Abstract
Expression of the ciliary neurotrophic factor (CNTF) receptor essential ligand binding subunit, CNTF receptor α (CNTFRα), is induced in motor neurons and skeletal muscle following peripheral nerve lesion. We previously found muscle CNTFRα promotes motor neuron axon regeneration post-lesion. Both nerve lesion and CNTF administration activate motor neuron signal transducer and activator of transcription 3 (STAT3), a transcription factor implicated in axon growth, suggesting CNTF receptors may contribute to the lesion-induced STAT3 activation. However, many receptor types signal through STAT3, and if CNTF receptors contribute, motor neuron receptors seemed most likely to regulate motor neuron STAT3. To determine the role played by muscle CNTFRα, we used in vivo, muscle-specific CNTFRα depletion in mice and report here that this selectively impairs the second phase, sustained motor neuron STAT3 activation post-lesion. Thus, muscle CNTFRα makes an essential contribution to motor neuron STAT3 activation during axon regeneration and may thereby promote axon regeneration through such signaling. We also report CNTFRα quantitative PCR suggesting involvement of many denervated muscle types, as well as muscle damaged at the lesion site. The present data add to the evidence suggesting that enhancing muscle CNTFRα expression may promote motor neuron regeneration in trauma and disease.
Keywords: mouse, sciatic nerve, mlc1f-Cre, regeneration
Graphical Abstract
Using muscle-specific CNTFRα gene disruption and the mouse sciatic nerve crush model, this study shows muscle CNTFRα contributes to the pSTAT3 response in lesion motor neurons. Image shows pSTAT3 immunohistochemistry of spinal cord ventral horns from control (A,B) and CNTFRα-knockdown (C,D) mice with lesioned side on right (B,D).
INTRODUCTION
CNTF receptors contain CNTFRα, leukemia inhibitory factor receptor β (LIFRβ) and gp130 (Davis et al., 1993a). Unlike LIFRβ and gp130, CNTFRα is unique to CNTF receptors and required for all CNTF receptor signaling (Davis et al., 1993a; Elson et al., 2000). Therefore, CNTFRα disruption best reveals endogenous CNTF receptor function. Unconditional CNTFRα knockout mice die perinatally with motor neuron (MN) loss (DeChiara et al., 1995), precluding adult studies needed to identify neuroregenerative mechanisms to target in adult MN diseases and neurotrauma.
Adult MNs survive and regenerate axons following nerve crush (e.g., Lee et al., 2013), and are therefore used to identify mechanisms supporting adult MN maintenance and axon regeneration. Indirect data suggest CNTF receptor involvement. Neuromuscular CNTFRα expression is restricted to MNs and skeletal muscle (MacLennan et al., 1996; Lee et al., 1997) and nerve crush increases expression in both cell types during axon regeneration (Davis et al., 1993b; Helgren et al., 1994; MacLennan et al., 1999). CNTF administration accelerates post-lesion axonal regeneration (Sahenk et al., 1994).
Moreover, CNTF administration and nerve lesion both activate MN STAT3 through Tyr705 phosphorylation (MacLennan et al., 2000; Kirsch et al., 2003; Lee et al., 2004) and phospho-Tyr705 STAT3 (pSTAT3) activation promotes axon growth (Shin et al., 2012; Pernet et al., 2013; Luo et al., 2016). However, many receptor types signal through STAT3 (Levy and Darnell, 2002). Therefore, it was not known whether CNTF receptors contribute to STAT3 activation in regenerating MNs. Moreover, it seemed most likely that if they did contribute to MN STAT3 activation, involvement would be limited to MN CNTF receptors.
In the studies here, muscle-specific in vivo gene disruption and pSTAT3 immunohistochemistry were used to directly determine the contribution of muscle CNTFRα to post-lesion MN STAT3 activation. The work also includes CNTFRα quantitative real-time PCR (qRT-PCR) to access the potential involvement of different classes of denervated muscles, as well as muscle damaged at the lesion site.
MATERIALS and METHODS
Floxed CNTFRα (Lee et al., 2008) and mlc1f-Cre (Bothe et al., 2000; from Dr. Steven Burden [NYU]) mice were genotyped by tail biopsy PCR and maintained on a 129SvEvBrd background. CNTFRα-depleted/knockdown mice (mlc1f-Cre+/−/flxCNTFRα+/+) and non-floxed littermate controls (mlc1f-Cre+/−/flxCNTFRα−/−) were generated by mlc1f-Cre+/−/flxCNTFRα+/− X mlc1f-Cre−/−/flxCNTFRα+/− breeding (both sexes; 2.5–4 months old; housed 4/pressurized individually ventilated cage) and processed in parallel (blind to genotype) through all procedures (total mice used in experiments=37).
For the unilateral sciatic nerve crush, mice were anesthetized with 100 mg/kg ketamine; 20 mg/kg xylazine. Mid-thigh skin was opened and a 5 mm longitudinal cut in the biceps femoris exposed the underlying nerve which was freed from surrounding connective tissue where it passes superficial to the tendon of the obturator internus. The nerve was crushed for 10 seconds with Dumont #5 Biologic Tip forceps (Fine Science Tools). The biceps femoris muscle and overlying skin were then closed with 4–0 suture. The mice were then monitored until completely recovered from anesthetic, and daily thereafter.
University of Cincinnati IACUC approved all procedures (06–09-04–01) as conducted in accordance with U.S. NIH/OLAW regulations.
Statistics:
In each pSTAT3 immunohistochemistry experiment Student’s t-tests were used to compare the knockdown and littermate control mice. For each CNTFRα qRT-PCR analyses, an ANOVA was conducted to evaluate the main effects of knockdown and lesion. All statistics performed with GraphPad Prism 5 software.
TaqMan CNTFRα qRT-PCR (GAPDH normalization) was performed with an Applied Biosystems StepOnePlus Real-Time PCR system and ThermoFisher primers (CNTFRα; cat.#4331182 [Mm00516693_m1], GAPDH; cat.# 4331182 [Mm99999915_g1]).
Following overdose with avertin (20 mg/ml; IP), mice were perfused with 4°C saline and then 4°C 4% paraformaldehyde. Spinal cords were post-fixed in 4% paraformaldehyde overnight at 4°C and then cryoprotected for at least 48 hours in 30% sucrose with 2.5mM sodium azide before sectioning. Every sixth 30um coronal cryostat section was collected throughout the complete L5 cord, which contains the sciatic nerve projecting MNs (Janjua and Leong, 1984), and processed slide mounted with previously described immunohistochemistry details (Lee et al., 2004) using an antiserum (1:1,000) specifically recognizing Tyr705-phosphorylated STAT3 (Cell Signaling Technology; #9131), ABC amplification (Vector Laboratories; #PK-6100) and cyanine-3 tyramide (Perkin Elmer; #SAT704B).
Individuals unaware of genotype stereologically quantified (Hyman et al., 1998) MNs displaying a lesion-induced pSTAT3 response (lesion side MNs with nuclear pSTAT3 label much greater than any contralateral MNs; data in Figure 3). With very little contralateral label (e.g., Figure 4), this reliably measured relative pSTAT3 response, particularly given the substantial knockdown effect (Figure 3). This response was not observed outside the area of expected sciatic MNs, further indicating its specificity. MNs were identified by their characteristic location in the ventro-lateral horn and their large nuclei.
Figure 3. Muscle CNTFRα knockdown selectively reduces the second phase of lesion-induced STAT3 activation in motor neurons.

Muscle CNTFRα knockdown and littermate controls received a unilateral sciatic nerve crush. MNs with a lesion-induced pSTAT3 response were immunohistochemically quantified. The knockdown had no effect on STAT3 activation one day post-lesion (p=0.88, n=7), but substantially reduced activation one week post-lesion (**p=0.0014, n=5).
Figure 4. pSTAT3 immunohistochemistry examples.

Spinal cord ventral horns from control (A,B) and CNTFRα knockdown (C,D) mice 1 week post-lesion. Lesion on right (B,D); dorsal to top. The lesion-induced increase in MN nuclear pSTAT3 label was smaller with CNTFRα knockdown. Broken lines=grey matter borders not otherwise apparent since signal is highly specific to pSTAT3. Scale bar=100µm.
The pSTAT3 antiserum has been extensively characterized by western blot (e.g., Starr et al., 1997) and MN immunohistochemistry. In vivo CNTF-induced pSTAT3 MN labelling, under the immunohistochemical conditions used here, is: 1) blocked by preabsorption with the antigen peptide but not a non-phosphorylated control or a pseudorandomized phospho-Tyr peptide with the same residues (MacLennan et al., 2000), 2) induced by low picogram quantities of CNTF and leukemia inhibitory factor, which signal through STAT3 Tyr705-phosphorylation, but not much greater quantities of neurotrophin-3 and brain derived neurotrophic factor, which activate MN phospho-tyrosine-based signaling unrelated to pSTAT3 (MacLennan et al., 2000), and 3) blocked by a specific CNTF receptor antagonist (MacLennan et al., 2000).
Images (Figure 4) captured with a 12 megapixel DXM1200 camera and Nikon E800 microscope with a 10X (NA=0.45) lens were identically adjusted with CorelDRAW.
RESULTS
To specifically deplete muscle CNTFRα we crossed floxed CNTFRα mice (Lee et al., 2008) with mice carrying a Cre recombinase (Cre) gene inserted into the myosin light chain 1f locus (mlc1f-Cre) (Bothe et al., 2000), a locus expressed very selectively in skeletal muscle cells (Lyons et al., 1990). Mlc1f-Cre excises floxed sequence in all adult skeletal muscles tested, with no excision in brain, spinal cord, sciatic nerve, liver, heart or stomach (Bothe et al., 2000; Lee et al., 2013).
CNTFRα-depleted/knockdown mice (mlc1f-Cre+/−/flxCNTFRα+/+) were compared with non-floxed, littermate controls (mlc1f-Cre+/−/flxCNTFRα−/−). We have shown by semi-quantitative PCR that extensor digitorum longus (EDL) muscle CNTFRα expression is increased by sciatic nerve crush and inhibited in the muscle CNTFRα-depleted mice (Lee et al., 2013). Here we used real time PCR to quantitatively compare EDL, soleus and tibialis anterior muscles, which are denervated by the lesion but have different functions and fiber type compositions (Augusto et al., 2004).
All muscles displayed a dramatic lesion-induced increase in CNTFRα expression (Figure 1A-C; tibialis, F1,8=248.1, p<0.0001; EDL, F1,8=133.0, p<0.0001; soleus, F1,8=54.4, p<0.0001) with CNTFRα gene excision greatly reducing CNTFRα expression (Figure 1A-C; tibialis, F1,8=162.0, p<0.0001; EDL, F1,8=979.1, p<0.0001; soleus, F1,8=30.72, p=0.0005). The large but incomplete decrease is consistent with previous mlc1f-Cre work (Bothe et al., 2000) and may result from Cre expression in most but not all the many nuclei in each myofiber.
Figure 1. Muscle CNTFRα gene disruption greatly reduces CNTFRα expression in a wide range of denervated muscles.
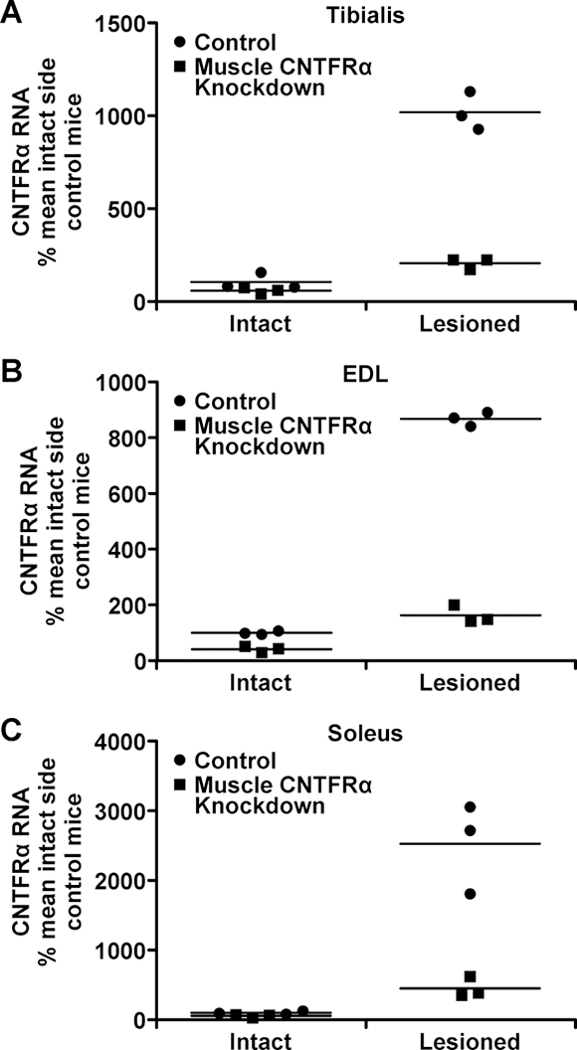
CNTFRα knockdown and littermate controls received a unilateral sciatic nerve crush. Twenty-four hours later, ipsilateral and contralateral tibialis anterior (A), EDL (B), and soleus (C) muscles were dissected for CNTFRα qRT-PCR. n=3/condition. All lesion and knockdown effects were significant (P<0.0005; text for statistics).
We also quantified CNTFRα expression in biceps femoris muscle which was longitudinally cut during the nerve crush procedure. It displayed an ~2-fold increase in CNTFRα expression (Figure 2A; F1,10=10.1, p=0.0099). The gene excision again greatly reduced CNTFRα expression (Figure 2A; F1,10=58.2, p<0.0001).
Figure 2. CNTFRα is induced in muscle damaged at the lesion site.
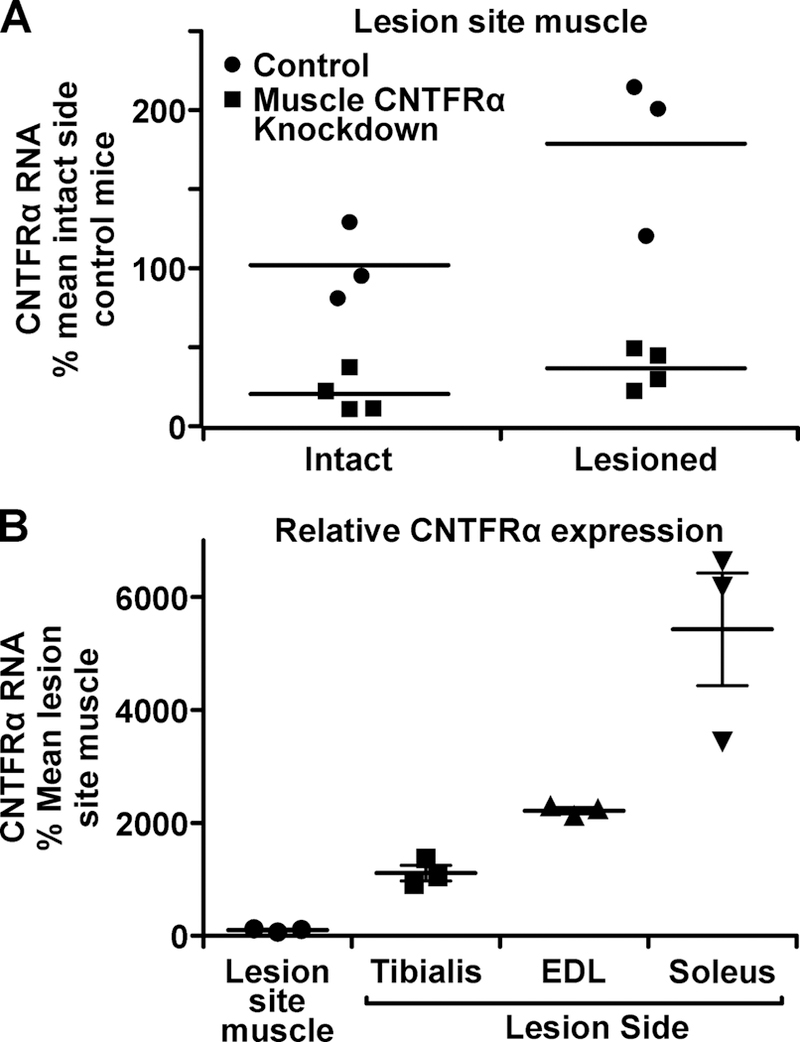
Muscle CNTFRα knockdown and littermate controls received a unilateral sciatic nerve crush. Twenty-four hours later biceps femoris muscles ipsilateral and contralateral to the lesion were dissected. CNTFRα RNA, quantified by qRT-PCR, is presented relative to control mouse intact side biceps femoris (A) and denervated muscles (B). Biceps femoris damage increases CNTFRα expression (p=0.0099) which is reduced by the knockdown (p<0.0001). n=4 knockdown, 3 controls.
We next determined whether muscle CNTFRα depletion affects the MN STAT3 activation which occurs during the ~2-week period of axon regeneration following sciatic nerve crush (Lee et al., 2004). This activation involves STAT3 Try705 phosphorylation and nuclear translocation (Levy and Darnell, 2002). To track this at a subcellular level in vivo, we used an extensively characterized (see Methods) phospho-Tyr705-STAT3-specific antiserum and quantified MNs displaying a lesion-induced increase in nuclear phospho-Try705-STAT3 (see Methods). As previously found (Lee et al., 2004), this pSTAT3 activation was restricted to MNs, like the pSTAT3 response seen after intraparenchymal CNTF injection (MacLennan et al., 2000). The muscle CNTFRα depletion had no effect on MN pSTAT activation at one day post-lesion (Figure 3; t=0.15, p=0.88, n=7, df=6) but substantially decreased the MN pSTAT3 activation 1 week post-lesion (Figures 3,4; t=7.83, p=0.0014, n=5 df=4).
DISCUSSION
We previously found muscle-specific CNTFRα disruption surprisingly has no effect on muscle itself, but instead impairs MN axon regrowth and motor recovery following nerve lesion (Lee et al., 2013) indicating muscle CNTFRα makes an essential/non-redundant contribution to these functions. Here we show the same muscle CNTFRα depletion decreases lesion-induced MN STAT3 activation. This form of STAT3 activation has been implicated in axon growth (Shin et al., 2012; Pernet et al., 2013; Luo et al., 2016). Therefore, together the data raise the possibility that muscle CNTFRα’s promotion of axon regeneration and motor recovery may at least partly result from its contribution to MN STAT3 activation.
Sciatic nerve lesion induces an essentially immediate pSTAT3 increase in lesion site axons that reaches spinal MN nuclei 24 hrs later (Lee et al., 2004) as pSTAT3 acts as a retrograde transcription factor (Ben-Yaakov et al., 2012). While this transient, variable lesion site response does not lend itself to quantification, we did not observe any reliable decrease in it with muscle CNTFRα depletion (data not shown). Although not conclusive, this is consistent with the lack of CNTFRα depletion effect on MN nuclear pSTAT3 1 day post-lesion (Figure 3) since the axonal pSTAT3 response, while maximal at 1 day post-lesion, is gone by 3 days post-lesion (Lee et al., 2004). In other words, the data suggest the axonal response contributes to the 1 day post-lesion MN pSTAT3 increase but does not directly contribute to the 1 week post-lesion increase. Here we find muscle CNTFRα depletion does not affect MN STAT3 activation 1 day post-lesion but substantially reduces activation at 1 week (Figure 3). Together the data suggest a rapid pSTAT3 response starting in lesion site axons contributes to an early MN nuclear pSTAT3 increase independent of muscle CNTFRα expression, but a second phase, longer lasting MN nuclear pSTAT3 response is substantially dependent on muscle CNTFRα.
Unconditional knockout work indicates the ligand CNTF is essential for the initial phase of post-lesion MN STAT3 activation but is not required for the second longer lasting phase (Kirsch et al., 2003). It has been suggested that Schwann cell CNTF released by the lesion produces an initial surge of CNTF receptor activation mediating early MN responses (Sendtner et al., 1992b). Therefore, current data suggest two phases of MN STAT3 response with the initial phase at least partly resulting from CNTF activation of STAT3 in lesion site axons leading to the MN nuclei response 1 day post-lesion. This phase is independent of muscle CNTFRα and presumably involves MN expressed CNTFRα. In contrast, the second phase STAT3 response involves muscle CNTFRα and a CNTF receptor ligand other than CNTF. The complex of cardiotrophin-like cytokine/cytokine-like factor-1 (CLC/CLF) also serves as a CNTF receptor ligand (Elson et al., 2000) and may participate in this second phase response. Unfortunately, conditional genetic disruption techniques will need to be developed to address this because complete unconditional knockout of CLF (Elson et al., 2000) or CLC (Zou et al., 2009) leads to perinatal death.
Muscle CNTFRα may promote muscle CNTF receptor signaling leading to release of unidentified muscle factor(s) acting directly or indirectly on MNs to activate STAT3. Moreover, following nerve lesion, muscle CNTFRα is released in a soluble, functional form (Davis et al., 1993b) that could potentially, over time, enhance MN CNTF receptor signaling, including STAT3 activation, by diffusing to regenerating axons for retrograde transport to MN soma, or diffusing directly to the soma. In vitro data also suggest muscle CNTFRα is released as a CNTFRα/CLC complex able to activate CNTF receptor signaling including STAT3 (Plun-Favreau et al , 2001).
The separation between the mid-thigh lesion and lower limb denervated muscles may seem to argue against factors from these muscles acting directly on axotomized MNs. However, the rapid, dramatic increase in muscle CNTFRα expression is sustained for at least 10 days (Helgran et al., 1994) suggesting CNTFRα and/or CNTFRα-dependent factors from denervated muscles could act on MNs, particularly with several days to diffuse to regenerating axons and contribute to the second phase of STAT3 activation.
The CNTFRα qRT-PCR data suggest a wide variety of denervated muscles are involved. They also identify, for the first time, the damaged lesion site muscle as another potential source of CNTFRα contributing to MN STAT3 activation and the muscle CNTFRα-dependent axon regeneration and motor recovery (Lee et al., 2013). This muscle is analogous to lesion site muscle invariably damaged clinically with accidental nerve lesion. Although the CNTFRα expression is much less than in denervated muscles (Figure 2B), the damage-induced increase in expression, and the muscles’ close proximity to the crushed axons, both suggest it may participate.
While CNTFRα depletion substantially reduces the second phase of MN STAT3 activation it does not eliminate it. The activation may be completely dependent on muscle CNTFRα with the remaining 20% of muscle CNTFRα expression sustaining the STAT3 activation observed. Alternatively, CNTFRα expressed by MNs may contribute, as could other STAT3-signaling receptor types.
Sciatic nerve lesion also increases pSTAT3 in dorsal root ganglion neurons (Lee et al., 2004). Unfortunately, we were unable to establish a quantitative assay for mouse dorsal root ganglion pSTAT3 (likely due to variability in fix, antibody penetration, etc. with such small tissues). However, it certainly remains possible that muscle CNTFRα contributes to this pSTAT3 activation.
The present data and our previous work (Lee et al., 2013) suggest muscle CNTFRα upregulation promotes MN regeneration by indirectly regulating MN signaling. Therefore, enhancing muscle CNTFRα expression may be worth considering as a therapeutic to promote MN regeneration in trauma and disease. In contrast to MN gene expression, skeletal muscle gene expression can be enhanced in humans with gene therapy techniques currently approved for market (Ylä-Herttuala, 2012).
Acknowledgements:
Supported by NIH grants NS35224 and NS052700 to A.J.M.
Abbreviations:
- CLC
cardiotrophin-like cytokine
- CLF
cytokine-like factor-1
- CNTF
ciliary neurotrophic factor
- CNTFRα
ciliary neurotrophic factor receptor α
- Cre
Cre recombinase
- EDL
extensor digitorum longus
- flxCNTFRα
floxed CNTFRα gene
- LIFRβ
leukemia inhibitory factor receptor β
- MN
motor neuron
- PFA
paraformaldehyde
- pSTAT3
phospho-Tyr705 STAT3
- STAT3
signal transducer and activator of transcription 3
- qRT-PCR
quantitative real-time PCR
Footnotes
Conflict of interest: The authors declare no conflicts.
Data accessibility statement: All data generated or analyzed during this study are included in this published article.
References
- Augusto V, Padovani CR & Campos GER (2004) Skeletal muscle fiber types in C57BL6J mice. Braz. J. Morphol. Sci, 21(2), 89–94. [Google Scholar]
- Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, Blesch A, Pilpel Y, Twiss JL & Fainzilber M (2012) Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J, March 21;31(6), 1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe GWM, Haspel JA, Smith CL, Wiener HH & Burden SJ (2000) Selective expression of Cre recombinase in skeletal muscle fibers. Genesis, 26, 165–166. [PubMed] [Google Scholar]
- Davis S, Aldrich TH, Stahl N, Pan L, Taga T, Kishimoto T, Ip NY & Yancopoulos GD (1993a) LIFRβ and gp130 as heterodimerizing signal transducers of the tripartite CNTF receptor. Science, 260, 1805–1808. [DOI] [PubMed] [Google Scholar]
- Davis S, Aldrich TH, Ip NY, Stahl N, Scherer S, Farruggella T, DiStefano PS, Curtis R, Panayotatos N, Gascan H, Chevalier S & Yancopoulos GD (1993b) Released form of CNTF receptor α component is a soluble mediator of CNTF responses. Science, 259, 1736–1739. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Vejsada R, Poueymirou WT, Acheson A, Suri C, Conover JC, Friedman B, McClain J, Pan L, Stahl N, Ip NY, Kato A & Yancopoulos GD (1995) Mice lacking the CNTF receptor, unlike mice lacking CNTF, exhibit profound motor neuron deficits at birth. Cell, 83, 313–322. [DOI] [PubMed] [Google Scholar]
- Elson GCA, Lelievre E, Guillet C, Chevalier S, Plun-Favreau H, Froger J, Suard I, Benoit de Coignac A, Delneste Y, Bonnefoy J–Y, Gauchat J-F & Gascan H (2000) CLF associates with CLC to form a functional heteromeric ligand for the CNTF receptor complex. Nature Neurosci, 3, 867–872. [DOI] [PubMed] [Google Scholar]
- Helgren ME, Squinto SP, Davis HL, Parry DJ, Boulton TG, Heck CS, Zhu Y, Yancopoulos GD, Lindsay RM & DiStefano PS (1994) Trophic effect of ciliary neurotrophic factor on denervated skeletal muscle. Cell, 76, 493–504. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Gomez-Isla T & Irizarry MC (1998) Stereology: A practical primer for neuropathology. J. Neuropath. Exp. Neurol, 57, 305–310. [DOI] [PubMed] [Google Scholar]
- Janjua MZ & Leong SK (1984) Organization of neurons forming the femoral, sciatic, common peroneal and tibial nerves in rats and monkeys. Brain Res, 310, 311–323. [DOI] [PubMed] [Google Scholar]
- Kirsch M, Terheggen U & Hofmann HD (2003) Ciliary neurotrophic factor is an early lesion-induced retrograde signal for axotomized facial motoneurons. Mol. Cell. Neurosci, 24(1), 130–8. [DOI] [PubMed] [Google Scholar]
- Lee MY, Hofmann HD & Kirsch M (1997) Expression of ciliary neurotrophic factor receptor-alpha messenger RNA in neonatal and adult rat brain: an in situ hybridization study. Neurosci, 77, 233–246. [DOI] [PubMed] [Google Scholar]
- Lee N, Neitzel KL, Devlin BK & MacLennan AJ (2004) STAT3 phosphorylation in injured axons before sensory and motor neuron nuclei: potential role for STAT3 as a retrograde signaling transcription factor. J. Comp. Neurol, July 5;474(4), 535–45. [DOI] [PubMed] [Google Scholar]
- Lee N, Robitz R, Zurbrugg RJ, Karpman AM, Mahler AM, Cronier SA, Vesey R, Spearry RP, Zolotukhin S & MacLennan AJ (2008) Conditional, genetic disruption of ciliary neurotrophic factor receptors reveals a role in adult MN survival. Eur. J. Neurosci, 27, 2830–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Spearry RP, Leahy KM, Robitz R, Trinh DS, Mason CO, Zurbrugg RJ, Batt MK, Paul RJ & MacLennan AJ (2013) Muscle ciliary neurotrophic factor receptor α promotes axonal regeneration and functional recovery following peripheral nerve lesion. J. Comp. Neurol, 521(13), 2947–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE & Darnell JE (2002) STATS: transcriptional control and biological impact. Nature Rev, 3, 651–662. [DOI] [PubMed] [Google Scholar]
- Luo X, Ribeiro M, Bray ER, Lee DH, Yungher BJ, Mehta ST, Thakor KA, Diaz F, Lee JK, Moraes CT, Bixby JL, Lemmon VP & Park KK (2016) Enhanced Transcriptional Activity and Mitochondrial Localization of STAT3 Co-induce Axon Regrowth in the Adult Central Nervous System. Cell Rep, 15(2), 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons GE, Ontell M, Cox R, Sassoon D & Buckingham M (1990) The expression of myosin genes in developing skeletal muscle in the mouse embryo. J. Cell. Biol, 111, 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan AJ, Vinson EN, Marks L, McLaurin DL, Pfeifer M & Lee N (1996) Immunohistochemical localization of ciliary neurotrophic factor receptor α expression in the rat nervous system. J. Neurosci, 16, 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan AJ, Devlin BK, Neitzel KL, McLaurin DL, Anderson KJ & Lee N (1999) Regulation of ciliary neurotrophic factor receptor alpha in sciatic MNs following axotomy. Neuroscience, 91(4), 1401–1413. [DOI] [PubMed] [Google Scholar]
- MacLennan AJ, Neitzel KL, Devlin BK, Garcia J, Hauptman GA, Gloaguen I, Di Marco A, Laufer R & Lee N (2000) In vivo localization and characterization of functional ciliary neurotrophic factor receptors which utilize JAK-STAT signaling. Neuroscience, 99(4), 761–72. [DOI] [PubMed] [Google Scholar]
- Pernet V, Joly S, Jordi N, Dalkara D, Guzik-Kornacka A, Flannery JG & Schwab ME (2013) Misguidance and modulation of axonal regeneration by Stat3 and Rho/ROCK signaling in the transparent optic nerve. Cell Death Dis, 18;4:e734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plun-Favreau H, Elson G, Chabbert M, Froger J, deLapeyrière O, Lelièvre E, Guillet C, Hermann J, Gauchat JF, Gascan H & Chevalier S (2001) The ciliary neurotrophic factor receptor alpha component induces the secretion of and is required for functional responses to cardiotrophin-like cytokine. EMBO J, 20(7), 1692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahenk Z, Seharaseyon J & Mendell JR (1994) CNTF potentiates peripheral nerve regeneration. Brain Res, 655, 246–250. [DOI] [PubMed] [Google Scholar]
- Sendtner M, Stockli KA & Thoenen H (1992b) Synthesis and localization of ciliary neurotrophic factor in the sciatic nerve of the adult rat after lesion and during regeneration. J Cell Biol, 118, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Cho Y, Beirowski B, Milbrandt J, Cavalli V & DiAntonio A (2012) Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron, 74(6):1015–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA & Hilton DJ (1997) A family of cytokine-inducible inhibitors of signalling. Nature, 387, 917–921. [DOI] [PubMed] [Google Scholar]
- Ylä-Herttuala S (2012) Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther, 20(10), 1831–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X,Bolon B, Pretorius JK, Kurahara C, McCabe J, Christiansen KA, Sun N, Duryea D, Foreman O, Senaldi G, Itano AA & Siu G (2009) Neonatal death in mice lacking cardiotrophin-like cytokine is associated with multifocal neuronal hypoplasia. Vet. Pathol, 46(3):514–9. [DOI] [PubMed] [Google Scholar]


