In Bacillus subtilis, the Spx transcription factor is proteolytically unstable, and protein stabilization figures prominently in the induction of the Spx regulon in response to oxidative and cell envelope stresses. ClpXP is largely, but not entirely, responsible for Spx instability. Here, we identify ClpCP as the protease that degrades Spx under conditions that antagonize the ClpXP pathway. Spx itself contributes to activation of the ctsR operon, which encodes ClpC as well as the McsB arginine kinase and protease adaptor, thereby providing a negative feedback mechanism. Genetic studies reveal that dysregulation of the CtsR regulon or inactivation of the YwlE phosphoarginine phosphatase decreases Spx activity through mechanisms involving both protein degradation and posttranslational modification.
KEYWORDS: Bacillus subtilis, Clp protease, Spx, proteolysis, stress response
ABSTRACT
In Bacillus subtilis, the Spx transcription factor controls a large regulon in response to disulfide, heat, and cell wall stresses. The regulatory mechanisms that activate the Spx regulon are remarkably complex and involve changes in transcription, proteolysis, and posttranslational modifications. To identify genes involved in Spx regulation, we performed a transposon screen for mutations affecting expression of trxB, an Spx-dependent gene. Inactivation of ctsR, encoding the regulator of the Clp proteases, reduced trxB expression and lowered Spx levels. This effect required ClpP, but involved ClpC rather than the ClpX unfoldase. Moreover, cells lacking McsB, a dual function arginine kinase and ClpCP adaptor, largely reverted the ctsR phenotype and increased trxB expression. The role of McsB appears to involve its kinase activity, since loss of the YwlE phosphoarginine phosphatase also led to reduced trxB expression. Finally, we show that Spx is itself a regulator of the ctsR operon. Altogether, this work provides evidence for a role of CtsR regulon members ClpC, ClpP, and McsB in Spx regulation and identifies a new feedback pathway associated with Spx activity in B. subtilis.
IMPORTANCE In Bacillus subtilis, the Spx transcription factor is proteolytically unstable, and protein stabilization figures prominently in the induction of the Spx regulon in response to oxidative and cell envelope stresses. ClpXP is largely, but not entirely, responsible for Spx instability. Here, we identify ClpCP as the protease that degrades Spx under conditions that antagonize the ClpXP pathway. Spx itself contributes to activation of the ctsR operon, which encodes ClpC as well as the McsB arginine kinase and protease adaptor, thereby providing a negative feedback mechanism. Genetic studies reveal that dysregulation of the CtsR regulon or inactivation of the YwlE phosphoarginine phosphatase decreases Spx activity through mechanisms involving both protein degradation and posttranslational modification.
INTRODUCTION
Spx belongs to the ArsC family of transcriptional regulators and is best known as the master regulator of the disulfide stress response in Bacillus subtilis (1). Spx consists of two major domains: one is formed by the N-terminal and C-terminal parts of the protein, and one is formed by its central region. The first domain contains a Cys-X-X-Cys (CXXC) redox-sensing switch that modulates Spx activity upon formation of an intramolecular disulfide bond (2). The central domain is involved in binding to the α-C-terminal domain (CTD) of RNA polymerase (3). Spx controls the expression of well more than 200 genes, including those involved in the synthesis of bacillithiol and cysteine and the thioredoxin system (1, 4).
Spx is encoded in a bicistronic operon, along with a putative acetyltransferase, and its expression is regulated by at least three different promoters (PA, PM1, and PB) that are dependent on different holoforms of the RNA polymerase (5–7). Expression of spx from the PA promoter, dependent on σA, is sufficient to complement an Δspx mutant and for normal regulation in response to disulfide stress (7). The activity of the PA promoter is regulated by two protein repressors, PerR and YodB, and is therefore induced by redox or electrophile stress (8). The σM-controlled PM1 promoter was recently shown to be critical for induction of the Spx regulon in response to cell wall stress (5). Little is known, however, about the functional role of the PB promoter, which is induced as part of the general stress response, as documented in the specific case of phosphate starvation (6).
Under unstressed conditions, Spx levels remain low since the protein is actively proteolyzed via ClpXP; this process is assisted by the adaptor protein YjbH, which is itself under Spx control (9–11). Stabilization of Spx plays a critical role in the induction of the Spx regulon in response to diamide, an electrophilic compound used to generate disulfide stress. Under disulfide stress, Spx is stabilized by aggregation of YjbH (12) and a decrease in ClpXP activity (13). Diamide also triggers oxidation of the redox-sensitive CXXC motif which activates Spx (2). Spx stabilization is also required for the full activation of the Spx regulon in response to cell wall stress. However, this stabilization is mediated by the anti-adaptor protein YirB, which binds YjbH and prevents Spx degradation through ClpXP (14, 15). Interestingly, under cell wall stress, Spx remains in the reduced state; therefore, the contribution of the redox-active disulfide switch appears to be limited under these conditions (5).
Although the in vivo mechanisms of Spx activation by stress are fairly well understood, some questions regarding Spx stability and activity remain to be answered. For instance, cells harboring a SpxC10A or SpxC10AC13A protein, both unable to form the intramolecular disulfide switch, display different profiles of activation of Spx-controlled genes in response to cell wall and disulfide stress (5). Moreover, Spx accumulates in response to cell wall stress in a YirB-independent manner, which therefore implicates other stabilization mechanisms (14).
In this work, we sought to identify additional regulatory pathways affecting Spx activity. We demonstrate that ClpCP degrades Spx under conditions that antagonize the ClpXP pathway, and this pathway is further activated when the CtsR repressor is inactive. Moreover, Spx itself contributes to activation of the ctsR operon, which encodes ClpC as well as the McsB arginine kinase and protease adaptor, thereby providing a negative feedback mechanism that likely involves both protein degradation and posttranslational modification.
RESULTS AND DISCUSSION
Dysregulation of the CtsR regulon leads to reduced induction of trxB.
We carried out mariner transposon mutagenesis to identify novel pathways involved in Spx regulation. For this, we used cells harboring a PtrxB-lacZ fusion, which is positively regulated by Spx and serves as a readout of Spx activity. The transposon library was plated on LB plus X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) medium, and light blue or white colonies were selected for further analysis. Transposon-generated mutations that decreased Spx activity included iolR (8 independent insertions), ctsR (5), galK (3), menH (3), and ywlE (1) (see Table S1 in the supplemental material). In this study, we focus on the ctsR gene, as it encodes the master regulator of proteolysis in B. subtilis (16, 17) and hence is a potential regulator of Spx stability. Additionally, CtsR was also previously reported to interact with YjbH in yeast two-hybrid experiments (15), and its regulon, similar to the Spx regulon, is induced in response to disulfide stress (4, 18, 19).
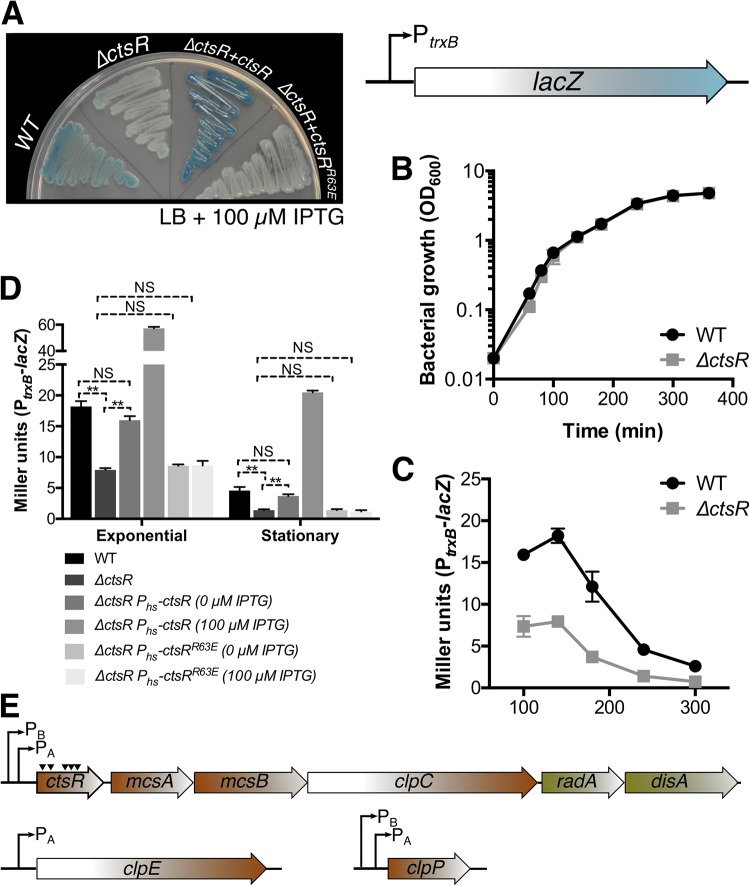
To validate the results obtained with the ctsR::mTn insertion, we used the BKE collection (20) to construct a strain harboring a clean deletion of the ctsR gene (ΔctsR), which also contained the PtrxB-lacZ reporter fusion. As expected, the ΔctsR null strain displayed whiter colonies than the wild type on LB plates supplemented with X-Gal (Fig. 1A), and ectopic complementation of the ΔctsR null strain with a conditional allele of ctsR (i.e., Phs-ctsR) restored the wild-type phenotype (Fig. 1A and D). In contrast, complementation with the ctsRR63E allele, which encodes a CtsR protein unable to bind DNA (21), was unable to restore the phenotype (Fig. 1A and D). Deletion of ctsR did not result in changes in growth (Fig. 1B) but did result in reduced expression of the PtrxB-lacZ fusion all along the growth curve (Fig. 1C). These results indicate that PtrxB activity is reduced when the CtsR regulon (Fig. 1E) is derepressed.
FIG 1.
Dysregulation of the CtsR regulon leads to reduced induction of the trxB promoter, a reporter of Spx activity. (A) Cells with a clean deletion in the ctsR gene (i.e., ΔctsR) display reduced induction of the Spx-controlled gene trxB on LB plates supplemented with X-Gal. The deletion mutant was complemented by ectopic expression of ctsR but not ctsRR63E. (B) Growth curves of WT and ΔctsR strains in LB were monitored by measuring optical density at 600 nm. (C) Induction of the PtrxB-lacZ transcriptional fusion throughout the exponential and early stationary phases. (D) Complementation of ΔctsR with an ectopic ctsR allele (but not ctsRR63E) driven from the Phs promoter restores the WT phenotype during exponential (t = 100 min) and stationary (t = 240 min) phases. **, P < 0.01; NS, not significant. (E) Diagram of the CtsR regulon in B. subtilis. The triangles illustrate the sites of mariner insertion.
ctsR inactivation leads to increased proteolysis of Spx by ClpCP.
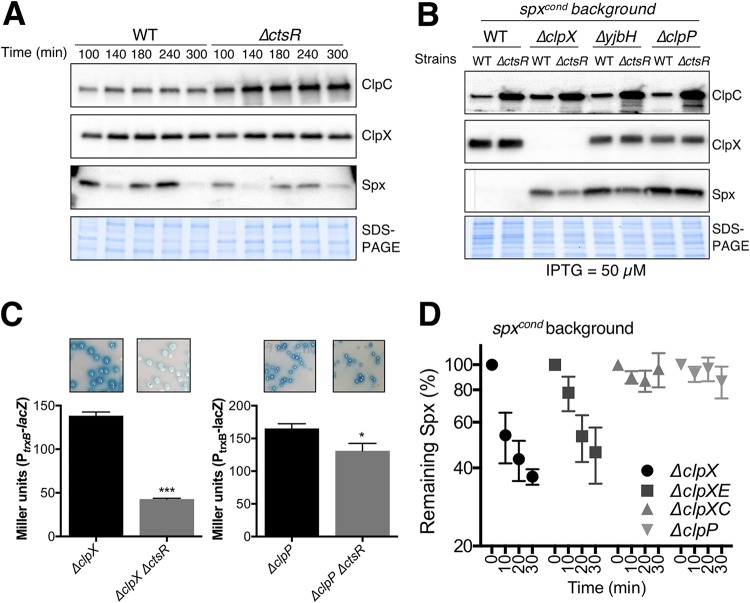
CtsR represses genes encoding two Clp unfoldases (ClpC and ClpE) as well as the ClpP protease (Fig. 1E). We therefore suspected that Spx levels would be reduced by inactivation of ctsR. Indeed, decreased Spx levels were observed in the ΔctsR strain (Fig. 2A), which correlated with the reduced activity of the PtrxB reporter fusion (Fig. 1C). For reasons not clear, we consistently observed that the lowest levels of Spx protein were seen as cells enter transition phase (∼140 min) and again in stationary phase (∼300 min) (Fig. 1B), and this effect is the same in both wild-type (WT) and ΔctsR cells (Fig. 2A). In parallel, we monitored the levels of ClpX and ClpC by Western blotting. ClpC, but not ClpX, was significantly elevated in the ΔctsR strain compared to that in the WT (Fig. 2A). This is consistent with CtsR acting as a repressor of clpC transcription (Fig. 2A and 1E). We therefore hypothesized that the reduction in Spx in the ΔctsR strain likely involved ClpP proteolysis through the ClpCP or ClpEP proteases, rather than ClpXP, which is generally considered the primary pathway for Spx degradation.
FIG 2.
The phenotype associated with ΔctsR is largely explained by increased ClpCP proteolysis. (A) Spx, ClpX, and ClpC levels in both WT and ΔctsR cells were surveyed during exponential and early stationary phases by Western blotting. The presented blot is representative of four biological replicates. (B) WT, ΔclpX (clpX::spec), ΔyjbH (yjbH::kan), and ΔclpP (clpP::tet) cells in an spxcond background were assessed for Spx levels during exponential phase (t = 100 min); 50 µM IPTG was added to the bacterial cultures (OD600 of ∼0.1) to induce spx expression. When cells reached an OD600 of 0.4 to 0.6, samples were taken for Western blot analyses. ClpC levels were included in panels A and B to illustrate the derepression of the CtsR regulon in ΔctsR cells. This blot is representative of three independent replicates. (C) PtrxB activity was studied on 48-h-old colonies of ΔclpX (clpX::spec) and ΔclpP (clpP::tet) strains growing on LB plus X-Gal plates. The colonies were derived from transformations of the clpX::spec and clpP::tet cassettes into the WT and ΔctsR strains. A t test was performed to compare the mean values of each pair of strains. *, P < 0.05; ***, P < 0.001. (D) A chloramphenicol chase was performed in spxcond cells lacking clpX, clpX clpE, clpX clpC, or clpP to determine Spx stability. Cells were grown with IPTG (100 µM) to induce spx expression and then treated with 100 µg ml−1 of chloramphenicol to stop protein synthesis. ΔclpX denotes clpX::spec; ΔclpE and ΔclpC denote clpE::ery and clpC::ery, respectively; ΔclpP denotes clpP::tet. The protein levels were quantified using Image Lab 5.2.1 software (Bio-Rad) as described in Materials and Methods. Error bars represent standard errors of the means (SEMs) from at least three independent replicates.
To explore in further detail the basis for the ΔctsR phenotypes, we used a strain with conditional expression of spx (i.e., Δspx amyE::Phs-spx), which we called spxcond, in order to maintain a fixed spx transcription rate. To determine if ClpXP was involved in degradation of Spx in the ΔctsR strain, we monitored Spx levels in spxcond cells lacking either ClpX or the ClpX adaptor protein, YjbH (Fig. 2B). As expected, these cells had significantly elevated levels of Spx, but this level was still reduced in the ΔctsR background. However, Spx levels were unaffected by the ctsR mutation in cells lacking ClpP (Fig. 2B). These results are consistent with a role for the ClpP protease in the degradation of Spx. Since a ΔctsR mutation can still modestly reduce PtrxB activity, even in cells lacking ClpP, this suggests that another member of the CtsR regulon can reduce Spx activity (Fig. 2C; see also Fig. S1), as explored in more detail below.
Next, we used a chloramphenicol chase assay to quantify the contribution of ClpCP or ClpEP to Spx degradation (Fig. 2D). In the WT strain, Spx was rapidly degraded with a half-life of ∼2 min, as shown previously (14) and confirmed here (see below). Although Spx was much more stable in ΔclpX spxcond cells, it was still degraded, with an approximately ∼60% decrease over 20 min (Fig. 2D). A similar rate of Spx turnover was noted in ΔclpX spxcond cells lacking clpE but not in spxcond cells lacking clpC clpX or clpP (Fig. 2D). Interestingly, although Spx is stable in both the ΔclpX ΔclpC double mutant and the ΔclpP background (Fig. 2D), the level of Spx was higher in the latter strain (see Fig. S2). The basis for this difference is not clear but might involve a backup role for ClpEP under these conditions. In sum, we conclude that ClpCP contributes to Spx degradation in vivo, and this pathway is likely to be important under conditions where ClpXP degradation is impeded. Indeed, previous biochemical experiments demonstrated that ClpCP is competent to degrade Spx in vitro (11).
The McsB arginine kinase affects Spx activity.
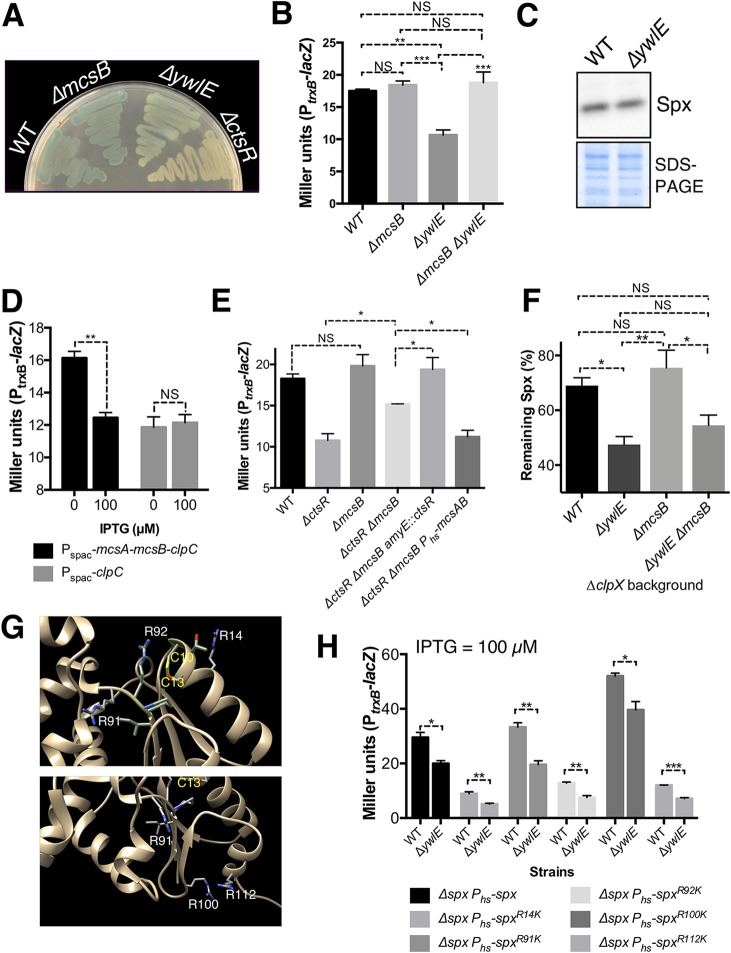
The activity of ClpC relies on adaptor proteins, which help assemble ClpCP and deliver substrates (22). We therefore reasoned that one of those adaptors might account for the ClpP-dependent and/or ClpP-independent phenotypes of ΔctsR cells. In fact, the gene encoding the McsB arginine kinase, one of the ClpC adaptors, lies within the clpC operon and was thus also upregulated in the ΔctsR strain (Fig. 1E). McsB activity requires the product of the adjacent gene, mcsA (18, 23), and its activity has been shown to be important to both modulate protein activity (21) and mark proteins for ClpCP degradation (24). Our attention was drawn to the possible role of McsB-mediated phosphoarginine modification in Spx regulation due to our recovery of a mariner ywlE::mTn insertion in our screen for altered expression of trxB (Fig. 3A and B; Table S1). YwlE functions as a phosphoarginine phosphatase (23, 25), suggesting that an increase in protein phosphorylation likely leads to a decrease in Spx stability and/or activity. As observed for the ΔctsR strain, the phenotype due to inactivation of ywlE was independent of spx transcriptional control and ClpXP-mediated Spx proteolysis (see Fig. S3B). Indeed, Western blotting showed no significant differences of Spx levels between WT and ΔywlE strains (Fig. 3C), suggesting that the observed effects (Fig. 3A and B) are the result of changes in Spx activity. The decrease in Spx activity in the ΔywlE mutant reverted to WT levels in a ΔywlE ΔmcsB double mutant, which additionally lacks the McsB arginine kinase (Fig. 3B). The effect of a ΔywlE mutation was less dramatic than that for the ΔctsR strain (Fig. 3A) and was complemented by an ectopic copy integrated at amyE (Fig. S3A). The impact of arginine phosphorylation on the expression of the Spx regulon was previously noted in transcriptomic studies of ΔmcsB and ΔywlE mutants (26).
FIG 3.
Elevated arginine phosphorylation affects Spx activity. (A and B) Cells lacking ywlE display reduced induction of the trxB promoter, and this phenotype was reversed by deletion of mcsB. (C) Spx levels were assessed on WT and ΔywlE cells at mid-exponential phase. This experiment is representative of three biological replicates. (D) PtrxB activity was studied to assess whether altering clpC and/or mcsA-mcsB-clpC expression levels were sufficient to replicate the difference between WT and ΔctsR cells (Fig. 1D). (E) PtrxB activity was measured to determine the effect of mcsB deletion on Spx activity in WT and ΔctsR cells. (F) Spx levels were measured 30 min after treatment of mid-exponential cells with chloramphenicol as described for Fig. 2D. (G) Predicted Spx R-phosphorylation sites according to Trentini et al. (24). The residues that form the redox-sensing switch are included for reference. (H) The contribution of each potentially phosphorylated Arg residue to the ΔywlE phenotype was assessed by replacing them by Lys (i.e., R to K) and measuring PtrxB activity. The ectopic WT or mutant allele of spx was induced with IPTG (100 µM), and PtrxB activity measured at mid-exponential phase. Error bars represent SEMs from at least three independent replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant. For panels B, E, and F, one-way analyses of variance (ANOVAs) and Tukey’s honestly significant difference (HSD) tests were used to compare the different bacterial strains. For panels D and H, t tests were performed to compare the effects of either IPTG addition or deletion of ywlE, respectively.
In light of the above-described results, we postulated that the reduction in Spx activity in the ΔctsR strain likely reflects a combined effect of increased ClpCP activity and increased activity of the McsA/McsB pathway for protein phosphorylation. Indeed, Spx was shown previously by phosphoproteomics to be phosphorylated on multiple Arg residues (24). We therefore placed either clpC alone or the mcsA, mcsB, and clpC genes together, under IPTG (isopropyl-β-d-thiogalactopyranoside) control at the native locus using the pMUTIN system. In the Pspac-clpC strain, induction of ClpC (under conditions where McsA and McsB are expressed from the native ctsR promoter) (Fig. 1E) did not lead to a decrease in Spx activity, as judged from the PtrxB-lacZ reporter. When all three genes were placed downstream of the Pspac promoter (in the Pspac-mcsA-mcsB-clpC strain), Spx activity was decreased upon induction, although there was also a higher basal level of activity (Fig. 3D). These observations are consistent with the idea that the negative effect of the ΔctsR mutation likely requires the concerted action of McsA, McsB, and the ClpCP-protease. Furthermore, deletion of mcsB in the wild-type strain had no effect on PtrxB activity (Fig. 3E), which is consistent with McsB being largely inactive in unstressed cells (27). In contrast, its inactivation in the ΔctsR strain (i.e., the ΔctsR ΔmcsB double mutant) partially reversed the large decrease in trxB expression noted in a ΔctsR strain, and this effect was complemented by ectopic expression of mcsA and mcsB (Fig. 3E). Taking these results together, we conclude that McsB modulates Spx activity through arginine phosphorylation and may also contribute to a reduction in stability by acting as an adaptor for Spx proteolysis. The regulatory control of McsA/McsB on Spx thus appears to be critical under conditions that result in induction of the CtsR regulon.
Spx was shown previously to be phosphorylated on four Arg residues: R14, R91, R100, and R112 (24). Inspection of the Spx structure reveals that the first two (R14 and R91) are near the disulfide switch, and the latter two (R100 and R112) are near the putative sites of binding of the YjbH adaptor protein for ClpXP proteolysis (Fig. 3G) (28, 29). The Spx paralog MgsR was phosphorylated on R17 and R95 (24), which align with Spx R14 and R92 in the protein structures. To determine which of these arginines might be relevant to the observed phenotypes of the mcsB and ywlE mutants, we individually replaced each of these Arg residues (and an additional arginine in the disulfide switch region, R92) with lysine (SpxR14K, SpxR91K, SpxR92K, SpxR100K, and SpxR112K). Induction of some of the Spx variants decreased trxB expression compared to that in an isogenic strain harboring the wild-type spx allele (Fig. 3H). In all cases, however, cells lacking ywlE still displayed reduced induction of the reporter fusion, suggesting that no single residue is absolutely required for the reduction in activity of the ywlE deletion (Fig. 3H).
Analysis of representative Firmicutes species further showed that mcsB cooccurs with spx in some Bacillales species, including B. subtilis, Listeria monocytogenes, and Staphylococcus aureus but not in representative Lactobacillales. mcsB is, however, also present in some species lacking Spx homologs such as Veillonella parvula or Coprococcus catus (see Table S4). The arginine residues at the positions described in Fig. 3 were highly conserved in the species containing McsB and slightly less conserved in the species lacking McsB (see Fig. S5), which might suggest a potential functional role.
Phosphorylation of arginine residues in some proteins is important for their degradation via ClpCP (24). We therefore hypothesized that increased arginine phosphorylation of Spx might lead to increased Spx degradation in cells lacking ClpX. This was indeed the case, as chloramphenicol chase experiments in a clpX strain (to eliminate the major pathway of Spx degradation by ClpXP) lacking YwlE showed a modest increase in Spx degradation (Fig. 3F). Unexpectedly, deletion of mcsB did not prevent this effect, perhaps suggesting the presence of other pathways for Arg phosphorylation in vivo or other roles for YwlE. We note that in the absence of McsB other ClpC adaptor proteins (i.e., MecA and YpbH) might play a more prominent role in Spx turnover. In support of this hypothesis, both MecA and McsB were previously shown to interact at the same ClpC site, and competition between these adaptors for ClpCP proteolysis of α-casein was previously demonstrated in vitro (30). Additionally, MecA can accelerate ClpCP-catalyzed degradation in vitro (11).
The toxicity due to Spx accumulation is partially alleviated by deletion of ctsR or ywlE.
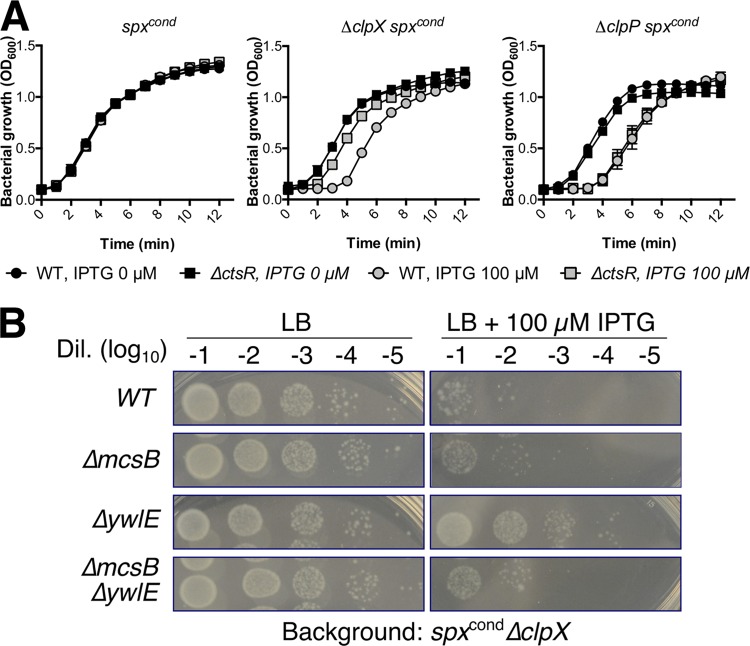
Spx accumulation in B. subtilis, as observed in ΔyjbH, ΔclpX, or ΔclpP mutants, results in reduced growth, sporulation, and competence (9, 31). Consistently, overexpression of an IPTG-inducible spxDD allele, which is resistant to proteolysis, as the only source of Spx for the cells, prevented bacterial growth upon induction (see Fig. S4). We therefore reasoned that if deletion of ctsR results in reduced Spx levels, ΔclpX cells lacking CtsR should display improved growth. This was indeed the case, since deletion of ctsR improved growth (as evidenced by a marked reduction in lag phase) in spxcond ΔclpX cells, but only when spx was induced (Fig. 4A). As expected, no improvement in growth was observed upon inactivation of ctsR in spxcond ΔclpP cells. These results suggest that derepression of the CtsR regulon alleviates the toxicity imposed by abnormally high Spx levels, a result correlated with reduced Spx levels (Fig. 4A), and also that this effect is due to increased ClpCP-mediated Spx proteolysis. Next, we assessed the effects of deletion of ΔywlE in spot dilution assays. Induction of Spx reduced the plating efficiency of the ΔclpX strain by ∼103, as observed in a spot dilution assay on LB with 100 µM IPTG (Fig. 4B), consistent with the increased lag seen in growth studies (Fig. 4A). Using this assay as a measure of Spx toxicity, we note that introduction of a ΔywlE mutation increased plating efficiency, and this effect was largely dependent on McsB (Fig. 4B), which correlated with our prior observations (Fig. 3B). Thus, the McsB arginine kinase reduces Spx toxicity, presumably by Spx phosphorylation.
FIG 4.
Spx toxicity is alleviated by deletion of ctsR or ywlE. (A) Growth curves of spxcond, spxcond ΔclpX, and spxcond ΔclpP (genotypes as for Fig. 2B) in absence and presence of 100 µM IPTG. (B) WT, ΔmcsB, ΔywlE, and ΔmcsB ΔywlE cells in a spxcond ΔclpX genetic background were surveyed on plates with or without 100 µM IPTG. The results are representative of two independent replicates.
ClpCP-mediated Spx degradation under disulfide stress.
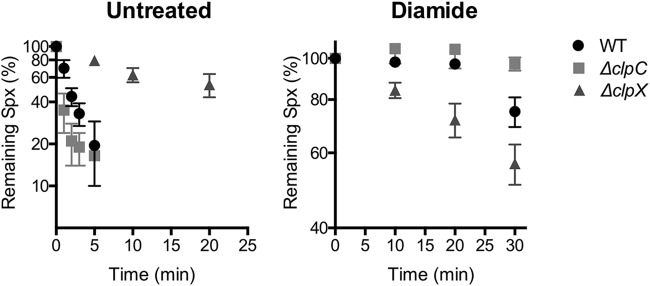
The present results suggest that ClpCP might be important for Spx degradation under conditions that result in upregulation of the CtsR regulon, such as diamide treatment (19). Importantly, some of the conditions that lead to CtsR derepression, such as disulfide, heat, or ethanol stress, also result in reduced ClpXP-mediated Spx proteolysis (12, 13). We therefore hypothesized that ClpCP-mediated Spx degradation provides a mechanism to detoxify Spx under conditions where the ClpXP pathway for Spx degradation is inactive. To determine if this is the case, we monitored Spx degradation in WT, ΔclpX, and ΔclpC cells in the presence and absence of diamide. In the WT and ΔclpC strains, under unstressed conditions, Spx was rapidly degraded, while degradation occurred slowly in ΔclpX cells (Fig. 5). These results confirm the primary role of ClpX in Spx degradation in unstressed cells. In the presence of diamide, Spx degradation was reduced in the WT, which is consistent with inactivation of ClpX and YjbH. Degradation was fully abolished in ΔclpC cells, confirming that under disulfide stress, ClpCP is responsible for the residual slow degradation of Spx.
FIG 5.
ClpCP-mediated Spx degradation under disulfide stress. Spx stability was assessed in the absence or presence of diamide in the WT, ΔclpC, and ΔclpX strains. Exponentially growing cells were treated or not with 0.5 mM diamide to induce Spx accumulation, protein synthesis was stopped with chloramphenicol, and Spx levels determined by Western blotting as described above for Fig. 2D. Each point represents the average from three biological replicates. Bars indicate the SEMs from biological replicates.
Spx regulates the expression of the ctsR operon.
Using chromatin immunoprecipitation (ChIP)-tilling microarray technology, Rochat et al. showed that Spx binds to and regulates the ctsR promoter (4). This interaction, however, was not explored in further detail. To assess the contribution of Spx for induction of the autoregulated ctsR operon, we studied the clpC mRNA levels in WT, ΔctsR, Δspx, and ΔctsR Δspx cells in response to vancomycin, a potent inducer of the Spx regulon (14). Deletion of ctsR resulted in derepression of the ctsR operon, which was observed as a more intense band at the initial time point and is consistent with negative autoregulation. This deletion, however, did not abolish the activation of the operon in response to cell envelope stress (Fig. 6). Conversely, deletion of Spx largely suppressed this activation, suggesting that both CtsR and Spx both regulate the ctsR operon. As a control, we studied another CtsR-regulated gene (i.e., clpE), which was also reported to be under Spx control; however, no contribution of Spx to clpE expression was observed (Fig. 6). Taken together, the present evidence demonstrates that Spx, which is activated by cell envelope and disulfide stress, can contribute to increased expression of the CtsR regulon, which, in turn, includes critical mediators of Spx proteolysis and activity (Fig. 7).
FIG 6.
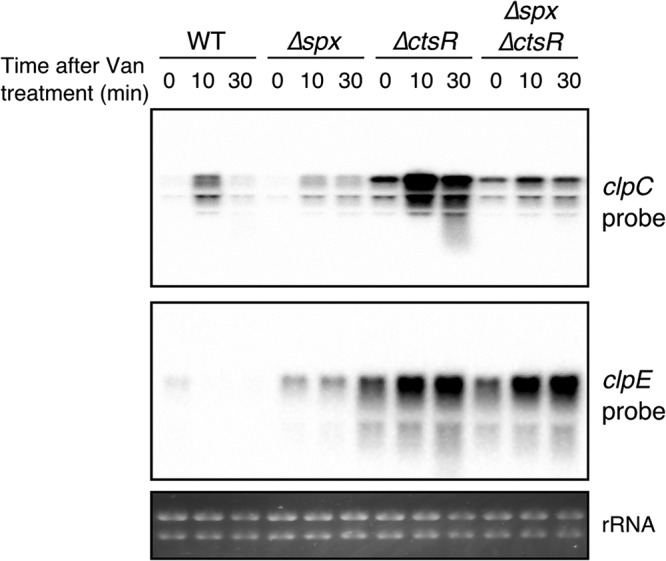
Both Spx and CtsR drive the expression of the ctsR operon. Northern blot showing the expression of two CtsR-regulated genes in response to vancomycin. Exponentially growing cells were treated with 1 µg ml−1 vancomycin, and RNA was harvested before vancomycin treatment (t = 0 min) and after 10 min and 30 min. RNA was run in a denaturing agarose gel, transferred to a Z probe membrane, and detected with biotin-labeled RNA probes. This experiment was performed in duplicates with identical results.
FIG 7.
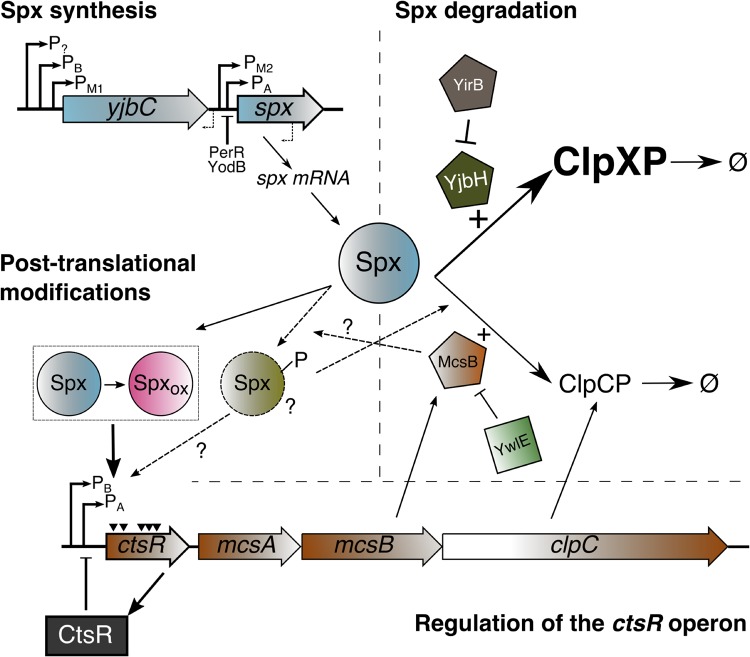
Model of Spx regulation in B. subtilis including pathways for Spx synthesis, degradation, and posttranslational modifications. Several promoters, controlled by different RNA polymerase holoforms, and two repressors regulate the transcription of spx. In unstressed cells, Spx is rapidly degraded through the ClpXP-YjbH system. Under cell wall stress, the YirB antiadaptor is induced and antagonizes YjbH, thus preventing Spx proteolysis. The ClpCP protease also contributes to Spx degradation. ClpCP-mediated Spx proteolysis appears to be at least in part mediated by McsB, an arginine kinase that acts as an adaptor for ClpCP. Spx activity is also modulated by changes in the oxidation state of the redox-sensing switch and also perhaps through Arg phosphorylation. The inactivation of Spx in cells lacking the YwlE arginine phosphatase suggests a negative role for Arg phosphorylation. Activation of the ctsR operon by Spx can increase levels of ClpCP and the arginine phosphorylation system, thereby providing a negative feedback loop.
Concluding remarks.
Spx is a pleiotropic transcription factor in B. subtilis, which is activated in response to proteotoxic and cell wall stress conditions (1, 5, 12, 14, 19, 32). The regulatory pathways that coordinate Spx activity are intricate and involve many cellular partners (5–7, 9, 11, 14, 33, 34). Here, using an unbiased screen, we identify two genes whose deletion leads to reduced Spx stability and activity: (i) ctsR, the master regulator of proteolysis gene (17), and (ii) ywlE, the B. subtilis arginine phosphatase gene (23, 25). Characterization of these genes uncovered novel molecular mechanisms associated with Spx regulation. First, by studying the ΔctsR phenotype, we found that ClpCP is also capable of degrading Spx in vivo (Fig. 2), particularly under conditions that potentially lead to inactivation of the ClpXP-YjbH system (Fig. 5). Moreover, McsB, a ClpC adaptor and arginine kinase, appears to be important for this process (Fig. 3). Both clpC and mcsB are members of the CtsR regulon (35). A second gene identified in our transposon screening encodes YwlE, an arginine phosphatase that antagonizes the action of the McsB protein arginine kinase (27). Interestingly, the ywlE phenotype was fully reverted in the double ΔywlE ΔmcsB mutant, thus implicating the kinase activity of McsB in the regulation of Spx (23, 25). Inactivation of ywlE had no effect on Spx levels, leading us to conclude that elevated arginine phosphorylation as observed in the ΔywlE strain primarily affects Spx activity (Fig. 3). Indeed, phosphorylation of several arginine residues on Spx and its paralog, MgsR, was previously observed in phosphoproteomic studies, which appears to support a direct effect of McsB on Spx (24). Analysis of Spx turnover in cells lacking the ClpXP pathway also revealed increased Spx proteolysis in ΔywlE cells, therefore suggesting that increased arginine phosphorylation might also affect Spx degradation.
The relationship between the CtsR and Spx regulons is, however, not unidirectional. In this work, we further show that Spx, together with CtsR, regulates the expression of the ctsR operon (Fig. 6); the positive correlation observed between the expression profile of CtsR- and Spx-regulated genes (i.e., nfrA and msrA) further strengthens this connection (19). The biological significance of this connection lies in the fact that both regulons are critical for degradation and/or refolding of damaged or misfolded proteins; this effect might be achieved through upregulation of the Clp proteases, the arginine phosphorylation system, the thioredoxin system, and other redox-associated proteins (1, 4, 35). Oxidative stress, which can lead to activation of both the Spx and CtsR operons, also leads to the inactivation of the arginine phosphatase activity of YwlE (36). These results thus suggest that Spx and CtsR participate in a feedback loop in which Spx contributes to the activation of the CtsR regulon and that the latter mediates the inactivation of Spx (Fig. 7). This regulatory circuit might be beneficial for the cells, since it enables the simultaneous activation of the Spx and CtsR regulons while preventing the negative effects of Spx overaccumulation (9, 31, 37). Interestingly, those proteotoxic stresses that lead to activation of the Spx regulon, as well as the CtsR regulon, also inactivate Spx proteolysis through ClpXP (12, 13). Consistent with a role for the CtsR regulon in Spx inactivation, we observed that in cells lacking the ClpXP protease, either CtsR derepression, which leads to increased ClpCP proteolysis, or deletion of ywlE, which leads to elevated arginine phosphorylation, prevents the deleterious effects of Spx accumulation (Fig. 4). ClpCP proteolysis, by contrast, does not appear to be affected by diamide treatment (Fig. 5) and, based on its transcription profile, is not predicted to be inactivated under other proteotoxic stress conditions (19). Altogether, these observations suggest a critical role of Spx in activation of the CtsR regulon and of the CtsR regulon in preventing Spx toxicity. This role appears to involve both arginine phosphorylation profiles and increased ClpCP-mediated degradation. The present model of Spx regulation, however, does not account for all the observed phenotypes, suggesting that even further mechanisms are at play. The intrinsic complexity of B. subtilis Spx regulation is reminiscent of the convoluted regulation of Escherichia coli RpoS (38, 39).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All bacterial strains are listed in Table S2 in the supplemental material. Bacillus subtilis strains (all based on the B. subtilis 168 wild type) were grown under standard conditions: lysogeny broth (LB; 10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) at 37°C with vigorous shaking, unless otherwise stated. Escherichia coli DH5α was used for plasmid construction. Antibiotics were added to the growth medium when appropriate: 100 µg ml−1 ampicillin for E. coli, and 1 µg ml−1 erythromycin plus 25 µg ml−1 of lincomycin (macrolide-lincomycin-streptogramin B [MLS] resistance), 10 µg ml−1 chloramphenicol, 100 µg ml−1 spectinomycin, and 10 µg ml−1 kanamycin for B. subtilis.
Transformation of Bacillus subtilis.
For genetic transformation of B. subtilis, a suspension of cells (optical density at 600 nm [OD600] of ∼0.1) was prepared in 5.0 ml of medium A [1.0 g yeast extract, 0.2 g Casamino Acids, 5.0 g glucose, 2.0 g (NH4)SO4, 18.3 g K2HPO4·3H2O, 6 g KH2PO4, 1 g Na citrate, and 0.2 g MgSO4·7H2O per liter] using an overnight culture of cells growing on LB plates as initial culture. This suspension was then incubated at 37°C with vigorous shaking for ∼4 h, which corresponded to 1.5 h after the end of the exponential phase, as determined by OD600 measurements. Then, 100 µl of this culture was transferred to a 5-ml tube containing 400 µl of prewarmed medium B (i.e., medium A plus 250 µM MgCl2 plus 250 µM CaCl2) and incubated for a further 90 min. Next, 200 ng of genomic DNA or 500 ng of plasmid DNA was added to the bacterial culture, unless otherwise stated, and cells were incubated for a further 2 h. Cells were plated on LB plates supplemented with the corresponding antibiotic. For transformation of pMUTIN-based constructs, the concentrations of erythromycin and spectinomycin were reduced to 0.5 µg ml−1 and 50 µg ml−1, respectively.
Cloning and site-directed mutagenesis.
For cloning purposes, the PCRs were performed using the high-fidelity Phusion DNA polymerase (NEB) according to the manufacturer’s instructions and genomic DNA of B. subtilis HB18501 unless otherwise stated. The primers used in this study are listed in Table S3. The details regarding strain constructions are described in the supplemental material.
Transposon library.
B. subtilis cells with the appropriate genotype were transformed with the pMarA plasmid, plated on LB plus 0.3 mg ml−1 erythromycin, and incubated at 28°C for 48 h. The resulting transformants were stored at −80°C (host for transposition). Cells containing the pMarA plasmid were then grown overnight at 28°C in LB supplemented with kanamycin and erythromycin. A new culture (1:40) was started in LB plus kanamycin (to select for the transposition events) and incubated for 4 h at 28°C; the cells were then transferred at 37°C and incubated for three more hours, and finally plated on LB amended with 15 µg ml−1 kanamycin and 0.2 mg ml−1 X-Gal. Plates were incubated at 42°C for loss of the plasmid, and then candidate mutant colonies were selected on the basis of the intensity of its blue color. Whiter colonies on the plates were chosen and subjected to one more round of selection; finally, 30 clones from each library were saved for further studies. To determine the site of mariner insertion, chromosomal DNA was isolated using the DNeasy kit (Qiagen) and digested with the Taqα1 restriction enzyme, and the products ligated using the T4 ligase. The resulting DNA was used as the template for an inverse PCR using the primers 6299 and 6300 annealing the mariner transposon. The PCR products were in-column cleaned and analyzed by sequencing using the 6301 internal primer. The sequencing information was then used to map the transposon insertion site.
β-Galactosidase activity.
The cells were grown until the OD600 reached ∼0.5. Then, cells were treated with different chemicals or not treated and incubated at 37°C with agitation. At specific time points, samples were taken, washed twice in phosphate-buffered saline (PBS), and finally resuspended in 900 µl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4·7H2O) supplemented with 400 µM dithiothreitol (DTT). Alternatively, several colonies were recovered from the plate and resuspended in PBS at an OD600 of ∼0.5, washed in PBS, and resuspended in 900 µl of Z buffer plus 400 µM DTT for further experiments. Optical density at 600 nm was measured, and then the cells were lysed using 100 µg ml−1 lysozyme at 37°C for 30 min. Next, 200 µl of 4 mg ml−1 o-nitrophenyl-β-d-galactopyranoside (ONPG) was added to the lysate, and the reaction mixture was incubated at 28°C until the samples produced a visible yellow color. The reaction was stopped by adding 500 µl of 1.0 M Na2CO3. The absorbance was then measured at 420 nm and 550 nm, and β-galactosidase activity was determined using the following equation: Miller units = 1,000 × (OD420 − 1.75 × OD550)/(t × v × OD600), where t is time in minutes and v is the volume of culture used in the reaction.
Western blots.
A total of 5 ml of cells was collected, washed in PBS, and resuspended in 150 µl of disruption buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 5% glycerol) supplemented with the cOmplete EDTA-free protease inhibitor cocktail. The cells were disrupted by sonication and then centrifuged for 15 min at 13,500 rpm at 4°C. The soluble fraction was collected and quantified using the Bradford assay. Reducing sample buffer was added to the protein extract, and then 5 µg of protein was loaded in a 4% to 20% SDS-PAGE gel. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane using the TransBlot Turbo transfer system (Bio-Rad, USA). The membrane was blocked using 5% protein blotting blocker dissolved in Tris-buffered saline with Tween 20 (TTBS) for 1 h at room temperature (RT). Then, the primary antibodies were resuspended in 0.5% protein blotting blocker dissolved in TTBS and incubated for 16 h at 4°C. Finally, an anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody was added and incubated for 2 h at RT. The membrane was revealed using the Clarity Western ECL substrate and visualized in a gel documenter. For quantification of Spx, the intensity of the bands was measured using Image Lab 5.2.1 software (Bio-Rad, USA)
Chloramphenicol chase assay.
Cells (50 ml) were grown under standard conditions up to an optical density at 600 nm of 0.500. Then, the culture was divided and left untreated or treated with 0.5 mM diamide. Next, chloramphenicol was added to the culture medium at a 100 µg ml−1 final concentration to stop protein synthesis, using as stock a 20 mg ml−1 chloramphenicol solution dissolved in 95% ethanol. Protein degradation was stopped by treatment with cold trichloroacetic acid (TCA; final concentration 10% TCA), and the amount of protein was normalized by optical density. Cells were then washed twice with ice-cold acetone and left to air dry for 10 min at room temperature. The pellet was resuspended into 100 µl of 1× Laemmli buffer, and 10-µl samples of the lysate were loaded in a 4% to 20% acrylamide gel. Protein degradation was then studied by Western blotting. For quantification of protein degradation, the Western blot bands were measured using Image Lab 5.2.1 software (Bio-Rad, USA) and normalized against the SDS-PAGE gel. For normalization, a total of 10 bands from the SDS-PAGE gel were quantified by densitometry and used to determine the amount of total protein loaded in each lane of the gel.
RNA isolation and Northern blots.
RNA isolation and Northern blotting were carried out as previously described (5, 14). RNA probes were synthesized from PCR products using in vitro transcription as previously described (5, 14), with the exception that biotin-16-UTP was used for labeling the probes. The generated RNA blots were developed using the North2South chemiluminescent hybridization and detection kit as per the manufacturer’s instructions (Thermo Scientific). The template PCR product for the clpC RNA probe was generated using the primers DR309 and DR310, while for the clpE RNA probe, we used DR302 and DR303.
Growth curves and spot dilution assays.
For growth curves, cells (5 ml) were grown under standard conditions to an optical density at 600 nm of ∼0.500. Then, cells were resuspended at a final OD600 of ∼0.01 in fresh sterile LB supplemented or not with IPTG, and samples of 150 µl were placed in a 96-well plate. Optical density at 600 nm was monitored for 20 h at 37°C with continuous shaking in a Synergy H1 microplate reader (BioTek). For spot dilution assays, cells (5 ml) were grown under standard conditions up to an optical density at 600 nm of ∼0.500 and serially diluted in sterile LB. The different dilutions were plated on LB plates supplemented or not with IPTG.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ulf Gerth for the anti-ClpX and anti-ClpC serum and Peter Zuber for the anti-Spx serum. We thank Camila Bustos, Manuela Alvarado, and Hye-Rim Hong for their valuable contributions to this work.
This work was supported by a grant from the National Institutes of Health (R35GM122461) to J.D.H.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00151-19.
REFERENCES
- 1.Nakano S, Küster-Schöck E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A 100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakano S, Erwin KN, Ralle M, Zuber P. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol 55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- 3.Newberry KJ, Nakano S, Zuber P, Brennan RG. 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci U S A 102:15839–15844. doi: 10.1073/pnas.0506592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rochat T, Nicolas P, Delumeau O, Rabatinova A, Korelusova J, Leduc A, Bessieres P, Dervyn E, Krasny L, Noirot P. 2012. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res 40:9571–9583. doi: 10.1093/nar/gks755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rojas-Tapias DF, Helmann JD. 2018. Induction of the Spx regulon by cell wall stress reveals novel regulatory mechanisms in Bacillus subtilis. Mol Microbiol 107:659–674. doi: 10.1111/mmi.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antelmann H, Scharf C, Hecker M, Hecker M. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J Bacteriol 182:4478–4490. doi: 10.1128/JB.182.16.4478-4490.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leelakriangsak M, Zuber P. 2007. Transcription from the P3 promoter of the Bacillus subtilis spx gene is induced in response to disulfide stress. J Bacteriol 189:1727–1735. doi: 10.1128/JB.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuber P. 2009. Management of oxidative stress in Bacillus. Annu Rev Microbiol 63:575–597. doi: 10.1146/annurev.micro.091208.073241. [DOI] [PubMed] [Google Scholar]
- 9.Larsson JT, Rogstam A, Wachenfeldt von C. 2007. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol Microbiol 66:669–684. doi: 10.1111/j.1365-2958.2007.05949.x. [DOI] [PubMed] [Google Scholar]
- 10.Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. 2009. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol 191:1268–1277. doi: 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano S, Zheng G, Nakano MM, Zuber P. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J Bacteriol 184:3664–3670. doi: 10.1128/JB.184.13.3664-3670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engman J, Wachenfeldt von C. 2015. Regulated protein aggregation: a mechanism to control the activity of the ClpXP adaptor protein YjbH. Mol Microbiol 95:51–63. doi: 10.1111/mmi.12842. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Zuber P. 2007. Requirement of the zinc-binding domain of ClpX for Spx proteolysis in Bacillus subtilis and effects of disulfide stress on ClpXP activity. J Bacteriol 189:7669–7680. doi: 10.1128/JB.00745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojas-Tapias DF, Helmann JD. 2018. Stabilization of Bacillus subtilis Spx under cell wall stress requires the anti-adaptor protein YirB. PLoS Genet 14:e1007531. doi: 10.1371/journal.pgen.1007531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kommineni S, Garg SK, Chan CM, Zuber P. 2011. YjbH-enhanced proteolysis of Spx by ClpXP in Bacillus subtilis is inhibited by the small protein YirB (YuzO). J Bacteriol 193:2133–2140. doi: 10.1128/JB.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruger E, Hecker M. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J Bacteriol 180:6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derré I, Rapoport G, Msadek T. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol Microbiol 31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 18.Elsholz AKW, Hempel K, Pöther D-C, Becher D, Hecker M, Gerth U. 2011. CtsR inactivation during thiol-specific stress in low GC, Gram+ bacteria. Mol Microbiol 79:772–785. doi: 10.1111/j.1365-2958.2010.07489.x. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 20.Koo B-M, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann A-B, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291.e7–305.e7. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuhrmann J, Schmidt A, Spiess S, Lehner A, Turgay K, Mechtler K, Charpentier E, Clausen T. 2009. McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science 324:1323–1327. doi: 10.1126/science.1170088. [DOI] [PubMed] [Google Scholar]
- 22.Kirstein J, MoliEre N, Dougan DA, Turgay K. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol 7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 23.Kirstein J, Zuhlke D, Gerth U, Turgay K, Hecker M. 2005. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J 24:3435–3445. doi: 10.1038/sj.emboj.7600780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trentini DB, Suskiewicz MJ, Heuck A, Kurzbauer R, Deszcz L, Mechtler K, Clausen T. 2016. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature 539:48–53. doi: 10.1038/nature20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuhrmann J, Mierzwa B, Trentini DB, Spiess S, Lehner A, Charpentier E, Clausen T. 2013. Structural basis for recognizing phosphoarginine and evolving residue-specific protein phosphatases in Gram-positive bacteria. Cell Rep 3:1832–1839. doi: 10.1016/j.celrep.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Elsholz AKW, Turgay K, Michalik S, Hessling B, Gronau K, Oertel D, Mader U, Bernhardt J, Becher D, Hecker M, Gerth U. 2012. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc Natl Acad Sci U S A 109:7451–7456. doi: 10.1073/pnas.1117483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elsholz AKW, Hempel K, Michalik S, Gronau K, Becher D, Hecker M, Gerth U. 2011. Activity control of the ClpC adaptor McsB in Bacillus subtilis. J Bacteriol 193:3887–3893. doi: 10.1128/JB.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Eryani Y, Ib Rasmussen M, Kjellström S, Højrup P, Emanuelsson C, Wachenfeldt von C. 2016. Exploring structure and interactions of the bacterial adaptor protein YjbH by crosslinking mass spectrometry. Proteins 84:1234–1245. doi: 10.1002/prot.25072. [DOI] [PubMed] [Google Scholar]
- 29.Chan CM, Hahn E, Zuber P. 2014. Adaptor bypass mutations of Bacillus subtilis spx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP. Mol Microbiol 93:426–438. doi: 10.1111/mmi.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirstein J, Dougan DA, Gerth U, Hecker M, Turgay K. 2007. The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J 26:2061–2070. doi: 10.1038/sj.emboj.7601655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol Microbiol 42:383–394. doi: 10.1046/j.1365-2958.2001.02639.x. [DOI] [PubMed] [Google Scholar]
- 32.Runde S, MoliEre N, Heinz A, Maisonneuve E, Janczikowski A, Elsholz AKW, Gerth U, Hecker M, Turgay K. 2014. The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis. Mol Microbiol 91:1036–1052. doi: 10.1111/mmi.12521. [DOI] [PubMed] [Google Scholar]
- 33.Leelakriangsak M, Kobayashi K, Zuber P. 2007. Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis. J Bacteriol 189:1736–1744. doi: 10.1128/JB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jervis AJ, Thackray PD, Houston CW, Horsburgh MJ, Moir A. 2007. SigM-responsive genes of Bacillus subtilis and their promoters. J Bacteriol 189:4534–4538. doi: 10.1128/JB.00130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsholz AKW, Gerth U, Hecker M. 2010. Regulation of CtsR activity in low GC, Gram+ bacteria. Adv Microb Physiol 57:119–144. doi: 10.1016/B978-0-12-381045-8.00003-5. [DOI] [PubMed] [Google Scholar]
- 36.Fuhrmann J, Subramanian V, Kojetin DJ, Thompson PR. 2016. Activity-based profiling reveals a regulatory link between oxidative stress and protein arginine phosphorylation. Cell Chem Biol 23:967–977. doi: 10.1016/j.chembiol.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schäfer H, Heinz A, Sudzinová P, Voß M, Hantke I, Krásný L, Turgay K. 2019. Spx, the central regulator of the heat and oxidative stress response in B. subtilis, can repress transcription of translation-related genes. Mol Microbiol 111:514–533. doi: 10.1111/mmi.14171. [DOI] [PubMed] [Google Scholar]
- 38.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. 2008. Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol 68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 39.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.