The bacterial flagellum has evolved as one of the most sophisticated self-assembled molecular machines, which confers locomotion and is often associated with virulence of bacterial pathogens. Variation in species-specific features of the flagellum, as well as in flagellar number and placement, results in structurally distinct flagella that appear to be adapted to the specific environments that bacteria encounter. Here, we used cutting-edge imaging techniques to determine high-resolution in situ structures of polar flagella in Pseudomonas aeruginosa and peritrichous flagella in Salmonella enterica serovar Typhimurium, demonstrating substantial variation between flagella in these organisms. Importantly, we observed novel flagellar subassemblies and provided additional insight into the structural basis of flagellar assembly and loss in both P. aeruginosa and S. Typhimurium.
KEYWORDS: bacterial cell motility, bacterial envelope, flagellar assembly, nanomachine, protein export, type III secretion
ABSTRACT
The bacterial flagellum is a sophisticated self-assembling nanomachine responsible for motility in many bacterial pathogens, including Pseudomonas aeruginosa, Vibrio spp., and Salmonella enterica. The bacterial flagellum has been studied extensively in the model systems Escherichia coli and Salmonella enterica serovar Typhimurium, yet the range of variation in flagellar structure and assembly remains incompletely understood. Here, we used cryo-electron tomography and subtomogram averaging to determine in situ structures of polar flagella in P. aeruginosa and peritrichous flagella in S. Typhimurium, revealing notable differences between these two flagellar systems. Furthermore, we observed flagellar outer membrane complexes as well as many incomplete flagellar subassemblies, which provide additional insight into mechanisms underlying flagellar assembly and loss in both P. aeruginosa and S. Typhimurium.
IMPORTANCE The bacterial flagellum has evolved as one of the most sophisticated self-assembled molecular machines, which confers locomotion and is often associated with virulence of bacterial pathogens. Variation in species-specific features of the flagellum, as well as in flagellar number and placement, results in structurally distinct flagella that appear to be adapted to the specific environments that bacteria encounter. Here, we used cutting-edge imaging techniques to determine high-resolution in situ structures of polar flagella in Pseudomonas aeruginosa and peritrichous flagella in Salmonella enterica serovar Typhimurium, demonstrating substantial variation between flagella in these organisms. Importantly, we observed novel flagellar subassemblies and provided additional insight into the structural basis of flagellar assembly and loss in both P. aeruginosa and S. Typhimurium.
INTRODUCTION
The bacterial flagellum is a complex, multiprotein nanomachine that allows bacteria to swim in liquid environments. Flagella from different species contain structurally conserved components. The cytoplasmic C-ring, which integrates chemotaxis signals, abuts the inner membrane MS-ring and interacts with stator proteins that anchor in the peptidoglycan and transform chemical energy into flagellar rotation (1). A dedicated flagellar export apparatus secretes proteins that assemble into the rod, hook, and filament to form the rotating axial “propeller” (2). L- and P-rings in the outer membrane and peptidoglycan, respectively, serve as a “bushing” for the flagellar rod as it traverses the cell envelope (3, 4).
Variation in specific features of the flagellum, as well as in flagellar number and placement, results in structurally distinct flagella that appear to be adapted to the specific environments that bacteria encounter (5–7). Escherichia coli and Salmonella enterica serovar Typhimurium assemble several external flagella at locations all over the cell (i.e., peritrichous). The counterclockwise rotation of these filaments causes them to form a bundle that powers a bacterial “run” toward a chemoattractant, while clockwise rotation results in filament unbundling and a reorienting bacterial “tumble” (8). Other bacteria restrict flagellar assembly to polar sites, among them Vibrio spp., which assemble one flagellum at a single pole; Campylobacter jejuni, with one flagellum at each pole; and Helicobacter pylori, with several flagella at one pole. Some bacteria, such as Vibrio parahaemolyticus and Vibrio alginolyticus, separately carry both lateral and polar flagellar systems (9). These marine Vibrio spp. also express two sets of stator proteins that use Na+ or H+ gradients to power rotation of the polar versus lateral flagella, respectively, in different environments (10, 11). The filament can even be assembled internally, as is the case for spirochetes that assemble periplasmic flagella between their cytoplasmic and outer membranes (12). This variety of flagellar structures, number, position, and regulation are reflected in the genetic loci that encode flagellar structural genes and regulators, which show clear evidence of evolution and diversification across bacterial phyla (13, 14). Cryo-electron tomography (cryo-ET) studies have revealed a corresponding variation in the macromolecular structures of the flagellar motor (5, 6). It is proposed that some of these unique structural features are related to the amount of torque the flagellum can generate or to its ability to interact with H+-versus Na+-driven stator systems (15, 16).
The opportunistic human pathogen Pseudomonas aeruginosa assembles a single polar flagellum that powers a run-and-reverse form of motility similar to that described for Vibrio spp. (17, 18). Although core structural features of the flagellum are well conserved and flagellar gene expression is regulated in similar stepwise fashions in P. aeruginosa and E. coli (19, 20), the P. aeruginosa flagellum appears to have several unusual features. Two distinct stators can drive rotation of the flagellum, and in contrast to V. alginolyticus, both appear to use proton motive force to generate torque (21). One of these stators can support swarming of monotrichous P. aeruginosa through media of increased viscosity (21, 22), in contrast to V. parahaemolyticus, which induces the expression of multiple lateral flagella in order to swarm (9). The P. aeruginosa flagellum propels bacteria in liquid media in response to signals sensed by more than 26 chemoreceptors (23). However, the flagellum also mediates bacterial attachment to surfaces (24) and initiates the process leading to biofilm formation (25). Despite the central importance of the flagellum in P. aeruginosa biology, a detailed structure of this nanomachine is not available.
We applied cryo-ET to visualize the P. aeruginosa polar flagellum in situ at high resolution and observed structural features that distinguish this flagellum from that of the model organism S. Typhimurium. We also observed “incomplete” flagellar assemblies in both P. aeruginosa and S. Typhimurium and investigated their possible roles in flagellar assembly and/or loss.
RESULTS
In situ structure of the intact P. aeruginosa flagellar motor and “incomplete” assemblies.
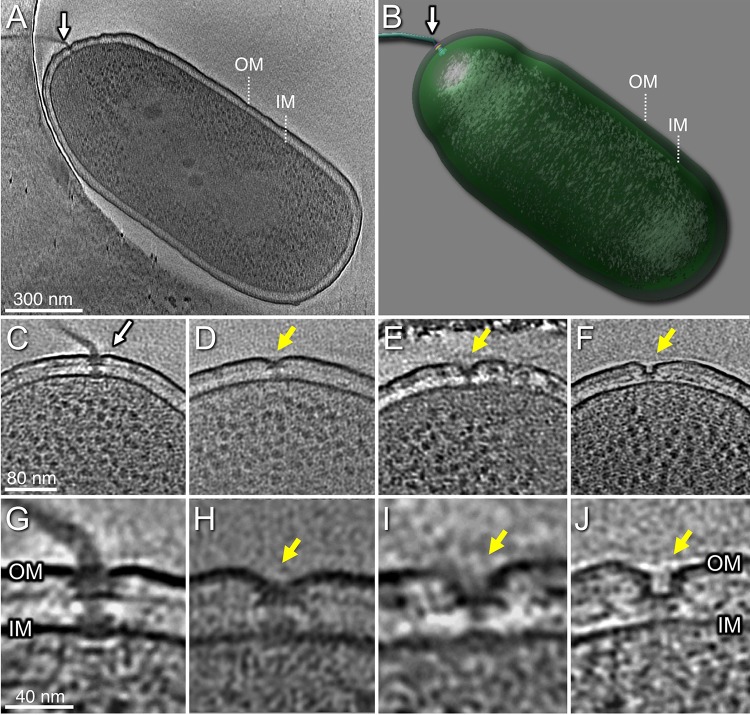
We used cryo-ET to visualize the single flagellum located at the pole of wild-type (WT) P. aeruginosa cells. Its basal body substructures, as well as the inner membrane, peptidoglycan layer, and outer membrane, were well defined in the tomograms (Fig. 1). In addition, we observed structures suggestive of “incomplete” flagellar assemblies at a subset of cell poles. These included the following: a flagellum lacking the hook and filament (Fig. 1D and H); a flagellum lacking the rod, hook, and filament (Fig. 1E and I); and an outer membrane complex without associated MS- or C-rings (Fig. 1F and J). Although these structures were much less common than complete flagellar motors in WT bacteria, the number of these “incomplete” assemblies increased in the hyperflagellated ΔfleN mutant (see below).
FIG 1.
Tomograms of P. aeruginosa PAK cells show the intact polar flagellum and subcomplexes. (A) One slice from a whole-cell tomogram shows a single polar flagellum. (B) A surface rendering of the tomogram shown in panel A. (C) A slice from one tomogram shows the intact flagellar motor. (D) A slice from one tomogram shows a flagellum lacking the hook and filament. (E) A tomogram slice shows a flagellum without the rod, hook, and filament. (F) A tomogram slice shows a novel structure with a large pore in outer membrane (OM). (G) A zoomed-in view of the flagellar motor shows major components embedded in OM and inner membrane (IM). (H) A zoomed-in view of the motor in panel D. (I) A zoomed-in view of the motor in panel E. (J) A zoomed-in view of the outer membrane pore in panel F.
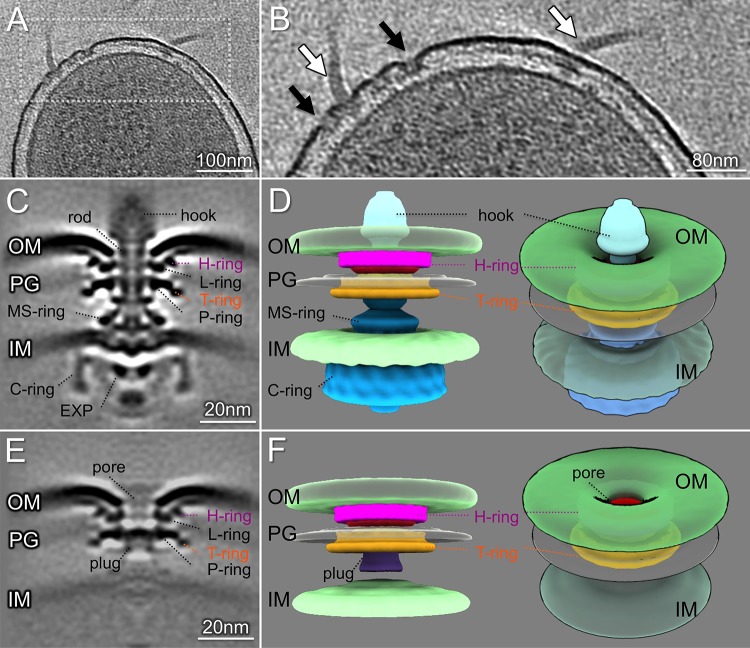
To determine the in situ structures of the P. aeruginosa flagellar motor at higher resolution, we made use of a hyperflagellated mutant, the ΔfleN mutant, which lacks the FleN/FlhG protein required for restriction of polar flagellar number in P. aeruginosa, Vibrio spp., and Shewanella spp. (26–30). This allowed us to visualize 1,170 flagellar motors and determine an in situ structure at higher resolution by subtomogram averaging (Fig. 2). A comparison of the P. aeruginosa flagellar motor structure with those of S. Typhimurium and V. alginolyticus (see Fig. S1 in the supplemental material) identified features common to all of these motors, namely the MS-ring, C-ring, export apparatus, rod, and P- and L-rings (Fig. 2C and E; see Fig. S2 in the supplemental material). The polar flagellar motors of P. aeruginosa and V. alginolyticus shared additional features not seen in the S. Typhimurium structure. Prominent densities adjacent to the P- and L-rings were observed in the P. aeruginosa flagellar motor, occupying a similar location to the T- and H-rings of the V. alginolyticus structure (31).
FIG 2.
Structures of the P. aeruginosa flagellar motor and subassembly. (A) A representative slice of a cell pole reconstruction from P. aeruginosa ΔfleN showing multiple flagella. (B) A zoomed-in view of the slice shown in panel A. Flagellar motors and hooks are indicated by white arrows. The flagellar outer membrane complexes (FOMCs) are shown by black arrows. (C) A slice of an averaged structure of the intact flagellar motor. Note the striking curvature of the outer membrane. (D) Side and tilted views of the three-dimensional (3D) surface rendering of the motor show the curvature of the outer membrane and the FOMC. (E) A slice of an averaged structure of the FOMC. (F) A 3D surface rendering of the FOMC. The novel “plug” is colored purple. A tilted view of the 3D surface rendering shows the membrane pore in the FOMC. EXP, export apparatus; OM, outer membrane; PG, peptidoglycan layer; IM, inner membrane.
The Vibrio T-ring is composed of two proteins, MotY and MotX, while FlgT is a component of the H-ring (31–33). Homologs of MotX and FlgT are not identified in P. aeruginosa, but a MotY (PA3526) homolog is encoded (21). We constructed the deletion mutant ΔmotY in P. aeruginosa and used cryo-ET and subtomogram averaging to determine its motor structure. The density adjacent to the P-ring was not visible in ΔmotY (see Fig. S3 in the supplemental material), suggesting that MotY contributes to the assembly of a “T-ring”-like structure in P. aeruginosa. Further work is required to determine if any proteins in addition to MotY participate in the formation of this ring and to identify proteins that contribute to the densities that are seen adjacent to the L-ring.
The interface between the flagellar motor and outer membrane in P. aeruginosa differed markedly from that seen in either S. Typhimurium or V. alginolyticus (Fig. S1). Compared with S. Typhimurium, the P. aeruginosa outer membrane appeared to undergo significant deflection at its interface with the L-ring (Fig. 2C; Fig. S1A). Although the spacing between the inner and outer membranes of S. Typhimurium and P. aeruginosa was quite similar (∼33 nm), the distance between the L-ring and the inner membrane was 24.6 nm in P. aeruginosa, much less than that observed for the motors of S. Typhimurium and V. alginolyticus (∼29.1 nm) (Fig. S1). Thus, the P. aeruginosa motor is both shorter and wider than that of S. Typhimurium.
In situ structure of a flagellar outer membrane complex in P. aeruginosa.
The cell poles of ΔfleN bacteria contained multiple intact flagellar motors, as well as numerous examples of a flagellar outer membrane complex (FOMC) (Fig. 2A and B) that was rarely observed in WT cells (Fig. 1F and J; Table 1). Subtomogram averaging of these abundant FOMCs allowed us to determine the FOMC in situ structure at high resolution (Fig. 2C and E). The FOMCs closely resembled the outer membrane portion of the fully assembled flagellar motor, possessing both P/L-rings and the adjacent T- and H-ring-like densities (Fig. 2C, D, F, and G; see Fig. S4 in the supplemental material). The outer membrane again exhibited significant deflection at its interface with the FOMC L-ring (Fig. 2), suggesting that this deformation was due to outer membrane interactions with the L/H-ring alone. The FOMC formed a large outer membrane pore of ∼12 nm in diameter and had a plug-like density within the P-ring, whereas the rod occupied this axial region in the flagellar motor (Fig. 2C, F, and H). Lastly, the inner membrane and MS-ring were clearly resolved in the intact motor structure, while the MS-ring was absent and the inner membrane poorly resolved in the structure of the FOMC. Classification of FOMC structures and averaging of these subgroups allowed us to observe variation in the distance between the outer membrane surrounding the FOMC and the inner membrane (see Fig. S5 in the supplemental material). In contrast, the distance between the inner and outer membranes in the vicinity of intact flagellar motors was less variable (Fig. S5), consistent with the notion that flagellar rod length is tightly controlled (34).
TABLE 1.
Cryo-ET data and analysis
| Sample name | Microscope | No. of tomograms | No. of subtomograms | No. of motors | No. of FOMCs | No. of intermediates |
|---|---|---|---|---|---|---|
| PAK (WT) | Titan Krios | 217 | 112 | 103 | 6 | 3 |
| PAK ΔfleN | Polara | 340 | 1,746 | 1,170 | 566 | 11 |
| PAK ΔfleN ΔflgG | Titan Krios | 46 | 121 | 121 | 0 | 0 |
| PAK ΔflgG | Titan Krios | 193 | 42 | 42 | 0 | 0 |
| PAK ΔmotY | Titan Krios | 65 | 26 | 26 | 0 | 0 |
| S. Typhimurium SB1780 | Polara | 1,470 | 3,695 | 3,669 | 6 | 20 |
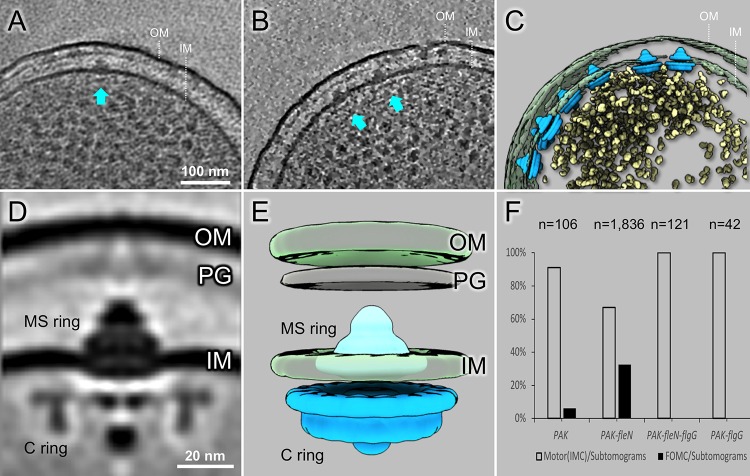
FOMCs as well as intact flagellar motors are observed in S. Typhimurium.
Our examination of P. aeruginosa revealed FOMCs composed of L/P- and T/H-rings forming plugged pores in the outer membrane, as well as “incomplete” flagellar assemblies (Fig. 1). To determine whether FOMCs were restricted to bacteria with polar flagella, we examined minicells prepared from peritrichously flagellated S. Typhimurium bacteria. We observed many fully assembled flagellar motors (Fig. 3A) and determined their structure with sufficient detail to resolve the well-ordered bilayers of both the inner and outer membranes and the close apposition of the L-ring to both the inner and outer leaflets of the outer membrane (Fig. 3D). After screening tomograms of 1,470 S. Typhimurium minicells (Table 1), we identified six FOMCs and determined their low-resolution structure (Fig. 3B and C). The overall shape and dimensions of the S. Typhimurium FOMC were well matched to those of the L/P rings of the fully assembled flagellar motor (Fig. 3E and H). As in P. aeruginosa, the S. Typhimurium FOMC formed a pore of approximately 12 nm, with a plug-like density visible below the P-ring. This membrane pore would be large enough to accommodate the rod assembly, as shown in the fully assembled flagellar motor (Fig. 3D and G).
FIG 3.
Intact flagellar motor, FOMC, and a subcomplex revealed in S. Typhimurium minicells. (A) A representative tomogram slice of an S. Typhimurium minicell shows a complete flagellar motor. (B, C) Two representative slices show FOMCs in S. Typhimurium. (D) A slice of a subtomogram average of the complete S. Typhimurium flagellar motor. (E) A slice of the averaged structure of the FOMC. (F) One class average shows the FOMC assembly aligned with the MS-ring in the absence of a distal rod. (G) Surface view of the intact flagellar motor. (H) A 3D surface rendering of the FOMC structure. (I) Surface rendering of a subcomplex in which the MS-ring is associated with the FOMC.
Subtomogram classification was used to analyze over 3,600 S. Typhimurium flagellar assemblies. Most class averages showed fully assembled motors, but others revealed “incomplete” assemblies. In addition to the class containing the six FOMCs, we observed a class with 20 particles that clearly differed from either the intact motor or the FOMC (Fig. 3F and I). In this class, a FOMC appeared axially aligned with a MS/C-ring structure in the absence of a rod density (Fig. 3F and I), raising the question of whether the FOMC can assemble independently of the rod and provide a conduit through which the rod reaches the outer membrane (Fig. 3F and I).
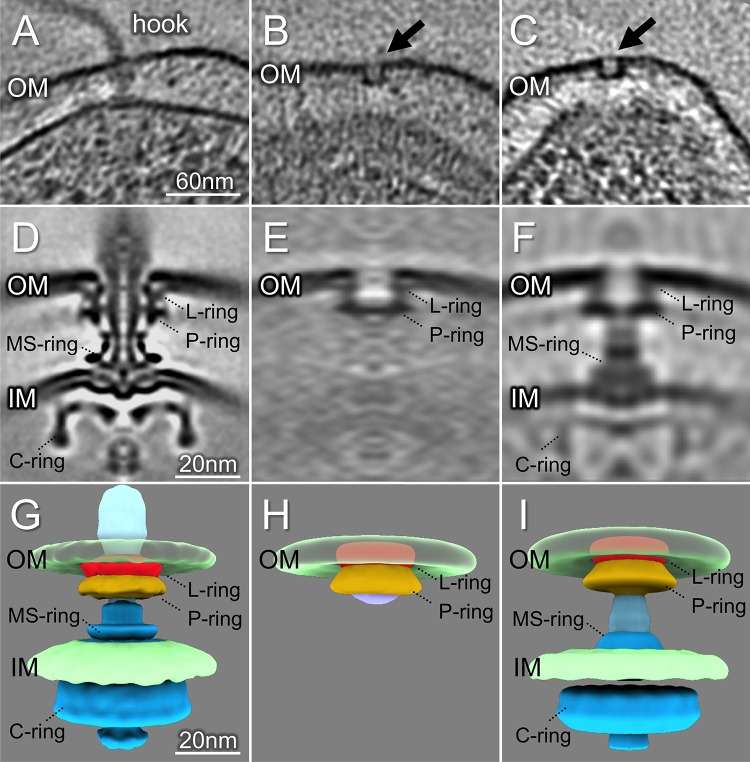
The FOMC is not observed in P. aeruginosa mutants lacking the distal rod protein FlgG.
To test whether the FOMC represents a flagellar assembly intermediate that can form in the absence of the flagellar rod, we constructed an in-frame, unmarked deletion of the gene encoding the predicted distal rod protein flgG in the PAK and PAK ΔfleN backgrounds. PAK ΔflgG was nonmotile in soft-agar swimming assays, but motility was fully complemented by expression of plasmid-encoded FlgG (see Fig. S6 in the supplemental material). Cryo-ET reconstructions from both PAK ΔflgG and PAK ΔfleN ΔflgG cells showed flagellar subcomplexes corresponding to the MS-ring and C-ring (Fig. 4). In 193 PAK ΔflgG cell pole tomograms, we observed 42 MS/C-ring complexes but no FOMC (Table 1). Likewise, we observed 121 MS/C-ring complexes from 46 PAK ΔfleN ΔflgG cell pole tomograms but no FOMC (Fig. 4B and C). The averaged structure of these complexes clearly shows the C-ring, MS-ring, and proximal rod, while the distal rod is absent (Fig. 4D and E). These results strongly argue that the FOMC cannot form in the absence of the distal rod protein, in agreement with previous models which propose that the P- and L-rings form around the assembled rod (35–37).
FIG 4.
P. aeruginosa ΔflgG and ΔfleN ΔflgG fail to assemble FOMCs. (A) A representative slice from a ΔflgG tomogram shows the MS/C-rings (arrow). (B) A representative slice from a ΔfleN ΔflgG tomogram shows two MS/C-rings (arrows). (C) A surface view of the tomogram in (B) shows seven MS/C-rings. (D) A central section from the averaged structure of the MS/C-rings. The distal rod is absent, while both MS/C-rings and expert apparatus are clearly resolved. (E) A surface view of the averaged structure shows the MS-ring (colored in cyan) embedded in inner membrane (IM) and the C-ring in blue. (F) Number of motors (or MS/C-rings) and FOMC observed in different strains. White bars represent motors (MS-ring and C-ring) found in cells; black bars indicate FOMCs. No FOMCs were seen in ΔflgG and ΔfleN ΔflgG cells.
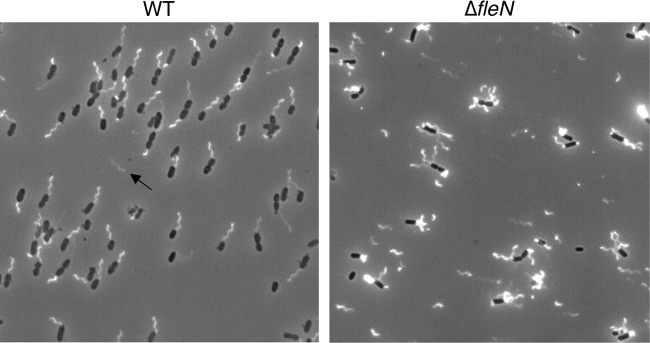
If the FOMC is not an assembly intermediate, it may be a residual structure left after a flagellum detaches. We measured the position of FOMCs relative to the cell pole and found that most were within 60 nm of the pole, with a distribution that matched that of intact flagellar motors in both WT and ΔfleN cells (see Fig. S7A and B in the supplemental material). This overlapping distribution is consistent with FOMCs being residual structures generated from flagellar motors, as the only known signal for polar flagellar placement is provided by the cytoplasmic protein FlhF (38). FOMCs were much more common in ΔfleN bacteria than in WT cells, accounting for 32% of all analyzed structures in ΔfleN cells (566/1747) compared with 5.5% in WT PAK (6/109) (Table 1). We examined WT and ΔfleN cells for evidence of increased flagellar loss in mutant bacteria and observed many detached flagellar filaments in samples of ΔfleN bacteria (Fig. 5). In contrast, detached flagella were rarely seen in samples prepared from WT bacteria, which is consistent with the rare observation of FOMCs in these cells.
FIG 5.
Detached flagella are frequently seen for ΔfleN bacteria. Live P. aeruginosa WT and ΔfleN bacteria expressing fliC(T394C) were stained with Alexa Fluor 488-maleimide and visualized by fluorescence microscopy. Single polar flagella are clearly visible on most WT cells, and one detached flagellum is indicated by an arrow. ΔfleN cells show multiple polar flagella, and many detached flagellar filaments are observed.
DISCUSSION
Flagella are the principal organelles powering motility in many bacterial species. These organelles share a conserved core structure but also demonstrate marked structural variation that is thought to underlie the ability of bacteria to swim through a wide range of environments (5, 6). We have used cryo-ET to visualize polar flagella in P. aeruginosa and lateral flagella in S. Typhimurium, revealing notable differences between these two flagellar systems. Specifically, the polar flagellum of P. aeruginosa possesses additional T- and H-rings that assemble outside the P- and L-rings, respectively. This wider stator would allow the P. aeruginosa motor to generate a greater maximum torque than S. Typhimurium (15), allowing P. aeruginosa to swim at higher velocities and swarm through more viscous media, as observed recently (39).
MotY is likely involved in the formation of the T-ring, as motY mutants lack this feature in both P. aeruginosa and V. alginolyticus (31). The protein(s) responsible for H-ring formation remains unknown. However, the H-ring proteins likely cause the marked deflection of the outer membrane seen at the motor-membrane interface (Fig. S2), which distinguishes the P. aeruginosa flagellum from those of V. alginolyticus and S. Typhimurium.
Kaplan et al. recently used cryo-ET to examine the polar motors of Shewanella oneidensis, Legionella pneumophila, and P. aeruginosa (16). The structure that they present for the P. aeruginosa motor lacks the H-ring-like densities that we clearly observe in both the complete flagellar motor and the FOMC and instead resembles the low-pass filtered structure shown in Fig. S3D. This difference in resolution between their structure and ours may account for the additional structures and detail that we observe; it is also possible that their use of a different P. aeruginosa strain (PAO1) grown overnight in minimal media contributes to these differences.
Flagellar gene expression in both S. Typhimurium and P. aeruginosa is tightly regulated and coupled to the completion of flagellar intermediates, with the transcription of genes encoding hook-basal body (HBB) components preceding transcription of the flagellar filament subunit, stator proteins, or chemotaxis receptors (19, 20). The “early” flagellar genes encoding HBB components are under the control of master regulators, such as FlhDC in S. Typhimurium and FleQ in P. aeruginosa, and are expressed at the same time (19, 20). The Sec-dependent assembly of the inner membrane MS-ring, with which the cytoplasmic C-ring and export apparatus associate, creates a flagellar type III secretion system responsible for secreting proximal and distal rod proteins, as well as subunits of the hook and filament. The rod passes through the peptidoglycan layer and outer membrane within P- and L-rings, respectively. FlgI and FlgH, the proteins that form the P- and L-rings, also use the Sec secretory pathway to reach the periplasmic space (40). In addition to forming an outer membrane “bushing” for flagellar rod rotation, the L-ring appears to catalyze removal of the FlgJ rod cap, allowing assembly of the hook cap protein FlgD and the transition to extracellular hook assembly (41). The long-accepted model of flagellar assembly proposes that P/L-rings form around a completed rod, with assembly of the L-ring catalyzing the removal of the FlgJ rod cap and coupling the formation of the outer membrane pore to the rod-to-hook transition (36, 37). We observed isolated FOMCs in both S. Typhimurium and P. aeruginosa, showing that the L/P-ring outer membrane plugged “pore” can stably exist in the absence of the flagellar inner membrane or rod components. We considered the possibility that the FOMCs are intermediates that assemble independently of the MS-ring, C-ring, and rod proteins. However, the absence of FOMCs in bacteria lacking FlgG provides compelling evidence that the FOMC cannot form in the absence of this distal rod protein. This is in contrast to the assembly of structurally related type 3 secretion systems, in which the outer membrane secretin pore can assemble independently and dock with the inner membrane rings (42).
FOMCs are likely to be remnants of flagella that have detached from the cell, although this process must be relatively uncommon under conditions of exponential growth, given the low frequency of these structures in WT P. aeruginosa cells (Fig. 6). A recent paper describes similar-appearing flagellar “relics” in V. cholerae and Plesiomonas shigelloides (43). The latter is a member of the family Enterobacteriaceae which is found in fresh or brackish waters and is associated with diarrheal disease following the consumption of raw oysters (44). P. shigelloides produces polar flagella when grown in liquid media and assembles lateral flagella when grown in solid or semisolid media (45). The polar flagellum of Plesiomonas (formerly Aeromonas) spp. is powered by Na+-conducting stators (46) as is the case for marine Vibrio spp. The polar flagella of both P. shigelloides and V. cholerae are proposed to be ejected under conditions of nutrient deprivation, leaving numerous flagellar “relics” at the P. shigelloides pole (43). Reduced swimming speeds and decreased numbers of surface flagella are observed as P. shigelloides and V. cholerae enter stationary phase, while S. Typhimurium swimming speed and flagellar number increase with culture density. Although Ferreira et al. argue that P. aeruginosa must undergo a similar process of flagellar “ejection” based on their observation of flagellar relics in this species, no data supporting this hypothesis are presented in their study. Our results suggest that P. aeruginosa FOMCs result when flagella detach, as happens frequently in the ΔfleN mutant and rarely in WT P. aeruginosa. As many ΔfleN bacteria still possess multiple polar flagella at a time when both detached flagellar filaments (Fig. 5) and FOMCs (Fig. 2B) are also observed, we hypothesize that detachment and/or shearing of entangled flagella is more likely to account for our findings than programmed flagellar ejection. Similar mechanical loss may also lead to the rare FOMCs that we observed for S. Typhimurium.
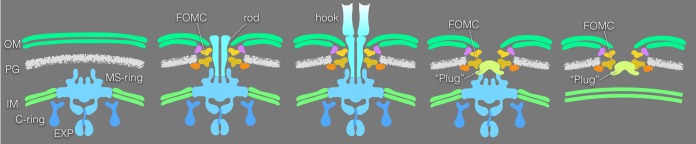
FIG 6.
A schematic model of flagellar assembly. The first panel shows an early stage of flagellar assembly with the MS-ring in the inner membrane. The second panel shows a FOMC assembled around the rod. The third panel shows an intermediate transitioning from rod to hook assembly. The fourth panel shows a subcomplex lacking the rod and hook. The last panel shows an FOMC. The final two structures are likely products of flagellar disassembly. EXP, export apparatus; OM, outer membrane; IM, inner membrane; PG, peptidoglycan layer.
In summary, we have used cryo-ET to observe in situ structures associated with the assembly of P. aeruginosa and S. Typhimurium flagella. These in situ structures have revealed the features of the P. aeruginosa polar flagellar motor in unprecedented detail. Our work has also demonstrated that MotY contributes to the formation of the T-ring in P. aeruginosa, but additional components that form the “H-ring” structure of this envelope spanning nanomachine still await discovery.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains used in this study are listed in Table 2. P. aeruginosa, E. coli, and S. Typhimurium strains were cultured in Luria broth (LB) with shaking at 37°C unless otherwise indicated. The following antibiotics were added as indicated: chloramphenicol (12.5 μg/ml) and gentamicin (15 µg/ml, E. coli; 100 µg/ml, P. aeruginosa).
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAK | WT | 60 |
| PAK ΔfleN | Unmarked, in-frame deletion of fleN | This study |
| PAK ΔflgG | Unmarked, in-frame deletion of flgG | This study |
| PAK ΔfleN ΔflgG | Unmarked, in-frame deletion of fleN and flgG | This study |
| PAK ΔmotY | Unmarked, in-frame deletion of motY | This study |
| PA14 | WT | 61 |
| PA14 fliC(T394C) | Replacement of chromosomal fliC in PA14 | This study |
| PA14 ΔfleN | Unmarked, in-frame deletion of fleN in PA14 | This study |
| PA14 fliC(T394C) ΔfleN | Replacement of chromosomal fliC in PA14 ΔfleN | This study |
| S. Typhimurium SB1780 | minD::cat (Cmr); minicell producer | 51 |
| E. coli | ||
| DH5α | Invitrogen | |
| S17.1 | thi pro hsdR recA RP4-2 (Tc::Mu) (Km::Tn7) | 62 |
| Plasmids | ||
| pEX-GW | pEX-based deletion vector, Gmr | 63 |
| pEX-fleNKO | pDONRX with ∼1-kb region upstream and downstream of fleN; Gmr | This study |
| pEX-flgGKO | pDONRX with ∼1-kb region upstream and downstream of flgG; Gmr | This study |
| pEX-motYKO | pDONRX with ∼1-kb region upstream and downstream of motY; Gmr | This study |
| pMQ72 | Expression vector, Gmr | 49 |
| pMQ72-FlgG | Plasmid expressing FlgG under pBAD control, Gmr | This study |
Cmr, chloramphenicol resistant; Gmr, gentamicin resistant.
Strain construction.
All PCR primers used in this study are listed in Table 3 and were synthesized by the Keck Facility (Yale University). Unmarked, in-frame deletion of P. aeruginosa genes was carried out by allelic exchange (47). Briefly, ∼1-kb regions upstream and downstream of fleN, flgG, or motY were PCR amplified from WT genomic DNA with Phusion polymerase (New England BioLabs [NEB]) using gene-specific N1/N2 and C1/C2 primer pairs and then spliced by overlap extension using primers N1/C2. The resulting fragment was integrated into a Gateway-adapted pEX-based deletion vector and confirmed by Sanger sequencing. The pEX-knockout (KO) plasmid was transformed into chemically competent E. coli S17.1 and then mated into P. aeruginosa. Exconjugant merodiploids were selected on Vogel-Bonner medium (VBM)-gentamicin and then plated on VBM and 10% (wt/vol) sucrose to select for excision of plasmid backbone sequences. Knockout candidates were screened by PCR and confirmed by motility phenotype and microscopy.
TABLE 3.
Primers
| Name | Sequence (5′ to 3′) |
|---|---|
| fleN N1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGCGCATGCTGTTGGCGCACCTGT |
| fleN N2 | CCGCTGTCATACGGCCGAACCACCCATCTGCTTCATACCTTG |
| fleN C1 | CAAGGTATGAAGCAGATGGGTGGTTCGGCCGTATGACAGCGG |
| fleN C2 | GGGGACCACTTTGTACAAGAAAGCTGGGTTCTTGTCCAAGTAAACCTCCGTACAGA |
| flgG N1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGGAGGATTCGATGGACAAGAT |
| flgG N2 | GACGAAGGACTTGCTGACCCACAGTGCCG |
| flgG C1 | GTCAGCAAGTCCTTCGTCACCCAGAATCTTTG |
| flgG C2 | GGGGACCACTTTGTACAAGAAAGCTGGGTGGATGCTGGCGATATCCTTCA |
| motY N1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGAGTCGGTGCCGAGGAAGA |
| motY N2 | CGCGACTCAAACCGTGGGTTTAAGGCGCGGCTGCACGACGGC |
| motY C1 | GCCGTCGTGCAGCCGCGCCTTAAACCCACGGTTTGAGTCGCG |
| motY C2 | GGGGACCACTTTGTACAAGAAAGCTGGGTCAGCTCGATGTAGGTCGGGGT |
| fliC(T394C) N1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCGACCTGGCCCTGCAATC |
| fliC(T394C) N2 | CGCCGTCTGCGCAGGAGATGTCGACGCTGGCAAC |
| fliC(T394C) C1 | CATCTCCTGCGCAGACGGCGCCCAGAACGCCA |
| fliC(T394C) C2 | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTCGATAGGGCAAGCACCATG |
| EcoRI-flgG | GGAGAATCCTTGCAAATCAGCTAATCACTTGAG |
| flgG-HindIII | GGACAAGCTTCTACCTCGCGGCGTACA |
The flagellin-encoding gene fliC was mutated to replace threonine with cysteine at amino acid position 394 to allow staining of flagellar filaments in live, unfixed cells with Alexa Fluor 488 C5 maleimide, as previously described (48). Gene-specific N1/N2 and C1/C2 primer pairs incorporating the mutation were used to PCR amplify fliC from PA14; these amplicons were spliced by overlap extension using primers N1/C2 and integrated into pDONRX. This construct was confirmed by Sanger sequencing and then employed as described above to replace the WT fliC genes of PA14 and PA14 ΔfleN.
The full-length flgG gene was PCR amplified from WT genomic DNA using Phusion polymerase (NEB) and cloned into pMQ72 (49) under the control of an arabinose-inducible pBAD promoter.
Swimming assays.
Two microliters of an exponential-phase bacterial culture was spotted onto LB solidified with 0.3% agar and incubated overnight at 30°C. Antibiotics and arabinose (0.2%) were added to plates carrying pMQ72 vectors.
Flagellar staining.
Flagellar filaments were stained as previously described (48). Briefly, PA14 fliC(T394C) and PA14 ΔfleN fliC(T394C) bacteria were grown to log phase (optical density at 600 [OD600], ∼0.5) in LB at 37°C. One milliliter of cell culture was gently pelleted (5,000 × g for 5 min), washed once with phosphate-buffered saline (PBS; pH 8), resuspended in 50 µl of PBS containing 50 µg/ml Alexa Fluor 488 C5 maleimide (Thermo Fisher Scientific), and incubated for 3 min at room temperature. Cells were washed with PBS and resuspended in 30 µl of PBS. Four microliters of suspension was spotted to a clean microscope slide, immobilized with a poly-l-lysine-treated coverslip, and imaged using a Nikon Eclipse Ti-E inverted microscope equipped with an Andor Zyla 5.5MP 10-tap scientific complementary metal-oxide semiconductor (sCMOS) camera.
Sample preparation for cryo-ET.
Two microliters of an overnight P. aeruginosa LB culture (grown at 37°C with shaking) was spotted onto a 1% LB agar plate and incubated for 12 hours at 37°C. The cells at the edges of colonies were collected and checked for motility. Collected cells were then washed once with PBS buffer and resuspended. Colloidal gold particles (10 nm) were added to the cell suspension to yield a 10× dilution and then deposited on a freshly glow-discharged, holey carbon grid (copper 200 mesh; Quantifoil) for 1 min. The grid with a 5-μl mixture was blotted with filter paper (Waterman qualitative filter paper, grade 1) and rapidly plunge-frozen in liquid ethane using a gravity-driven plunger apparatus, as described previously (31, 50).
S. Typhimurium SB1780 was grown overnight at 37°C in LB containing 0.3 M NaCl. Bacterial subcultures were prepared the following day and enriched for minicells, as previously described (51). Minicell deposition and freezing were carried out as described above for P. aeruginosa.
Cryo-ET data collection and image processing.
The data from P. aeruginosa PAK ΔfleN and S. Typhimurium were collected using a Polara G2 electron microscope (FEI). The data from P. aeruginosa WT cells were collected using a Titan Krios electron microscope (Thermo Fisher Scientific). Both microscopes were equipped with a 300-kV field emission gun and a direct electron detector (Gatan K2 Summit). Images collected by the Polara G2 electron microscope were observed at ×15,000 magnification and at 4- to 6-μm defocus, resulting in 0.25 nm/pixel. The images taken by the Titan Krios electron microscope were collected at focus using a Volta phase plate and energy filter with a 20-eV slit. The resulting physical resolution is 0.27 nm/pixel. All data were acquired with SerialEM software (52). A total dose of 50 e−/Å2 is distributed among 35 tilt images covering angles from −51° to +51° at tilt steps of 3° and starting to collect tilts series at −30°. For every single tilt series collection, the dose-fractionated mode was used to generate 8 to 10 frames per projection image. Collected dose-fractionated data were first subjected to the motion correction program to generate drift-corrected stack files (53). The stack files were aligned using gold fiducial markers and volumes reconstructed by the weighted back-projection method, using Tomoauto (54), IMOD (55), and Tomo3d (56).
Subtomogram analysis with i3 package.
Bacterial flagellar motors were detected manually and analyzed using the i3 program (57, 58). In total, 1,746 subtomograms of P. aeruginosa motors from PAK ΔfleN and 3,695 subtomograms of S. Typhimurium motors from SB1780 were used for subtomogram analysis. The i3 tomographic package was used on the basis of the “alignment by classification” method with missing wedge compensation for generating the averaged structure of the motor, as described previously (58). Detailed information about tomograms and subtomograms in this study is provided in Table 1.
Three-dimensional visualization.
Tomographic reconstructions were visualized using IMOD (55). The segmentation was generated using IMOD and surface rendering was done with UCSF chimera (59).
Determination of flagellar motor and FOMC distance from the P. aeruginosa cell pole.
The distance of motors and FOMCs from the cell pole was extracted from tomograms for 38 motors and 29 FOMCs in ΔfleN cells and for 103 motors and 6 FOMCs in WT PAK cells, and data were plotted using Microsoft Excel.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants GM107629 and R01AI087946 (to J.L.), AI075051 and AI117333 (to B.I.K.), and AI079022 (to J.E.G.) from National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00117-19.
REFERENCES
- 1.Minamino T, Imada K. 2015. The bacterial flagellar motor and its structural diversity. Trends Microbiol 23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181:1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones CJ, Homma M, Macnab RM. 1987. Identification of proteins of the outer (L and P) rings of the flagellar basal body of Escherichia coli. J Bacteriol 169:1489–1492. doi: 10.1128/jb.169.4.1489-1492.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones CJ, Homma M, Macnab RM. 1989. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol 171:3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao X, Norris SJ, Liu J. 2014. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaban B, Hughes HV, Beeby M. 2015. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol 46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 9.McCarter L. 2004. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 10.Sato K, Homma M. 2000. Functional reconstitution of the Na(+)-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem 275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 11.Atsumi T, McCarter L, Imae Y. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 12.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. 2012. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu R, Ochman H. 2007. Origins of flagellar gene operons and secondary flagellar systems. J Bacteriol 189:7098–8104. doi: 10.1128/JB.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pallen MJ, Penn CW, Chaudhuri RR. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol 13:143–149. doi: 10.1016/j.tim.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. 2016. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci U S A 113:E1917–E1926. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan M, Ghosal D, Subramanian P, Oikonomou CM, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ. 2019. The presence and absence of periplasmic rings in bacterial flagellar motors correlates with stator type. Elife 8:e43487. doi: 10.7554/eLife.43487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai Q, Li Z, Ouyang Q, Luo C, Gordon VD. 2016. Singly flagellated Pseudomonas aeruginosa chemotaxes efficiently by unbiased motor regulation. mBio 7:e00013. doi: 10.1128/mBio.00013-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie L, Altindal T, Chattopadhyay S, Wu XL. 2011. Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc Natl Acad Sci U S A 108:2246–2251. doi: 10.1073/pnas.1011953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 20.Chevance FF, Hughes KT. 2017. Coupling of flagellar gene expression with assembly in Salmonella enterica. Methods Mol Biol 1593:47–71. doi: 10.1007/978-1-4939-6927-2_4. [DOI] [PubMed] [Google Scholar]
- 21.Doyle TB, Hawkins AC, McCarter L. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol 186:6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchma SL, Delalez NJ, Filkins LM, Snavely EA, Armitage JP, O'Toole GA. 2015. Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J Bacteriol 197:420–430. doi: 10.1128/JB.02130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortega DR, Fleetwood AD, Krell T, Harwood CS, Jensen GJ, Zhulin IB. 2017. Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:12809–12814. doi: 10.1073/pnas.1708842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad JC, Gibiansky ML, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GC. 2011. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys J 100:1608–1616. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 26.Dasgupta N, Arora SK, Ramphal R. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 182:357–364. doi: 10.1128/JB.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusumoto A, Kamisaka K, Yakushi T, Terashima H, Shinohara A, Homma M. 2006. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem 139:113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
- 30.Schuhmacher JS, Rossmann F, Dempwolff F, Knauer C, Altegoer F, Steinchen W, Dorrich AK, Klingl A, Stephan M, Linne U, Thormann KM, Bange G. 2015. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci U S A 112:3092–3097. doi: 10.1073/pnas.1419388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu S, Nishikino T, Hu B, Kojima S, Homma M, Liu J. 2017. Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus. Proc Natl Acad Sci U S A 114:10966–10971. doi: 10.1073/pnas.1712489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terashima H, Koike M, Kojima S, Homma M. 2010. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J Bacteriol 192:5609–5615. doi: 10.1128/JB.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu S, Nishikino T, Kojima S, Homma M, Liu J. 2018. The Vibrio H-ring facilitates the outer membrane penetration of the polar sheathed flagellum. J Bacteriol 200:e00387-18. doi: 10.1128/JB.00387-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii T, Kato T, Hiraoka KD, Miyata T, Minamino T, Chevance FF, Hughes KT, Namba K. 2017. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun 8:14276. doi: 10.1038/ncomms14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol 226:433–446. doi: 10.1016/0022-2836(92)90958-M. [DOI] [PubMed] [Google Scholar]
- 36.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 37.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray TS, Kazmierczak BI. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol 188:6995–7004. doi: 10.1128/JB.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schniederberend M, Johnston JF, Shine E, Shen C, Jain R, Emonet T, Kazmierczak BI. 2019. Modulation of flagellar rotation in surface-attached bacteria: a circuit for rapid surface-sensing. bioRxiv doi: 10.1101/567438. [DOI] [PMC free article] [PubMed]
- 40.Karlinsey JE, Pease AJ, Winkler ME, Bailey JL, Hughes KT. 1997. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J Bacteriol 179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen EJ, Hughes KT. 2014. Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the L-ring-dependent rod scaffold removal. J Bacteriol 196:2387–2395. doi: 10.1128/JB.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira JL, Gao FZ, Rossmann FM, Nans A, Brenzinger S, Hosseini R, Wilson A, Briegel A, Thormann KM, Rosenthal PB, Beeby M. 2019. Gamma-proteobacteria eject their polar flagella under nutrient depletion, retaining flagellar motor relic structures. PLoS Biol 17:e3000165. doi: 10.1371/journal.pbio.3000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Chen Y, Yang Q, Kong H, Yu F, Han D, Zheng S, Cui D, Li L. 2013. Plesiomonas shigelloides infection in Southeast China. PLoS One 8:e77877. doi: 10.1371/journal.pone.0077877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merino S, Aquilini E, Fulton KM, Twine SM, Tomas JM. 2015. The polar and lateral flagella from Plesiomonas shigelloides are glycosylated with legionaminic acid. Front Microbiol 6:649. doi: 10.3389/fmicb.2015.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilhelms M, Vilches S, Molero R, Shaw JG, Tomas JM, Merino S. 2009. Two redundant sodium-driven stator motor proteins are involved in Aeromonas hydrophila polar flagellum rotation. J Bacteriol 191:2206–2217. doi: 10.1128/JB.01526-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 48.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 49.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, Picking WD, Liu J. 2015. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A 112:1047–1052. doi: 10.1073/pnas.1411610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu B, Lara-Tejero M, Kong Q, Galán JE, Liu J. 2017. In situ molecular architecture of the Salmonella type III secretion machine. Cell 168:1065–1067. doi: 10.1016/j.cell.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morado DR, Hu B, Liu J. 2016. Using Tomoauto: a protocol for high-throughput automated cryo-electron tomography. J Vis Exp 107:e53608. doi: 10.3791/53608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 56.Agulleiro JI, Fernandez JJ. 2015. Tomo3D 2.0–exploitation of advanced vector extensions (AVX) for 3D reconstruction. J Struct Biol 189:147–152. doi: 10.1016/j.jsb.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 57.Winkler H. 2007. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol 157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 58.Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. 2009. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol 165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Frost LS, Paranchych W. 1977. Composition and molecular weight of pili purified from Pseudomonas aeruginosa K. J Bacteriol 131:259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 62.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 63.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.