Streptococcus pneumoniae is an opportunistic human pathogen responsible for over a million deaths every year. Although both vaccination programs and antibiotic therapies have been effective in prevention and treatment of pneumococcal infections, respectively, the sustainability of these solutions is uncertain. The pneumococcal genome is highly flexible, leading to vaccine escape and antibiotic resistance. This flexibility is predominantly facilitated by competence, a state allowing the cell to take up and integrate exogenous DNA. Thus, it is essential to obtain a detailed overview of gene expression during competence. This is stressed by the fact that administration of several classes of antibiotics can lead to competence. Previous studies on the competence regulon were performed with microarray technology and were limited to an incomplete set of known genes. Using RNA sequencing combined with an up-to-date genome annotation, we provide an updated overview of competence-regulated genes.
KEYWORDS: BlpR, CiaR, ComE, ComX, RNA-seq, Streptococcus pneumoniae, transcriptomics, VraR, genetic competence
ABSTRACT
Competence for genetic transformation allows the opportunistic human pathogen Streptococcus pneumoniae to take up exogenous DNA for incorporation into its own genome. This ability may account for the extraordinary genomic plasticity of this bacterium, leading to antigenic variation, vaccine escape, and the spread of antibiotic resistance. The competence system has been thoroughly studied, and its regulation is well understood. Additionally, over the last decade, several stress factors have been shown to trigger the competent state, leading to the activation of several stress response regulons. The arrival of next-generation sequencing techniques allowed us to update the competence regulon, the latest report on which still depended on DNA microarray technology. Enabled by the availability of an up-to-date genome annotation, including transcript boundaries, we assayed time-dependent expression of all annotated features in response to competence induction, were able to identify the affected promoters, and produced a more complete overview of the various regulons activated during the competence state. We show that 4% of all annotated genes are under direct control of competence regulators ComE and ComX, while the expression of a total of up to 17% of all genes is affected, either directly or indirectly. Among the affected genes are various small RNAs with an as-yet-unknown function. Besides the ComE and ComX regulons, we were also able to refine the CiaR, VraR (LiaR), and BlpR regulons, underlining the strength of combining transcriptome sequencing (RNA-seq) with a well-annotated genome.
IMPORTANCE Streptococcus pneumoniae is an opportunistic human pathogen responsible for over a million deaths every year. Although both vaccination programs and antibiotic therapies have been effective in prevention and treatment of pneumococcal infections, respectively, the sustainability of these solutions is uncertain. The pneumococcal genome is highly flexible, leading to vaccine escape and antibiotic resistance. This flexibility is predominantly facilitated by competence, a state allowing the cell to take up and integrate exogenous DNA. Thus, it is essential to obtain a detailed overview of gene expression during competence. This is stressed by the fact that administration of several classes of antibiotics can lead to competence. Previous studies on the competence regulon were performed with microarray technology and were limited to an incomplete set of known genes. Using RNA sequencing combined with an up-to-date genome annotation, we provide an updated overview of competence-regulated genes.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is a mostly harmless human commensal found in the nasopharynx. However, when the pneumococcus leaves the nasopharynx and ends up in other niches, it can cause severe diseases, such as sepsis, pneumonia, and meningitis (1). Especially among individuals with an underdeveloped or weakened immune system, these diseases lead to over a million deaths per year (2). Although both vaccination and antibiotic therapy have been used successfully for prevention and treatment of infections, respectively, the pneumococcus remains a threat to human health. This persistence is largely due to the remarkable genomic plasticity of the pneumococcus, allowing the acquisition of antibiotic resistance and evasion of the host immune response. Horizontal gene transfer, underlying the vast majority of such diversification strategies, is facilitated by pneumococcal competence. The competent state allows cells to take up exogenous DNA and integrate it into their own genome (i.e., transformation). During competence, various functionalities are activated, including DNA repair, bacteriocin production, and activities of several stress-response regulons (3, 4). This diversity of activated functions is relevant in light of the fact that a broad spectrum of antimicrobial compounds (causing various forms of stress) can induce competence development (5–7) through at least three distinct mechanisms: HtrA substrate competition (8, 9), oriC-proximal gene dosage increases (6), and chaining-mediated autocrine-like signaling (7). Other parameters that affect competence development include pH, oxygen, phosphate, and diffusibility of the growth medium (10–12). The fact that various forms of stress induce competence, leading to the activation of several stress-response regulons, has led to the hypothesis that competence in the pneumococcus may function as a general stress response mechanism (13, 14).
Among the genes activated during competence are the CiaR and VraR (LiaR) regulons. Although the underlying molecular mechanisms of activation are unknown, both regulons have been associated with cell wall damage control (3, 15). Indeed, a growth lag during competence (4) and the reduced fitness of both ciaR and vraR mutants (3, 15) indicate that competence represents a significant burden for a pneumococcal cell. It seems plausible that the production and insertion of the DNA uptake machinery (16) into the rigid cell wall have a significant impact on cell wall integrity. The CiaR regulon seems to be responsible for resolving such issues and preventing subsequent lysis (3, 17). An additional dose of competence-related cell wall stress is caused by fratricide, where competent cells kill and lyse noncompetent sister cells and members of closely related species. Specifically during competence, pneumococci produce a fratricin, CbpD, and the corresponding immunity protein, ComM (18). Secreted CbpD, aided by the action of autolysins LytA and LytC, can kill noncompetent, neighboring cells, which then release their DNA and other potentially valuable resources. Eldholm et al. showed that the VraR regulon represents a second layer of protection, beyond that represented by ComM, by which competent cells prevent CbpD-mediated lysis (15). ComM is also crucial in causing cell division arrest during competence by inhibiting initiation of division and by interfering with the activity of StkP (19).
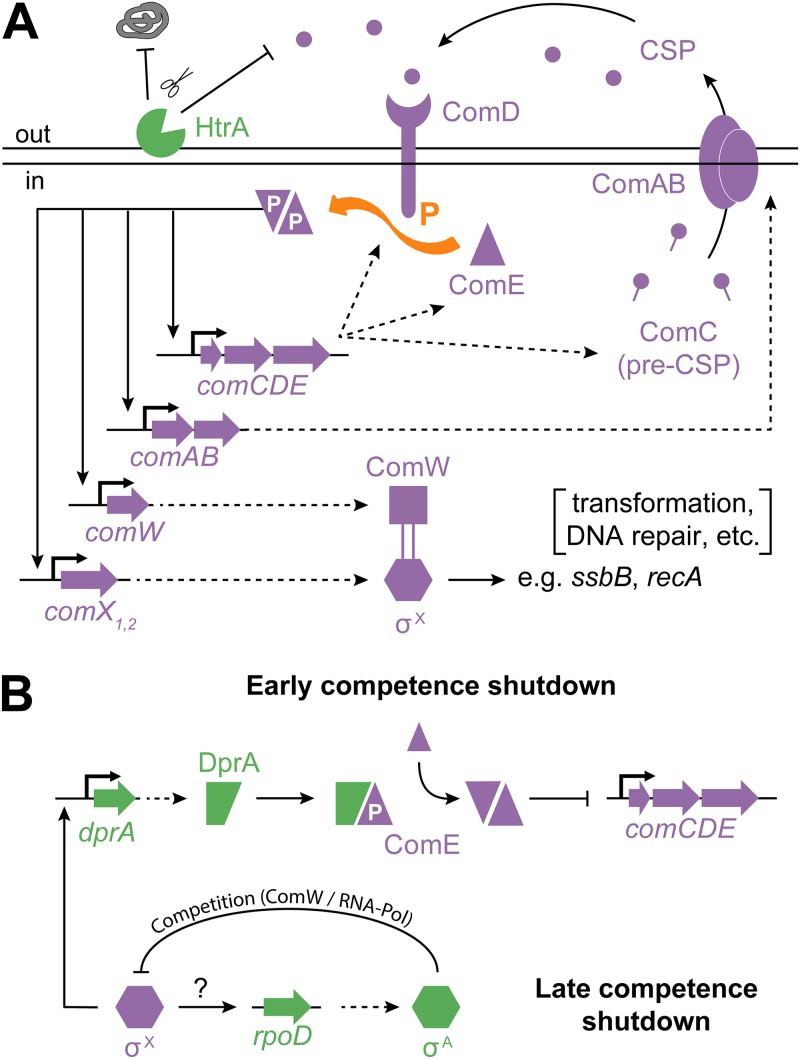
The activation of competence (Fig. 1A) depends on the action of two key transcriptional regulators, ComE and ComX. The competence regulon is divided into groups of early-competence (early-com) (i.e., ComE-dependent) genes and late-competence (late-com) (i.e., ComX-dependent) genes. Specifically, early competence involves, among others, the comCDE and comAB operons. A basal expression level of comCDE (20) ensures the production of the small peptide ComC, which contains a double-glycine leader and is processed and exported into the extracellular milieu by the bipartite transporter ComAB (21, 22). The resulting 17-residue matured peptide is referred to as competence-stimulating peptide (CSP) (23) and can interact with ComD, the membrane histidine kinase component of the two-component system (TCS) ComDE (24). Upon CSP binding, ComD autophosphorylates and, subsequently, transfers its phosphate group to its cognate response regulator, ComE (25). Finally, phosphorylated ComE dimerizes and binds specific recognition sequences to activate the members of the early-com regulon (26, 27). This regulon contains both the aforementioned comAB and comCDE operons, creating a positive-feedback loop that self-amplifies once a certain threshold of extracellular CSP is reached. Since CSP can interact with ComD on the producer cell as well as on other pneumococcal cells, competence represents a quorum-sensing system. Although cell-to-cell contact is not required for the spread of competence in a population (7, 28), it does lead to more efficient signaling (29) and to more-extensive recombination events (30).
FIG 1.
Overview of the regulatory network controlling competence in Streptococcus pneumoniae. (A) Positive-feedback loop responsible for activation of competence. HtrA-mediated degradation of CSP is relevant to the competence-inducing effect of certain antibiotics (8). (Adapted from reference 6 with permission of the publisher.) (B) Early competence is shut down through the action of late-competence protein DprA. Binding of DprA to phosphorylated ComE leads to an increase in the levels of unphosphorylated ComE dimers repressing comCDE expression. Late-competence shutdown is the result of competition for ComW and RNA polymerase binding between sigma factors ComX (σX) and RpoD (σA). Although expression of rpoD is strongly upregulated during competence, the underlying mechanism is unknown.
Additional members of the early-com regulon are comX1 and comX2, two identical genes that encode the alternative sigma factor ComX (σX) (31). The rapid accumulation of ComX during early competence leads to the activation of promoters with a ComX-binding motif, resulting in the expression of the late-com regulon (3, 4, 32, 33). Finally, within 20 min after the initiation of competence, the process is largely shut down through a combination of different mechanisms (Fig. 1B) (34–36). Early-com genes are repressed by late-competence protein DprA and unphosphorylated ComE (27). As a second layer of control, sigma factor competition between ComX and RpoD (σA; induced during late competence) is probably responsible for the shutdown of late-com genes (36).
While the backbone of this regulatory system is quite well understood, there are many other factors that complicate the matter, including the system’s sensitivity to growth medium acidity and potential small RNA (sRNA)-mediated control of ComC expression (37).
To fully understand the implications of competence activation in the pneumococcus, it is important to know which genes are (directly or indirectly) differentially expressed during competence. Several comprehensive studies, based on DNA microarray technology, have been performed to determine the competence regulon, resulting in the identification of more than 100 reported competence-associated genes (3, 4, 38). All of those studies showed a high level of agreement on a certain core regulon, but discrepancies remained. Moreover, the recent identification of early-competence protein BriC (39, 40) illustrates that the description of the competence regulon can still be refined. In order to generate a more complete and nuanced overview of the competence regulon, we first utilized data from PneumoExpress (39), a resource containing data on the pneumococcal transcriptome under various infection-relevant conditions. We used transcriptome sequencing (RNA-seq) data sets from S. pneumoniae D39V (41) cells just prior to (t = 0) and 3, 10, and 20 min after the addition of exogenous CSP. More importantly, our data set has higher sensitivity and precision and has a larger dynamic range than the data sets used with previous genome-wide assays of the competence regulon, which were based on DNA microarrays (42). Second, the recent reannotation of the pneumococcal genome revealed previously nonannotated protein-encoding sequences and small RNAs (41). DNA microarray studies are limited to the target sequences present on the array, and a new data set was therefore required to obtain information on the expression of these new elements. Finally, the new annotation also contains information on transcription start sites (TSSs) and terminators (41), which allows both a more accurate search of transcription regulatory motifs (e.g., ComE- or ComX-binding sites) and the integration of operon information into the interpretation of transcriptome data.
As expected, our results largely confirmed those of previous microarray-based studies and we observed distinct time-dependent expression patterns of ComE- and ComX-regulated genes. In addition, we provide an overview of the transcription start sites most likely to be responsible for the observed transcriptome changes, adding up to, among others, 15 ComE-regulated, 19 ComX-regulated, 18 CiaR-regulated, and 4 VraR-regulated operons. We identified 7 new noncoding RNAs (ncRNAs), affected by several regulators, among the differentially expressed genes, but elucidation of their role in competence requires future studies.
RESULTS
Competence induction disrupts the pneumococcal transcriptional landscape.
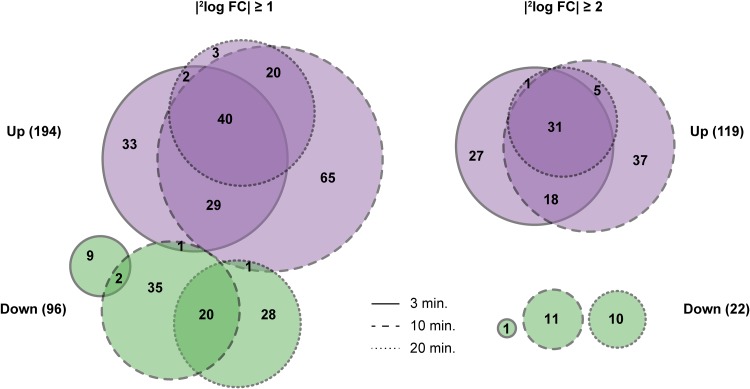
Differential gene expression analysis revealed that many genes (13% and 17%, corresponding to data from gene-based and promoter-based analysis, respectively) are affected by the induction of competence (Fig. 2): a total of 288 genes undergo a change in expression of more than 2-fold. Among these, 192 genes were exclusively upregulated, 94 were exclusively downregulated, and 2 genes were upregulated at one time point and downregulated at another. Using a stricter fold change cutoff value of 4-fold, 141 genes were still significantly affected, 119 of which were upregulated and 22 were downregulated. As can be seen in Fig. 2, upregulated genes tended to be affected more strongly and consistently, while not a single gene was found to be significantly downregulated at all three time points.
FIG 2.
Venn diagrams of differentially expressed genes, created with http://eulerr.co. Diagrams show how many genes were significantly upregulated (purple) or downregulated (green), using a cutoff fold change value of 2 (left) or 4 (right). Differential expression results seen 3, 10, and 20 min after addition of CSP are indicated by outlines.
Identification of ComE-, ComX-, and CiaR/VraR-regulated WGCNA clusters.
Weighted gene coexpression network analysis (WGCNA) (see Materials and Methods) of all of the genes, performed on the basis of their regularized log (rlog) expression levels across the 22 conditions included in PneumoExpress (39), yielded 36 clusters (see Table S2 in the supplemental material). Using these results, we verified whether some of the clusters corresponded to specific regulons known to be affected during competence. Indeed, one of these clusters (cluster [cl.] 29, n = 26) contained 20 of the 25 genes that had been reported previously to be regulated by ComE (3, 4, 38), including briC (SPV_0391), which was identified as a member of the competence regulon only recently (39, 40). Aggarwal et al. showed that the double-glycine peptide encoded by briC is conserved only in pneumococci and related streptococci and is important for late biofilm development (40). Those authors also found that BriC is secreted (in part by ComAB), but its exact mode of action has yet to be elucidated.
The five ComE-regulated genes that did not end up in this cluster include blpA, blpY, blpZ, and pncP (SPV_0472 to SPV_0475, respectively), which are part of the BlpR regulon and whose promoters are likely to have lower affinity for ComE (43, 44). The fifth off-cluster ComE-regulated gene is ybbK (SPV_1984). A second cluster (cl. 11, n = 56) contained 41 of 51 members of the reported ComX regulon (3, 4, 38), confirming the power of the WGCNA approach while simultaneously highlighting the general reliability of previous descriptions of the competence regulon. Less clearly, 13 of 32 known CiaR-regulated genes (37, 45, 46) and 5 of 14 VraR-regulated genes (15) clustered together (cl. 33, n = 22). The fact that genes from the CiaR and VraR regulon cluster less clearly may, in part, be explainable by the more diverse nature of their regulation. For example, the heat shock hrcA-grpE operon is regulated not only by VraR but also by HrcA itself, accounting for a different expression dynamic across the diverse conditions sampled for PneumoExpress (39). Additionally, the two TSSs of tarIJ-licABC (SPV_1127 to SPV_1123) (37) and the downregulatory effect of CiaR on the manLMN operon (SPV_0264 to SPV_0262) (46) may prevent clear clustering.
Time-resolved expression profiles of several regulons during competence.
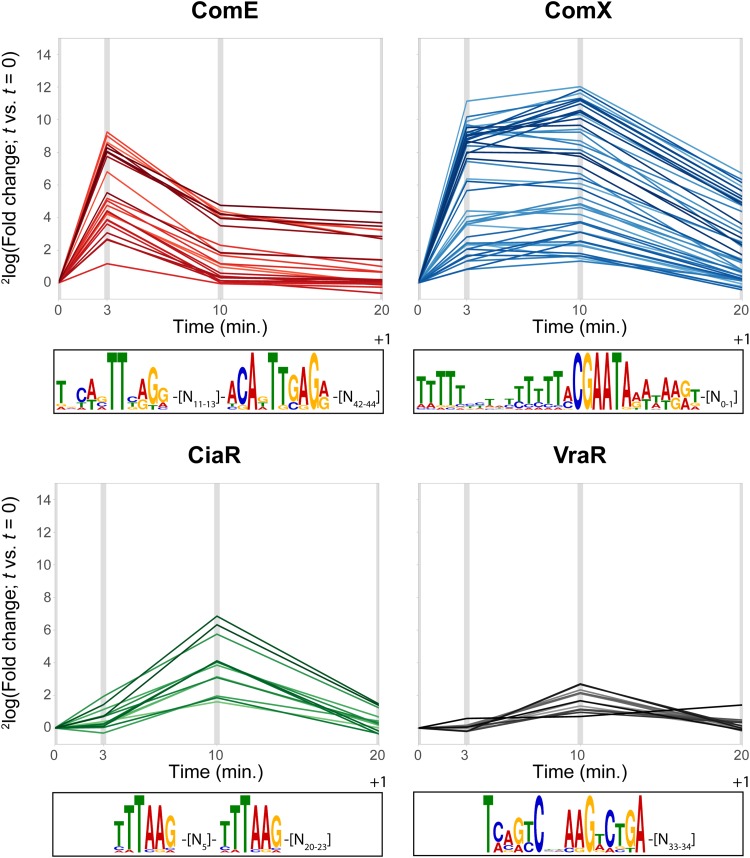
We visualized the typical time-resolved expression patterns of the various regulons by plotting the fold expression changes, relative to t = 0, of all genes that (i) were previously reported to be activated by the corresponding regulator and (ii) fell into the associated WGCNA cluster (see above). We refer to these sets of genes as “high-confidence” (HC) members of their respective regulons. It is clear from these plots that ComE-regulated genes peaked early and rapidly dropped in expression level afterward (Fig. 3, top left). This is in line with previous studies which showed that early competence is actively shut down through the action of late-competence protein DprA. By specifically binding to active, phosphorylated ComE, DprA causes a shift toward a state where regulated promoters are, instead, bound by dephosphorylated ComE, leading to a shutdown of transcription (Fig. 1B) (36, 47).
FIG 3.
Regulons affected during competence. (Top panels) Expression profiles of genes previously reported to be ComE, ComX, CiaR, or VraR activated. Each line shows the fold changes (versus t = 0) for a single gene from the corresponding regulon. Except for the VraR regulon (all genes shown), only genes that fell into the appropriate WGCNA cluster (cl. 29 for ComE, cl. 11 for ComX, cl. 33 for CiaR) were included. (Bottom panels) Position weight matrices of the recognition sites for ComE, ComX, CiaR, and VraR, as determined with MEME (94), from the promoters of the HC members of the corresponding regulons (ComE, ComX, and CiaR) or from three pneumococcal and six L. lactis promoters (VraR [15]).
Similarly to the ComE regulon results, the level of expression of ComX-regulated genes also increased rapidly, with high fold changes after 3 min. However, the expression of these genes remained stable for a longer period of time than was seen with the ComE regulon, with most still increasing their level of expression until 10 min after CSP addition (Fig. 3, top right). The decrease in expression level that followed was, in part, indirectly linked to the shutdown of early competence, since both production and stabilization by ComW (34) of ComX depend on the activity of phosphorylated ComE. However, a recent modeling approach suggested that another shutdown mechanism was required to explain the observed rate of late-competence shutdown (36). The authors argued, convincingly, that competition between sigma factors ComX (σX) and RpoD (σA) for interaction with RNA polymerase and/or stabilizing factor ComW would be a suitable explanation for the discrepancies between the model data and the experimental data. Indeed, the fact that rpoD transcription is upregulated up to 10-fold during competence (3, 4) would make this a credible hypothesis, although rpoD upregulation could also simply serve to restore the expression levels of RpoD-controlled genes.
Constituting a more indirect consequence of competence induction, the CiaR-mediated response was generally weaker than and delayed in comparison to the responses seen with the ComE and ComX regulons (Fig. 3, bottom left). Interestingly, the activation of this regulon also seems to be quite transient, with a fast drop in expression from 10 to 20 min after CSP addition.
Finally, the expression profile of all reported VraR-regulated genes (15), regardless of their clustering behavior under the infection-relevant conditions, was similar to that of the CiaR regulon (Fig. 3, bottom right).
Shared expression trends within operons allow switching from gene level analysis to promoter level analysis of transcriptional regulation.
Transcriptome studies are typically performed on a per-gene level, reporting for each individual gene whether or not it is differentially expressed under the different studied conditions. That type of information is certainly relevant in assessments of what changes occur in a cell or population under conditions of confrontation with a certain change in environment or identity. However, to find out how these changes come about, it is also interesting to consider which transcripts or, rather, which promoters have been affected. To this end, we used our previously created map of the pneumococcal transcriptional landscape (39, 41), where possible, to identify which promoters are responsible for the observed differential levels of expression of individual genes. As an example, we highlight a cluster of 22 genes (SPV_0192 to SPV_0213), encoding 21 ribosomal proteins and protein translocase subunit SecY (Table 1). Although Peterson et al. reported previously that expression of the entire operon was downregulated (4), we observed significant upregulation (just above the 2-fold cutoff) for only 10 genes and no significant effects for the rest of the operon. However, it is clear both from visual inspection and from the fact that all genes cluster together in the WGCNA analysis that the entire operon behaves as a single transcriptional unit, with modest upregulation occurring 3 min after CSP addition, followed by a drop in expression at 10 min. Therefore, regulation of a single TSS, at position 195877 [positive strand (+)], would suffice to explain the behavior of these 22 genes. Note that the observed gradient in fold changes across this operon was observed in several other operons as well (see Table S5). Although we do not fully understand this phenomenon, we speculate that differences in the rates of mRNA degradation might play a role. Since (exo)ribonucleases have a specific directionality (i.e., 5ʹ to 3ʹ or 3ʹ to 5ʹ in Gram-positive bacteria), such differences could explain the observed gradients.
TABLE 1.
Expression trend of a 22-gene operon (SPV_0192 to SPV_0213) transcribed from TSS 195877 (+)a
| Locus tag | Gene | TPM at 0 min | Log2 fold change at: |
||
|---|---|---|---|---|---|
| 3 min | 10 min | 20 min | |||
| SPV_0192 | rpsJ | 4,585 | 0.7 | 0.0 | −0.1 |
| SPV_0193 | rplC | 2,992 | 0.9 | −0.1 | 0.0 |
| SPV_0194 | rplD | 2,181 | 0.8 | −0.1 | −0.3 |
| SPV_0195 | rplW | 2,919 | 1.0 | −0.2 | 0.2 |
| SPV_0196 | rplB | 2,611 | 1.0 | −0.2 | −0.1 |
| SPV_0197 | rpsS | 4,134 | 1.0 | −0.5 | 0.1 |
| SPV_0198 | rplV | 3,838 | 1.0 | −0.5 | 0.1 |
| SPV_0199 | rpsC | 3,158 | 1.0 | −0.4 | 0.0 |
| SPV_0200 | rplP | 4,329 | 1.0 | −0.6 | 0.0 |
| SPV_0201 | rpmC | 3,060 | 0.9 | −0.7 | 0.0 |
| SPV_0202 | rpsQ | 4,348 | 1.1 | −0.7 | −0.1 |
| SPV_0203 | rplN | 3,640 | 1.1 | −0.7 | 0.1 |
| SPV_0204 | rplX | 3,917 | 1.1 | −0.7 | 0.0 |
| SPV_0205 | rplE | 2,848 | 1.1 | −0.7 | −0.2 |
| SPV_0206 | rpsN | 2,717 | 1.1 | −0.6 | −0.3 |
| SPV_0207 | rpsH | 4,013 | 1.3 | −0.8 | 0.0 |
| SPV_0208 | rplF | 4,411 | 1.1 | −0.8 | −0.2 |
| SPV_0209 | rplR | 3,598 | 1.1 | −0.9 | −0.2 |
| SPV_0210 | rpsE | 3,477 | 1.1 | −0.9 | −0.2 |
| SPV_0211 | rpmD | 4,120 | 1.1 | −0.9 | −0.4 |
| SPV_0212 | rplO | 2,247 | 1.2 | −0.7 | −0.2 |
| SPV_0213 | secY | 1,920 | 1.0 | −0.7 | −0.3 |
Boldface data indicate significance (P < 0.001; log2 fold change [FC] > 1; DESeq [91]).
Early-competence genes: the ComE and BlpR regulons.
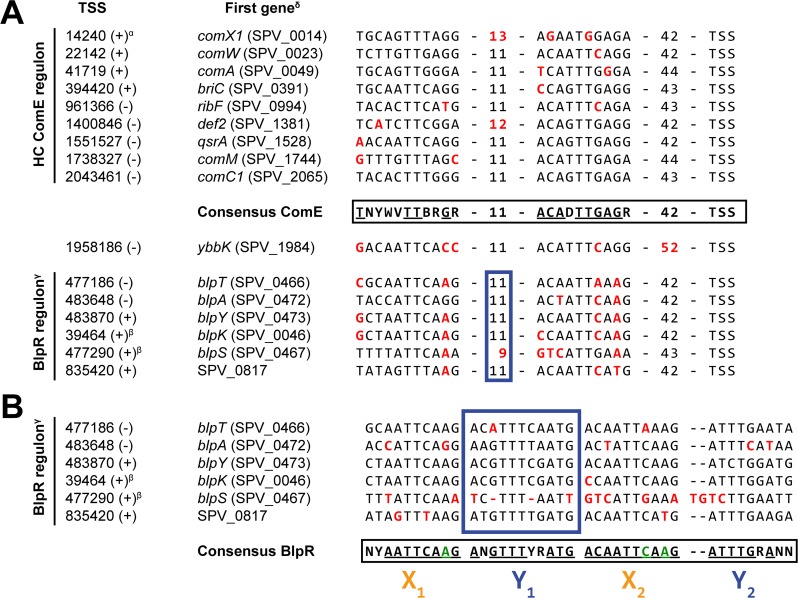
We reasoned that confining the analysis of promoter regions to only those upstream of HC members (defined above) of a regulon was likely to yield a more accurate set of data representing the consensus binding site of the corresponding regulator than analysis of such a consensus on the basis of all known regulated genes, as is the common procedure. Therefore, combining transcription start site (TSS) data on these selected genes (Table S3) as reported previously (41) and known characteristics of regulatory sites (e.g., typical distances from TSS), we redefined the binding motifs for ComE, ComX, and CiaR (see Materials and Methods for details). The identified ComE-binding sequence (Fig. 3, top left; see also Fig. 4A) strongly resembles sequences described in previous reports (26, 27) and consists of two imperfect repeats. The spacing between these repeats is 11 to 13 nucleotides (nt) in length (mode = 11), while the spacing between the right motif and the TSS is 42 to 44 nt (mode = 42). Clearly, the second repeat is the more highly conserved repeat and is likely to be most important for ComE recognition. In summary, this yields the following consensus ComE-binding motif: [TNYWVTTBRGR]-[N11]-[ACADTTGAGR]-[N42]-[TSS] (Fig. 4A).
FIG 4.
(A) ComE-binding sequences on the S. pneumoniae D39V genome. The consensus sequence was determined as described in Materials and Methods. Multiple possible nucleotides are indicated, according to IUPAC nomenclature, by R (A or G [A/G]), Y (C/T), W (A/T), B (C/G/T), D (A/G/T), V (A/C/G), or N (any). Nucleotides and spacings colored red deviate from this consensus. HC, high confidence. (B) Putative BlpR-binding site consensus. The blue box corresponds to the internal spacer indicated in panel A. The consensus sequence (IUPAC nomenclature used as described for panel A) was determined as described in Materials and Methods, where letters colored green indicate where the BlpR consensus is incompatible with the ComE consensus. A superscript alpha indicates that the promoters of comX1 (SPV_0014) and comX2 (SPV_1818) are identical. A superscript beta indicates that the operons were not differentially expressed in response to CSP addition. A superscript gamma indicates that the genes encoding the export machinery of signaling peptide BlpC and bacteriocins BlpK and PncW and the blpA and blpB genes were frameshifted, eliminating the regulatory positive-feedback loop of the Blp system. A superscript delta indicates the first gene annotated in both D39V and D39W (97).
As Martin et al. previously reported (27), the internal spacing and the right arm of the binding sequence in PcomX deviate from the consensus (Fig. 4A). Indeed, from our data, this deviation seemed to lead to a somewhat lower level of expression from PcomX after induction (Table 2), relative to other members of the ComE regulon. This reduction is partially compensated by the presence of two copies of comX on the chromosome. In contrast, we found no indication that mismatches with the consensus ComE-binding sequence found in the promoter region of comAB resulted in lower expression of those genes (Table 2), which was suggested by Martin et al. (27) to explain an earlier observation that comAB transcript levels were rate-limiting in the development of competence (48). Indeed, although higher ComAB levels may indeed accelerate competence development, the fact that a duplication of comC (22, 25) leads to competence upregulation suggests that ComAB is not exporting CSP at maximum capacity in wild-type cells.
TABLE 2.
ComE-regulated genes, distributed over 15 operonsa
| Gene | Product | TPM at 0 min | Log2 fold change at: |
Note(s) | ||
|---|---|---|---|---|---|---|
| 3 min | 10 min | 20 min | ||||
| comX1 | Competence-specific sigma factor | 8 | 9.1 | 3.8 | 3.1 | |
| tRNA-Glu-1 | tRNA-Glu-UUC | 387 | 2.8 | 0.1 | −0.6 | Secondary |
| comW | Competence positive regulator | 17 | 9.1 | 4.7 | 3.0 | |
| purA | Adenylosuccinate synthetase | 619 | 4.4 | 1.0 | 0.0 | Secondary |
| ccnC | csRNA3 | 155 | 4.4 | 1.2 | 0.0 | Secondary; also CiaR regulon |
| srf-03 | ncRNA of unknown function | 39 | 7.1 | 2.0 | 1.5 | |
| comA | CSP ABC transporter ATP-binding protein | 21 | 9.3 | 4.1 | 3.4 | |
| comB | CSP ABC transporter permease | 24 | 9.5 | 4.4 | 3.3 | |
| briC | Biofilm-regulating peptide | 97 | 4.9 | 1.7 | 0.7 | |
| ydiLb | Putative membrane peptidase | 67 | 5.3 | 2.4 | 1.0 | |
| blpT | BlpT protein | 8 | 3.9 | 0.7 | 1.5 | BlpR regulon |
| blpAb | Peptide ABC transporter permease/ATP-binding protein | 2 | 5.1 | 2.1 | 2.5 | BlpR regulon |
| blpBb | Peptide ABC transporter permease | 3 | 4.5 | 2.2 | 2.4 | BlpR regulon |
| blpC | Peptide pheromone | 3 | 4.4 | 2.7 | 1.9 | BlpR regulon |
| pncW | Putative bacteriocin | 13 | 4.0 | 1.1 | 2.0 | BlpR regulon |
| blpY | Bacteriocin immunity protein | 17 | 4.3 | 1.4 | 2.2 | BlpR regulon |
| blpZ | Immunity protein | 13 | 4.3 | 1.5 | 1.9 | BlpR regulon |
| pncP | Putative protease | 14 | 4.2 | 1.3 | 1.9 | BlpR regulon |
| SPV_2249 | Hypothetical protein | 50 | 1.2 | 0.3 | 0.8 | BlpR regulon |
| SPV_0817 | CAAX amino terminal protease family protein | 45 | 1.3 | 0.2 | 0.4 | BlpR regulon |
| ribF | FMN adenylyltransferase/riboflavin kinase | 147 | 5.2 | 1.2 | 0.7 | |
| def2 | Peptide deformylase | 177 | 3.7 | 0.0 | −0.1 | |
| SPV_1380 | Cell shape-determining protein | 276 | 3.9 | 0.1 | 0.0 | Secondary |
| SPV_1379 | Hypothetical protein | 183 | 3.3 | 0.0 | −0.1 | Secondary |
| yaaA | UPF0246 protein | 85 | 1.2 | 0.0 | −0.3 | Secondary |
| qsrA | ABC transporter ATP-binding protein-Na+ export | 223 | 4.5 | 0.6 | 0.2 | Secondary |
| qsrB | ABC transporter permease-Na+ export | 216 | 4.5 | 0.4 | −0.2 | Secondary |
| srf-22 | ncRNA of unknown function | 54 | 4.1 | 0.2 | −0.7 | Secondary |
| comM | Immunity factor | 9 | 8.0 | 4.4 | 2.8 | |
| tsaE | tRNA processing protein | 280 | 3.1 | 0.3 | 0.2 | Secondary |
| SPV_1742 | Acetyltransferase | 319 | 2.7 | 0.3 | 0.2 | Secondary |
| lytR | LytR-CpsA-Psr family protein | 355 | 2.8 | 0.1 | 0.1 | Secondary |
| comX2 | Competence-specific sigma factor | 8 | 9.1 | 3.8 | 3.1 | |
| tRNA-Glu-3 | tRNA-Glu-UUC | 387 | 2.8 | 0.1 | −0.6 | Secondary |
| ybbK | Putative membrane protease subunit | 795 | 2.1 | 0.6 | 0.6 | TSS too far from ComE-binding site |
| comC1 | Competence-stimulating peptide precursor | 36 | 8.8 | 5.1 | 4.6 | |
| comD | Two-component system sensor histidine kinase | 27 | 8.4 | 4.3 | 3.8 | |
| comE | Two-component system response regulator | 38 | 8.3 | 4.0 | 3.5 | |
| tRNA-Glu-5 | tRNA-Glu-UUC | 63 | 5.8 | 1.9 | 1.5 | Secondary |
| tRNA-Asn-2 | tRNA-Asn-GUU | 181 | 5.1 | 0.8 | 0.4 | Secondary |
The 15 operons are indicated by the eight blocks of data highlighted in gray and the seven blocks of data highlighted in white. Members of the BlpR regulon are included as indicated in the Note(s) column. “Secondary” indicates either read-through after incomplete termination or the influence of an additional TSS. For complete supplemental information, including TSS positions, see Table S5. Boldface data indicate significance (P < 0.001; log2 FC > 1; DESeq [91]). FMN, flavin mononucleotide; ncRNA, noncoding RNA.
Pseudogene.
As reported before, the comAB genes are preceded by a BOX element (49), an imperfectly repeated DNA element occurring 127 times in the D39V genome (41). Interestingly, the BOX element is located downstream of the ComE-regulated TSS and is therefore part of the comAB transcript. It was shown by Knutsen et al. that the BOX element upstream of comAB is important for the fine-tuning of competence (50), but the underlying mechanism is unknown. We previously detected several RNA fragments that terminated between the BOX element and the start codon of comA, leading us to annotate it as an ncRNA, srf-03 (41). Although some BOX elements were reported to contain putative protein-encoding sequences, we did not find any uninterrupted coding sequence in this specific case. It seems unlikely that the BOX element functions as an RNA switch, since srf-03 and comAB displayed the same time-dependent expression patterns, with relatively low expression at t = 0 (Table 2). Since removing this BOX element from PcomA leads to reduced comAB expression (50), it is tempting to speculate that the prematurely terminated transcript corresponding to srf-03 plays a role in competence regulation. However, a similar effect on transcription was observed when the BOX element in front of qsrAB (also ComE regulated) was removed (50) and we did not find any evidence for premature termination between that BOX element and the start of qsrA.
Analysis of all upregulated promoters resulted in the detection of five additional operons putatively regulated by ComE (the complete proposed ComE regulon is listed in Table 2), including ybbK, a known early-com gene. The weaker induction of this gene and the fact that its expression did not cluster with that of other early-com genes can be explained by the fact that the ComE site is located 10 nt too far from the TSS, compared to a canonical ComE-regulated gene (Fig. 4A).
Three other ComE-induced operons (blpT, blpABC, and pncW-blpYZ-pncP) are known to be part of the BlpR regulon, and their activation is the result of cross talk, where ComE can recognize the binding sites of BlpR but with lower efficiency (43, 44). Indeed, close inspection of the corresponding promoter regions shows marked differences with those of other ComE-regulated genes, consistently deviating from the consensus ComE-binding site at specific positions (Fig. 4A). The same discrepancies were observed in the promoter regions of blpK and, to a lesser extent, blpSRH, the two remaining blp operons. These operons were not differentially expressed during competence, probably due to the poorer resemblance to the ComE-binding consensus. Additionally, both blpK and blpSRH are constitutively expressed, such that any minor inducing effect by ComE would be negligible. A multiple-sequence alignment of the promoter regions of the five known blp operons in strain D39V revealed a conserved sequence very similar to, but slightly more extended than, that of the putative BlpR-binding site postulated by de Saizieu et al. (51). The binding site reported here can be seen as the following imperfect tandem 19- to 21-bp repeat sequence: [NYAATTCAAGANGTTTYRATG]-[ACAATTCAAG(NN)ATTTGRANN]-[N33]-[TSS]. More specifically, the region can be written as X1-Y1-X2-Y2, where X (resembling the ComE-binding site) and Y are 10 and 9 to 11 bp in length, respectively, having a highly conserved “TT” sequence (or a TTT sequence in Y) at their centers (Fig. 4B). Interestingly, the promoter region of the final operon putatively regulated by ComE, SPV_2249 to SPV_0817, resembles the putative BlpR recognition site (Fig. 4B) and we speculate that these genes are actually part of the BlpR regulon. This idea is supported by the very modest levels of induction (2.3-fold and 2.5-fold, respectively) of this operon during competence. Additionally, SPV_0817 encodes a probable CAAX protease (41) that could be speculated to be involved in immunity against self-produced bacteriocins (52, 53).
Finally, only one feature from the WGCNA cluster associated with ComE regulation remained that could not be directly linked to a ComE-binding site. This feature, a pseudogene (SPV_2414), is part of an ISSpn7 insertion sequence (IS) (54) and represents a truncated version of the gene encoding the corresponding transposase. Since the D39V genome contains eight additional sites with ≥95% sequence identity to this insertion sequence, clearly undermining mapping fidelity, and since no significant differential expression was observed at any competence time point, we ruled out SPV_2414 as a member of the ComE regulon.
Late-competence genes: the ComX regulon.
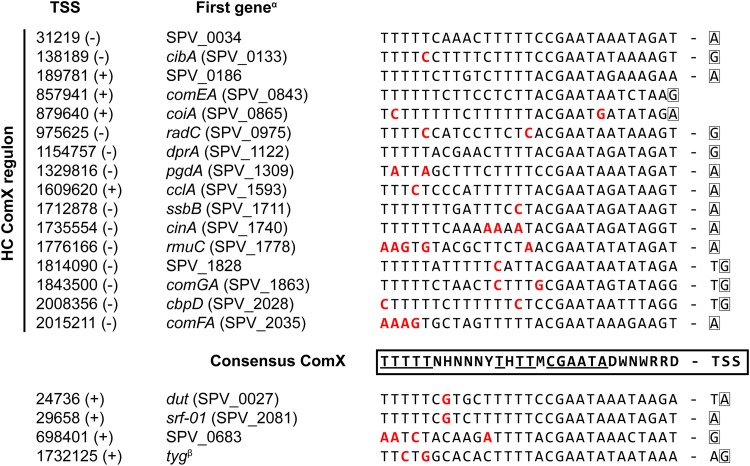
Directly following the strong, ComE-mediated increase in comX expression, the late-competence regulon is activated. On the basis of the promoter sequences of HC members of the late-com regulon (Fig. 5; see also Table S3), the ComX recognition sequence was reevaluated. Not surprisingly, the identified motif (Fig. 3, top right) did contain a near-perfect match (TMCGAATA) to the previously reported 8-nucleotide consensus sequence (3, 32, 33):. However, our analysis showed that the region relevant to ComX binding is likely much wider than that; with a thymine-rich stretch upstream and a (less extensively conserved) adenine-rich stretch downstream of the reported 8 nucleotides, the actual recognition site is extended to 20 to 30 bp. In summary, this yields the following consensus ComX-binding motif: [TTTTTNHNNNYTHTTMCGAATADWNWRRD]-[TSS] (where the underlined sequence represents the previously reported consensus sequence) (Fig. 5).
FIG 5.
ComX-binding sequences on the S. pneumoniae D39V genome. The consensus sequence was determined as described in Materials and Methods. Multiple possible nucleotides are indicated, according to IUPAC nomenclature, by R (A/G), Y (C/T), W (A/T), M (A/C), D (A/G/T), H (A/C/T), or N (any). HC, high confidence. A superscript alpha indicates the first gene annotated in both D39V and D39W (97), if available. A superscript beta indicates the tyg gene described by Campbell et al. (32). Its TSS was detected in PneumoBrowse (41), but no coding DNA sequence (CDS) or ncRNA has been reported (41, 59).
In addition to the 16 HC ComX-regulated operons, we identified three promoters containing the motif reported here (Table 3). First, SPV_0027 to SPV_0030, an operon encoding, among others, a dUTP pyrophosphatase (dut) and DNA repair protein RadA (radA), was already previously reported to be part of the late-com regulon (3, 4) but was not found to cluster with the HC ComX regulon. A secondary TSS, 11 nucleotides downstream of the ComX-regulated TSS, might be responsible for the lower correlation with other ComX-regulated genes. Similarly, a second previously reported late-com gene, SPV_0683, might be under the control of a secondary, as-yet-unidentified TSS, besides the ComX-activated TSS reported here (Table 3), a speculation supported by its relatively high expression level prior to CSP addition and sequencing coverage observed in PneumoBrowse (41). A third ComX-binding site (SPV_0033) was found downstream of prs1 and immediately upstream of a novel ncRNA, srf-01 (SPV_2081), which we identified recently (41). While this addition to the known competence regulon seemed interesting at first, the partial overlap between the ncRNA and a nearby IS element (containing pseudogene SPV_0034) led us to question the functionality of this novel element (see Fig. S1 in the supplemental material). Additionally, another pseudogene (SPV_2082) was located on the other side of the IS element and was also found to be under the control of ComX (Table 3). A multiple-genome alignment of several pneumococcal strains (not shown) revealed that the ComX-binding element downstream of prsA1 (i.e., prs1) was conserved in, among other strains, S. pneumoniae INV200 (GenBank accession number FQ312029.1) but was followed in that strain by a set of pseudogenes (Fig. S1). A BLASTX search showed that the two pseudogenes were probably derived from a protein-encoding gene mostly annotated as encoding a recombination-promoting nuclease or transposase. Interestingly, this gene was highly similar to SPV_2082 and to an additional pseudogene, SPV_2340, located elsewhere on the D39V chromosome and also ComX regulated. We speculate that the presence of a repeat unit of the pneumococcus (RUP) (55) upstream of SPV_2082, in combination with the action of IS elements, might have enabled several events of duplication and/or reorganization of the SPV_2082 locus. While these findings suggest that, in pneumococcal strains with an intact copy of this gene, it might be relevant to transformation and horizontal gene transfer, we do not expect srf-01 (or pseudogenes SPV_2082 and SPV_2340, for that matter) to have a role in competence.
TABLE 3.
ComX-regulated genes, distributed over 19 operonsa
| Gene | Product | TPM at 0 min | Log2 fold change at: |
Note | ||
|---|---|---|---|---|---|---|
| 3 min | 10 min | 20 min | ||||
| dut | Deoxyuridine 5ʹ-triphosphate nucleotidohydrolase | 78 | 2.7 | 1.5 | −0.2 | |
| SPV_0028 | Hypothetical protein | 94 | 2.6 | 1.6 | −0.2 | |
| radA | DNA repair protein | 96 | 2.5 | 2.4 | −0.1 | |
| SPV_0030 | Carbonic anhydrase | 388 | 0.8 | 1.1 | 0.1 | Secondary |
| srf-01 | ncRNA of unknown function | 71 | 3.7 | 3.3 | 0.9 | Putative pseudogene |
| SPV_2082b | Hypothetical protein | 22 | 6.6 | 6.3 | 2.6 | |
| SPV_0034b | IS1167 transposase | 3 | 3.6 | 3.7 | 1.1 | |
| cibA | Two-peptide bacteriocin peptide | 8 | 12.0 | 12.9 | 7.4 | |
| cibB | Two-peptide bacteriocin peptide | 11 | 10.2 | 11.0 | 5.5 | |
| cibC | CibAB immunity factor | 10 | 10.6 | 12.3 | 6.5 | |
| SPV_2121 | Hypothetical protein | 124 | 4.6 | 4.4 | 0.6 | |
| SPV_0186 | Competence-damage induced protein | 283 | 2.4 | 2.3 | 0.1 | Secondary |
| SPV_0683 | Hypothetical protein | 136 | 4.2 | 4.9 | 0.8 | |
| comEA | Late-competence DNA receptor | 4 | 9.8 | 9.2 | 3.7 | |
| comEC | Late-competence DNA transporter | 3 | 9.5 | 9.8 | 5.0 | |
| SPV_2256 | Hypothetical protein | 156 | 4.0 | 4.8 | 1.4 | Secondary |
| SPV_2257b | ABC transporter ATP-binding protein | 91 | 3.7 | 5.0 | 1.3 | Secondary |
| SPV_0846 | Hypothetical protein | 68 | 3.9 | 5.4 | 1.5 | Secondary |
| coiA | Competence protein | 1 | 8.2 | 7.4 | 1.2 | |
| pepF1 | Oligoendopeptidase F | 252 | 1.3 | 1.2 | 0.0 | Secondary |
| SPV_0867 | O-Methyltransferase family protein | 136 | 1.4 | 1.5 | 0.3 | Secondary |
| radC | DNA repair protein | 3 | 9.8 | 9.8 | 2.6 | |
| dprA | DNA protecting protein | 6 | 10.1 | 8.8 | 4.6 | |
| SPV_2340b | Hypothetical protein | 222 | 2.5 | 2.6 | 0.2 | Secondary |
| pgdA | Peptidoglycan N-acetylglucosamine deacetylase | 397 | 1.4 | 1.7 | 0.1 | Secondary |
| SPV_1308 | Oxidoreductase of aldo/keto reductase family, subgroup 1 | 182 | 0.9 | 1.4 | 0.3 | Secondary |
| cclA | Type IV prepilin peptidase | 2 | 9.3 | 9.0 | 3.7 | |
| ssbB | Single-stranded DNA-binding protein | 9 | 10.7 | 11.9 | 6.8 | |
| cinA | ADP-ribose pyrophosphatase/nicotinamide-nucleotide amidase | 78 | 5.8 | 6.5 | 2.5 | |
| recA | DNA recombination/repair protein | 391 | 3.0 | 3.8 | 0.7 | Secondary |
| dinF | MATE efflux family protein | 288 | 2.2 | 3.8 | 0.4 | Secondary |
| lytA | Autolysin/N-acetylmuramoyl-l-alanine amidase | 703 | 1.4 | 3.2 | 0.4 | Secondary |
| tygc | NA | <1 | 6.7 | 6.8 | 4.2 | |
| rmuC | DNA recombination protein | 314 | 2.1 | 1.8 | −0.3 | |
| yhaM | 3ʹ→5ʹ exoribonuclease | 312 | 1.7 | 1.7 | −0.4 | |
| SPV_1828 | Hypothetical protein | 142 | 6.5 | 6.0 | 1.4 | |
| nadC | Quinolinate phosphoribosyltransferase | 72 | 1.7 | 3.2 | 0.3 | Secondary |
| SPV_1825b | IS630-Spn1 transposase | 162 | 0.5 | 1.7 | 0.0 | Secondary |
| SPV_1824 | ABC transporter, permease | 34 | 0.9 | 2.7 | 0.2 | Secondary |
| comGA | Late-competence protein | 14 | 9.4 | 10.4 | 5.1 | |
| comGB | Late-competence protein | 8 | 9.4 | 10.8 | 5.4 | |
| comGC | Late-competence protein | 6 | 9.3 | 11.2 | 5.3 | |
| comGD | Late-competence protein | 4 | 10.1 | 12.1 | 6.3 | |
| comGE | Late-competence protein | 3 | 10.3 | 12.3 | 6.5 | |
| comGF | Late-competence protein | 2 | 9.9 | 12.2 | 6.5 | |
| comGG | Late-competence protein | 8 | 8.7 | 11.0 | 5.5 | |
| SPV_2427b | S-Adenosylmethionine-dependent methyltransferase | 2 | 10.3 | 13.0 | 7.2 | |
| srf-29 | ncRNA of unknown function | 3 | 9.3 | 8.8 | 3.0 | |
| cbpD | Choline-binding protein D | 6 | 9.9 | 10.0 | 3.8 | |
| SPV_2027 | Cytoplasmic thiamin-binding component of thiamin ABC transporter | 89 | 1.0 | 1.2 | −0.7 | Secondary |
| thiX | Thiamin ABC transporter transmembrane component | 88 | 0.8 | 1.2 | −1.1 | Secondary |
| thiY | Thiamin ABC transporter substrate-binding component | 134 | 0.8 | 0.9 | −1.1 | Secondary |
| thiZ | Thiamin ABC transporter ATPase component | 128 | 0.6 | 1.0 | −1.2 | Secondary |
| comFA | DNA transporter ATPase | 3 | 9.0 | 8.1 | 2.9 | |
| comFC | Phosphoribosyltransferase domain protein | 3 | 8.6 | 8.5 | 2.7 | |
| hpf | Ribosome hibernation promotion factor | 624 | 0.9 | 1.4 | −0.3 | Secondary |
The 19 operons are indicated by the 9 blocks of data in a gray background and the 10 blocks of data in a white background. “Secondary” indicates either read-through after incomplete termination or the influence of an additional TSS. For complete supplemental information, including TSS positions, see Table S5. Boldface data indicate significance (P < 0.001; |log2 FC| > 1; DESeq [91]). NA, data not available.
Pseudogene.
The tyg TSS was previously found to be ComX regulated (32). An artificial 250-nucleotide transcript starting at this TSS was added to the annotation file to allow differential expression analysis.
A second ncRNA, srf-29, is located upstream of and partially overlaps cbpD (SPV_2028), a known late-com gene. A high-confidence terminator (41) inside the coding region of cbpD marks the 3ʹ end of srf-29. It is not clear whether srf-29 represents an uncharacterized RNA switch regulating cbpD expression, produces a functional sRNA, or simply is an artifact produced by a premature terminator [see PneumoBrowse coordinates 2008356 to 2008242 (−)].
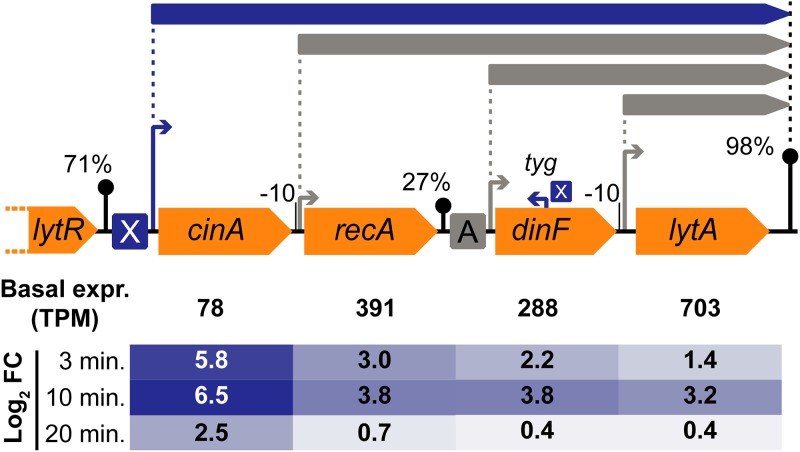
Since TSS and terminator information permits a promoter-based interpretation of our data, we observed examples of complex operon structures wherein TSSs or imperfect terminators inside the operon can lead to differences in the expression levels of different genes in the same operon (41, 56, 57). A striking example is the cinA-recA-dinF-lytA operon (SPV_1740 to SPV_1737), shown in Fig. 6, which is under the control of ComX, with only an inefficient (27%) terminator between recA and dinF. However, the presence of three internal TSSs, upstream of recA, dinF, and lytA, respectively, led to very different basal expression levels at t = 0 (Table 3). Due to these differences, the extent to which competence induction affects the expression of the four genes decreases from the 5ʹ end to the 3ʹ end of the operon (Fig. 6). Finally, Campbell et al. identified a transcription start site inside and antisense to dinF that was induced during competence and provisionally named the associated hypothetical gene tyg (Fig. 6) (32). Although not discussed by Campbell and coworkers, both Håvarstein (58) and Claverys and Martin (59) argued that the peculiar positioning of the tyg TSS represents reason for doubts regarding the functionality of any transcript produced. However, as Claverys and Martin concede, the possibility cannot be ruled out that tyg has a role in mRNA stability of the cinA-recA-dinF-lytA operon. With Cappable-seq (60), we did indeed detect a transcription start site (41), accompanied by a consensus ComX recognition site (Fig. 6). Since we did not detect a clearly demarcated associated transcript, we artificially annotated a 250-nucleotide-long transcript, starting at the tyg TSS, to allow differential expression analysis. The time-dependent trend of expression of this transcript during competence seemed to follow that of other late-com genes (39). However, the extremely low expression level detected prior to (and even after) CSP addition precluded any further statistical analysis regarding differential expression or clustering. We reiterate that TSS and terminator data regarding other competence-specific operons can be retrieved from PneumoBrowse (41).
FIG 6.
(Top) Overview of the complex cinA-recA-dinF-lytA operon, with an imperfect internal terminator and TSSs upstream of each gene, leading to the presence of four overlapping operons. The TSS upstream of cinA is preceded by a ComX-binding site, and addition of CSP indeed affected expression of all four genes in the operon. (Bottom) Log2 fold change relative to t = 0 (i.e., basal expression [expr.]). However, the effect size decreased with every gene, due to differences in basal expression from the internal TSSs. Additionally, a ComX-regulated TSS was found inside and antisense to dinF, giving rise to the hypothetical transcript tyg (32), with an unknown 3ʹ end.
The expression pattern of 44 of the 56 members of the ComX-associated WGCNA cluster (cl. 11) can now be linked to a ComX-regulated TSS. Bearing in mind that the clustering was performed based on expression under all 22 infection-relevant conditions studied in PneumoExpress (39), only five other cluster members (SPV_0553, SPV_0957 to SPV_0959, and SPV_2317) showed an expression pattern similar to that seen with ComX-regulated genes specifically under competence conditions. While the TSS for SPV_0553 has not been determined, this gene is surrounded by two repeat units of the pneumococcus (55) and one BOX element (49) and nothing resembling a ComX-binding site was found near it. Second, SPV_2317 represents a novel ncRNA (srf-19), potentially an RNA switch, that is preceded by a predicted RpoD site, rather than a ComX site. The last cluster member, operon SPV_0957 to SPV_0959, contains rpoD (SPV_0958). In light of the proposed role of RpoD in the shutdown of late competence (see above), it would be interesting if its upregulation was directly induced by ComX. However, analysis of the promoter region of the operon yielded no indication of a ComX-binding site and the mechanism of rpoD induction in competence continues to elude us.
The CiaR regulon is induced during competence and contains a novel noncoding RNA.
Besides early-com genes and late-com genes, terminology reserved for the ComE and ComX regulons, respectively, many other genes are more indirectly affected by the addition of CSP. Although a small portion of these genes can already be seen to be affected after 3 min, here we refer to all of these genes collectively as “delayed,” as these changes occur at least after the activation of the ComE regulon and most likely also downstream of the ComX regulon. Among the delayed genes are nearly all members of the CiaR regulon (Table 4), and promoter analysis of the HC members of the CiaR regulon (Table S3; see also Fig. S2) identified the following CiaR recognition site as previously reported (37, 45, 46): [TTTAAG]-[N5]-[TTTAAG]-[N22]-[TSS] (Fig. 3, bottom left). Analysis of other affected promoters turned up three additional monocistronic operons (ccnC, SPV_0098, and SPV_0775), all of which have already previously been reported to be regulated by CiaR. While SPV_0098 is expressed from two different TSSs (45) and ccnC (SPV_2078) expression is affected by transcriptional read-through from the upstream ComE-regulated comW operon, it is not clear why SPV_0775 does not cluster with other CiaR-regulated genes.
TABLE 4.
CiaR-regulated genes, distributed over 18 operonsa
| Gene | Product | TPM at 0 min | Log2 fold change at: |
Note | ||
|---|---|---|---|---|---|---|
| 3 min | 10 min | 20 min | ||||
| ccnC | csRNA3 | 155 | 4.4 | 1.2 | 0.0 | Also ComE regulon |
| SPV_0098 | Glycosyltransferase, group 2 family | 218 | −0.5 | 1.0 | 0.3 | |
| ccnE | csRNA5 | 32 | 0.6 | 0.8 | 0.2 | |
| ccnA | csRNA1 | 27 | −2.0 | −1.4 | −1.0 | |
| ccnB | csRNA2 | 2 | −0.6 | 2.8 | −1.5 | |
| ccnD | csRNA4 | 25 | 0.4 | 1.9 | 0.0 | |
| manL | Mannose-specific PTS IIAB components | 3,379 | 0.0 | −3.3 | −1.0 | Also CcpA-binding site |
| manM | Mannose-specific PTS IIC component | 3,296 | 0.1 | −3.1 | −1.1 | |
| manN | Mannose-specific PTS IID component | 4,750 | 0.2 | −3.3 | −1.0 | |
| rimP | Bacterial ribosome SSU maturation protein | 228 | −0.2 | 1.0 | 0.8 | CiaR-binding motif on opposite strand |
| nusA | Transcription termination/antitermination protein | 206 | 0.1 | 1.0 | 0.9 | |
| SPV_0480 | Putative transcription termination protein | 117 | 0.5 | 1.2 | 1.1 | |
| SPV_0481 | L7Ae family ribosomal protein | 103 | 0.6 | 1.4 | 1.4 | |
| infB | Translation initiation factor 2 | 277 | 0.8 | 0.7 | 0.9 | |
| rbfA | Ribosome-binding factor A | 194 | 1.0 | 0.5 | 0.8 | |
| ciaR | Two-component system response regulator | 225 | 0.2 | 3.2 | 0.3 | |
| ciaH | Two-component system sensor histidine kinase | 164 | 0.3 | 3.2 | 0.2 | |
| SPV_0775 | Acetyltransferase | 28 | 0.4 | 4.8 | 1.0 | |
| prsA | Putative parvulin type peptidyl-prolyl isomerase | 267 | 1.3 | 3.9 | 0.6 | Potentially also ComX regulon |
| SPV_0913 | Extracellular protein | 55 | 2.0 | 5.9 | 1.3 | |
| rlmCD | 23S rRNA [uracil(1939)-C(5)]-methyltransferase | 38 | −0.4 | 2.0 | 0.4 | Secondary |
| tarIb | Ribitol-5-phosphate cytidylyltransferase | 308 | 0.6 | 0.4 | 0.2 | |
| tarJb | Ribulose-5-phosphate reductase | 362 | 0.4 | 0.3 | 0.1 | |
| licAb | Choline kinase | 319 | 0.6 | 0.2 | 0.1 | |
| licBb | Choline permease | 397 | 0.6 | 0.0 | −0.1 | |
| licCb | Cholinephosphate cytidylyltransferase | 422 | 0.8 | 0.0 | 0.0 | |
| srf-21 | ncRNA of unknown function | 325 | 0.7 | 3.2 | 0.3 | |
| axe1 | Acetyl xylan esterase 1/cephalosporin-C deacetylase | 58 | 0.8 | 4.2 | 0.2 | Secondary |
| SPV_1769 | Membrane protein | 497 | 0.0 | 1.9 | −0.4 | |
| malQ | 4-Alpha-glucanotransferase (amylomaltase) | 69 | 0.1 | 4.2 | −0.2 | |
| malP | Maltodextrin phosphorylase | 83 | 0.2 | 4.2 | −0.4 | |
| dltX | d-Alanyl-lipoteichoic acid biosynthesis protein | 277 | −0.6 | 1.9 | 0.2 | CiaR-binding motif on opposite strand |
| dltA | d-Alanine-poly(phosphoribitol) ligase subunit 1 | 392 | −0.3 | 1.6 | 0.1 | |
| dltB | d-Alanyl transfer protein | 322 | 0.0 | 1.6 | 0.0 | |
| dltC | d-Alanine-poly(phosphoribitol) ligase subunit 2 | 429 | 0.0 | 1.5 | 0.1 | |
| dltD | Poly(glycerophosphate chain) d-alanine transfer protein | 326 | 0.3 | 1.5 | 0.0 | |
| htrA | Serine protease | 49 | 1.5 | 7.0 | 1.5 | |
| parB | Chromosome partitioning protein | 57 | 0.9 | 6.5 | 1.4 | Secondary |
The 18 operons are indicated by the 9 blocks of data in a gray background and the 9 blocks of data in a white background. “Secondary” indicates either read-through after incomplete termination or the influence of an additional TSS. For complete supplemental information, including TSS positions, see Table S5. Boldface data indicate significance (P < 0.001; |log2 FC| > 1; DESeq [91]). PTS, phosphotransferase system; SSU, small subunit.
The indicated operon has two different detected TSSs. The TSS at 1159217 (−) is under the control of CiaR.
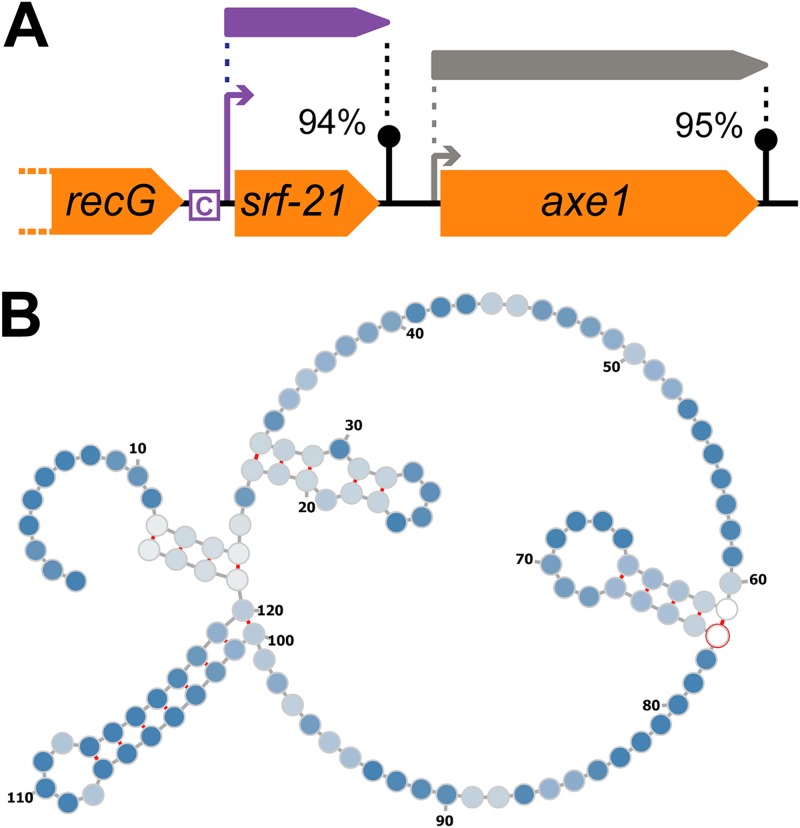
Since CiaR-binding sites were found on the opposite strand for dltXABCD (SPV_2006 to SPV_2002; upregulated) and manLMN (downregulated), we speculate that the operon containing, among others, rimP, infB, nusA, and rbfA (SPV_0478 to SPV_0483), which encode several proteins involved in translation, is also regulated by CiaR (Fig. S2). Intriguingly, another new member of the CiaR regulon is srf-21 (SPV_2378), a novel, uncharacterized noncoding RNA (41). The TSS from which this ncRNA is expressed was already part of the reported CiaR regulon but was linked to the overexpression of the downstream axe1 gene (SPV_1506). Inspection of the transcriptional layout of the region (Fig. 7A) showed that srf-21 and axe1 are separated by a relatively efficient terminator and a second TSS. Nonetheless, axe1 overexpression might still be attributable to read-through from srf-21. We did not find any similar ncRNAs in the RFAM and BSRD databases (61, 62), and, since axe1 is expressed from its own TSS, it seems unlikely that srf-21 functions as an RNA switch. Preliminary minimum free energy (MFE) secondary structure prediction performed with RNAfold (63) and target prediction performed with TargetRNA2 (64) did not provide us with any clear hints with regard to the function of this ncRNA. First, the predicted MFE structure (Fig. 7B) might represent only a transient conformation, since it makes up less than 1% of the modelled ensemble. Second, sRNA target prediction produced many potential targets (Table S4). Some candidate regions are less likely to be targeted, because they are located more than 20 nucleotides upstream of the start codon or even upstream of the TSS, ruling out a possible interaction between srf-21 and the transcript in question. However, future work will be necessary to reveal whether any of the remaining genes (e.g., queT, pezA, and cps2H) are regulated by srf-21 or, indeed, whether this ncRNA might have a completely different mode of action.
FIG 7.
Noncoding RNA srf-21 (SPV_2378) is part of the CiaR regulon. (A) Overview of the genomic context of srf-21. CiaR-dependent upregulation of downstream gene axe1 might be due to read-through from srf-21. The CiaR-binding sequence is indicated by a boxed “C.” (B) MFE secondary structure of srf-21, as predicted by RNAfold (63).
It is noteworthy that four reported promoters of the CiaR regulon were not found to be significantly affected during competence. First, the tarIJ-licABC operon is under the control of two TSSs and is thereby apparently less sensitive to CiaR control. Finally, three of five cia-dependent small RNAs (csRNAs), described by Halfmann et al. (37) did not appear to be significantly affected (Table 4). Note, however, that the quality of the statistical data on these short transcripts is rather poor and that the current data can neither support nor disprove their upregulation in competence. However, we do believe that the data presented by Halfmann et al. regarding CiaR-regulation seem perfectly convincing and the promoter regions of each of the five csRNAs contain a clear match with the consensus CiaR-binding site (Fig. S2).
Other known regulons affected during competence.
Regardless of whether or not competence should be regarded as a stress response mechanism in itself, it is clear that the activation of competence, at least indirectly, leads to a multifactorial stress response. Still unknown in previous descriptions of the competence regulon, the VraR (LiaR) regulon has been described by Eldholm et al. to be activated in response to competence-induced cell wall damage (15). On the basis of three pneumococcal promoters and six Lactococcus lactis promoters (Table S3; see also Fig. S3), we rebuilt the consensus motif described by Eldholm et al. (Fig. 3, bottom right) and observed that all of these motifs are located 32 to 34 nucleotides upstream of the corresponding TSS. In contrast, we were able to confirm the reported presence of a VraR-binding site upstream of hrcA (15), but this site is 81 nucleotides removed from its target TSS. However, the fact that this promoter region also carries two HrcA-binding sequences could account for this difference in spacing. Finally, we suggest that SPV_1057 (spr1080 in R6) and SPV_1160 (spr1183) are not regulated by VraR, in contrast to the report by Eldholm and coworkers. First, both of these genes lacked a detected TSS and neither was found to be differentially expressed in our study. Second, the reported recognition site for SPV_1057 is actually located downstream of the gene, inside a repeat region (ISSpn7 element). Finally, as recognized by Eldholm et al., SPV_1160 represents a 5ʹ-truncated version of a gene putatively encoding the ATP-binding component of an ABC transporter. These observations, combined with the fact that the reported VraR-binding site is located only 24 nucleotides upstream of the annotated start of SPV_1160, led us to conclude that SPV_1160, like SPV_1057, is not regulated by VraR. This limits the VraR regulon to 15 genes, distributed over 4 operons (Table 5). Eldholm et al. showed that specifically CbpK (PcpC; SPV_0357) and a putative phage shock protein (SPV_0803) are important in preventing competence-mediated lysis, forming an additional layer of protection besides ComM (15).
TABLE 5.
Competence-induced genes that are part of the VraR, CtsR, and/or HrcA regulonsa
| Gene | Product | TPM at 0 min | Log2 fold change at: |
Regulon(s) | Note | ||
|---|---|---|---|---|---|---|---|
| 3 min | 10 min | 20 min | |||||
| spxA2 | Transcriptional regulator SpxA2 | 772 | 0.0 | 1.4 | 0.0 | VraR | |
| SPV_0179 | Hypothetical protein | 163 | −0.2 | 1.7 | 0.1 | VraR | Secondary |
| clpL | Clp protease ATP-binding subunit ClpL | 24 | −0.2 | 6.6 | 3.7 | CtsR | |
| vraT | Cell wall-active antibiotic response protein VraT | 62 | 0.1 | 2.4 | −0.1 | VraR | |
| vraS | Two-component system sensor histidine kinase VraS | 52 | 0.0 | 2.2 | 0.0 | VraR | |
| vraR | Two-component transcriptional regulator VraR | 48 | 0.1 | 2.2 | −0.3 | VraR | |
| alkDb | DNA alkylation repair enzyme AlkD | 43 | 0.0 | 2.2 | 0.0 | VraR | |
| SPV_0355 | Hypothetical protein | 118 | −0.2 | 1.2 | 0.5 | VraR | Secondary |
| cbpGa | Choline-binding protein CbpG | 156 | −0.2 | 1.1 | 0.4 | VraR | Secondary |
| cbpK | Choline-binding protein CbpK | 151 | 0.1 | 1.0 | 0.3 | VraR | Secondary |
| hrcA | Heat-inducible transcription repressor HrcA | 650 | −0.2 | 2.7 | 0.1 | VraR, HrcA | |
| grpE | Heat shock protein GrpE | 600 | −0.3 | 2.7 | −0.1 | VraR, HrcA | |
| dnaK | Chaperone protein DnaK | 1,005 | −0.6 | 2.5 | −0.2 | VraR, HrcA | |
| SPV_2171 | Hypothetical protein | 679 | −0.8 | 2.3 | −0.1 | VraR, HrcA | |
| dnaJ | Chaperone protein DnaJ | 551 | −0.6 | 2.1 | −0.1 | VraR, HrcA | |
| clpE | Clp protease ATP-binding subunit ClpE | 168 | 0.2 | 1.4 | 0.9 | CtsR | |
| SPV_0803 | Putative phage shock protein C | 316 | 0.1 | 1.7 | −0.1 | VraR, HrcA | |
| groES | Heat shock protein 60 family cochaperone GroES | 1,099 | 0.5 | 2.5 | 0.6 | CtsR, HrcA | |
| groEL | Heat shock protein 60 family chaperone GroEL | 1,146 | 0.3 | 2.2 | 0.6 | CtsR, HrcA | |
| ctsR | Transcriptional regulator CtsR | 162 | −0.2 | 1.3 | 1.2 | CtsR | |
| clpC | Clp protease ATP-binding subunit ClpC | 257 | −0.1 | 1.0 | 0.9 | CtsR | |
| SPV_2021 | Hypothetical protein | 220 | 0.3 | 1.1 | 1.0 | CtsR? | |
| SPV_2020 | Two-component system response regulator | 167 | 0.1 | 0.9 | 1.1 | CtsR? | |
| SPV_2019 | Two-component system sensor histidine kinase | 86 | 0.4 | 1.1 | 1.2 | CtsR? | |
The different operons are indicated by blocks of data in a gray background and blocks of data in a white background. “Secondary” indicates either read-through after incomplete termination or the influence of an additional TSS. For complete supplemental information, including TSS positions, see Table S5. Boldface data indicate significance (P < 0.001; |log2 FC| > 1; DESeq [91]).
Pseudogene.
Besides VraR and CiaR, the well-characterized HrcA (3, 65) and CtsR (3, 66) regulons are also activated in competent cells, as previously reported (3). The only addition to be made here regarding the HrcA regulon is the annotation of a gene encoding a protein of unknown function (SPV_2171) that had not been annotated in D39 or R6 strains previously. This gene is located between dnaK (SPV_0460) and dnaJ (SPV_0461) and is therefore regulated by both VraR and HrcA (Table 5). Expression of almost the entire CtsR regulon was found to be upregulated. The observed upregulation was below the employed cutoff only for clpP (SPV_0650), possibly because its basal expression level (t = 0) is 4-fold to 40-fold higher than that of other clp genes. Finally, the upregulation of an uncharacterized two-component regulatory system (SPV_2020 and SPV_2019), with unknown consequences, could be attributed to transcriptional read-through from the upstream ctsR-clpC operon.
Together, CtsR and HrcA account for the overproduction of several subunits of the Clp protease system and of several chaperone proteins (e.g., DnaK, DnaJ, and GroES-GroEL) during competence (Table 5). Both of these protein functions are aimed at the reduction of stress caused by misfolded proteins and might be required to ensure proper folding of the many competence proteins.
Two other regulons seemed overrepresented in the set of differentially expressed genes (P < 10−4, hypergeometric test). First, transcription of all six genes predicted to be regulated by GntR (SPV_1524), on the basis of homology to Streptococcus pyogenes Spy_1285 (67), was found to be upregulated 10 and 20 min after addition of CSP. These six genes are distributed over two operons (SPV_0686 to SPV_0688 and SPV_1524 to SPV_1526). Second, expression of a significant number (n = 31) of RpoD-regulated genes was downregulated, mostly after 10 and 20 min, which may readily be explained by the competition for RNA polymerase of RpoD (σA) with the alternative sigma factor ComX (σX).
Other differentially expressed genes.
A total of 367 genes (i.e., 17% of all annotated genes) were either found to be differentially expressed or, at least, to be under the control of a TSS that appears to be differentially regulated at some point during competence (Table S5). The response of a large portion (204 genes) of these can be ascribed to the action of one of the transcriptional regulators discussed above. While 56% of the members of the latter group displayed a maximum absolute change in expression of more than 4-fold, only 29 of the remaining 163 genes (16%), distributed over 14 operons, met the same criterion. These data show that the bulk of strong induction or repression can be explained by the activity of a small set of regulators. Among the 29 strongly differentially expressed genes with no known regulators, our data confirmed the upregulation of rpoD, in line with the role that RpoD might play in late-competence shutdown (36).
Functional analysis did not reveal any gene ontology (GO) or KEGG classes overrepresented among upregulated genes. Only classes related to ribosomes and translation seemed overrepresented, but the realization that all affected genes from these classes were part of a single operon (shown in Table 1) led us to discard them due to lack of evidence (see Materials and Methods). Similarly, most of the potential hits among downregulated genes were discarded. Only classes related to thiamine metabolism (GO 0009228, KEGG ko00730) remained. The five genes in question are distributed over four operons: adk (SPV_0214), thiM-1-thiE-1 (SPV_0623 and SPV_0624), thiD (SPV_0632), and sufS (SPV_0764). Two of these four operons are regulated by a TPP riboswitch, an RNA element that, when bound to thiamine pyrophosphate (TPP), prevents transcription of the downstream operon (68). We suspect, therefore, that the temporary growth lag accompanying competence development (3) leads to a transient accumulation of TPP, which then represses transcription of operons under the control of a TPP riboswitch. Indeed, both of the other D39V operons led by a TPP riboswitch showed a similar expression trend 10 to 20 min after competence induction. First, ykoEDC-tenA-thiW-thiM-2-thiE-2 (SPV_0625 to SPV_0631) is already minimally expressed prior to CSP addition, preventing significant downregulation (not shown). The second operon, SPV_2027-thiXYZ (SPV_2027 to SPV_2024), was excluded from gene enrichment analysis since it was part of the ComX regulon (Table 3). Since the hypothesized accumulation of TPP seems to occur with a delay, relative to the activation of the ComX regulon, expression of these genes is first upregulated (3 to 10 min) and then downregulated (20 min) even relative to the basal expression level.
Comparison to previous reports of the competence regulon.
Finally, we compared our findings with those reported from previous, microarray-based studies by Peterson et al. (4) and Dagkessamanskaia et al. (3). Although both reports give a remarkably complete overview, our approach allowed us to refine and nuance the description of the competence regulon even further (Table S5). The higher levels of sensitivity and accuracy of Illumina sequencing, the improved genome annotation, and the application of a promoter-based (rather than gene-based) analysis allowed us to expand the set of genes identified as under direct control of ComE and ComX to 40 and 55 genes, respectively (combined, 4% of all genes). In particular, several genes with putative BlpR-binding sites (Table 2) and, therefore, a generally weaker response were missing in previous reports. Additionally, the previously reported briC operon (40) is now included in the ComE regulon and we confirmed that, while undetected by Dagkessamanskaia et al., ybbK and def2 (early) and radC (late) are indeed part of the com regulon (Tables 2 and 3). Remaining discrepancies could be explained either by transcriptional read-through or by the absence of certain elements (e.g., ncRNAs) from the TIGR4 and R6 genome annotation files used by Peterson et al. and Dagkessamanskaia et al., respectively.
Not surprisingly, larger discrepancies were found for genes displaying delayed differential expression, since their fold changes are (mostly) considerably smaller and therefore more sensitive to technical variation and differences in experimental conditions (e.g., growth medium, pH, or time of induction). In contrast with the previous studies, we identified nearly the entire CiaR regulon as differentially expressed in our data, confirming the higher sensitivity of RNA-seq than of microarray-based technology.
DISCUSSION
Competence for genetic transformation is defined as a state in which a bacterial cell can take up exogenous DNA and incorporate it into its own genome, either in the form of a plasmid or via homologous recombination. Since the very first demonstrations of bacterial transformation were provided by Griffith and Avery et al. (69, 70) in S. pneumoniae, the pneumococcal competence system was widely studied as soon as the required tools became available. Therefore, much knowledge has been assembled about how this state is regulated, which environmental triggers affect its development, and what downstream consequences it has. Indeed, several studies have been performed to compile a comprehensive list of all competence-regulated pneumococcal genes. The most recent of these studies (3, 4), although of very high quality and invaluable to the research field, date from nearly 15 years ago. Since then, the fields of transcriptome analysis and genome sequencing and annotation have been revolutionized by second-generation (e.g., Illumina) and third-generation (e.g., PacBio) sequencing techniques. Therefore, we have analyzed Illumina-based RNA-seq data (39), using the recently sequenced and deeply annotated S. pneumoniae D39V strain (41), to refine the previously reported pneumococcal competence regulon. Additionally, rather than just reporting affected individual genes, we used previously determined transcript boundaries (TSSs and terminators [41]) to identify the affected promoters, which may be used to gain more insights into the regulatory processes at work during competence.
In short, we report that ComE directly regulates 15 early-com transcripts (40 genes), including 4 transcripts (10 genes) that are part of the BlpR regulon (Table 2; see also Fig. 4). Alternative sigma factor ComX (σX) was found to control 19 late-com transcripts (55 genes), in addition to the previously described tyg TSS, inside and antisense to dinF (Table 3). We should note that four genes from the early-com and late-com regulons (e.g., blpC) did not meet the requirements with respect to fold change and/or statistical cutoff values but were part of operons that were clearly regulated. For each of these genes, the observed expression trends correlated with those of other operon members.
Our data confirmed that, as shown in the previous studies (3, 4), the activation of the early-competence and late-competence regulons indirectly resulted in the activation of several other regulons, most of which are implicated in pneumococcal stress response. First, in the newly compiled CiaR regulon (18 transcripts, 38 genes), based on the work of Halfmann et al. (37), 4 operons were not found to be significantly affected (Table 4). Other affected regulons were those under the control of VraR (LiaR), HrcA, CtsR, and GntR (see Table S5 in the supplemental material). Additionally, transcription of genes led by a TPP riboswitch was downregulated, suggesting a transient increase in intracellular thiamine pyrophosphate levels in competent cells.
Expression of many other genes, including rpoD, is upregulated or downregulated through unknown mechanisms. In total, approximately 140 transcripts (containing 367 genes, 17% of all genes) undergo some extent of differential expression. For several reasons (e.g., fold changes near cutoff, expression from multiple TSSs, or poor statistics due to low expression), 79 genes from these operons did not individually meet the detection criteria, leaving 288 differentially expressed genes (13%) following the traditional, gene-based analysis approach (Fig. 2). Among the affected genes are several small, noncoding transcripts. Some of these ncRNAs, like the CiaR-activated csRNAs (see below), have been characterized, and we showed that srf-01 is unlikely to be functional. For others, e.g., srf-03 (upstream of comAB) and srf-21 (upstream of axe1), future work is required to determine their role, if any, during competence.
Given the fact that so many different functionalities are activated during competence, including stress response systems such as the Clp protease and several chaperone proteins, Claverys et al. proposed to refer to the system more neutrally as “X-state” (for ComX). We would argue, however, that the primary response to a high extracellular CSP level is the activation of the ComE and ComX regulons, which mostly encode proteins relevant to transformation. First, the expression of fratricin CbpD (SPV_2028), along with immunity protein ComM (SPV_1744), allows the lysis of neighboring noncompetent cells, which may offer access to both nutrients and DNA (71, 72). The upregulation of the gene encoding autolysin LytA (lytA) would fit in nicely here, since the simultaneous deletion of lytC and lytA abolishes competence-induced lysis completely (71). However, basal-level lytA expression was reported to be already sufficient for the observed lysis rate (73). Next, the DNA uptake machinery comes into play, involving many proteins encoded by the late-com genes, as reviewed by Claverys et al. (16); most of ComGC to ComGG (SPV_1861 to SPV_1857), ComEA/C (SPV_0843 and SPV_0844), and ComFA (SPV_2035) are, together with constitutively expressed endonuclease EndA (SPV_1762) and through the action of prepilin peptidase CclA (SPV_1593) and proteins ComGA/B (SPV_1863 and SPV_1862), assembled into a pilus-like structure (74). The importation of DNA is followed by DNA processing and recombination and involves DNA protection protein DprA (SPV_1122), recombinase RecA (SPV_1739), single-stranded DNA (ssDNA)-binding SsbB (SPV_1711), and other late-competence proteins (75–78). For the reasons discussed above, combined with the fact that the vast majority of other differentially expressed genes display a more modest change in expression (Table S5), we decided to continue calling the system competence for genetic transformation. We do, however, agree that the downstream effects of competence development should not be ignored, as discussed below.
The genome of Streptococcus pneumoniae D39V contains 13 two-component systems (TCSs) for regulation (41, 79). Interestingly, the regulons of two TCSs (ComDE and BlpRH) are activated during competence, while the regulons of another two TCSs (CiaRH and VraRS) are activated shortly after. Expression of a fifth, uncharacterized TCS (SPV_2020 and SPV_2019) is also slightly upregulated, possibly due to transcriptional read-through from the ctsR-clpC operon. The additional activation of stress-related regulons of HrcA and CtsR gives support to the hypothesis, as proposed by Prudhomme at al. (5), that competence activation serves as a general stress response in the pneumococcus, which lacks a LexA-mediated SOS response such as is common in many other bacteria. The activation of competence in response to various types of stress (5–8) provides even more relevance to this idea. On the other hand, Dagkessamanskaia et al. showed that a deletion of the CiaR regulon caused an extended growth lag, as well as stronger activation of the HrcA and CtsR regulons, after competence induction (3). Both observations are in line with the notion that the development of competence is accompanied by a significant burden to the cell. To our knowledge, the exact mechanism underlying CiaR-mediated prevention of autolysis is unknown. However, Halfmann et al. showed that the deletion of CiaR-induced noncoding csRNA4 and csRNA5 led to an enhanced-lysis phenotype (37). Furthermore, d-alanylation of lipoteichoic acids by the DltXABCD proteins (45) might contribute to cell wall integrity during or after the transient block in cell division that accompanies competence (19). Finally, HtrA can play a role in the cleaning up of potential misfolded or excess proteins that cause stress to the pneumococcus (9).
We consider it likely that the main benefit of the activation of the CiaR and other stress response regulons during competence is that of dealing with the stress invoked by competence itself, by building the large membrane- and cell wall-protruding DNA uptake machinery and the production of cell wall hydrolases such as the fratricin CbpD. It is not unthinkable, however, that the activation of the many different stress response regulons renders it beneficial to a pneumococcal cell to become competent even under specific stressful conditions that do not require DNA uptake or recombination machineries. Whether or not such conditions played a role in the (co)evolution of competence and downstream processes is open to speculation. In this respect, it is interesting that the production of competence-induced bacteriocins was found to be important to prevent “intruder” pneumococci from colonizing (80).
The apparent severity of the stress imposed on a competent cell emphasizes the need to shut down competence after a short transformation-permissive time window. In addition to the known role of DprA in early-competence shutdown, Weyder et al. proposed that the upregulation of rpoD is responsible for the (less efficient) shut-down of late competence (36). Related to this, RpoD-regulated genes are, to some extent, overrepresented among the downregulated genes during competence. While this explains the need for upregulation of rpoD, the underlying mechanism is still unknown. Other aspects that might play a role in the shutting down of competence are the CiaR-mediated upregulation of HtrA, which has been shown to degrade extracellular CSP (9), and the recently discovered CiaR-regulated noncoding csRNAs (ccnA to ccnE) (37, 81–83), which were shown to repress ComC translation in an additive fashion.
Finally, although several of the activated regulons are quite well understood, a large portion of affected genes are still differentially expressed through unknown mechanisms. It seems plausible that the expression of many of these genes is due to the sudden and severe shift in metabolic state. For example, the higher translational demands present during competence could lead to the upregulation of genes encoding ribosomal proteins (Table 1). Similarly, the hypothetical transient increase in TPP concentrations, leading to riboswitch-mediated downregulation of four operons, could be accompanied by the accumulation or depletion of other, unknown metabolites, with potential transcriptional consequences.
MATERIALS AND METHODS
The samples studied here represent a subset of the data set presented in PneumoExpress (samples C+Y; CSP, 3 min; CSP, 10 min; CSP, 20 min [39]), and detailed procedures regarding bacterial growth, RNA isolation, and sequencing and mapping of reads are reported there. The key points of these methods are summarized below.
Culturing and harvesting of S. pneumoniae D39V.
Eight tubes with 2 ml C+Y medium (pH 6.8; nonpermissive for natural competence [39]) without antibiotics were each inoculated with wild-type S. pneumoniae D39V cells (initial optical density at 600 nm [OD600], ∼0.004) and incubated at 37°C (standing culture in ambient air). When the cultures reached an OD600 of 0.05, two cultures were harvested for RNA isolation (t = 0). To the other six, 100 ng/ml synthetic competence-stimulating peptide (CSP-1), purchased from GenScript (Piscataway, NJ), was added. Duplicate samples were harvested 3, 10, and 20 min after CSP-1 addition. Before harvesting, cultures were pretreated with a saturated ammonium sulfate solution as described before (84), to prevent protein-dependent RNA production and degradation (85 [patent]). Afterward, cells were harvested by centrifugation (20 min, 4°C, 10,000 × g) and cell pellets were snap-frozen with liquid nitrogen and stored at −80°C.
Total RNA isolation, library preparation, and sequencing and mapping of reads.
RNA was isolated using phenol-chloroform extraction, followed by DNase treatment and another round of phenol-chloroform extraction (39). The quantity and quality of total RNA were estimated by the use of NanoDrop technology, while a 1% bleach gel (86) was employed to confirm the presence of rRNA bands (23S, 2.9 kbp; 16S, 1.5 kbp) and the absence of genomic DNA. Subsequently, RNA quality was again checked using chip-based capillary electrophoresis (Agilent Bioanalyzer). Stranded cDNA library preparation was performed, without depletion of rRNA, using a TruSeq stranded total RNA sample preparation kit (Illumina, USA). Sequencing was performed on an Illumina NextSeq 500 system, in 75-nucleotide single-end mode. The raw FASTQ data are accessible at https://www.ncbi.nlm.nih.gov/geo/ with accession number GSE108031 (samples B05 to B11).
After a quality check performed with FastQC v0.11.5 (87), reads were trimmed using Trimmomatic 0.36 (88). Alignment of trimmed reads to the reference S. pneumoniae D39V genome (GenBank CP027540 [41]) was performed with STAR (89).
Read quantification and differential gene analysis.
The aligned reads were then counted (90) according to the D39V annotation file (GenBank CP027540 [41]) in a strand-specific fashion, allowing mapping to multiple sites (-M), for which fractional counts are reported (–fraction), and allowing reads to overlap multiple features (-O) to account for polycistronic operons.
Subsequently, we analyzed the libraries in R-studio (R v3.4.2). We performed differential gene expression analysis on rounded raw counts by DESeq2 (91). Normalized expression levels are presented as TPM (transcripts per million) (92) and can be found in Table S1 in the supplemental material. Genes with an absolute change of expression of more than 2-fold and a corresponding P value of below 0.001 were considered to be significantly differentially expressed.
When possible, PneumoBrowse (https://veeninglab.com/pneumobrowse [41]) was used to trace back differential expression levels of individual genes to specific TSSs and promoter regions. As a starting point, the operon prediction from PneumoBrowse was used to define groups of genes differentially expressed in competence. Note that strong transcriptional responses such as those observed during competence may have significant downstream effects. Even in the presence of highly efficient transcriptional terminators, which were defined to be operon boundaries in PneumoBrowse, such read-through effects may be visible. Therefore, these coexpressed groups were refined by inspection of the raw data in PneumoBrowse and in accordance with the consideration that minor read-through from a highly expressed gene can still be significant if the expression level of the downstream gene is sufficiently lower.
Clustering and creation of position weight matrices.
Using the weighted gene coexpression network analysis (WGCNA) R software package (93), genes were clustered based on their rlog (regularized log) expression value (Table S2), as output by DESeq2, across all 22 infection-relevant conditions analyzed in PneumoExpress (39). We noticed that the reported members of the ComE (26, 27), ComX (3, 4, 38), and CiaR (37, 45, 46) regulons each largely ended up in specific clusters (here, clusters 29, 11, and 33 for ComE, ComX, and CiaR, respectively). Previously reported regulon members that properly clustered in these three identified main clusters, which are referred to here as “training sets” (Table S3), were used to define the recognition motifs of these three regulators, in the form of position weight matrices (PWMs) and to determine the most common distance of such a motif from the TSS. Using the MEME suite (94), we analyzed the upstream regions in each training set for enriched sequence motifs. First, since earlier work had shown slightly different consensus sequences for the two tandem ComE boxes that make up the ComE site (27), we extracted the left ComE motif (CEML) by scanning the regions from 77 to 63 bp upstream and the right motif (CEMR) by scanning the regions from 56 to 42 bp upstream of TSSs in the training set. The ComX-binding motif (CXM) was determined from the regions 35 bp upstream to the +1 site (TSS). In building the CEML, CEMR, and CXM PWMs, each sequence in the training set was required to have exactly one match to the motif, in the transcription direction (i.e., on the locally defined “plus” strand).
CiaR has been described to bind to a direct repeat (37), and we scanned the regions 41 to 19 bp upstream, allowing multiple hits per sequence in the training set. While some members of the CiaR regulon have binding sites on the opposite strand, no such genes were part of the training set and the CiaR-binding motif (CRM) was therefore also limited to the plus strand. Genes reported to belong to the VraR (LiaR) regulon (15) did not cluster together throughout the 22 sets of conditions, and the TSS was unknown for some of these genes. To enable extraction of a VraR-binding motif (VRM), we combined upstream regions of pneumococcal genes spxA2 (SPV_0178), vraT (SPV_0350), and SPV_0803 with those of six Lactococcus lactis genes that were reported to be regulated by the close VraR homolog CesR (15, 95) as follows: llmg_0165, llmg_0169, llmg_1115, llmg_1155, llmg_1650, and llmg_2164. Cappable-seq (60) was used to identify L. lactis TSSs (S. B. van der Meulen and O. P. Kuipers, unpublished data). Importantly, we did not use the standard “0-order model of sequences” as a background model for motif discovery but instead created background models based on the corresponding regions upstream of all known TSSs in the pneumococcal genome (e.g., −35 to +1 for ComX). Additionally, we defined summary consensus sequences using IUPAC nucleotide coding. Since the CiaR-binding motif reportedly consists of two perfect repeats, we determined the consensus based on the 16 motif occurrences in the CiaR training set (8 promoter sequences). Single-base codes (A, C, G, and T) were called when 75% (rounded up) of all promoters matched. Double-base codes (R, Y, S, W, K, and M) were called when 8/9 (ComE and VraR), 15/16 (ComX), or 5/5 (BlpR) promoters matched either of the two encoded bases. Triple-base codes (B, D, H, and V) were called when all promoters matched either of the three encoded bases. Note that, due to its degenerate appearance, the blpRS promoter was excluded from determinations of the BlpR-binding consensus.
Assigning putative regulons.
After creating PWMs for ComE-, ComX-, CiaR-, and VraR-binding sites, we used FIMO (96) to scan the 100 bp upstream of all known pneumococcal TSSs for matches to these motifs. Here, too, we used the appropriate background models (see above). A cutoff false-discovery rate (q) value of 0.01 was used for hits with ComX- and VraR-binding motifs. We defined a reliable ComE-binding site as CEML-[N11-13]-CEMR, using a cutoff P value of 0.01 for each motif. Similarly, we defined a CiaR-binding site as CRM-[N5-6]-CRM. Additionally, to assign a gene cluster to a given putative regulon, we also applied a constraint to the position of the motif relative to the corresponding TSS, based on the typical spacing observed in the training sets. Thus, the allowed first nucleotide positions were as follows: −77, −76, −75, −74, or −73 for ComE; −30, −29, or −28 for ComX; −40, −39, −38, −37, or −36 for CiaR; and −51, −50, −49, −48, or −47 for VraR.
Putative binding sites for other regulatory proteins were copied from the propagated S. pneumoniae D39 regulons, as found in the RegPrecise database (67) and annotated in PneumoBrowse (41). RNA switches, annotated in D39V, were also taken into consideration as putatively responsible regulatory mechanisms.
Functional analysis of differentially expressed genes.
Differentially expressed genes that could not be ascribed to the action of ComE, ComX, CiaR, or VraR were subjected to gene set enrichment analysis (functional analysis). For this, gene ontology and KEGG classifications (Table S6) were extracted from the GenBank file corresponding to the latest annotation of S. pneumoniae D39V (41). Additionally, predicted transcription factor binding sites were used to assign genes to their putative regulons (Table S6). A total of 448 hypergeometric tests were performed, and a Bonferroni-corrected cutoff P value of 0.0001 (i.e., 0.05 divided by 448) was used to determine whether certain regulons or gene ontology or KEGG classes were overrepresented among differentially expressed genes. We excluded overrepresented classes for all affected genes belonging to the same operon, since the activation of a single promoter does not confer any statistical evidence.
Data availability.
The data analyzed here can be extracted from PneumoExpress (https://veeninglab.com/pneumoexpress-app). Raw RNA-seq data used to build PneumoExpress were deposited to the GEO repository under accession number GSE108031.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to A. de Jong and S. Holsappel for (bio)informatics support and to S. B. van der Meulen for sharing L. lactis TSS data.
Work in the Veening laboratory is supported by the Swiss National Science Foundation (project grant 31003A_172861), by a JPIAMR grant (50-52900-98-202) from the Netherlands Organization for Health Research and Development (ZonMW), and by ERC consolidator grant 771534-PneumoCaTChER.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00780-18.
For a commentary on this article, see https://doi.org/10.1128/JB.00238-19.
REFERENCES
- 1.Henriques-Normark B, Tuomanen EI. 2013. The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb Perspect Med 3:a010215. doi: 10.1101/cshperspect.a010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2016 Lower Respiratory Infections Collaborators. 19 September 2018, posting date. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis doi: 10.1016/S1473-3099(18)30310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagkessamanskaia A, Moscoso M, Hénard V, Guiral S, Overweg K, Reuter M, Martin B, Wells J, Claverys J-P. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol 51:1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- 4.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 5.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys J-P. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 6.Slager J, Kjos M, Attaiech L, Veening J-W. 2014. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 7.Domenech A, Slager J, Veening J-W. 2018. Antibiotic-induced cell chaining triggers pneumococcal competence by reshaping quorum sensing to autocrine-like signaling. Cell Rep 25:2390–2400.e3. doi: 10.1016/j.celrep.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens KE, Chang D, Zwack EE, Sebert ME. 2011. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2:e00071-11. doi: 10.1128/mBio.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassone M, Gagne AL, Spruce LA, Seeholzer SH, Sebert ME. 2012. The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J Biol Chem 287:38449–38459. doi: 10.1074/jbc.M112.391482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J-D, Morrison DA. 1987. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J Gen Microbiol 133:1959–1967. doi: 10.1099/00221287-133-7-1959. [DOI] [PubMed] [Google Scholar]
- 11.Claverys J-P, Håvarstein LS. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front Biosci 7:d1798–d1814. doi: 10.2741/claverys. [DOI] [PubMed] [Google Scholar]
- 12.Echenique JR, Chapuy-Regaud S, Trombe MC. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol Microbiol 36:688–696. [DOI] [PubMed] [Google Scholar]
- 13.Claverys J-P, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu Rev Microbiol 60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 14.Turlan C, Prudhomme M, Fichant G, Martin B, Gutierrez C. 2009. SpxA1, a novel transcriptional regulator involved in X-state (competence) development in Streptococcus pneumoniae. Mol Microbiol 73:492–506. doi: 10.1111/j.1365-2958.2009.06789.x. [DOI] [PubMed] [Google Scholar]
- 15.Eldholm V, Gutt B, Johnsborg O, Brückner R, Maurer P, Hakenbeck R, Mascher T, Håvarstein LS. 2010. The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J Bacteriol 192:1761–1773. doi: 10.1128/JB.01489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claverys J-P, Martin B, Polard P. 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev 33:643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 17.Mascher T, Heintz M, Zahner D, Merai M, Hakenbeck R. 2006. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in β-lactam resistance. J Bacteriol 188:1959–1968. doi: 10.1128/JB.188.5.1959-1968.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei H, Håvarstein LS. 2012. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl Environ Microbiol 78:5897–5905. doi: 10.1128/AEM.01343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergé MJ, Mercy C, Mortier-Barrière I, VanNieuwenhze MS, Brun YV, Grangeasse C, Polard P, Campo N. 2017. A programmed cell division delay preserves genome integrity during natural genetic transformation in Streptococcus pneumoniae. Nat Commun 8:1621. doi: 10.1038/s41467-017-01716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin B, Granadel C, Campo N, Hénard V, Prudhomme M, Claverys J-P. 2010. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol Microbiol 75:1513–1528. doi: 10.1111/j.1365-2958.2010.07071.x. [DOI] [PubMed] [Google Scholar]
- 21.Hui FM, Zhou L, Morrison DA. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153:25–31. doi: 10.1016/0378-1119(94)00841-F. [DOI] [PubMed] [Google Scholar]
- 22.Alloing G, Martin B, Granadel C, Claverys JP. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol Microbiol 29:75–83. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 23.Håvarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Håvarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol 21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 25.Pestova EV, Håvarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 26.Ween O, Gaustad P, Håvarstein LS. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol 33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 27.Martin B, Soulet A-L, Mirouze N, Prudhomme M, Mortier-Barrière I, Granadel C, Noirot-Gros M-F, Noirot P, Polard P, Claverys J-P. 2013. ComE/ComE∼P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol Microbiol 87:394–411. doi: 10.1111/mmi.12104. [DOI] [PubMed] [Google Scholar]
- 28.Moreno-Gámez S, Sorg RA, Domenech A, Kjos M, Weissing FJ, van Doorn GS, Veening J-W. 2017. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun 8:854. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prudhomme M, Berge M, Martin B, Polard P. 2016. Pneumococcal competence coordination relies on a cell-contact sensing mechanism. PLoS Genet 12:e1006113. doi: 10.1371/journal.pgen.1006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowley LA, Petersen FC, Junges R, Jimson D Jimenez M, Morrison DA, Hanage WP. 2018. Evolution via recombination: cell-to-cell contact facilitates larger recombination events in Streptococcus pneumoniae. PLoS Genet 14:e1007410. doi: 10.1371/journal.pgen.1007410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell EA, Choi SY, Masure HR. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol 27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 33.Pestova EV, Morrison DA. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J Bacteriol 180:2701–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piotrowski A, Luo P, Morrison DA. 2009. Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW. J Bacteriol 191:3359–3366. doi: 10.1128/JB.01750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng L, Piotrowski A, Morrison DA. 2013. Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS One 8:e64197. doi: 10.1371/journal.pone.0064197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weyder M, Prudhomme M, Bergé M, Polard P, Fichant G. 2018. Dynamic modeling of Streptococcus pneumoniae competence provides regulatory mechanistic insights into its tight temporal regulation. Front Microbiol 9:1637. doi: 10.3389/fmicb.2018.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halfmann A, Kovács M, Hakenbeck R, Brückner R. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol 66:110–126. doi: 10.1111/j.1365-2958.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 38.Rimini R, Jansson B, Feger G, Roberts TC, de Francesco M, Gozzi A, Faggioni F, Domenici E, Wallace DM, Frandsen N, Polissi A. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol Microbiol 36:1279–1292. [DOI] [PubMed] [Google Scholar]
- 39.Aprianto R, Slager J, Holsappel S, Veening J-W. 2018. High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res 46:9990–10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aggarwal SD, Eutsey R, West-Roberts J, Domenech A, Xu W, Abdullah IT, Mitchell AP, Veening J-W, Yesilkaya H, Hiller NL. 2018. Function of BriC peptide in the pneumococcal competence and virulence portfolio. PLoS Pathog 14:e1007328. doi: 10.1371/journal.ppat.1007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slager J, Aprianto R, Veening J-W. 2018. Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Res 46:9971–9989. doi: 10.1093/nar/gky725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kjos M, Miller E, Slager J, Lake FB, Gericke O, Roberts IS, Rozen DE, Veening J-W. 2016. Expression of Streptococcus pneumoniae bacteriocins is induced by antibiotics via regulatory interplay with the competence system. PLoS Pathog 12:e1005422. doi: 10.1371/journal.ppat.1005422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wholey W-Y, Kochan TJ, Storck DN, Dawid S. 2016. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog 12:e1005413. doi: 10.1371/journal.ppat.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denapaite D, Brückner R, Hakenbeck R, Vollmer W. 2012. Biosynthesis of teichoic acids in Streptococcus pneumoniae and closely related species: lessons from genomes. Microb Drug Resist 18:344–358. doi: 10.1089/mdr.2012.0026. [DOI] [PubMed] [Google Scholar]
- 46.Mascher T, Zähner D, Merai M, Balmelle N, de Saizieu AB, Hakenbeck R. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J Bacteriol 185:60–70. doi: 10.1128/JB.185.1.60-70.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirouze N, Bergé MA, Soulet A-L, Mortier-Barrière I, Quentin Y, Fichant G, Granadel C, Noirot-Gros M-F, Noirot P, Polard P, Martin B, Claverys J-P. 2013. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A 110:E1035–E1044. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol 38:867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 49.Martin B, Humbert O, Camara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res 20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knutsen E, Johnsborg O, Quentin Y, Claverys J-P, Håvarstein LS. 2006. BOX elements modulate gene expression in Streptococcus pneumoniae: impact on the fine-tuning of competence development. J Bacteriol 188:8307–8312. doi: 10.1128/JB.00850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Saizieu A, Gardès C, Flint N, Wagner C, Kamber M, Mitchell TJ, Keck W, Amrein KE, Lange R. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol 182:4696–4703. doi: 10.1128/JB.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. 2010. The Abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol 192:2068–2076. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pei J, Grishin NV. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci 26:275–277. doi: 10.1016/S0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- 54.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oggioni MR, Claverys JP. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145:2647–2653. doi: 10.1099/00221287-145-10-2647. [DOI] [PubMed] [Google Scholar]
- 56.Warrier I, Ram-Mohan N, Zhu Z, Hazery A, Echlin H, Rosch J, Meyer MM, van Opijnen T. 2018. The transcriptional landscape of Streptococcus pneumoniae TIGR4 reveals a complex operon architecture and abundant riboregulation critical for growth and virulence. PLoS Pathog 14:e1007461. doi: 10.1371/journal.ppat.1007461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosinski-Chupin I, Sauvage E, Sismeiro O, Villain A, Da Cunha V, Caliot M-E, Dillies M-A, Trieu-Cuot P, Bouloc P, Lartigue M-F, Glaser P. 2015. Single nucleotide resolution RNA-seq uncovers new regulatory mechanisms in the opportunistic pathogen Streptococcus agalactiae. BMC Genomics 16:419. doi: 10.1186/s12864-015-1583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Håvarstein LS. 1998. Identification of a competence regulon in Streptococcus pneumoniae by genomic analysis. Trends Microbiol 6:297–300. doi: 10.1016/S0966-842X(98)01328-6. [DOI] [PubMed] [Google Scholar]
- 59.Claverys J-P, Martin B. 1998. Competence regulons, genomics and streptococci. Mol Microbiol 29:1126–1127. doi: 10.1046/j.1365-2958.1998.01005.x. [DOI] [PubMed] [Google Scholar]
- 60.Ettwiller L, Buswell J, Yigit E, Schildkraut I. 2016. A novel enrichment strategy reveals unprecedented number of novel transcription start sites at single base resolution in a model prokaryote and the gut microbiome. BMC Genomics 17:199. doi: 10.1186/s12864-016-2539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L, Huang D, Cheung MK, Nong W, Huang Q, Kwan HS. 2013. BSRD: a repository for bacterial small regulatory RNA. Nucleic Acids Res 41:D233–D238. doi: 10.1093/nar/gks1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kery MB, Feldman M, Livny J, Tjaden B. 2014. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res 42:W124–W129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narberhaus F. 1999. Negative regulation of bacterial heat shock genes. Mol Microbiol 31:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- 66.Derré I, Rapoport G, Msadek T. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol 31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 67.Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, Kazanov MD, Riehl W, Arkin AP, Dubchak I, Rodionov DA. 2013. RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 69.Griffith F. 1928. The significance of pneumococcal types. J Hyg (Lond) 27:113–159. doi: 10.1017/S0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Håvarstein LS, Martin B, Johnsborg O, Granadel C, Claverys J-P. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol 59:1297–1037. doi: 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 72.Veening J-W, Blokesch M. 2017. Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat Rev Microbiol 15:621–629. doi: 10.1038/nrmicro.2017.66. [DOI] [PubMed] [Google Scholar]
- 73.Moscoso M, Claverys J-P. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol Microbiol 54:783–794. doi: 10.1111/j.1365-2958.2004.04305.x. [DOI] [PubMed] [Google Scholar]
- 74.Laurenceau R, Péhau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le Cam E, Claverys J-P, Fronzes R. 2013. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog 9:e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergé M, Mortier-Barrière I, Martin B, Claverys J-P. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol Microbiol 50:527–536. doi: 10.1046/j.1365-2958.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 76.Desai BV, Morrison DA. 2006. An unstable competence-induced protein, CoiA, promotes processing of donor DNA after uptake during genetic transformation in Streptococcus pneumoniae. J Bacteriol 188:5177–5186. doi: 10.1128/JB.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burghout P, Bootsma HJ, Kloosterman TG, Bijlsma JJE, de Jongh CE, Kuipers OP, Hermans P. 2007. Search for genes essential for pneumococcal transformation: the RadA DNA repair protein plays a role in genomic recombination of donor DNA. J Bacteriol 189:6540–6550. doi: 10.1128/JB.00573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Attaiech L, Olivier A, Mortier-Barrière I, Soulet A-L, Granadel C, Martin B, Polard P, Claverys J-P. 2011. Role of the single-stranded DNA–binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity. PLoS Genet 7:e1002156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, Caspers P, Kamber M, Keck W, Amrein KE. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223–234. doi: 10.1016/S0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 80.Shen P, Lees JA, Bee GCW, Brown SP, Weiser JN. 2019. Pneumococcal quorum sensing drives an asymmetric owner–intruder competitive strategy during carriage via the competence regulon. Nat Microbiol 4:198. doi: 10.1038/s41564-018-0314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schnorpfeil A, Kranz M, Kovács M, Kirsch C, Gartmann J, Brunner I, Bittmann S, Brückner R. 2013. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol 89:334–349. doi: 10.1111/mmi.12277. [DOI] [PubMed] [Google Scholar]
- 82.Laux A, Sexauer A, Sivaselvarajah D, Kaysen A, Brückner R. 2015. Control of competence by related non-coding csRNAs in Streptococcus pneumoniae R6. Front Genet 6:246. doi: 10.3389/fgene.2015.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsui H-C, Mukherjee D, Ray VA, Sham L-T, Feig AL, Winkler ME. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol 192:264–279. doi: 10.1128/JB.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aprianto R, Slager J, Holsappel S, Veening J-W. 2016. Time-resolved dual RNA-seq reveals extensive rewiring of lung epithelial and pneumococcal transcriptomes during early infection. Genome Biol 17:198. doi: 10.1186/s13059-016-1054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Korfhage C, Wyrich R, Oelmüller U. August 2002. Ammonium sulfate for neutralization of inhibitory effects. European patent WO2002059360A2.
- 86.Aranda PS, LaJoie DM, Jorcyk CL. 2012. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis 33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 88.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 91.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 93.Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 95.Martínez B, Zomer AL, Rodríguez A, Kok J, Kuipers OP. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol Microbiol 64:473–486. doi: 10.1111/j.1365-2958.2007.05668.x. [DOI] [PubMed] [Google Scholar]
- 96.Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lanie JA, Ng W-L, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed here can be extracted from PneumoExpress (https://veeninglab.com/pneumoexpress-app). Raw RNA-seq data used to build PneumoExpress were deposited to the GEO repository under accession number GSE108031.