Abstract
Background: Mycosis fungoides (MF) is indolent, but may disseminate to leukemia. We reported that C-C motif chemokine ligand 21 (CCL21) is associated with MF invasion and progression. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long noncoding RNA, is associated with several cancer types, however, how it interacts with CCL21 to regulate MF progression, remains unclear. Materials and Methods: Expression of long noncoding RNAs MALAT1, antisense noncoding RNA in the INK4 locus (ANRIL), Hox antisense intergenic RNA (HOTAIR), highly up-regulated in liver cancer RNA (HULC), and leukemia-associated non-coding insulin-like growth factor 1 receptor activator RNA 1 (LUNAR1) in tissues from MF was studied using polymerase chain reaction and RNA interference in MF cell line MyLa were used to address this question. Results: Expression of MALAT1 was selectively increased in MF tissues. C-C Chemokine receptor type 7 (CCR7) expression was found to be increased in MyLa cells. CCL21 was found not only to mediate migration, but also to enhance MALAT1 and mammalian target of rapamycin (mTOR) activation in MyLa cells. Knockdown of MALAT1 abrogated CCL21-mediated migration, but not mTOR activation. In contrast, mTOR inhibition reduced CCL21-mediated migration and MALAT1 expression. Conclusion: CCL21 induced mTOR activation in MyLa cells, followed by expression of MALAT1, causing cell migration. MALAT1 and mTOR are potential therapeutic targets for MF.
Keywords: Mycosis fungoides, MALAT1, lncRNA, mTOR, migration
Primary cutaneous lymphomas belong to a heterogenous group of extra-nodal non-Hodgkin lymphomas which are usually confined to skin at initial presentation (1). Whereas many nodal lymphomas originate from B-cells, about 75% of primary cutaneous lymphomas derive from T-cells. The incidence of cutaneous T-cell lymphomas (CTCL) has been increasing and is estimated to be 6.4 per million persons, more in males and in the elderly (2,3). Among these T-cell-derived primary cutaneous lymphoma, two-thirds are mycosis fungoides (MF) (2,4). MF usually runs an indolent course for several decades with confinement to the skin, however, in advanced MF, malignant lymphocytes may disseminate to lymph nodes and metastasize to peripheral blood and visceral organs (Sézary syndrome). A variety of MF tumor cells have been shown to express chemokine receptors, which have been demonstrated to be involved in organ-specific cancer metastasis. The role of chemokines and chemokine receptors in the pathogenesis of MF and other CTCLs has been reviewed by us and others (5,6). Moreover, our previous study showed that C-C chemokine receptor type 7 (CCR7) was expressed in 62% of tissue specimens of MF, and its expression correlated with subcutaneous extension of lymphoma cells (6).
In the human genome, only 2-3% out of 3 billion bases actually encode protein-related transcriptional messages (7). More than 90% of these bases are transcribed to non-protein coding RNA (8,9), which was initially considered a non-functional part of the genome (10). However, exons with transcripts do not exclusively explain the pathophysiology and progression of several diseases, such as cancer metastasis (11,12). Epigenetic regulation includes regulation by means other than the traditional paradigm of mRNA transcription and protein production. It may involve chromatin changes, histone modifications, DNA methylation, microRNAs (miRs), and other non-coding RNA types. In CTCL, the treatment efficacy of histone deacetylase inhibitor (HDACi) suggests the involvement of histone modification in the progression of CTCL (13). Microarray data from tumorous MF tissue indicated that miR-34a is a candidate oncogenic molecule and miR-29a acts as a tumor suppressor, highlighting their regulatory role in the progression of MF (14). DNA methylation analysis has shown that Sézary syndrome is characterized by widespread yet distinct DNA methylation alterations, and that promoter hypermethylation of a single gene, chemokine-like factor chemokine-like factor-like MARVEL transmembrane domain containing 2 (CMTM2), was sufficient to accurately distinguish it from other erythrodermic inflammatory diseases (15). Together this evidence suggests epigenetic regulation may play a significant role on the pathogenesis and progression of MF.
Long noncoding RNA (lncRNA), a specific type of RNA with long noncoding domains, has recently aroused research interest due to their multi-functional and pluripotential role in many biological processes. LncRNAs actively regulate gene expression in carcinogenesis. In many cancer types, hundreds of lncRNAs become dysregulated, among which some act as tumor suppressors or promoters. LncRNAs contribute to various epigenetic processes, including dynamic coordination of chromatin, regulation of DNA methylation, modulation of RNA stability, and coordination of altered tumor metabolism [reviewed in (16)]. In 2014, Xing et al. reported the role of lncRNA breast cancer anti-estrogen resistance 4 (BCAR4) in breast cancer metastasis, showing that CCL21 activates BCAR4 by releasing SMAD nuclear interacting protein 1 (SNIP1) inhibition of p300-dependent histone acetylation, enabling BCAR4-recruited protein phosphatase 1 regulatory subunit 10 to bind H3K18ac and relieve inhibition of RNA Pol II to facilitate gene transcription (17). In mesenchymal stem cells, chemokine (C-X-C motif) ligand 13 (CXCL13) was shown to mediate the positive regulation of AK028326, a lncRNA, in osteogenic gene expression (18). In cholangiocarcinoma, lncRNAs H19 and highly up-regulated in liver cancer RNA (HULC) were shown to regulate cell migration and invasion by targeting CXCR4 via miR-372/miR-373 (19). Methylation of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), through CXCL5, was found to regulate cell migration and invasion by non-small-cell lung cancer cells (20). In Sezary syndrome, transcriptome assembly analysis by multiple algorithms identified 13 differentially expressed unannotated transcripts that include 12 predicted lncRNAs (21). These data suggest that lncRNAs might play a role in the pathogenesis and progression of MF. In this study, we aimed to investigate the role of selected lncRNAs in the progression of MF and determine how chemokine CCL21 was involved in the process.
Materials and Methods
Identification of patients with MF. Five patients with biopsy-proven MF and three age- and gender-matched healthy controls were identified from the Kaohsiung Chang Gung Memorial Hospital in southern Taiwan. None of them had received phototherapy, radiotherapy, nor chemotherapy before skin biopsy. The study was approved by the Institutional Review Board (approval number 104-9782A3) of the hospital and conducted according to the Declaration of Helsinki.
Real-time polymerase chain reaction (PCR) study of MF skin lesions. Skin tissue was obtained and processed to isolate tissue RNA using RNeasy Mini Kit (Qiagen, Germantown, MD, USA). Quantification and purity of the RNA were assessed by A260/A280 absorption (Nanodrop spectrophotometer; Thermo Fisher (Waltham, MA, USA), and RNA samples with ratios greater than 1.7 were stored at –70˚C for further analysis. Extracted RNA (1 μl) was then subjected to PCR amplification using MPCR kits (Maxim Biotech, San Francisco, CA, USA) according to the manufacturer’s instructions. The primer sequences used for MALAT1, antisense noncoding RNA in the INK4 locus (ANRIL), Hox antisense intergenic RNA (HOTAIR), HULC, and leukemia-associated non-coding insulin-like growth factor 1 receptor activator RNA 1 (LUNAR1) are listed in Table I. The reactions were carried out under following conditions: 96˚C for 1 min and 60˚C for 4 min; 30 cycles of 94˚C for 1 min and 60˚C for 3 min; and extension at 70˚C for 10 min.
Table I. Primer sequences of long noncoding RNAs studied.
F: Forward; R: reverse.
Cell line and cell cultures. The human MF cell line MyLa obtained from European Collection of Cell Cultures (Porton Down, Salisbury, UK) was grown in RPMI 1640 medium with 10% fetal bovine serum (FBS; Gibco, Waltham, MA, USA). All cells were maintained at 37˚C in a humid atmosphere with 5% CO2. Jurkat cells (ATCC® TIB-152) were acquired from the American Type Culture Collection (Germantown, DC, USA). Peripheral blood mononuclear cells (PBMCs) were obtained from healthy volunteers and used as controls in experiments to measure cell migration and expression of chemokine receptors.
Transfection of small interfering RNA (siRNA). Cells were seeded to be 70-90% confluent before transfection. Cells were transfected with MALAT1 siRNA or negative control (NC) siRNA (both from Ambion, Waltham, MA, USA) using Lipofectamine® 3000 Transfection Reagent according to instructions from the manufacturer (ThermoFisher, Waltham, MA, USA). Lipofectamine® 3000 Reagent was diluted in siRNA-containing serum-free medium for 15 min at room temperature. The siRNA–lipid complex was then added to the cells for 6 h. Thereafter, the siRNA-lipid complex was replaced and incubated with FBS at 37˚C in a humid atmosphere with 5% CO2 before the subsequent western blot and quantitative PCR.
Western blotting. Cells were grown in 6 cm dishes until near confluence. MyLa cells were treated for 10, 20, 40 or 60 min with CCL21 at 10 ng/ml. In order to investigate whether CCL21-induced MALAT1 expression was dependent on mammalian target of rapamycin (mTOR), cells were pre-treated with 20 ng/ml rapamycin (Calbiochem, Billerica, MA, USA) for 2 h before the addition of CCL21. Following treatment, the cells were harvested and the protein content was measured by the protein assay dye (BIO-RAD, Hercules, CA, USA) The proteins were then extracted based on the manufacturer's instructions. Equivalent amounts of protein per sample were electrophoretically resolved on 10% polyacrylamide gels and transferred onto 0.2 mm nitrocellulose membranes. Proteins were probed overnight with primary antibodies against mTOR (1:500), phospho-(p)mTOR (1:1,000), extracellular-regulated protein kinases (ERK) (1:2,000), p-ERK (1:2,000), signal transducer and activator of transcription 3 (STAT3)(1:1,000), p-STAT3 (1:1,000) (all from Cell Signaling, Danvers, MA, USA) and actin (1:1,000; Millipore, Burlington, MA, USA). The nitrocellulose membranes were then incubated with an appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature, and the immunoreactivity was observed by enhanced chemiluminescence detection, a semiquantitative assay through blot dosimetry. Anti-β-actin (Epitomics, Cambridge, MA, USA) was used to check for equal loading of protein between wells.
Cell migration assay. Migration was measured in a 96-well double-chamber transwell chemotaxis assay using filters with 8-mm pore membranes (Neuro Probe, Gaithersburg, MD, USA). The upper chamber was seeded with 106 cells/ml (in 29 ml of medium). In the lower chamber, either vehicle or chemokine (CCL21) was added to the culture medium (RPMI1640). In some wells, cells were pre-treated with rapamycin (Calbiochem, Billerica, MA, USA) for 2 h to determine the effect of mTOR inhibition on the migratory response. Plates were then incubated for 8 h at 37˚C. After incubation for 2 and 4 h, non-migrating cells were collected from the membrane using a cotton-tipped swab. Cells migrating through the filter to the lower surface were counted at 100× magnification.
Statistical analysis. The data are presented as means±SD. The two-sample t-test was used to compare continuous data from two groups. For comparisons of continuous data from multiple groups, one-way analysis of variance (ANOVA) was performed. For western blot densitometric analysis, Dunnett’s test was used to compare different treatment groups with an untreated control. Statistical analysis was assessed using Prism software (Graphpad, La Jolla, CA, USA). Differences were regarded as statistically significant when the p-value was less than 0.05.
Results
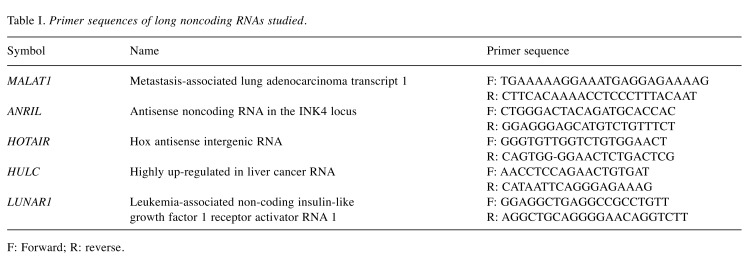
MALAT1 was up-regulated in skin tissues from patients with MF. We first asked whether the expression of lncRNAs MALAT1, ANRIL, HOTAIR, HULC, and LUNAR1 would be dysregulated in the tissues from patients with MF (Figure 1). Skin tissue samples were obtained from healthy controls (n=3, age range=45-70 years) and patients with MF (n=5, age range=56-72 years). The expression of MALAT1 in tissues from patients with MF was approximately 5-fold that of the controls. In contrast, the expressions of HOTAIR and ANRIL were similar in skin tissues from the two groups. The expressions of LUNAR1 and HULC were low in both groups. The result showed that there was selectively and significantly increased expression of MALAT1 in MF tissue.
Figure 1. Selectively and significantly increased expression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in skin tissues from patients with mycosis fungoides (MF). Skin tissues were taken from lesional skin in patients with MF and from healthy controls (n=5 and n=3, respectively). Extracted tissue mRNA was processed for real-time polymerase chain reaction. The relative expression of MALAT1 (A), Hox antisense intergenic RNA (HOTAIR) (B), leukemia-associated non-coding insulin-like growth factor 1 receptor activator RNA 1 (LUNAR1) (C), antisense noncoding RNA in the INK4 locus (ANRIL) (D), and highly up-regulated in liver cancer RNA (HULC) (E) long noncoding RNA was then measured. Data are presented as means±SD from two independent experiments with triplicates. *Significantly different at p<0.05.
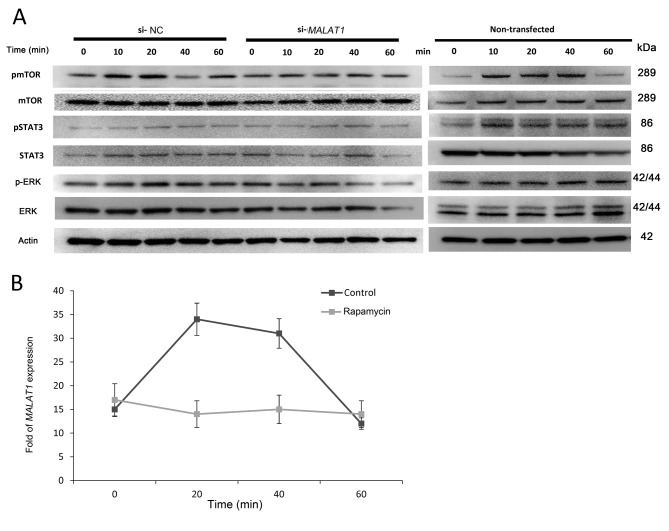
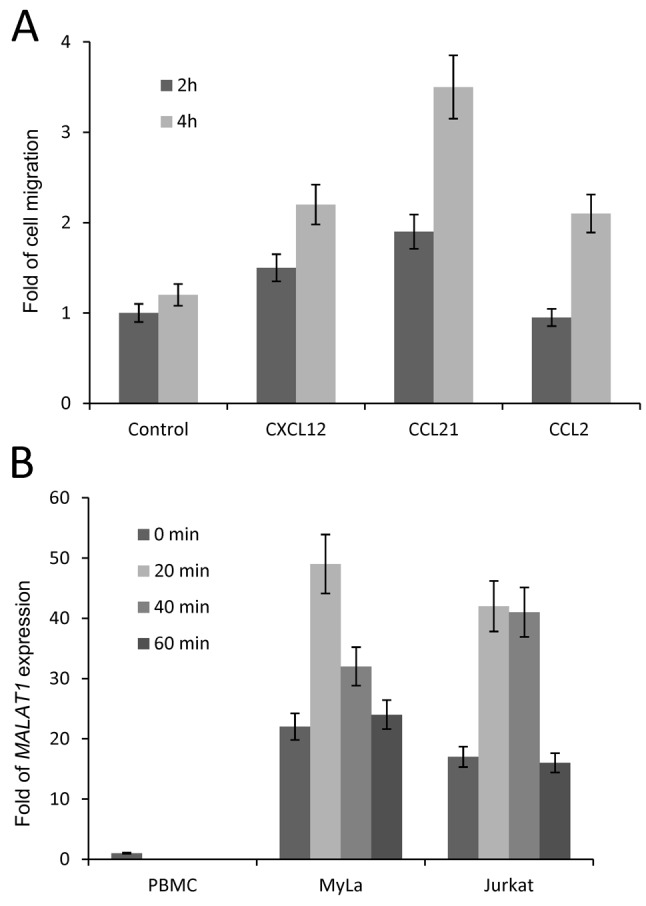
CCL21 enhances MALAT1 expression and cell migration in MyLa cells. In previous work, we demonstrated that MyLa cells migrated toward CCL21 in a transwell migration assay (6). In the current study, we showed that CCL21 increased cell migration almost 4-fold in 4 h compared to the control (Figure 2A). Next, we were curious whether MALAT1 up-regulation was increased in vitro in MyLa and Jurkat cells as that in MF tissues ex vivo. Using PBMC cells as controls, we demonstrated that in both MyLa cells and Jurkat cells, the basal expression of MALAT1 was significantly higher by approximately 20-fold (Figure 2B). More importantly, treatment of both MyLa and Jurkat cells with CCL21 induced significant up-regulation of MALAT1 after 20 min (Figure 2B).
Figure 2. C-C Motif chemokine ligand 21 (CCL21) enhanced expression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and cell migration in MyLa cells. A: MyLa cells were incubated in a transwell migration plate with bottom chamber nourished with chemokine (C-X-C motif) ligand 12 (CXCL12), CCL21, and CCL2 for 2 and 4 h. Cell migration was measured as the number of cells migrating from upper chamber to lower chamber as standardized by the data from the group without any chemokines (three independent experiments with triplicates). B: MyLa cells were incubated with CCL21 for 0, 20, 40, and 60 min and their expression of MALAT1 was then measured by real-time polymerase chain reaction. Data are presented as means±SD from three independent experiments with triplicates.
CCL21 induced cell migration through MALAT1 expression. Since CCL21 induced MALAT1 expression, we next asked whether CCL21-induced cell migration was mediated by the MALAT1 expression. To address this, we transfected MyLa cells with siRNA targeting MALAT1 or negative control siRNA and performed a 4-h transwell migration assay. Efficiency of si-MALAT1 was demonstrated by significant down-regulation of MALAT1 in si-MALAT1-transfected MyLa cells but no significant change in NC-si-transfected MyLa cells (Figure 3A). In addition, the increased expression of MALAT1 at 30 min induced by CCL21 was practically abolished in si-MALAT1-transfected MyLa cells but not in NC-si-transfected MyLa cells. In fact, migration which was increased by CCL21 in MyLa cells at 4 h was inhibited in MALAT1-siRNA-transfected MyLa cells but not in NC-siRNA-transfected MyLa cells (Figure 3B). The data indicate that CCL21-induced cell migration in MyLa cells is mediated by MALAT1 expression.
Figure 3. C-C Motif chemokine ligand 21 (CCL21) induced cell migration through expression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1). A: MyLa cells were transfected with siRNA targeting MALAT1 or with mock negative control (NC) siRNA. Peripheral blood mononuclear cells (PBMCs), MyLa cells, MyLa cells transfected with MALAT1-siRNA and MyLa cells transfected with NC siRNA were treated with CCL21 for 30 and 60 min. The expression of MALAT1 was then measured by real-time polymerase chain reaction (two independent experiments with triplicates). B: MyLa cells transfected with MALAT1- siRNA or NC siRNA were used for cell migration assay. CCL21 was used to stimulate MyLa cell migration for 4 h. Data are presented as means±SD from two independent experiments with triplicates. *Significantly different at p<0.05.
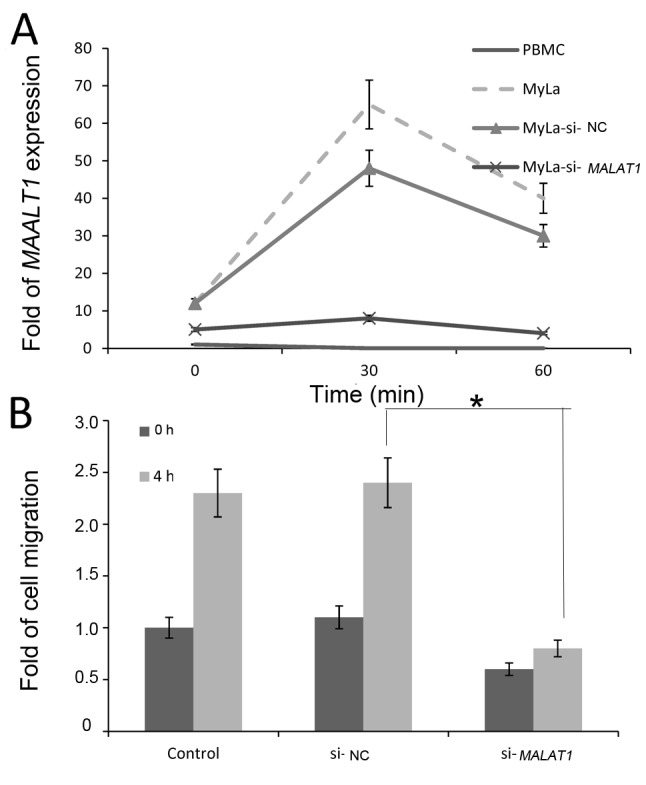
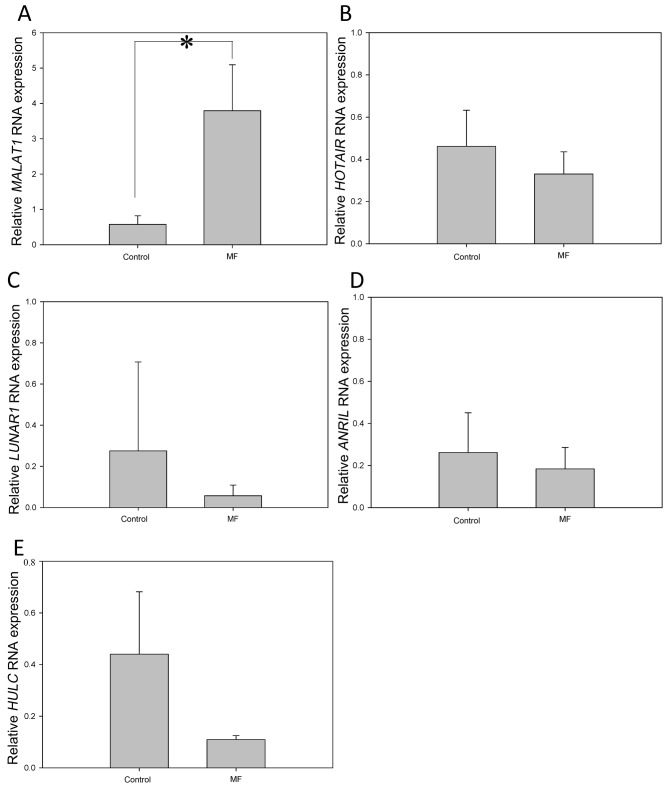
CCL21-induced MALAT1 expression is mediated by mTOR phosphorylation. We previously demonstrated that CCL21 induced mTOR phosphorylation, which led to cell migration (6). We wanted to determine whether CCL21 induced mTOR phosphorylation through MALAT1 expression or vice versa. To do so, MyLa cells transfected with NC-siRNA or MALAT1-siRNA were treated with CCL21, and the expression and activation of mTOR, STAT3, and ERK at different times was determined by western blot. The results showed that while CCL21 induced minimal changes of phophorylation of ERK and STAT3, CCL21 induced mTOR phosphorylation biphasically at 10-20 min and at 60 min in both NC-si- and MALAT1-si-transfected cells (Figure 4A). On the other hand, when cells were pretreated with rapamycin, which inhibits mTOR phosphorylation, induction of MALAT1 expression by CCL21 was abolished, indicating that CCL21-induced mTOR phosphorylation was followed by MALAT1 up-regulation (Figure 4B).
Figure 4. A: Blocking expression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) did not alter mammalian target of rapamycin (mTOR) activation by C-C motif chemokine ligand 21 (CCL21). CCL21 at 10 ng/ml was used to treat MyLa cells for 0, 10, 20, 40, and 60 min. Protein lysates were obtained for western blot of mTOR, phospho-(p)-mTOR, extracellular-regulated protein kinases (ERK), p-ERK, signal transducer and activator of transcription 3 (STAT3), pSTAT and actin. A representative blot is shown from two independent experiments. B: mTOR inhibition disrupted CCL21-mediated MALAT1 expression. MyLa cells were pretreated with or without mTOR inhibitor rapamycin for 2 h. The expression of MALAT1 was then measured by real time polymerase chain reaction at 0, 20, 40, and 60 min after CCL21 treatment. Data are presented as means±SD from three independent experiments with triplicates.
Discussion
In this study, we showed that MALAT1 expression is selectively increased in tissues from patients with MF. In vitro study showed that CCL21-mediated migration in MyLa cells is dependent on mTOR activation and MALAT1 expression. Blocking MALAT1 expression by RNA interference did not affect CCL21-mediated mTOR activation, however, blocking mTOR by rapamycin, inhibited CCL21-mediated MALAT1 expression.
Several reports have demonstrated that MALAT1 mediates the migration of different cancer cell lines. Zhang et al. showed that MALAT1 mediates migration and invasion of ovarian cancer cell in vitro (22) and several other reports showed that MALAT1 is overexpressed and promotes cell invasion and migration in cervical (23), esophageal (24), and prostate (25) cancer. However, whether increased migration or invasion was chemokine- or cytokine-specific was not clear from these publications. Specifically for lymphoma, only one study has been reported showing that inhibition of MALAT1 can improve chemotherapy sensitivity of diffuse large B-cell lymphoma by enhancing expression of autophagy-related proteins (26).
In this study, we showed that mTOR is an upstream molecule regulating CCL21-induced MALAT1 expression. The exact mechanism by which CCL21 mediates the activation of MALAT1 is now known. A previous study showed in vitro and in vivo that BCAR4, another lncRNA, binds to CCL21, directing delicate cooperative downstream epigenetic regulation in breast cancer metastasis (17). CCL5, another chemokine, was also found to induce MALAT1 expression, leading to the subsequent expression of snail family transcriptional repressor 1 (SNAI) (27), a molecule related to epithelial–mesenchymal transformation.
In this study, we showed that mTOR mediated CCL21-induced MALAT1 expression. An immunohistochemical study in follicular thyroid carcinoma showed parallel increases of expression of mTOR and MALAT1 (28). In hepatocellular carcinoma, MALAT1 was shown to modulate serine/arginine-rich splicing factor 1 (SRSF1) splicing targets, enhancing the production of antiapoptotic splicing isoforms and activating the mTOR pathway by modulating the alternative splicing of S6K1. Inhibition of SRSF1 expression or mTOR activity abolished the oncogenic properties of MALAT1, suggesting that mTOR activation is essential for MALAT1-induced transformation (29).
Several studies have shown that cytokines and chemokines mediate their pro-inflammatory actions through lncRNAs. For example, ANRIL silencing abrogated the tumor necrosis factor-induced expression of interleukin-6 and -8 (30). In terms of cancer progression, it has been demonstrated that MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells (31). A subsequent study showed that MALAT1 silencing reduced the expression of CXCL5 in lung cancer cells (20). Long intergenic non-protein coding RNA 963LINC00963 was found to interact with glycolytic phosphoglycerate kinase 1 (PGK1) and prevent its ubiquitination, leading to activation of the oncogenic protein kinase B (AKT)/mTOR signaling pathway (32). In other chronic inflammatory diseases, down-regulation of nuclear paraspeckle assembly transcript 1 (NEAT1) was shown to inhibit the progression of non-alcoholic fatty liver disease through mTOR/S6K1 signaling pathway in rats (33). Moreover, miR-503 was found to modulate epithelial–mesenchymal transition in silica-induced pulmonary fibrosis by targeting phosphoinositide 3-kinase p85 (34).
This study has several limitations. Firstly, the sample numbers were low. Whether the findings might be extrapolated requires further investigation. Secondly, we did not include any animal study to show the regulatory effect of MALAT1 in tumor formation in vivo. However, the establishment of a xenotopic model of MF in mice remains a challenge (35) and the heterogeneous and polyclonal features of cell lines used for implantation remain an unsolved issue. Nevertheless, in this study, the human data clearly show that among the five lncRNAs measured, expression of MALAT1 was selectively increased.
In conclusion, we showed that MALAT1 expression is selectively increased in tissues from patients with MF. We also showed that CCL21-mediated migration in MF MyLa cells is dependent on mTOR activation followed by MALAT1 expression. MALAT1 or mTOR could be a potential therapeutic target for MF.
Conflicts of Interest
The Authors declare that they have no competing interests in regard to this study.
Authors’ Contributions
CHH collected the tissue samples and performed most of the in vitro experiments. SHL helped collect tissue samples and perform chemotaxis assay. CHL proposed the research strategy, supervised the study, and provided laboratory facilities.
Acknowledgements
The Authors are grateful for the funding supported by the Ministry of Science and Technology in Taiwan (104-2314-B-182A-073-MY3, CH Lee; 105-2314-B-010-054-MY3, CH Hong; 105-2628B-182A-004-MY3, SH Lin). The Authors thank the Chang Gung Memorial Foundation for their generous laboratory support. The funding agency had no role in the design of the study and collection, analysis, interpretation of data, nor in writing the article.
References
- 1.Wilcox RA. Cutaneous t-cell lymphoma: 2017 update on diagnosis, risk-stratification, and management. Am J Hematol. 2017;92(10):1085–1102. doi: 10.1002/ajh.24876. PMID: 28872191, 10.1002/ajh.24876. [DOI] [PubMed] [Google Scholar]
- 2.Bradford PT, Devesa SS, Anderson WF, Toro JR. Cutaneous lymphoma incidence patterns in the united states: A population-based study of 3,884 cases. Blood. 2009;113(21):5064–5073. doi: 10.1182/blood-2008-10-184168. PMID: 19279331, 10.1182/blood-2008-10-184168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, Robson A, Calonje E, Stefanato CM, Wain EM, Wilkins B, Fields PA, Dean A, Webb K, Scarisbrick J, Morris S, Whittaker SJ. Survival outcomes and prognostic factors in mycosis fungoides/sezary syndrome: Validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer Staging proposal. J Clin Oncol. 2010;28(31):4730–4739. doi: 10.1200/JCO.2009.27.7665. PMID: 20855822, 10.1200/JCO.2009.27.7665. [DOI] [PubMed] [Google Scholar]
- 4.Criscione VD, Weinstock MA. Incidence of cutaneous T-cell lymphoma in the United States, 1973-2002. Arch Dermatol. 2007;143(7):854–859. doi: 10.1001/archderm.143.7.854. PMID: 17638728, 10.1001/archderm. 143.7.854. [DOI] [PubMed] [Google Scholar]
- 5.Sarvaiya PJ, Guo D, Ulasov I, Gabikian P, Lesniak MS. Chemokines in tumor progression and metastasis. Oncotarget. 2013;4(12):2171–2185. doi: 10.18632/oncotarget.1426. PMID: 24259307, 10.18632/oncotarget.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu SC, Lin CL, Hong CH, Yu HS, Chen GS, Lee CH. Ccr7 expression correlates with subcutaneous involvement in mycosis fungoides skin lesions and promotes migration of mycosis fungoides cells (MyLa) through mtor activation. J Dermatol Sci. 2014;74(1):31–38. doi: 10.1016/j.jdermsci.2013.12.003. PMID: 24411580, 10.1016/j.jdermsci.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Consortium IHGS Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. PMID: 15496913, 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 8.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306(5705):2242–2246. doi: 10.1126/science.1103388. PMID: 15539566, 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 9.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. PMID: 17486121, 10.1038/ nrg2083. [DOI] [PubMed] [Google Scholar]
- 10.Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10(9):1106–1113. doi: 10.1038/ncb1771. PMID: 19160492, 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 11.Croce CM. Causes and consequences of microrna dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. PMID: 19763153, 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negrini M, Nicoloso MS, Calin GA. Micrornas and cancer–new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21(3):470–479. doi: 10.1016/j.ceb.2009.03.002. PMID: 19411171, 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Moskowitz AJ, Horwitz SM. Targeting histone deacetylases in t-cell lymphoma. Leuk Lymphoma. 2017;58(6):1306–1319. doi: 10.1080/10428194.2016.1247956. PMID: 27813438, 10428194.2016.1247956. [DOI] [PubMed] [Google Scholar]
- 14.Papadavid E, Braoudaki M, Bourdakou M, Lykoudi A, Nikolaou V, Tounta G, Ekonomidi A, Athanasiadis E, Spyrou G, Antoniou C, Kitsiou-Tzeli S, Rigopoulos D, Kolialexi A. Aberrant microrna expression in tumor mycosis fungoides. Tumour Biol. 2016;37(11):14667–14675. doi: 10.1007/s13277-016-5325-2. PMID: 27623940, 10.1007/s13277-016-5325-2 [pii]. [DOI] [PubMed] [Google Scholar]
- 15.van Doorn R, Slieker RC, Boonk SE, Zoutman WH, Goeman JJ, Bagot M, Michel L, Tensen CP, Willemze R, Heijmans BT, Vermeer MH. Epigenomic analysis of Sezary syndrome defines patterns of aberrant DNA methylation and identifies diagnostic markers. J Invest Dermatol. 2016;136(9):1876–1884. doi: 10.1016/j.jid.2016.03.042. PMID: 27113428, 10.1016/j.jid.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 16.Forrest ME, Khalil AM. Review: Regulation of the cancer epigenome by long non-coding RNAs. Cancer Lett. 2017;407:106–112. doi: 10.1016/j.canlet.2017.03.040. PMID: 28400335, 10.1016/j.canlet.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 17.Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, Hung MC, Lin C, Yang L. LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–1125. doi: 10.1016/j.cell.2014.10.013. PMID: 25416949, 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao B, Liu N, Wang W. High glucose prevents osteogenic differentiation of mesenchymal stem cells via lncRNA AK028326/CXCL13 pathway. Biomed Pharmacother. 2016;84:544–551. doi: 10.1016/j.biopha.2016.09.058. PMID: 27693963, 10.1016/j.biopha.2016.09.058. [DOI] [PubMed] [Google Scholar]
- 19.Wang WT, Ye H, Wei PP, Han BW, He B, Chen ZH, Chen YQ. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9(1):117–117. doi: 10.1186/s13045-016-0348-0. PMID: 27809873, 10.1186/s13045-016-0348-0 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo F, Guo L, Li Y, Zhou Q, Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol. 2015;8(12):15903–15910. PMID: 26884862, [PMC free article] [PubMed] [Google Scholar]
- 21.Lee CS, Ungewickell A, Bhaduri A, Qu K, Webster DE, Armstrong R, Weng WK, Aros CJ, Mah A, Chen RO, Lin M, Sundram U, Chang HY, Kretz M, Kim YH, Khavari PA. Transcriptome sequencing in Sezary syndrome identifies sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts. Blood. 2012;120(16):3288–3297. doi: 10.1182/blood-2012-04-423061. PMID: 22936659, 10.1182/blood-2012-04-423061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, Sheng X. The long noncoding RNA MALAT-1 is highly expressed in ovarian cancer and induces cell growth and migration. PLoS One. 2016;11(5):e0155250–e0155250. doi: 10.1371/journal.pone.0155250. PMID: 27227769, 10.1371/jouRNAl.pone.0155250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Wang T, Huang HQ, Li W, Cheng XL, Yang J. Human MALAT-1 long non-coding RNA is overexpressed in cervical cancer metastasis and promotes cell proliferation, invasion and migration. J BUON. 2015;20(6):1497–1503. PMID: 26854446. [PubMed] [Google Scholar]
- 24.Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y, Qi B, Liu S, Qin X, Zhao B. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37(4):4305–4312. doi: 10.1007/s13277-015-4223-3. PMID: 26493997, 10.1007/s13277-015-4223-3 [pii]. [DOI] [PubMed] [Google Scholar]
- 25.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Up-regulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8(9):2289–2294. doi: 10.1039/c2mb25070e. PMID: 22722759, 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 26.Li LJ, Chai Y, Guo XJ, Chu SL, Zhang LS. The effects of the long non-coding RNA malat-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed Pharmacother. 2017;89:939–948. doi: 10.1016/j.biopha.2017.02.011. PMID: 28292022, 10.1016/j.biopha.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Kan JY, Wu DC, Yu FJ, Wu CY, Ho YW, Chiu YJ, Jian SF, Hung JY, Wang JY, Kuo PL. Chemokine (C-C motif) ligand 5 is involved in tumor-associated dendritic cell-mediated colon cancer progression through non-coding RNA MALAT-1. J Cell Physiol. 2015;230(8):1883–1894. doi: 10.1002/jcp.24918. PMID: 25546229, 10.1002/ jcp.24918. [DOI] [PubMed] [Google Scholar]
- 28.Covach A, Patel S, Hardin H, Lloyd RV. Phosphorylated mechanistic target of rapamycin (p-mTOR) and noncoding RNA expression in follicular and hurthle cell thyroid neoplasm. Endocr Pathol. 2017;28(3):207–212. doi: 10.1007/s12022-017-9490-7. PMID: 28660408, 10.1007/ s12022-017-9490-7. [DOI] [PubMed] [Google Scholar]
- 29.Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, Karni R. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 up-regulation and mTOR activation. Cancer Res. 2017;77(5):1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. PMID: 27993818, 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, Gan LM, Cao H, Liang Z. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-kappaB pathway. RNA Biol. 2016;13(1):98–108. doi: 10.1080/15476286.2015.1122164. PMID: 26618242, 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. PMID: 23243023, 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, Li Z, Chen B, Zhang X, Pan H, Li S, Lin H, Liu L, Yan M, He X, Yao M. Metalnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. 2017;77(21):5782–5794. doi: 10.1158/0008-5472.CAN-17-0671. PMID: 28923857, 10.1158/0008-5472.CAN-17-0671. [DOI] [PubMed] [Google Scholar]
- 33.Wang X. Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J Cell Biochem. 2018;119(2):1567–1574. doi: 10.1002/jcb.26317. PMID: 28771824, 10.1002/jcb.26317. [DOI] [PubMed] [Google Scholar]
- 34.Yan W, Wu Q, Yao W, Li Y, Liu Y, Yuan J, Han R, Yang J, Ji X, Ni C. Mir-503 modulates epithelial–mesenchymal transition in silica-induced pulmonary fibrosis by targeting PI3K p85 and is sponged by lncRNA MALAT1. Sci Rep. 2017;7(1):11313–11313. doi: 10.1038/s41598-017-11904-8. PMID: 28900284, 10.1038/s41598-017-11904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thaler S, Burger AM, Schulz T, Brill B, Bittner A, Oberholzer PA, Dummer R, Schnierle BS. Establishment of a mouse xenograft model for mycosis fungoides. Exp Dermatol. 2004;13(7):406–412. doi: 10.1111/j.0906-6705.2004.00201.x. PMID: 15217360, 10.1111/j.0906-6705. [DOI] [PubMed] [Google Scholar]