Abstract
Throughout the past decade, studying ancient genomes has provided unique insights into human prehistory, and differences between modern humans and other branches like Neanderthals can enrich our understanding of the molecular basis of unique modern human traits. Modern human variation and the interactions between different hominin lineages are now well studied, making it reasonable to go beyond fixed genetic changes and explore changes that are observed at high frequency in present-day humans. Here, we identify 571 genes with non-synonymous changes at high frequency. We suggest that molecular mechanisms in cell division and networks affecting cellular features of neurons were prominently modified by these changes. Complex phenotypes in brain growth trajectory and cognitive traits are likely influenced by these networks and other non-coding changes presented here. We propose that at least some of these changes contributed to uniquely human traits, and should be prioritized for experimental validation.
Subject terms: Evolutionary genetics, Evolutionary biology
Introduction
Homo sapiens appears to be a “very special primate”1. Our position among animal species stands out largely thanks to the composite complexity of our cultures, social structures and communication systems. It seems reasonable that this “human condition” is rooted, at least in part, in the properties of our brain, and that these can be traced to changes in the genome on the modern human lineage. This phenotype in the population called “anatomically modern humans” emerged in Africa likely before the deepest divergence less than 100,000–200,000 years ago2,3, although complex population structure may reach back up to 300,000 years ago4–6. Except for some early dispersals7,8, humans most likely peopled other parts of the world than Africa and the Middle East permanently only after around 65,000 years ago. It has been claimed that the brain of modern humans adopted a specific, apomorphic growth trajectory early in life that gave rise to the skull shape difference between modern humans and extinct branches of the genus Homo9. Importantly, the growth pattern might differ between the populations10,11, with Neanderthal alleles influencing the endocranial shape in modern humans12, while the brain size and encephalization of humans and Neanderthals is similar9,13,14. This ontogenic trajectory, termed the “globularization phase”, might have contributed to cognitive changes that underlie behavioral traits in which humans differ from their extinct relatives, despite mounting evidence for their cognitive sophistication9,15–18.
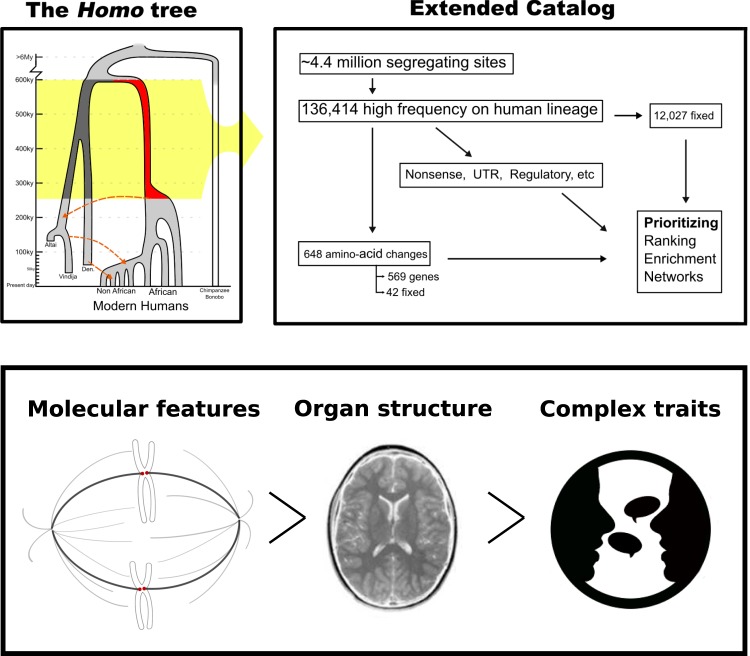
We are now in a favorable position to examine the evolution of human biology with the help of the fossil record, in particular thanks to breakthroughs in paleogenomics: The recent reconstruction of the high quality genomes of members of archaic Homo populations19–21 has opened the door to new comparative genomic approaches and molecular analyses. The split of the lineages leading to modern humans and other archaic forms (Neanderthals and Denisovans) is estimated to around 600,000 years ago2, setting the timeframe for truly modern human-specific changes after this split, but before the divergence of modern human populations (Fig. 1). Together with efforts to explore present-day human diversity22, this progress has allowed to narrow down the number of candidate point mutations from ~35 million differences since the split from chimpanzee when comparing only reference genomes23 to 31,389 fixed human-specific changes in a previous seminal study1. Other types of more complex changes like structural variants most likely contributed to human-specific traits. For example, it is well known that since the split from chimpanzees functional differences arose through gene duplications in ARHGAP11B and other genes24,25, copy number variants in SRGAP2 and other genes26–28 or regulatory deletions29. In these cases, the variants arose before the split between humans and Neanderthals, but the differences in structural variation that exist between the hominin lineages30 need to be explored in more detail, with advancement of technologies in ancient DNA sequencing and computational methods. This will result in complementary lists of changes for understanding the human condition outside the scope of this study.
Figure 1.
Conceptual summary of this study.
Some of the single nucleotide changes have been linked to putative functional consequences1,20,31, and evidence is mounting that several molecular changes affecting gene expression in the brain were subject to selective pressures32–36. Furthermore, the genomic impact of interbreeding events is not evenly distributed across the genome. Genes expressed in regions of the brain regarded as critical for certain cognitive functions such as language are depleted in introgressed archaic genetic material37–40, and introgressed alleles are downregulated in some of these brain regions, suggesting natural selection acting on tissue-specific gene regulation41. Thus, it seems reasonable to conclude that there were differences between anatomically modern human and Neanderthal brains, and that these underlie at least some of the characteristics of our lineage42. We want to emphasize that such recent differences are likely to be subtle when compared to those after the split from our closest living relatives on a scale of 6–10 million years43, where fundamental changes arose since the divergence from chimpanzees and bonobos44,45. The observation of recurrent gene flow between modern human and archaic populations also implies a broad overall similarity, yet, such subtle differences may still have contributed to the evolutionary outcome18. This does not imply a superiority of humans, but specific changes that might have facilitated survival under the given environmental conditions. Obviously, not all human-specific changes are beneficial: while most mutations may be rather neutral and have little effect on the phenotype, some may have had deleterious effects or side-effects, possibly increasing the risks for neurodevelopmental or neurodegenerative disorders in humans46–48.
The goal of this paper is to provide a revised, extended set of recent single nucleotide changes in humans since their split from Neanderthals that could enrich our understanding of the molecular basis of the recent human condition. The previous focus on fixed alleles was reasonable given limited data1, but having a better grasp of the magnitude of modern human variation and the interaction between different hominin lineages seems a good reason to cast a wider net, and take into account not only fixed differences but also high-frequency changes shared by more than 90% of present-day individuals. Here, we present a revised list of 36 genes that carry missense substitutions that are fixed across thousands of human individuals and for which all archaic hominin individuals sequenced so far carry the ancestral state. In total, 647 protein-altering changes in 571 genes reached a frequency of at least 90% in the present-day human population. We attempt to interpret this list, as well as some regulatory changes, since it seems very likely that some of these genes would have contributed to the human condition. We discuss some of their known functions, and how these relate to pathways that might have been modified during human evolution from the molecular level to cellular features and more complex phenotypic traits (Fig. 1). We restrict our attention to genes where the literature allows reasonably firm conclusions and predictions about functional effects, since many genes likely have pleiotropic functions49. Obviously, it cannot be emphasized enough that ultimately, experimental validation will be needed to confirm our hypotheses concerning alterations in specific functions. For example, transcription factors or enzymatically active proteins can be tested using cell cultures or in vitro assays, while brain organoids could be used to test differences in neuronal functions50, especially in combination with single-cell RNA sequencing51–53. Ultimately, these variants can be introduced into model organisms like mice to test complex features related to cognitive abilities or behavior54. Still, given existing limitations concerning the amount of changes that can be tested at once, networks that are modified by multiple changes cannot be tested with current technologies, making synthesizing attempts like ours valuable.
Results
Genetic differences between present-day humans and archaic hominins
Using publicly available data on one Denisovan and two Neanderthal individuals and present-day human variation (Methods), we calculated the numbers of single nucleotide changes (SNCs) which most likely arose recently on the respective lineages after their split from each other, and their functional consequences (Table 1). Previously, a number of 31,389 sites has been reported as recently fixed derived in present-day humans, while being ancestral in archaics1,20. We find a smaller number of only 12,027 positions in the genome, in part due to the inclusion of another archaic individual and different filters, but mainly because of a richer picture of present-day human variation. The 1,000 Genomes Project as well as other sources contributing to the dbSNP database now provide data for thousands of individuals, which results in very high allele frequencies for many loci, instead of full fixation. Indeed, 29,358 positions show allele frequencies larger than 0.995, demonstrating that the level of near-fixation is similar to the level of previously presented fixation. The number of loci with high frequency (HF) changes of more than 90% in present-day humans is an order of magnitude larger than the number of fixed differences. This cutoff is somewhat arbitrary and based on previous work20. However, when increasing the frequency cutoff, the number declines sharply, while decreasing it results in a near-linear increase of sites (Fig. S1). The three archaic individuals carry more than twice as many changes than present-day humans; however, we emphasize that much of this difference is not due to more mutations in archaics, but rather the fact that data for only three individuals is available, compared to thousands of humans. The variation across the archaic population is not represented equally well, which makes these numbers not directly comparable. On the other hand, much less variation is found by the sequencing of each additional Neanderthal individual compared to humans due to the low diversity of Neanderthals (Fig. S36 in reference 22). This low diversity across their geographic range suggests that most alleles observed as ancestral here will be the same state in other individuals. Furthermore, we take variability into account due to gene flow or errors, decreasing the possibility that positions ancestral in the archaic individuals studied to date turn out to be derived in most archaic individuals, leading to the prediction that this extended catalog will likely not undergo drastic changes. However, changes in structural variants or regions of the genome that are not accessible by current sequencing technologies will most likely complement our results45.
Table 1.
Summary of single nucleotide changes.
| Fixed human | HF human | Extended human | Fixed archaic | HF archaic | Extended archaic | |
|---|---|---|---|---|---|---|
| All | 12,027 | 136,435 | 83,254 | 33,498 | 380,756 | 983 |
| Non-synonymous | 42 | 647 | 327 | 167 | 1,921 | 13 |
| Synonymous | 41 | 843 | 363 | 193 | 2,123 | 14 |
| Start/stop | 1 | 14 | 10 | 3 | 48 | 2 |
| Splice site | 4 | 23 | 8 | 4 | 54 | 0 |
| TFBS | 28 | 226 | 126 | 87 | 914 | 1 |
| Upstream | 1,935 | 19,599 | 11,235 | 4,920 | 55,188 | 289 |
| 5′ UTR | 180 | 1,853 | 1,012 | 195 | 2,016 | 7 |
| 3′ UTR | 77 | 702 | 334 | 506 | 5,303 | 19 |
| Downstream | 1,922 | 19,704 | 11,673 | 4,956 | 55,832 | 281 |
| miRNA | 0 | 1 | 2 | 0 | 4 | 0 |
| Regulatory element | 1,952 | 20,971 | 12,320 | 5,125 | 59,248 | 195 |
TFBS: Transcription factor binding sites. UTR: Untranslated Region. HF: High frequency. Fixed changes are a subset of HF changes.
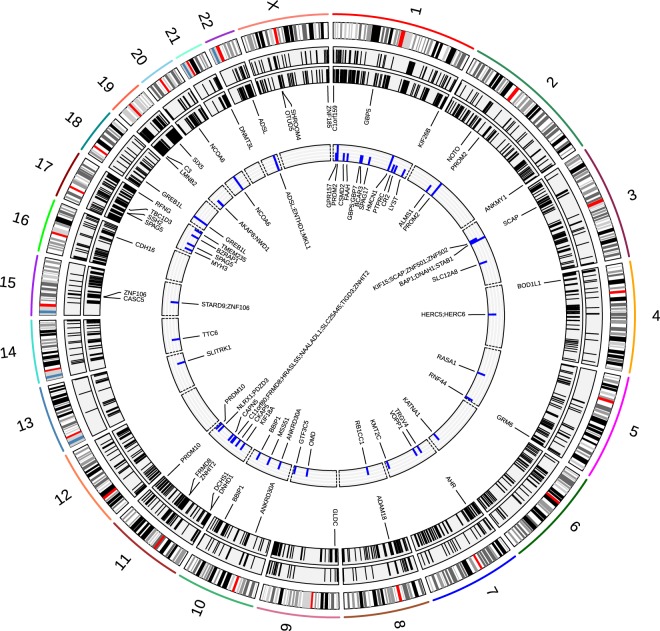
Present-day humans carry 42 fixed amino acid-changes in 36 genes (Table 2, Fig. 2), while Neanderthals carry 159 such changes. Additionally, modern humans carry 605 amino acid-changes at high frequency (human-lineage high-frequency missense changes, referred to as HHMCs), amounting to a total of 647 such changes in 571 genes (Table S1). Together with 323 SNCs on the human lineage with low confidence (Methods, Table S2), almost 1,000 putative protein-altering changes were found across most present-day humans. Generally, synonymous changes are found at a similar magnitude as missense changes, but only few SNCs altering start and stop codons, and thousands of changes in putative regulatory and untranslated regions. We admit that some of the loci presented here are variable across the phylogenetic tree, or less reliable due to low coverage in the archaics, but we accept this since our intention is retrieve an inclusive picture of possibly functional recent changes. The 42 protein-altering changes for which the ancestral allele has not been observed in any present-day human, most of which have been presented before1, constitute without doubt the strongest entry points into a molecular understanding of the human condition, and should be prime candidates for experimental validation. Only one gene, SPAG5, carries three such SNCs, and four genes (ADAM18, CASC5, SSH2 and ZNHIT2) carry two fixed protein-coding changes in all modern humans. We identified 15 SNCs (in AHR, BOD1L1, C1orf159, C3, DNHD1, DNMT3L, FRMD8, OTUD5, PROM2, SHROOM4, SIX5, SSH2, TBC1D3, ZNF106, ZNHIT2) that have not been previously described as fixed differences between humans and archaics. We note that another 12 previously described1 protein-altering substitutions were not found among the genotypes analyzed here (in C21orf62, DHX29, FAM149B1, FRRS1L, GPT, GSR, HERC5, IFI44L, KLF14, PLAC1L, PTCD2, SCAF11). These genotype calls are absent from the files provided for the three archaic genomes due to different genotype calling and filtering procedures compared to the original publication of the Altai Neanderthal genome20,21. Hence, some potentially relevant candidate changes were not included here, and future research is necessary to evaluate these as well. Despite attempting an extended interpretation, our data is thus not fully exhaustive.
Table 2.
Genes with fixed non-synonymous changes on the human lineage, genes under positive selection with HHMCs, and deleterious candidate HHMCs.
| Fixed HHMCs | ADAM18, ADSL, AHR, ANKMY1, ANKRD30A, BBIP1, BOD1L1, C1orf159, C3, CASC5, CDH16, DCHS1, DNHD1, DNMT3L, FRMD8, GBP5, GLDC, GREB1L, GRM6, KIF26B, LMNB2, NCOA6, NOTO, OTUD5, PRDM10, PROM2, RFNG, SCAP, SHROOM4, SIX5, SPAG5, SSH2, TBC1D3, ZNF106, ZNF185, ZNHIT2 |
| Selection 2014 | C11orf80, CKAP5, GREB1L, HMCN1, NLRX1, PDZD3, PRDM2, RB1CC1 |
| Selection 2015 | MSS51, NCOA6, OMD, SPAG17, SPAG5 |
| Selection 2016 | ACE, ADSL, ALMS1, ANKRD30A, BZRAP1, DNAH1, GREB1L, KMT2C, NWD1, PROM2, RASA1, STAB1, STARD9, ZNF106 |
| Selection 2017 | ADSL, AKAP8, BAP1, BBIP1, BCAR3, CAPN5, CR2, CSMD2, DNAH1, ENTHD1, FAAH, FRMD8, GBP5, GBP7, GPR157, GTF3C5, HERC5, HERC6, HMCN1, HRASLS5, KATNA1, KIF15, KIF18A, LYST, MKL1, MYH3, NAALADL1, NCOA6, PRDM10, PRDM2, PROM2, PTPRC, RNF44, SCAP, SLC12A8, SLC25A45, SLITRK1, TIGD3, TMEM235, TRGV4, TTC6, VOPP1, ZNF501, ZNF502, ZNHIT2 |
| Grantham | ABHD14A-ACY1, ACY1, ABHD14A, CCDC158, CCDC30, DNHD1, EML2, ERI1, GBA3, GREB1, OR1K1, TTC6, UBQLN3, UIMC1, ZBP1, ZNF510, ZNHIT2 |
| SIFT | BEND2, CCT6B, COPA, CUL4B, GBP7, KRTAP10-10, MEPE, NHEJ1, OR1K1, SLC6A15, TPO, ZNF510 |
| PolyPhen-2 | FSHR, NLN, TPO |
| CADD | C11orf80, C5orf66, CCT6B, CDH15, CEP128, CPM, FGF21, FMN2, FUT1, H2AFY, HERC6, KCNK5, KPNA4, KRT33A, KRT8P12, MUM1, NR1H2, OPRM1, PDSS2, ROCK1, RPS15P9, SLC22A31, SUCLG2P4, TMPRSS7, UNC5D |
| GWAVA | ANK2, COPA, CTRC, CYP2B6, MAPK10, MCTP1, SLC38A6, SYT1, YTHDC1 |
Figure 2.
Features discussed in this study. From inside to outside: Genes with HHMCs and signatures of positive selection (compare Table 2), genes with fixed non-synonymous SNCs on the human lineage, HHMCs, AHMCs, karyogram of human chromosomes.
It is noteworthy that the number of fixed SNCs decreased substantially, and it is possible that single individuals will be found to carry some of the ancestral alleles for the remaining fixed sites. Hence, it is important to focus not only on fixed differences, but also consider variants at high frequency. When analyzing the 647 HHMCs, 68 genes carry more than one amino acid-altering change. Among these, TSGA10IP (Testis Specific 10 Interacting Protein) and ABCC12 (ATP Binding Cassette Subfamily C Member 12) carry four such changes, and seven more genes (MUC5B, NPAP1, OR10AG1, OR5M9, PIGZ, SLX4, VCAN) carry three HHMCs. 1,542 genes carry at least one HF missense change on the archaic lineage (archaic-lineage high-frequency missense change, referred to as AHMC, Tables S3, S4). We find an overlap of 122 genes with HHMCs and AHMCs, which is more than expected considering that among 1,000 sets of random genes of a similar length distribution, no overlap of this extent was observed. The same genes seem to have acquired missense changes on both lineages since their divergence more often than expected. We find a high ratio of HHMCs over synonymous changes for chromosome 21 (1.75-fold), and a very small ratio (0.18-fold) for chromosome 13. We do not find such extreme ratios for AHMCs and corresponding synonymous changes, suggesting differences in the distribution of amino acid changes between both lineages (Fig. S2).
Ranking and enrichment
We assessed the impact of mutations for different deleteriousness scores (Table 2), finding 12 genes with deleterious HHMCs according to SIFT, 3 according to PolyPhen, and 16 when using the Grantham score (>180), measuring the physical properties of amino acid changes. The C-score and GWAVA can be used to rank all mutation classes, including non-coding changes, and we present the top candidates.
Then, we attempted a ranking of genes by the density of lineage-specific changes in the dataset. As expected, the total number of segregating sites is correlated with gene length (Pearsons’ R = 0.93). This correlation is weaker for HF human SNCs (R = 0.73) and fixed human-specific SNCs (R = 0.25), as well as for fixed (R = 0.37) and HF (R = 0.82) SNCs in archaics. We conclude that some genes with a large number of human-specific changes might carry these large numbers by chance, while others are depleted. Indeed, 17,453 (88.9%) of these genes do not carry any fixed human-specific change, and 80.5% do not carry fixed archaic-specific changes. Of note, genes that have attracted attention in the context of traits related to the “human condition” like CNTNAP2 and AUTS2 are among the longest genes in the genome, hence changes in these genes should be interpreted with caution as they are not unexpected. We ranked the genes by the number of HF changes in either modern humans or archaics, divided by their genomic lengths, and categorize the top 5% of this distribution as putatively enriched for changes on each lineage (Table S5). We note that 191 genes (30.9%) fall within this category for both human HF changes and archaic HF changes, as a result of differences in mutation density. In order to distinguish a truly lineage-specific enrichment, we calculated the ratios of HF changes for humans and archaics, defining the top 10% of genes in this distribution as putatively enriched (Table S5). Among the genes enriched for changes on the modern human lineage, 18 carry no HF changes on the archaic lineage, and ten of these also fall within the 5% of genes carrying many changes considering their length (ARSJ, CLUAP1, COL20A1, EPPIN, KLHL31, MKNK1, PALMD, RIC3, TDRD7, UBE2H). These might be candidates for an accumulation of changes, even though this is not identical to selective sweep signals. Among these, the collagen COL20A1 and the Epididymal Peptidase Inhibitor EPPIN carry HHMCs. ACAD10, DST and TTC40, which carry two HHMCs, might be other notable genes with a human-specific enrichment.
Gene Ontology (GO) categories are neither enriched for HHMCs on the human lineage in a hypergeometric test, nor for genes carrying AHMCs, HF changes in UTRs or transcription factor binding sites. However, instead of singular changes that might be observed more often in long genes, or genes that are more prone to mutations in hominins, the density of HF changes in a gene might yield a better picture of lineage-specific changes, possibly for cumulative changes. We applied a test for the ratio of the number of gene-wise HF changes on one lineage over the other lineage, finding an enrichment for 12 GO categories on the human lineage (Table S6), with “soft palate development”, “negative regulation of adenylate cyclase activity”, “collagen catabolic process” and “cell adhesion” in the biological process category. Among the cellular components category, the “postsynaptic membrane”, “spermatoproteasome complex”, “collagen trimer”, “dendrite” and “cell junction” show enrichment, as well as the molecular functions “calcium ion binding”, “histone methyltransferase activity (H3-K27 specific)” and “metallopeptidase activity”. We find no GO enrichment for genes with an excess of changes on the archaic lineage. In order to approach a deeper exploration of genes with associated complex traits in humans, we explored the NHGRI-EBI GWAS Catalog55, containing 2,385 traits. We performed a systematic enrichment screen, finding 17 unique traits enriched for genes with HHMCs, and 11 for genes with AHMCs (Table S7). Changes in genes associated to “Cognitive decline (age-related)”, “Rheumatoid arthritis” or “Major depressive disorder” might point to pathways that could have been influenced by protein-coding changes on the human lineage. In archaics, genes are enriched, among others, for associations to traits related to body mass index or cholesterol levels, which might reflect differences in their physiology.
We find a significant enrichment of protein-protein interactions (P = 0.006) among the gene products of HHMC genes (Fig. S3), meaning that these proteins interact with each other more than expected. Functional enrichment is found for the biological process “cellular component assembly involved in morphogenesis”, most strongly for the cellular components cytoskeleton and microtubule, as well as the molecular function “cytoskeletal protein binding”. Three proteins have at least 20 interactions in this network and might be considered important nodes: TOP2A, PRDM10 and AVPR2 (Table S8). However, proteins encoded by genes with synonymous changes on the modern human lineage seem to be enriched for interactions as well (P = 0.003), as are proteins encoded by genes with AHMCs (P = 1.68 × 10−14), with an enrichment in GO categories related to the extracellular matrix and the cytoskeleton, and proteins with more than 40 interactions (Table S8). We caution that these networks might be biased due to more mutations and possibly more interactions in longer, multi-domain genes.
Regulatory changes might have been important during our evolution56, hence we tested for an overrepresentation of transcription factors. We find 78 known or putative transcription factors among the HHMC genes (Table S9) on the modern human lineage57, which is no overrepresentation among genes with HHMCs (with 49.2% of random genes sets containing fewer HHMCs). Despite no enrichment as a category, single transcription factors on the modern human lineage might have been important, particularly those with an excess of modern human over archaic HF changes (AHR, MACC1, PRDM2, TCF3, ZNF420, ZNF516). Others, like RB1CC120 or PRDM10 and NCOA633 have been found in selective sweep screens, suggesting contributions of individual transcription factors, rather than the whole class of proteins. We also tested for an enrichment of gene expression in different brain regions and developmental stages58,59, using the HF synonymous changes on each lineage as background sets. We find an enrichment of gene expression in the orbital frontal cortex at infant age (0–2 years) for genes with HHMCs, but no enrichment for genes with AHMCs. Furthermore, when testing the genes with HHMCs and using the set of genes with AHMCs as background, “gray matter of forebrain” at adolescent age (12–19 years) is enriched, while no enrichment was found for genes with AHMCs.
Discussion
The enrichment of broad categories above suggests traits prominently represented by HHMCs, some of which are possibly brain-related. It should be noted that such results would be less clear if we just focused on completely fixed changes, given the drastically reduced number of genes harboring such changes. It seems likely that many human-specific traits will rather be the consequence of cumulative changes than a single change60. Hence, we suggest that the “full modernity” of modern humans is constituted from a network of changes, where the presence of single ancestral alleles in some individuals would not lead to “partially modern” phenotype. Here, we will further examine the possible impact on the brain that some of these changes might have, paying special attention to hypotheses formulated in earlier work on modern human-specific changes. Our extended catalog of changes appears to provide additional support for some of these hypotheses.
Cell division and the brain growth trajectory
It has been proposed previously that protein-coding changes in cell cycle-related genes are highly relevant candidates for human-specific traits1,20, with the brain being specifically sensitive to such changes61. Indeed, three genes (CASC5, SPAG5, and KIF18A) have been singled out as involved in spindle pole assembly during mitosis1. Other genes with protein-coding SNCs (NEK6, STARD9/KIF16A and CDK5RAP2) turn out to be implicated in the regulation of spindle pole assembly as well62–64. Among the 15 fixed protein-coding changes identified here but absent from previous analyses1,20, some might have also contributed to complex modifications of pathways in cell division, like AHR65 or DNHD166 (Supplementary Information 1), as well as other genes with HHMCs, like CHEK167 or the gene encoding for the protein TOP2A68, which shows the largest number of interactions with other HHMC-carrying proteins, suggesting a function as interaction hub in the cell division complex (Supplementary Information 1). Taken together, these changes suggest that the cell cycle machinery might have been modified in a specific way in humans compared to other hominins.
It has been claimed20 that genes with fixed non-synonymous changes in humans are also more often expressed in the ventricular zone of the developing neocortex, compared to fixed synonymous changes. Since the kinetochore-associated genes CASC5, KIF18A and SPAG5 are among these genes, it has been emphasized that this “may be relevant phenotypically as the orientation of the mitotic cleavage plane in neural precursor cells during cortex development is thought to influence the fate of the daughter cells and the number of neurons generated69,20”. Several fixed SNCs on the modern human lineage are observed for CASC5 (two changes) and SPAG5 (three changes), which is also among genes with a relatively high proportion of HF changes (Table S5). The changes in KIF18A, KIF16A and NEK6 can no longer be considered as fixed, but occur at very high frequencies (>99.9%) in present-day humans. We attempted to determine whether an enrichment of genes with HHMCs on the human lineage can be observed in the ventricular zone59, but instead find an enrichment in the intermediate zone, where less than 5% of random gene sets of the same size are expressed. However, synonymous HF changes also show an enrichment in this layer, as well as genes with AHMCs (Table S10), suggesting an overrepresentation of genes that carry mutations in the coding regions rather than lineage-specific effects. We were able to broadly recapitulate the observation of an enrichment of expression in the ventricular zone if restricting the test to genes with non-synonymous changes at a frequency greater than 99.9% in present-day humans, which is not observed for corresponding synonymous and archaic non-synonymous changes (Table S10). Among the 28 genes expressed in the ventricular zone that carry almost fixed HHMCs, four might be enriched for HF changes in humans (HERC5, LMNB2, SPAG5, VCAM1), and one shows an excess of HF changes on the human compared to the archaic lineage (AMKMY1). Other notable genes discussed in this study include ADSL, FAM178A, KIF26B, SLC38A10, and SPAG17.
The centrosome-cilium interface is known to be critical for early brain development, and centrosome-related proteins are overrepresented in studies on the microcephaly phenotype in humans70. We find 126 genes (Table S9) with 143 HHMCs that putatively interact with proteins at the centrosome-cilium interface71. Some of the genes listed here and discussed in this study, such as FMR1, KIF15, LMNB2, NCOA6, RB1CC1, SPAG5 and TEX2, harbor not only HHMCs, but an overall high proportion of HF changes on the human lineage. Although an early analysis suggested several candidate genes associated to microcephaly, not all of these could be confirmed by high-coverage data. Among eleven candidate genes32, only two (PCNT, UCP1) are among the HHMC gene list presented here, while most of the other changes are not human-specific, and only PCNT has been related to microcephaly72. Nevertheless, more changes related to microcephaly are found on both lineages, for example in ATRX73 or CASC574 (Supplementary Information 3).
Changes in genes associated with brain growth trajectory differences lead not necessarily to a decrease but also an increase of brain size75, suggesting that the disease phenotype of macrocephaly might point to genes relevant in the context of brain growth as well. One of the few genes with several HHMCs, CASC5, has been found to be associated with gray matter volume differences76. It has been claimed that mutations in PTEN alter the brain growth trajectory and allocation of cell types through elevated Beta-Catenin signaling77. This well-known gene, critical for brain development78, has not been highlighted in the context of human-specific changes, while we find that PTEN falls among the genes with an excess on the modern human over the archaic lineage, suggesting that regulatory changes in this gene might have contributed to human-specific traits. This is also the case for the HHMC-carrying transcription factor TCF3, which is known to repress Wnt-Beta-Catenin signaling and maintain the neural stem cell population during neocortical development79. Changes in these and other genes (Supplementary Information 3) like CCND280, GLI381, or RB1CC182, for which a regulatory SNC has been suggested to modify transcriptional activity83 and which carries a signature of positive selection20, could have contributed to the brain growth trajectory changes hypothesized to give rise to the modern human-specific globular braincase shape during the past several 100,000 years9,11,15. Finally, we find changes that might have affected the size of the cerebellum, a key contributor to our brain shape11,84, possibly even since the split from Neanderthals85: For example, HF regulatory SNCs in ZIC1 and ZIC486, an excess of HF mutations in AHI187, and a deleterious HHMC in ABHD14A, which is a target of ZIC188.
Cellular features of neurons
To form critical networks during the early development of the brain, axonal extensions of the neurons in the cortical region must be sent and guided to eventually reach their synaptic targets. Studies conducted on avian vocal learners89,90 have shown a convergent differential regulation of axon guidance genes of the SLIT-ROBO families in the pallial motor nucleus of the learning species, allowing for the formation of connections virtually absent in the brains of vocal non-learners. In modern humans, genes with axon-guidance-related functions such as FOXP2, SLIT2 and ROBO2 have been found to lie within deserts of archaic introgression39,40,91, suggesting incompatibilities between modern humans and archaics for these regions. Even though these particular genes do not carry protein-coding changes, but potentially relevant regulatory changes92, our dataset contains a fair amount of genes known to impact brain wiring: Some of the aforementioned microtubule-related genes, specifically those associated with axonal transport and known to play a role in post-mitotic neural wiring and plasticity93, are associated with signals of positive selection, such as KIF18A94 or KATNA195,96. Furthermore, an interactor of KIF18A, KIF1597, might have been under positive selection in modern humans33, and contains two HHMCs. Versican (VCAN), which promotes neurite outgrowth98, carries three HHMCs, and SSH2 (two HHMCs) might be involved in neurite outgrowth99. PIEZO1, which carries a non-synonymous change that is almost fixed in modern humans, is another factor in axon guidance100, as well as NOVA1101, which is an interactor of ELAVL4102, a gene that codes for a neuronal-specific RNA-binding protein and might have been under positive selection in humans33,36. Furthermore, we find one of the most deleterious regulatory SNCs in the Netrin receptor UNC5D, which is critical for axon guidance103.
We also detect changes in genes associated with myelination and synaptic vesicle endocytosis, critical to sustain a high rate of synaptic transmission, including DCX104, SCAP105, RB1CC1106, ADSL107 and PACSIN1108 among others (Supplementary Information 2). It is noteworthy that among traits associated with cognitive functions such as language or theory of mind, the timing of myelination appears to be a good predictor of computational abilities109,110. Computational processing might have been facilitated by some of the changes presented here, at least in some of the circuits that have expanded in our lineage111,112, since subtle maturational differences early in development113 may have had a considerable impact on the phenotype. In this context, it is worth mentioning that in our dataset, several genes carrying HHMCs and associated with basal ganglia functions (critical for language and cognition) stand out, like SLITRK1114 and NOVA136,115–118 (Supplementary Information 4). Finally, in the broader context of traits potentially related to cognition, we find an enrichment of HHMCs in genes associated to “Alzheimer’s disease (cognitive decline)” and “Cognitive decline (age-related)”, with seven associated genes (COX7B2, BCAS3, DMXL1, LIPC, PLEKHG1, TTLL2 and VIT). Among genes influencing behavioral traits (Supplementary Information 4) are GPR153119, NCOA6120, or the Adenylosuccinate Lyase (ADSL)121, for which the ancestral Neanderthal-like allele has not been observed in 1,000 s of modern human genomes and which has been pointed out before as under positive selection31,33,34,122. We know that archaic hominins likely had certain language-like abilities123,124, and hybrids of modern and archaic humans must have survived in their communities125, underlining the large overall similarity of these populations. However, genes associated with axon guidance functions, which are important for the refinement of neural circuits including those relevant for speech and language, are found in introgression deserts126,127. We suggest that modifications of a complex network in cognition or learning took place in modern human evolution128, possibly related to other brain-related9,16,129,130, vocal tract131 or neural changes132.
The craniofacial phenotype
In previous work on ancient genomes changes related to craniofacial morphology have been highlighted31,131, and we find an enrichment of genes with an excess of HF SNCs on the modern human lineage for soft palate development (Table S6). Among genes harboring an excess of HF SNCs associated with specific facial features, we find RUNX2, EDAR, and GLI3133, NFATC1134, SPOP135, DDR2136 and NELL1137, possibly carrying changes in regulatory regions, while mutations in the HHMC-carrying gene encoding for the transcription factor ATRX cause facial dysmorphism138. In addition, genes with HHMCs such as PLXNA2139, EVC2140, MEPE141, OMD142, and SPAG17143 are known to affect craniofacial bone and tooth morphologies. These genes appear to be important in determining bone density, mineralization and remodeling, hence they may underlie differences between archaic and modern human facial growth144. Some of these facial properties may have been present in the earliest fossils attributed to H. sapiens, like the Jebel Irhoud fossils4, deviating from craniofacial features which emerged in earlier forms of Homo145, and may have become established before some brain-related changes discussed here11,146. The gene encoding the transcription factor PRDM10 stands out for carrying HHMCs, being found in selective sweep regions and the second-most interacting protein within the HHMC dataset. Although little is known about PRDM10, it may be related to dendrite growth147 and neural crest related changes that contributed to the formation of our distinct modern face148. Other craniofacial morphology-related genes, such as DCHS2133, HIVEP2149, HIVEP3150, FREM1151, and FRAS1152 harbor AHMCs, while another bone-related gene, MEF2C153, shows an excess of HF changes on the archaic lineage. These changes may underlie some of the prominent derived facial traits of Neanderthals154,155.
Life history and other phenotypic traits
Apart from their consequences for cognitive functions, it has been suggested that changes involved in synaptic plasticity might be interpreted in a context of neoteny33,156–158, with the implication of delayed maturation in humans159 and a longer timeframe for brain development. However, given their similar brain sizes160, humans and Neanderthals might both have needed a long overall maturation time161,162. Accordingly, notions like neoteny and heterochrony are unlikely to be fine-grained enough to capture differences between these populations, but early differences in infant brain growth between humans and Neanderthals9,10 could have rendered our maturational profile distinct during limited developmental periods and within specific brain regions, imposing different metabolic requirements163. One of the brain regions where such differences are found is the orbitofrontal cortex (OFC)129, and we find that the OFC at infant age (0–2 years) is enriched for the expression of genes that carry HHMCs compared to synonymous SNCs. We suggest that the development of the OFC in infants might have been subject to subtle changes since the split from Neanderthals rather than a general developmental delay, which is particularly interesting given that this brain region has been implied in social cognition164 and learning165.
Genes carrying HHMCs are enriched for expression in the gray matter of the forebrain at the adolescent age compared to AHMC-carrying genes, hence additional human-specific modifications during this period might have taken place, possibly linked to changes in myelination described above. It has been suggested that differences in childhood adolescence time existed between humans and Neanderthals, after a general developmental delay in the hominin lineage166,167. Dental evidence suggests an earlier maturation in Neanderthals than modern humans168, and it has been claimed that Neanderthals might have reached adulthood earlier169. Furthermore, an introgressed indel from Neanderthals causes an earlier onset of menarche in present-day humans30, supporting at least the existence of alleles for earlier maturation in the Neanderthal population. Among the genes carrying fixed HHMCs, NCOA6 has also been linked to age at menarche and onset of puberty170, as well as placental function171. This putative transcription factor is enriched in HF changes and has been suggested to have been under positive selection on the modern human lineage33,122. The HHMC is located nearby and three 5′-UTR variants within a putatively selected region36, with an estimated time of selection at around 150 kya (assuming a slow mutation rate). Even though this gene carries an AHMC as well, it remains possible that modern humans acquired subtle differences in their reproductive system through lineage-specific changes in this gene. A delay in reproductive age may influence overall longevity, another trait for which our data set yields an enrichment of genes with HHMCs (SLC38A10, TBC1D22A and ZNF516).
The male reproductive system might have been subject to changes as well, since we find that several proteins in spermatogenesis seem to carry two HHMCs: Sperm Specific Antigen 2 (SSFA2), Sperm Associated Antigen 17 (SPAG17), ADAM18172 and WDR52173, out of which ADAM18 and SPAG17 also carry AHMCs. Lineage-specific differences in genes related to sperm function or spermatogenesis might have been relevant for the genetic compatibility between humans and Neanderthals. Another gene harboring a HHMC with similar functions is EPPIN174, which shows no HF changes on the archaic, but 27 such SNCs on the modern human lineage. The gene encoding for the Testis Expressed 2 protein (TEX2) is enriched for HF changes in both humans and archaics, with one HHMC and five AHMCs, but its function is not yet known. Another possible SNC that might be relevant in this context is a splice site change in IZUMO4, since proteins encoded by the IZUMO family form complexes on mammalian sperm175. The adjacent exon is not present in all transcripts of this gene, suggesting a functional role of this splice site SNC. Finally, genes in the GO category “spermatoproteasome complex” are enriched for an excess of HF changes on the human lineage.
It has been found that Neanderthal alleles contribute to addiction and, possibly, pain sensitivity in modern humans176,177. In this context, an interesting protein-truncating SNC at high frequency in humans is the loss of a stop codon in the opioid receptor OPRM1 (6:154360569), potentially changing the structure of the protein encoded by this gene in some transcripts. Other mutations in this gene are associated to heroin addiction178, and pain perception179, but also sociality traits180. Interestingly, a recent study found a pain insensitivity disorder caused by a mutation in ZFHX2181, which carries an AHMC, and three HHMCs are observed in NPAP1, which might be associated with the Prader-Willi syndrome, involving behavioral problems and a high pain threshold182. Such changes may point to differences in levels of resilience to pain between Neanderthals and modern humans.
Conclusion
The long-term evolutionary processes that led to the human condition1 are still subject to debate and investigation, and the high-quality genomes from archaic humans provide opportunities to explore the recent evolution of our species. We want to contribute to an attempt to unveil the genetic basis of specific molecular events in the time-window after the split from these archaic populations and before the emergence of most of the present-day diversity. It needs to be emphasized again that this does not imply a superiority of humans over other populations, but rather small differences after a long shared evolutionary history and their genetic underpinnings. We sought to combine different sources of information, from genome-wide enrichment analyses to functional information available for specific genes, to identify threads linking molecular needles in this expanded haystack. In doing so, we have mainly built on existing proposals concerning brain-related changes, but we have divided the observations into different biological levels, from cellular changes through brain organization differences to complex phenotypic traits. Only future experimental work will determine which of the changes highlighted here contributed significantly to making us “fully human”. We hope that our characterization and presentation of some new candidate genes will help prioritize inquiry in this area, since the specific type of validation depends on each candidate gene or network.
Methods
We used the publicly available high-coverage genotypes for three archaic individuals: One Denisovan19, one Neanderthal from the Denisova cave in Altai mountains20, and another Neanderthal from Vindija cave, Croatia21. The data is publicly available under http://cdna.eva.mpg.de/neandertal/Vindija/VCF/, with the human genome version hg19 as reference, covering ~1.8 billion base pairs of the genome21. We applied further filtering to remove sites with less than 5-fold coverage and more than 105-fold coverage in the Altai Neanderthal or 75-fold coverage in the other archaic individuals, if such cases occurred. We also removed sites with genotype quality smaller than 20, and heterozygous sites with strong allele imbalance (<0.2 minor allele frequency). Although these permissive filters increase power compared to previous studies, we caution that in some cases genotypes might be incorrect. We added the genotype and coverage for the exome and chromosome 21 sequences of the Vindija and El Sidrón Neanderthals from previous studies2,31, with 75-fold and 50-fold coverage cutoffs, respectively. These studies provided data for the same Vindija individual21.
We applied the Ensembl Variant Effect Predictor VEP183 in order to obtain inferences for protein-coding and regulatory mutations, scores for SIFT184, PolyPhen185, CADD186 and GWAVA187, and allele frequencies in the 1000 Genomes and ExAC human variation databases22,188. We used the inferred ancestral allele from published data on multiple genome alignments189, and at positions where this information was not available, the macaque reference allele, rheMac3190. We determined the allele frequencies in present-day humans using the dbSNP database build 147191. We retrieved the counts for each allele type, and summarized the counts of non-reference alleles at each position. Grantham scores192 were calculated for missense mutations.
Data processing and database retrieval was performed using bcftools/samtools v1.0193, bedtools v2.16.2194, and R/Bioconductor195, with rtracklayer196 and biomaRt197 packages, and plotting with Rcircos198. We analyzed all positions where at least two alleles (human reference and alternative allele) were observed among the human reference and at least one out of three of the high-coverage archaic individuals, in at least one archaic chromosome. The 22 autosomal chromosomes and the X chromosome were analyzed, in the absence of Y chromosome data for the three female archaic individuals. The data for 4,437,803 segregating sites is available under http://cbl.ub.edu/index.php/resources and on Figshare under 10.6084/m9.figshare.8184038. The following subsets were created:
Fixed differences: Positions where all present-day humans carry a derived allele, while at least two out of three archaics carry the ancestral allele, accounting for potential human gene flow into Neanderthals.
High-frequency (HF) differences: Positions where more than 90% of present-day humans carry a derived allele, while at least the Denisovan and one Neanderthal carry the ancestral allele, accounting for different types of errors and bi-directional gene flow.
Extended high-frequency differences: Positions where more than 90% of present-day humans carry a derived allele, while one of the following conditions is true: (a) Not all archaics have reliable genotypes, but those that have carry the ancestral allele. (b) Some archaics carry an alternative genotype that is not identical to either the human or the ancestral allele. (c) The Denisovan carries the ancestral allele, while one Neanderthal carries a derived allele, which allows for gene flow from humans into Neanderthals. (d) The ancestral allele is missing in the EPO alignment, but the macaque reference sequence is identical to the allele in all three archaics.
We also created corresponding lists of archaic-specific changes. Fixed changes were defined as sites where the three archaics carry the derived allele, while humans carry the ancestral allele at more than 99.999%. High-frequency changes occur at less than 1% in present-day humans, while at least two archaic individuals carry the derived allele. An extended list presents high-frequency changes where the ancestral allele is unknown, but the macaque allele is identical to the present-day human allele.
A ranking of mutation density was performed for genes with protein-coding sequences and their genomic regions as retrieved from Ensembl. For each gene, unique associated changes as predicted by VEP were counted. A ranking on the number of HF changes per gene length was performed for all genes that span at least 5,000 bp in the genome and carry at least 25 segregating sites in the dataset (at any frequency in humans or in archaics), in order to remove genes which are very short or poor in mutations. The top 5% of the empirical distribution was defined as putatively enriched for changes on each lineage. The ratio of lineage-specific HF changes was calculated for the subset of genes where at least 20 lineage-specific HF changes were observed on the human and the archaic lineages combined. The top 10% of the empirical distribution was defined as putatively enriched for lineage-specific changes.
We performed enrichment tests using the R packages ABAEnrichment58 and DescTools199. We used the NHGRI-EBI GWAS Catalog55, and overlapped the associated genes with protein-coding changes on the human and archaic lineages, respectively. We performed an enrichment test as described elsewhere200: We counted the number of HF missense changes on each lineage and the subset of those associated to each trait (“Disease trait”), and performed a significance test (G-test) against the number of genes associated to each trait, and all genes in the genome, with a P value cutoff at 0.1. This suggests a genome-wide enrichment of changes for each trait. We then performed a G-test between the numbers of HF missense changes on each lineage, and the subset of each associated to each trait (P-value cutoff at 0.1), to determine a difference between the two lineages. We then performed an empirical test by creating 1,000 random sets of genes with similar length as the genes associated to each trait, and counting the overlap with the lineage-specific missense changes. At least 90% of these 1,000 random sets were required to contain fewer missense changes than the real set of associated genes. Only traits were considered for which at least 10 associated loci were annotated.
Gene Ontology (GO) enrichment was performed using the software FUNC201, with a significance cutoff of the adjusted p-value < 0.05 and a family-wise error rate <0.05. When testing missense changes, a background set of synonymous changes on the same lineage was used for the hypergeometric test. When testing genes with relative mutation enrichment, the Wilcoxon rank test was applied. Enrichment for sequence-specific DNA-binding RNA polymerase II transcription factors and transcription factor candidate genes from57, and genes interacting at the centrosome-cilium interface71 was tested with an empirical test in which 1,000 random sets of genes were created that matched the length distributions of the genes in the test list. The same strategy was applied for genes expressed in the developing brain (Table S10)59. Protein-protein interactions were analyzed using the STRING online interface v10.5202 with standard settings (medium confidence, all sources, query proteins only) as of January 2018. The overlap with selective sweep screens considers HHMCs within 50,000 bp of the selected regions20,33,36.
Supplementary information
Acknowledgements
We thank S. Han and T. Marques-Bonet for helpful discussions, and A. G. Andirkó, Thomas O'Rourke and P.T. Martins for help with figures. M.K. was supported by a Deutsche Forschungsgemeinschaft (DFG) fellowship (KU 3467/1-1) and the Postdoctoral Junior Leader Fellowship Programme from “la Caixa” Banking Foundation (LCF/BQ/PR19/11700002). C.B. acknowledges research funds from the Spanish Ministry of Economy and Competitiveness/FEDER (grant FFI2016-78034-C2-1-P), Marie Curie International Reintegration Grant from the European Union (PIRG-GA-2009-256413), research funds from the Fundació Bosch i Gimpera, MEXT/JSPS Grant-in-Aid for Scientific Research on Innovative Areas 4903 (Evolinguistics: JP17H06379), and Generalitat de Catalunya (Government of Catalonia) – 2017-SGR-341.
Author Contributions
M.K. and C.B. analyzed data and wrote the manuscript.
Data Availability
Variants used were publicly available. Variant annotation data generated for this article can be found at 10.6084/m9.figshare.8184038. All other data analyzed are included in this published article and its Supplementary Information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44877-x.
References
- 1.Pääbo S. The Human Condition—A Molecular Approach. Cell. 2014;157:216–226. doi: 10.1016/j.cell.2013.12.036. [DOI] [PubMed] [Google Scholar]
- 2.Kuhlwilm M, et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 2016;530:429–433. doi: 10.1038/nature16544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlebusch CM, et al. Genomic Variation in Seven Khoe-San Groups Reveals Adaptation and Complex African History. Science (80-.). 2012;338:374 LP–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hublin J-J, et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature. 2017;546:289. doi: 10.1038/nature22336. [DOI] [PubMed] [Google Scholar]
- 5.Schlebusch, C. M. et al. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science (80-.) (2017). [DOI] [PubMed]
- 6.Skoglund P, et al. Reconstructing Prehistoric African Population Structure. Cell. 2017;171:59–71.e21. doi: 10.1016/j.cell.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabett RJ. The success of failed Homo sapiens dispersals out of Africa and into Asia. Nat. Ecol. Evol. 2018;2:212–219. doi: 10.1038/s41559-017-0436-8. [DOI] [PubMed] [Google Scholar]
- 8.Posth, C. et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. 8, 16046 (2017). [DOI] [PMC free article] [PubMed]
- 9.Hublin, J.-J., Neubauer, S. & Gunz, P. Brain ontogeny and life history in Pleistocene hominins. Philos. Trans. R. Soc. B Biol. Sci. 370 (2015). [DOI] [PMC free article] [PubMed]
- 10.Gunz P, Neubauer S, Maureille B, Hublin JJ. Brain development after birth differs between Neandertals and modern humans. Curr. Biol. 2010;20:921–922. doi: 10.1016/j.cub.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Neubauer, S., Hublin, J.-J. & Gunz, P. The evolution of modern human brain shape. Sci. Adv. 4 (2018). [DOI] [PMC free article] [PubMed]
- 12.Gunz P, et al. Neandertal Introgression Sheds Light on Modern Human Endocranial Globularity. Curr. Biol. 2019;29:120–127.e5. doi: 10.1016/j.cub.2018.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trinkaus E, Howells WW. The Neanderthals. Sci. Am. 1979;241:118–133. doi: 10.1038/scientificamerican1279-118. [DOI] [Google Scholar]
- 14.Schoenemann PT. Brain Size Scaling and Body Composition in Mammals. Brain. Behav. Evol. 2004;63:47–60. doi: 10.1159/000073759. [DOI] [PubMed] [Google Scholar]
- 15.Gunz P, et al. A uniquely modern human pattern of endocranial development. Insights from a new cranial reconstruction of the Neandertal newborn from Mezmaiskaya. J. Hum. Evol. 2012;62:300–313. doi: 10.1016/j.jhevol.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Boeckx C. The language-ready head: Evolutionary considerations. Psychon. Bull. Rev. 2017;24:194–199. doi: 10.3758/s13423-016-1087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann, D. L., Angelucci, D. E., Villaverde, V., Zapata, J. & Zilhão, J. Symbolic use of marine shells and mineral pigments by Iberian Neandertals 115,000 years ago. Sci. Adv. 4 (2018). [DOI] [PMC free article] [PubMed]
- 18.Wynn T, Overmann K, Coolidge F. The false dichotomy: a refutation of the Neandertal indistinguishability claim. J. Anthropol. Sci.=Riv. di Antropol. JASS. 2016;94:201–221. doi: 10.4436/JASS.94022. [DOI] [PubMed] [Google Scholar]
- 19.Meyer M, et al. a High Coverage Genome Sequence From an Archaic Denisovan Individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–9. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prüfer, K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science (80-.) (2017). [DOI] [PMC free article] [PubMed]
- 22.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium TCS. and A. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 24.Ju X-C, et al. The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. Elife. 2016;5:e18197. doi: 10.7554/eLife.18197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–70. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 26.Dumas LJ, et al. DUF1220-Domain Copy Number Implicated in Human Brain-Size Pathology and Evolution. Am. J. Hum. Genet. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis MY, et al. Evolution of Human-Specific Neural SRGAP2 Genes by Incomplete Segmental Duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki IK, et al. Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell. 2018;173:1370–1384.e16. doi: 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chintalapati M, Dannemann M, Prüfer K. Using the Neandertal genome to study the evolution of small insertions and deletions in modern humans. BMC Evol. Biol. 2017;17:179. doi: 10.1186/s12862-017-1018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castellano S, et al. Patterns of coding variation in the complete exomes of three Neandertals. Proc. Natl. Acad. Sci. USA. 2014;111:6666–71. doi: 10.1073/pnas.1405138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green RE, et al. A Draft Sequence of the Neandertal Genome. Science (80-.). 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peyrégne S, Boyle MJ, Dannemann M, Prüfer K. Detecting ancient positive selection in humans using extended lineage sorting. Genome Res. 2017;27:1563–1572. doi: 10.1101/gr.219493.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Racimo F. Testing for Ancient Selection Using Cross-population Allele Frequency Differentiation. Genetics. 2016;202:733 LP–750. doi: 10.1534/genetics.115.178095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somel M, Liu X, Khaitovich P. Human brain evolution: transcripts, metabolites and their regulators. Nat. Rev. Neurosci. 2013;14:1–16. doi: 10.1038/nrn3372. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, H. et al. A Chronological Atlas of Natural Selection in the Human Genome during the Past Half-million Years. bioRxiv (2015).
- 37.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vernot B, Akey JM. Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science (80-.). 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 39.Vernot B, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science (80-.). 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sankararaman S, Mallick S, Patterson N, Reich D. The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans. Curr. Biol. 2016;26:1241–1247. doi: 10.1016/j.cub.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy RC, Wakefield J, Akey JM. Impacts of Neanderthal-Introgressed Sequences on the Landscape of Human Gene Expression. Cell. 2017;168:916–927.e12. doi: 10.1016/j.cell.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynn T, Coolidge FL. The expert Neandertal mind. J. Hum. Evol. 2004;46:467–487. doi: 10.1016/j.jhevol.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Langergraber KE, et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. …. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varki A, Altheide TK. Comparing the human and chimpanzee genomes: Searching for needles in a haystack. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 45.O’Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat. Rev. Genet. 2012;13:853–866. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bufill E, Blesa R, Augusti J. Alzheimer’s disease: an evolutionary approach. J. Anthropol. Sci.=Riv. di Antropol. JASS. 2013;91:135–157. doi: 10.4436/jass.91001. [DOI] [PubMed] [Google Scholar]
- 47.Bufill E, Agustí J, Blesa R. Human neoteny revisited: The case of synaptic plasticity. Am. J. Hum. Biol. 2011;23:729–739. doi: 10.1002/ajhb.21225. [DOI] [PubMed] [Google Scholar]
- 48.Bruner E, Jacobs HIL. Alzheimer’s disease: the downside of a highly evolved parietal lobe? J. Alzheimers. Dis. 2013;35:227–240. doi: 10.3233/JAD-122299. [DOI] [PubMed] [Google Scholar]
- 49.Gratten J, Visscher PM. Genetic pleiotropy in complex traits and diseases: implications for genomic medicine. Genome Med. 2016;8:78. doi: 10.1186/s13073-016-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giandomenico SL, Lancaster MA. Probing human brain evolution and development in organoids. Curr. Opin. Cell Biol. 2017;44:36–43. doi: 10.1016/j.ceb.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Camp JG, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA. 2015;112:15672–7. doi: 10.1073/pnas.1508055112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camp JG, Treutlein B. Human organomics: a fresh approach to understanding human development using single-cell transcriptomics. Development. 2017;144:1584 LP–1587. doi: 10.1242/dev.150458. [DOI] [PubMed] [Google Scholar]
- 53.Pollen AA, et al. Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell. 2019;176:743–756.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enard W, et al. A Humanized Version of Foxp2 Affects Cortico-Basal Ganglia Circuits in Mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 55.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 57.Chawla K, Tripathi S, Thommesen L, Lægreid A, Kuiper M. TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics. 2013;29:2519–2520. doi: 10.1093/bioinformatics/btt432. [DOI] [PubMed] [Google Scholar]
- 58.Grote S, Prüfer K, Kelso J, Dannemann M. ABAEnrichment: an R package to test for gene set expression enrichment in the adult and developing human brain. Bioinformatics. 2016;32:3201–3203. doi: 10.1093/bioinformatics/btw392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller JA, et al. Transcriptional Landscape of the Prenatal Human Brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Boer, B., Thompson, B., Ravignani, A. & Boeckx, C. Evolutionary Dynamics Do Not Motivate a Single-Mutant Theory of Human Language. bioRxiv517029, 10.1101/517029 (2019). [DOI] [PMC free article] [PubMed]
- 61.Omer Javed A, et al. Microcephaly Modeling of Kinetochore Mutation Reveals a Brain-Specific Phenotype. Cell Rep. 2018;25:368–382.e5. doi: 10.1016/j.celrep.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Regan L, Fry AM. The Nek6 and Nek7 Protein Kinases Are Required for Robust Mitotic Spindle Formation and Cytokinesis. Mol. Cell. Biol. 2009;29:3975–3990. doi: 10.1128/MCB.01867-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torres JZ, et al. The STARD9/Kif16a Kinesin Associates with Mitotic Microtubules and Regulates Spindle Pole Assembly. Cell. 2017;147:1309–1323. doi: 10.1016/j.cell.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barr AR, Kilmartin JV, Gergely F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J. Cell Biol. 2010;189:23 LP–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puga A, Xia Y, Elferink C. Role of the aryl hydrocarbon receptor in cell cycle regulation. Chem. Biol. Interact. 2002;141:117–130. doi: 10.1016/S0009-2797(02)00069-8. [DOI] [PubMed] [Google Scholar]
- 66.Bader JR, et al. Polo-like Kinase1 Is Required for Recruitment of Dynein to Kinetochores during Mitosis. J. Biol. Chem. 2011;286:20769–20777. doi: 10.1074/jbc.M111.226605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zachos G, et al. Chk1 Is Required for Spindle Checkpoint Function. Dev. Cell. 2017;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida MM, Azuma Y. Mechanisms behind Topoisomerase II SUMOylation in chromosome segregation. Cell Cycle. 2016;15:3151–3152. doi: 10.1080/15384101.2016.1216928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fietz SA, Huttner WB. Cortical progenitor expansion, self-renewal and neurogenesis—a polarized perspective. Curr. Opin. Neurobiol. 2011;21:23–35. doi: 10.1016/j.conb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 70.Megraw TL, Sharkey JT, Nowakowski RS. Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol. 2011;21:470–480. doi: 10.1016/j.tcb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta GD, et al. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li F-F, et al. Identification of two novel critical mutations in PCNT gene resulting in microcephalic osteodysplastic primordial dwarfism type II associated with multiple intracranial aneurysms. Metab. Brain Dis. 2015;30:1387–1394. doi: 10.1007/s11011-015-9712-y. [DOI] [PubMed] [Google Scholar]
- 73.Ritchie K, Watson LA, Davidson B, Jiang Y, Bérubé NG. ATRX is required for maintenance of the neuroprogenitor cell pool in the embryonic mouse brain. Biol. Open. 2014;3:1158 LP–1163. doi: 10.1242/bio.20148730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Genin A, et al. Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Hum. Mol. Genet. 2012;21:5306–5317. doi: 10.1093/hmg/dds386. [DOI] [PubMed] [Google Scholar]
- 75.Montgomery SH, Capellini I, Venditti C, Barton RA, Mundy NI. Adaptive Evolution of Four Microcephaly Genes and the Evolution of Brain Size in Anthropoid Primates. Mol. Biol. Evol. 2011;28:625–638. doi: 10.1093/molbev/msq237. [DOI] [PubMed] [Google Scholar]
- 76.Shi L, et al. Regional selection of the brain size regulating gene CASC5 provides new insight into human brain evolution. Hum. Genet. 2017;136:193–204. doi: 10.1007/s00439-016-1748-5. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y, Huang W-C, Séjourné J, Clipperton-Allen AE, Page DT. Pten Mutations Alter Brain Growth Trajectory and Allocation of Cell Types through Elevated β-Catenin Signaling. J. Neurosci. 2015;35:10252 LP–10267. doi: 10.1523/JNEUROSCI.5272-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, et al. Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell. 2017;20:385–396.e3. doi: 10.1016/j.stem.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuwahara A, et al. Tcf3 Represses Wnt–β-Catenin Signaling and Maintains Neural Stem Cell Population during Neocortical Development. PLoS One. 2014;9:e94408. doi: 10.1371/journal.pone.0094408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mirzaa GM, et al. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat. Genet. 2014;46:510. doi: 10.1038/ng.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jamsheer A, et al. Expanded mutational spectrum of the GLI3 gene substantiates genotype–phenotype correlations. J. Appl. Genet. 2012;53:415–422. doi: 10.1007/s13353-012-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang C, Liang C-C, Bian ZC, Zhu Y, Guan J-L. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat. Neurosci. 2013;16:532. doi: 10.1038/nn.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weyer S, Pääbo S. Functional Analyses of Transcription Factor Binding Sites that Differ between Present-Day and Archaic Humans. Mol. Biol. Evol. 2016;33:316–322. doi: 10.1093/molbev/msv215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kochiyama T, et al. Reconstructing the Neanderthal brain using computational anatomy. Sci. Rep. 2018;8:6296. doi: 10.1038/s41598-018-24331-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller IF, Barton RA, Nunn CL. Quantitative uniqueness of human brain evolution revealed through phylogenetic comparative analysis. Elife. 2019;8:e41250. doi: 10.7554/eLife.41250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blank MC, et al. Multiple developmental programs are altered by loss of Zic1 and Zic4 to cause Dandy-Walker malformation cerebellar pathogenesis. Development. 2011;138:1207 LP–1216. doi: 10.1242/dev.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng Y-Z, et al. Investigating Embryonic Expression Patterns and Evolution of AHI1 and CEP290 Genes, Implicated in Joubert Syndrome. PLoS One. 2012;7:e44975. doi: 10.1371/journal.pone.0044975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoshino J, Aruga J, Ishiguro A, Mikoshiba K. Dorz1, a novel gene expressed in differentiating cerebellar granule neurons, is down-regulated in Zic1-deficient mouse. Mol. Brain Res. 2003;120:57–64. doi: 10.1016/j.molbrainres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 89.Pfenning, A. R. et al. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science (80-.). 346 (2014). [DOI] [PMC free article] [PubMed]
- 90.Wang R, et al. Convergent Differential Regulation of SLIT-ROBO Axon Guidance Genes in the Brains of Vocal Learners. J. Comp. Neurol. 2015;523:892–906. doi: 10.1002/cne.23719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuhlwilm Martin. The evolution of FOXP2 in the light of admixture. Current Opinion in Behavioral Sciences. 2018;21:120–126. doi: 10.1016/j.cobeha.2018.04.006. [DOI] [Google Scholar]
- 92.Fisher SE. Human Genetics: The Evolving Story of FOXP2. Curr. Biol. 2019;29:R65–R67. doi: 10.1016/j.cub.2018.11.047. [DOI] [PubMed] [Google Scholar]
- 93.Lüders Jens., editor. The Microtubule Cytoskeleton. Vienna: Springer Vienna; 2016. [Google Scholar]
- 94.McVicker DP, et al. Transport of a kinesin-cargo pair along microtubules into dendritic spines undergoing synaptic plasticity. Nat. Commun. 2016;7:12741. doi: 10.1038/ncomms12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ahmad FJ, Yu W, McNally FJ, Baas PW. An Essential Role for Katanin in Severing Microtubules in the Neuron. J. Cell Biol. 1999;145:305 LP–315. doi: 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karabay A, Yu W, Solowska JM, Baird DH, Baas PW. Axonal Growth Is Sensitive to the Levels of Katanin, a Protein That Severs Microtubules. J. Neurosci. 2004;24:5778 LP–5788. doi: 10.1523/JNEUROSCI.1382-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kevenaar JT, et al. Kinesin-Binding Protein Controls Microtubule Dynamics and Cargo Trafficking by Regulating Kinesin Motor Activity. Curr. Biol. 2017;26:849–861. doi: 10.1016/j.cub.2016.01.048. [DOI] [PubMed] [Google Scholar]
- 98.Wu Y, et al. Versican V1 Isoform Induces Neuronal Differentiation and Promotes Neurite Outgrowth. Mol. Biol. Cell. 2004;15:2093–2104. doi: 10.1091/mbc.e03-09-0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cuberos H, et al. Roles of LIM kinases in central nervous system function and dysfunction. FEBS Lett. 2015;589:3795–3806. doi: 10.1016/j.febslet.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 100.Koser DE, et al. Mechanosensing is critical for axon growth in the developing brain. Nat. Neurosci. 2016;19:1592. doi: 10.1038/nn.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jensen KB, et al. Nova-1 Regulates Neuron-Specific Alternative Splicing and Is Essential for Neuronal Viability. Neuron. 2000;25:359–371. doi: 10.1016/S0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 102.Ratti Antonia, Fallini Claudia, Colombrita Claudia, Pascale Alessia, Laforenza Umberto, Quattrone Alessandro, Silani Vincenzo. Post-transcriptional Regulation of Neuro-oncological Ventral Antigen 1 by the Neuronal RNA-binding Proteins ELAV. Journal of Biological Chemistry. 2008;283(12):7531–7541. doi: 10.1074/jbc.M706082200. [DOI] [PubMed] [Google Scholar]
- 103.Takemoto M, et al. Laminar and Areal Expression of Unc5d and Its Role in Cortical Cell Survival. Cereb. Cortex. 2011;21:1925–1934. doi: 10.1093/cercor/bhq265. [DOI] [PubMed] [Google Scholar]
- 104.Yap CC, et al. Doublecortin (DCX) Mediates Endocytosis of Neurofascin Independently of Microtubule Binding. J. Neurosci. 2012;32:7439 LP–7453. doi: 10.1523/JNEUROSCI.5318-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Verheijen MHG, et al. SCAP is required for timely and proper myelin membrane synthesis. Proc. Natl. Acad. Sci. 2009;106:21383–21388. doi: 10.1073/pnas.0905633106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015;16:345. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 107.Jurecka A, Jurkiewicz E, Tylki-Szymanska A. Magnetic resonance imaging of the brain in adenylosuccinate lyase deficiency: a report of seven cases and a review of the literature. Eur. J. Pediatr. 2012;171:131–138. doi: 10.1007/s00431-011-1503-9. [DOI] [PubMed] [Google Scholar]
- 108.Widagdo J, Fang H, Jang SE, Anggono V. PACSIN1 regulates the dynamics of AMPA receptor trafficking. Sci. Rep. 2016;6:31070. doi: 10.1038/srep31070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Skeide MA, Friederici AD. The ontogeny of the cortical language network. Nat. Rev. Neurosci. 2016;17:323. doi: 10.1038/nrn.2016.23. [DOI] [PubMed] [Google Scholar]
- 110.Grosse Wiesmann C, Schreiber J, Singer T, Steinbeis N, Friederici AD. White matter maturation is associated with the emergence of Theory of Mind in early childhood. Nat. Commun. 2017;8:14692. doi: 10.1038/ncomms14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mars RB, Eichert N, Jbabdi S, Verhagen L, Rushworth MFS. Connectivity and the search for specializations in the language-capable brain. Curr. Opin. Behav. Sci. 2018;21:19–26. doi: 10.1016/j.cobeha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ardesch DJ, et al. Evolutionary expansion of connectivity between multimodal association areas in the human brain compared with chimpanzees. Proc. Natl. Acad. Sci. 2019;116:7101 LP–7106. doi: 10.1073/pnas.1818512116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dubois J, et al. Exploring the Early Organization and Maturation of Linguistic Pathways in the Human Infant Brain. Cereb. Cortex. 2016;26:2283–2298. doi: 10.1093/cercor/bhv082. [DOI] [PubMed] [Google Scholar]
- 114.Abelson JF, et al. Sequence Variants in SLITRK1 are Associated with Tourette’s Syndrome. Science (80-.). 2005;310:317 LP–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 115.Jelen N, Ule J, Živin M. Cholinergic regulation of striatal Nova mRNAs. Neuroscience. 2010;169:619–627. doi: 10.1016/j.neuroscience.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 116.Alsiö JM, Tarchini B, Cayouette M, Livesey FJ. Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc. Natl. Acad. Sci. 2013;110:E716–E725. doi: 10.1073/pnas.1215707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Popovitchenko T, et al. The RNA binding protein HuR determines the differential translation of autism-associated FoxP subfamily members in the developing neocortex. Sci. Rep. 2016;6:28998. doi: 10.1038/srep28998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Konopka G, et al. Human-Specific Transcriptional Networks in the Brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sreedharan S, et al. The G protein coupled receptor Gpr153 shares common evolutionary origin with Gpr162 and is highly expressed in central regions including the thalamus, cerebellum and the arcuate nucleus. FEBS J. 2011;278:4881–4894. doi: 10.1111/j.1742-4658.2011.08388.x. [DOI] [PubMed] [Google Scholar]
- 120.Takata A, et al. Integrative Analyses of De Novo Mutations Provide Deeper Biological Insights into Autism Spectrum Disorder. Cell Rep. 2018;22:734–747. doi: 10.1016/j.celrep.2017.12.074. [DOI] [PubMed] [Google Scholar]
- 121.Fon EA, et al. Adenylosuccinate lyase (ADSL) and infantile autism: Absence of previously reported point mutation. Am. J. Med. Genet. 1995;60:554–557. doi: 10.1002/ajmg.1320600614. [DOI] [PubMed] [Google Scholar]
- 122.Racimo F, Kuhlwilm M, Slatkin M. A test for ancient selective sweeps and an application to candidate sites in modern humans. Mol. Biol. Evol. 2014;31:3344–3358. doi: 10.1093/molbev/msu255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dediu D, Levinson SC. On the antiquity of language: The reinterpretation of neandertal linguistic capacities and its consequences. Front. Psychol. 2013;4:1–17. doi: 10.3389/fpsyg.2013.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dediu D, Levinson SC. Neanderthal language revisited: not only us. Curr. Opin. Behav. Sci. 2018;21:49–55. doi: 10.1016/j.cobeha.2018.01.001. [DOI] [Google Scholar]
- 125.Fu Q, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lei H, et al. Axon guidance pathways served as common targets for human speech/language evolution and related disorders. Brain Lang. 2017;174:1–8. doi: 10.1016/j.bandl.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 127.Jeong J-W, Sundaram S, Behen ME, Chugani HT. Relationship between genotype and arcuate fasciculus morphology in six young children with global developmental delay: Preliminary DTI stuy. J. Magn. Reson. Imaging. 2016;44:1504–1512. doi: 10.1002/jmri.25306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Boeckx C, Benítez-Burraco A. Globularity and language-readiness: generating new predictions by expanding the set of genes of interest. Frontiers in Psychology. 2014;5:1324. doi: 10.3389/fpsyg.2014.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bastir M, et al. Evolution of the base of the brain in highly encephalized human species. Nat. Commun. 2011;2:588. doi: 10.1038/ncomms1593. [DOI] [PubMed] [Google Scholar]
- 130.Bryant Katherine L., Preuss Todd M. Digital Endocasts. Tokyo: Springer Japan; 2017. A Comparative Perspective on the Human Temporal Lobe; pp. 239–258. [Google Scholar]
- 131.Gokhman, D. et al. Extensive Regulatory Changes in Genes Affecting Vocal and Facial Anatomy Separate Modern from Archaic Humans. bioRxiv (2017).
- 132.Belyk M, Brown S. The origins of the vocal brain in humans. Neurosci. Biobehav. Rev. 2017;77:177–193. doi: 10.1016/j.neubiorev.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 133.Adhikari K, et al. A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat. Commun. 2016;7:11616. doi: 10.1038/ncomms11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim JH, Kim N. Regulation of NFATc1 in Osteoclast Differentiation. J Bone Metab. 2014;21:233–241. doi: 10.11005/jbm.2014.21.4.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cai H, Liu A. Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc. Natl. Acad. Sci. 2016;113:14751–14756. doi: 10.1073/pnas.1612520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang Y, et al. An essential role of discoidin domain receptor 2 (DDR2) in osteoblast differentiation and chondrocyte maturation via modulation of Runx2 activation. J. Bone Miner. Res. 2011;26:604–617. doi: 10.1002/jbmr.225. [DOI] [PubMed] [Google Scholar]
- 137.Zhang Xinli, Ting Kang, Pathmanathan Dharmini, Ko Theodore, Chen Weiwei, Chen Feng, Lee Haofu, James Aaron W., Siu Ronald K., Shen Jia, Culiat Cymbeline T., Soo Chia. Calvarial Cleidocraniodysplasia-Like Defects With ENU-Induced Nell-1 Deficiency. Journal of Craniofacial Surgery. 2012;23(1):61–66. doi: 10.1097/SCS.0b013e318240c8c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moncini S, et al. ATRX mutation in two adult brothers with non-specific moderate intellectual disability identified by exome sequencing. Meta Gene. 2013;1:102–108. doi: 10.1016/j.mgene.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oh J-E, et al. PlexinA2 mediates osteoblast differentiation via regulation of Runx2. J. Bone Miner. Res. 2012;27:552–562. doi: 10.1002/jbmr.1471. [DOI] [PubMed] [Google Scholar]
- 140.Kwon EK, et al. The Role of Ellis-Van Creveld 2(EVC2) in Mice During Cranial Bone Development. Anat. Rec. 2018;301:46–55. doi: 10.1002/ar.23692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gullard A, et al. MEPE Localization in the Craniofacial Complex and Function in Tooth Dentin Formation. J. Histochem. Cytochem. 2016;64:224–236. doi: 10.1369/0022155416635569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tashima T, Nagatoishi S, Sagara H, Ohnuma S, Tsumoto K. Osteomodulin regulates diameter and alters shape of collagen fibrils. Biochem. Biophys. Res. Commun. 2015;463:292–296. doi: 10.1016/j.bbrc.2015.05.053. [DOI] [PubMed] [Google Scholar]
- 143.Teves ME, et al. Spag17 Deficiency Results in Skeletal Malformations and Bone Abnormalities. PLoS One. 2015;10:e0125936. doi: 10.1371/journal.pone.0125936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lacruz RS, et al. Ontogeny of the maxilla in Neanderthals and their ancestors. Nat. Commun. 2015;6:8996. doi: 10.1038/ncomms9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lacruz RS, et al. Facial Morphogenesis of the Earliest Europeans. PLoS One. 2013;8:e65199. doi: 10.1371/journal.pone.0065199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stringer, C. The origin and evolution of Homo sapiens. Philos. Trans. R. Soc. B Biol. Sci. 371 (2016). [DOI] [PMC free article] [PubMed]
- 147.Siegel DA, Huang MK, Becker SF. Ectopic dendrite initiation: CNS pathogenesis as a model of CNS development. Int. J. Dev. Neurosci. 2002;20:373–389. doi: 10.1016/S0736-5748(02)00055-2. [DOI] [PubMed] [Google Scholar]
- 148.Park J-A, Kim KC. Expression patterns of PRDM10 during mouse embryonic development. BMB Rep. 2010;43:29–33. doi: 10.5483/BMBRep.2010.43.1.029. [DOI] [PubMed] [Google Scholar]
- 149.Jones DC, et al. Uncoupling of growth plate maturation and bone formation in mice lacking both Schnurri-2 and Schnurri-3. Proc. Natl. Acad. Sci. 2010;107:8254–8258. doi: 10.1073/pnas.1003727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Imamura K, et al. Human Immunodeficiency Virus Type 1 Enhancer-binding Protein 3 Is Essential for the Expression of Asparagine-linked Glycosylation 2 in the Regulation of Osteoblast and Chondrocyte Differentiation. J. Biol. Chem. 2014;289:9865–9879. doi: 10.1074/jbc.M113.520585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee MK, et al. Genome-wide association study of facial morphology reveals novel associations with FREM1 and PARK2. PLoS One. 2017;12:e0176566. doi: 10.1371/journal.pone.0176566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Talbot JC, et al. Pharyngeal morphogenesis requires fras1-itga8-dependent epithelial-mesenchymal interaction. Dev. Biol. 2016;416:136–148. doi: 10.1016/j.ydbio.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Verzi MP, et al. The Transcription Factor MEF2C Is Required for Craniofacial Development. Dev. Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rak Y. The Neanderthal: A new look at an old face. J. Hum. Evol. 1986;15:151–164. doi: 10.1016/S0047-2484(86)80042-2. [DOI] [Google Scholar]
- 155.Stelzer S, Neubauer S, Hublin J-J, Spoor F, Gunz P. Morphological trends in arcade shape and size in Middle Pleistocene Homo. Am. J. Phys. Anthropol. 2019;168:70–91. doi: 10.1002/ajpa.23721. [DOI] [PubMed] [Google Scholar]
- 156.Somel M, et al. Transcriptional neoteny in the human brain. Proc. Natl. Acad. Sci. USA. 2009;106:5743–8. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu X, et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012;22:611–622. doi: 10.1101/gr.127324.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sherwood CC, Gómez-Robles A. Brain Plasticity and Human Evolution. Annu. Rev. Anthropol. 2017;46:399–419. doi: 10.1146/annurev-anthro-102215-100009. [DOI] [Google Scholar]
- 159.Bednarik Robert. From Human Past to Human Future. Humanities. 2013;2(1):20–55. doi: 10.3390/h2010020. [DOI] [Google Scholar]
- 160.Hofman Michel A. Encephalization in Hominids: Evidence for the Model of Punctuationalism. Brain, Behavior and Evolution. 1983;22(2-3):102–117. doi: 10.1159/000121511. [DOI] [PubMed] [Google Scholar]
- 161.Ponce de León MS, Bienvenu T, Akazawa T, Zollikofer CPE. Brain development is similar in Neanderthals and modern humans. Curr. Biol. 2017;26:R665–R666. doi: 10.1016/j.cub.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 162.Rosas A, et al. The growth pattern of Neandertals, reconstructed from a juvenile skeleton from El Sidrón (Spain) Science (80-.). 2017;357:1282 LP–1287. doi: 10.1126/science.aan6463. [DOI] [PubMed] [Google Scholar]
- 163.Bruner E, de la Cuétara JM, Masters M, Amano H, Ogihara N. Functional craniology and brain evolution: from paleontology to biomedicine. Front. Neuroanat. 2014;8:19. doi: 10.3389/fnana.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Beer JS, John OP, Scabini D, Knight RT. Orbitofrontal Cortex and Social Behavior: Integrating Self-monitoring and Emotion-Cognition Interactions. J. Cogn. Neurosci. 2006;18:871–879. doi: 10.1162/jocn.2006.18.6.871. [DOI] [PubMed] [Google Scholar]
- 165.Miller, K. J., Botvinick, M. M. & Brody, C. D. Value Representations in Orbitofrontal Cortex Drive Learning, but not Choice. bioRxiv (2018). [DOI] [PMC free article] [PubMed]
- 166.Smith BH, Tompkins RL. Toward A Life History of the Hominidae. Annu. Rev. Anthropol. 1995;24:257–279. doi: 10.1146/annurev.an.24.100195.001353. [DOI] [Google Scholar]
- 167.Bock J, Sellen DW. Childhood and the evolution of the human life course. Hum. Nat. 2002;13:153–159. doi: 10.1007/s12110-002-1006-5. [DOI] [PubMed] [Google Scholar]
- 168.Smith TM, et al. Dental evidence for ontogenetic differences between modern humans and Neanderthals. Proc Natl Acad Sci USA. 2010;107:20923–20928. doi: 10.1073/pnas.1010906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ramirez Rozzi, F. V. & de Castro, J. M. surprisingly rapid growth in Neanderthals. Nature428 (2004). [DOI] [PubMed]
- 170.Day FR, et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat. Genet. 2017;49:834–841. doi: 10.1038/ng.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Antonson P, et al. Inactivation of the Nuclear Receptor Coactivator RAP250 in Mice Results in Placental Vascular Dysfunction. Mol. Cell. Biol. 2003;23:1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhu G-Z, Lin Y, Myles DG, Primakoff P. Identification of four novel ADAMs with potential roles in spermatogenesis and fertilization. Gene. 1999;234:227–237. doi: 10.1016/S0378-1119(99)00208-5. [DOI] [PubMed] [Google Scholar]
- 173.Tang S, et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang Z, Widgren EE, Sivashanmugam P, O’Rand MG, Richardson RT. Association of Eppin with Semenogelin on Human Spermatozoa1. Biol. Reprod. 2005;72:1064–1070. doi: 10.1095/biolreprod.104.036483. [DOI] [PubMed] [Google Scholar]
- 175.Ellerman DA, et al. Izumo is part of a multiprotein family whose members form large complexes on mammalian sperm. Mol. Reprod. Dev. 2009;76:1188–1199. doi: 10.1002/mrd.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Simonti CN, et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science (80-.). 2016;351:737 LP–741. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dannemann M, Prüfer K, Kelso J. Functional implications of Neandertal introgression in modern humans. Genome Biol. 2017;18:61. doi: 10.1186/s13059-017-1181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shi J, et al. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum. Mutat. 2002;19:459–460. doi: 10.1002/humu.9026. [DOI] [PubMed] [Google Scholar]
- 179.Tan E, et al. Ethnicity and OPRM variant independently predict pain perception and patient-controlled analgesia usage for post-operative pain. Mol. Pain. 2009;5:32. doi: 10.1186/1744-8069-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pearce E, Wlodarski R, Machin A, Dunbar RIM. Variation in the β-endorphin, oxytocin, and dopamine receptor genes is associated with different dimensions of human sociality. Proc. Natl. Acad. Sci. 2017;114:5300 LP–5305. doi: 10.1073/pnas.1700712114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Habib, A. M. et al. A novel human pain insensitivity disorder caused by a point mutation in ZFHX2. Brain awx326-awx326 (2017). [DOI] [PMC free article] [PubMed]
- 182.Buiting K, et al. C15orf2 and a novel noncoding transcript from the Prader–Willi/Angelman syndrome region show monoallelic expression in fetal brain. Genomics. 2007;89:588–595. doi: 10.1016/j.ygeno.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 183.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 185.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ritchie GRS, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat. Methods. 2014;11:294–6. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Paten B, et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18:1829–1843. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yan G, et al. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat. Biotechnol. 2011;29:1019. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 191.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Grantham R. Amino Acid Difference Formula to Help Explain Protein Evolution. Science. 1974;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 193.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Meth. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 198.Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Signorell, A. DescTools: Tools for Descriptive Statistics. (2017).
- 200.Han Sojung, Andrés Aida M, Marques-Bonet Tomas, Kuhlwilm Martin. Genetic Variation in Pan Species Is Shaped by Demographic History and Harbors Lineage-Specific Functions. Genome Biology and Evolution. 2019;11(4):1178–1191. doi: 10.1093/gbe/evz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Prüfer K, et al. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Variants used were publicly available. Variant annotation data generated for this article can be found at 10.6084/m9.figshare.8184038. All other data analyzed are included in this published article and its Supplementary Information files.