobble GU pairs have their thermodynamic stabilities at 37 °C more accurately predicted as a 4×4 nt internal loop by current models5, 56 rather than as a 2×2 nt internal loop (Table 4). This is not surprising because current models use the nearest neighbor parameters measured for wobble GU pairs.45 As expected, all loops studied here that were flanked by wobble GU pairs are more accurately predicted as 2×2 nt internal loops. In the current work, the most dramatic absence of wobble GU pairing was discovered in the GUCU duplex (Figure 9D). In that case, a considerable fraction of duplexes form two GC pairs and a UU pair, instead of two GU and two UC pairs. This was accompanied by flipping of two (or possibly three) U’s outside of the helix instead of forming GU pairs. Presumably, the two unexpected GC pairs allow formation of a 5’-GUC/3’-CUG motif, which can have a favorable free energy between −0.5 and −1.8 kcal/mol depending on location in a duplex.51 A related motif was discovered for the major structure of the 5’-GAGU/3’-UGAG internal loop where two GG pairs form and two U’s are flipped out.38 Evidently, flipping out of the U and alternative pairing of the G with a loop nucleotide should also be considered when searching for potentially unusual structures that are more favorable than GU pairs.
Table 4.
Free energy increments for 2×2 nt internal loops along with whether NMR detected wobble GU pairs are present.
| Internal Loop7 | 2×2 Measured ΔG°37 (kcal/mol)7 | 2×2 Predicted ΔG°37 (kcal/mol)5 | 2×2 |ΔΔG| (kcal/mol) | 4×4 Measured ΔG°37 (kcal/mol)7 | 4×4 Predicted ΔG°37 (kcal/mol)56 | 4×4 |ΔΔG| (kcal/mol) | Wobble GU |
|---|---|---|---|---|---|---|---|
| 5’-GCAU/3’-UACG | 3.6 | 3.0 | 0.6 | 0.0 | 2.2 | 2.2 | Y |
| 5’-UUCG/3’-GCUU | 4.3 | 3.0 | 1.3 | 2.9 | 3.6 | 0.7 | Y |
| 5’-GCUU/3’-UUCG | 5.4 | 3.0 | 2.4 | 4.6 | 3.6 | 1.0 | N |
| 5’-GUCU/3’-UCUG | 4.1 | 3.0 | 1.1 | 3.4 | 3.6 | 0.2 | N |
| 5’-GCCU/3’-UCCG | 5.1 | 3.0 | 2.1 | 4.4 | 3.6 | 0.8 | N |
| 5’-GAAU/3’-UAAG | 5.4 | 3.0 | 2.4 | 4.5 | 3.6 | 0.9 | Na |
| 5’-GACU/3’-UCAG | 3.6 | 3.0 | 0.6 | 2.8 | 3.6 | 0.8 | Na |
Lack of a GU wobble pairing is inferred from 1D spectra.7
3D Structure Prediction Accuracy by ROSIE
The accuracy of the de novo structural predictions by the FARFAR26, 29 algorithm was remarkable. Nearly all non-canonical interactions were predicted by the lowest energy structures for 5’-GCAU/3’-UACG, 5’-UUCG/3’-GCUU, and 5’-GCUU/3’-UUCG with or without chemical shift restraints (Table 3). Introduction of chemical shifts into the predictions fixed an unusual G5-U20 pairing within 5’-GCUU/3’-UUCG (Figure S11). Calculations for the GUCU duplex were not attempted because the chemical shifts are averages of more than one structure.
The accuracy of 3D predictions within the internal loop region suggests that using chemical shift based predictions can further validate structures modeled with NMR derived distance and torsion restraints. Furthermore, the accuracy of the predictions also indicates that chemical shift based restraints in addition to distance/torsion restraints in modeling could further aid structural determination from NMR. A weighted combination of distance restraints from NOEs, angular data from scalar coupling and/or RDCs, and chemical shifts could help the force field refinement yield more accurate 3D models.
Implications for benchmarking forcefields
Benchmarking the accuracy of RNA force fields is often done primarily using hairpins.57–60 Stable hairpins such as UNCG or GNRA have historically been used to test RNA force fields because the systems are well defined. However, it is being realized that many RNAs exist as ensembles of structures.61–63 Moreover, RNA structure and dynamics depend on subtle competition between hydrogen bonding, stacking, hydration, and electrostatics.60 Thus, benchmarking of RNA force fields requires many different types of systems. The duplexes discussed here provide several new subjects for testing force fields. For example, the GUCU duplex provides a particularly challenging system for testing whether force fields can reproduce the flipping out of U8, U20, and/or U21 while forming G5-C19, G17-C7, and U6-U18 pairs (Figure 9D).
Potential implications for biological function
Two unexpected pairing schemes were revealed by the NMR spectra: (1) bifurcated GU pairs couple with a narrow minor groove in the GCUU duplex (Figure 8B and C), and (2) GC pairs replacing GU pairs in the GUCU duplex (Figure 9). Both provide potential recognition elements for binding to RNA. While the duplexes studied here have sequence symmetric internal loops, it is likely that the nearest neighbor interactions drive these structures. Thus, such structures should be considered at each end of an asymmetric loop, especially when a potential GU pair has the G 5’ of the loop. One example is the 5’-GGAAGG/3’-CUAAUU loop from some group I self-splicing introns.55
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Susan Schroeder for comments and for suggesting comparisons to ROSIE predictions.
Funding: This work was supported by NIH grants R01GM22939 to D. H. T. Additional support for K. D. B. was provided by the NIH training grant T32GM68411-10.
Footnotes
Notes: The authors declare no competing financial interest
ASSOCIATED CONTENT:
Supporting information:
The Supporting Information is available free of charge on the ACS Publications website.
The supporting information contains tables of chemical shifts for all studied duplexes, restraints used in modeling, statistics for NMR derived structures, structural parameters for base pairs related to the observed bifurcated GU in GCUU, and additional figures of NMR spectra/structures.
Accession Codes:
Structural coordinates were deposited in the RCSB Protein Data Bank as entries 6N8F, 6N8H, and 6N8I.
REFERENCES
- (1).Miao Z, Adamiak RW, Blanchet MF, Boniecki M, Bujnicki JM, Chen SJ, Cheng C, Chojnowski G, Chou FC, Cordero P, Cruz JA, Ferre-D’Amare AR, Das R, Ding F, Dokholyan NV, Dunin-Horkawicz S, Kladwang W, Krokhotin A, Lach G, Magnus M, Major F, Mann TH, Masquida B, Matelska D, Meyer M, Peselis A, Popenda M, Purzycka KJ, Serganov A, Stasiewicz J, Szachniuk M, Tandon A, Tian S, Wang J, Xiao Y, Xu X, Zhang J, Zhao P, Zok T, and Westhof E (2015) RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures, Rna 21, 1066–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Leontis NB, Stombaugh J, and Westhof E (2002) The non-Watson-Crick base pairs and their associated isostericity matrices, Nucleic Acids Res 30, 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wu M, McDowell JA, and Turner DH (1995) A periodic table of symmetric tandem mismatches in RNA, Biochemistry 34, 3204–3211. [DOI] [PubMed] [Google Scholar]
- (4).Christiansen ME, and Znosko BM (2008) Thermodynamic characterization of the complete set of sequence symmetric tandem mismatches in RNA and an improved model for predicting the free energy contribution of sequence asymmetric tandem mismatches, Biochemistry 47, 4329–4336. [DOI] [PubMed] [Google Scholar]
- (5).Christiansen ME, and Znosko BM (2009) Thermodynamic characterization of tandem mismatches found in naturally occurring RNA, Nucleic Acids Res 37, 4696–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Turner DH, and Mathews DH (2010) NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure, Nucleic Acids Res 38, D280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Berger KD, Kennedy SD, Schroeder SJ, Znosko BM, Sun H, Mathews DH, and Turner DH (2018) Surprising Sequence Effects on GU Closure of Symmetric 2 × 2 Nucleotide RNA Internal Loops, Biochemistry 57, 2121–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Masquida B, and Westhof E (2000) On the wobble GoU and related pairs, Rna 6, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Varani G, and McClain WH (2000) The G·U wobble base pair: A fundamental building block of RNA structure crucial to RNA function in diverse biological systems, EMBO Reports 1, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Perrotta AT, and Been MD (1996) Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage, Nucleic Acids Research 24, 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ferre-D’Amare AR, Zhou K, and Doudna JA (1998) Crystal structure of a hepatitis delta virus ribozyme, Nature 395, 567–574. [DOI] [PubMed] [Google Scholar]
- (12).Correll CC, Freeborn B, Moore PB, and Steitz TA (1997) Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain, Cell 91, 705–712. [DOI] [PubMed] [Google Scholar]
- (13).Allain FH, and Varani G (1995) Structure of the P1 helix from group I self-splicing introns, J Mol Biol 250, 333–353. [DOI] [PubMed] [Google Scholar]
- (14).Ban N, Nissen P, Hansen J, Moore PB, and Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution, Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- (15).Shankar N, Xia T, Kennedy SD, Krugh TR, Mathews DH, and Turner DH (2007) NMR reveals the absence of hydrogen bonding in adjacent UU and AG mismatches in an isolated internal loop from ribosomal RNA, Biochemistry 46, 12665–12678. [DOI] [PubMed] [Google Scholar]
- (16).Sklenář V, and Bax A (1987) Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra, Journal of Magnetic Resonance (1969) 74, 469–479. [Google Scholar]
- (17).Grzesiek S, and Bax A (1993) The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements, Journal of the American Chemical Society 115, 12593–12594. [Google Scholar]
- (18).Piotto M, Saudek V, and Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions, J Biomol NMR 2, 661–665. [DOI] [PubMed] [Google Scholar]
- (19).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- (20).Lee W, Tonelli M, and Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy, Bioinformatics 31, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wijmenga SS, and van Buuren BNM (1998) The use of NMR methods for conformational studies of nucleic acids, Progress in Nuclear Magnetic Resonance Spectroscopy 32, 287–387. [Google Scholar]
- (22).Kauffmann AD, Kennedy SD, Zhao J, and Turner DH (2017) Nuclear Magnetic Resonance Structure of an 8 × 8 Nucleotide RNA Internal Loop Flanked on Each Side by Three Watson–Crick Pairs and Comparison to Three-Dimensional Predictions, Biochemistry 56, 3733–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bansal M, Bhattacharyya D, and Ravi B (1995) NUPARM and NUCGEN: software for analysis and generation of sequence dependent nucleic acid structures, Comput Appl Biosci 11, 281–287. [DOI] [PubMed] [Google Scholar]
- (24).Still WC, Tempczyk A, Hawley RC, and Hendrickson T (1990) Semianalytical treatment of solvation for molecular mechanics and dynamics, Journal of the American Chemical Society 112, 6127–6129. [Google Scholar]
- (25).The PyMOL Molecular Graphics System, Version 1.7.4.4, (2015) Schrödinger, LLC. [Google Scholar]
- (26).Das R, Karanicolas J, and Baker D (2010) Atomic accuracy in predicting and designing noncanonical RNA structure, Nature Methods 7, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lyskov S, Chou F-C, Conchúir SÓ, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ, and Das R (2013) Serverification of Molecular Modeling Applications: The Rosetta Online Server That Includes Everyone (ROSIE), PLOS ONE 8, e63906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sripakdeevong P, Cevec M, Chang AT, Erat MC, Ziegeler M, Zhao Q, Fox GE, Gao X, Kennedy SD, Kierzek R, Nikonowicz EP, Schwalbe H, Sigel RK, Turner DH, and Das R (2014) Structure determination of noncanonical RNA motifs guided by (1)H NMR chemical shifts, Nat Methods 11, 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sripakdeevong P, Kladwang W, and Das R (2011) An enumerative stepwise ansatz enables atomic-accuracy RNA loop modeling, Proc Natl Acad Sci U S A 108, 20573–20578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Johnston PD, and Redfield AG (1981) Nuclear magnetic resonance and nuclear Overhauser effect study of yeast phenylalanine transfer ribonucleic acid imino protons, Biochemistry 20, 1147–1156. [DOI] [PubMed] [Google Scholar]
- (31).Tanaka Y, Kojima C, Yamazaki T, Kodama TS, Yasuno K, Miyashita S, Ono A, Ono A, Kainosho M, and Kyogoku Y (2000) Solution Structure of an RNA Duplex Including a C−U Base Pair, Biochemistry 39, 7074–7080. [DOI] [PubMed] [Google Scholar]
- (32).Holbrook SR, Cheong C, Tinoco I Jr, and Kim S-H (1991) Crystal structure of an RNA double helix incorporating a track of non-Watson–Crick base pairs, Nature 353, 579. [DOI] [PubMed] [Google Scholar]
- (33).Spasic A, Kennedy SD, Needham L, Manoharan M, Kierzek R, Turner DH, and Mathews D (2018) Molecular Dynamics Correctly Models the Unusual Major Conformation of the GAGU RNA Internal Loop and with NMR Reveals an Unusual Minor Conformation, Rna. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chen X, McDowell JA, Kierzek R, Krugh TR, and Turner DH (2000) Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2), Biochemistry 39, 8970–8982. [PubMed] [Google Scholar]
- (35).Tolbert BS, Kennedy SD, Schroeder SJ, Krugh TR, and Turner DH (2007) NMR structures of (rGCUGAGGCU)2 and (rGCGGAUGCU)2: probing the structural features that shape the thermodynamic stability of GA pairs, Biochemistry 46, 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chen JL, Kennedy SD, and Turner DH (2015) Structural Features of a 3′ Splice Site in Influenza A, Biochemistry 54, 3269–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Jeffrey GA (1997) An Introduction to Hydrogen Bonding, Oxford University Press. [Google Scholar]
- (38).Kennedy SD, Kierzek R, and Turner DH (2012) Novel conformation of an RNA structural switch, Biochemistry 51, 9257–9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Keel AY, Rambo RP, Batey RT, and Kieft JS (2007) A general strategy to solve the phase problem in RNA crystallography, Structure 15, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Cate JH, and Doudna JA (1996) Metal-binding sites in the major groove of a large ribozyme domain, Structure 4, 1221–1229. [DOI] [PubMed] [Google Scholar]
- (41).Kieft JS, and Tinoco I Jr. (1997) Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexammine, Structure 5, 713–721. [DOI] [PubMed] [Google Scholar]
- (42).Gautheret D, Konings D, and Gutell RR (1995) G.U base pairing motifs in ribosomal RNA, Rna 1, 807–814. [PMC free article] [PubMed] [Google Scholar]
- (43).Lee JC, and Gutell RR (2004) Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs, J Mol Biol 344, 1225–1249. [DOI] [PubMed] [Google Scholar]
- (44).Stombaugh J, Zirbel CL, Westhof E, and Leontis NB (2009) Frequency and isostericity of RNA base pairs, Nucleic Acids Res 37, 2294–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chen JL, Dishler AL, Kennedy SD, Yildirim I, Liu B, Turner DH, and Serra MJ (2012) Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters, Biochemistry 51, 3508–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mizuno H, and Sundaralingam M (1978) Stacking of Crick Wobble pair and Watson-Crick pair: stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon-anticodon Wobble interaction, Nucleic Acids Res 5, 4451–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lu X-J, Bussemaker HJ, and Olson WK (2015) DSSR: an integrated software tool for dissecting the spatial structure of RNA, Nucleic Acids Research 43, e142–e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Popenda M, Szachniuk M, Blazewicz M, Wasik S, Burke EK, Blazewicz J, and Adamiak RW (2010) RNA FRABASE 2.0: an advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures, BMC Bioinformatics 11, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Yang J, Christianson LA, and Gellman SH (1999) Comparison of an HXH Three-Center Hydrogen Bond with Alternative Two-Center Hydrogen Bonds in a Model System, Organic Letters 1, 11–14. [Google Scholar]
- (50).Hammond NB, Tolbert BS, Kierzek R, Turner DH, and Kennedy SD (2010) RNA internal loops with tandem AG pairs: the structure of the 5’GAGU/3’UGAG loop can be dramatically different from others, including 5’AAGU/3’UGAA, Biochemistry 49, 5817–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kierzek R, Burkard ME, and Turner DH (1999) Thermodynamics of single mismatches in RNA duplexes, Biochemistry 38, 14214–14223. [DOI] [PubMed] [Google Scholar]
- (52).Davis AR, and Znosko BM (2010) Positional and neighboring base pair effects on the thermodynamic stability of RNA single mismatches, Biochemistry 49, 8669–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, and Zardecki C (2002) The Protein Data Bank, Acta Crystallogr D Biol Crystallogr 58, 899–907. [DOI] [PubMed] [Google Scholar]
- (54).Walter AE, Wu M, and Turner DH (1994) The stability and structure of tandem GA mismatches in RNA depend on closing base pairs, Biochemistry 33, 11349–11354. [DOI] [PubMed] [Google Scholar]
- (55).Znosko BM, Kennedy SD, Wille PC, Krugh TR, and Turner DH (2004) Structural features and thermodynamics of the J4/5 loop from the Candida albicans and Candida dubliniensis group I introns, Biochemistry 43, 15822–15837. [DOI] [PubMed] [Google Scholar]
- (56).Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, and Turner DH (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure, Proc Natl Acad Sci U S A 101, 7287–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Chen AA, and García AE (2013) High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations, Proceedings of the National Academy of Sciences 110, 16820–16825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Kührová P, Best RB, Bottaro S, Bussi G, Šponer J, Otyepka M, and Banáš P (2016) Computer Folding of RNA Tetraloops: Identification of Key Force Field Deficiencies, Journal of Chemical Theory and Computation 12, 4534–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kührová P, Banáš P, Best RB, Šponer J, and Otyepka M (2013) Computer Folding of RNA Tetraloops? Are We There Yet?, Journal of Chemical Theory and Computation 9, 2115–2125. [DOI] [PubMed] [Google Scholar]
- (60).Šponer J, Bussi G, Krepl M, Banáš P, Bottaro S, Cunha RA, Gil-Ley A, Pinamonti G, Poblete S, Jurečka P, Walter NG, and Otyepka M (2018) RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview, Chemical Reviews 118, 4177–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, and Assmann SM (2013) In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features, Nature 505, 696. [DOI] [PubMed] [Google Scholar]
- (62).Kutchko KM, Sanders W, Ziehr B, Phillips G, Solem A, Halvorsen M, Weeks KM, Moorman N, and Laederach A (2015) Multiple conformations are a conserved and regulatory feature of the RB1 5’ UTR, Rna 21, 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Cordero P, and Das R (2015) Rich RNA Structure Landscapes Revealed by Mutate-and-Map Analysis, PLOS Computational Biology 11, e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.