The induction of lipoprotein N-terminal remodeling in response to environmental copper in Gram-positive bacteria suggests a more general role in bacterial cell envelope physiology. N-terminal modification by lyso formation, in particular, simultaneously modulates the TLR2 response in direct comparison to their diacylglycerol-modified precursors. Thus, use of copper as a frontline antimicrobial control agent and ensuing selection raises the potential of diminished innate immune sensing and enhanced bacterial virulence.
KEYWORDS: Cell surface, copper, lipoproteins, Toll-like receptors
ABSTRACT
Bacterial lipoproteins are globular proteins anchored to the extracytoplasmic surfaces of cell membranes through lipidation at a conserved N-terminal cysteine. Lipoproteins contribute to an array of important cellular functions for bacteria, as well as being a focal point for innate immune system recognition through binding to Toll-like receptor 2 (TLR2) heterodimer complexes. Although lipoproteins are conserved among nearly all classes of bacteria, the presence and type of α-amino-linked acyl chain are highly variable and even strain specific within a given bacterial species. The reason for lyso-lipoprotein formation and N-acylation variability in general is presently not fully understood. In Enterococcus faecalis, lipoproteins are anchored by an N-acyl-S-monoacyl-glyceryl cysteine (lyso form) moiety installed by a chromosomally encoded lipoprotein intramolecular transacylase (Lit). Here, we describe a mobile genetic element common to environmental isolates of Listeria monocytogenes and Enterococcus spp. encoding a functional Lit ortholog (Lit2) that is cotranscribed with several well-established copper resistance determinants. Expression of Lit2 is tightly regulated, and induction by copper converts lipoproteins from the diacylglycerol-modified form characteristic of L. monocytogenes type strains to the α-amino-modified lyso form observed in E. faecalis. Conversion to the lyso form through either copper addition to media or constitutive expression of lit2 decreases TLR2 recognition when using an activated NF-κB secreted embryonic alkaline phosphatase reporter assay. While lyso formation significantly diminishes TLR2 recognition, lyso-modified lipoprotein is still predominantly recognized by the TLR2/TLR6 heterodimer.
IMPORTANCE The induction of lipoprotein N-terminal remodeling in response to environmental copper in Gram-positive bacteria suggests a more general role in bacterial cell envelope physiology. N-terminal modification by lyso formation, in particular, simultaneously modulates the TLR2 response in direct comparison to their diacylglycerol-modified precursors. Thus, use of copper as a frontline antimicrobial control agent and ensuing selection raises the potential of diminished innate immune sensing and enhanced bacterial virulence.
INTRODUCTION
Lipoproteins are highly conserved bacterial cell surface-bound proteins and consequently are a focal recognition point for innate immune pathways involved in microbial-pathogen recognition (1–4). Lipoproteins are structurally unique in having posttranslational acylation at a conserved N-terminal cysteine that anchors variable globular protein domains to the cell membrane surface, where they perform a myriad of functions (2, 3, 5–7). Preprolipoproteins are first exported across the cytoplasmic membrane, after which they become modified by lipoprotein diacylglyceryl transferase (Lgt). Lgt recognizes a short, conserved amino acid sequence called a lipobox containing an invariant cysteine residue that is modified by transferring a diacylglycerol (DA) moiety from a membrane phospholipid to the cysteine thiol to form a thioether (8, 9). Then, lipoprotein signal peptidase II (Lsp) cleaves the leader peptide, exposing the cysteine α-amino group (10). Beyond this step, further lipoprotein modification varies between species and in a way that does not simply correspond to Gram-negative versus Gram-positive bacteria, as had previously been accepted (6, 11–14).
In most Gram-negative bacteria, acylation of the α-amino cysteine terminus is catalyzed by the essential lipoprotein N-acyl transferase (Lnt) using a phospholipid acyl donor to create the mature triacylated lipoprotein (TA-LP) (15). This N-acyl chain is critical for lipoprotein recognition and transport to the outer membrane (16, 17). However, an lnt ortholog, and thus TA-LP, is also present in many high-GC Gram-positive Actinobacteria (6, 13, 18). More surprisingly, TA-LPs are found in some low-GC Gram-positive Firmicutes (Staphylococcus aureus and Staphylococcus epidermidis) that have no identifiable lnt sequence ortholog (11, 19, 20). Further studies by Kurokawa et al. among a panel of Firmicutes revealed several novel lipoprotein forms, all featuring novel N-terminal modifications: the N-acetyl form, which has an amide-linked acetyl group (Bacillus subtilis, among others); the lyso form (lyso-LP), with an N-acyl S-monoacyl-glyceryl (Enterococcus faecalis, Bacillus cereus, and others); and a peptidyl form, containing two additional amino acids before the lipid-modified cysteine (Mycoplasma fermentans) (11). More recently, Nguyen et al. identified the N-acetyl form in Staphylococcus carnosus, as well (21). The widespread phylogenetic conservation of N-terminally modified lipoprotein forms, when considered with the seemingly random intraspecies lipoprotein type, suggests strong selective pressure favoring N-terminal modifications that appeared postspeciation. The underlying physiological purpose of N modification, though, remains unclear, especially as some Gram-positive species continue to elaborate unmodified DA-LP (diacylglycerol-modified lipoprotein), as in Listeria monocytogenes (11). In either case, however, lipoprotein N-terminal structural diversity is accommodated by the innate immune response through cognate Toll-like receptor 2 (TLR2) heterodimerization (TLR2/TLR1 and TLR2/TLR6 for TA-LP and DA-LP ligands, respectively) (4, 21–23).
Capitalizing on the shared N-acyl chain of TA-LP and lyso-LP as means for rescue of the otherwise lethal lnt-null phenotype in Escherichia coli, we identified the previously uncharacterized lipoprotein intramolecular transacylase gene (lit) as responsible for the synthesis of lyso-LP in Firmicutes (12). Homology searches among type strains revealed a relatively narrow phylogenomic distribution in comparison to the highly conserved Lgt and Lsp proteins. Here, however, we report the surprising distribution of a distinct Lit-type protein (named Lit2) encoded on a mobile genetic element embedded within a copper resistance operon. We investigate the effect of Lit2 on lipoprotein formation and subsequent TLR2 signaling using two environmental isolates, Enterococcus faecalis TX1342 and Listeria monocytogenes CFSAN023459. We show that Lit2 expression is induced by copper, in turn converting the lipoprotein profile from DA-LP to a lyso form in L. monocytogenes. We also compare the TLR2 responses of the lyso-LP form, using both synthetic lipopeptide standards and heat-inactivated whole bacterial cells, and demonstrate that lyso-LP signals preferentially through the TLR2/TLR6 heterodimer. Overall, however, the lyso form is shown to be a markedly weaker ligand than either the conventional DA-LP or TA-LP form. Taken together, the connection between copper resistance, lyso form lipoproteins, and altered TLR2 innate immune recognition suggests a greater role of N-terminal lipoprotein modification in Gram-positive bacteria in copper resistance, and potentially virulence, through muted TLR2 recognition.
RESULTS
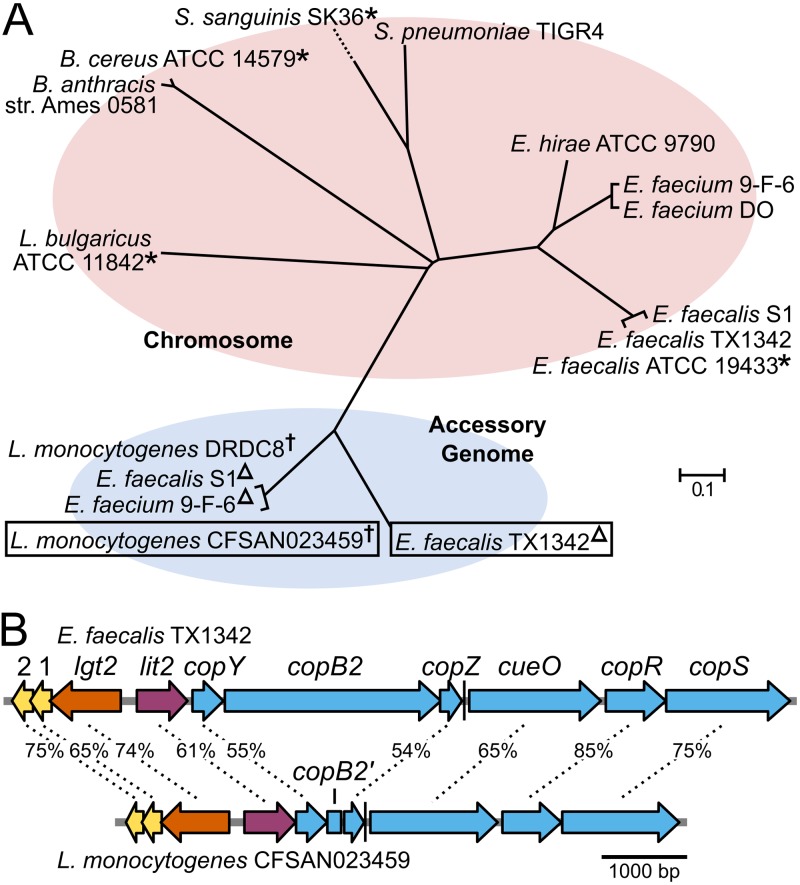
Phylogenetic analyses reveal a lit sequence ortholog colocalized within a putative copper resistance operon.
When the E. faecalis ATCC 19433 Lit protein sequence is queried using a BLASTp search, Lit orthologs bifurcate into two distinct clades (Fig. 1A). In the first clade are species known to produce lyso-LP (11), as well as clinically relevant organisms, such as E. faecium, Bacillus anthracis, and Streptococcus pneumoniae. All the genes are chromosomally located, though there is no shared cross-genus genomic synteny. This is in stark contrast to the second clade of Lit orthologs, where all are flanked by highly similar genes within a larger putative mobile element either on a freely replicating plasmid (L. monocytogenes) or integrated into the chromosome (Enterococcus spp.). In Enterococcus sp. isolates, these strains therefore contain two copies of lit, while isolates of L. monocytogenes harboring the plasmid contain a single copy. To differentiate between the two, here, we refer to the chromosomal copy as “lit1” and the accessory genome copy as “lit2.”
FIG 1.
Phylogenomic distribution of Lit. (A) A nonexhaustive phylogenetic tree was generated using the amino acid sequences of Lit from select strains. Sequences of Lit1 and Lit2 separate into two clades based on their location in the chromosome or the accessory genome, respectively. Strains previously demonstrated to make lyso form lipoproteins are indicated with asterisks (11, 12). Other strains were assumed to make the lyso form based on the presence of lit1. The strains E. faecalis TX1342 and L. monocytogenes CFSAN023459, characterized in this study, are boxed. Plasmid-borne copies of lit2 are indicated with a dagger and chromosomally integrated copies with a triangle. (B) Schematic representations of the E. faecalis TX1342 and L. monocytogenes CFSAN023459 lit2-copper resistance operons with percent ortholog sequence identities indicated. The proteins with unknown functions are designated “1” and “2” for clarity, and the incomplete CopB2 fragment is designated copB2′. A predicted rho-independent terminator following copZ is indicated by a vertical line.
The overall genetic architecture and gene similarity between E. faecalis TX1342 and L. monocytogenes CFSAN023459 indicates a common origin and transmission potential between Enterococcus and Listeria spp. (Fig. 1B). Intriguingly, lit2 appears to be the first gene in a polycistronic operon with well-characterized copper resistance determinants (24, 25). CopB is a P-type ATPase that exports copper across the cell membrane, thereby reducing intracellular copper concentrations (26). While L. monocytogenes CFSAN023459 does not carry an intact copB, a copB′ fragment is present, suggesting a gene deletion event. The metallochaperone CopZ binds cytoplasmic copper and shuttles it to CopB to be extruded from the cell or to CopY, a copper-responsive transcriptional regulator that derepresses expression of target copper resistance-related genes (27–29). These genes are followed by the genes encoding CueO, a multicopper oxidase that helps detoxify copper by oxidizing Cu+ to Cu2+ (30), and a two-component signal transduction system, CopRS, that senses extracellular copper and induces expression of copper resistance genes (31–33). Together, these proteins maintain copper homeostasis and provide resistance to elevated levels of copper. Divergently transcribed from the lit2-copper resistance operon is a second copy of another lipoprotein-biosynthetic gene, lgt, here known as “lgt2,” again to differentiate it from the chromosomal copy, “lgt1.”
Lit2 is a lipoprotein intramolecular transacylase.
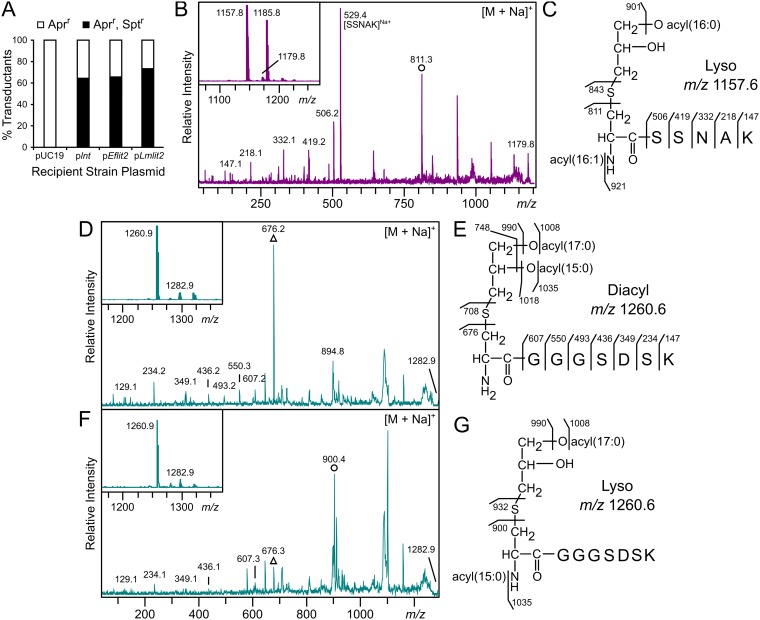
The colocalization of lgt2 with lit2 strongly suggests Lit2 is indeed a lipoprotein intramolecular transacylase, and the Lit2 orthologs are more similar to each other than to the experimentally characterized E. faecalis ATCC 19433 Lit1 (Fig. 1A). To confirm this, a P1vir cotransduction linkage analysis assay was performed to assess whether lit2 could functionally replace the lipoprotein N-acyltransferase lnt gene in E. coli. If functional, lyso-LP formation would allow proper lipoprotein trafficking to the outer membrane (12). A donor lysate of E. coli strain TXM541 (lnt::Sptr chiQ::Aprr) was transduced into recipient strains harboring the experimental plasmid pUC19, plnt, pEflit2, or pLmlit2. Apramycin-resistant (Aprr) colonies marked successful transduction events, while cotransduction of spectinomycin resistance (Sptr) demonstrated that lnt can be functionally replaced (Fig. 2A). While lnt could not be deleted from the control pUC19 strain, the lnt::Sptr allele could be established at frequencies comparable to the lnt-expressing positive control when either E. faecalis TX1342 lit2 or L. monocytogenes CFSAN023459 lit2 was present (12). This confirms Lit2 can functionally substitute for lnt.
FIG 2.
Lit2 is a functional lipoprotein intramolecular transacylase. (A) The linked lnt::Sptr and chiQ::Aprr markers were cotransduced by P1vir into recipient strains carrying pUC19, plnt, pEflit2, or pLmlit2. The percentage of transductants resistant to apramycin alone versus both apramycin and spectinomycin are shown (n = 24 to 40) and compared to negative (pUC19) and positive (plnt) controls (12). (B) Trypsinized N-terminal lipopeptides of Lpp from the lnt-null strain KA811 expressing E. faecalis lit2 were analyzed by MALDI-TOF MS. The protonated m/z 1,157.8 and sodiated m/z 1,179.8 parent ions are shown (inset), with the latter further fragmented by MS-MS. (C) This spectrum was used to assign Lpp structure from KA811 as the lyso form. Note that the m/z 1,185.8 peak in the parent spectrum is consistent with a C16:0, C18:1 acyl chain combination. (D and F) Trypsinized N-terminal lipopeptides of the lipoprotein KO07_11695 from wild-type L. monocytogenes ATCC 19115 (D) and the corresponding L. monocytogenes lit2-expressing strain KA849 (F) were analyzed by MALDI-TOF MS. The parent spectra (insets) display the protonated m/z 1,260.9 and sodiated m/z 1,282.9 parent ions. (E and G) The sodiated ions were fragmented by MS-MS (D and F), revealing production of DA-LP in KA847 (E) and the lyso-LP when L. monocytogenes lit2 was expressed (G). (D and F) The structurally diagnostic dehydroalanyl ions for the diacylglycerol-modified (triangle) and the lyso (circle) lipopeptides are indicated.
To determine the lipoprotein form, Lpp(K58A) from E. coli strain KA811 (lnt::Sptr pEflit2) was analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Similar to when Lpp was modified by Lit1 (12), the parent spectrum revealed ions at m/z 1,157.8 and 1,179.8, consistent with the predicted mass of the N-terminal CSSNAK tryptic lipopeptide possessing two acyl chains (C16:0 and C16:1) and ionizing as protonated and sodium adducts, respectively (Fig. 2B, inset). Fragmentation of the sodium adduct by tandem MS (MS-MS) (Fig. 2B), which fragments preferentially toward the dehydroalanyl peptide (see Fig. S1A in the supplemental material) (34), exhibited Lpp’s y series ions and a prominent ion at m/z 811.3. This ion is diagnostic of a monounsaturated C16:1 N-acyl chain on the lipopeptide (Fig. 2C), demonstrating that Lpp is converted to the lyso form when E. faecalis TX1342 lit2 is expressed in E. coli.
To show that L. monocytogenes CFSAN023459 Lit2 similarly functions as an intramolecular transacylase, L. monocytogenes lit2 was cloned under a xylose-inducible promoter into L. monocytogenes ATCC 19115, a type strain of L. monocytogenes that lacks any lit sequence orthologs. Lipoproteins were extracted from cultures grown with 2% xylose and wild-type ATCC 19115 cells and then separated by SDS-PAGE (see Fig. S2 in the supplemental material). A predicted peptide ABC transporter substrate-binding lipoprotein, KO07_11695, previously studied by Kurokawa et al. (11), was chosen for further analysis by MALDI-TOF MS. The parent spectra of both strains, regardless of L. monocytogenes lit2 expression, contained peaks at m/z 1,260.9 and 1,282.9 corresponding to the protonated and sodiated ions of the diacylated N-terminal CGGGSDSK tryptic lipopeptide (Fig. 2D and E, insets). To differentiate between the DA- and lyso-lipopeptides, which have identical masses, the sodiated ion at m/z 1,282 from each sample was further fragmented. While the MS-MS spectra of both strains displayed the same CGGGSDSK y series peptide ions, the dehydroalanyl m/z ion differed depending on lit2 expression. The type strain spectrum had a prevalent ion at m/z 676, corresponding to the dehydroalanyl CGGGSDSK peptide with a free α-amino terminus (Fig. 2D and E), while introduction of lit2 resulted in a shift to m/z 900.4. This mass is consistent with an added C15:0 N-acyl chain (Fig. 2F and G) and is further supported by MS-MS spectra of the protonated parent m/z 1,260 ion (see Fig. S1B and C). Thus, lipoproteins in the L. monocytogenes type strain ATCC 19115 are DA-LPs, and expression of L. monocytogenes CFSAN023459 lit2 alone is sufficient for lipoprotein conversion.
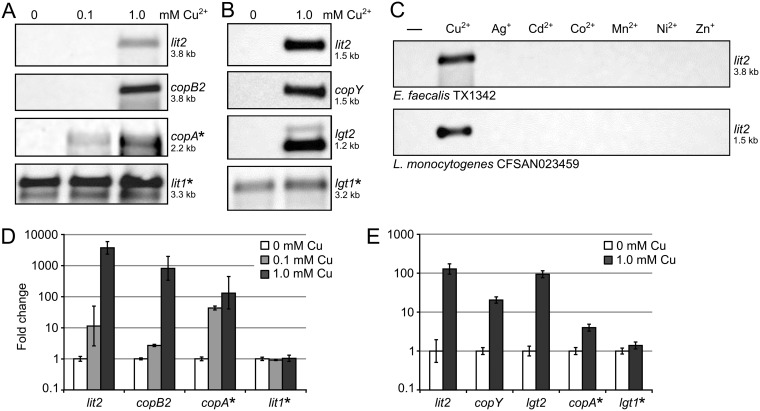
Copper induces expression of lit2.
As the lit2-copper resistance operon (Fig. 1B) contains orthologs of CopY and CopR, two known copper-responsive transcriptional regulators (27, 31), we hypothesized that lit2 expression is induced by excess copper. To test this, we performed a series of Northern blots in E. faecalis TX1342 (Fig. 3A) and L. monocytogenes CFSAN023459 (Fig. 3B). Copper concentrations were chosen so that there was no effect on growth rates (data not shown). In both strains, lit2 transcripts were detected only when cells were grown with 1 mM copper, indicating tight basal regulation. The observed lit2 transcript sizes in both strains were consistent with a polycistronic operon beginning with lit2 and terminating after copZ at the predicted rho-independent terminator (Fig. 1B). Copper-induced expression of select downstream genes (copB2 for E. faecalis TX1342 and copY for L. monocytogenes CFSAN023459) produced identical transcript lengths, confirming coexpression on a single transcript. We also probed for expression of the divergently oriented lipoprotein-related gene lgt2 in L. monocytogenes CFSAN023459 and likewise observed copper-dependent expression. To check whether copper-induced expression of all lipoprotein-related genes is a general response, we probed the expression of either chromosomal lit1 (in L. monocytogenes CFSAN023459) or lgt1 (in E. faecalis TX1342). Transcripts were constitutively expressed at similar levels regardless of the presence of copper in both cases. Induction was specific to copper, as neither strain upregulated lit2 when grown with alternative metals (silver, cadmium, cobalt, magnesium, nickel, or zinc) (Fig. 3C). Quantification of the copper-induced transcriptional response by reverse transcription-quantitative PCR (RT-qPCR) was consistent with Northern blotting (Fig. 3D and E). Collectively, the data indicate that lit2 and lgt2 are integral parts of the copper-responsive regulon.
FIG 3.
Copper induces expression of the lit2-copper resistance operon. (A and B) RNA was extracted from E. faecalis TX1342 cells grown with 0, 0.1, or 1.0 mM CuCl2 (A) and L. monocytogenes CFSAN023459 cells grown with 0 or 1.0 mM CuCl2 (B) and probed for expression of the indicated genes. Chromosomal genes are indicated by asterisks. (C) RNA was extracted from E. faecalis TX1342 (top) and L. monocytogenes CFSAN023459 (bottom) cells induced with various metal ions and probed for lit2 expression. Estimated transcript lengths are indicated. (D and E) Expression of the indicated target genes was measured by RT-qPCR from E. faecalis TX1342 (D) and L. monocytogenes CFSAN023459 (E) cells grown with 0, 0.1, or 1.0 mM copper. Expression was normalized in each strain against the internal control gene gyrA. The data are shown as the means ± standard deviations of the results of three replicates.
Copper induces conversion of lipoproteins from the diacylglycerol-modified to the lyso form in L. monocytogenes CFSAN023459.
While E. faecalis TX1342 already encodes a constitutively expressed chromosomal lit gene and thus addition of a second copy in lit2 would not be expected to alter the lipoprotein form, L. monocytogenes normally makes DA-LP lacking N-terminal modifications (Fig. 2D). Therefore, environmental isolates of L. monocytogenes, like L. monocytogenes CFSAN023459, that have acquired the lit2-copper resistance operon may elaborate lyso-LP, specifically when exposed to elevated copper levels.
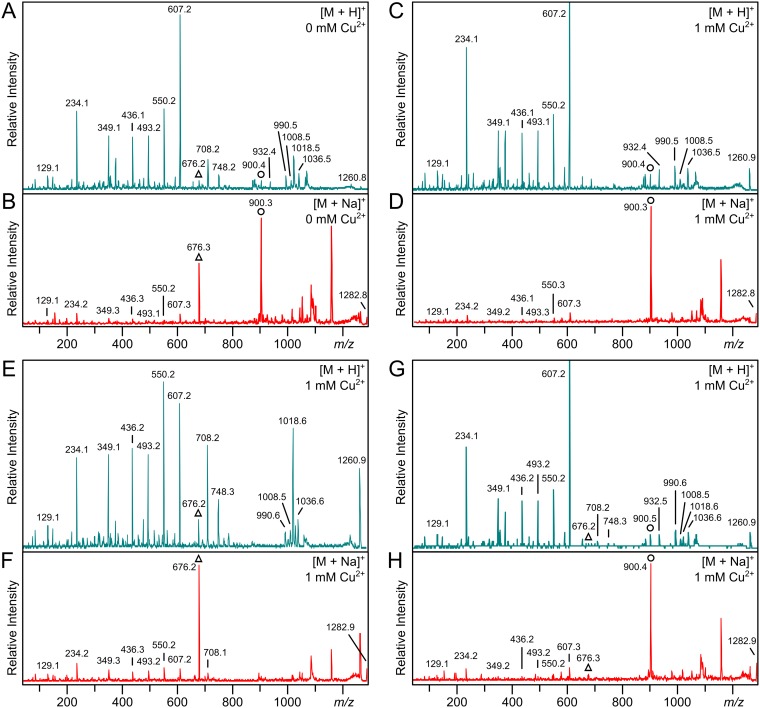
Lipoproteins were extracted following growth with or without 1 mM copper, which did not alter the overall lipoprotein profile or relative abundance according to SDS-PAGE (see Fig. S2). The lipoprotein KO07_11695 was again chosen for analysis in L. monocytogenes CFSAN023459, and the two samples had identical parent spectra by MALDI-TOF MS analysis (see Fig. S3 in the supplemental material). Prevalent ions at m/z 1,260 and m/z 1,282, along with the same corresponding y series ions, could likewise be assigned to the diacylated N-terminal CGGGSDSK peptide whether copper was added or not. However, MS-MS analysis revealed pronounced differences in acylation of the product ions (Fig. 4; Fig. 2E and G shows KO07_11695’s N-terminal lipopeptide structures). When the strain was grown without copper, fragmentation of the protonated parent m/z 1,260 ion yielded DA-LP-related ions at m/z 676, 708, 748, and 1,018, corresponding to dehydroalanyl CGGGSDSK, thiolated peptide, and the peptide with either both fatty acids (C32:0) or a single C15:0 fatty acid (C14H28COOH) lost (Fig. 4A). Additional ions at m/z 900 and 932 were diagnostic of the lyso form and correspond to the N-acyl(C15:0)-dehydroalanyl peptide and the thiolated N-acyl(C15:0)-peptide. Additional ions at m/z 990, 1,008, and 1,035 can be assigned to either the DA-LP or lyso-LP form and correspond to the parent ions with a C17:0 fatty acid (C16H32COOH), C17:0 ketene (C15H29CH=C=O), and C15:0 ketene (C13H27CH=C=O) lost, respectively. Fragmentation of the sodiated parent at m/z 1,282 also resulted in abundant peaks at m/z 676 and 900, evidence of both nonacylated and N-acylated dehydroalanyl forms (Fig. 4B; Fig. 2G shows the lipopeptide structure). From this, we conclude that a mixture of DA-LP and lyso-LP exists within a cell population even without added copper, suggesting some basal lit2 expression. We could not reliably determine the ratio of DA-LP to lyso-LP due to inherent differences in ionization efficiency and multiple ion adduct formation.
FIG 4.
Copper induces conversion of lipoproteins from DA-LP to lyso-LP. (A and B) MS-MS spectra of the protonated m/z 1,260 (A) and sodiated m/z 1,282 (B) parent ions of lipoprotein KO07_11695 from L. monocytogenes CFSAN023459 grown in the absence of copper. (C and D) MS-MS spectra of the protonated m/z 1,260 (C) and sodiated m/z 1,282 (D) parent ions of L. monocytogenes strain CFSAN023459 grown with 1 mM copper. (E and F) MS-MS spectra of the protonated m/z 1,260 (E) and sodiated m/z 1,282 (F) parent ions of the Δlit2 strain grown with 1 mM CuCl2. (G and H) MS-MS spectra of the protonated m/z 1,260 (G) and sodiated m/z 1,282 (H) parent ions of the Δlit2 strain back-complemented with pPxylLmlit2 grown with 1 mM CuCl2 and 2% xylose. The diagnostic dehydroalanyl ions for the diacylglycerol-modified (triangles) and the lyso (circles) lipopeptides are indicated. The parent spectra are shown in Fig. S3.
In contrast to the mixed lipoprotein profile from extracts grown without copper, only the lyso-LP-specific ions at m/z 900 and 932 were observed when L. monocytogenes CFSAN023459 was cultured with copper (from protonated [Fig. 4C] and sodiated [Fig. 4D] parent ions). There was also a marked absence of DA-LP-related ions. To see whether copper-induced lipoprotein conversion was global, two additional lipoproteins were analyzed (see Fig. S4 and S5 in the supplemental material). A clear enrichment in the lyso-LP population, as seen with KO07_11695, was likewise observed. To demonstrate that copper itself is not responsible for conversion of lipoproteins to the lyso form, we deleted lit2 from L. monocytogenes CFSAN023459 and grew the resulting strain with 1 mM copper. Even when grown with copper, only DA-LP could be detected (Fig. 4E and F). Back complementation with a plasmid-borne lit2 copy of the gene completely restored lyso-LP production (Fig. 4G and H), consistent with global conversion of lipoproteins to the lyso form upon induction of the lit2-copper resistance operon cassette.
TLR2 stimulation by synthetic lyso form lipopeptides.
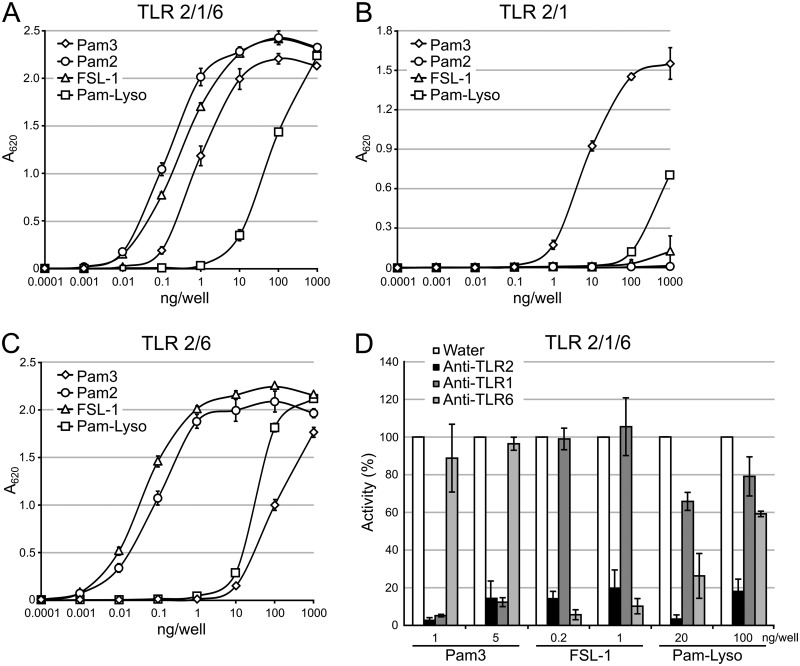
As lipoproteins are converted from DA-LP to the lyso form in L. monocytogenes CFSAN023459, we questioned if this alteration affects recognition by TLR2. While it has been established that DA-LPs are sensed by the TLR2/TLR6 heterodimer and TA-LPs are sensed by TLR2/TLR1 (4, 6, 22, 23, 35), it is not clear which heterodimer senses the lyso form and with what sensitivity. Since little is known about the specific TLR2 response to lyso-LP (11), we first measured TLR2 signaling activity using a set of defined synthetic lipopeptides and a HEK-Blue-TLR2/1/6 reporter cell line that secretes alkaline phosphatase (SEAP) upon activation of the TLR2-responsive NF-κB transcription factor. TLR2 activation was measured using Pam3CSK4, Pam2CSK4, and PamC(Pam)SK4, representing the TA-LP, DA-LP, and lyso-LP forms, respectively, as well as the Mycoplasma salivarium-derived DA-LP FSL-1 ligand (Pam2CGDPKHPKSF). The DA-LP Pam2CSK4 and FSL-1 ligands elicited signal at the lowest concentration (0.01 ng/well), with an approximately 10-fold-higher concentration (0.1 ng/well) needed for half-maximal activation (50% effective concentration [EC50]) (Fig. 5A). Both the detection limit (0.1 ng/well) and the EC50 (1 ng/well) were shifted to slightly less than 10-fold higher for the TA-LP Pam3CSK4 in comparison to the DA-LP ligand set. The lyso-LP PamC(Pam)SK4 was by far, however, the weakest TLR2/TLR1/TLR6 ligand, with a shift higher in the detection limit and EC50 of well over 2 log units in comparison to the DA-LP standards (Fig. 5A).
FIG 5.
TLR2 response to synthetic lipopeptides. (A) HEK-Blue-TLR2/1/6 cells were exposed to 10-fold dilutions of the synthetic lipopeptides Pam3CSK4 (Pam3) and PamC(Pam)SK4 (Pam-Lyso), representing the TA-LP and lyso-LP, respectively, as well as Pam2CSK4 (Pam2) and FSL-1, both DA-LP ligands. (B and C) The same lipopeptides were exposed to HEK-Blue cells expressing only TLR2/TLR1 (B) or TLR2/TLR6 (C). (D) HEK-Blue-TLR2/1/6 cells pretreated with 10 μg/ml TLR-neutralizing antibodies were then exposed to two optimized concentrations of Pam3CSK4, PamC(Pam)SK4, and FSL-1. The percent activity when normalized to the water control wells is indicated. The data shown are the means ± standard deviations of the results of three biological replicates.
Structural and biochemical studies of TLR2/TLR1 and TLR2/TLR6 have shown that conventional DA-LP lipopeptides signal through TLR2/TLR6 while TA-LP lipopeptides signal through TLR2/TLR1 (22, 23). Since TLR2 has a larger, diffuse binding pocket for the thioether-linked diacylglycerol moiety, ligand specificity is predominantly imparted by TLR1 accommodating the extra N-acyl chain of TA-LP in a hydrophobic binding pocket that is inaccessible in TLR6 (23, 35). At least two distinct binding modes could be envisioned for lyso-LP: one where the monoacyl-glycerol acyl chain remains in TLR2 while the N-acyl is bound within TLR1 in a TLR2/TLR1 heterocomplex, and a second whereby both acyl chains bind within TLR2 in a TLR2/TLR6 complex. Therefore, we used TLR2/TLR1- and TLR2/TLR6-specific reporter cell lines to probe specificity. Using HEK-Blue-TLR2/1 cells, Pam3CSK4 was the highest-affinity ligand, while the DA-LPs Pam2CSK4 and FSL-1 elicited little signal even at the highest concentration tested (1,000 ng/well), as expected (Fig. 5B). The lyso form PamC(Pam)SK4 ligand was 2 log units weaker than TA-LP with respect to the detection limit and EC50, albeit still a slightly better ligand than the DA-LPs at the highest concentration tested (Fig. 5B). In HEK-Blue-TLR2/6 cells, the PamC(Pam)SK4 detection limit and EC50 were also over 2 log units weaker than the canonical DA-LP ligand (Fig. 5C). Collectively, the data are consistent with the lyso form PamC(Pam)SK4 lipopeptide being predominantly sensed by the TLR2/TLR6 heterodimer, with limited TLR2/TLR1 activation when present at high concentrations. Engagement of TLR2/TLR6, however, remained much weaker than the cognate DA-LP standards.
To support this finding, we treated HEK-Blue-TLR2/1/6 cells with neutralizing antibodies specific for each TLR component and then challenged with an optimized concentration of lipopeptide standard. Two separate ligand concentrations, each chosen to achieve robust knockdown of TLR2 signaling (greater than 80%) and within the linear range of the dose-response curve (Fig. 5A), were utilized with equal amounts of neutralizing antibody (Fig. 5D). A more pronounced signal knockdown of the lyso form PamC(Pam)SK4 standard was observed when TLR6 was neutralized than with TLR1 at both ligand concentrations tested. Taken together, these results suggest that while the lyso form can indeed be sensed by TLR2/TLR1, the bulk of signaling activity observed in HEK-Blue-TLR2/1/6 cells is due to TLR2/TLR6.
TLR2 stimulation by whole bacteria.
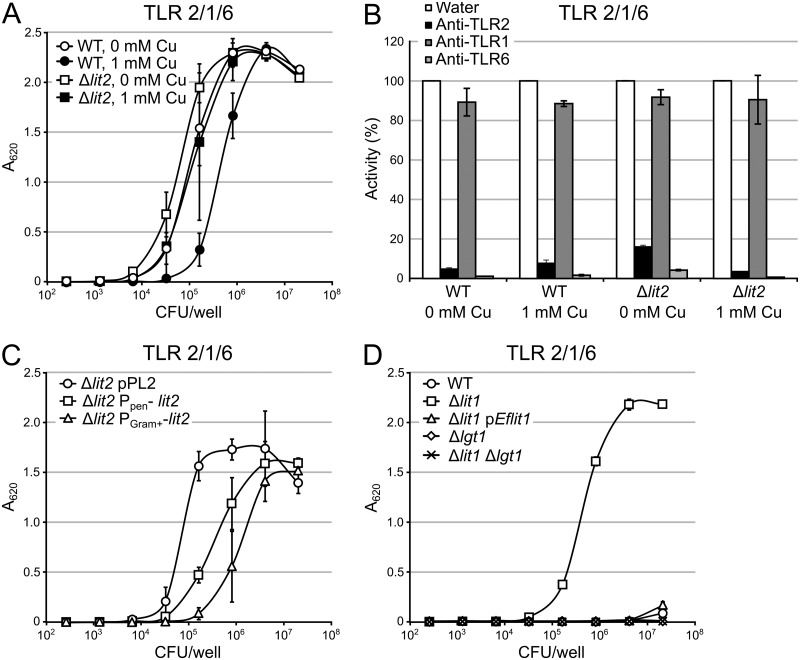
As copper-induced expression of lit2 converts lipoproteins to the lyso form in L. monocytogenes CFSAN023459 (Fig. 4) and lyso-LP is a weaker TLR2 agonist than the cognate DA-LP (Fig. 5), we hypothesized that copper-rich growth environments could modulate TLR2 detection, as well. Thus, we repeated the previous TLR2 assays using heat-inactivated whole L. monocytogenes CFSAN023459 cells grown with or without 1 mM copper (Fig. 6A). Using HEK-Blue-TLR2/1/6 reporter cells, L. monocytogenes CFSAN023459 cells elaborating mostly DA-LP (wild type grown without copper and Δlit2 grown with or without copper) are detected at approximately 5-fold-fewer CFU per milliliter with respect to the detection limit and EC50 than lit2-induced L. monocytogenes CFSAN023459 cells (wild type grown with copper). When the contributions of TLR2/TLR1 and TLR2/TLR6 were isolated using TLR2 heterodimer-specific reporter assays, there was a similar ligand potency trend using TLR2/TLR6 cells (see Fig. S6A in the supplemental material). Interestingly, none of the samples, including those with mostly lyso-LP compositions, were able to measurably induce NF-κB in the TLR2/TLR1 reporter cell line (see Fig. S6B). We next used neutralizing antibodies to probe specificity in TLR2/TLR1/TLR6 when using heat-inactivated bacterial preparations (Fig. 6B). There was little to no reduction in signal when TLR1 was neutralized, while TLR6 neutralization reduced TLR2 stimulation by more than 80%. This implicates the TLR2/TLR6 heterodimer as the critical TLR2 heterodimer for sensing lyso-LP.
FIG 6.
TLR2 response to whole bacteria. (A) HEK-Blue-TLR2/1/6 cells were exposed to 5-fold dilutions of heat-inactivated whole bacterial cells of L. monocytogenes CFSAN023459 (wild type [WT]) grown with or without 1 mM copper, as well as the derivative Δlit2 cells grown with or without 1 mM CuCl2. (B) HEK-Blue-TLR2/1/6 cells were pretreated with 10 μg/ml of TLR-neutralizing antibodies and then exposed to the same bacterial cell preparations as in panel A. The “WT + 1 mM Cu” sample was added to 8.0 × 105 CFU/ml, while the others were added to 3.2 × 104 CFU/ml. The percent activity normalized to the water control is indicated. (C and D) HEK-Blue-TLR2/1/6 cells were exposed to 5-fold dilutions of heat-inactivated, whole bacterial cells of strains L. monocytogenes CFSAN023459 (C) and E. faecalis ATCC 19433 (D). The data are shown as the means ± standard deviations of the results of three biological replicates.
Lipoprotein conversion in the wild-type L. monocytogenes CFSAN023459 strain even when grown with copper under our conditions is not complete (Fig. 4; see Fig. S4 and S5). Considering the difference in potency between the DA-LP and lyso-LP, even a small residual population of DA-LP would be expected to make an outsize contribution to total signal in TLR2/TLR1/TLR6 reporter cells. To measure the effect of lit2 expression independently of endogenous copper induction, L. monocytogenes CFSAN023459 lit2 was placed under the control of the constitutive promoters Ppen and PGram+, an optimized Gram-positive promoter (36). According to the MS-MS spectra for these strains, PGram+-lit2 (strain KA1179) realizes a more complete conversion to lyso-LP than PPen-lit2 (strain KA1178) (see Fig. S7 in the supplemental material). The lyso form-producing Ppen-lit2 and PGram+-lit2 strains elicited signals at approximately 10- and 25-fold higher loads (CFU per milliliter) than the parent Δlit2 strain (Fig. 6C). The TLR2 response thus correlates with the relative extent of lipoprotein population conversion from DA-LP to lyso-LP. This trend was consistent when measured with TLR2/TLR6 reporter cells (see Fig. S8A in the supplemental material), and once again, little to no signal was detected with TLR2/TLR1 cells (see Fig. S8B).
As lyso-LP formation in L. monocytogenes CFSAN023459 reduced detection by TLR2/TLR6, we sought to test whether this is true in other lyso-LP-producing bacteria where the lit gene is naturally present and chromosomally integrated. We thus measured TLR2 response to heat-inactivated E. faecalis ATCC 19433 cells and compared it to that to the isogenic Δlit1 derivative strain KA543 (12) using HEK-Blue-TLR2/1/6 reporter cells. This resulted in the most remarkable shift, with enhanced TLR2-mediated detection of the DA-LP Δlit1 strain at inputs (CFU per milliliter) 2 to 3 log units lower than the lyso-LP-producing wild type (Fig. 6D). Complementation of lit1 on a plasmid restored signal to that of wild-type cells. Deletion of lgt, thus abrogating production of lipoproteins altogether, resulted in no detectable signal and confirmed that the bulk of observed signal in fact originates from differences in lipoprotein acylation. These results were echoed in TLR2/TLR6 cells, while no signal was detected with TLR2/TLR1 cells for either DA-LP or lyso-LP (see Fig. S9 in the supplemental material).
DISCUSSION
We originally identified Lit1 in E. faecalis and noted the narrow distribution of similar sequences in the NCBI database to only those bacteria expressing lyso-LP (12). Further phylogenomic analysis here revealed two distinct clades: (i) lit1, located on the chromosome without synteny, and (ii) lit2, located on a mobile genetic element within a copper resistance operon (Fig. 1). We confirmed that Lit2 is indeed a functional lipoprotein transacylase (Fig. 2), that expression of lit2 is induced specifically by copper (Fig. 3), and that lit2 alone is sufficient to convert lipoproteins from DA-LP to lyso-LP in L. monocytogenes CFSAN023459 (Fig. 4). Finally, we demonstrate that the shift in lipoprotein N-terminal structure to lyso tangibly impacts TLR2 detection (Fig. 5 and 6).
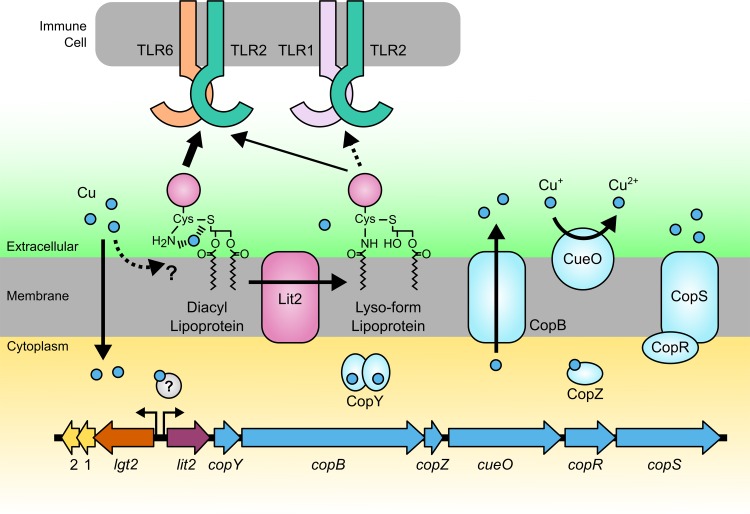
Although copper is an essential nutrient, it is highly toxic in excess and is often used as an antimicrobial in health care and agriculture settings (37–39). Bacteria have in turn evolved mechanisms to combat copper toxicity, many of which are subject to horizontal gene transfer (40, 41). Indeed, the presence of a lit2-copper resistance operon has been noted before in the L. monocytogenes dairy isolate DRDC8, where it is part of a freely replicating plasmid containing multiple heavy metal resistance determinants (42), similar to the L. monocytogenes CFSAN023459 plasmid studied here, and again in a comparative genomics study of copper resistance determinants in E. faecalis strains isolated from farms using copper-supplemented pig feed (43). In the latter, the lit2-copper resistance element is chromosomally integrated in a pathogenicity/fitness island, likely through transposition, as in E. faecalis TX1342. Copper resistance orthologs of CopY, a copper-sensing transcriptional regulator (27, 44), CopB, a copper efflux pump (26), CopZ, a metallochaperone (28), CueO, a copper oxidase (30), and CopRS, a two-component response regulator (33), are all associated with lit2 (Fig. 1B). Thus, selection for copper resistance may also lead to lipoprotein conversion and in turn to the unanticipated modulation of TLR2 signaling (Fig. 7).
FIG 7.
Model of transposon-mediated response to copper. Copper enters the cell nonspecifically or via transporters. The proposed high-affinity association of copper at the N terminus of DA-LP is shown. Elevated copper levels induce transcription through an undetermined copper-dependent regulator. Expression of genes downstream of lit2 contributes to copper resistance by various mechanisms, including copper efflux, copper oxidation, and regulation of additional genes. Coinduction of Lit2 simultaneously converts lipoproteins from the DA-LP form to the lyso-LP, which is proposed to reduce copper coordination at the lipoprotein N terminus. While both DA-LP and lyso-LP are sensed by the TLR2/TLR6 heterodimer, the lyso form is overall a less potent ligand at TLR2/TLR6 than DA-LP and a poor TLR2/TLR1 ligand.
There is a growing appreciation of the structural diversity found among the N termini of bacterial lipoproteins (6, 11, 19, 21). While understanding the physiological function and consequences of N-acylation has lagged, it is clear that N-acylation is not simply required for lipoprotein trafficking from the inner to the outer membrane of Gram-negative bacteria by the Lol transport system (16, 17, 45, 46). Rather, the Lol transporter substrate selectivity may be more of a quality control check to ensure that only mature N-acylated TA-LPs are transported to the outer membrane. Why lipoprotein N-acylation is advantageous in bacterial membranes is an outstanding question. The widespread occurrence of N-terminal modification, though, does suggest a more universal selective pressure in nature beyond Gram-negative-specific transport (14, 47). The genetic coregulation between lit2 and copper tolerance determinants in Firmicutes reported here, in tandem with the peculiar distribution of N-terminal modification machinery, may provide a clue. Bioavailable copper in the more soluble Cu2+ is thought to have accumulated largely after environmental oxygenation (48). Thus, a preexisting Lgt-Lsp lipoprotein pathway would need to be edited postspeciation on a strain-by-strain basis. Acute copper challenge, typical in farms and health care settings, may likewise be contributing to acquisition of N-acylation genes within DA-LP-producing bacteria. When lipoprotein N-terminal modification is viewed as a postpathway edit, the seemingly random species level N-terminal structural variation becomes less puzzling.
This is not the first experimental connection between lipoproteins and copper, as the lipoprotein-N-acylating E. coli lnt gene was originally named cutE due to accumulation of intracellular copper and enhanced susceptibility in cutE- and lnt-defective mutants (49). However, we have yet to observe comparable changes in copper sensitivity in Δlit1 and/or Δlit2 mutants in E. faecalis TX1342 or L. monocytogenes CFSAN023459 under standard growth conditions (unpublished data). Nevertheless, TA-LPs, including Braun's lipoprotein (Lpp) (50), are among the most abundant protein classes in Gram-negative bacteria (51). Thus, lipoprotein N-acylation appears to be a general defense strategy against copper. DA-LPs have a thioether sulfur atom, as well as a free α-amino group on the N-terminal cysteine residue, which can conceivably coordinate copper in a high-affinity complex (Fig. 7). Any structural modifications that weaken copper cation coordination, such as a decrease in the nucleophilicity of the amino terminus, would be expected to limit lipoprotein-copper interactions. Indeed, the common denominator among all lipoprotein N-terminal modifications discovered to date (TA-LP with N-acyl/N-acetyl, N-peptidyl, and lyso-LP) is α-amide formation, which delocalizes electron density on the α-amino nitrogen and the corresponding copper coordination potential. This would be advantageous, as copper binding at the cell membrane interface can increase copper uptake and disrupt metal homeostasis (52) or directly impact membrane integrity by promoting reactive oxygen species formation (53). Lipoprotein function itself could also be impacted by enhancing cysteine thioether oxidation. Further studies will be necessary to quantify copper binding to lipoproteins of various N-terminal structures.
What is clear is that induction by copper of the lit2-copper resistance operon leads to lipoprotein conversion from DA-LP to the lyso form, and this alters recognition by TLR2 in HEK-TLR2 reporter cell assays (Fig. 5 and 6). DA-LPs are sensed by the TLR2/TLR6 heterodimer, while TA-LPs are sensed by TLR2/TLR1 (4). Limited studies measuring the TLR2 response to the lyso form using single purified lipoproteins conducted thus far have revealed complex, mixed TLR2 heterodimer activation (11). Our results show that while the lyso form can be sensed by both the TLR2/TLR1 and TLR2/TLR6 heterodimers, lyso-LP is a less potent TLR2 agonist than either DA-LP or TA-LP. There was a clear inverse correlation between lyso-LP content and NF-κB reporter activation, whether lipopeptide standards or heat-inactivated bacteria were used. Once more, there was a strong preference for lyso-LP engaging TLR2/TLR6 over TLR2/TLR1, which is somewhat surprising given crystallographic structures clearly showing the TA-LP N-acyl chain binding to the hydrophobic channel of TLR1 (22). The fact that lyso-LP, which also contains an N-acyl chain, interacts preferentially with the TLR2/TLR6 heterodimer suggests that complete TLR2 ligand engagement with both acyl chains is likely the most critical determinant driving receptor heterodimerization and that lyso-LP acyl chain distribution is nonoptimal for this binding mode. This in turn may dampen the host innate immune response, particularly during the onset of infection, when TLR2 response is most crucial (4). It will be interesting to determine whether the results observed here in nonimmune HEK reporter cells directly translate to primary immune cells and ultimately infection models. Roles for other chaperones and adapter proteins in recognizing noncanonical or weak TLR2 ligands, which may not be expressed in HEK cells, have been proposed (4). Regardless, the potential of copper use in health care and agriculture not only to spread copper resistance-conferring genes (43, 54) but also to alter the bacterial lipoprotein structural landscape and hence the TLR2 response, needs to be considered.
MATERIALS AND METHODS
Phylogenetic analysis.
Strains of interest were identified in a BLAST search using the protein sequence of Enterococcus faecalis ATCC 19433 Lit as the query. To identify isolates from partially assembled genomes, a tBLASTn search was performed against the database of whole-genome shotgun (wgs) contigs. Sequences were aligned using Muscle. A phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA7 (55).
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1 and primers in Table S1 in the supplemental material. E. faecalis TX1342 was grown in tryptic soy broth (TSB) at 37°C with agitation. Strain KA666 was grown with chloramphenicol (5 μg/ml) and nisin (100 ng/ml) when appropriate. L. monocytogenes CFSAN023459 contains two plasmids, CFSAN023459_01 (12,949 bp; GenBank accession no. NZ_CP014253.1) and CFSAN023459_02 (52,687 bp; GenBank accession no. NZ_CP014254.1); the latter contains the lit2-copper resistance operon. Listeria strains were grown in modified Hsiang-Ning Tsai medium (HTM) at 37°C with agitation (56). To create HTM+, HTM was supplemented with 0.1 mg/ml each of alanine, arginine, asparagine, aspartic acid, glutamine, glutamate, glycine, histidine, isoleucine, leucine, lysine, phenylalanine, proline, serine, threonine, and valine. Antibiotic markers were selected with chloramphenicol (2.5 μg/ml). Cells were induced with a final concentration of 1 mM copper(II) chloride or 2% (wt/vol) xylose, each added when cultures had reached an optical density at 600 nm (OD600) of 0.1 unless otherwise noted.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Reference |
|---|---|---|
| Escherichia coli | ||
| S17-1 | recA pro hsdR RP42Tc::MuKm::Tn7 integrated into the chromosome | |
| BW25113 | K-12 wild type [Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514] | |
| TXM327 | lpp::Cmr | 12 |
| TXM541 | gut::Kanr-rrnB TT-araC-PBAD-lnt lnt::Sptr chiQ::Aprr | 12 |
| KA707 | TXM327 + pLmlit2 | This study |
| KA708 | TXM327 + pEflit2 | This study |
| KA808 | KA708 chiQ::Aprr lnt::Sptr | This study |
| KA811 | KA808 + pKA810 | This study |
| Listeria monocytogenes | ||
| KA694 | CFSAN023459 with plasmid CFSAN023459_02 (courtesy of Dwayne Roberson) | |
| KA738 | KA694 Δlit2 | This study |
| KA834 | KA834 + pPxylLmlit2 | This study |
| KA847 | L2 ATCC 19115 | |
| KA849 | KA847 + pPxylLmlit2 | This study |
| KA1171 | KA738 attB::pPL2 | This study |
| KA1178 | KA738 attB::PpenLmCFSANlit2 | This study |
| KA1179 | KA738 attB::PGram+LmCFSANlit2 | This study |
| Enterococcus faecalis | ||
| TXM465 | ATCC 19433 | |
| KA543 | TXM465 Δlit1 | 12 |
| KA666 | KA543 + pKA635 | 12 |
| KA693 | TX1342 (courtesy of Barbara E. Murray) | |
| GKM744 | KA543 Δlgt | This study |
| GKM760 | TXM465 Δlgt | This study |
| Plasmids | ||
| pKFC | Temperature-sensitive shuttle vector; Cmr | |
| pKFC (ts fix) | Cmr | This study |
| pPxylLmlit2 | pKFC (ts fixed)-PxylLmCFSANlit2; Cmr | This study |
| pKA635 | pMS3535-lit1; Eryr | 12 |
| pKA810 | pCL25-E. coli lppK58A-Strep tag; Trimr | This study |
| plnt | pUC19-E. coli lnt; Carr | 12 |
| pEflit2 | pUC19-E. faecalis TX1342 lit2; Carr | This study |
| pLmLit2 | pUC19-L. monocytogenes CFSAN023459 lit2; Carr | This study |
| pPL2 | Integration into Lm tRNAArg site; Cmr (courtesy of Richard Calendar) | 60 |
| pTXM1170 | pPL2-PpenLmCFSANlit2; Cmr | This study |
| PTXM1169 | pPL2-PGram+LmCFSANlit2; Cmr | This study |
Km, kanamycin; Kanr, kanamycin resistance; Cmr, chloramphenicol resistance; Sptr, spectinomycin resistance; Aprr, apramycin resistance; Eryr, erythromycin resistance; Trimr, trimethoprim resistance; Carr, carbenicillin resistance.
Construction of deletion strains and lit2 complementation plasmid.
An unmarked internal deletion of lit2 from plasmid CFSAN02359_02 was generated using the temperature-sensitive pKFC plasmid (57) and verified by PCR. To complement this deletion and for lit2 expression in L. monocytogenes ATCC 19115, the xylose-inducible promoter from pSPNprM-hp (a gift from Dieter Jahn; Addgene plasmid number 48120) and lit2 from CFSAN023459_02 were cloned into pKFC-ts fix that had been repaired for stable replication.
Transformation of L. monocytogenes.
Either electroporation or conjugation from E. coli S17-1 was employed to transform strains of L. monocytogenes. Electroporation was performed following the protocol described by Monk et al. (58). The target genes were cloned into the phage attachment site integrating vector pPL2 under the control of the indicated promoters (a gift from Richard Calendar [59]) by a 3-piece DNA fragment assembly (InFusion; Clontech). Constructs were introduced into recipient L. monocytogenes isolates through biparental conjugation.
Total RNA isolation.
RNA was extracted using an RNeasy Mini kit (Qiagen) with the following modifications. One-milliliter of cells at an OD600 of 1.0 was pelleted by centrifugation, treated with 1 ml of RNAlater for 20 min, and stored at −20°C until used. The cells were washed with 1 ml of TE buffer (10 mM Tris, pH 8.0, 1 mM EDTA), resuspended in 900 μl QIAzol lysis reagent, and combined with an equal volume of 0.1-mm zirconium beads. The cells were disrupted using a MagNA Lyser (Roche) at 7,000 rpm for two 20-s cycles with a 2-min rest on ice in between. Beads and unbroken cells were removed by centrifugation (3,000 × g for 1 min) and washed with an additional 300 μl of QIAzol lysis reagent, and the supernatants were pooled. After chloroform-induced phase separation, 500 μl of the aqueous phase was collected for subsequent steps. The optional centrifugation at maximum speed for 1 min was performed to dry the column. RNA was eluted with two 40-μl volumes of RNase-free water for a total of 80 μl. RNA was quantified by absorbance at 260 nm, and the rRNA integrity was assessed with a MOPS (morpholinepropanesulfonic acid)-formaldehyde-agarose gel.
Total RNA was prepared as described above for metal induction experiments. The MIC for each metal in TSB was determined for E. faecalis TX1342 (data not shown). Overnight cultures were then diluted in metal-supplemented TSB (1:100 [vol/vol]; 1 mM CuCl2, NiCl2, MnCl2, or ZnSO4; 0.015 mM AgNO3 or CdCl2; 0.25 mM CoCl2) and grown at 37°C until the OD600 reached 1 before harvesting cells for RNA isolation. For L. monocytogenes CFSAN023459, cells were grown to an OD600 of 0.3 and induced with 1 mM each metal for 1 h before harvesting.
Northern blotting.
Northern blots were performed using a NorthernMax kit (Ambion) according to the manufacturer’s instructions. Briefly, 500 ng of total RNA from L. monocytogenes and 2 μg from E. faecalis were separated on a MOPS-formaldehyde-agarose gel and transferred to a BrightStar-Plus positively charged nylon membrane (Invitrogen) using a Whatman Nytran SuPerCharge TurboBlotter kit (GE Healthcare Life Sciences) for 3.5 h. The membranes were cross-linked by baking at 80°C for 20 min. Biotin-labeled RNA probes were synthesized from DNA using a Maxiscript T7 transcription kit (Thermo Fisher), including the optional DNase digestion and cleanup with NucAway spin columns (Invitrogen), along with gene-T7-specific primer sets (see Table S1). Probes were added to 10 ng/ml in Ultrahyb ultrasensitive hybridization buffer (Invitrogen) and incubated at 72°C for 20 h. The membranes were washed as directed in the NorthernMax kit, with the two high-stringency washes performed at 68°C. RNA was visualized with a chemiluminescent nucleic acid detection kit (Thermo Fisher) according to the manufacturer’s instructions.
RT-qPCR.
A Maxima First Strand cDNA synthesis kit for RT-qPCR with double-stranded DNase (dsDNase) treatment (Thermo Fisher) was used to synthesize cDNA template from 150 ng of total RNA isolated as described above. PowerUp SYBR green master mix (Applied Biosystems) was used for RT-qPCRs, with gene-qPCR-specific primer sets (see Table S1) and measured using an Applied Biosystems 7300 real-time PCR machine. All data were measured in triplicate and normalized to an internal gyrA control, and relative expression levels were calculated using the 2−ΔΔCT method (60).
Purification of lpp(K58A)-Strep tag and lipoprotein extraction.
Lpp(K58A)-Strep tag was affinity column purified from E. coli strain KA811 and separated by SDS-PAGE using a 16.5% Tris-tricine gel as described previously (12). Unlabeled lipoproteins were extracted using the Triton X-114 phase-partitioning method and separated with a 12% Tris-glycine SDS-PAGE gel (12, 34).
Preparation of N-terminal tryptic lipopeptides for MALDI-TOF MS and MS-MS analysis.
Following transfer to a nitrocellulose membrane, bands were visualized by Ponceau S staining. The bands selected for mass spectrometric analysis were trypsinized overnight and eluted from the membrane as described previously (12, 34). Single or multiple 0.5-μl layers of lipopeptide in 10-mg/ml CHCA (α-cyano-4-hydroxycinnamic acid) were deposited onto a steel target plate. Mass spectra were collected using an Ultraflextreme (Bruker Daltonics) MALDI-TOF mass spectrometer in positive reflectron mode. MS-MS spectra were acquired on the same instrument in Lift mode.
TLR2 assays.
HEK293 NF-κB/SEAP-reporter cells (HEK-Blue) expressing human TLR2/TLR1/TLR6, TLR2/TLR1, and TLR2/TLR6 (InvivoGen, San Diego, CA) were cultivated in 75-cm2 culture flasks in 15 ml Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 U/ml penicillin, 50 mg/ml streptomycin, 100 mg/ml Normocin (InvivoGen), and 4 μl/ml HEK-Blue selection antibiotics at 37°C in 5% CO2. The triacylated Pam3CSK4 and diacylglycerol-modified Pam2CSK4 and FSL-1 (Pam2CGDPKHPKSF) synthetic lipopeptides were purchased from InvivoGen and the lyso form PamC(Pam)SK4 from EMC Microcollections (Tubingen, Germany). Cells were seeded in a 96-well plate at 25,000 cells per well in 180 μl of growth medium and stimulated for 20 h at 37°C in 5% CO2 with 20 μl of synthetic lipopeptide or heat-inactivated whole bacterial cells diluted in endotoxin-free water. Bacteria were grown to an OD600 of 0.5 in HTM+ for L. monocytogenes or TSB for E. faecalis, washed once with water, heat inactivated at 58°C for 1 h, and stored at −20°C. Heat inactivation of cells was verified by plating on solid medium. Where indicated, anti-hTLR2-IgA-, anti-TLR1-IgG-, and anti-hTLR6-IgG-neutralizing antibodies (InvivoGen) were added to a final concentration of 10 μg/ml and incubated for 1 h before challenge. NK-κB-dependent SEAP activity was measured using Quanti-Blue solution (InvivoGen) according to the manufacturer’s instructions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tatiana Laremore (Penn State Proteomics and Mass Spectrometry Core Facility, University Park, PA) for support with MALDI-TOF MS. We thank Barbara E. Murray (McGovern Medical School, University of Texas) for the gift of Enterococcus faecalis TX1342, Dwayne Roberson (Food and Drug Administration) for Listeria monocytogenes CFSAN023459, Dieter Jahn for pSPNprM-hp (Addgene plasmid no. 48120), and Richard Calendar (University of California Berkeley) for pPL2.
This work was funded by the National Institutes of Health (R01GM127482 to T.C.M.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00195-19.
REFERENCES
- 1.Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect Immun 79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buddelmeijer N. 2015. The molecular mechanism of bacterial lipoprotein modification—how, when and why?. FEMS Microbiol Rev 39:246–261. doi: 10.1093/femsre/fuu006. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen MT, Götz F. 2016. Lipoproteins of Gram-positive bacteria: key players in the immune response and virulence. Microbiol Mol Biol Rev 80:891–903. doi: 10.1128/MMBR.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliveira-Nascimento L, Massari P, Wetzler LM. 2012. The role of TLR2 in infection and immunity. Front Immunol 3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szewczyk J, Collet JF. 2016. The journey of lipoproteins through the cell: one birthplace, multiple destinations. Adv Microb Physiol 69:1–50. doi: 10.1016/bs.ampbs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J 279:4247–4268. doi: 10.1111/febs.12041. [DOI] [PubMed] [Google Scholar]
- 7.Sutcliffe IC, Russell R. 1995. Lipoproteins of Gram-positive bacteria. J Bacteriol 177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankaran K, Wu HC. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem 269:19701–19706. [PubMed] [Google Scholar]
- 9.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 188:2761–2773. doi: 10.1128/JB.188.8.2761-2773.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain M, Ichihara S, Mizushima S. 1982. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J Biol Chem 257:5177–5182. [PubMed] [Google Scholar]
- 11.Kurokawa K, Ryu K-H, Ichikawa R, Masuda A, Kim M-S, Lee H, Chae J-H, Shimizu T, Saitoh T, Kuwano K, Akira S, Dohmae N, Nakayama H, Lee BL. 2012. Novel bacterial lipoprotein structures conserved in low-GC content Gram-positive bacteria are recognized by Toll-like receptor 2. J Biol Chem 287:13170–13181. doi: 10.1074/jbc.M111.292235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armbruster KM, Meredith TC. 2017. Identification of the Lyso-form N-acyl intramolecular transferase in low-gc Firmicutes. J Bacteriol 199:e00099-17. doi: 10.1128/JB.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tschumi A, Nai C, Auchli Y, Hunziker P, Gehrig P, Keller P, Grau T, Sander P. 2009. Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J Biol Chem 284:27146–27156. doi: 10.1074/jbc.M109.022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutcliffe IC, Harrington DJ, Hutchings MI. 2012. A phylum level analysis reveals lipoprotein biosynthesis to be a fundamental property of bacteria. Protein Cell 3:163–170. doi: 10.1007/s13238-012-2023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta SD, Wu HC. 1991. Identification and subcellular localization of apolipoprotein N-acyltransferase in Escherichia coli. FEMS Microbiol Lett 62:37–41. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda A, Matsuyama S-I, Hara T, Nakayama J, Nagasawa H, Tokuda H. 2002. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J Biol Chem 277:43512–43518. doi: 10.1074/jbc.M206816200. [DOI] [PubMed] [Google Scholar]
- 17.Robichon C, Vidal-Ingigliardi D, Pugsley AP. 2005. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J Biol Chem 280:974–983. doi: 10.1074/jbc.M411059200. [DOI] [PubMed] [Google Scholar]
- 18.Widdick DA, Hicks MG, Thompson BJ, Tschumi A, Chandra G, Sutcliffe IC, Brülle JK, Sander P, Palmer T, Hutchings MI. 2011. Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol Microbiol 80:1395–1412. doi: 10.1111/j.1365-2958.2011.07656.x. [DOI] [PubMed] [Google Scholar]
- 19.Asanuma M, Kurokawa K, Ichikawa R, Ryu KH, Chae JH, Dohmae N, Lee BL, Nakayama H. 2011. Structural evidence of α-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J 278:716–728. doi: 10.1111/j.1742-4658.2010.07990.x. [DOI] [PubMed] [Google Scholar]
- 20.Kurokawa K, Lee H, Roh K-B, Asanuma M, Kim YS, Nakayama H, Shiratsuchi A, Choi Y, Takeuchi O, Kang HJ, Dohmae N, Nakanishi Y, Akira S, Sekimizu K, Lee BL. 2009. The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for Toll-like receptor 2. J Biol Chem 284:8406–8411. doi: 10.1074/jbc.M809618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen M-T, Uebele J, Kumari N, Nakayama H, Peter L, Ticha O, Woischnig A-K, Schmaler M, Khanna N, Dohmae N, Lee BL, Bekeredjian-Ding I, Götz F. 2017. Lipid moieties on lipoproteins of commensal and non-commensal staphylococci induce differential immune responses. Nat Commun 8:2246. doi: 10.1038/s41467-017-02234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik S-G, Lee H, Lee J-O. 2007. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Kang JY, Nan X, Jin MS, Youn S-J, Ryu YH, Mah S, Han SH, Lee H, Paik S-G, Lee J-O. 2009. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Solioz M, Stoyanov JV. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev 27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 25.Solioz M, Abicht HK, Mermod M, Mancini S. 2010. Response of Gram-positive bacteria to copper stress. J Biol Inorg Chem 15:3–14. doi: 10.1007/s00775-009-0588-3. [DOI] [PubMed] [Google Scholar]
- 26.Odermatt A, Krapf R, Solioz M. 1994. Induction of the putative copper ATPases, CopA and CopB, of Enterococcus hirae by Ag+ and Cu2+, and Ag+ extrusion by CopB. Biochem Biophys Res Commun 202:44–48. doi: 10.1006/bbrc.1994.1891. [DOI] [PubMed] [Google Scholar]
- 27.Strausak D, Solioz M. 1997. CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J Biol Chem 272:8932–8936. doi: 10.1074/jbc.272.14.8932. [DOI] [PubMed] [Google Scholar]
- 28.Wickramasinghe WA, Dameron CT, Weber T, Solioz M, Cobine P, Harrison MD. 1999. The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS Lett 445:27–30. [DOI] [PubMed] [Google Scholar]
- 29.Multhaup G, Strausak D, Bissig KD, Solioz M. 2001. Interaction of the CopZ copper chaperone with the CopA copper ATPase of Enterococcus hirae assessed by surface plasmon resonance. Biochem Biophys Res Commun 288:172–177. doi: 10.1006/bbrc.2001.5757. [DOI] [PubMed] [Google Scholar]
- 30.Grass G, Rensing C. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun 286:902–908. doi: 10.1006/bbrc.2001.5474. [DOI] [PubMed] [Google Scholar]
- 31.Mills SD, Lim CK, Cooksey DA. 1994. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol Gen Genet 244:341–351. [DOI] [PubMed] [Google Scholar]
- 32.Schelder S, Zaade D, Litsanov B, Bott M, Brocker M. 2011. The two-component signal transduction system coprs of Corynebacterium glutamicum is required for adaptation to copper-excess stress. PLoS One 6:e22143. doi: 10.1371/journal.pone.0022143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintana J, Novoa-Aponte L, Argüello JM. 2017. Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J Biol Chem 292:15691–15704. doi: 10.1074/jbc.M117.804492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armbruster KM, Meredith TC. 2018. Enrichment of bacterial lipoproteins and preparation of N-terminal lipopeptides for structural determination by mass spectrometry. J Vis Exp 136:e56842. doi: 10.3791/56842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manavalan B, Basith S, Choi S. 2011. Similar structures but different roles—an updated perspective on TLR structures. Front Physiol 2:41. doi: 10.3389/fphys.2011.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouloc P, Lartigue M-F, Glaser P, Trieu-Cuot P, Villain A, Sismeiro O, Dillies M-A, Sauvage E, Da Cunha V, Rosinski-Chupin I, Caliot M-E. 2015. Single nucleotide resolution RNA-seq uncovers new regulatory mechanisms in the opportunistic pathogen Streptococcus agalactiae. BMC Genomics 16:419. doi: 10.1186/s12864-015-1583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodgkinson V, Petris MJ. 2012. Copper homeostasis at the host-pathogen interface. J Biol Chem 287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djoko KY, Ong C-L, Walker MJ, McEwan AG. 2015. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290:18954–18961. doi: 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samanovic MI, Ding C, Thiele DJ, Darwin KH. 2012. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe 11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobman JL, Crossman LC. 2015. Bacterial antimicrobial metal ion resistance. J Med Microbiol 64:471–497. doi: 10.1099/jmm.0.023036-0. [DOI] [PubMed] [Google Scholar]
- 41.Silveira E, Freitas AR, Antunes P, Barros M, Campos J, Coque TM, Peixe L, Novais C. 2014. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages. J Antimicrob Chemother 69:899–906. doi: 10.1093/jac/dkt479. [DOI] [PubMed] [Google Scholar]
- 42.Bell FY, Adelaide B. 2010. Copper tolerance of Listeria monocytogenes strain DRDC8. PhD thesis. University of Adelaide, Adelaide, Australia. [Google Scholar]
- 43.Zhang S, Wang D, Wang Y, Hasman H, Aarestrup FM, Alwathnani HA, Zhu YG, Rensing C. 2015. Genome sequences of copper resistant and sensitive Enterococcus faecalis strains isolated from copper-fed pigs in Denmark. Stand Genomic Sci 10:35. doi: 10.1186/s40793-015-0021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Portmann R, Magnani D, Stoyanov JV, Schmechel A, Multhaup G, Solioz M. 2004. Interaction kinetics of the copper-responsive CopY repressor with the cop promoter of Enterococcus hirae. J Biol Inorg Chem 9:396–402. doi: 10.1007/s00775-004-0536-1. [DOI] [PubMed] [Google Scholar]
- 45.Grabowicz M, Silhavy TJ. 2017. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci U S A 114:4769–4774. doi: 10.1073/pnas.1702248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narita SI, Tokuda H. 2011. Overexpression of LolCDE allows deletion of the Escherichia coli gene encoding apolipoprotein N-acyltransferase. J Bacteriol 193:4832–4840. doi: 10.1128/JB.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabowicz M. 2018. Lipoprotein transport: greasing the machines of outer membrane biogenesis: re-examining lipoprotein transport mechanisms among diverse Gram-Negative bacteria while exploring new discoveries and questions. BioEssays 40:e1700187. doi: 10.1002/bies.201700187. [DOI] [PubMed] [Google Scholar]
- 48.Rodushkin I, El Albani A, Chi Fru E, Lalonde SV, Konhauser KO, Andersson P, Partin CA, Rodríguez NP, Weiss DJ. 2016. Cu isotopes in marine black shales record the Great Oxidation Event. Proc Natl Acad Sci U S A 113:4941–4946. doi: 10.1073/pnas.1523544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers SD, Bhave MR, Mercer JF, Camakaris J, Lee BT. 1991. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J Bacteriol 173:6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asmar AT, Collet JF. 2018. Lpp, the Braun lipoprotein, turns 50—major achievements and remaining issues. FEMS Microbiol Lett 365:fny199. doi: 10.1093/femsle/fny199. [DOI] [PubMed] [Google Scholar]
- 51.Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. 2014. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 28:1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grass G, Rensing C, Solioz M. 2011. Metallic copper as an antimicrobial surface. Appl Environ Microbiol 77:1541–1547. doi: 10.1128/AEM.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yazdankhah S, Rudi K, Bernhoft A. 2014. Zinc and copper in animal feed—development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb Ecol Heal Dis 25:10.3402/mehd.v25.25862. doi: 10.3402/mehd.v25.25862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai H-N, Hodgson DA. 2003. Development of a synthetic minimal medium for Listeria monocytogenes. Appl Environ Microbiol 69:6943–6945. doi: 10.1128/AEM.69.11.6943-6945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato F, Sugai M. 2011. A simple method of markerless gene deletion in Staphylococcus aureus. J Microbiol Methods 87:76–81. doi: 10.1016/j.mimet.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Monk IR, Gahan CGM, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lauer P, Chow MYN, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol 184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.