Abstract
There is a great deal of excitement around the concept of targeting proteins for degradation as an alternative to conventional inhibitory small molecules and antibodies. Protein degradation can be undertaken by bifunctional molecules that bind the target for ubiquitin mediated degradation by complexing them with Cereblon (CRBN), von Hippel-Lindau or other E-3 ligases. Alternatively, E-3 ligase receptors such as CRBN or DCAF15 can also be used as a ‘template’ to bind IMiD or sulphonamide like compounds to degrade multiple context specific proteins by the selected E-3 ligases. The ‘template approach’ results in the degradation of neo-substrates, some of which would be difficult to drug using conventional approaches. The chemical properties necessary for drug discovery, the rules by which neo-substrates are selected by E-3 ligase receptors and defining the optimal components of the ubiquitin proteasome for protein degradation are still to be fully elucidate. Theis review will aim to critically evaluate the different approaches and principles emerging for targted protein degradation.
Introduction
With the realisation that proteins are in a dynamic state of synthesis and degradation, the scene was set for the investigation of mechanisms associated with proteolysis (reviewed by Ref. [1]). Proteolysis is a highly regulated process associated with lysosomal as well as ubiquitin mediated protein degradation. The deep understanding of the molecular pathways associated with ubiquitin-proteasome (UP) regulation has resulted in a great deal of activity to target components of this system for drug discovery [2]. This has resulted in the successful development of proteasome inhibitors such as bortezimib, carfilzomib, marizomib and ixazomib [3], inhibitors of neddylation [4], and inhibitors of deubiquitinases [5]. Whilst inhibition of individual components has resulted in successful anticancer agents, so far this has made a clinical impact in a limited number of malignancies, most notably multiple myeloma. Research is still ongoing however and, for example, the deubiquitinase inhibitors are expected to have activity in a broader range of cancers and human disease. The broad potential of manipulating protein homeostasis was demonstrated by the discovery that clinical anticancer immunomodulatory drugs (IMiDs), such as thalidomide, pomalidomide and lenalidomide, induce the UPS-dependent degradation of a range of neo-substrate proteins [6].
This possibility of enhancing small molecule based drug discovery by exploitation of the ubiquitin-proteasome system (UPS) for targeted protein degradation has resulted in a great deal of excitement. The ability to degrade a target protein rather than modulate specific catalytic or functional domain activity has a number of advantages, including: the potential to degrade proteins that have been difficult to drug, e.g. transcription factors; removal of proteins which exert scaffolding functions independent of their catalytic/kinase activity [7]; and the potential to partially degrade target proteins that may be elevated in human disease to restore function and cellular homeostasis.
Proteolysis targeting chimeras (PROTACs)
The degradation of specific protein targets using heterobifunctional small molecules to hijack the UPS has been rapidly adopted as a tool for biological research and as a potential modality for the discovery of new drugs [2], [8], [9], [10], [11], [37]. These molecular constructs consist of two discrete pharmacophores, one capable of binding the protein to be degraded and the other with affinity for an E3 ubiquitin ligase complex, connected by a flexible linker. Together these components serve to recruit the protein of interest into proximity with the E3 ligase through formation of a ternary complex where ubiquitination occurs, thus entering the neo-substrate protein into the UPS pathway for degradation by the 26S proteasome. The field was led by the pioneering work of Deshaies and Crews in developing the first heterobifunctional inducers of degradation, the proteolysis targeting chimeras (PROTACs) that intercepted the Hrt1 (SCF) E3 ligase [12]. Subsequent progress was made by exploiting VHL-dependent degradation using peptide ligands to bind the E3 ligase, later replaced by more cell permeable peptidomimetics [13], [14], [15]. Other early examples in the field include the specific and non-genetic IAP-dependent protein erasers (SNIPERs) developed by Hashimoto and co-workers that hijack the cIAP E3 ligase [16]. Further research extended the range of E3 ligases recruited [17], most notably to include redirection of the cereblon-containing cullin4 (CUL4CRBN) E3 complex [18], [19]. Between them, the various heterobifunctional inducers of degradation have been used to deplete an impressive array of target proteins in human cells, including nuclear hormone receptors, chromatin binders and modulators, protein and lipid kinases, and transcriptional regulators [2], [20], [21].
The mechanism of heterobifunctional promoters of protein degradation involves reversible formation of a ternary complex (E3–heterobifunctional molecule–target). This exploits the catalytic activity of the E3 ligase so that one heterobifunctional molecule can induce the ubiquitination of many molecules of neo-substrate through turnover of the complex. Thus potent degradation (quantified as a DC50; the concentration of the heterobifunctional molecule that gives 50% degradation of the target relative to an untreated control) can be achieved with relatively low target occupancy in the cell, in contrast to the classical model for reversible inhibitors of protein function where high occupancy is generally required for potent efficacy [14], [22], [23]. As a result, a significant proportion of the unmodified E3 ligase complex can remain functioning in tandem with the hijacked enzyme, potentially leading to less perturbation of the endogenous substrate binding and providing a wider therapeutic window. The formation of the ternary complex may involve co-operative effects, due to formation of a sandwich structure in which interactions of the linker with the proteins or direct protein-protein interactions contribute to the binding affinity, such that stable complex formation can be seen with binding elements whose individual affinities for the E3 and/or target protein are only moderate [24], [25]. Techniques for measuring cooperativity and the thermodynamics of ternary complex formation have been developed to assess heterobifunctional molecules [24], [26]. Another common feature of ternary complex formation is the emergence of a bell-shaped concentration-response curve for degradation induced by potent binders, the so called ‘hook effect’, where high concentrations of the heterobifunctional molecule independently saturate the binding sites on the two components preventing complex formation [18].
An attractive feature of heterobifunctional inducers of degradation is the interchangeable nature of the binding domains. This allows for rational design of the molecules to target different neo-substrates to a given E3 ligase, or to recruit different E3 ligases to the same protein target [27]. While clearly generally applicable to a range of proteins once sufficiently potent binding groups can be identified, a number of factors determine the success or otherwise of the strategy [2]. Firstly, considering the biological mechanism, the initial single ubiquitination of the target protein requires that one or more lysine residues are accessible to the E3 ligase function in the ternary complex. The first ubiquitinated substrate must also be a substrate for polyubiquitination and subsequent steps in the down-stream UPS, for example decomplexation by enzymes such as valosin-containing protein (VCP)/p97 that process CUL4CRBN ubiquitinated proteins [28]. These requirements are mitigated by the generality of the UPS, which has evolved to handle many different proteins for endogenous degradation, but the ubiquitination events can be the major rate-limiting factors determining the efficiency of eventual target degradation [29]. A ubiquitination steady state is dependent on the balance of the E3 ligase activity against competing deubiquitinase (DUB) enzymes, which will determine the effectiveness of target degradation. The spatial and temporal expression of the E3 ligase within the cell, and how this is matched to the expression of the target protein is also critical. The majority of successful examples of heterobifunctional degraders so far involve soluble cytosolic or nuclear target proteins, but degradation of membrane associated proteins has also been reported [23]. The time-course of degradation induced by heterobifunctional molecules is typically of the order of minutes to several hours. The approach therefore offers advantages over genetic depletion of target proteins through RNA interference or CRISPR in terms of rapid onset and minimisation of the opportunity for cellular adaptation to loss of the protein. On wash-out in cellular assays, or following drug elimination in vivo, the duration of response and recovery of protein expression will depend in part on the re-synthesis rate of the target.
Considering chemical factors that determine the effectiveness of the heterobifunctional inducers of degradation, the points of attachment of the linker to the E3 binding and target protein binding motifs are critical, to ensure that each remains capable of interacting with its primary binding site. The selection of linker attachment points can be made empirically, guided by suitable biochemical or biophysical affinity assays, but is greatly aided by structural biology insights into the structure of the E3 ligase and target proteins. For the most widely used E3 ligases, VHL, CRBN, cIAP, MDM2, known potent binding motifs (e.g. thalidomide or lenalidomide derivatives for CRBN, peptidomimetics of the HIF1α binding peptide for VHL, derivatives of the cIAP binding small molecule bestatin, nutlin derivatives for MDM2) are available with well characterised attachment points and chemical routes for synthesis of the heterobifunctional molecules. Finding attachment points on the target binding motifs also benefits from structural biology information, as exemplified by the discovery of several recent heterobifunctional molecules that induce degradation of bromodomain-containing proteins [19], [15], [24], [30], protein kinases [27] and lysine deacetylases [32].
The choice of linker group to give an effective degrader is essential but less easily predetermined than selecting the attachment points. The linker is required to be long enough and flexible enough to permit binding of both protein components, in a relative orientation that is productive for ubiquitin transfer. Molecular modelling based on the separate structures of the E3 and target protein binding sites has been used to suggest minimal linker lengths for fruitful interactions [19], [31], [32]. While originally conceived as non-functional with respect to the two binding events, there is an emerging dichotomy in the experimentally determined role of the linkers in heterobifunctional molecules that induce degradation. Seminal work by Ciulli to elucidate the mechanism of a PROTAC (MZ1) promoting VHL-dependent degradation of BRD4 clearly demonstrated thermodynamic cooperativity in the binding of the PROTAC, favouring shorter linkers [24], [25]. This was supported by the first crystal structure of a VHL-PROTAC-target ternary complex, showing the linker folding within a sandwich complex to mediate interactions between the proteins, as well as highlighting direct positive interactions of the two protein surfaces. Using this information, a shorter and more efficient linker was designed for a second generation PROTAC. In contrast, for a recent PROTAC that promoted CUL4CRBN-dependent degradation of Bruton’s tyrosine kinase (BTK), a clear thermodynamic non-cooperative mechanism was defined [26]. Here, optimum linker length was associated with relief of steric clashes between the proteins, with shorter linkers that would require direct protein–protein contacts leading to negative thermodynamic cooperativity. While ternary complex formation was not excluded, interpretation of the biophysical data for binding favoured a looser and more transient interaction than seen for the crystallized VHL-PROTAC complex. It seems highly likely therefore, that both cooperative and uncooperative mechanisms are possible, depending on the E3 and target protein involved and the surface complementarity between them, as well as the length, rigidity and functionality of the linker in the heterobifunctional molecule.
Understanding the degree of cooperativity can inform the best optimization strategy for a given heterobifunctional molecule. It is notable that while many successful examples of promoters of degradation have now been reported, often a comprehensive set of linker lengths and functionalities is required to be explored, as well as different target binding and E3 recruitment motifs [33], [14], [15], [30]. Indeed, where direct comparisons have been made, significant variation in effectiveness and selectivity of target degradation can be seen through variation of these components [27]. Major increases in selectivity for degradation relative to the warhead kinase binding profile were achieved when promiscuous kinase binders were converted into heterobifunctional degraders [34], [35], [36] and altered selectivity was also seen for a multi-targeted bromodomain ligand incorporated into a PROTAC [15]. In a comparison of various matched ABL-kinase targeting PROTACs, CRBN-binding molecules more often induced degradation than the VHL-binding counterparts [27]. It has been suggested that this reflects the different molecular structures of the E3 complexes, where the large and flexible CUL4CRBN assembly provides a very permissive scaffold that can accommodate a larger range of substrates and orientations for ubiquitination than the more compact VHL- or cIAP-containing complexes [37]. Differences between the E3 ligases and the currently used small molecule binding motifs also lead to distinct off-target degradation profiles. Incorporation of thalidomide-based ligands that recruit CRBN into heterobifunctional molecules can retain the intrinsic neo-substrate degradation promoted by binding of the immunomodulatory imide drug (IMiD) element to CRBN, resulting in lower degradation selectivity than equivalent heterobifunctional degraders based on recruitment of VHL [26]. Off-target pharmacology due to the E3 targeting motif can also be seen for some bestatin-derived components of cIAP-targeting molecules [38], while recruitment of neo-substrates to MDM2 can reduce endogenous MDM2-dependent degradation of the tumour suppressor TP53 [39].
The high molecular weight and high polar surface area of heterobifunctional molecules requires consideration during optimization of the species, particularly for in vivo and ultimately clinical use. Solubility and membrane permeability may be reduced in the initial linking of two physicochemically well-behaved binding groups. However, the heterobifunctional molecules are amenable to the same rational approaches to increase solubility and permeability that apply to smaller molecules, especially through systematic variation of the linker lengths and functional group composition. This was demonstrated for heterobifunctional promoters of degradation of the protein pirin, where changes to linker polarity and functionality and the masking of hydrogen bond donors greatly improved the cellular potency [40]. The application of endogenous tagging methods to directly monitor ternary complex formation and target protein depletion in living cells promises to increase the speed and efficiency of the optimisation of the heterobifunctional species for cellular activity [29]. Using systematic approaches, heterobifunctional molecules suitable for in vivo studies have been generated [19], [22], [31], [30], [41], [42], [26], [65], [66], notably for molecules targeting degradation of nuclear receptors for the treatment of cancer. A clinical candidate heterobifunctional molecule (ARV-110, [43] that redirects E3 ligase activity to induce androgen receptor degradation has been recently announced (http://ir.arvinas.com/news-releases/news-release-details/arvinas-receives-authorization-proceed-its-ind-application; accessed 03.02.2019).
Targeted protein degradation through hydrophobic tagging
The targeting of specific E3 ligase complexes is not the only means to convey proteins into the UPS for degradation. Direct recruitment to the 20S proteasome has been reported for heterobifunctional molecules bearing a lipophilic tert-butyl carbamate-protected arginine group that promotes degradation without the need for ubiquitination [44], [45]. Hydrophobic tagging of proteins also leads to degradation through the UPS by recruitment of molecular chaperones that recognise unfolded proteins [46] and has been extended to heterobifunctional molecules for the degradation of the androgen receptor [47]. Based on the mode of action of the selective estrogen receptor degrader, Fulvestrant, that promotes ER degradation through the presence of a lipophilic alkyl sidechain, other hydrophobic analogues have been generated that can serve as modular components of heterobifunctional degraders [48].
E3 ligase receptors as templates for targeted degradation
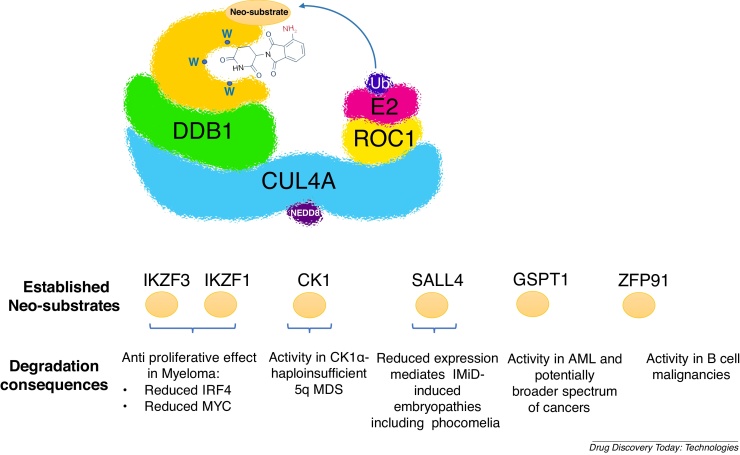
Another approach to targeted protein degradation is based on the observation that binding of the IMiD compounds to CRBN, the receptor for the CRL4CRBN E3 ligase family, results in a neomorphic activity causing degradation of two critical Zinc Finger (ZF)-associated B cell transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) in multiple myeloma cells, as well as T cells [49], [50], [6]. Subsequently it was shown that other proteins such as CSNK1A1 [51] and GSPT1 [52] are also degraded by these compounds in myeloid cells (Fig. 1, Fig. 2).
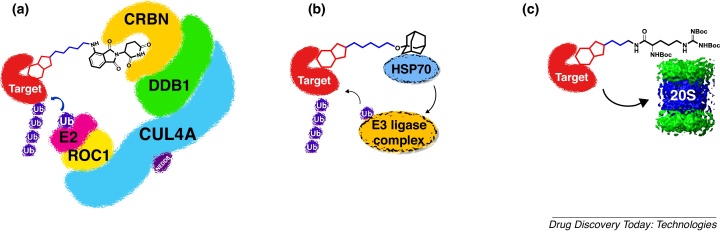
Fig. 1.
A schematic view of the mechanism of action of bifunctional molecules that induce targeted protein degradation. (A) PROteolysis TArgeting Chimeric (PROTAC) molecules consisting of a target affinity group (red), flexible linker (blue) and E3 ligase binding group (black) exemplified for the CUL4CRBN E3 ligase; (B) adamantyl-tagged (black) bifunctional molecules that bind to HSP70; (C) (Boc)3-arginine-tagged (black) bifunctional molecules that bind to the 20S proteasome. (For interpretation of the references to colour in the text, the reader is referred to the web version of this article.)
Fig. 2.
A schematic view of the mechanism of action of IMiD agents, with summary of degraded substrates that have defined clinical consequences.
Insights into the mechanism by which neosubstrates are degraded were defined by the elucidation of the target ‘degron’ found on proteins that had hitherto been shown to be degraded by the IMiD agents [52], [53]. These studies indicate that the IMiD agents interact with a hydrophobic tri-tryptophan cage which is the thalidomide-binding domain at the C-terminal end of CRBN. The glutarimide moiety of the IMiD agents binds into this highly conserved hydrophobic pocket, with the phthalamide ring exposed on the surface of the CRBN protein. The phthalamide portion of the IMiD compounds will interact with specific amino acids on CRBN to ‘create’ a thermo dynamically favourable hot-spot for neosubstrate interactions [52], [53]. These neosubstrates contain a β-hairpin loop with a sentinel glycine residue that engages the phthalamide moiety of the IMiDs. Initial data suggest that proteins containing a β-hairpin loop with a glycine motif found in ZF containing transcription factors (e.g. IKZF1 and IKZF3) or non-ZF containing proteins (e.g. CSNK1A1) can be potentially degraded by IMiD agents [53], [54].
The importance of the β-hairpin loop as a determinant of target degradation is further illustrated by the discovery of the compound, CC-885, a potent anti-cancer agent with activity in a broad range of cancer cell lines including primary acute myeloid leukemia (AML) cells. CC-885 has a tri-tryptophan binding glutaramide moiety and a phthalamide extended by a methylene urea group. This compound enhanced the degradation of IKZF3 and also triggered the degradation of GSPT1, a translation termination factor that binds eRF1 to mediate stop codon recognition and nascent protein release from the ribosome. The role of GSPT1 had not been described in cancer and highlights how these agents can also be exploited for discovering new biology [52], [55].
Refining the CRL4 CRBN degron and implications for teratogenicity
The observation that hitherto undruggable ZF transcription factors could be degraded, led the Ebert and Thoma groups to hypothesise that other ZF containing proteins could also be degraded [54]. IKZF1 and 3 contain a conserved Cys2-His2 (C2H2) motif and these groups generated different IKZF1 deletion constructs and measured their affinity for binding to CRBN in the presence of thalidomide analogues using a time resolved fluorescence energy transfer (TR-FRET) assay. They showed clearly that the minimal degron consisted of the second ZF (ZF2) but the optimal degron which gave the highest affinity was an IKZF1 construct spanning ZF2 and ZF3.
The minimal degron was used to define potential proteins that could be targeted for degradation and 6572 distinct CH2H2-containing ZFs were defined using PROSITE (www.prosite.expasy.org). The cDNAs from these ZFs were transduced into degradation reporter vectors in HEK293T cells in the presence of thalidomide, lenalidomide and pomalidomide. Pomalidomide was the most potent degrader but degraded only 6 out of the 6572 ZF containing proteins assayed (these included IKZF1/IKZF3, ZNF629, ZNF91, ZNF276, ZNF653 and ZNF827). When the next generation of thalidomide analogues were employed in the degradation assays a broader range of C2H2-containing ZFs were degraded. Using structural studies, including in silico docking, and combining the experimental data a number of principles can be elucidated:
-
1.
The sequence of the minimal degron associated with the early generation of thalidomide analogues consists of x(2)-C-x(2)-CG-x(2)-C-x(5). With the newer generation of substituted thalidomide analogues such as CC-885, CC-122 and CC-220, there is no additional consensus sequence apart from the residues that stabilise the ternary ZF fold. However, these chemically distinct analogues have an overlapping and additional repertoire of ZF degradation. Recruitment and degradation are therefore influenced not only by the complementarity of the ZF motifs to a drug–CRBN binding groove but also by the close contacts between the thalidomide analogues and specific ZF [54]. Altering the non-glutarimide moiety of the thalidomide analogues will therefore change the specificity for neosubstrates. The complete definition of ZF degrons will likely be further refined upon elucidation of the crystal structures of new CRBN binders. This is highlighted by the fact that the compound CC-122 degrades WIZ (a transcription factor involved in histone methylation chromatin regulation and cerebellar development) which has ZFs that are widely spaced, being separated by distances ranging from 16 to 263 amino acids in the longest human splice variant [56]. This compares to the typical C2H2-type ZF motifs, which are separated by seven amino acids. One interpretation of these data are that ZFs as such are not necessary for drug-CRBN mediated degradation but rather a stabilised β loop which can interact with both the compound and the substrate receptor. While the recent focus on defining degrons has been on ZF containing proteins, it should be noted that the degron in GSPT1 is similar to the one seen in IKZF1 and 3, and CSNK1A1. The precise patterns of degrons in non-ZF neosubstrates requires further consideration.
-
2.
The affinity of binding of neosubstrates to the drug-CRBN complex does not always predict for degradation in cells. This is due to a number of factors including the relative abundance of ZF substrates, their concentration and expression in cells, and, as these ZF are in epistasis, competition for CRBN binding sites. Thus cell context will be as important as the specific CRBN-binding compound in defining neosubstrate repertoires.
It should be pointed out that many ZF containing proteins do not have a defined function. However, SALL4, member of the spalt-like family of developmental transcription factors, contains a C2H2 domain, and germline mutations in SALL4 have been shown to cause Duane Radial Ray Syndrome (DRRS) and Holt-Oram Syndrome (ORS) [57]. Both these congenital syndromes have a phenotypic overlap with thalidomide associated embryopathy. Recently, both the Ebert/Fischer and Celgene groups have shown that known IMiD agents degrade SALL4 in human, primates and rabbits but not in rodents or fish [58], [59]. The inability of rodents to degrade SALL4 is explained by the fact that mouse CRBN contains an isoleucine at CRBN 388aa (human sequence) instead of valine [51], [58]. This substitution clashes with C2H2 neosubstrate engagement and and may explain why thalidomide associated teratogenicity was not detected in rodent species, although the poly-pharmacology of IMiDs presents the possibility that degradation of other targets may contribute to the teratogenicity in humans. The ability of different thalidomide analogues to degrade different ZF-containing proteins (Table 1) will enable development of novel thalidomide analogues for degradation of specific ZF proteins in specific cell contexts for therapeutic intervention.
Table 1.
Zinc Finger domain-containing proteins known to be degraded in cells. Data was assembled from the work of Thoma and co-workers [54] and Ebert/Fischer and co-workers [58].
| Zinc Fingers known to be degraded in cells | Known degrader compounds |
|---|---|
| IKZF3 | Pomalidomide, Lenalidomide, CC-885, CC-220 |
| IKZF1 | Pomalidomide, Lenalidomide, CC-885, CC-220, FPFT-2216 |
| ZFP91 | Pomalidomide, CC-220 |
| SALL4 | Thalidomide, Pomalidomide, Lenalidomide, dBET57, AR-825 |
| ZNF692 | Thalidomide, Pomalidomide, Lenalidomide |
| ZNF276 | Pomalidomide, Lenalidomide |
| ZNF827 | Pomalidomide, Lenalidomide |
| ZNF653 | Thalidomide, Pomalidomide, Lenalidomide, CC-220 |
| GZF1 | Pomalidomide |
| ZNF98 | CC-220, Pomalidomide |
Other DCAFs or E3 ligases as templates for targeted protein degradation
The powerful impact of the IMiD agents in hematological malignancies, and use of CRBN has spurred the activity to define novel E3 ligases that may be used as templates for drug discovery. Whilst studying mechanisms of cancer drug resistance to a sulfonamide, Indisulam, Nijahawan and co-workers discovered DCAF15 as a receptor for the CRL4 family that could be exploited for neosubstrate degradation [60]. Indisulam was shown to bind RBM39 (RNA binding motif protein 39) for UPS-mediated degradation. RBM39 is a known splicing factor and its degradation may have potential in haemopoietic lineages. Similar data have been described by the Eisai group [61].
So far, the DCAFs identified have been as a consequence of serendipity, but represent a great example of how the unexpected has been carefully studied to open a new area of therapeutics. More rational approaches to define additional DCAFs are being undertaken. For example, Cravatt and co-workers incorporated electrophilic “scout” fragments that covalently bind to cysteine-containing proteins [62] into heterobifunctional chimera containing the selective FKBP12 binder SLF to identify novel E3 ligase-mediated degradation pathways. Using FLAG-mediated affinity enrichment/proteomics, the authors showed that the substrate receptor DCAF16 was substantially enriched in FLAG-FKBP12 variants-expressing HEK293T when treated with the scout-SLF chimera. Importantly, they showed that DCAF16 was able to selectively bind the nuclear FLAG-FKBP12_NLS variant and not the cytosolic one which is consistent with DCAF16 predicted nuclear localisation [63].
CRBN and DCAF15 and 16 are part of the CRL4 E3 ligase family, which has a significant advantage for neosubstrate degradation in that the CUL4 ligase arm has a unique flexibility [53], with the ability to rotate 180° to create an exceptionally large ubiquitination zone that brings E2–ubiquitin bound to ROC1 into direct contact with the substrate. Other E3 ligases may however have other advantages, such as ability to target smaller and more specific neosubstrates and hence limit side effects.
Finally, it is becoming apparent some reversible small molecule inhibitors of protein function can also directly induce degradation of their targets, as exemplified by ligands for the transcription factor BCL6 [64]. Understanding the extent and generality of this phenomenon promises an exciting union of small molecule target inhibition and induced degradation in the future.
Comparison of the PROTAC and DCAF template approaches
The exploitation of CRBN as a template is different from the PROTAC approach and they are compared in Table 2. Both rely on establishing tractable ligand binding sites on the E3 ligase component that are “hotspots” for the formation of ternary complexes with other proteins, and the identification of small molecules that bind those sites. The construction of a PROTAC also requires the prior definition of the molecular target to be degraded, and a sufficiently potent ligand that can be incorporated into the PROTAC without perturbing target binding. It is not necessary that the ligand has a functional effect on the target protein, widening the scope beyond typical enzyme or receptor binding sites. This approach is well matched to the general paradigm for validating and prosecuting drug discovery against molecular targeted therapies for specific proteins, and tuning of the selectivity of PROTACs starting from promiscuous binding groups has been shown [35]. In contrast the exploitation of CRBN and other DCAFs as templates for the recruitment of neo-substrates for degradation, uses a screening approach to identify DCAF-binding small molecules that induce a defined, UPS-dependent phenotypic change. The targets degraded by these compounds are identified subsequently, typically using proteomic techniques. Effective DCAF reprogramming small molecules may be quite specific for degradation of one target, or may result from the induced degradation of a wide range of proteins. The discovery of DCAF-binding is an empirical process however, the emerging patterns of degron motifs within classes of neo-substrates degraded by CRBN binding small molecules suggest that more focussed approaches can also be applied.
Table 2.
Comparison of the PROTAC vs empirical exploitation of E3 ligase receptors to create a template for targeted protein degradation.
| PROTAC | DCAFs as templates |
|---|---|
| Requires a ligand binder for the target | Requires identification of ligand binding “hotspots” to attract neo-substrates |
| Linkers, E3-binding and target-binding components of the large bifunctional molecules need to be optimized | DCAF “hotspots” select for drug like properties in the ligands |
| Target known | Phenotypic screen defines target; Requires empirical definition of neo-substrates using proteomics or selecting targets based on degron motifs |
| Protein-protein interactions (ternary complexes) and E3-ligase binding “hotspots” are important | Protein-protein interactions (ternary complexes) and template ‘hotspots’ are important |
| Exploit known biology; molecular targeted approach | Explore new biology; targeted or poly-pharmacology |
| First examples reaching clinic | Clinical compounds |
Conclusion
Targeted protein degradation offers a great deal of promise for drug discovery and addition they raise a number of challenges, not least the ability to find binders and appropriate drug like properties for the PROTAC approach and the need to move beyond serendipity for selecting E3 ligases as templates for drug discovery. It is still unclear whether CRL4CRBN has unique properties or whether other E3 ligase receptors can also be similarly modulated, using small molecule approaches for degradation of a novel repertoire of neosubstrates.
Acknowledgements
The authors acknowledge support from The Institute of Cancer Research and Cancer Research UK program grant C309/A11566.
Section editors: Alessio Ciulli, FRSC – Professor of Chemical & Structural Biology, School of Life Sciences, University of Dundee, Division of Biological Chemistry and Drug Discovery, James Black Centre, Dow Street, Dundee DD1 5EH, United Kingdom. William Farnaby – Professor of Chemical & Structural Biology, School of Life Sciences, University of Dundee, Division of Biological Chemistry and Drug Discovery, University of Dundee, James Black Centre, Dow Street, Dundee DD1 5EH, United Kingdom.
References
- 1.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Hematology Am Soc Hematol Educ Program. 2006:505–506. doi: 10.1182/asheducation-2006.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Collins I., Wang H., Caldwell J.J., Chopra R. Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway. Biochem J. 2017;474:1127–1147. doi: 10.1042/BCJ20160762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lub S., Maes K., Menu E., De Bruyne E., Vanderkerken K., Van Valckenborgh E. Novel strategies to target the ubiquitin proteasome system in multiple myeloma. Oncotarget. 2016;7:6521–6537. doi: 10.18632/oncotarget.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y., Jia L. Neddylation pathway as a novel anti-cancer target: mechanistic investigation and therapeutic implication. Anticancer Agents Med Chem. 2015;15:1127–1133. doi: 10.2174/1871520615666150305111257. [DOI] [PubMed] [Google Scholar]
- 5.Harrigan J.A., Jacq X., Martin N.M., Jackson S.P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu G., Middleton R.E., Sun H., Naniong M., Ott C.J., Mitsiades C.S. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivanco I., Chen Z.C., Tanos B., Oldrini B., Hsieh W.Y., Yannuzzi N. A kinase-independent function of AKT promotes cancer cell survival. Elife. 2014;3(December):3. doi: 10.7554/eLife.03751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churcher I. Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J Med Chem. 2018;61:444–452. doi: 10.1021/acs.jmedchem.7b01272. [DOI] [PubMed] [Google Scholar]
- 9.Neklesa T.K., Winkler J.D., Crews C.M. Targeted protein degradation by PROTACs. Pharmacol Ther. 2017;174:138–144. doi: 10.1016/j.pharmthera.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 10.Lucas X., Ciulli A. Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr Opin Struct Biol. 2017;44:101–110. doi: 10.1016/j.sbi.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Ohoka N., Shibata N., Hattori T., Naito M. Protein knockdown technology: application of ubiquitin ligase to cancer therapy. Curr Cancer Drug Targets. 2016;16:136–146. doi: 10.2174/1568009616666151112122502. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto K.M., Kim K.B., Kumagai A., Mercurio F., Crews C.M., Deshaies R.J. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneekloth J.S., Jr., Fonseca F.N., Koldobskiy M., Mandal A., Deshaies R., Sakamoto K. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 14.Bondeson D.P., Mares A., Smith I.E.D., Ko E., Campos S., Miah A.H. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zengerle M., Chan K., Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh Y., Ishikawa M., Naito M., Hashimoto Y. Protein knockdown using methyl bestatin–ligand hybrid molecules: design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J Am Chem Soc. 2010;132:5820–5826. doi: 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- 17.Schneekloth A.R., Pucheault M., Tae H.S., Crews C.M. Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg Med Chem Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu J., Qian Y., Altieri M., Dong H., Wang J., Raina K. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter G.E., Buckley D.L., Paulk J., Roberts J.M., Souza A., Dhe-Paganon S. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang K., Song Y., Xie H., Wu H., Wu Y., Leisten E.D. Development of the first small molecule histone deacetylase 6 (HDAC6) degraders. Bioorg Med Chem Lett. 2018;28:2493–2497. doi: 10.1016/j.bmcl.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 21.McCoull W., Cheung T., Anderson E., Barton P., Burgess J., Byth K. Development of a novel B-cell lymphoma 6 (BCL6) PROTAC to provide insight into small molecule targeting of BCL6. ACS Chem Biol. 2018;13:3131–3141. doi: 10.1021/acschembio.8b00698. [DOI] [PubMed] [Google Scholar]
- 22.Raina K., Lu J., Qian Y., Altieri M., Gordon D., Rossi A.M. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burslem G.M., Smith B.E., Lai A.C., Jaime-Figueroa S., McQuaid D.C., Bondeson D.P. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol. 2018;18(25) doi: 10.1016/j.chembiol.2017.09.009. 67–77.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gadd M.S., Testa A., Lucas X., Chan K.H., Chen W., Lamont D.J. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes S.J., Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem. 2017;61:505–516. doi: 10.1042/EBC20170041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zorba A., Nguyen C., Xu Y., Starr J., Borzilleri K., Smith J. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc Natl Acad Sci U S A. 2018;115:E7285–E7292. doi: 10.1073/pnas.1803662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai A.C., Toure M., Hellerschmied D., Salami J., Jaime-Figueroa S., Ko E. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed Engl. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen T.V., Li J., Lu C.J., Mamrosh J.L., Lu G., Cathers B.E. p97/VCP promotes degradation of CRBN substrate glutamine synthetase and neosubstrates. Proc Natl Acad Sci U S A. 2017;114:3565–3571. doi: 10.1073/pnas.1700949114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riching K.M., Mahan S., Corona C.R., McDougall M., Vasta J.D., Robers M.D. Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem Biol. 2018;13(9):2758–2770. doi: 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- 30.Qin C., Hu Y., Zhou B., Fernandez-Salas E., Yang C., Liu L. Discovery of QCA570 as an exceptionally potent and efficacious proteolysis targeting chimera (PROTAC) degrader of the bromodomain and extra-terminal (BET) proteins capable of inducing complete and durable tumor regression. J Med Chem. 2018;61:6685–6704. doi: 10.1021/acs.jmedchem.8b00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai L., Zhou B., Yang C.Y., Ji J., McEachern D., Przybranowski S. Targeted degradation of BET proteins in triple-negative breast cancer. Cancer Res. 2017;77:2476–2487. doi: 10.1158/0008-5472.CAN-16-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiedel M., Herp D., Hammelmann S., Swyter S., Lehotzky A., Robaa D. Chemically induced degradation of sirtuin 2 (Sirt2) by a proteolysis targeting chimera (PROTAC) based on sirtuin rearranging ligands (SirReals) J Med Chem. 2018;61:482–491. doi: 10.1021/acs.jmedchem.6b01872. [DOI] [PubMed] [Google Scholar]
- 33.Cyrus K., Wehenkel M., Choi E., Swanson H., Kim K. Two-headed PROTAC: an effective new tool for targeted protein degradation. ChemBioChem. 2011;11:1531–1534. doi: 10.1002/cbic.201000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H.T., Dobrovolsky D., Paulk J., Yang G., Weisberg E.L., Doctor Z.M. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol. 2018;25:88–99. doi: 10.1016/j.chembiol.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bondeson D.P., Smith B.E., Burslem G.M., Buhimschi A.D., Hines J., Jaime-Figueroa S. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol. 2018;25:78–87. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson C.M., Jiang B., Erb M.A., Liang Y., Doctor Z.M., Zhang Z. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai A.C., Crews C.M. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekine K., Takubo K., Kikuchi R., Nishimoto M., Kitagawa M., Abe F. Small molecules destabilize cIAP1 by activating auto-ubiquitylation. J Biol Chem. 2008;283:8961–8968. doi: 10.1074/jbc.M709525200. [DOI] [PubMed] [Google Scholar]
- 39.Hines J., Lartigue S., Dong H., Qian Y., Crews C.M. MDM2-recruiting PROTAC offers superior, synergistic anti-proliferative activity via simultaneous degradation of BRD4 and stabilization of p53. Cancer Res. 2019;79:251–262. doi: 10.1158/0008-5472.CAN-18-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chessum N.E.A., Sharp S.Y., Caldwell J.J., Pasqua A.E., Wilding B., Colombano G. Demonstrating in-cell target engagement using a pirin protein degradation probe (CCT367766) J Med Chem. 2018;61:918–933. doi: 10.1021/acs.jmedchem.7b01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun B., Fiskus W., Qian Y., Rajapakshe K., Raina K., Coleman K.G. BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia. 2018;32:343–352. doi: 10.1038/leu.2017.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C., Han X.R., Yang X., Jiang B., Liu J., Xiong Y. Proteolysis targeting chimeras (PROTACs) of anaplastic lymphoma kinase (ALK) Eur J Med Chem. 2018;151:304–314. doi: 10.1016/j.ejmech.2018.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neklesa T., Snyder L.B., Willard R.R., Vitale N., Raina K., Pizzano J. ARV-110: an androgen receptor PROTAC degrader for prostate cancer. Cancer Res. 2018;78(Suppl. (3)) Abstract nr 5236. [Google Scholar]
- 44.Long M.C.J., Gollapalli D.R., Hedstrom L. Inhibitor mediated protein degradation. Chem Biol. 2012;19:629–637. doi: 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi Y., Long M.J.C., Rosenberg M.M., Li S., Kobjack A., Lessans P. Boc3Arg-linked ligands induce degradation by localizing target proteins to the 20S proteasome. ACS Chem Biol. 2016;11:3328–3337. doi: 10.1021/acschembio.6b00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neklesa T.K., Tae H.S., Schneekloth A.R., Stulberg M.J., Corson T.W., Sundberg T.B. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gustafson J.L., Neklesa T.K., Cox C.S., Roth A.G., Buckley D.L., Tae H.S. Small-molecule-mediated degradation of the androgen receptor through hydrophobic tagging. Angew Chem Int Ed Engl. 2015;54:9659–9662. doi: 10.1002/anie.201503720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Guillen V.S., Sharma N., Flessa K., Min J., Carlson K.E. New class of selective estrogen receptor degraders (SERDs): expanding the toolbox of PROTAC degrons. ACS Med Chem Lett. 2018;9:803–808. doi: 10.1021/acsmedchemlett.8b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandhi A.K., Kang J., Havens C.G., Conklin T., Ning Y., Wu L. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) Br J Haematol. 2014;164(March (6)):811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krönke J., Udeshi N.D., Narla A., Grauman P., Hurst S.N., McConkey M. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krönke J., Fink E.C., Hollenbach P.W., MacBeth K.J., Hurst S.N., Udeshi N.D. Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matyskiela M.E., Lu G., Ito T., Pagarigan B., Lu C.C., Miller K. A novel cereblon modulator recruits GSPT1 to the CRL4 (CRBN) ubiquitin ligase. Nature. 2016;535:252–257. doi: 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- 53.Petzold G., Fischer E.S., Thomä N.H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- 54.Sievers Q.L., Petzold G., Bunker R.D., Renneville A., Słabicki M., Liddicoat B.J. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362:eaat0572. doi: 10.1126/science.aat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen J.D., Condroski K., Correa M., Muller G., Man H.W., Ruchelman A. Protein degradation via CRL4CRBN ubiquitin ligase: discovery and structure-activity relationships of novel glutarimide analogs that promote degradation of aiolos and/or GSPT1. J Med Chem. 2018;61:492–503. doi: 10.1021/acs.jmedchem.6b01911. [DOI] [PubMed] [Google Scholar]
- 56.Simon J.M., Parker J.S., Liu F., Rothbart S.B., Ait-Si-Ali S., Strahl B.D. A role for widely interspaced zinc finger (WIZ) in retention of the G9a methyltransferase on chromatin. J Biol Chem. 2015;290:26088–26102. doi: 10.1074/jbc.M115.654459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohlhase J., Schubert L., Liebers M., Rauch A., Becker K., Mohammed S.N. Mutations at the SALL4 locus on chromosome 20 result in a range of clinically overlapping phenotypes, including Okihiro syndrome, Holt-Oram syndrome, acro-renal-ocular syndrome, and patients previously reported to represent thalidomide embryopathy. J Med Genet. 2003;40:473–478. doi: 10.1136/jmg.40.7.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donovan K.A., An J., Nowak R.P., Yuan J.C., Fink E.C., Berry B.C. Thalidomide promotes degradation of SAL, a transcription factor implicated in Duane Radial Ray syndrome. Elife. 2018;8430 doi: 10.7554/eLife.38430. pii: e38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matyskiela M.E., Couto S., Zheng X., Lu G., Hui J., Stamp K. SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat Chem Biol. 2018;14:981–987. doi: 10.1038/s41589-018-0129-x. [DOI] [PubMed] [Google Scholar]
- 60.Han T., Goralski M., Gaskill N., Capota E., Kim J., Ting T.C. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356:eaal3755. doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- 61.Uehara T., Minoshima Y., Sagane K., Sugi N.H., Mitsuhashi K.O., Yamamoto N. Selective degradation of splicing factor CAPERα by anticancer sulfonamides. Nat Chem Biol. 2017;13:675–680. doi: 10.1038/nchembio.2363. [DOI] [PubMed] [Google Scholar]
- 62.Backus K.M., Correia B.E., Lum K.M., Forli S., Horning B.D., González-Páez G.E. Proteome-wide covalent ligand discovery in native biological systems. Nature. 2016;534(June (7608)):570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X., Crowley V.M., Wucherpfennig T.G., Dix M.M., Cravatt B.F. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Biorxiv. 2018 doi: 10.1038/s41589-019-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kerres N., Steurer S., Schlager S., Bader G., Berger H., Caligiuri M. Chemically induced degradation of the oncogenic transcription factor BCL6. Cell Rep. 2017;20:2860–2875. doi: 10.1016/j.celrep.2017.08.081. [DOI] [PubMed] [Google Scholar]
- 65.Jiang Y., Deng Q., Zhao H., Xie M., Chen L., Yin F. Development of stabilized peptide-based PROTACs against estrogen receptor. ACS Chem Biol. 2018;13:628–635. doi: 10.1021/acschembio.7b00985. [DOI] [PubMed] [Google Scholar]
- 66.Zhou B., Hu J., Xu F., Chen Z., Bai L., Fernandez-Salas E. Discovery of a small-molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J Med Chem. 2018;61(January (2)):462–481. doi: 10.1021/acs.jmedchem.6b01816. [DOI] [PMC free article] [PubMed] [Google Scholar]