Abstract
Introduction:
Hypoxia is one of the intrinsic features of solid tumors and it is always associated with aggressive phenotypes, including resistance to radiation and chemotherapy, metastasis, and poor patient prognosis. Hypoxia manifests these unfavorable effects through activation of a family of transcription factors, Hypoxia- inducible factors (HIFs) play a pivotal role in the adaptation of tumor cells to hypoxic and nutrient-deprived conditions by upregulating the transcription of several pro-oncogenic genes. Several advanced human cancers share HIFs activation as a final common pathway.
Areas covered:
This review highlights the role and regulation of the HIF-1/2 in cancers and alludes on the biological complexity and redundancy of HIF-1/2 regulation. Moreover, this review summarizes recent insights into the therapeutic approaches targeting the HIF-1/2 pathway.
Expert opinion:
More studies are needed to unravel the extensive complexity of HIFs regulation and to develop more precise anticancer treatments. Inclusion of HIF-1/2 inhibitors to the current chemotherapy regimens has been proven advantageous in numerous reported preclinical studies. The combination therapy ideally should be personalized based on the type of mutations involved in the specific cancers and it might be better to include two drugs that inhibit HIF-1/2 activity by synergistic molecular mechanisms.
Keywords: Hypoxia, Hypoxia-inducible factors, HIF-1α, HIF-2α, HIF-3α, Hypoxia response elements, HIF-1 inhibitors, HIF-2 inhibitors, chemoresistance, radioresistance, angiogenesis
1. Introduction:
Hypoxia and necrosis are two characteristic features of solid tumors. Tumor cells develop hypoxia as a result of inadequate supply of oxygen (chronic hypoxia) or transient fluctuation in blood flow (acute hypoxia)1. The impairment in diffusion, the abnormalities in the tumor microvessels and the disturbed microcirculation, all lead to deficiency or even abolishment in oxygen supply in the tumor microenvironment2. Eventually, tumor cells become necrotic due to lack of oxygen. Hypoxia negatively influences the results of radiotherapy and chemotherapy and potentiates tumor metastasis. This is well supported by scientific evidence from early 1950s as in 1953, Gray and co-workers showed that the therapeutic response of tumor cells to radiation in a well-oxygenated medium is better than when the tumor cells are irradiated under anoxic conditions3. However, these findings could be explained based on the crucial role of oxygen in the success of radiation therapy. Oxygen interacts with the free electron in the DNA that formed because of the radiation therapy. This oxygen-free radicals interaction makes the radiation damage permanent, and this explains the radioresistance of hypoxic cells which have low level of oxygen4. The impact of hypoxia on chemoresistance can be attributed to several factors. First, low drug concentration in hypoxic cells as it accumulates in areas away from functioning blood vessels4. Second, most anticancer drugs target proliferating cells; however, hypoxic cells experience nutrient starvation and impaired cell proliferation compared to aerobic cells, and thus they have less effect on hypoxic cells4, 5. The unfavorable effects of hypoxia extend beyond its negative impact on the effectiveness of radiotherapy and chemotherapy, as hypoxic microenvironment is linked with genomic instability, genetic alterations, mutagenesis, and poor prognosis6. For instance, hypoxia is able to select for cells expressing mutations in both p53, a tumor suppressor gene, and Bcl-2, an apoptosis-inhibiting protein, in oncogenically transformed cells7. By doing so, hypoxia contributes to solid tumor malignancy and metastasis. Hypoxia either locally or systemically provokes acute (short-term) and long-term responses in a number of physiologically relevant genes and mediates the switch from aerobic to anaerobic energy metabolism. Hypoxia induces the transcription of over 40 genes involved in various cellular functions including vascular architecture and tone: vascular endothelial growth factor (VEGF), red blood cell production (erythropoietin, EPO), iron metabolism (transferrin), energy metabolism that mainly includes the glucose transport proteins (GLUT-1, GLUT-3), insulin like growth factor-2 (IGF-2), and pH regulation (carbonic anhydrase-9, CA IX)8, 9. The overall goal of these various activation mechanisms is to increase oxygen delivery and decrease oxygen consumption or activate alternative metabolic pathways that do not require oxygen10, 11. However, these oxygen-regulated genes bear a specific protein sequence known as hypoxia-responsive elements (HREs) that specifically bind to Hypoxia-inducible factors-1–3 (HIF-1–3). HIFs are intrinsic markers of tumor hypoxia in which their expression are increased in hypoxic cells as a means of an adaptive response to hypoxic environment and tumor progression and metastasis.
2. The three HIF isoforms: their structures and functions:
2.1. HIF-1α:
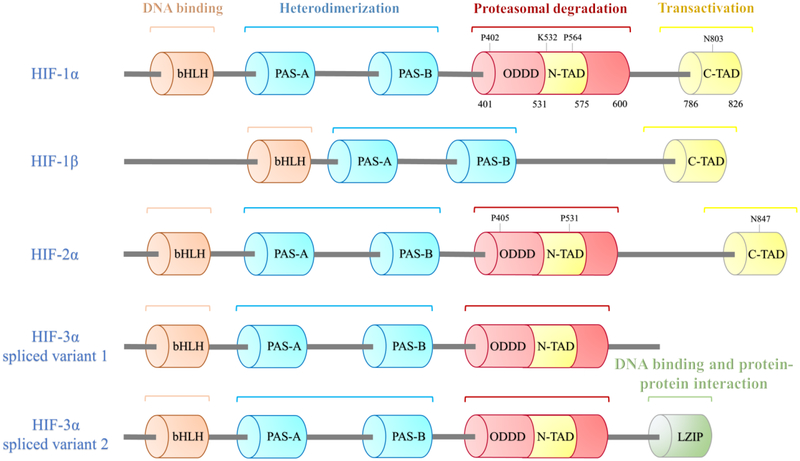
HIFs is a family of three members and they are heterodimers composed of an O2 sensitive α subunits (HIF-1α, or HIF-2α or HIF-3α) and an O2 insensitive HIF-1β subunit12. HIF-1α is the most well-characterized isoform of the HIFs, and most of the current knowledge on the structure and the regulation of these transcriptional factors are based on the studies of mammalian HIF-1α and to a lesser extent, HIF-2α. HIF-1α, a 120-kDa polypeptide subunit, heterodimerizes with HIF-1β which is a 91- to 94-kDa polypeptide subunit, to form the transcription factor HIF-1. Both are classified as a member of the basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) family of transcription factors because both HIF-1α and HIF-1β exhibit regions of amino acid sequence homology with Per (a Drosophila circadian rhythm protein) and Sim (a protein involved in Drosophila central nervous system development). However, they contain one bHLH domain and two PAS domains (PAS-A and PAS-B). The HLH and PAS domains mediate heterodimerization between HIF-1α and HIF-1β, while the basic regions, prior to the N terminus of the HLH domain are responsible for the binding of the HIF-1α/HIF-1β heterodimer to the HREs-DNA motif of target gene promoters (Figure 1)13, 14. In addition to these domains, HIF-1α has two transactivation domains (TAD), which are functionally distinct from the DNA-binding and heterodimerization domains. These TAD are located at the C-terminal half of the HIF-1α, not within their N-terminal portion, which contains their bHLH and PAS domains13. Both hypoxia responsiveness and transactivation capability of HIF-1α reside within its C-terminal region14, 15. These independent transactivation domains are: the NH2-terminal transactivation domain (N-TAD) localized to amino acids residues (531–575) in humans and the COOH terminal transactivation domain (C-TAD) localized to amino acids residues (786–826) in humans. Consistent with other previously described transactivation domains, both N-TAD and C-TAD are rich in acidic and hydrophobic residues16. The two transactivation domains are separated by (576–785) amino acid sequences in humans known as the inhibitory domain (ID) and this negative regulatory domain suppresses the transcriptional activity of N-TAD and C-TAD in normoxic conditions15, 16. The hypoxia-inducible transactivation capability of HIF-1 is mainly dependent on its HIF-1α subunit since HIF-1α is the oxygen labile subunit and contains the transactivation domains which are responsible for HIF-1α transcriptional activity. Whereas, HIF-1β is dispensable for the induction and serves as a dimerization partner14,17. The transcriptional activity of HIF-1α is regulated by the oxygen cellular tension16, 18. The C-TAD regulates the transactivation of target genes through co-activators recruitment, co-activators CBP and p30019. The N-TAD is located within a region known as the oxygen-dependent degradation domain (ODDD) which is localized to the human HIF-1α amino acid residues (400–600). The ODDD is recognized by von Hipple-Lindau (pVHL) only under normoxic conditions and pVHL is required for mediating the HIF-α degradation by ubiquitin-proteasome pathway15, 20. The ODDD contains two key proline residues targeted for hydroxylation in normoxic conditions, thus; the ODDD controls the activity and stability of the α subunit and the carboxyl terminal region of HIF-1α represents the protein stability domain15, 21, 22. The ODDD and N-TAD regions are present only in HIF-1α which is unlike HIF-1β where HIF-1β contains C-TAD only. Both HIF-1α and HIF-1β mRNAs are constitutively expressed in various human tissues, however; HIF-1α mRNA levels show steady-state expression regardless of cellular oxygen tension and are not induced by hypoxia in most tissue culture cell lines13, 23, 24. In contrast to what have been observed in vitro, HIF-1α mRNA levels increase significantly in response to hypoxia in brain, heart, kidney, lungs, and skeletal muscle in vivo25. Nonetheless, HIF-1α and HIF-1β are also differ in that HIF-1β protein is constitutively active and stable in both aerobic and hypoxic cells whereas HIF-1α protein is degraded rapidly under normoxic conditions by the ubiquitin-proteasome system11, 26. Thus, the stability of HIF-1α is the primary determinant for the regulation of HIF-1 activity23. Hence, HIF-1α is a conditionally regulated transcription factor whereas HIF-1β is a constitutively active transcription factor. HIF-1β is also different from HIF-1α in its ability to heterodimerize with other proteins such as Aryl hydrocarbon receptor (AhR) and SIM and this is owed to the fact that HIF-1β is identical to the Aryl hydrocarbon receptor nuclear translocator (ARNT) protein, which is required for the dioxin-AhR function13. In addition, HIF-1β can homodimerize in vitro unlike HIF-1α.
Figure 1.
Functional domain structures of HIF isoforms and their potential function. Columns represent different function domains. The hydroxylation sites are shown above the domain. HIF isoforms are bHLH–PAS proteins, they all have a bHLH motif, two PAS domains (PAS-A and PAS-B) for the heterodimerization between HIF-α and HIF-1β. Unlike HIF-1β, HIF-α subunits have an ODDD that mediates hydroxylation of two proline (P) residues and the acetylation of a lysine (K) followed by proteasomal degradation, a N-TAD within the ODDD and a C-TAD, which involved in transcriptional activation. The proline residues are conserved in HIF-1/2α subunits. Multiple HIF-3α splice variants exist, such as HIF-3α variant 1 without C-TAD and HIF-3α variant 2 with a LZIP, which mediates DNA binding and protein-protein interaction.
2.2. HIF-2α:
HIF-2α and HIF-3α are two closely related homologues of HIF-1α (Figure 1). HIF-2α was reported by groups of researchers around the same time and it was previously denoted by different names: Endothelial PAS domain protein 1 (EPAS1), HIF-1α-like factor (HLF), HIF-1α related factor (HRF) and member of the PAS superfamily-1 (MOP-1)27–30. HIF-2α shows 48% amino acid sequence homology overall with HIF-1α and it has a similar domain arrangement21, 27, 28. Although HIF-1α and HIF-2α share very similar characteristics including their abilities to heterodimerize with HIF-1β, binding to hypoxia inducible genes bearing HREs motif, and transcriptional activation, they are different in their expression levels in different tissues during different developmental stages21, 27–30. HIF-2α is expressed most abundantly in embryonic development stage and adult vascular endothelial cells, lungs, placenta and heart, whereas; HIF-1α has a ubiquitous expression in all analyzed mammalian tissues and cell types, specifically heart and kidney25, 28, 30, 31. HIF-1α and HIF-2α show different specificity in their transcriptional targets. For instance, HIF-1α effectively stimulates the expression of glycolytic enzymes, such as Lactate dehydrogenase-A (LDH-A) and CA IX. In contrast, HIF-2α acts more effectively on EPO gene and genes involved in iron metabolism while another group of genes, including VEGF and GLUT-1, are regulated by both HIF-1α and HIF-2α32, 33.
2.3. HIF-3α:
HIF-3α (long HIF-3α variant) was firstly reported as a new bHLH-PAS protein in mice with 662 amino acids and a molecular weight of 73 kDa34. In the same paper, Gu and co-authors showed that HIF-3α has 57% and 53% amino acid sequences identity in the bHLH-PAS domain with HIF-1α and HIF-2α respectively, and 61% identity in the ODDD with HIF-1α. The first human HIF-3α (667 amino acid sequence) (Figure 1) was reported in 2001 with a high similarity with human HIF-1α and HIF-2α in the bHLH and PAS domains, and it contains N-TAD but lacks C-TAD transactivation domain. Interestingly, another HIF-3α was showed to contain a leucine zipper (LZIP) domain in the place of the C-TAD, which mediates DNA binding and protein-protein interaction35, 36. The expression pattern of HIF-3α is distinct from that of HIF-1α and HIF-2α. HIF-3α is expressed in adult mice thymus, lung, brain, heart and kidney. Similar to HIF-1α and HIF-2α, HIF-3α is shown to heterodimerize with HIF-1β in vitro and in vivo34. A lot of evidences suggest that the expression and the activity of the mouse, rat and human HIF-3α are enhanced in response to hypoxia37, 38. Moreover, other evidence suggests that HIF-2α plays a role in upregulating the expression and the transcriptional activity of the HIF-3α mRNA39. Another spliced variant of HIF-3α (short HIF-3α variant) in mice was reported as a negative regulator of HIF mediated expression of hypoxia inducible genes and it was designated as Inhibitory PAS domain protein (IPAS)40. Nevertheless, IPAS is N-TAD and C-TAD deficient and it is unique in its ability to act as repressor of hypoxia induced transactivation function of HIF-1α and HIF-2α, which adds to the complexity in the regulation of hypoxia-inducible genes by the HIFs. IPAS exerts its inhibitory action through physical interaction with the bHLH/PAS domain of HIF-1α and this interaction results in the formation of dysfunctional complex with HIF-1α in the nucleus41. In contrast to HIF-1α, IPAS shows a restricted pattern of tissue expression with rigorous expression in Purkinje cells of the cerebellum and corneal epithelium of the mice. This extensive expression of IPAS in the cornea has its biological significance where IPAS counteracts the function of HIF-1α in the cornea and suppresses the corneal epithelium VEGF gene activation and neovascularization, and this is important in maintaining corneal transparency which is required for clear visions. Interestingly, IPAS is preferentially singled out HIFs without affecting other members of the bHLH/PAS transcription factor family such as AhR40. The selective negatively regulation of IPAS on HIF-1α could be exploited in compromising tumor angiogenesis and other pathological conditions associated with elevated activation of HIF-1α. Yuichi Makino et al. have shown that IPAS-expressing hepatoma cells produced tumors with slower growth rate and lower vascular density relative to the wild type cells40. However, it was shown that HIF-1α binds to the HREs in the promoter region of the IPAS gene and mediates hypoxia-dependent activation of gene transcription42. Likewise, Neonatal and embryonic PAS (NEPAS) is another splice variant of mouse HIF-3α, which is expressed predominantly during late embryonic and early postnatal stages. It acts as a negative regulator of HIF-1α and HIF-2α mediated gene expression where it can dimerize with HIF-1β and indirectly inhibit HIF-1α and HIF-2α activity43. Other multiple splice variants of the human HIF-3α have also been reported36, 44, 45, however; it was evident that all human HIF-3α variants are induced by hypoxia and the induction is mediated by HIF-1α but not by HIF-2α45. In contrast, it worth mentioning that contradicting evidence was published concerning the expression of both the mRNA and protein levels of adult and embryos zebrafish hif-3α where its expression was increased by hypoxia and it was not directly regulated by HIF-1α46. Moreover, the hypoxic regulation of HIF-3α mRNA levels is tissue-specific in zebrafish unlike in the mammals46.
4. HIF-1/2α regulation pathways:
4.1. Canonical mechanisms regulating HIF-1/2α:
4.1.1: Hydroxylation:
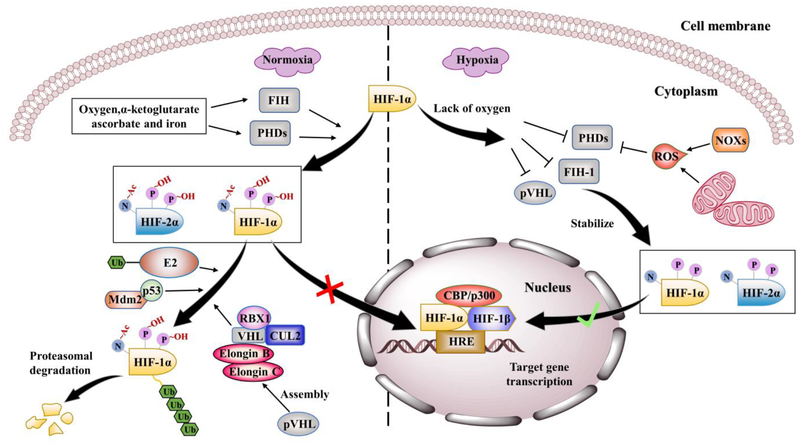
It is well established that transcriptional activity and stability of HIF-1/2α are tightly regulated by oxygen-dependent hydroxylation of their α subunits, where normoxia leads to quick degradation of HIF-1/2α transcript. Conversely, Hypoxia stabilizes HIF-1/2α via inactivation of pVHL, thus decreases HIF-α ubiquitination and proteasomal degradation (Figure 2)47, 48. pVHL mediates the assembly of a complex composed of VHL, Elongin B, Elongin C and a catalytic RING subunit (RBX1), which binds ubiquitin-conjugated E2 component, and it is organized on a cullin scaffold protein (CUL2) to accomplish ubiquitination of VHL-bound HIF-1/2α proteins. However, this ubiquitination step requires a posttranslational hydroxylation step of two separate consensus proline residues (P402 and P564) within the ODDD of the human HIF-1α and (P405 and P531) with in the ODDD of human HIF-2α subunits49, 50. Here, prolyl hydroxylases domain enzymes (PHD1–4), most prominently PHD2, represent an essential component of the canonical regulation pathways of HIF-1/2. They utilize molecular oxygen, α-ketoglutarate, ascorbate, and iron as substrates and generate CO2 and succinate as by-products. Beside the prolyl hydroxylases, Factor inhibiting HIF (FIH), an asparaginyl hydroxylase, regulates HIF-1/2α transcriptional activity under normoxic conditions by hydroxylating the asparagine 803 and asparagine 847 residues within the C-TADs of HIF-1α and HIF-2α respectively and blocking the interaction between HIF-1/2α and transcriptional co-activators CBP/p30019, 51. FIH is also an iron and α-ketoglutarate dependent dioxygenase and is activated only in the presence of molecular oxygen. Therefore, hypoxia inhabits the functions of PHDs and FIH, stabilizes HIF-1α and HIF-2α and their transcriptional activity. Parallel to limited oxygen availability caused by hypoxia, inhibition of PHDs and FIH, and inactivation of pVHL are also owed to the oxidation of PHDs and FIH, which contain Fe (II) in their active sites, by reactive oxygen species (ROS) generated in the mitochondria through electron transport Complex III52. However, other ROS generating mechanisms that might stabilize HIF-1/2α have been reported such as NADPH oxidase systems and specifically the Nox family of NADPH oxidases. NADPH oxidase 1 (also known as NOX1) mediates ROS production as a mechanism to upregulate HIF-1α53. Whereas, NOX1 and NADPH oxidase 4 (also known as NOX4) help maintain HIF-2α expression in renal carcinoma via ROS generation and therefore, contribute to renal carcinogenesis54.
Figure 2.
Schematic diagram of canonical mechanisms regulating HIF-1/2. Under normoxic conditions (left panel), PHDs and FIH hydroxylate HIF-1/2α on proline residues and asparagine residue and trigger formation of hydroxylated HIF-1/2α. In the meantime, pVHL mediates the assembly of a complex containing VHL, Elongin B, Elongin C, CUL2 and RBX1, which binds ubiquitin-conjugated E2 component to accomplish ubiquitination of HIF-1/2α proteins. P53 is able to recruit an E3 ubiquitin–protein ligase, Mdm2, to help the proteasomal degradation of HIF-1α mediated by an E2 and E3 ubiquitin ligase–pVHL complex. Besides, hydroxylation of the asparagine residue in the C-TAD of HIF-1/2, FIH blocks the essential interaction between HIF-1α and co-activators such as CBP/p300. However, under hypoxic conditions (right panel), pVHL, PHDs and FIH activities are inhibited by limited oxygen, and ROS generation mechanisms in mitochondria and others such as NADPH oxidases (NOXs), leading to the escape of HIF-1/2α from proteasomal degradation. Thus, the HIF transcriptional complex binds to the HREs motif on the DNA of target genes and activates the target gene transcription.
4.2. Non-canonical mechanisms regulating HIF-1/2:
Although the oxygen dependent pVHL pathway provides a major regulatory mechanism for HIF-1/2α protein stability, additional mechanisms do exist to fine-tune the HIF-1/2α protein stability, synthesis, and transcriptional activity in both hypoxia and normoxia. However, because our understanding for the biology of the oxygen-independent control of HIF-1α is becoming clearer than its isoform HIF-2α, the oxygen-independent control of HIF-2α is needed to be investigated in more details.
4.2.1: Acetylation and deacetylation:
Many posttranslational acetylation and deacetylation events have been reported to play a role in regulating both HIF-1/2α protein stability and transcriptional activity. However, conflicting data bring into question about the foundations of these regulation mechanisms and their roles in HIF-1/2α regulation require clarification. Multiple sites of the HIF-1α protein can be modified by lysine acetylation leading to different downstream effects. For example, acetylation within the ODDD is related to the pVHL-dependent HIF-1α degradation where Jeong et al. showed that pVHL binding is also promoted by acetylation of lysine (K532) residue of HIF-1α by direct binding of Arrest defective-1(ARD1), a protein acetyltransferase55. Thereafter, the ubiquitinated HIF-1α serves as the signal for degradation mediated by the 26S proteasome. However, the acetylation function of ARD1 is counteracted by the action of Metastasis-associated protein 1 (MTA1), where MTA1 induces the deacetylation of HIF-1α at K532R by increasing the expression of Histone deacetylase 1 (HDAC1) and thus enhances the transcriptional activity and stability of HIF-1α protein. In addition, the expression of MTA1 is strongly induced under hypoxic conditions and it is physically associated with HIF-1α when they are co-expressed56. Therefore, both MTA1 and HIF-1α are expected to have important roles in tumor metastasis and progression. Similarly, Zhu et al. showed that Metastasis-associated protein 2 (MTA2), another member of the MTA family, deacetylates HIF-1α and enhances its stability through interacting with HDAC1 in pancreatic carcinoma57. Yet, Arnesen et al. and Murray-Rust et al. reported that K532R mutation did not affect the interaction between the HIF-1α ODDD and human ARD1 (hARD1), and they concluded that hARD1 did not acetylate and destabilize HIF-1α58. Moreover, Fisher et al. showed that inhibition and overexpression of ARD1 did not affect basal HIF-1α levels or its response to hypoxia59. Whereas, acetylations of lysine (K709) and lysine (K674) in the carboxy terminal region of HIF-1α are related to HIF-1α/p300 interaction and HIF-1 transactivation. For example, Geng et al. demonstrated that p300, a component of the HIF-1 transcriptional complex, stabilizes HIF-1α via acetylating lysine (K709) residue in both normal and hypoxic conditions, and they showed that this acetylation is opposed by HDAC160. However, K674 in HIF-1α was shown to be acetylated primarily by the CBP/p300 -associated factor (PCAF) leading to the increase of HIF-1α protein levels and binding of p30061. Moreover, in the same study, Lim et al. showed that Sirtuin 1 (SIRT1), a NAD-dependent deacetylase, binds to HIF-1α, deacetylates it at K674 position, blocks p300 recruitment and consequently represses HIF-1 target genes. Conversely, Dioum et al. and Chen et al. demonstrated that HIF-2α can be acetylated at K385, K685, and K741 positions within its C terminus by CBP and selectively deacetylated by SIRT1 to augment HIF-2 signaling62, 63.
4.2.2: PI3K/AKT and MAPK/ERK pathways:
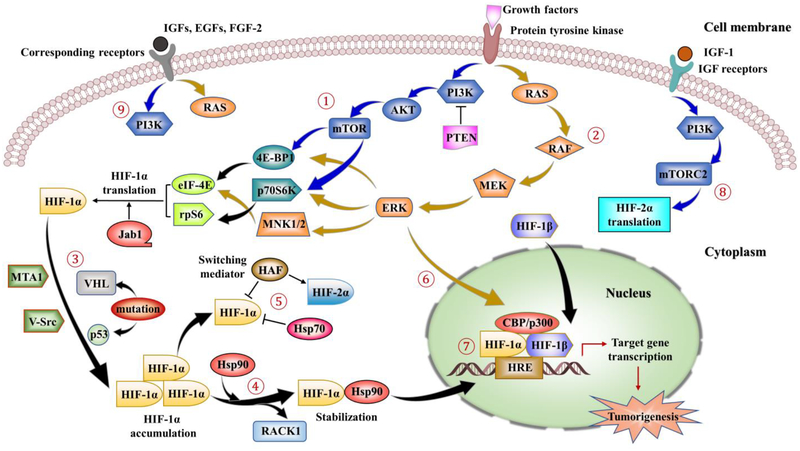
In non-hypoxic conditions, overexpression of HIF-1α could be achieved by growth factors stimulation where they are able to increase HIF-1α protein synthesis in a cell type-specific manner via activation of protein tyrosine kinases (PTKs) by mutation or ligand binding. This activation leads to signaling via the phosphatidylinositol 3- kinase (PI3K)/protein kinase B (AKT) pathway or mitogen-activated protein kinase (MAPK/ERK) pathway (Figure 3). The two pathways are affected by the tumor microenvironment favorable selection of cells with somatic mutations that activates oncogenes and inactivate tumor suppressor genes. These mutations drive cells through the cell cycle in an uncontrolled manner and prevent apoptosis. PI3K regulates HIF-1α protein synthesis via its target downstream serine threonine kinases, AKT and rapamycin-associated protein (FRAP/FKBP), which is also known as mammalian target of rapamycin (mTOR). FRAP/mTOR mediates its action via phosphorylation of two downstream effectors, the translational regulatory proteins eIF-4E binding protein 1 (4E-BP1) and p70 ribosomal protein S6 kinase (p70S6K). Phosphorylation of p70S6K phosphorylates its substrate, the ribosomal protein S6 (rpS6) and phosphorylation of 4E-BP1 disrupts its inhibitory interaction with eukaryotic translation initiation factor 4E (eIF-4E)64 and eventually these phosphorylation actions result in enhanced translation of HIF-1α mRNA into protein. While HIF-1α expression is dependent on both mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) in renal carcinoma cells, HIF-2α expression is solely dependent on mTORC265. The mTORC1 and mTORC2 represent two functionally distinct mTOR-containing signaling complexes, where mTORC1 phosphorylates p70S6K and 4E-BP1 and mTORC2 phosphorylates AKT. Similarly, the activation of the RAS-RAF-MEK-ERK pathway, where activated ERK1 and ERK2 phosphorylate and activate the MAP kinase-interacting kinases 1 and 2 (MNK1 and MNK2), which directly phosphorylate and stimulate the activity of eIF-4E and thus enhancing HIF-1 α translation. Accordingly, IGF-166, IGF-2, Epidermal growth factor (EGF), and Fibroblast growth factor 2 (FGF-2) as well as autocrine activation of the IGF-1 and IGF-2 receptors induce HIF-1α expression66, 67 and protein synthesis via activation of both the PI3K/AKT and MAPK pathways66, 68. Likewise, IGF-1 was reported to induce the transcription of HIF-2α via PI3K-mTORC2 system and promote vascularization in neuroblastoma69. Interestingly, HIF-1 is a transcriptional activator of IGF-2 which is the most highly upregulated gene in colon cancer70. However, increased activity of the HER-2 (also known as neu) tyrosine kinase receptor in breast cancer is associated with increased HIF-1α protein and VEGF mRNA expression via activation of a signal transduction pathway of PI3K/AKT71, 72. In prostate carcinoma and glioblastoma, mutation and inactivation of the tumor suppressor PTEN, which acts as a negative regulator of the PI3K via dephosphorylating the 3 position of phosphoinositides, is evident with elevated HIF-1α protein expressions73–75. In addition to PTEN, VHL and p53 are two tumor suppressor genes in which their loss of function mutations results in increased expression of HIF-1α. p53 is known to recruit the murine double minute 2 (Mdm2), an E3 ubiquitin–protein ligase to degrade HIF-1α76. Jun activating binding protein 1 (Jab1), also known as constitutive photomorphogenic-9 (COP9) signalosome subunit 5 (CSN5), counteracts the effect of p53 by competing with p53 to bind directly to the ODDD of HIF-1α. Jab1/CSN5 stabilizes HIF-1α by blocking hypoxia dependent p53-mediated degradation and promotes the transcriptional activity of HIF-1α77. However, activating mutations in oncogenes such as the previously mentioned MTA1 is associated with increased HIF-α protein level and VEGF protein level, progression and metastasis of the pancreatic cancer78. V-Src is another oncogene where its overexpression increases the expression of HIF-1α protein level without affecting its transactivation domain function, and thus increases the transcriptional activation of genes encoding VEGF and enolase 1 (ENO1)79 via increased activity of both the PI3K/AKT pathway and HIF-1α80.
Figure 3.
Main signaling involved in non-canonical pathways regulating HIF-1/2. ①-②: HIF-1α regulation could be initiated by growth factors stimulation via activation of protein tyrosine kinases (PTKs). This stimulation leads to downstream signaling activations via the PI3K–AKT–mTOR pathway (indicated by the blue arrows) and MAPK/ERK pathway (indicated by dark yellow arrows). mTOR and ERK signaling further mediate phosphorylation and activation of three downstream effectors, 4E-BP1, p70S6K and MNK1/2, followed by the activation of eIF-4E and rpS6. Finally, these phosphorylation actions result in enhanced translation of HIF-1α mRNA into protein. In addition, Jab1 may promote the transcriptional activity of HIF-1α, while PTEN, a negative regulator of the PI3K, could downregulate the HIF-1α expression. ③: Mutations of VHL, PTEN and p53 result in increased expression of HIF-1α, as well as activations of oncogenes, such as MTA1 and V-Src. ④: Hsp90 competes with the RACK1 for binding to the HIF-1α PAS domain since RACK1 destabilizes HIF-1α via proteasomal degradation pathway. HIF-1α further accumulates in the cytoplasm with the help of Hsp90 which assists the protein folding, prevents the degradation of HIF-1α by the proteasome and contributes its nuclear translocation. ⑤: HIF-1α is also regulated by HAF by inducing the degradation of HIF-1α, while increasing HIF-2α transactivation. Compared with Hsp90, Hsp70 could mediate HIF-1α degradation in the prolonged hypoxia but not HIF-2α. ⑥: ERK phosphorylates the co-activator CBP/p300 and increases HIF-1α/p300 complex formation. ⑦: In the nucleus, the HIF complex binds to the HREs motif on the DNA of target genes and activates the transcription, causing the upregulation of genes involved in cell proliferation, cell survival, angiogenesis and tumorigenesis. ⑧: IGF-1 activation could induce the transcription of HIF-2α via PI3K-mTORC2 system. ⑨: Stimulations of IGFs, EGFs and FGF-2 on corresponding receptors induce HIF-1α expression and protein synthesis through activation of both PI3K/AKT and MAPK pathways.
4.2.3: HIF-1/2α and other phosphorylation events:
In addition to regulating HIF-1α protein synthesis, the MAPK/ERK pathway is also implicated in HIF-1α transcriptional activation, where ERK is reported to phosphorylate the co-activator CBP/p300 and increase HIF-1α/p300 complex formation81. Moreover, ERK2 was reported to directly phosphorylate the C-terminal domain of HIF-1α at two distinct serine residues (S641 and S643) and by doing so it blocks HIF-1α nuclear exclusion by Chromosomal maintenance 1 (CRM1 also known as Exportin 1, XPO1) and enhances the nuclear accumulation and activity of HIF-1α82. However, Gradin et al. showed that HIF-1α and HIF-2α were phosphorylated at threonine 796 and threonine 844, respectively, under hypoxic conditions. This phosphorylation step increased the affinity of the interaction between HIF-1/2α and the transcriptional co-activator CBP/p30083. Moreover, Lancaster et al. showed that phosphorylation of HIF-1α at threonine 796 prevented the hydroxylation of asparagine 803 by FIH84. However, HIF-1α can be phosphorylated at several threonine and serine residues including: threonine 63 and serine 692 by Protein kinase A (PKA)85, serine 696 by Ataxia telangiectasia mutated serine/threonine kinase (ATM)86, and serine 668 by Cyclin-dependent kinase 1 (CDK1)87. Whereas these modifications by PKA, ATM and CDK1 resulted in increased HIF-1α stability, phosphorylation by Glycogen synthase kinase 3β (GSK3β) at serine 551, threonine 555, and serine 58988, or by polo-like kinase 3 (PLK3) at serine 576 and serine 657 increases HIF-1α degradation89. HIF-2α is phosphorylated at by casein kinase 1delta (CK1δ) at serine 383 and threonine 528, and as a result, CK1δ enhances the nuclear accumulation of HIF-2α via blocking the CRM1-dependent export of HIF-2α from the nucleus90. On the contrary, CK1δ phosphorylates HIF-1α at serine 247 and inhibits its activity via inhibiting its ability to associate with HIF-1β91,92.
4.2.4: Hsp90, HAF and Hsp70:
HIF-1α, like other bHLH-PAS protein such as AhR and Sim, is stably associated with the Heat shock protein 90 (Hsp90), a 90 kDa molecular chaperone that assists the covalent folding and assembly and controls stabilization of several client proteins. Hsp90 is involved in HIF-α subunit stabilization against non-pVHL mediated ubiquitination and proteasomal degradation and it aids HIF-1α accumulation in the nucleus. It binds to the PAS domain and stabilizes the HIF-α subunit predominantly under normoxic conditions, and it is displaced by HIF-β subunit primarily under hypoxia following nuclear translocation93, 94. Hsp90 competes with the Receptor of activated protein kinase C1 (RACK1) for binding to the HIF-1α PAS domain since the interaction between HIF-1α and RACK1 destabilizes HIF-1α via proteasomal degradation pathway in an oxygen-independent manner95. Moreover, Hsp90 appears to chaperone a proper conformation of the HIF-1 and facilitates its binding to HREs motif on the DNA of target genes96. Interestingly, whereas acute exposure to hypoxia resulted in increased HIF-1α accumulation, hypoxia could cause degradation and decrease of the accumulation of HIF-1α, but not HIF-2α protein levels in cell culture systems upon prolonged exposure. Mei Koh and colleagues have shown that selective oxygen-independent degradation of HIF-1α and promotion of HIF-2α transactivation are controlled by Hypoxia-associated factor (HAF; also known as SART1). HAF is an E3 ubiquitin ligase targets HIF-1α specifically for proteasomal degradation following prolonged hypoxia. HAF also binds to a different site on HIF-2α although it increases HIF-2α transactivation without causing its degradation. Thus, HAF represents a switching mediator for the hypoxic response of the cancer cell from HIF-1α-dependent to HIF-2α-dependent and provides an elucidation for enhanced tumor progression under prolonged hypoxia97, 98. However, Heat shock protein 70 (Hsp70) was reported to have a role in the prolonged hypoxia mediated HIF-1α degradation but not HIF-2α. Hsp70 recruits the ubiquitin ligase, carboxyl terminus of Hsp70-interacting protein (CHIP) to selectively promote the ubiquitination and proteasomal degradation of HIF-1α but not HIF-2α99. Consequently, the literatures suggest the opposite roles of Hsp70 and Hsp90 in regulating HIF-1α where Hsp70 and Hsp90 may be involved in degradation and stabilization of HIF-1α, respectively.
5. HIF-1/2 transcriptional co-activators:
Hypoxia inducible transcription of various hypoxia-responsive genes is not solely dependent on HIF-1/2 binding; however, optimal gene transcription requires HIF-1/2 to interact with adjacent, or sometimes distant, transcription factors and co-activating proteins to form multiprotein complexes100, 101. These multiple interacting transcription factor binding sites are different in each oxygen-regulated gene and each cell type, which convey some degree of tissue selectivity and contribute to the unique regulation of that gene with respect to its level of induction by hypoxia100. Moreover, they play a role in HIF-1/2 target gene specificity since many reported HIFs co-activators exhibit specific physical interaction with only one HIF isoform, and thus preferentially enhancing only one HIF isoform’s activity in the correct gene promoter context. Interestingly, being an essential part in HIF-1α or HIF-2α transcriptional complexes, they may provide additional therapeutic targets for anticancer treatments. However, several co-activators have been previously reported to enhance HIF-1/2 ability to activate target genes expressions through stabilizing the interaction of HIF-1/2 with the transcriptional co-activator CBP or p300. The activation of Endothelin-1 (ET-1) promoter expression by hypoxia in endothelial cells is one example where it has a three adjacent transcription factor binding sites: Activator Protein-1 (AP-1), GATA-binding factor 2 (GATA-2) and Nuclear Factor-1(NF-1), and they form a functional hypoxia responsive complex with HIF-1. AP-1, GATA-2, and NF-1 to stabilize the binding of HIF-1 and promote recruitment of CBP/p300 to the HIF-1 hypoxia response complex102. Hypoxia, along with anemia, induces the synthesis of the EPO in liver and kidney. The function of EPO enhancer in hypoxic liver tissue is shown to be modulated by sequences lying 3′ downstream of HREs which contains the HIF binding site. These 3′ sequences co-operate to permit the action of the inducible hypoxia at a distance from EPO enhancer and they have been identified as being necessary for hypoxic induction101, 103. Hepatocyte nuclear factor-4 (HNF4) was reported to bind to the region of EPO enhancer and was shown to physically interact with the HIF-1β subunit of the dimeric HIF-1 and cooperate with HIF-1 in the induction of the EPO gene under hypoxia104. Likewise, Upstream stimulatory factor-2 (USF2) was reported to be specifically essential in the hypoxic induction of HIF-2 target genes including EPO in hepatic and renal cancer cell lines. USF2 activates HIF-2 target genes by binding to HIF-2 target gene promoters/enhancers, interacting physically and functionally with the N-TAD of HIF-2α and recruiting co-activators CBP and p300 to form a multi-factoral transcription complex on HIF-2 target gene promoters105. Signal transducer and activator of transcription-3 (STAT3) has been shown to be involved in the HIF-1 and activation of HIF-1 target genes during hypoxia in renal and breast cancer cell lines, but not HIF-2-mediated hypoxic transcriptional response. STAT3 physically and functionally interacts with the N-TAD of HIF-1α, but not with HIF-2α, and increases the recruitment of CBP and p300 co-activators to HIF-1 target promoters such as VEGF and haptoglobin promoters106. Steroid receptor coactivator-1 (SRC-1) has been shown to form a complex with CBP and interacts with the transactivation domains of HIF-1α. Thus, SRC-1 functions as a co-activator for HIF-1α and enhances HIF-1α hypoxia-inducible transactivation potential107. Apurinic/apyrimidinic endonuclease-1 (APE1), also known as Redox effector factor 1 (Ref-1), has been shown to act as a co-activator for HIF-1α through its redox activity, mediating the DNA-binding activity of HIF-1 and the expression of HIF-1 downstream genes, including VEGF and CA IX23. The enhanced expression of the Lactate dehydrogenase A (LDH-A) promoter in hypoxic conditions is dependent on two domains or sequences close to HIF-1 binding sites. One sequence is located in an analogous position to one of the crucial regions in the EPO 3′ hypoxic enhancer, and the other domain has the motif of a cAMP response element (CRE)108. Although all these domains are crucial for oxygen-regulated expression of LDH-A promoter, they are not capable of driving hypoxic induction in isolation108. In similar fashion, the induction of VEGF and EPO enhancers and hence synthesis are governed by the participation of CBP/p300–HIF-1/2 complexes109. In general, the induction of hypoxia-responsive genes is not solely dependent on HIF-1 recognition, but rather dependent on a tripartite array of sites participation100. Hypoxia activates the expression of Tyrosine hydroxylase (TH), the rate-limiting enzyme in the biosynthesis of dopamine, via increasing the rate of transcription of the TH gene and it is regulated by a region of the proximal promoter that contains a number of cis-acting regulatory elements including AP-1, Activating protein 2 (AP-2) and HIF-1110, 111.
6. Targeting HIF-1/2 in cancer:
Taken together, the fact that solid tumors create their own characteristic hypoxic environment and the fact that hypoxia mediates the aggressive, metastatic and resistant forms of the tumors, it was rational to exploit the hypoxic tumor microenvironment to design and develop targeted therapies for cancers. Several approaches have been pursued including gene therapy, recombinant anaerobic bacteria, and the use of hypoxia-activated prodrugs112, 113. Another important route is to take advantage of the selective induction of the transcription factors HIF-1/2 under hypoxia. Ideally, selectivity could be achieved, and normal cells would remain unaffected. However, HIF-1/2 regulation pathways are highly complex and contain interconnected signaling cascades and overlapping mechanisms. This convolution manifests itself in two facts: 1) designing selective inhibitors of HIF-1/2 becomes highly challenging task, thus a growing number of HIF-1/2 targeting compounds that have been developed and classified based on their inhibitory mechanisms do not appear to inhibit the HIF-1/2 pathway as their specific target. 2) Considering, the regulatory mechanism of the transcriptional activation of HIF-1/2 which is dependent on a series of interrelated events, including elevated steady-state levels of HIF-1/2 α subunit via physiological stimulation as well as genetic alterations, HIF-1/2α nuclear translocation, heterodimerization with the β subunit, HREs-DNA binding, co-activators recruitment and formation of an active transcriptional complex, lots of efforts have been made to modulate and intervene each step of these HIF-1/2 regulating events. Many of the synthesized HIF-1/2 inhibitors act on HIF-1α, or HIF-2α, or both through direct mechanisms, while many other compounds and approved drugs have been shown to indirectly inhibit HIF-1/2 activity due to the connection between HIF-1/2 signaling and other cellular pathways such as the upstream pathways: the PI3K/AKT/mTOR pathways or the downstream cellular pathways: anti-VEGF-therapy. So far, many reviews on hypoxia- targeted anticancer agents have been reported114, 115. In this review, we describe a more updated set of new small molecule HIF inhibitors as potential anticancer agents listed in Table 1 and Figure 4.
Table 1.
Newly reported HIF inhibitors
| Inhibitor | Mechanism of Inhibition |
|---|---|
| Benzopyranyl triazole (1) | Increased HIF-1α hydroxylation |
| BIX01294 (2) | Increased HIF-1α hydroxylation/HIF-1α protein degradation |
| Cardenolides (3) | Decreased HIF-1 transcriptional activity |
| CRLX-101 (4) | Decreased HIF-1α protein accumulation |
| EZN-2208 (5) | Decreased HIF-1α protein expression and transcriptional activity |
| Glyceollins (6) | Decreased HIF-1α translation/stability |
| IDF-1174 (7) | Increased HIF-1α degradation |
| LBH589 (8) | Increased HIF-1α degradation |
| MPT0G157 (9) | Increased HIF-1α degradation |
| Vorinostat (10) | Decreased HIF-1/2α translation/nuclear localization/stability |
| NNC 55–0396 (11) | Decreased HIF-1α protein expression/translation/Increased HIF-1α hydroxylation |
| Kresoxim-methyl analogues (12) | Decreased HIF-1α stability |
| PT2385 (13) and PT2399 (14) | Decreased HIF-2 heterodimerization |
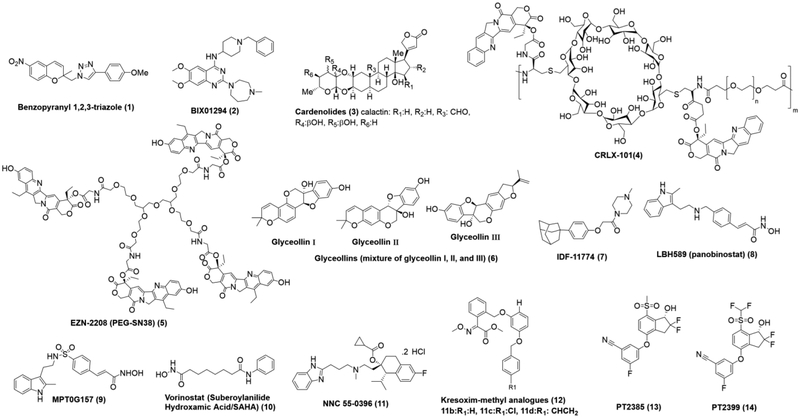
Figure 4.
Recently reported chemical structures of molecules inhibiting HIF-1/2.
Benzopyranyl 1,2,3-triazole (1) is a novel anticancer agent that was identified during in house chemical library screening and reported to work as HIF-1 inhibitor via increasing HIF-1α hydroxylation and subsequent ubiquitination and proteasomal degradation. It has an IC50 value of 2–24 nM in cancer cells. However, it decreases VEGF expression and angiogenesis in a dose-dependent manner. It has a synergistic effect with gefitinib, an Epidermal growth factor receptor (EGFR) inhibitor that is used clinically in lung and breast cancer with mutated and overactive EGFR, because the combination treatment inhibits tumor growth and angiogenesis in allograft model significantly116.
BIX01294 (2) is a diazepinquinazolin-amine derivative that was originally identified as an Euchromatic histone-lysine N-methyltransferase 2 (EHMT2)/G9a inhibitor during a chemical library screening of small molecules117. EHMT2 is an essential enzyme that catalyzes the methylation of histone H3 at lysine residue 9 to form H3K9me2, which is an epigenetic marker118, 119. EHMT2 is highly expressed in human cancer cells such as in neuroblastoma and glioblastoma brain cancers and BIX01294 was reported to decrease the proliferation of neuroblastoma cells. BIX01294 was also reported to decrease HIF-1 expression in HepG2 human hepatocellular carcinoma cells via increasing the hydroxylation of HIF-1α by increasing PHD2 and pVHL expressions and thus diminishing HIF-1α stability (at 1 μM range)120. However, it is noteworthy that G9a inhibition was reported to upregulate the HIF-1α and HIF-2α in breast cancer cells after using higher concentrations of BIX01294121.
Cardenolides (3) were isolated from the latex and fruits of Calotropis gigantea, a medicinal plant native to Southeast Asia used traditionally for the treatment of several diseases including cancers. Cardenolides are steroids with 25–26 carbon atoms and a five or six-membered lactone ring at C-17. Among them, Calactin inhibits HIF-1 transcriptional activity with an IC50 value of 21.8 nM and it shows cytotoxic potency on human breast cancer cell line MCF-7 with an IC50 value of 45.2 nM122.
CRLX-101(4) is a 20- to 30-nm diameter nanoparticle consisting of water-soluble cyclodextrins-based polymers that contain pendant carboxylate groups, camptothecin (CPT) and alternating repeat polyethylene glycol (PEG) blocks123. It was designed to accumulate into solid tumors and slowly release CPT in tumors over an extended period. In addition, it has the advantage of reducing the gastrointestinal (GI) toxicity related to CPT and improving patients’ compliance due to its favorable safety profile124. It was demonstrated that CRLX-101 has improved the efficacy of the chemoradiotherapy for locally advanced rectal cancer and when use as monotherapy or in combination therapy125. However, CRLX101 showed improved efficacy when used alone or in combination therapy with Bevacizumab in metastatic triple negative breast cancer mouse models126. CPT, the active moiety in CRLX101, is a potent inhibitor of Topoisomerase I (Topo-I) and HIF-1α. It was originally identified as HIF-1 inhibitor in 2002 using a cell-based high-throughput screening of approximately 2000 compounds. It inhibits HIF-1α protein accumulation, hypoxic induction of VEGF mRNA and protein expression in hypoxic U251 human glioma cells in a dose-dependent manner. CRLX101 showed higher efficacy and good tolerability in preclinical mouse model of gastric adenocarcinoma compared to its parent compound CPT and its synthetic analogues Irinotecan (CPT-11) and Topotecan (TPT)127. Currently, CRLX101 is being evaluated in phase II clinical trials as in combination therapy with other anticancer drugs for several tumor types124. The established dose for CRLX101 concluded from phase I clinical trial is 15 mg/m2 to be given intravenously every 2 weeks124. The preferential safety profile of CRLX101 was also confirmed in a pilot trial using the same recommended dose in patients with chemotherapy-refractory gastroesophageal cancer125, 128.
EZN-2208 (PEG-SN38, 5) is the multiarmed PEG backbone linked form of SN38. SN38 (7-ethyl-10-hydroxy-camptothecin) is the active metabolite of CPT-11 and it has anti Topo-I activity129. EZN-2208 was made to address the poor solubility of SN38 and it has the same anticancer potency as the native SN38 compound. Furthermore, it showed significantly greater antitumor activity than CPT-11 in several human tumor xenograft models including a CPT-111 resistant model130. EZN-2208 reduced the expression and transcriptional activity of HIF-1α in preclinical models131. Moreover, EZN-2208 in combination therapy with All-trans retinoic acid-arsenic trioxide (ATRA-ATO), the current standard of care for patients with acute promyelocytic leukemia (APL), was highly effective in treating patients with APL who develop resistance to ATO or patients carrying the PLZF-RARα fusion protein132. This synergistic effect was foreseeable since PML-RARα, the oncogenic fusion proteins found in 95% of the (APL) cases, behave as transcriptional co-activators of HIF-1α in leukemic promyelocytes of APL patients, and HIF-1α regulates leukemia-initiating cells (LICs) maintenance and leukemia progression133. In addition, ATRA increases HIF-1α levels in APL cells via induction of the transcriptional activation of PML-RARα134
Glyceollins (a mixture of glyceollin I, II, and III, 6) are a group of phytoalexins that are synthesized de novo in soybean via the stimulatory action of elicitors as a protective mechanism against microbial invasion, ultraviolet radiation, and chemical stressors135, 136. Glyceollins exhibit various biological functions including anti-estrogenic activity137, anti-contractile activity in vascular smooth muscle138, enhanced insulin sensitivity139 and anti-melanin synthesis activity140. Most importantly, Glyceollins regulate tumor growth and inhibit the expression of VEGF in cancer cells through regulation of HIF-1α141, 142. They perform their HIF-1 inhibitory action by two means: 1) they block HIF-1α translation via inhibiting the PI3K/AKT/mTOR pathway under hypoxic conditions. 2) They interfere with Hsp90 binding activity and thus decrease HIF-1α stability.
IDF-11774 (7) is another aryloxyacetylaminobenzoic acid analogue like LW6, which have been reported to inhibit the accumulation of HIF-1α via regulation of VHL expression143. LW6 was discovered based on a high-throughput cell-based reporter assay for in house chemical library in human hepatocellular carcinoma Hep3B cells144. IDF-11774 is an (E)-phenoxyacrylic amide derivative of LW6 where the oxyacetylamide linker has been replaced with an oxyacrylic amide to provide a more constrained conformation. In comparison to LW6, IDF-11774 showed superior potency in colorectal carcinoma HCT116 cells and improved aqueous solubility. IDF-11774 promotes HIF-1α degradation and inhibits its accumulation in colorectal cancer cells in vitro and in vivo145, 146. IDF-11774 was shown to significantly inhibit mitochondrial respiration and increase the intracellular oxygen tension, and thus promote the proteasomal degradation of HIF-1α146. Intriguingly, IDF-11774 was reported to act as Hsp70 inhibitor by binding to the allosteric pocket of Hsp70147. Moreover, IDF-11774 suppresses the hypoxia-induced mRNA expression of hypoxia target genes including VEGF and EPO145. Currently, IDF-11774 is being evaluated in phase I clinical trial for cancer therapy by the Korean Food and Drug Administration (KFDA)146.
LBH589, also known as (panobinostat, 8), is a pan histone deacetylase (HDAC) inhibitor that exhibits antitumor effects in vitro and in vivo in various hematologic malignancies and solid tumors, including diffuse large B-cell lymphoma, Hodgkin lymphoma, multiple myeloma, hepatocellular carcinoma, pancreatic cancer and non-small cell lung cancer148–151. Moreover, LBH589 showed anticancer activity against glioblastoma both in vitro and in vivo through interfering with HIF-1α stability and inducing its degradation. LBH589 disrupts Hsp90/Histone deacetylase 6 (HDAC6) complex because Hsp90 chaperone activity is regulated by its interaction and reversible acetylation by HDAC6 and inactivation of HDAC6 leads to Hsp90 hyperacetylation, its dissociation from p23 co-chaperone, and a loss of Hsp90 activity152. However, the newly synthesized HIF-1α molecules need to interact with the chaperone Hsp90 to complete its maturation and LBH589 treatment causes disruption of Hsp90-mediated folding of HIF-1α leading to its subsequent degradation by proteasome153, 154 However, LBH589 attenuates hypoxia in a much higher level of complexity due to its influence on multiple HDACs. For instance, in virtue of its pan inhibitory activity LBH589 could disrupt HIF-1α stability via inhibiting HDAC 1, 3, and 4 because they reported to bind directly to the ODDD of HIF-1α and induce deacetylation of lysine residues, and thus invoke HIF-1α stability and HIF-1 transactivation function in hypoxic conditions155, 156. Moreover, LBH589 could simultaneously target Histone deacetylase 7 (HDAC7) since HDAC7 was reported to form a complex with HIF-1α and p300 after co-translocation to the nucleus under hypoxic conditions and thus increases the transcriptional activity of HIF-1α157. However, it worthy to mention that a recently reported phase II clinical trial of combined LBH589 with Bevacizumab in patients with recurrent high-grade glioma (HGG) did not significantly improve the 6-month progression-free survival (PFS6) compared with historical controls of Bevacizumab monotherapy158.
MPT0G157 (9) an indole-3-ethylsulfamoylphenylacrylamide compound was developed based on the core structure of PXD101 (Belinostat) and LBH589, HDAC inhibitors159. MPT0G157 exerted potent inhibition against HDAC1, 2, 3 and 6 and subsequently resulted in hyper-acetylation of Hsp90 and of HIF-1α, leading to its subsequent degradation. In addition to its anti-inflammatory effect, MPT0G157 showed anticancer activity in vitro and in vivo particularly in human colorectal cancer (HCT116) cells160.
Vorinostat also known as (Suberoylanilide Hydroxamic Acid/SAHA, 10) is an FDA approved HDAC pan inhibitor used clinically for the treatment of cutaneous T cell lymphoma (CTCL). It displayed anti HIF-1/2 activity in hepatocellular carcinoma (HCC) (both in vitro and in vivo), osteosarcoma (OS), and glioblastoma (GBM) cell lines in vitro161. Different hypotheses are proposed to elucidate the suppression of hypoxia signaling mediated by Vorinostat. Although SAHA was shown to inhibit HIF-1/2 stabilization via direct acetylation of Hsp90 and increase HIF-1/2 degradation through a ubiquitin-dependent mechanism, other reports support that SAHA and other class I and II HDAC inhibitors enhance HIF-1/Hsp70 interactions and mediated HIF-1α degradation via non-ubiquitin-mediated degradation by the 20S proteasome162. Moreover, Vorinostat inhibits the interaction between HIF-1/2α and Importin and hence blocks HIF-1/2 α nuclear translocation161. However, Hutt et al have demonstrated that Vorinostat inhibits HIF-1α translation through Histone deacetylase 9 (HDAC9) and indirectly interferes with Eukaryotic translation initiation factor 3subunit G (eIF3G) in hepatocellular carcinoma163.
NNC 55–0396 (11) is a derivative of Mibefradil and it is a selective T-type Ca2+ channel blocker. It was identified based on a library screening study and cell growth assay with glucose or galactose medium, where it exhibited stronger inhibitory effects on cell growth in galactose medium than in glucose, an indication of modulating the mitochondrial function. NNC 55–0396 was found to significantly suppress the hypoxia-induced mitochondrial ROS generation and thus blocks HIF-1 activation and tumor growth and angiogenesis in vitro and in vivo164. Increased mitochondrial ROS generation in hypoxic cells triggers the HIF-1 induction which in turn activates the transcription of the genes encoding Pyruvate dehydrogenase kinase 1 (PDK1) and LDH-A in order to decrease the production of the ROS. This represents a pivotal regulating mechanism that functions to prevent excessive ROS production165. By the virtue of inducing PDK1 and LDH-A, HIF-1 can limit the delivery of the reducing equivalents NADH and FADH2 to the electron-transport chain, thus blocks the mitochondrial respiration. In addition, NNC 55–0396 increased the hydroxylation of HIF-1α and its subsequent ubiquitination and degradation. Moreover, NNC 55–0396 suppressed the de novo HIF-1 α synthesis via inhibition the phosphorylation of mTOR and p70S6K.
Kresoxim-methyl analogues (12) were designed and showed to inhibit hypoxia-induced HIF-1 transcriptional activation with IC50 values of 0.60–0.94 μM in human colorectal cancer (HCT116) cells. They work by increasing the intracellular oxygen tension under hypoxic condition, and thus promoting the ubiquitin-dependent proteasomal degradation of HIF-1α and impairing its accumulation166. Kresoxim-methyl is marketed synthetic analogue of strobilurin fungicide with the representative toxophore methyl methoxy-imino-acetate. It is a mitochondrial inhibitor specifically targets the cytochrome bc1 complex (complex III) of the mitochondrial electron transport chain by binding to its Q0 site, therefore; perturbs the ROS mitochondrial production responsible for hypoxia-dependent stabilization of HIF-1.
PT2385 (13) and PT2399 (14) are selective HIF-2α inhibitors that allosterically blocks its dimerization with HIF-1β. They are reported to have efficacy in clear cell renal cell carcinomas (ccRCC) cell lines and tumor xenografts. However, they exhibited preferable safety profile in xenograft models, with no adverse cardiovascular side effects, which have been associated with anti-VEGF receptor chemotherapeutic agents. They were identified using structure-based drug design using the PAS-B domains of HIF-2α and ARNT, and it was proved that they bind to the internal cavity of the HIF-2α PAS-B domain167. However, phase I clinical trial demonstrated that PT2385 has a favorable safety profile and is active in patients with heavily pretreated ccRCC168, 169.
7. Conclusion:
From the date of HIF-1 discovery by Semena and co-workers170, and from the first published report by Zhong et al171that linked HIF-1α overexpression in primary human cancers and their metastases, and this field has since expanded into a major area of cancer research. This partially due to the large number of clinical data that demonstrate an association between increased levels of HIF-1/2α proteins with increased both radiotherapy and chemotherapy resistance, cancer progression and patient mortality in many different human cancers. Beside intratumoral hypoxia, diverse sets of mechanisms have been reported to contribute to HIF-1/2 signaling and regulations, including, low–molecular weight signaling molecules such as ROS, cytokines and growth factors, tumor suppressor loss of function and oncogene gain of function. This complexity in the regulation pathways has made the process of rational design of HIF-1/2 α inhibitors very challenging. A growing number of HIF-1/2 α inhibitors have been identified, although; no selective HIF-1/2 α inhibitor has been clinically approved. In addition, several approved drugs have been reported to indirectly affect the HIF-1/2 α pathway, which reflects the extended connectivity between HIF-1/2 signaling and other cellular pathways. Therefore, targeting these pathways in cancer leads to secondary (indirect) inhibition of HIF activity. Future directions would be directed towards developing selective HIF-1/2 α inhibitors and addressing those combinations of drugs to which addition of a HIF-1/2 inhibitor will have additive or synergistic effects.
8. Expert opinion:
The reported literature regarding hypoxia and its playmakers HIFs provide a good insight into the complexity that characterize HIFs regulation pathways. The stability and degradation of HIF-1α is differentially affected by short or prolonged exposure to hypoxia, and the cross talks between HIF-1/2 and its affecter genes such as autocrine growth factors and other cellular stress response mediators are complicated. However, further research is needed to unravel the extensive complexity of HIF-1/2 regulation and to develop a more precise anticancer treatment. Many types of cancer such as breast, brain, cervical, colorectal, acute lymphoid, myeloid leukemias, melanoma, gastric cancer as well as liver, lung, ovarian, and pancreatic have been shown to respond poorly to chemotherapeutic agents and radiation therapy because of increased HIF-1/2 activation. HIF-1/2 activation contributes to cancer patients’ mortality through activation of genes encoding proteins that mediate vascularization, immune evasion, metabolic reprogramming, growth factor signaling, invasion, tumor progression, and metastasis. Consequently, there have been great interests in developing inhibitors targeting HIF-1/2. Even though there are large number of chemical compounds and approved drugs that have been shown to inhibit HIF-1/2 activation via a variety of molecular mechanisms, HIF-1/2 inhibition selectivity is far from fulfillment. This makes developing selective HIF-1/2 inhibitors as the ultimate goal for this area of research. Recurrent and metastatic triple-negative breast cancer tumors where paclitaxel or carboplatin is the established therapy are associated with overexpression of HIF-1α mRNA and protein and HIF-1 target genes. Consistently, co-administration of a HIF-1 inhibitor counteracts these negative pathological responses172. The addition of HIF-1/2 inhibitors to the current treatment have been proven advantageous in a wide range of preclinical studies including, acute and chronic myeloid leukemia, glioblastoma, colon, and liver cancer. Taken together, it is reasonable to argue that adding HIF-1/2 inhibitors to the cancer treatment regimens is a necessity. Ideally, the combination therapy using two or more chemotherapeutic agents should exert additive or synergistic effects and could preferably include multiple drugs that inhibit HIF-1/2 activity by different molecular mechanisms for long-term efficacy. For instances, large number of ccRCC show genetic mutation in VHL and TCEB1, which encodes Elongin C. These defects impair the classical regulation pathway of HIF-1/2, cause failure to degrade HIF-α subunits and result in its accumulation under normoxic conditions173. Furthermore, inhibition of HIF-2α has proven to be therapeutically useful in in mice bearing pVHL-defective clear-cell renal carcinoma174, and HIF-2α knockdown studies phenocopy the effects of pVHL reintroduction with respect to decreased expression of hypoxia-inducible genes and tumor growth suppression175. In contrast, HIF-1α is often deleted or mutated in a subtype of ccRCC characterized by chromosome 14q loss of function because HIF-1α gene (HIF1A) resides in this chromosome176. These data along with others suggest that HIF-2α might be the pathogenic driver in ccRCC and underscore the therapeutic potential of HIF-2α inhibitor for ccRCC treatment. PT2385 may particularly effective in the treatment of renal cell carcinomas especially with the positive results of its phase I clinical trial. However, PT2399 prolonged treatment causes cross-resistance in xenograft models, probably due to mutations in the binding site in HIF-2α177. Thus, co-administration of multiple drugs that inhibit HIF-2 activity by different molecular mechanisms may be necessary. However, drug-drug interaction, side effects and toxicity should be considered upon choosing the HIF-1/2 inhibitor as some of the currently available pan-HIF-1/2 inhibitors such as echinomycin and YC-1 have dose-limiting side effects. Overall, while it is still very challenging to develop more specific and thus selective HIF-1/2 inhibitors, with increased understanding of the HIFs structures, molecular mechanisms, and interactions with other signal transduction pathways, we certainly are seeing more and more lights in the tunnel for developing more efficacious HIF-1/2 inhibitors in the near future.
Article Highlights:
Solid tumors develop hypoxia because of deficiency or abolishment in oxygen supply in the tumor microenvironment. Hypoxia has negative impacts on radiotherapy and chemotherapy, and it potentiates tumor metastasis, genomic instability, and poor prognosis.
Hypoxia induces short and long-term responses in hypoxia-responsive elements bearing genes through its transcriptional factors HIFs, heterodimeric proteins that consist of two proteins, HIF-α and HIF-β. HIF-α stability is the primary determinant for the regulation of HIF activity. HIF-α has three closely related homologues, HIF-1α, HIF-2α and HIF-3α.
In normoxia, HIF-α is regulated by O2-dependent prolyl hydroxylation which facilitates its ubiquitylation by E3 ubiquitin-protein ligases containing the von Hippel–Lindau. Thereafter, the ubiquitinated HIF-α is signaled for proteasomal degradation. HIF-1/2α is overexpressed in human cancers as a result of intratumoral hypoxia, low–molecular weight signaling molecules such as reactive oxygen species, gain-of-function mutations in oncogenes and loss-of-function mutations in tumor-suppressor genes. HIF-1/2 regulation pathway is highly complex and contains interconnected signaling cascades and overlapping mechanisms.
The regulatory mechanism of the transcriptional activation of HIF-1/2 is dependent on a series of interrelated events, which includes elevated steady-state levels of HIF-α subunit, localization of HIF-α inside the nucleus, heterodimerization with the β subunit, hypoxia-responsive elements-DNA binding, increased transcriptional activity mediated by C-TAD and N-TAD of HIFα, as well as co-activators recruitments.
HIF-1/2α overexpression is linked with primary human cancers and their metastases, radiotherapy and chemotherapy resistance, and patient mortality. In preclinical and clinical studies, inhibition of HIF-1/2 activity has marked effects on angiogenesis and tumor growth. Thus, efforts are directed towards developing selective HIF-1/2 inhibitors.
Funding:
This research is supported by National Institutes of Health/National Cancer Institute grant R01CA193609 (to W Li). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH/NCI.
Reviewer Disclosures
One referee has received research funding and has consulted for Peloton. Peer reviewers on this manuscript have no other relevant financial relationships or otherwise to disclose.
List of Abbreviations used:
- 4E-BP1
eIF4E-binding protein 1
- AP-1
Activator Protein-1
- AP-2
Activating protein 2
- APL
Acute promyelocytic leukemia
- ATRA-ATO
All-trans retinoic acid-arsenic trioxide
- APE1
Apurinic/apyrimidinic endonuclease-1
- AhR
Aryl hydrocarbon receptor
- ARD1
Arrest defective-1
- ARNT
Aryl hydrocarbon receptor nuclear translocator
- ATM
Ataxia telangiectasia mutated
- CPT
Camptothecin
- CA IX
Carbonic anhydrase-9
- CHIP
Carboxyl terminus of Hsp70-interacting protein
- CK1δ
Casein kinase 1delta
- CRM1
Chromosomal maintenance 1
- CRE
cAMP response element
- ccRCC
Clear cell renal cell carcinomas
- COP9
Constitutive photomorphogenic-9
- CSN5
signalosome subunit 5
- CUL2
Cullin scaffold protein
- CTCL
Cutaneous T cell lymphoma
- CDK1
Cyclin-dependent kinase 1
- EGF
Epidermal growth factor
- EHMT2
Euchromatic histone-lysine N-methyltransferase 2
- eIF3G
Eukaryotic translation initiation factor 3subunit G
- eIF-4E
Eukaryotic translation initiation factor 4E
- EPAS1
Endothelial PAS domain protein 1
- EPO
Erythropoietin
- ET-1
Endothelin-1
- ENO1
Enolasen1
- EGFR
Epidermal growth factor receptor
- XPO1
Exportin 1
- FGF-2
Fibroblast growth factor 2
- FIH
Factor inhibiting HIF
- FRAP
Rapamycin-associated protein
- GATA-2
GATA-binding factor 2
- GBM
Glioblastoma
- GLUT-1
Glucose transport-1
- GLUT-3
Glucose transport-3
- GSK3β
Glycogen synthase kinase 3β
- HAF
Hypoxia-associated factor
- HNF4
Hepatocyte nuclear factor-4
- HCC
Hepatocellular carcinoma
- HGG
High-grade glioma
- HDAC
Histone deacetylase
- HDAC1
Histone deacetylase 1
- HDAC6
Histone deacetylase 6
- HDAC7
Histone deacetylase 7
- HDAC9
Histone deacetylase 9
- HIFs
Hypoxia- inducible factors
- HIF-1
Hypoxia-inducible factor-1
- HIF-2
Hypoxia-inducible factor-2
- HIF-3
Hypoxia-inducible factor-3
- HIF-1α
Hypoxia-inducible factor-1 α subunit
- HIF-2α
Hypoxia-inducible factor-2 α subunit
- HIF-3α
Hypoxia-inducible factor-3 α subunit
- HIF-1β
Hypoxia-inducible factor-1 β subunit
- HIF1A
HIF-1α gene
- HLF
HIF-1α-like factor
- HREs
Hypoxia-responsive elements
- HRF
HIF-1α related factor
- Hsp70
Heat shock protein 70
- Hsp90
Heat shock protein 90
- IGF-1
Insulin-like growth factor-1
- IGF-2
Insulin-like growth factor-2
- IPAS
Inhibitory PAS domain protein
- CPT-11
Irinotecan
- Jab1
Jun activating binding protein 1
- LDH-A
Lactate dehydrogenase A
- MAPK/ERK
Mitogen-activated protein kinase
- Mdm2
Murine double minute 2
- MNK1
MAP kinase interacting kinase 1
- MNK2
MAP kinase interacting kinase 2
- MOP-1
Member of the PAS superfamily-1
- MTA1
Metastasis-associated protein 1
- MTA2
Metastasis-associated protein 2
- mTOR
Mammalian target of rapamycin
- mTORC1
mTOR complex 1
- mTORC2
mTOR complex 2
- NEPAS
Neonatal and embryonic PAS
- NF-1
Nuclear Factor-1
- OS
Osteosarcoma
- PCAF
CBP/p300-associated factor
- p70S6K
p70 ribosomal protein S6 kinase
- PI3K
Phosphatidylinositol 3- kinase
- PHD 1–4
Prolyl hydroxylases domain enzymes 1–4
- PKA
Protein kinase A
- AKT
Protein kinase B
- PLK3
Polo-like kinase 3
- PDK1
Pyruvate dehydrogenase kinase 1
- PEG
polyethylene glycol
- ROS
Reactive oxygen species
- RACK1
Receptor of activated protein kinase C 1
- Ref-1
Redox effector factor 1
- rpS6
Ribosomal protein S6
- STAT3
Signal transducer and activator of transcription-3
- SIRT1
Sirtuin 1
- SRC-1
Steroid receptor coactivator-1
- SAHA
Suberoylanilide hydroxamic acid
- TH
Tyrosine hydroxylase
- Topo-I
Topoisomerase I
- TPT
Topotecan
- USF2
Upstream stimulatory factor-2
- VEGF
Vascular endothelial growth factor
- pVHL
von Hipple-Lindau
Footnotes
Declaration of Interest:
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References
- 1.Brown JM. Evidence for acutely hypoxic cells in mouse tumours, and a possible mechanism of reoxygenation. Br J Radiol 1979;52(620):650–6. [DOI] [PubMed] [Google Scholar]
- 2.ckel M, Vaupel P. Tumor Hypoxia: Definitions and Current Clinical, Biologic, and Molecular Aspects. JNCI: Journal of the National Cancer Institute 2001;93(4):266–76. [DOI] [PubMed] [Google Scholar]
- 3.Gray LH, Conger AD, Ebert M, Hornsey S, Scott OCA. The Concentration of Oxygen Dissolved in Tissues at the Time of Irradiation as a Factor in Radiotherapy. 1953;26(312):638–48. [DOI] [PubMed] [Google Scholar]
- 4.Brown JM. Tumor microenvironment and the response to anticancer therapy. Cancer Biol Ther 2002;1(5):453–8. [DOI] [PubMed] [Google Scholar]
- 5.Doktorova H, Hrabeta J, Khalil AM, Eckschlager TJBp. Hypoxia-induced chemoresistance in cancer cells: The role of not only HIF-1. 2015;159(2):166–77. [DOI] [PubMed] [Google Scholar]
- 6.Keysar SB, Trncic N, Larue SM, Fox MH. Hypoxia/reoxygenation-induced mutations in mammalian cells detected by the flow cytometry mutation assay and characterized by mutant spectrum. Radiation research 2010;173(1):21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 1996;379(6560):88–91. [DOI] [PubMed] [Google Scholar]
- 8.Fang J, Yan L, Shing Y, Moses MA. HIF-1α-mediated Up-Regulation of Vascular Endothelial Growth Factor, Independent of Basic Fibroblast Growth Factor, Is Important in the Switch to the Angiogenic Phenotype during Early Tumorigenesis. 2001;61(15):5731–35. [PubMed] [Google Scholar]
- 9.Semenza GL. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med 2001;7(8):345–50. [DOI] [PubMed] [Google Scholar]
- 10.Maxwell P HIF-1: an oxygen response system with special relevance to the kidney. J Am Soc Nephrol 2003;14(11):2712–22. [DOI] [PubMed] [Google Scholar]
- 11.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proceedings of the National Academy of Sciences of the United States of America 1995;92(12):5510–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev 2012;92(3):967–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. The Journal of biological chemistry 1996;271(30):17771–8. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Ko HP, Whitlock JP. Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1alpha. J Biol Chem 1996;271(35):21262–7. [DOI] [PubMed] [Google Scholar]
- 15.Pugh CW, O’Rourke JF, Nagao M, Gleadle JM, Ratcliffe PJ. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the alpha subunit. J Biol Chem 1997;272(17):11205–14. [DOI] [PubMed] [Google Scholar]
- 16.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. The Journal of biological chemistry 1997;272(31):19253–60. [DOI] [PubMed] [Google Scholar]
- 17.Wood SM, Gleadle JM, Pugh CW, Hankinson O, Ratcliffe PJ. The role of the aryl hydrocarbon receptor nuclear translocator (ARNT) in hypoxic induction of gene expression. Studies in ARNT-deficient cells. J Biol Chem 1996;271(25):15117–23. [DOI] [PubMed] [Google Scholar]
- 18.Luo G, Gu YZ, Jain S, Chan WK, Carr KM, Hogenesch JB, et al. Molecular characterization of the murine Hif-1 alpha locus. Gene expression 1997;6(5):287–99. [PMC free article] [PubMed] [Google Scholar]
- 19.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 2002;16(12):1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science (New York, NY) 2001;294(5545):1337–40. [DOI] [PubMed] [Google Scholar]
- 21.Iyer NV, Leung SW, Semenza GL. The human hypoxia-inducible factor 1alpha gene: HIF1A structure and evolutionary conservation. Genomics 1998;52(2):159–65. [DOI] [PubMed] [Google Scholar]
- 22.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A 1998;95(14):7987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem 1996;271(50):32253–9. [DOI] [PubMed] [Google Scholar]
- 24.Wenger RH, Rolfs A, Marti HH, Guenet JL, Gassmann M. Nucleotide sequence, chromosomal assignment and mRNA expression of mouse hypoxia-inducible factor-1 alpha. Biochemical and biophysical research communications 1996;223(1):54–9. [DOI] [PubMed] [Google Scholar]
- 25.Wiener CM, Booth G, Semenza GL. In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem Biophys Res Commun 1996;225(2):485–8. [DOI] [PubMed] [Google Scholar]
- 26.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. The Journal of biological chemistry 1995;270(3):1230–7. [DOI] [PubMed] [Google Scholar]
- 27.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes & development 1997;11(1):72–82. [DOI] [PubMed] [Google Scholar]
- 28.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U S A 1997;94(9):4273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flamme I, Frohlich T, von Reutern M, Kappel A, Damert A, Risau W. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1 alpha and developmentally expressed in blood vessels. Mechanisms of development 1997;63(1):51–60. [DOI] [PubMed] [Google Scholar]
- 30.Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, et al. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem 1997;272(13):8581–93. [DOI] [PubMed] [Google Scholar]
- 31.Wiesener MS, Jurgensen JS, Rosenberger C, Scholze CK, Horstrup JH, Warnecke C, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J 2003;17(2):271–3. [DOI] [PubMed] [Google Scholar]
- 32.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol 2005;25(13):5675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol 2003;23(24):9361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr 1998;7(3):205–13. [PMC free article] [PubMed] [Google Scholar]
- 35.Hara S, Hamada J, Kobayashi C, Kondo Y, Imura N. Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: suppression of HIF-mediated gene expression by HIF-3alpha. Biochem Biophys Res Commun 2001;287(4):808–13. [DOI] [PubMed] [Google Scholar]
- 36.Maynard MA, Qi H, Chung J, Lee EH, Kondo Y, Hara S, et al. Multiple splice variants of the human HIF-3 alpha locus are targets of the von Hippel-Lindau E3 ubiquitin ligase complex. J Biol Chem 2003;278(13):11032–40. [DOI] [PubMed] [Google Scholar]
- 37.Heidbreder M, Frohlich F, Johren O, Dendorfer A, Qadri F, Dominiak P. Hypoxia rapidly activates HIF-3alpha mRNA expression. FASEB J 2003;17(11):1541–3. [DOI] [PubMed] [Google Scholar]
- 38.Li QF, Wang XR, Yang YW, Lin H. Hypoxia upregulates hypoxia inducible factor (HIF)-3alpha expression in lung epithelial cells: characterization and comparison with HIF-1alpha. Cell Res 2006;16(6):548–58. [DOI] [PubMed] [Google Scholar]
- 39.Hatanaka M, Shimba S, Sakaue M, Kondo Y, Kagechika H, Kokame K, et al. Hypoxia-inducible factor-3alpha functions as an accelerator of 3T3-L1 adipose differentiation. Biol Pharm Bull 2009;32(7):1166–72. [DOI] [PubMed] [Google Scholar]
- 40.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 2001;414(6863):550–4. [DOI] [PubMed] [Google Scholar]
- 41.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J Biol Chem 2002;277(36):32405–8. [DOI] [PubMed] [Google Scholar]
- 42.Makino Y, Uenishi R, Okamoto K, Isoe T, Hosono O, Tanaka H, et al. Transcriptional up-regulation of inhibitory PAS domain protein gene expression by hypoxia-inducible factor 1 (HIF-1): a negative feedback regulatory circuit in HIF-1-mediated signaling in hypoxic cells. J Biol Chem 2007;282(19):14073–82. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita T, Ohneda O, Nagano M, Iemitsu M, Makino Y, Tanaka H, et al. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol 2008;28(4):1285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravenna L, Salvatori L, Russo MA. HIF3alpha: the little we know. FEBS J 2016;283(6):993–1003. [DOI] [PubMed] [Google Scholar]
- 45.Pasanen A, Heikkila M, Rautavuoma K, Hirsila M, Kivirikko KI, Myllyharju J. Hypoxia-inducible factor (HIF)-3alpha is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol 2010;42(7):1189–200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang P, Lu L, Yao Q, Li Y, Zhou J, Liu Y, et al. Molecular, functional, and gene expression analysis of zebrafish hypoxia-inducible factor-3alpha. Am J Physiol Regul Integr Comp Physiol 2012;303(11):R1165–74. [DOI] [PubMed] [Google Scholar]
- 47.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999;399(6733):271–5. [DOI] [PubMed] [Google Scholar]
- 48.Semenza GL. Involvement of hypoxia-inducible factor 1 in human cancer. Intern Med 2002;41(2):79–83. [DOI] [PubMed] [Google Scholar]
- 49.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science (New York, NY) 2001;292(5516):468–72. [DOI] [PubMed] [Google Scholar]
- 50.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J 2001;20(18):5197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pappalardi MB, McNulty DE, Martin JD, Fisher KE, Jiang Y, Burns MC, et al. Biochemical characterization of human HIF hydroxylases using HIF protein substrates that contain all three hydroxylation sites. Biochem J 2011;436(2):363–9. [DOI] [PubMed] [Google Scholar]
- 52.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem 2000;275(33):25130–8. [DOI] [PubMed] [Google Scholar]
- 53.Goyal P, Weissmann N, Grimminger F, Hegel C, Bader L, Rose F, et al. Upregulation of NAD(P)H oxidase 1 in hypoxia activates hypoxia-inducible factor 1 via increase in reactive oxygen species. Free Radic Biol Med 2004;36(10):1279–88. [DOI] [PubMed] [Google Scholar]
- 54.Block K, Gorin Y, Hoover P, Williams P, Chelmicki T, Clark RA, et al. NAD(P)H oxidases regulate HIF-2alpha protein expression. J Biol Chem 2007;282(11):8019–26. [DOI] [PubMed] [Google Scholar]
- 55.Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, et al. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell 2002;111(5):709–20. [DOI] [PubMed] [Google Scholar]
- 56.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO J 2006;25(6):1231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu S, Deng S, He C, Liu M, Chen H, Zeng Z, et al. Reciprocal loop of hypoxia-inducible factor-1alpha (HIF-1alpha) and metastasis-associated protein 2 (MTA2) contributes to the progression of pancreatic carcinoma by suppressing E-cadherin transcription. J Pathol 2018;245(3):349–60. [DOI] [PubMed] [Google Scholar]
- 58.Arnesen T, Kong X, Evjenth R, Gromyko D, Varhaug JE, Lin Z, et al. Interaction between HIF-1 alpha (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1 alpha. FEBS Lett 2005;579(28):6428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisher TS, Etages SD, Hayes L, Crimin K, Li B. Analysis of ARD1 function in hypoxia response using retroviral RNA interference. J Biol Chem 2005;280(18):17749–57. [DOI] [PubMed] [Google Scholar]
- 60.Geng H, Liu Q, Xue C, David LL, Beer TM, Thomas GV, et al. HIF1alpha protein stability is increased by acetylation at lysine 709. J Biol Chem 2012;287(42):35496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell 2010;38(6):864–78. [DOI] [PubMed] [Google Scholar]
- 62.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science 2009;324(5932):1289–93. [DOI] [PubMed] [Google Scholar]
- 63.Chen R, Xu M, Hogg RT, Li J, Little B, Gerard RD, et al. The acetylase/deacetylase couple CREB-binding protein/Sirtuin 1 controls hypoxia-inducible factor 2 signaling. J Biol Chem 2012;287(36):30800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes & development 2001;15(7):807–26. [DOI] [PubMed] [Google Scholar]
- 65.Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Biol Chem 2008;283(50):34495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukuda R, Hirota K, Fan F, Jung YD, Ellis LM, Semenza GL. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. The Journal of biological chemistry 2002;277(41):38205–11. [DOI] [PubMed] [Google Scholar]
- 67.Feldser D, Agani F, Iyer NV, Pak B, Ferreira G, Semenza GL. Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer research 1999;59(16):3915–8. [PubMed] [Google Scholar]
- 68.Agani F, Semenza GL. Mersalyl is a novel inducer of vascular endothelial growth factor gene expression and hypoxia-inducible factor 1 activity. Mol Pharmacol 1998;54(5):749–54. [DOI] [PubMed] [Google Scholar]
- 69.Mohlin S, Hamidian A, von Stedingk K, Bridges E, Wigerup C, Bexell D, et al. PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res 2015;75(21):4617–28. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, et al. Gene expression profiles in normal and cancer cells. Science (New York, NY) 1997;276(5316):1268–72. [DOI] [PubMed] [Google Scholar]
- 71.Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer research 2000;60(6):1541–5. [PubMed] [Google Scholar]
- 72.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol 2001;21(12):3995–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giri D, Ittmann M. Inactivation of the PTEN tumor suppressor gene is associated with increased angiogenesis in clinically localized prostate carcinoma. Hum Pathol 1999;30(4):419–24. [DOI] [PubMed] [Google Scholar]
- 74.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes & development 2000;14(4):391–6. [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A 1998;95(26):15587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes & development 2000;14(1):34–44. [PMC free article] [PubMed] [Google Scholar]
- 77.Bae MK, Ahn MY, Jeong JW, Bae MH, Lee YM, Bae SK, et al. Jab1 interacts directly with HIF-1alpha and regulates its stability. J Biol Chem 2002;277(1):9–12. [DOI] [PubMed] [Google Scholar]
- 78.Sun X, Zhang Y, Li B, Yang H. MTA1 promotes the invasion and migration of pancreatic cancer cells potentially through the HIF-alpha/VEGF pathway. J Recept Signal Transduct Res 2018;38(4):352–58. [DOI] [PubMed] [Google Scholar]
- 79.Jiang BH, Agani F, Passaniti A, Semenza GL. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer research 1997;57(23):5328–35. [PubMed] [Google Scholar]
- 80.Datta K, Bellacosa A, Chan TO, Tsichlis PN. Akt is a direct target of the phosphatidylinositol 3-kinase. Activation by growth factors, v-src and v-Ha-ras, in Sf9 and mammalian cells. The Journal of biological chemistry 1996;271(48):30835–9. [DOI] [PubMed] [Google Scholar]
- 81.Sang N, Stiehl DP, Bohensky J, Leshchinsky I, Srinivas V, Caro J. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J Biol Chem 2003;278(16):14013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mylonis I, Chachami G, Samiotaki M, Panayotou G, Paraskeva E, Kalousi A, et al. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1alpha. J Biol Chem 2006;281(44):33095–106. [DOI] [PubMed] [Google Scholar]
- 83.Gradin K, Takasaki C, Fujii-Kuriyama Y, Sogawa K. The transcriptional activation function of the HIF-like factor requires phosphorylation at a conserved threonine. J Biol Chem 2002;277(26):23508–14. [DOI] [PubMed] [Google Scholar]
- 84.Lancaster DE, McNeill LA, McDonough MA, Aplin RT, Hewitson KS, Pugh CW, et al. Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. Biochem J 2004;383(Pt. 3):429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bullen JW, Tchernyshyov I, Holewinski RJ, DeVine L, Wu F, Venkatraman V, et al. Protein kinase A-dependent phosphorylation stimulates the transcriptional activity of hypoxia-inducible factor 1. Sci Signal 2016;9(430):ra56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol Cell 2010;40(4):509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warfel NA, Dolloff NG, Dicker DT, Malysz J, El-Deiry WS. CDK1 stabilizes HIF-1alpha via direct phosphorylation of Ser668 to promote tumor growth. Cell Cycle 2013;12(23):3689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flugel D, Gorlach A, Michiels C, Kietzmann T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1alpha and mediates its destabilization in a VHL-independent manner. Mol Cell Biol 2007;27(9):3253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu D, Yao Y, Lu L, Costa M, Dai W. Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1alpha. J Biol Chem 2010;285(50):38944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pangou E, Befani C, Mylonis I, Samiotaki M, Panayotou G, Simos G, et al. HIF-2alpha phosphorylation by CK1delta promotes erythropoietin secretion in liver cancer cells under hypoxia. J Cell Sci 2016;129(22):4213–26. [DOI] [PubMed] [Google Scholar]
- 91.Kourti M, Ikonomou G, Giakoumakis NN, Rapsomaniki MA, Landegren U, Siniossoglou S, et al. CK1delta restrains lipin-1 induction, lipid droplet formation and cell proliferation under hypoxia by reducing HIF-1alpha/ARNT complex formation. Cell Signal 2015;27(6):1129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kalousi A, Mylonis I, Politou AS, Chachami G, Paraskeva E, Simos G. Casein kinase 1 regulates human hypoxia-inducible factor HIF-1. J Cell Sci 2010;123(Pt 17):2976–86. [DOI] [PubMed] [Google Scholar]
- 93.Katschinski DM, Le L, Heinrich D, Wagner KF, Hofer T, Schindler SG, et al. Heat induction of the unphosphorylated form of hypoxia-inducible factor-1alpha is dependent on heat shock protein-90 activity. The Journal of biological chemistry 2002;277(11):9262–7. [DOI] [PubMed] [Google Scholar]
- 94.Katschinski DM, Le L, Schindler SG, Thomas T, Voss AK, Wenger RH. Interaction of the PAS B domain with HSP90 accelerates hypoxia-inducible factor-1alpha stabilization. Cell Physiol Biochem 2004;14(4–6):351–60. [DOI] [PubMed] [Google Scholar]
- 95.Liu YV, Baek JH, Zhang H, Diez R, Cole RN, Semenza GL. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol Cell 2007;25(2):207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hur E, Kim HH, Choi SM, Kim JH, Yim S, Kwon HJ, et al. Reduction of hypoxia-induced transcription through the repression of hypoxia-inducible factor-1alpha/aryl hydrocarbon receptor nuclear translocator DNA binding by the 90-kDa heat-shock protein inhibitor radicicol. Mol Pharmacol 2002;62(5):975–82. [DOI] [PubMed] [Google Scholar]
- 97.Koh MY, Darnay BG, Powis G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1alpha, leading to its oxygen-independent degradation. Mol Cell Biol 2008;28(23):7081–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koh MY, Lemos R Jr., Liu X, Powis G. The hypoxia-associated factor switches cells from HIF-1alpha- to HIF-2alpha-dependent signaling promoting stem cell characteristics, aggressive tumor growth and invasion. Cancer research 2011;71(11):4015–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luo W, Zhong J, Chang R, Hu H, Pandey A, Semenza GL. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1alpha but Not HIF-2alpha. The Journal of biological chemistry 2010;285(6):3651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ebert BL, Bunn HF. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol Cell Biol 1998;18(7):4089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Semenza GL, Koury ST, Nejfelt MK, Gearhart JD, Antonarakis SE. Cell-type-specific and hypoxia-inducible expression of the human erythropoietin gene in transgenic mice. Proc Natl Acad Sci U S A 1991;88(19):8725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamashita K, Discher DJ, Hu J, Bishopric NH, Webster KA. Molecular regulation of the endothelin-1 gene by hypoxia. Contributions of hypoxia-inducible factor-1, activator protein-1, GATA-2, AND p300/CBP. The Journal of biological chemistry 2001;276(16):12645–53. [DOI] [PubMed] [Google Scholar]
- 103.Pugh CW, Ebert BL, Ebrahim O, Ratcliffe PJ. Characterisation of functional domains within the mouse erythropoietin 3’ enhancer conveying oxygen-regulated responses in different cell lines. Biochimica et biophysica acta 1994;1217(3):297–306. [DOI] [PubMed] [Google Scholar]
- 104.Huang LE, Ho V, Arany Z, Krainc D, Galson D, Tendler D, et al. Erythropoietin gene regulation depends on heme-dependent oxygen sensing and assembly of interacting transcription factors. Kidney Int 1997;51(2):548–52. [DOI] [PubMed] [Google Scholar]
- 105.Pawlus MR, Wang L, Ware K, Hu CJ. Upstream stimulatory factor 2 and hypoxia-inducible factor 2alpha (HIF2alpha) cooperatively activate HIF2 target genes during hypoxia. Mol Cell Biol 2012;32(22):4595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene 2014;33(13):1670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carrero P, Okamoto K, Coumailleau P, O’Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol Cell Biol 2000;20(1):402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Firth JD, Ebert BL, Ratcliffe PJ. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. The Journal of biological chemistry 1995;270(36):21021–7. [DOI] [PubMed] [Google Scholar]
- 109.Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A 1996;93(23):12969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Millhorn DE, Raymond R, Conforti L, Zhu W, Beitner-Johnson D, Filisko T, et al. Regulation of gene expression for tyrosine hydroxylase in oxygen sensitive cells by hypoxia. Kidney international 1997;51(2):527–35. [DOI] [PubMed] [Google Scholar]
- 111.Norris ML, Millhorn DE. Hypoxia-induced protein binding to O2-responsive sequences on the tyrosine hydroxylase gene. The Journal of biological chemistry 1995;270(40):23774–9. [DOI] [PubMed] [Google Scholar]
- 112.Baran N, Konopleva M. Molecular Pathways: Hypoxia-Activated Prodrugs in Cancer Therapy. Clin Cancer Res 2017;23(10):2382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Interesting review paper on prodrug approach in targeting hypoxia for cancer therapy.
- 113.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer 2011;11(6):393–410. [DOI] [PubMed] [Google Scholar]; **Highly interesting review paper on targeting hypoxia in cancer therapy.
- 114.Masoud GN, Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta pharmaceutica Sinica B 2015;5(5):378–89. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Highly interesting review paper on targeting hypoxia in cancer therapy.
- 115.Xia Y, Choi HK, Lee K. Recent advances in hypoxia-inducible factor (HIF)-1 inhibitors. Eur J Med Chem 2012;49:24–40. [DOI] [PubMed] [Google Scholar]; **Highly interesting review paper on targeting hypoxia in cancer therapy.
- 116.Park K, Lee HE, Lee SH, Lee D, Lee T, Lee YM. Molecular and functional evaluation of a novel HIF inhibitor, benzopyranyl 1,2,3-triazole compound. Oncotarget 2017;8(5):7801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 2007;25(3):473–81. [DOI] [PubMed] [Google Scholar]
- 118.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & development 2002;16(14):1779–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes & development 2011;25(8):781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oh SY, Seok JY, Choi YS, Lee SH, Bae JS, Lee YM. The Histone Methyltransferase Inhibitor BIX01294 Inhibits HIF-1alpha Stability and Angiogenesis. Mol Cells 2015;38(6):528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ho JC, Abdullah LN, Pang QY, Jha S, Chow EK, Yang H, et al. Inhibition of the H3K9 methyltransferase G9A attenuates oncogenicity and activates the hypoxia signaling pathway. PLoS One 2017;12(11):e0188051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parhira S, Zhu GY, Chen M, Bai LP, Jiang ZH. Cardenolides from Calotropis gigantea as potent inhibitors of hypoxia-inducible factor-1 transcriptional activity. J Ethnopharmacol 2016;194:930–36. [DOI] [PubMed] [Google Scholar]
- 123.Cheng J, Khin KT, Jensen GS, Liu A, Davis ME. Synthesis of linear, beta-cyclodextrin-based polymers and their camptothecin conjugates. Bioconjug Chem 2003;14(5):1007–17. [DOI] [PubMed] [Google Scholar]
- 124.Weiss GJ, Chao J, Neidhart JD, Ramanathan RK, Bassett D, Neidhart JA, et al. First-in-human phase 1/2a trial of CRLX101, a cyclodextrin-containing polymer-camptothecin nanopharmaceutical in patients with advanced solid tumor malignancies. Invest New Drugs 2013;31(4):986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Clark AJ, Wiley DT, Zuckerman JE, Webster P, Chao J, Lin J, et al. CRLX101 nanoparticles localize in human tumors and not in adjacent, nonneoplastic tissue after intravenous dosing. Proc Natl Acad Sci U S A 2016;113(14):3850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pham E, Yin M, Peters CG, Lee CR, Brown D, Xu P, et al. Preclinical Efficacy of Bevacizumab with CRLX101, an Investigational Nanoparticle-Drug Conjugate, in Treatment of Metastatic Triple-Negative Breast Cancer. Cancer research 2016;76(15):4493–503. [DOI] [PubMed] [Google Scholar]
- 127.Gaur S, Chen L, Yen T, Wang Y, Zhou B, Davis M, et al. Preclinical study of the cyclodextrin-polymer conjugate of camptothecin CRLX101 for the treatment of gastric cancer. Nanomedicine 2012;8(5):721–30. [DOI] [PubMed] [Google Scholar]
- 128.Chao J, Lin J, Frankel P, Clark AJ, Wiley DT, Garmey E, et al. Pilot trial of CRLX101 in patients with advanced, chemotherapy-refractory gastroesophageal cancer. J Gastrointest Oncol 2017;8(6):962–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao H, Rubio B, Sapra P, Wu D, Reddy P, Sai P, et al. Novel prodrugs of SN38 using multiarm poly(ethylene glycol) linkers. Bioconjug Chem 2008;19(4):849–59. [DOI] [PubMed] [Google Scholar]
- 130.Sapra P, Zhao H, Mehlig M, Malaby J, Kraft P, Longley C, et al. Novel delivery of SN38 markedly inhibits tumor growth in xenografts, including a camptothecin-11-refractory model. Clin Cancer Res 2008;14(6):1888–96. [DOI] [PubMed] [Google Scholar]
- 131.Sapra P, Kraft P, Pastorino F, Ribatti D, Dumble M, Mehlig M, et al. Potent and sustained inhibition of HIF-1alpha and downstream genes by a polyethyleneglycol-SN38 conjugate, EZN-2208, results in anti-angiogenic effects. Angiogenesis 2011;14(3):245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Coltella N, Valsecchi R, Ponente M, Ponzoni M, Bernardi R. Synergistic Leukemia Eradication by Combined Treatment with Retinoic Acid and HIF Inhibition by EZN-2208 (PEG-SN38) in Preclinical Models of PML-RARalpha and PLZF-RARalpha-Driven Leukemia. Clin Cancer Res 2015;21(16):3685–94. [DOI] [PubMed] [Google Scholar]
- 133.Lee KE, Simon MC. From stem cells to cancer stem cells: HIF takes the stage. Curr Opin Cell Biol 2012;24(2):232–5. [DOI] [PubMed] [Google Scholar]
- 134.Coltella N, Percio S, Valsecchi R, Cuttano R, Guarnerio J, Ponzoni M, et al. HIF factors cooperate with PML-RARalpha to promote acute promyelocytic leukemia progression and relapse. EMBO Mol Med 2014;6(5):640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Graham TL. Role of Constitutive Isoflavone Conjugates in the Accumulation of Glyceollin in Soybean Infected with Phytophthora megasperma, 1990.
- 136.Bhattacharyya MK, Ward EWB. Resistance, susceptibility and accumulation of glyceollins I–III in soybean organs inoculated with Phytophthora megasperma f. sp. glycinea. Physiological and Molecular Plant Pathology 1986;29(2):227–37. [Google Scholar]
- 137.Salvo VA, Boue SM, Fonseca JP, Elliott S, Corbitt C, Collins-Burow BM, et al. Antiestrogenic glyceollins suppress human breast and ovarian carcinoma tumorigenesis. Clin Cancer Res 2006;12(23):7159–64. [DOI] [PubMed] [Google Scholar]
- 138.Song MJ, Baek I, Jeon SB, Seo M, Kim YH, Cui S, et al. Effects of glyceollin I on vascular contraction in rat aorta. Naunyn Schmiedebergs Arch Pharmacol 2010;381(6):517–28. [DOI] [PubMed] [Google Scholar]
- 139.Park S, Ahn IS, Kim JH, Lee MR, Kim JS, Kim HJ. Glyceollins, one of the phytoalexins derived from soybeans under fungal stress, enhance insulin sensitivity and exert insulinotropic actions. J Agric Food Chem 2010;58(3):1551–7. [DOI] [PubMed] [Google Scholar]
- 140.Lee YS, Kim HK, Lee KJ, Jeon HW, Cui S, Lee YM, et al. Inhibitory effect of glyceollin isolated from soybean against melanogenesis in B16 melanoma cells. BMB Rep 2010;43(7):461–7. [DOI] [PubMed] [Google Scholar]
- 141.Lee SH, Lee J, Jung MH, Lee YM. Glyceollins, a novel class of soy phytoalexins, inhibit angiogenesis by blocking the VEGF and bFGF signaling pathways. Mol Nutr Food Res 2013;57(2):225–34. [DOI] [PubMed] [Google Scholar]
- 142.Lee SH, Jee JG, Bae JS, Liu KH, Lee YM. A group of novel HIF-1alpha inhibitors, glyceollins, blocks HIF-1alpha synthesis and decreases its stability via inhibition of the PI3K/AKT/mTOR pathway and Hsp90 binding. J Cell Physiol 2015;230(4):853–62. [DOI] [PubMed] [Google Scholar]
- 143.Lee K, Kang JE, Park SK, Jin Y, Chung KS, Kim HM, et al. LW6, a novel HIF-1 inhibitor, promotes proteasomal degradation of HIF-1alpha via upregulation of VHL in a colon cancer cell line. Biochem Pharmacol 2010;80(7):982–9. [DOI] [PubMed] [Google Scholar]
- 144.Lee K, Lee JH, Boovanahalli SK, Jin Y, Lee M, Jin X, et al. (Aryloxyacetylamino)benzoic acid analogues: A new class of hypoxia-inducible factor-1 inhibitors. J Med Chem 2007;50(7):1675–84. [DOI] [PubMed] [Google Scholar]
- 145.Naik R, Won M, Kim BK, Xia Y, Choi HK, Jin G, et al. Synthesis and structure-activity relationship of (E)-phenoxyacrylic amide derivatives as hypoxia-inducible factor (HIF) 1alpha inhibitors. J Med Chem 2012;55(23):10564–71. [DOI] [PubMed] [Google Scholar]
- 146.Ban HS, Kim BK, Lee H, Kim HM, Harmalkar D, Nam M, et al. The novel hypoxia-inducible factor-1alpha inhibitor IDF-11774 regulates cancer metabolism, thereby suppressing tumor growth. Cell Death Dis 2017;8(6):e2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ban HS, Naik R, Kim HM, Kim BK, Lee H, Kim I, et al. Identification of Targets of the HIF-1 Inhibitor IDF-11774 Using Alkyne-Conjugated Photoaffinity Probes. Bioconjug Chem 2016;27(8):1911–20. [DOI] [PubMed] [Google Scholar]
- 148.Assouline SE, Nielsen TH, Yu S, Alcaide M, Chong L, MacDonald D, et al. Phase 2 study of panobinostat with or without rituximab in relapsed diffuse large B-cell lymphoma. Blood 2016;128(2):185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wahaib K, Beggs AE, Campbell H, Kodali L, Ford PD. Panobinostat: A histone deacetylase inhibitor for the treatment of relapsed or refractory multiple myeloma. Am J Health Syst Pharm 2016;73(7):441–50. [DOI] [PubMed] [Google Scholar]
- 150.Jin N, Lubner SJ, Mulkerin DL, Rajguru S, Carmichael L, Chen H, et al. A Phase II Trial of a Histone Deacetylase Inhibitor Panobinostat in Patients With Low-Grade Neuroendocrine Tumors. Oncologist 2016;21(7):785–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lachenmayer A, Toffanin S, Cabellos L, Alsinet C, Hoshida Y, Villanueva A, et al. Combination therapy for hepatocellular carcinoma: additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J Hepatol 2012;56(6):1343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 2005;18(5):601–7. [DOI] [PubMed] [Google Scholar]
- 153.Aoyagi S, Archer TK. Modulating molecular chaperone Hsp90 functions through reversible acetylation. Trends in Cell Biology 2005;15(11):565–67. [DOI] [PubMed] [Google Scholar]
- 154.Yao ZG, Li WH, Hua F, Cheng HX, Zhao MQ, Sun XC, et al. LBH589 Inhibits Glioblastoma Growth and Angiogenesis Through Suppression of HIF-1alpha Expression. J Neuropathol Exp Neurol 2017;76(12):1000–07. [DOI] [PubMed] [Google Scholar]
- 155.Kim SH, Jeong JW, Park JA, Lee JW, Seo JH, Jung BK, et al. Regulation of the HIF-1alpha stability by histone deacetylases. Oncol Rep 2007;17(3):647–51. [PubMed] [Google Scholar]
- 156.Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. The Journal of biological chemistry 2011;286(44):38095–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kato H, Tamamizu-Kato S, Shibasaki F. Histone deacetylase 7 associates with hypoxia-inducible factor 1alpha and increases transcriptional activity. The Journal of biological chemistry 2004;279(40):41966–74. [DOI] [PubMed] [Google Scholar]
- 158.Lee EQ, Reardon DA, Schiff D, Drappatz J, Muzikansky A, Grimm SA, et al. Phase II study of panobinostat in combination with bevacizumab for recurrent glioblastoma and anaplastic glioma. Neuro Oncol 2015;17(6):862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mehndiratta S, Hsieh YL, Liu YM, Wang AW, Lee HY, Liang LY, et al. Indole-3-ethylsulfamoylphenylacrylamides: potent histone deacetylase inhibitors with anti-inflammatory activity. Eur J Med Chem 2014;85:468–79. [DOI] [PubMed] [Google Scholar]
- 160.Huang YC, Huang FI, Mehndiratta S, Lai SC, Liou JP, Yang CR. Anticancer activity of MPT0G157, a derivative of indolylbenzenesulfonamide, inhibits tumor growth and angiogenesis. Oncotarget 2015;6(21):18590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang C, Yang C, Feldman MJ, Wang H, Pang Y, Maggio DM, et al. Vorinostat suppresses hypoxia signaling by modulating nuclear translocation of hypoxia inducible factor 1 alpha. Oncotarget 2017;8(34):56110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1alpha. Mol Cell Biol 2006;26(6):2019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hutt DM, Roth DM, Vignaud H, Cullin C, Bouchecareilh M. The histone deacetylase inhibitor, Vorinostat, represses hypoxia inducible factor 1 alpha expression through translational inhibition. PLoS One 2014;9(8):e106224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim KH, Kim D, Park JY, Jung HJ, Cho YH, Kim HK, et al. NNC 55–0396, a T-type Ca2+ channel inhibitor, inhibits angiogenesis via suppression of hypoxia-inducible factor-1alpha signal transduction. J Mol Med (Berl) 2015;93(5):499–509. [DOI] [PubMed] [Google Scholar]
- 165.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J 2007;405(1):1–9. [DOI] [PubMed] [Google Scholar]
- 166.Lee S, Kwon OS, Lee C-S, Won M, Ban HS, Ra CS. Synthesis and biological evaluation of kresoxim-methyl analogues as novel inhibitors of hypoxia-inducible factor (HIF)-1 accumulation in cancer cells. Bioorganic & Medicinal Chemistry Letters 2017;27(13):3026–29. [DOI] [PubMed] [Google Scholar]
- 167.Wallace EM, Rizzi JP, Han G, Wehn PM, Cao Z, Du X, et al. A Small-Molecule Antagonist of HIF2alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma. Cancer research 2016;76(18):5491–500. [DOI] [PubMed] [Google Scholar]
- 168.Courtney KD, Infante JR, Lam ET, Figlin RA, Rini BI, Brugarolas J, et al. Phase I Dose-Escalation Trial of PT2385, a First-in-Class Hypoxia-Inducible Factor-2alpha Antagonist in Patients With Previously Treated Advanced Clear Cell Renal Cell Carcinoma. J Clin Oncol 2018;36(9):867–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature 2016;539(7627):107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3’ to the human erythropoietin gene. Proc Natl Acad Sci U S A 1991;88(13):5680–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer research 1999;59(22):5830–5. [PubMed] [Google Scholar]
- 172.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci U S A 2014;111(50):E5429–38. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 173.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature Genetics 2013;45:860. [DOI] [PubMed] [Google Scholar]
- 174.Kondo K, Kim WY, Lechpammer M, Kaelin WG, Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol 2003;1(3):E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zimmer M, Doucette D, Siddiqui N, Iliopoulos O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol Cancer Res 2004;2(2):89–95. [PubMed] [Google Scholar]
- 176.Monzon FA, Alvarez K, Peterson L, Truong L, Amato RJ, Hernandez-McClain J, et al. Chromosome 14q loss defines a molecular subtype of clear-cell renal cell carcinoma associated with poor prognosis. Mod Pathol 2011;24(11):1470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016;539(7627):112–17. [DOI] [PMC free article] [PubMed] [Google Scholar]