Abstract
AT-rich interaction domain 1A gene (ARID1A) encodes for a subunit of the switch/sucrose non-fermentable (SWI/SNF) complex, a chromatin remodeling complex, and it has been implicated in the pathogenesis of various cancer types. In this review, we discuss how ARID1A is linked to endometrial cancer and what molecular pathways are affected by mutation or inhibition of ARID1A. We also discuss the potential use of ARID1A not only as a prognostic biomarker, but also as a target for therapeutic interventions.
Keywords: ARID1A, endometrial cancer, molecular pathways, review
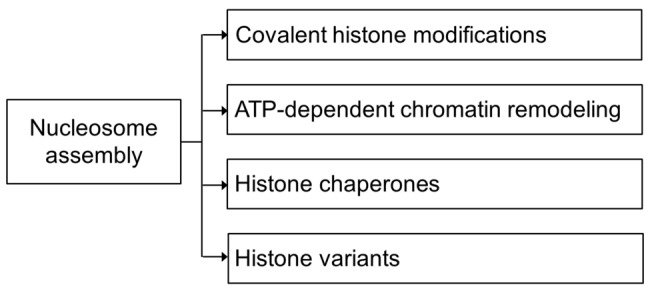
The dynamic modification of chromatin structure in a temporal- and spatial-specific manner determines cell fate by regulating expression levels of specific genes. The complexity of this process is further highlighted when considering all the endogenous and exogenous signals received by each cell during development and throughout its life. Numerous molecules (proteins and RNA) and macromolecular complexes are responsible for the organization of nucleosomes (Figure 1), epigenetic modifications, the dynamic change between the ‘relaxed’ or ‘tight’ conformation of chromatin (euchromatin and heterochromatin, respectively) and the accessibility of gene promoters determining cellular activities such as gene transcription, DNA repair and cell differentiation. Thus, disruption of normal chromatin remodeling impairs cellular development and homeostasis, and it has been associated extensively with tumorigenesis [reviewed in (1)].
Figure 1. Levels of nucleosome assembly. Nucleosome assembly depends on histone modifications, ATP-dependent chromatin remodeling, the action of histone chaperons and the involvement of different histone variants.
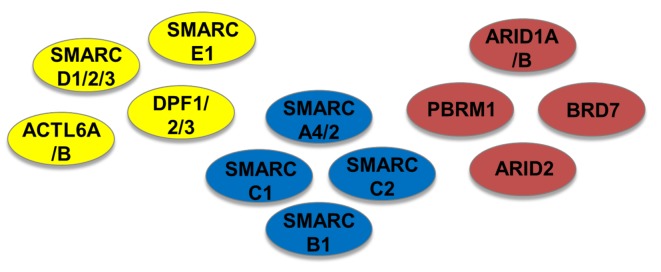
The switch/sucrose non-fermentable (SWI/SNF) complex is a nucleosome-remodeling factor found in both eukaryotes and prokaryotes. It is involved in gene expression through transcriptional regulation and plays a pivotal role in carcinogenesis (2). This complex changes the DNA conformation in nucleosomes, allowing recruitment of transcription factors or other complexes responsible for DNA repair, replication and proliferation. Thus, when the SWI/SNF complex is disrupted, aberrant cell cycling is observed, as well as a loss of control of proliferation (3). SWI/SNF is a multi-subunit complex and many of its subunits, such AT-rich interaction domain 1A (ARID1A), ARID1B, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 2 (SMARCA2) and SMARCA4 (Figure 2), have been incriminated as driving mutations in various cancer types due to the high mutation frequencies observed (4). In particular, when considering human primary cancer cases with mutations in the SWI/SNF complex, most of the mutations seen are encountered in the gene encoding ARID1A (5-7). ARID1A and ARID1B genes encode DNA-targeting subunits, while SMARCA2 and SMARCA4 encode ATPase enzymes. The mutation frequency of these subunits in different cancer types seems to be tumor type-specific indicating that there is probably differential participation of the complex in gene regulation in different tissues (4). Loss of ARID1A has been shown in numerous human malignancies, such as uterine endometrioid carcinoma (8-10), ovarian endometrioid carcinoma (11), gastric cancer (12,13), esophageal adenocarcinoma (14), pancreatic cancer (15) and colorectal cancer (16).
Figure 2. Switch/sucrose non-fermentable (SWI/SNF) complex consists of the core elements SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily A, member 4/2 (SMARCA4/2), SMARCC1, SMARCC2 and SMARCB1 (shown in blue); with the additional subunits SMARCD1/2/3, SMARCE1, actin-like 6A/B (ACTL6A/B) and double PHD fingers 1/2/3 (DPF1/2/3) (shown in yellow), and the variant components AT-rich interaction domain 1A/B (ARID1A/B), polybromo 1 (PBRM1), bromodomain-containing 7 (BRD7) and ARID2 (shown in red).
ARID1A
ARID1A gene has been characterized as a tumor suppressor, and the majority of the mutations seen in human cases are frame-shift or nonsense mutations (17). It encodes a protein with both nuclear and cytoplasmic localization, however, it was shown that nuclear ARID1A is unstable, as it is rapidly degraded by the nuclear ubiquitin-proteasome system (17,18). It is known that ARID1A expression is regulated by somatic mutations, copy number and methylation of its promoter (19). In-frame deletions disrupting the nuclear export signal lead to reduced ARID1A levels, due to the nuclear retention of the protein and its subsequent degradation (17,18). As the majority of ARID1A mutations result in truncated proteins and proteins prone to degradation, its mutation in cancer highly correlates with loss of its protein expression, which can be assessed by immunohistochemistry (12,15,20). In addition, missense mutations in the DNA-binding domain of ARID1A impaired the activity of the SWI/SNF complex by reducing its genomic occupancy in mouse embryos, which resulted in embryonic lethality due to cardiovascular development defects (21). Furthermore, loss of ARID1A promoted colon cancer in a mouse model of human colorectal cancer (16) indicating its involvement in different tissues. According to next-generation sequencing data, mutations in ARID1A were not only found in endometrial cancer, but at different frequencies in a variety of human malignancies, such as 3.2-3.5% in breast carcinoma (22), 9.1-15% in esophageal adenocarcinoma (14,23), 8-27% in gastric carcinoma (24), 8% in pancreatic carcinoma (25), 10-13% in hepatocellular carcinoma (26), 13% in transitional cell carcinoma of the bladder (27), 6% in neuroblastoma (28), and 17% in Burkitt lymphoma (29).
The extensive list of human malignancies found to have mutated ARID1A highlights the importance of this molecule and suggests that regulating its expression might be a viable therapeutic strategy. In fact, restoring ARID1A expression in ovarian cancer cells expressing mutant ARID1A resulted in suppression of proliferation and tumor growth in mice, whereas ARID1A silencing in a mouse tumor xenograft model increased cellular proliferation and tumorigenicity (8). Similar findings were also shown in cell lines of esophageal, breast and gastric cancer, with silencing of ARID1A leading to increased proliferation and restoration of its expression in cells carrying the mutant form leading to suppression of proliferation (23,24). In addition, further characterization in a murine preosteoblast cell line showed that ARID1A inhibition enhanced proliferation by preventing cell-cycle arrest through the regulation of c-MYC expression (30). Furthermore, using a high-throughput genetic screening approach, it was shown that inhibition of ARID1A not only enhanced proliferation, but also reduced apoptosis of Jurkat leukemia cells by inhibiting FAS-mediated cell death (31). When considering tumor progression, it was shown that loss of ARID1A was associated with reduced progression-free survival in ovarian clear cell carcinoma (32), as well as deep myometrial invasion in endometrial carcinoma (33). A role of ARID1A in tumor initiation was also supported by the identification of ARID1A mutations and expression loss in precancerous lesions of esophageal adenocarcinoma, with higher frequency of loss in lesions with severe dysplasia (14). All these studies suggest that ARID1A acts as a tumor suppressor and has an important role in tumor initiation, disease progression and apoptosis through its regulation of cellular proliferation in multiple cancer types, including endometrial cancer (32).
In direct contrast to cancer cells, where Arid1a loss promoted proliferation and halted cellular differentiation, its loss in healthy embryonic stem cells had the opposite effect by impairing the maintenance of the pluripotent characteristics of the cells, as well as cell proliferation (34). In particular, ablation of ARID1A in early mouse embryos resulted in developmental arrest and failure of mesodermal layer formation. In addition to defective development, loss of pluripotency, inhibition of self-renewal and promotion of differentiation into primitive-endoderm-like cells were seen. This study suggested that ARID1A has a key role in differentiation with lineage-specific characteristics, as in vitro development of neurons and skeletal muscle was permitted, but development of cardiomyocytes and adipocytes was inhibited. Thus, ARID1A function and expression should be considered in a spatial and temporal context in relation to therapeutic and diagnostic potential applications.
Endometrial Cancer and ARID1A
One of the most common and invasive types of gynecological cancer is endometrial cancer, which in 2015 accounted for 40% of all gynecological cancer cases worldwide (35). Endometrial cancer has been divided into two major groups according to clinical, pathological and molecular features: type I and type II. Type I encompasses endometrioid and estrogen-related cancer cases, and is the most frequent form, as it appears in 80% of patients suffering from endometrial cancer. It is characterized by unopposed estrogen stimulation, early onset of symptoms (young women) and relatively favorable prognosis. In contrast, type II endometrial cancer (non-endometrioid and non-estrogen-related) usually presents as serous and clear cell carcinomas, and this type of cancer is characterized by reduced association with hormonal risk factors, late onset symptoms and aggressive clinical course (36).
Epigenetic defects have been identified in endometrial carcinomas and have been implicated in the pathogenesis of these carcinomas (37). In particular, ARID1A has been found mutated in all types of endometrium-associated tumors, including undifferentiated and dedifferentiated endometrial carcinomas (38-40). More specifically, ARID1A mutations were found in 57% of ovarian clear-cell carcinomas (41), 40% of uterine endometrioid carcinomas (42), and between 20% and 36% of uterine carcinosarcomas (43); however, they were found to be rare in endometrial serous carcinoma (44). Furthermore, a study investigating the ARID1A loss in patients with endometrioid carcinoma reported that no ARID1A loss was seen in complex atypical hyperplasia, with loss increasing to 25% and 44% of patients with low-grade and high-grade endometrioid carcinomas, respectively (9). The differential mutation rates and expression levels of ARID1A in endometrial and ovarian cancer may denote that distinct processes might be involved in their tumorigenesis or disease progression. Thus, it is of great importance to study the role of ARID1A more not only in the pathogenesis of endometrial cancer, but also in its effect on the downstream molecular pathways in order to pinpoint the clinical relevance of the gene for both diagnostic and therapeutic interventions.
ARID1A has also been used as a prognostic marker in endometrial cancer. A significant association of reduced ARID1A expression has been found with shorter progression-free survival in patients suffering from endometrium-related cancer and, especially from ovarian clear-cell carcinoma, as well as with higher FIGO stage of both endometrial and ovarian cancer (45). Furthermore, the very rare occurrence of ARID1A mutations in complex atypical hyperplasia cases compared to endometrioid endometrial carcinoma (9,46), in conjunction with evidence linking ARID1A to tumor initiation and progression suggest that it might play a significant role in the transition from complex atypical hyperplasia to carcinoma through somatic inactivating mutations (47).
Molecular Pathways
ARID1A mutations affect the activity of SWI/SNF complexes, which, in turn, lead to transcriptional dysfunction resulting in aberrant transcriptional signatures in diverse pathways (6). Loss of components of SWI/SNF complex, including ARID1A, was found to disrupt not only the physical assembly of the complex, but also its association with genomic loci and the recruitment of interacting partners (co-activators or co-repressors), thereby impeding its remodeling activity (48). For example, under normal conditions, ARID1A protein interacts with topoisomerase IIα, which plays an important role in chromosome segregation during mitosis (49,50). Furthermore, it was shown that ARID1A is essential in maintaining chromatin accessibility through histone modifications at enhancers and this function is shared with ARID1B, a recently found homolog of ARID1A (51), highlighting the importance of enhancer architecture in health and disease (52). Loss of ARID1A was shown to induce neoplastic transformation in non-tumorigenic endometriotic cells due to alterations in histone modifications around AP-1 motifs at enhancers, indicating that ARID1A loss could be an early stage event in oncogenic transformation (53). When association was sought with other components of the SWI/SNF complex and ARID1A, reduced SMARCD3 expression was significantly associated with human tumors also exhibiting ARID1A loss, such as lung, pancreatic and kidney carcinomas and sarcomas (54-58).
In endometrioid endometrial carcinogenesis, activation of WNT signaling pathway has been observed, potentially induced by various molecular events such as inhibition of phosphatase and tensin homolog (PTEN), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene (PIK3CA) mutations, KRAS proto-oncogene GTPase gene (KRAS) mutations, microsatellite instability (MSI), tumor protein P53 gene (TP53) mutations or combinations of these defects (59). Apart from the high percentage of cases presenting with ARID1A mutations (40% of low-grade endometrioid carcinomas), loss of ARID1A expression was observed in 26-29% of low-grade and up to 39% of high-grade endometrioid carcinomas (20,46). In addition, co-occurrence of ARID1A mutations has been found in carcinomas exhibiting mutations of PTEN and PIK3CA, and ARID1A-deficient cancer cells exhibited significantly increase sensitivity to PI3K and protein kinase B (AKT) inhibition (60). Furthermore, impairment in the PI3K pathway is a very common feature of endometrioid carcinoma, accounting for more than 80% of cases, and ARID1A regulates this pathway through the phosphorylation of AKT (61). Due to the important effect of ARID1A deregulation in endometrial carcinogenesis, ARID1A has been used in addition to eight other genes (protein phosphatase 2 scaffold subunit Aalpha (PPP2R1A) gene, PTEN, PIK3CA, KRAS, catenin beta 1 (CTNNB1) gene, TP53, B-Raf proto-oncogene, serine/threonine kinase (BRAF) gene and PPP2R5C) to sub-classify endometrial carcinomas based on mutational profile (62). ARID1A has also been associated with MSI and mismatch repair (MMR) deficiency due to silencing of MMR genes in gastric carcinoma and uterine endometrioid carcinoma (13,59,63). In particular, loss of ARID1A was seen in sporadic MSI in significantly more cases than in other conditions, and ARID1A-regulated epigenetic silencing of mutL homolog 1 (MLH1) gene through promoter hypermethylation has been suggested to be the cause of MSI (59). Lately, converging pathways were identified for endometrial and ovarian carcinogenesis, through genetic and epigenetic analyses in Lynch syndrome, which is associated with hereditary defects in DNA repair resulting in increased MSI (64), while ARID1A loss seems to arise in early stages of carcinogenesis (65). Such a link of ARID1A and MSI was also described in gastric cancer, further supporting the hypothesis of mutated ARID1A-driven MSI (12,13,66). When considering the MMR pathways, ARID1A has been found to interact with MMR protein mutS homolog 2 (MSH2) and ARID1A deficiency contributes significantly to MMR impairment (67). Furthermore, Niskakoski et al. found a final diagnosis of endometrial cancer was given to all patients that presented with loss of either ARID1A gene or any of the four MMR genes MLH1, PMS2, MSH6 or MSH2 (64).
On the other hand, the molecular landscape in non-endometrioid cancer (type II) is characterized by high frequency of TP53 mutations, up-regulation of p16, human epidermal growth factor receptor 2 (HER2) gene amplification, global loss of heterozygosity and alterations in e-cadherin signaling (68-70). It has been shown that p53 protein directly interacts with the ARID1A–SMARCA4 complex and this interaction regulates the transcription of various genes including cyclin-dependent kinase inhibitor 1A (CDKN1A) gene and SMAD family member 3 (SMAD3) (8). It has been proposed that when ARID1A is lost, the CDKN1A and SDMA3 pathways are disrupted and PI3K/AKT pathway is aberrantly activated (59). Although loss of ARID1A and aberrant p53 expression have been described extensively in clear cell cancer (71-73), examination of ovarian clear-cell carcinoma, uterine carcinoma and endometrial carcinoma samples showed that mutations in both ARID1A and TP53 were mutually exclusive (59,63). ARID1A, p53 and β-catenin can be used as prognostic biomarkers in both clear-cell and endometrioid carcinoma, however, a significant correlation has been established only between ARID1A and β-catenin expression in endometrioid tumors (74). In a recent study, the estrogen receptor cofactor nuclear receptor interacting protein 1 (NRIP1) was found to be mutated in 12% of patients with metastatic endometrial cancer and these mutations were driving oncogenesis. In the same study, ARID1A mutations were not in agreement with the finding that driver events might be common in primary and metastatic tumors, demonstrating considerable heterogeneity within endometrial cancer cases and relative homogeneity in metastatic tissues (75).
Since tumors with ARID1A mutations are seen frequently in hormone-responsive tissues, it is worth considering the role of ARID1A in the regulation of hormonal signaling. Increased transcriptional activation through the glucocorticoid, estrogen, and androgen receptors was seen when ARID1A expression was restored in ARID1A-deficient breast cancer cells (76). Regarding other pathways affected by ARID1A, in ARID1A-deficient endometrial cancer cells, a significant increase in the level of forkhead box O1 (FOXO1) was observed along with down-regulation of the mitogen-activated protein kinase pathway and the insulin-like growth factor-1 signaling pathway (77). Although a systematic study investigating all pathways affected by mutant ARID1A is not yet available, the importance of such a study is emphasized by the fact that synthetic lethality was generated when enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) inhibitors were used as therapeutic agents in ARID1A-mutated cancer, through targeting the phosphoinositide-3-kinase interacting protein 1 (PIK3IP1), an ARID1A-regulated molecule of the PI3K/AKT pathway (78), while a new inhibitor of PI3Kα, showed positive results in human trials (79). Through synthetic lethality, suppression of cells growth and promotion of apoptosis are achieved, whereas ARID1B, might have potential as a therapeutic target in ARID1A-mutated cancer cells in a molecular-targeted therapeutic approach (51).
Conclusion
Endometrial cancer is one of the most common types of gynecological cancers, with carcinoma incidence increasing in every year worldwide. ARID1A loss has been found in every type of endometrial cancer, indicating its important role in initiation and progression of this disease. ARID1A, as member of SWI/SNF chromatin remodeling complex, interacts with numerous of transcription factors and other signaling molecules, while its association with several molecular pathways has been shown. Due to the fact that defects in ARID1A can affect the expression levels of various downstream genes, a better understanding is needed to elucidate which of the pathways affected are important in the pathogenesis and progression of cancer, and how these pathways can be targeted. Thus, the mechanism of action of ARID1A in initiation and progression of cancer cells needs to be clarified in order to develop new diagnostic tools and design new, more effective, therapeutic approaches.
Conflicts of Interest
All Authors declare that no conflict of interest exists in regard to this study.
Authors’ Contributions
CT and ENK researched and wrote the article. AL, IT, TT, SK, LI, XA and GT helped with the writing of the article, offered scientific advice and proof-read the article.
References
- 1.Nebbioso A, Tambaro FP, Dell’Aversana C, Altucci L. Cancer epigenetics: Moving forward. PLoS Genet. 2018;14:e10073622018–e10073622018. doi: 10.1371/journal.pgen.1007362. PMID: 29879107. DOI: 10.1371/ journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pulice JL, Kadoch C. Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb Symp Quant Biol. 2016;81:53–60. doi: 10.1101/sqb.2016.81.031021. PMID: 28408647. DOI: 10.1101/sqb.2016.81.031021. [DOI] [PubMed] [Google Scholar]
- 3.Nagl NG, Patsialou A, Haines DS, Dallas PB, Beck GR Jr., Moran E. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell-cycle arrest. Cancer Res. 2005;65:9236–9244. doi: 10.1158/0008-5472.CAN-05-1225. PMID: 16230384. DOI: 10.1158/0008-5472.CAN-05-1225. [DOI] [PubMed] [Google Scholar]
- 4.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. PMID: 23644491. DOI: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. PMID: 21654818. DOI: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 6.Wu JN, Roberts CW. ARID1A mutations in cancer: Another epigenetic tumor suppressor. Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. PMID: 23208470. DOI: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119–e55119. doi: 10.1371/journal.pone.0055119. PMID: 23355908. DOI: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan B, Wang TL, Shih IeM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. PMID: 21900401. DOI: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao TL, Ardighieri L, Ayhan A, Kuo KT, Wu CH, Wang TL, Shih IeM. Loss of ARID1A expression correlates with stages of tumor progression in uterine endometrioid carcinoma. Am J Surg Pathol. 2013;37:1342–1348. doi: 10.1097/PAS.0b013e3182889dc3. PMID: 24076775. DOI: 10.1097/ PAS.0b013e3182889dc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. PMID: 23636398. DOI: 10.1038/ nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih IeM, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. PMID: 20942669. DOI: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, Chan KH, Chan AS, Tsui WY, Ho SL, Chan AK, Man JL, Foglizzo V, Ng MK, Chan AS, Ching YP, Cheng GH, Xie T, Fernandez J, Li VS, Clevers H, Rejto PA, Mao M, Leung SY. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–852. doi: 10.1038/ng.2983. PMID: 24816253. DOI: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 13.Abe H1, Maeda D, Hino R, Otake Y, Isogai M, Ushiku AS, Matsusaka K, Kunita A, Ushiku T, Uozaki H, Tateishi Y, Hishima T, Iwasaki Y, Ishikawa S, Fukayama M. ARID1A expression loss in gastric cancer: Pathway-dependent roles with and without Epstein-Barr virus infection and microsatellite instability. Virchows Arch. 2012;461:367–377. doi: 10.1007/s00428-012-1303-2. PMID: 22915242. DOI: 10.1007/s00428-012-1303-2. [DOI] [PubMed] [Google Scholar]
- 14.Streppel MM, Lata S, DelaBastide M, Montgomery EA, Wang JS, Canto MI, Macgregor-Das AM, Pai S, Morsink FH, Offerhaus GJ, Antoniou E, Maitra A, McCombie WR. Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett’s esophagus. Oncogene. 2014;33:347–357. doi: 10.1038/onc.2012.586. PMID: 23318448. DOI: 10.1038/onc.2012.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA, Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM. Australian Pancreatic Cancer Genome Initiative. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. PMID: 25719666. DOI: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, Roberts CW. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302. doi: 10.1038/ng.3744. PMID: 27941798. DOI: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao TL, Shih IeM. The roles of ARID1A in gynecologic cancer. J Gynecol Oncol. 2013;24:376–381. doi: 10.3802/jgo.2013.24.4.376. PMID: 24167674. DOI: 10.3802/jgo.2013.24.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan B, Gao M, Wu CH, Wang TL, Shih IeM. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia. 2012;14:986–993. doi: 10.1593/neo.121218. PMID: 23097632. DOI: 10.1593/ neo.121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Sun Q, Shan M, Niu M, Liu T, Xia B, Liang X, Wei W, Sun S, Zhang Y, Liu XS, Song Q, Yang Y, Ma Y, Liu Y, Yang L, Ren Y, Zhang G, Pang D. Promoter hypermethylation of ARID1A gene is responsible for its low mRNA expression in many invasive breast cancers. PLoS ONE. 2013;8:e53931–e53931. doi: 10.1371/journal.pone.0053931. PMID: 23349767. DOI: 10.1371/journal.pone.0053931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiegand KC, Lee AF, Al-Agha OM, Chow C, Kalloger SE, Scott DW, Steidl C, Wiseman SM, Gascoyne RD, Gilks B, Huntsman DG. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol. 2011;224:328–333. doi: 10.1002/path.2911. PMID: 21590771. DOI: 10.1002/path.2911. [DOI] [PubMed] [Google Scholar]
- 21.Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. ARID1A-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 2013;33:265–280. doi: 10.1128/MCB.01008-12. PMID: 23129809. DOI: 10.1128/MCB.01008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornen S, Adelaide J, Bertucci F, Finetti P, Guille A, Birnbaum DJ, Birnbaum D, Chaffanet M. Mutations and deletions of ARID1A in breast tumors. Oncogene. 2012;31:4255–4256. doi: 10.1038/onc.2011.598. PMID: 22249247. DOI: 10.1038/onc.2011.598. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, Wang LD, Wang L, Yang W, Velculescu VE, Vogelstein B, Papadopoulos N, Kinzler KW, Meltzer SJ. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. PMID: 22877736. DOI: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. PMID: 22484628. DOI: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 25.Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, Maitra A, Pollack JR. Convergent structural alterations defne switch/ sucrose nonfermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci USA. 2012;109:E252–E259. doi: 10.1073/pnas.1114817109. PMID: 22233809. DOI: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. PMID: 22634756. DOI: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 27.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, He M, Li Z, Sun X, Jia W, Chen J, Yang S, Zhou F, Zhao X, Wan S, Ye R, Liang C, Liu Z, Huang P, Liu C, Jiang H, Wang Y, Zheng H, Sun L, Liu X, Jiang Z, Feng D, Chen J, Wu S, Zou J, Zhang Z, Yang R, Zhao J, Xu C, Yin W, Guan Z, Ye J, Zhang H, Li J, Kristiansen K, Nickerson ML, Theodorescu D, Li Y, Zhang X, Li S, Wang J, Yang H, Wang J, Cai Z. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. PMID: 21822268. DOI: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA Jr., Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE, Hogarty MD. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. PMID: 23202128. DOI: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, Palmer G, Lucena-Silva N, Pedrosa F, Pedrosa M, Teruya-Feldstein J, Bhagat G, Alobeid B, Leoncini L, Bellan C, Rogena E, Pinkney KA, Rubin MA, Ribeiro RC, Yelensky R, Tam W, Stephens PJ, Cesarman E. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–5184. doi: 10.1182/blood-2012-06-437624. PMID: 23091298. DOI: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagl NG, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. PMID: 17255939. DOI: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. PMID: 19091943. DOI: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katagiri A, Nakayama K, Rahman MT, Rahman M, Katagiri H, Nakayama N, Ishikawa M, Ishibashi T, Iida K, Kobayashi H, Otsuki Y, Nakayama S, Miyazaki K. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol. 2012;25:282–288. doi: 10.1038/modpathol.2011.161. PMID: 22101352. DOI: 10.1038/modpathol. 2011.161. [DOI] [PubMed] [Google Scholar]
- 33.Werner HM, Berg A, Wik E, Birkeland E, Krakstad C, Kusonmano K, Petersen K, Kalland KH, Oyan AM, Akslen LA, Trovik J, Salvesen HB. ARID1A loss is prevalent in endometrial hyperplasia with atypia and low-grade endometrioid carcinomas. Mod Pathol. 2013;26:428–434. doi: 10.1038/modpathol.2012.174. PMID: 23080032. DOI: 10.1038/modpathol.2012.174. [DOI] [PubMed] [Google Scholar]
- 34.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germlayer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. PMID: 18448678. DOI: 10.1073/ pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. PMID: 25559415. DOI: 10.3322/ caac.21254. [DOI] [PubMed] [Google Scholar]
- 36.Prat J, Gallardo A, Cuatrecasas M, Catasus L. Endometrial carcinoma: pathology and genetics. Pathology. 2007;39:72–87. doi: 10.1080/00313020601136153. PMID: 17365824. DOI: 10.1080/00313020601136153. [DOI] [PubMed] [Google Scholar]
- 37.Bartosch C, Lopes JM, Jerónimo C. Epigenetics in endometrial carcinogenesis – part 2: histone modifications, chromatin remodeling and noncoding RNAs. Epigenomics. 2017;9:873–892. doi: 10.2217/epi-2016-0167. PMID: 28523964. DOI: 10.2217/epi-2016-0167. [DOI] [PubMed] [Google Scholar]
- 38.Köbel M, Hoang LN, Tessier-Cloutier B, Meng B, Soslow RA, Stewart CJR, Lee CH. Undifferentiated endometrial carcinomas show frequent loss of core switch/sucrose nonfermentable complex proteins. Am J Surg Pathol. 2018;42:76–83. doi: 10.1097/PAS.0000000000000941. PMID: 28863077. DOI: 10.1097/PAS.0000000000000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coatham M, Li X, Karnezis AN, Hoang LN, Tessier-Cloutier B, Meng B, Soslow RA, Blake Gilks C, Huntsman DG, Stewart CJ, Postovit LM, Köbel M, Lee CH. Concurrent ARID1A and ARID1B inactivation in endometrial and ovarian dedifferentiated carcinomas. Mod Pathol. 2016;29:1586–1593. doi: 10.1038/modpathol.2016.156. PMID: 27562491. DOI: 10.1038/modpathol.2016.156. [DOI] [PubMed] [Google Scholar]
- 40.Espinosa I, De Leo A, D’Angelo E, Rosa-Rosa JM, Corominas M, Gonzalez A, Palacios J, Prat J. Dedifferentiated endometrial carcinomas with neuroendocrine features: A clinicopathologic, immunohistochemical, and molecular genetic study. Hum Pathol. 2018;72:100–106. doi: 10.1016/j.humpath.2017.11.006. PMID: 29133142. DOI: 10.1016/j.humpath.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. PMID: 28055103. DOI: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton CA, Cheung MK, Osann K, Chen L, Teng NN, Longacre TA, Powell MA, Hendrickson MR, Kapp DS, Chan JK. Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br J Cancer. 2006;94:642–646. doi: 10.1038/sj.bjc.6603012. PMID: 16495918. DOI: 10.1038/sj.bjc.6603012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boruta DM II, Gehrig PA, Fader AN, Olawaiye AB. Management of women with uterine papillary serous cancer: A Society of Gynecologic Oncology (SGO) review. Gynecol Oncol. 2009;115:142–153. doi: 10.1016/j.ygyno.2009.06.011. PMID: 19592079. DOI: 10.1016/ j.ygyno.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Fadare O, Roma AA, Parkash V, Zheng W, Walavalkar V. Does a p53 "wild-type" immunophenotype exclude a diagnosis of endometrial serous carcinoma. Adv Anat Pathol. 2018;25:61–70. doi: 10.1097/PAP.0000000000000171. PMID: 28945609. DOI: 10.1097/PAP.0000000000000171. [DOI] [PubMed] [Google Scholar]
- 45.Liu G, Xu P, Fu Z, Hua X, Liu X, Li W, Zhang M, Wu J, Wen J, Xu J, Jia X. Prognostic and clinicopathological significance of ARID1A in endometrium-related gynecological cancers: a meta-analysis. J Cell Biochem. 2017;118:4517–4525. doi: 10.1002/jcb.26109. PMID: 28466574. DOI: 10.1002/jcb.26109. [DOI] [PubMed] [Google Scholar]
- 46.Guan B, Mao TL, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL, Shih IeM. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. PMID: 21412130. DOI: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yen TT, Miyamoto T, Asaka S, Chui MH, Wang Y, Lin SF, Stone RL, Fader AN, Asaka R, Kashima H, Shiozawa T, Wang TL, Shih I, Tanner EJ 3rd. Loss of ARID1A expression in endometrial samplings is associated with the risk of endometrial carcinoma. Gynecol Oncol. 2008;150:426–431. doi: 10.1016/j.ygyno.2018.06.025. PMID: 30126589. DOI: 10.1016/j.ygyno.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 48.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. PMID: 10078207. DOI: https://doi.org/10.1016/S1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 49.Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nat Struct Mol Biol. 2005;12:589–593. doi: 10.1038/nsmb953. PMID: 15965487. DOI: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- 50.Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, Pfister S, Cho Y-J, Zhao K, Crabtree GR. mSWI/SNF (BAF) complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;497:624–627. doi: 10.1038/nature12146. PMID: 23698369. DOI: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, Kim Y, Kryukov GV, Ghandi M, Aguirre AJ, Jagani Z, Wang Z, Garraway LA, Hahn WC, Roberts CW. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20:251–254. doi: 10.1038/nm.3480. PMID: 24562383. DOI: 10.1038/ nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelso TWR, Porter DK, Amaral ML, Shokhirev MN, Benner C, Hargreaves DC. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. Elife. 2017;6:e30506–e30506. doi: 10.7554/eLife.30506. PMID: 28967863. DOI: 10.7554/ eLife.30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lakshminarasimhan R, Andreu-Vieyra C, Lawrenson K, Duymich CE, Gayther SA, Liang G, Jones PA. Down-regulation of ARID1A is sufficient to initiate neoplastic transformation along with epigenetic reprogramming in non-tumorigenic endometriotic cells. Cancer Lett. 2017;401:11–19. doi: 10.1016/j.canlet.2017.04.040. PMID: 28483516. DOI: 10.1016/j.canlet.2017.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medina PP, Sanchez-Cespedes M. Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer. Epigenetics. 2008;3:64–68. doi: 10.4161/epi.3.2.6153. PMID: 18437052. DOI: 10.4161/epi.3.2.6153. [DOI] [PubMed] [Google Scholar]
- 55.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. PMID: 12566296. [PubMed] [Google Scholar]
- 56.Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland AM, Smith R, Salada G, Carillo A, Laity K, Gupte J, Swedlund B, Tavtigian SV, Teng DH, Lees E. BRG1, a component of the SWI–SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–6177. PMID: 11085541. [PubMed] [Google Scholar]
- 57.Dal MM, Hong SM, Hebbar S, Sharma R, Scrimieri F, de Wilde RF, Mayo SC, Goggins M, Wolfgang CL, Schulick RD, Lin MT, Eshleman JR, Hruban RH, Maitra A, Matthaei H. Loss of expression of the SWI/SNF chromatin remodeling subunit BRG1/ SMARCA4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum Pathol. 2012;43:585–591. doi: 10.1016/j.humpath.2011.06.009. PMID: 21940037. DOI: 10.1016/j.humpath.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hollmann TJ, Hornick JL. INI1-deficient tumors: Diagnostic features and molecular genetics. Am J Surg Pathol. 2011;35:e47–e63. doi: 10.1097/PAS.0b013e31822b325b. PMID: 21934399. DOI: 10.1097/PAS.0b013e31822b325b. [DOI] [PubMed] [Google Scholar]
- 59.Bosse T, ter Haar NT, Seeber LM, v Diest PJ, Hes FJ, Vasen HF, Nout RA, Creutzberg CL, Morreau H, Smit VT. Loss of ARID1A expression and its relationship with PI3K-AKT pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod Pathol. 2013;26:1525–35. doi: 10.1038/modpathol.2013.96. PMID: 23702729. DOI: 10.1038/modpathol.2013.96. [DOI] [PubMed] [Google Scholar]
- 60.Samartzis EP, Gutsche K, Dedes KJ, Fink D, Stucki M, Imesch P. Loss of ARID1A expression sensitizes cancer cells to PI3K and AKT inhibition. Oncotarget. 2014;5:5295–5303. doi: 10.18632/oncotarget.2092. PMID: 24979463. DOI: 10.18632/oncotarget.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, Guo W, Scherer SE, Carter H, Westin SN, Dyer MD, Verhaak RG, Zhang F, Karchin R, Liu CG, Lu KH, Broaddus RR, Scott KL, Hennessy BT, Mills GB. Whole exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. PMID: 23028188. DOI: 10.1101/ gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McConechy MK, Ding J, Cheang MC, Wiegand K, Senz J, Tone A, Yang W, Prentice L, Tse K, Zeng T, McDonald H, Schmidt AP, Mutch DG, McAlpine JN, Hirst M, Shah SP, Lee CH, Goodfellow PJ, Gilks CB, Huntsman DG. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012;228:20–30. doi: 10.1002/path.4056. PMID: 22653804. DOI: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allo G, Bernardini MQ, Wu RC, Shih IeM, Kalloger S, Pollett A, Gilks CB, Clarke BA. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod Pathol. 2014;27:255–261. doi: 10.1038/modpathol.2013.144. PMID: 23887303. DOI: 10.1038/modpathol.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niskakoski A, Pasanen A, Porkka N, Eldfors S, Lassus H, Renkonen-Sinisalo L, Kaur S, Mecklin JP, Bützow R, Peltomäki P. Converging endometrial and ovarian tumorigenesis in Lynch syndrome: Shared origin of synchronous carcinomas. Gynecol Oncol. 2018;150:92–98. doi: 10.1016/j.ygyno.2018.04.566. PMID: 29716739. DOI: 10.1016/ j.ygyno.2018.04.566. [DOI] [PubMed] [Google Scholar]
- 65.Niskakoski A, Pasanen A, Lassus H, Renkonen-Sinisalo L, Kaur S, Mecklin JP, Bützow R, Peltomäki P. Molecular changes preceding endometrial and ovarian cancer: a study of consecutive endometrial specimens from Lynch syndrome surveillance. Mod Pathol. 2018;31:1291–1301. doi: 10.1038/s41379-018-0044-4. PMID: 29588532. DOI: 10.1038/ s41379-018-0044-4. [DOI] [PubMed] [Google Scholar]
- 66.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, Hruban RH, Eshleman JR, Iacobuzio-Donahue CA, Goggins M, Maitra A, Malek SN, Powell S, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. PMID: 22009941. DOI: 10.1002/ humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, Nagel ZD, Zou J, Wang C, Kapoor P, Ma X, Ma D, Liang J, Song S, Liu J, Samson LD, Ajani JA, Li GM, Liang H, Shen X, Mills GB, Peng G. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–562. doi: 10.1038/s41591-018-0012-z. PMID: 29736026. DOI: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng W, Xiang L, Fadare O, Kong B. A proposed model for endometrial serous carcinogenesis. Am J Surg Pathol. 2011;35:e1–e14. doi: 10.1097/PAS.0b013e318202772e. PMID: 21164282. DOI: 10.1097/PAS.0b013e318202772e. [DOI] [PubMed] [Google Scholar]
- 69.Mariani A, Sebo TJ, Webb MJ, Riehle D, Katzmann JA, Keeney GL, Roche PC, Lesnick TG, Podratz KC. Molecular and histopathologic predictors of distant failure in endometrial cancer. Cancer Detect Prev. 2003;27:434–441. doi: 10.1016/j.cdp.2003.09.005. PMID: 14642551. DOI: 10.1016/j.cdp.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 70.Lax SF. Molecular genetic changes in epithelial, stromal and mixed neoplasms of the endometrium. Pathology. 2007;39:46–54. doi: 10.1080/00313020601146822. PMID: 17365822. DOI: 10.1080/00313020601146822. [DOI] [PubMed] [Google Scholar]
- 71.Hoang LN, McConechy MK, Meng B, McIntyre JB, Ewanowich C, Gilks CB, Huntsman DG, Köbel M, Lee CH. Targeted mutation analysis of endometrial clear cell carcinoma. Histopathology. 2015;66:664–674. doi: 10.1111/his.12581. PMID: 25308272. DOI: 10.1111/his.12581. [DOI] [PubMed] [Google Scholar]
- 72.Fadare O, Gwin K, Desouki MM, Crispens MA, Jones HW 3rd, Khabele D, Liang SX, Zheng W, Mohammed K, Hecht JL, Parkash V. The clinicopathologic significance of p53 and BAF-250a (ARID1A) expression in clear cell carcinoma of the endometrium. Mod Pathol. 2013;26:1101–1110. doi: 10.1038/modpathol.2013.35. PMID: 23524907. DOI: 10.1038/modpathol.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.An HJ, Logani S, Isacson C, Ellenson LH. Molecular characterization of uterine clear cell carcinoma. Mod Pathol. 2004;17:530–537. doi: 10.1038/modpathol.3800057. PMID: 14976538. DOI: 10.1038/modpathol. 3800057. [DOI] [PubMed] [Google Scholar]
- 74.Heckl M, Schmoeckel E, Hertlein L, Rottmann M, Jeschke U, Mayr D. The ARID1A, p53 and β-Catenin statuses are strong prognosticators in clear-cell and endometrioid carcinoma of the ovary and the endometrium. PLoS One. 2018;13:e0192881–e0192881. doi: 10.1371/journal.pone.0192881. PMID: 29451900. DOI: 10.1371/journal.pone.0192881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gibson WJ, Hoivik EA, Halle MK, Taylor-Weiner A, Cherniack AD, Berg A, Holst F, Zack TI, Salvesen HB. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet. 2016;48:848–855. doi: 10.1038/ng.3602. PMID: 27348297. DOI: 10.1038/ng.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem. 2002;277:41674–41685. doi: 10.1074/jbc.M205961200. PMID: 12200431. DOI: 10.1074/jbc.M205961200. [DOI] [PubMed] [Google Scholar]
- 77.Yang Y, Bao W, Sang Z, Yang Y, Lu M, Xi X. Microarray pathway analysis indicated that mitogen-activated protein kinase/extracellular signal-regulated kinase and insulin growth factor 1 signaling pathways were inhibited by small interfering RNA against AT-rich interactive domain 1A in endometrial cancer. Oncol Lett. 2018;15:1829–1838. doi: 10.3892/ol.2017.7489. PMID: 29399196. DOI: 10.3892/ol.2017.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih Ie M, Conejo-Garcia JR, Speicher DW, Zhang R. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. PMID: 25686104. DOI: 10.1038/ nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Juric D, Rodon J, Tabernero J, Janku F, Burris HA, Schellens JHM, Middleton MR, Berlin J, Schuler M, Gil-Martin M, Rugo HS, Seggewiss-Bernhardt R, Huang A, Bootle D, Demanse D, Blumenstein L, Coughlin C, Quadt C, Baselga J. Phosphatidylinositol 3-kinase α-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: Results From the first-in-human study. J Clin Oncol. 2018;36:1291–1299. doi: 10.1200/JCO.2017.72.7107. PMID: 29401002. DOI: 10.1200/JCO.2017.72.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]