Abstract
Purpose:
To design and implement a multi-slice MRSI method for fast spectroscopic imaging, employing a modified version of Echo Planar Spectroscopic Imaging (EPSI) that offers higher spectral width (SW) and/or shorter scan time.
Methods:
EPSI suffers from inconsistencies between readout lines acquired with gradients of opposite signs, which has typically been addressed by reconstructing the “positive” and “negative” datasets separately and averaging the two. Nevertheless, consistency between the readout lines of each phase encode (PE) can be achieved by interposing the EPSI readouts with alternating “blipped” PE gradients. This method exchanges inconsistencies along the temporal dimension with inconsistencies along the PE dimension, which are straightforward to correct, as is conventionally done in various EPI reconstruction schemes. Such consistent k-t-space EPSI doubles the SW in comparison to EPSI, or – in an alternative realization, yields the same SW as EPSI, but at half the acquisition time. In this work, multi-band CAIPIRINHA slice selection was integrated with consistent k-t-space EPSI to further accelerate the measurement two-fold.
Results:
The feasibility of a consistent k-t-space EPSI was demonstrated in both phantoms and in-vivo brain imaging at 3T and four pulse scheme variants were evaluated. It was demonstrated to be useful in optimizing the SW and scan acceleration, both of which are limiting factors in-vivo. Dual-band implementation was shown to shorten the duration of the scan 4-fold.
Conclusion:
The consistent k-t-space EPSI can be used to accelerate MRSI or, alternatively, double its spectral width. Adding dual-band CAIPIRINHA further accelerates the acquisition by factor two.
Keywords: Echo planar spectroscopic imaging, EPSI, fast magnetic resonance spectroscopic imaging, proton spectroscopy, MRSI
Introduction
In-vivo proton magnetic resonance spectroscopy (1H-MRS) conveys important physiological information that is beneficial in contemporary structural and functional research1–5, as well as the clinical assessment of multiple disorders6–10 including Alzheimer’s disease, schizophrenia, depression and cancer11–12. Magnetic Resonance Spectroscopic Imaging (MRSI) provides spatial and spectral information, capable of simultaneously visualizing the spatial extent of disease. Many common methods of MRSI are based on the phase-encoded chemical shift imaging method13 and require long acquisition times, especially when high spatial resolutions are required. To address this shortcoming, several methods using time-varying readout gradients have been proposed, including, among others, Echo Planar Spectroscopic Imaging (EPSI)14, spiral15, concentric rings16 and rosette17 schemes. While efficient, these methods still often require additional acceleration when very high resolution (~0.5 cm3 voxels), large coverage, “whole-brain” 3D acquisitions are desired. Thus, these schemes have been combined with k-space undersampling schemes, including parallel imaging, compressed sensing18–20, and multiband schemes (SMS) including the method of “controlled aliasing in parallel imaging results in higher acceleration” (CAIPIRINHA)20,21.
EPSI14,22,23, mentioned above, is an established fast spectroscopic imaging technique, in which an alternating readout gradient is used to simultaneously acquire 1D spatial and 1D spectral information within a single shot. Additional spatial dimensions are encoded using “slow” phase encoding (PE) gradients. The EPSI acquisition traces a zigzag trajectory in k-t space, implemented with alternating positive and negative readout gradients (Fig 1a). One drawback of EPSI is its restricted spectral-width6 (SW) – limited by the available gradient amplitude and slew-rate – making it difficult to cover the full spectral range of protons, especially at ultra-high fields (≥7T)24–27, where the full SW exceeds 2 kHz, or when imaging hyperpolarized 13C compounds, which span a wide spectral range of several dozen ppm28–30. Furthermore, the positive and negative lines in the nonuniform zigzag sampling pattern are often analyzed separately and eventually averaged, which doubles the effective echo spacing (TES) and halves the already-limited SW – an approach we refer to as “even/odd sum” (E+O) EPSI. Even if this zigzag behavior is accounted in the reconstruction (e.g., by a nonuniform Fourier transform), any misalignment between the positive and negative gradient echoes – e.g., due to B0 inhomogeneity and gradient hardware imperfections – would lead to ghosting artifacts along the spectral dimension. These ghosting artifacts are analogous to those typically observed in non-spectral Echo Planar Imaging (EPI) along the uniformly-sampled phase encoding axis31, where simple solutions to correct them exist31–33. Such solutions are only partially applicable to EPSI, due to its non-uniform sampling pattern.
Figure 1:
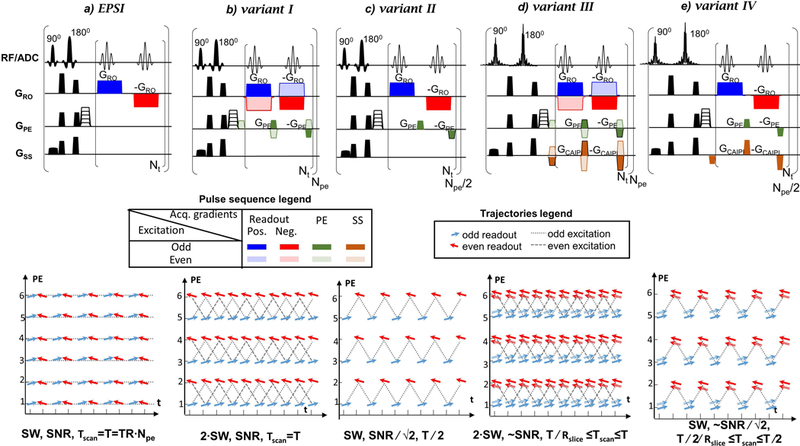
Sequences (top) and PE/t trajectories (bottom) : a) EPSI, b)-e) consistent k-t space EPSI variants I-IV. The relative SW, SNR and total scan time (Tscan) for each sequence (a-e) are noted relative to E+O EPSI (a). The blue and red arrows correspond to positive and negative readout gradient lobes, respectively. The dual-band slice-selection has dual-arrows representing two slices. The odd/even excitations are marked differently as shown in the legends. Variant II and IV (d) and ( e) use only the odd excitations of the full schemes variants I and III (b) and (d), respectively. Rslice is the acceleration factor of the multi-band excitation (Rslice=2 for dual-band).
Several solutions to EPSI’s limited SW exist. One such solution, called “flyback” EPSI 28,34, acquires data solely during the positive gradient lobes, while minimizing the time spent during the negative lobe by using the maximum slew rates and gradient amplitudes available. Its main drawback is lack of efficiency, since 30% or more of the readout time may be wasted during the reverse lobes, resulting in sub-optimal SNR. Another solution is temporal interleaving, in which the EPSI scan is repeated a number of times (N), delaying the acquisition’s onset by an amount The combined dataset offers a smaller effective echo-spacing1, given by TES/N, increasing the SW N-fold but at the cost of a concomitant N-fold increase in scan time. A third solution consists of repeating the acquisition with reversed readout gradients 35,36, from which one can extract two full datasets – one with only positive gradient readouts and one with only negative gradient readouts – thus doubling the SW, but at the cost of a two-fold increase in total scan time. The increased scan times are challenging when large coverage acquisitions are desired, as is often the case. Additional studies explored optimization of the SW and artifacts reduction by pulse schemes development and post-processing methods. The pulse schemes approach includes center-out EPSI with gradient-echo phase and time shifts corrections37. The post-processing methods include interlaced FT and gridding techniques38–39.
One of the pulse scheme solutions that doubles the SW of EPSI without requiring any additional scan time was published by Webb et al. in 198940. This approach has been demonstrated in phantoms40 but not in-vivo. In this EPSI implementation, blipped PE gradients are added in between positive and negative readouts (Fig.1b), such that time consistent acquisitions are acquired for each PE line. This comes at the expense of introducing misalignment between echoes in the PE dimension. However, as mentioned earlier, such inconsistencies are straightforward to correct, as routinely done for EPI31–33.
In the current work, the feasibility of the consistent k-t space EPSI for in-vivo 3D MRSI of the human brain at 3T is demonstrated. An additional variation demonstrated by Webb40 (Fig. 1c) is also examined. This variant only acquires the odd excitation lines of the above scheme, yielding the same SW as E+O EPSI at half the acquisition time (and with equal sensitivity – i.e., same SNR∕√T, where T is the scan time). In addition, since multi-slice MRSI imaging requires relatively long times, the consistent k-t space EPSI was combined with multi-band CAIPIRINHA for a multi-slice accelerated acquisition introducing two more pulse scheme variants. The combined sequences provide either a higher bandwidth or additional acceleration beyond those of EPSI. The four variants were evaluated and compared to more standard implementation of EPSI.
Methods
Sequences
Fig. 1 shows the pulse sequences compared in this study: E+O EPSI and four variants of the consistent k-t space EPSI – variant I) with maximal SW, variant II) with shorter scan duration, variant I and variant II combined with dual-band CAIPIRINHA into variants III and IV. The pulse scheme includes slice-selective excitation and refocusing, pre-phasing gradients and acquisition block. The acquisition block includes repeating positive and negative readout gradients with blipped PE gradients for the consistent k-t space EPSI schemes. No sampling occurred during gradient ramp up and down times. The different sequences are contrasted by their 2D sampling patterns in the PE-time plane, with the slice and readout axes omitted for brevity (a different graphical representation of the same sampling path can be found in Ref. 40). Left-to-right (blue) arrows and right-to-left (red) arrows indicate positive and negative readout gradients, respectively. The alternating arrows along the temporal axis in EPSI (Fig.1a) are replaced in variant I (Fig.1b) by readouts that are represented by unidirectional arrows. Variant II (Fig.1c) consists of only the odd excitations of Variant I, while enabling acquisition of the full spatial dataset with a halved-SW, the same as in E+O EPSI, but at half the number of excitations. Dual-band variants (III and IV) pulse schemes are shown in Figures 1d, 1e. In these schemes, the slice-selection pulses are replaced by dual-band pulses, while blipped gradients in the slice direction are integrated into the blipped PE gradients units as done in the blipped CAIPIRINHA method41. Blipped gradients in the slice direction for two-fold acceleration include 50% FOV shift41. Note that, when combining the blipped PE gradients with CAIPIRINHA, the blipped gradients in the slice direction need to be alternated in unison with the alternating PE blips (as shown in Fig.1d).
The water suppression that was used in this study is based on an in-house 256 ms long optimized VAPOR-like model consisting of six frequency selective pulses. The multi-slice acquisition includes slice selective single-band and dual-band excitation and refocusing pulses, in variants I,II and variants III,IV, respectively. The single-band excitation and refocusing pulses used in Fig.1-c were SLR pulses with a bandwidth-time product of 18 and 12 respectively and a duration of 3.8 ms and 8 ms respectively; whereas the dual-band excitation and refocusing pulses in Fig. 1d,e were SLR pulses with a bandwidth-time product of 12 and a duration of 6 ms and 8 ms, respectively.
Data Reconstruction and Evaluation
The data reconstruction steps employed for the consistent k-t space EPSI variants I-IV are shown in Figure 2. They consist of: i) reordering of the raw data, as each excitation step in variant I mixes information from two phase encodes; ii) applying a first order phase correction (with constant and linear terms) along the readout in image space to the even PE lines (due to the odd and even readout hardware inconsistencies, as in EPI31); iii) for variant II only, adding a linear phase to the even PE lines to correct for the even-to-odd echo spacing (Δteo) temporal shift relative to the odd PE lines; and iv) for dual-band sequences only, separating the two simultaneously excited slices using the coils’ sensitivity profiles41. The hardware inconsistencies of step ii) can be corrected using a range of methods developed for EPI sequences31–33,42. In this work we used the central region of the water reference data to extract a phase correction (see Fig.2 for formulas used) for the even PE lines (negative readout gradient). The correction consisted of a linear phase along the readout direction, including a constant component, and was applied to the even PE lines prior to the Fourier transform along PE. In addition, a MATLAB program (The Mathworks, Natick MA, USA) was used to interactively phase the spectra. The EPSI reconstruction included a separate FT of the even and odd line (along the temporal dimension), a FT along the k-space axes, and finally addition of the “even” and “odd” spectral images. Common to all EPSI based methods, the odd acquired lines were reversed before the reconstruction steps application.
Figure 2:
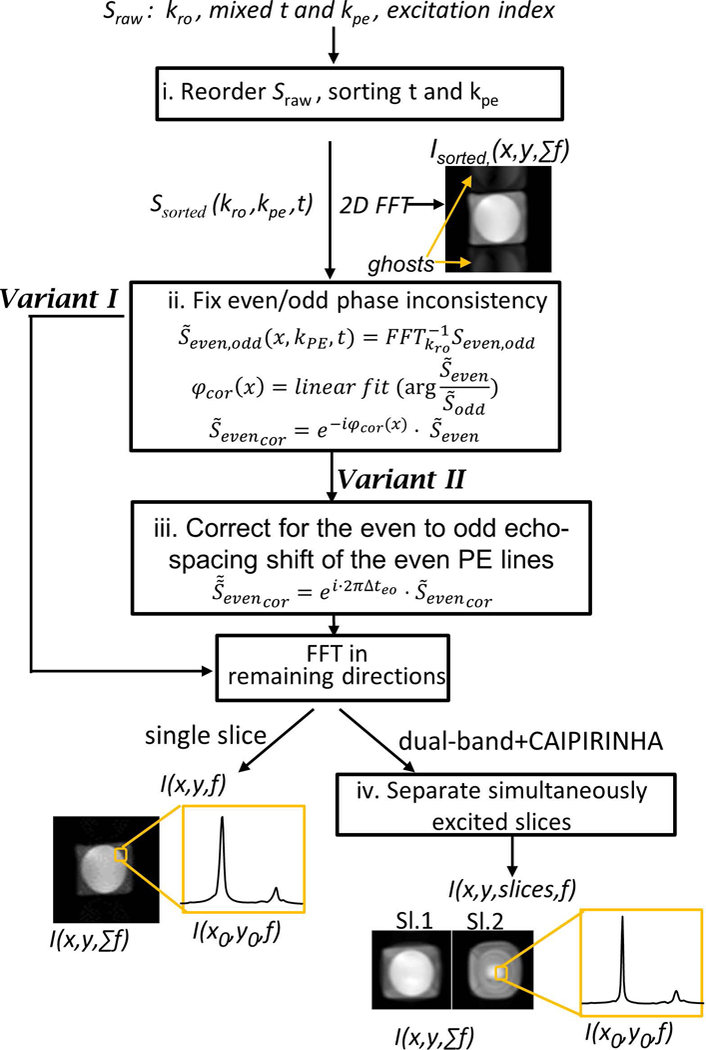
Reconstruction steps employed for variants I and II and their dual-band (variants III, IV) extensions. Sraw is the input raw data signal and the I(x,y,f) for each slice is the spectral spatial final reconstructed output. The images after the first and last steps show representative results, demonstrating an image with ghosts that will appear after the first step and the final images that include spectroscopic and spatial information.
The contribution of the coils to the signal was evaluated using a water reference scan43 and the final signal was estimated as where Si is the signal of ith channel and wi is the weighting calculated as . The final result shows real component spectra after zero- and first-order phase corrections. These spectra were fit in LCModel44 (v6.3) using simulated basis functions obtained by solving the full Liouville equations using in-house MATLAB routines, which was used to estimate linewidths and relative concentrations. Metabolite images were created using straightforward integration over each of the metabolite spectra (the integration range is specified in the figure captions). Unlike LCModel fitting, direct integration is more sensitive to lipid contamination and ghosting artifacts and highlights their presence, and was therefore deemed more appropriate for visual presentation.
Phantom Experiments
All experiments were performed on a 3T system (Prisma, Siemens, Erlangen) using a commercial 20-channel head coil. Two phantoms were used to validate and compare the sequences shown in Fig. 1. For metabolites spectroscopic measurements an homogeneous spherical metabolite phantom was used, with a solution of 10mM L-Glutamic acid, 10mM Creatine 8mM myo-Inositol, 2mM GABA, 2mM Choline chloride, 5mM Sodium lactate 12.5mM N-acetylaspartate (NAA) and 0.5mL Gd, all dissolved in water (T1,water= 625 ms). To evaluate the method’s voxel bleed we also examined, qualitatively, the spectroscopic imaging of an oil/water phantom, consisting of white cheese balls immersed in olive oil.
For SNR measurements, the consistent k-t space EPSI variant I was compared with EPSI (combined odd and even lines) and variant II with EPSI using only the odd lines, since these two sets represent equivalent SNR. The SNR ratios were calculated for both water and NAA images. The regions for signal and noise estimation in the phantom are shown in Figure 3a. In addition, the effect of the dual-band CAIPIRINHA acquisition on SNR was also examined. The geometry factor maps were calculated for two setups of FOV and object dimensions. In addition, the ACR MRI phantom45 was used to examine the reconstruction of high-resolution images for simultaneous acquisition of adjacent slices. The results of dual-band CAIPIRINHA comparisons are summarized in the Supporting Information Figure S1.
Figure 3:
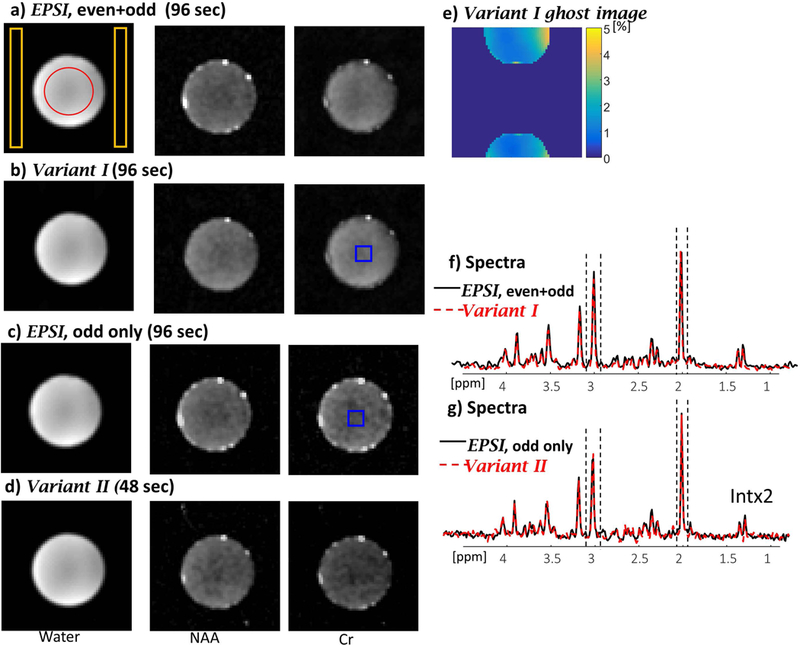
Phantom results comparing EPSI with combined odd and even lines, variant I, EPSI with odd lines only (Tscan=96 sec), and variant II (Tscan=48 sec). The images from left to right show water image (from a scan without water suppression), NAA and Cr images (from a separate scan with water-suppression) for EPSI (a), variant I (b), EPSI odd lines only (c) and variant II (d). e) Ghost artifacts evaluation in percent compared the image intensity. f) Spectra of EPSI (black) and variant I (red-dashed) and g) spectra of EPSI (black) and variant II (red dashed) for a central region shown by blue overlay (averaging 5×5 pixels). The dashed lines in f) and g) shows the region for NAA and Cr integration (0.14ppm and 0.16 ppm, respectively).
The scan parameters employed for the metabolic phantom were: TR/TE 1500/15 ms, FOV and thickness 300×300×15 mm3, in-plane resolution 4.7×4.7 mm2, 0.33 cc voxel, spectral resolution 0.7 Hz (final time vector-size 512 for EPSI and variant II and 1024 for variant I), ramp time 50μs (slew rate 180T/m/s), readout duration 570ms, echo spacing 0.7 ms, SW =700 Hz for EPSI and variant II and SW = 1400 Hz for variant I, and a single average. The scan duration of EPSI and variant I was 96 seconds, while variant II scan duration was 48 seconds. An additional non-water suppressed dataset was acquired for coil combination, phasing and signal normalization.
A two-fold accelerated variant IV was demonstrated on a metabolites phantom. Scan parameters were: TR/TE 1500/15 ms, FOV and thickness 300×300×15 mm3, 5.4cm gap between slices, in-plane resolution 4.7×4.7mm2, 0.33 cc voxel, spectral resolution 0.7 Hz, echo spacing 0.7 ms, SW=700 Hz, and a single average. The same parameters were also used for the oil/water phantom, apart from: TR/TE 500/15 ms, FOV 200×200 mm2, 2 Hz spectral resolution.
In-Vivo Experiments
In-vivo applicability of the consistent k-t space EPSI variants was examined by scanning three human volunteers, who provided written informed consent and following procedures approved by the Internal Review Board of the Wolfson Medical Center (Holon, Israel). The protocol included a localizer (0:30 min), a Turbo Spin Echo (TSE) for anatomical reference (5 min) and the fast MRSI methods described above: EPSI and the variants I-IV. The MRSI scans imaged six slices.
The scan parameters common to all the in-vivo MRSI scans were: TR/TE 2000/20 ms, FOV and thickness 300×145×15 mm3, 1.8cm gap between slices, spatial resolution 5.5×5.5 mm2, 0.45 mL voxel, and a single average. Additional scan parameters for EPSI, variant II and variant IV were spectral resolution 5.8Hz (final time vector-size 128 for EPSI and variants II, IV and 256 for variants I,III), ramp time 40μs (slew rate 180T/m/s), readout duration 590ms, echo spacing 0.7ms and SW=750 Hz. For variants I and III these were: spectral resolution 4Hz, echo spacing 1ms and SW 1000 Hz. The latter parameters demonstrate the flexibility of the variant I which can achieve a higher SW than EPSI, despite a longer echo-spacing. The dual-band acquisition excited adjacent slices, which is beneficial for reducing lipid contamination artifacts (see schematic explanation in Supporting Information Figure S2). The scan duration of EPSI, variant I and III were 1:48 min, while variant II and IV took 54 seconds. Scan parameters of the TSE scan were TR/TE 4000/77 ms, FOV and thickness 256×256×5 mm3 and in-plane resolution 1×1.4 mm2.
Despite the high in-plane resolution which minimized voxel bleed, lipid contamination persisted in the reconstructed datasets. To further mitigate it, in-plane k-space data was apodized using a 2D Hamming filter (Hamming window a0= 0.5) to reduce the PSF side-bands. The linewidth of the NAA peak was compared between the three representative ROIs, for all five sequences (EPSI and four discussed variants).
Results
Phantom Experiments
Figure 3 shows a representative example comparing SNR and spectra quality between EPSI and the consistent k-t EPSI variant I and II. Supporting Information Table S1 includes a comparison of the SNR of variant I and II, each relative to EPSI (as a reference). The SNR ratio was calculated as 1.01, SNREPSIddlines/SNRvariant II with variant II acquired in half the time of the EPSI scan duration. The results confirm that the same SNR is provided by the two sequences – for example, comparing EPSI using odd lines only (equivalent to “flyback” EPSI) and the variant II. Figure 3 also shows that the variant II spectrum has the same linewidth as in the EPSI spectrum (5.3 Hz for NAA). The ghost artifacts that occur in the PE direction due to the inaccuracies of the even/odd PE lines reconstruction in variant I sequence are quantified in Figure 3e. The ghost artifacts reach a maximum of 4.24%, with an average±standard deviation of 1.5%±0.7%.
Figure 4a shows results of 1H MRSI on a brain mimicking phantom, reconstructing two slices acquired by the dual-band variant IV method. The Supporting Information Figure S1 expands the analysis of the dual-band CAIPIRINHA implementation. Geometry factor maps of the two examined setups are shown and the values are ≤1.2 for both adjacent and non-adjacent slices. The details of the high-resolution ACR phantom are reconstructed faithfully for adjacent slices.
Figure 4:
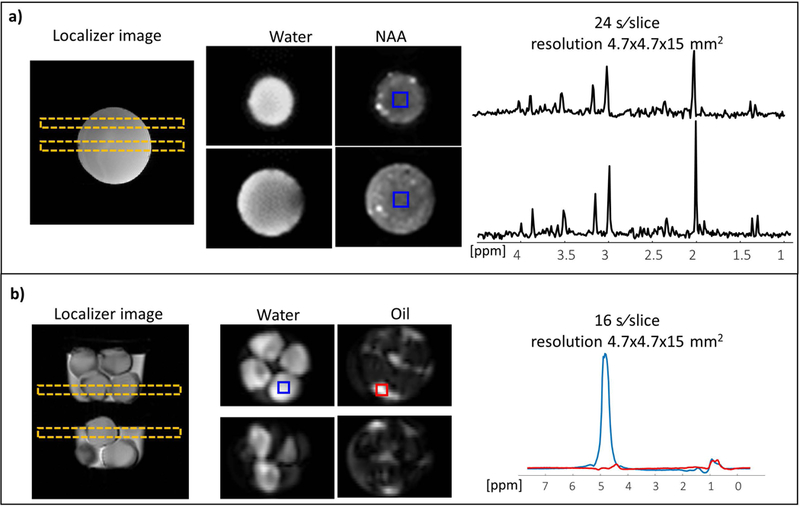
Dual-band variant IV – a) Metabolites and b) Water/Oil phantoms. The figure includes from left to right: a localizer image with the planned slices, images of the slices for selected peaks and spectra for the selected regions shown by overlays (blue overlay box on NAA image for metabolites and blue/red box for water/oil spectra, respectively).
In addition, an experiment to separate oil and water peaks is demonstrated using a phantom consisting of white cheese balls immersed in oil. Figure 4b shows two slices acquired using faster variant IV, where water and oil images were reconstructed. Spectra from the regions containing white cheese consisted of both water and lipid peaks (blue rectangle), while spectra from the surrounding oil consisted mainly of lipid peaks (red rectangle).
In-vivo experiments
Figure 5 shows a representative example of the human brain scanning results. The figure includes an anatomical TSE reference scan and results of the variant I (with higher SW) and variant IV (with faster acquisition) compared to EPSI (variants II and III are shown in the Supporting Information Figure S3). For each sequence the spectra of three regions, close to the edges, are shown. The results show comparable spectra between EPSI and the consistent k-t space EPSI sequences, including the dual-band slice acquisition. The images of NAA metabolite are also shown. The Supporting Information Figure S4–S6 includes Cr and Cho images as well as NAA/tCr ratio maps estimated by the LCModel fitting and single voxels spectra. The measured NAA linewidth fitted by LCModel for the blue ROI was 13.7±2.1, 12.9±2.2, 12.8±2.1, 13.1±3.4, 11.9±1.9 Hz for each of the methods above, respectively. The estimated deviation of the linewidth of NAA peak for variants I-IV sequences was 3±5% compared to EPSI. It can be seen that without averaging the full dataset can be acquired within one minute, using the dual-band variant IV acquisition scheme. Note that, in the dual-band version, lipid contamination and ghost artifacts were observed in the second slice (in a region with relatively high B0 inhomogeneity) and in fifth slice (which has relative high lipid content).The ghost artifacts for dual-band CAIPIRINHA for this in-vivo case are estimated in the Supporting Information Figure S7.
Figure 5:
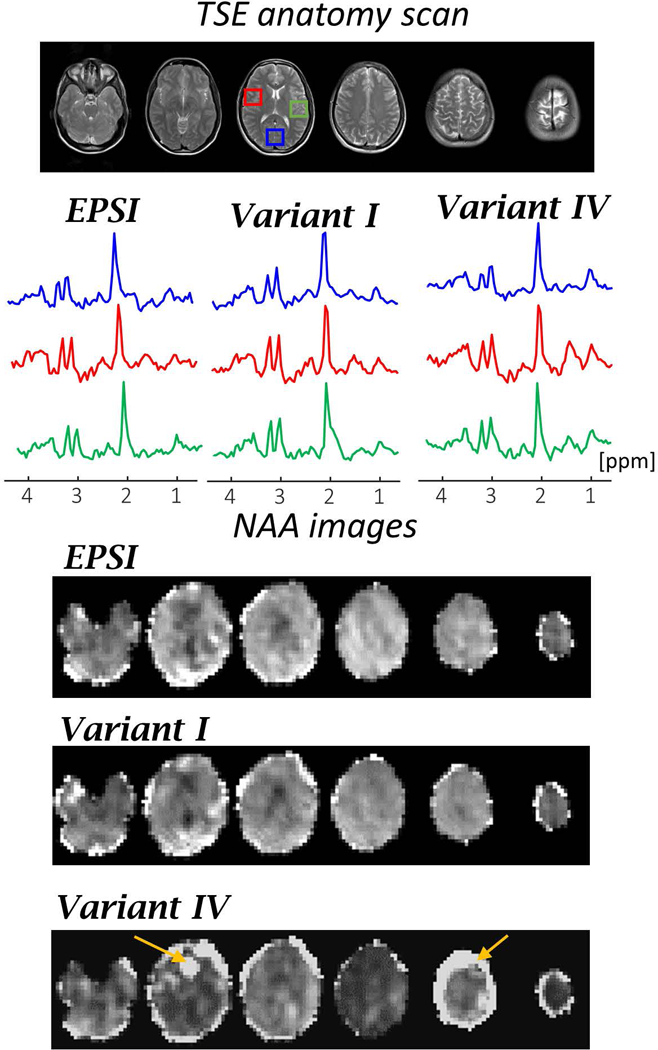
In-vivo results of EPSI and variants I (with higher SW) and variant IV (with faster acquisition). From top to bottom: TSE anatomy images of the scanned slices; real spectra for each MRSI method at three regions (each averaging over 5×5 voxels) in the central slice (shown in red, blue and green rectangle overlays); and images of six slices for NAA using spectra integration in the range of 0.4 ppm. The image intensities of NAA is normalized to the NAA intensity in each of the methods.
Discussion and Conclusions
In this work, we implemented a multi-slice consistent k-t space EPSI sequence combined with dual-band slice excitation for fast MRSI, and demonstrated its feasibility of accelerating in-vivo MRSI at 3T. The results from the metabolite phantom show EPSI and the examined variants I and II produce similar quality spectra, with equal sensitivity. The in-vivo results demonstrated the capability of the dual-band variant IV for producing spectroscopic images of the human brain within approximately one-minute, using a 54×26×6 matrix size (5.5×5.5 mm2 in-plane resolution, 15 mm slice thickness). The variant II enabled halving the acquisition time of EPSI, while maintaining its sensitivity and SW. On the other hand, the full acquisition version in variant I (Fig.1b) offered greater flexibility in optimizing the SW, as demonstrated in Fig. 5, where we increased the SW in variant I acquisition to 1000 Hz compared to 750 Hz in EPSI, by increasing the physical dwell time from 20 µs 33.3 µs. This flexibility can be beneficial in reducing the maximal readout gradients, since variant I can achieve the same SW as EPSI with lower readout gradient amplitudes (approximately half). This can also yield shorter ramp times that can contribute to the efficiency of the net acquisition time. Additional studies to further increase the SW in EPSI for in-vivo imaging are required, to ensure no aliased spectrally signal will arise.
The increased SNR due to recent advances in multi-channel receive coil technology, as well as the availability of ultra-high field MRI scanners, make large-coverage, fast MRSI pulse sequences particularly attractive. In comparison to EPSI, the consistent k-t space EPSI allows for faster acquisitions or reduced gradient demands. At 3T, where EPSI usually achieves the desired SWs, variant II can be harnessed for scan time reduction, which is important for achieving clinically relevant scan times of large imaging volumes. The ability of Variant I to extend the SW without resorting to time-consuming temporal interleaving can be used to address the ever-increasing SW requirements beyond 3T. Incorporating simultaneous multi-slice encoding into the scheme by using a dual-band excitation, as shown in the current study, can also play a significant role in multi-slice acquisitions at higher fields, improving the applicability of fast MRSI techniques. Even greater speedups can be achieved when using variant II, especially when integrated with CAIPIRINHA, which could even make it a potential candidate for functional spectroscopic brain imaging.
It should be pointed out that moving the time-inconsistencies in EPSI to inconsistencies in the PE encoding direction in the consistent k-t space EPSI results in artifacts which look like residual ghosts. In the current study, the PE inconsistencies were corrected using the water reference data and extracting a linear phase correction between the odd and even PE lines. This correction was found to produce acceptable spectroscopic images with less than 5% ghost intensity. However, when this scheme is combined with a dual-band CAIPIRINHA, the ghost artifacts will also lead to lipid contamination, as in the upper slices of the in-vivo example in Fig. 5. The lipid ghosts of up to 5% is still relatively high signal compared to the metabolites. A range of studies have developed more extensive corrections for similar artifacts in EPI, and may be used to further reduce the ghosts33,42. In applications that target superior slices, some form of lipid suppression is recommended. An additional concern that requires further analysis is the sensitivity of the proposed methods to motion. This analysis lies beyond the scope of the current work. However, the EPSI sequence topology can be easily expanded by adding navigators to each of the acquisition blocks, which can track and assist in correction motion artifacts46–47.
Although in the current study lipid suppression was not used, in studies that require better coverage of metabolites, including peaks like lactate that are in close proximity to lipids, a lipid suppression method can be added to the sequences implemented here. In addition, the multi-band CAIPIRINHA reconstruction can also be further improved 48,49, which is essential for reducing inter-slice lipid contamination. Moreover, one of the current drawbacks of EPSI-like acquisition and of other fast 3D acquisitions is the B0 inhomogeneity that varies between the slices. As a next step in the development of the above pulse sequences, one can further reduce the susceptibility to B0 inhomogeneity by implementing per-slice B0 shimming50.
Supplementary Material
Figure S1: Dual-band CAIPIRINHA variant III - g-factor maps and high resolution ACR phantom reconstruction of simultaneous excitation of adjacent (isl, isl+1) and not adjacent (isl, isl+3) slices. Set 1 and Set 2 examples show two different FOV and object dimensions. The scan parameters for set 1 were: TR/TE 1500/15 ms, FOV and thickness 270×192×8 mm3, in-plane resolution 3×3 mm2, SW =1000 Hz, for set 2: FOV and thickness 192×192×8 mm3, in-plane resolution 3×3 mm2, SW =1000 Hz.
Figure S2: Schematic illustration of the ghost artifacts for dual-band CAIPIRINHA case with adjacent (isl, isl+1) and not adjacent (isl, isl+3) slices, shown in a) and b) respectively. The simulation shows a spherical-shape phantom with signal inside mimicking metabolites equal to one and a ring mimicking lipid 10x higher. The ghost is simulated having 4% from the original signal. The adjacent slices have the same radius and the non-adjacent slices have different radius. The scale of the images is from 0 to 2.
Figure S3: In-vivo results of variants II and III. From top to bottom: TSE anatomy images of the scanned slices; real spectra for each MRSI method at three regions (each averaging over 5×5 voxels) in the central slice (shown in red, blue and green rectangle overlays); and images of six slices for NAA using spectra integration in the range of 0.4 ppm. The image intensities of NAA is normalized to the NAA intensity in each of the methods.
Figure S4: Creatine and Choline images of the in-vivo results of EPSI and variants I-IV acquisitions. Images of six slices are shown for each of the methods.
Figure S5. NAA/tCr ratio maps estimated by the LCModel fitting for EPSI and variant I-IV.
Figure S6. Single voxel spectra for each MRSI method at the blue ROI (central voxel) – EPSI and variants I-IV, respectively.
Figure S7. Lipid contamination estimation dual-band CAIPIRINHA variant III compared to variant I (in-vivo example from Fig.5). a) Ratio of the NAA images dual-band CAIPIRINHA variant III and variant I. b) histogram of the ratio values in a).
Table S1. Comparison of the SNR of variant I and II (in Figure 3), each relative to EPSI (as a reference).
Acknowledgment
We are grateful to Dr. Edna Haran and the Weizmann MRI technician team, for assistance in the human imaging scans, and to Osnat Volovyk for preparation of the phantom mimicking brain metabolites. Assaf Tal acknowledges the support of the Monroy-Marks Career Development Fund and the historic generosity of the Harold Perlman Family. This work was funded by the Minerva Foundation and NIH grant R01-NS097494.
Footnotes
Supporting Information
Additional supporting information may be found online in the Supporting information section at the end of the article.
References
- 1.Burlina AP, Aureli T, Bracco F, Conti F, & Battistin L (2000). MR spectroscopy: a powerful tool for investigating brain function and neurological diseases. Neurochemical Research, 25(9–10), 1365–1372. [DOI] [PubMed] [Google Scholar]
- 2.Mangia S, Tkác I, Gruetter R, Van De Moortele P-F, Giove F, Maraviglia B, & Uğurbil K (2006). Sensitivity of single-voxel 1H-MRS in investigating the metabolism of the activated human visual cortex at 7 T. Magnetic Resonance Imaging, 24(4), 343–348. 10.1016/j.mri.2005.12.023 [DOI] [PubMed] [Google Scholar]
- 3.Apšvalka D, Gadie A, Clemence M, & Mullins PG (2015). Event-related dynamics of glutamate and BOLD effects measured using functional magnetic resonance spectroscopy (fMRS) at 3T in a repetition suppression paradigm. NeuroImage, 118, 292–300. [DOI] [PubMed] [Google Scholar]
- 4.Puts NAJ, & Edden RAE (2012). In vivo magnetic resonance spectroscopy of GABA: a methodological review. Progress in Nuclear Magnetic Resonance Spectroscopy, 60, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogner W, Gagoski B, Hess AT, Bhat H, Tisdall MD, van der Kouwe AJW, … Andronesi OC (2014). 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. NeuroImage, 103, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Posse S, Otazo R, Dager SR, & Alger J (2013). MR spectroscopic imaging: principles and recent advances. Journal of Magnetic Resonance Imaging: JMRI, 37(6), 1301–1325. [DOI] [PubMed] [Google Scholar]
- 7.Soares DP, & Law M (2009). Magnetic resonance spectroscopy of the brain: review of metabolites and clinical applications. Clinical Radiology, 64(1), 12–21. [DOI] [PubMed] [Google Scholar]
- 8.Jack CR, Garwood M, Wengenack TM, Borowski B, Curran GL, Lin J, … Poduslo JF (2004). In vivo visualization of Alzheimer’s amyloid plaques by magnetic resonance imaging in transgenic mice without a contrast agent. Magnetic Resonance in Medicine, 52(6), 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keshavan MS, Stanley JA, & Pettegrew JW (2000). Magnetic resonance spectroscopy in schizophrenia: methodological issues and findings—part II. Biological Psychiatry, 48(5), 369–380. [DOI] [PubMed] [Google Scholar]
- 10.Auer DP, Pütz B, Kraft E, Lipinski B, Schill J, & Holsboer F (2000). Reduced glutamate in the anterior cingulate cortex in depression: an in vivo proton magnetic resonance spectroscopy study. Biological Psychiatry, 47(4), 305–313. [DOI] [PubMed] [Google Scholar]
- 11.Gillies RJ, & Morse DL (2005). In vivo magnetic resonance spectroscopy in cancer. Annual Review of Biomedical Engineering, 7, 287–326. [DOI] [PubMed] [Google Scholar]
- 12.Cheng LL, Burns MA, Taylor JL, He W, Halpern EF, McDougal WS, & Wu C-L (2005). Metabolic characterization of human prostate cancer with tissue magnetic resonance spectroscopy. Cancer Research, 65(8), 3030–3034. [DOI] [PubMed] [Google Scholar]
- 13.Brown TR, Kincaid BM, & Ugurbil K (1982). NMR chemical shift imaging in three dimensions. Proceedings of the National Academy of Sciences, 79(11), 3523–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansfield P (1984). Spatial mapping of the chemical shift in NMR. Magnetic Resonance in Medicine, 1(3), 370–386. [DOI] [PubMed] [Google Scholar]
- 15.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, & Spielman DM (1998). Volumetric spectroscopic imaging with spiral-basedk-space trajectories. Magnetic Resonance in Medicine, 39(6), 889–898. [DOI] [PubMed] [Google Scholar]
- 16.Furuyama JK, Wilson NE, & Thomas MA (2012). Spectroscopic imaging using concentrically circular echo-planar trajectories in vivo. Magnetic Resonance in Medicine, 67(6), 1515–1522. [DOI] [PubMed] [Google Scholar]
- 17.Schirda CV, Tanase C, & Boada FE (2009). Rosette spectroscopic imaging: optimal parameters for alias-free, high sensitivity spectroscopic imaging. Journal of Magnetic Resonance Imaging: JMRI, 29(6), 1375–1385. [DOI] [PubMed] [Google Scholar]
- 18.Dydak U, Weiger M, Pruessmann KP, Meier D, & Boesiger P (2001). Sensitivity-encoded spectroscopic imaging. Magnetic Resonance in Medicine, 46(4), 713–722. [DOI] [PubMed] [Google Scholar]
- 19.Nassirpour S, Chang P, Avdievitch N, & Henning A (2018). Compressed sensing for high-resolution nonlipid suppressed 1 H FID MRSI of the human brain at 9.4T. Magnetic Resonance in Medicine [DOI] [PubMed]
- 20.Strasser B, Považan M, Hangel G, Hingerl L, Chmelik M, Gruber S, … Bogner W (2017). (2 + 1)D-CAIPIRINHA accelerated MR spectroscopic imaging of the brain at 7T: (2 + 1)D-CAIPIRINHA Accelerated MRSI. Magnetic Resonance in Medicine, 78(2), 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breuer FA, Blaimer M, Mueller MF, Seiberlich N, Heidemann RM, Griswold MA, & Jakob PM (2006). Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magnetic Resonance in Medicine, 55(3), 549–556. [DOI] [PubMed] [Google Scholar]
- 22.Mulkern RV, & Panych LP (2001). Echo planar spectroscopic imaging: Echo Planar Spectroscopic Imaging. Concepts in Magnetic Resonance, 13(4), 213–237. [Google Scholar]
- 23.Posse S, DeCarli C, & Le Bihan D (1994). Three-dimensional echo-planar MR spectroscopic imaging at short echo times in the human brain. Radiology, 192(3), 733–738. [DOI] [PubMed] [Google Scholar]
- 24.Uğurbil K, Adriany G, Andersen P, Chen W, Garwood M, Gruetter R, … Zhu X-H (2003). Ultrahigh field magnetic resonance imaging and spectroscopy. Magnetic Resonance Imaging, 21(10), 1263–1281. [DOI] [PubMed] [Google Scholar]
- 25.Tkáč I, Öz G, Adriany G, Uğurbil K, & Gruetter R (2009). In vivo 1 H NMR spectroscopy of the human brain at high magnetic fields: Metabolite quantification at 4T vs. 7T. Magnetic Resonance in Medicine, 62(4), 868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiew M, Jiang W, Burns B, Larson P, Steel A, Jezzard P,… Emir UE (2018). Density-weighted concentric rings k -space trajectory for 1 H magnetic resonance spectroscopic imaging at 7 T. NMR in Biomedicine, 31(1), e3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hangel G, Strasser B, Považan M, Heckova E, Hingerl L, Boubela R, … Bogner W (2018). Ultra-high resolution brain metabolite mapping at 7 T by short-TR Hadamard-encoded FID-MRSI. NeuroImage, 168, 199–210. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham CH, Vigneron DB, Chen AP, Xu D, Nelson SJ, Hurd RE, … Pauly JM (2005). Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magnetic Resonance in Medicine, 54(5), 1286–1289. [DOI] [PubMed] [Google Scholar]
- 29.Mayer D, Yen Y-F, Takahashi A, Josan S, Tropp J, Rutt BK, … Pfefferbaum A (2011). Dynamic and high-resolution metabolic imaging of hyperpolarized [1–13C]-pyruvate in the rat brain using a high-performance gradient insert. Magnetic Resonance in Medicine, 65(5), 1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson PEZ, Hu S, Lustig M, Kerr AB, Nelson SJ, Kurhanewicz J, … Vigneron DB (2011). Fast dynamic 3D MR spectroscopic imaging with compressed sensing and multiband excitation pulses for hyperpolarized 13C studies. Magnetic Resonance in Medicine, 65(3), 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jesmanowicz A, Wong EC, Hyde JS. (1993). Phase correction for EPI using internal reference lines. Proc., SMRM, 12th Annual Meeting, New York, (p. 1239). [Google Scholar]
- 32.Hu X, & Kim SG (1994). Reduction of signal fluctuation in functional MRI using navigator echoes. Magnetic Resonance in Medicine, 31(5), 495–503. [DOI] [PubMed] [Google Scholar]
- 33.Chen N, & Wyrwicz AM (2004). Removal of EPI Nyquist ghost artifacts with two-dimensional phase correction. Magnetic Resonance in Medicine, 51(6), 1247–1253. [DOI] [PubMed] [Google Scholar]
- 34.Feinberg DA, Turner R, Jakab PD, & von Kienlin M (1990). Echo-planar imaging with asymmetric gradient modulation and inner-volume excitation. Magnetic Resonance in Medicine, 13(1), 162–169. [DOI] [PubMed] [Google Scholar]
- 35.Morgan PS, Bowtell RW, McIntyre DJO, & Worthington BS (2004). Correction of spatial distortion in EPI due to inhomogeneous static magnetic fields using the reversed gradient method. Journal of Magnetic Resonance Imaging: JMRI, 19(4), 499–507. [DOI] [PubMed] [Google Scholar]
- 36.An Z, Tiwari V, Ganji SK, Baxter J, Levy M, Pinho MC, … Choi C (2018). Echo-planar spectroscopic imaging with dual-readout alternated gradients (DRAG-EPSI) at 7 T: Application for 2-hydroxyglutarate imaging in glioma patients. Magnetic Resonance in Medicine, 79(4), 1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labadie C, Hetzer S, Schulz J, Mildner T, Aubert-Frécon M, & Möller HE (2013). Center-out echo-planar spectroscopic imaging with correction of gradient-echo phase and time shifts. Magnetic Resonance in Medicine, 70(1), 16–24. [DOI] [PubMed] [Google Scholar]
- 38.Du W, Du YP, Fan X, Zamora MA, & Karczmar GS (2003). Reduction of spectral ghost artifacts in high-resolution echo-planar spectroscopic imaging of water and fat resonances. Magnetic Resonance in Medicine, 49(6), 1113–1120 [DOI] [PubMed] [Google Scholar]
- 39.Hanson LG, Schaumburg K, & Paulson OB (2000). Reconstruction strategy for echo planar spectroscopy and its application to partially undersampled imaging. Magnetic Resonance in Medicine, 44(3), 412–417. [DOI] [PubMed] [Google Scholar]
- 40.Webb P, Spielman D, & Macovski A (1989). A fast spectroscopic imaging method using a blipped phase encode gradient. Magnetic Resonance in Medicine, 12(3), 306–315. [DOI] [PubMed] [Google Scholar]
- 41.Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, & Wald LL (2012). Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magnetic Resonance in Medicine, 67(5), 1210–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zur Y (2011). Two-dimensional phase correction method for single and multi-shot echo planar imaging. Magnetic Resonance in Medicine, 66(6), 1616–1626. [DOI] [PubMed] [Google Scholar]
- 43.Hall EL, Stephenson MC, Price D, & Morris PG (2014). Methodology for improved detection of low concentration metabolites in MRS: optimised combination of signals from multi-element coil arrays. NeuroImage, 86, 35–42. [DOI] [PubMed] [Google Scholar]
- 44.Provencher SW (1993). Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magnetic Resonance in Medicine, 30(6), 672–679. [DOI] [PubMed] [Google Scholar]
- 45. https://www.acraccreditation.org/-/media/ACRAccreditation/Documents/MRI/LargePhantomGuidance.pdf?la=en.
- 46.Ebel A, & Maudsley AA (2005). Detection and correction of frequency instabilities for volumetric1H echo-planar spectroscopic imaging. Magnetic Resonance in Medicine, 53(2), 465–469. [DOI] [PubMed] [Google Scholar]
- 47.Heckova E, Považan M, Strasser B, Krumpolec P, Hnilicová P, Hangel GJ, … Bogner W (2018). Real-time Correction of Motion and Imager Instability Artifacts during 3D γ-Aminobutyric Acid-edited MR Spectroscopic Imaging. Radiology, 286(2), 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyu M, Barth M, Xie VB, Liu Y, Ma X, Feng Y, & Wu EX (2018). Robust SENSE reconstruction of simultaneous multislice EPI with low-rank enhanced coil sensitivity calibration and slice-dependent 2D Nyquist ghost correction. Magnetic Resonance in Medicine, 80(4), 1376–1390. [DOI] [PubMed] [Google Scholar]
- 49.Rosenzweig S, Holme HCM, Wilke RN, Voit D, Frahm J, & Uecker M (2018). Simultaneous multi-slice MRI using cartesian and radial FLASH and regularized nonlinear inversion: SMS-NLINV: SMS Reconstruction Using Nonlinear Inversion. Magnetic Resonance in Medicine, 79(4), 2057–2066. [DOI] [PubMed] [Google Scholar]
- 50.Boer VO, Klomp DWJ, Juchem C, Luijten PR, & de Graaf RA (2012). Multislice 1H MRSI of the human brain at 7 T using dynamic B0 and B1 shimming. Magnetic Resonance in Medicine, 68(3), 662–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Dual-band CAIPIRINHA variant III - g-factor maps and high resolution ACR phantom reconstruction of simultaneous excitation of adjacent (isl, isl+1) and not adjacent (isl, isl+3) slices. Set 1 and Set 2 examples show two different FOV and object dimensions. The scan parameters for set 1 were: TR/TE 1500/15 ms, FOV and thickness 270×192×8 mm3, in-plane resolution 3×3 mm2, SW =1000 Hz, for set 2: FOV and thickness 192×192×8 mm3, in-plane resolution 3×3 mm2, SW =1000 Hz.
Figure S2: Schematic illustration of the ghost artifacts for dual-band CAIPIRINHA case with adjacent (isl, isl+1) and not adjacent (isl, isl+3) slices, shown in a) and b) respectively. The simulation shows a spherical-shape phantom with signal inside mimicking metabolites equal to one and a ring mimicking lipid 10x higher. The ghost is simulated having 4% from the original signal. The adjacent slices have the same radius and the non-adjacent slices have different radius. The scale of the images is from 0 to 2.
Figure S3: In-vivo results of variants II and III. From top to bottom: TSE anatomy images of the scanned slices; real spectra for each MRSI method at three regions (each averaging over 5×5 voxels) in the central slice (shown in red, blue and green rectangle overlays); and images of six slices for NAA using spectra integration in the range of 0.4 ppm. The image intensities of NAA is normalized to the NAA intensity in each of the methods.
Figure S4: Creatine and Choline images of the in-vivo results of EPSI and variants I-IV acquisitions. Images of six slices are shown for each of the methods.
Figure S5. NAA/tCr ratio maps estimated by the LCModel fitting for EPSI and variant I-IV.
Figure S6. Single voxel spectra for each MRSI method at the blue ROI (central voxel) – EPSI and variants I-IV, respectively.
Figure S7. Lipid contamination estimation dual-band CAIPIRINHA variant III compared to variant I (in-vivo example from Fig.5). a) Ratio of the NAA images dual-band CAIPIRINHA variant III and variant I. b) histogram of the ratio values in a).
Table S1. Comparison of the SNR of variant I and II (in Figure 3), each relative to EPSI (as a reference).


