Abstract
Background
While the pathogenic nature of CNVs on chromosome 22q11.2 have been recognized for decades, unbiased estimates of their population prevalence, mortality, disease risks, and diagnostic trajectories are lacking. Hence, we aim to provide the true population prevalence, trajectory of disease risk, and mortality of 22q11.2 CNVs utilizing the unbiased, representative Danish iPSYCH population case-cohort.
Methods
We use epidemiological methods in conjunction with nationwide hospital registers to analyze the iPSYCH case-control cohort i.e. cases born from 1981 to 2005 (n=57,377) with Attention-Deficit/Hyperactivity Disorder, Major Depressive Disorder, Schizophrenia, Autism or Bipolar Disorder as well as 30,000 randomly drawn individuals – to provide unbiased, population-adjusted estimates and 30-year long disease trajectories for major neuropsychiatric disorders.
Findings
Population prevalence in the Danish population was 1:3672 and 1:1606 for deletions and duplications, respectively, the mortality rate was zero and hazard ratios for neuropsychiatric disorders ranged from 1 to 82 comparably for both rearrangements. By age 32, 10% developed Attention-Deficit/Hyperactivity Disorder, Autism or Intellectual Disability; and deletion carriers had higher probability than duplication carriers of co-occurring Intellectual Disability and of epilepsy.
Interpretation
The significantly different prevalence of 22q11.2 duplications and deletions indicates distinct selective pressures on these rearrangements. While risk for congenital abnormalities, developmental delay, and Intellectual Disability is elevated in deletion carriers, the overall prevalence of neuropsychiatric disorders is higher in duplication carriers, which implies that identification and clinical monitoring should extend beyond congenital traits and into child and adolescent psychiatry.
Introduction
In recent years, large-scale genotyping and sequencing efforts have identified many loss-of-function (LoF) variants in the human genome, including frameshift mutations, truncations, and copy number variants (CNVs),1 greatly expanding our understanding of genetic contributions to neuropsychiatric and developmental disorders. However, the rarity of LoF variants in conjunction with sampling strategies typically used for gene discovery compromise estimates of prevalence and disease risk at the population level. The rarity of these mutations has promoted alternative analytical strategies that cluster together variants based on shared characteristics, e.g. genes impacting early brain development or under evolutionary constraint. While these approaches have successfully uncovered disease risk in groups of variants and implicated specific biological mechanisms,2 they are limited in their ability to estimate risk in individual patients for use in genetic counseling and clinical decision-making. This is largely because most studies are not designed to provide population-unbiased estimates of disease risk, but rather involve patient recruitment focused on specific clinical features or genetic disorders.
CNVs that map to the chromosome 22q11.2 locus have been known for over 25 years, and are among the most frequent and clinically best-characterized mutations in humans.3 These primarily de novo deletions typically involve 1·5–3 Megabases (Mb), and were first identified in patients with DiGeorge or Velo-CardioFacial syndromes, characterized by congenital heart-defects, recurrent infections, velopharyngeal insufficiency, and facial dysmorphism often in combination with Developmental Delay and Intellectual Disability.4 More recently, 22q11.2 deletions have been shown to account for 0·1–1% of patients in clinical cohorts with Autism,5–7 Developmental Delay and/or Intellectual Disability,8 and Schizophrenia,1,9 and have been associated with neurological disorders like epilepsy.10 In contrast, the reciprocal duplications are less frequent in clinical cohorts with Developmental Delay (1:350; unpublished data11) and Intellectual Disability (1:700)12, and whereas the deletions have been formally associated with high risk (OR ≥ 16)1 and penetrance (range: 0·12–0·55) for Schizophrenia13 the duplication has been proposed to protect against this disorder.1,9
Despite substantial clinical evidence regarding 22q11.2 aberrations, we remain largely uninformed at the population level of prevalence, global burden of disease, and mortality. Prevalence estimates of 22q11.2 deletions vary considerably from 1:2,000 to 1:9,70014,15 in newborns, possibly due to ascertainment biases or mortality due to somatic complications, particularly during infancy.16 The reciprocal duplications are less well characterized but appear to segregate more often within families than deletions17 and have higher frequency in healthy controls (1:1176).18 However, population prevalence estimates are lacking and there is a dearth of comprehensive knowledge of age-dependent mortality and morbidity. Recently, we performed the first population-based analysis to provide neuropsychiatric and developmental disorder risk estimates in a nationwide, clinical cohort of patients with 22q11.2 deletions and duplications.19 We found an elevated risk for Schizophrenia (OR = 6·64; 95% CI:2·64–13·46) but considerably lower than expected from other studies,1 presumably reflecting an ascertainment bias affecting the latter. Due to the use of clinically ascertained subjects, our former study could not estimate population prevalence or deletion/duplication ratio, compare risk of mental disease between clinically ascertained and non-ascertained individuals, or examine disease trajectories of individuals without 22q11 rearrangements, with 22q11 deletions and with 22q11 duplications.
In this study, we aim to explore the clinical manifestations and provide the true population prevalence, trajectory of disease risk, and mortality of 22q11.2 CNVs, leveraging a uniquely large and unbiased sample of the Danish population, the iPSYCH Initiative.20 The iPSYCH case-cohort is designed to optimize genetic studies of mental disorders in conjunction with nationwide electronic health records. The iPSYCH casecohort includes (a) the case-sample with all individuals diagnosed with at least one diagnosis of AttentionDeficit/Hyperactivity Disorder, Major Depressive Disorder, Autism Spectrum Disorder, Bipolar Disorder, or Schizophrenia, and (b) the cohort, i.e. a 2% random and thus truly representative sample of the Danish population, which includes individuals with diagnoses of mental disorders. Therefore, we provide prevalence of psychiatric disorders as a function of 22q11.2 rearrangement status and map populationunbiased disease trajectories from ages one to thirty-two.
Material and Methods
Study Population
The study population includes the iPSYCH case-cohort of 86,189 individuals selected among the study base of 1,472,762 singletons born in Denmark between May 1, 1981, and December 31, 2005, who have a known mother from the Danish Civil Registration System, were residents in Denmark, alive at age one year, and enrolled in the iPSYCH Initiative.21 Selection of cases was based on in- and out-patient discharge diagnoses from all Danish hospitals up until December 31, 2012, obtained as ICD-10 codes from the Danish Psychiatric Central Research Register (PCR). The case-cohort includes a population random sample of 30,000 subjects (of which 1,188 overlap with cases) and all individuals clinically diagnosed with AttentionDeficit/Hyperactivity Disorder, Major Depressive Disorder, Schizophrenia (Schizophrenia), Autism Spectrum Disorder, or Bipolar Disorder by December 31, 2012 (Supplementary Methods and eTable 1). Intellectual Disability is defined as IQ below 70; Mild Intellectual Disability: IQ 50–69; Moderate Intellectual Disability: IQ 35–49; Severe Intellectual Disability: IQ<35. Neonatal blood spots were retrieved from the Danish Neonatal Screening Biobank (DNSB) and DNA extraction, genetic analyses and quality control were performed as described in Supplementary Methods. With cross-reference to the clinically-diagnosed 22q11.2DS case-cohort (n=188 individuals), the ability to identify 22q11.2 rearrangements in whole genome amplified (WGA) and genomic DNA were compared (Supplementary Methods).
Statistical Analysis
Statistical analyses were performed in R version 3.3.1. Using the survival22 and mstate packages,23 we employed survival analysis stratified by sex and with inverse-probability of sampling (IPS) weights and a robust estimator of variance24 to assess risk of the 22q11.2 rearrangements for neuropsychiatric and developmental disorders. Since probability of being sampled is accurately known in iPSYCH, risk estimates are unbiased for the Danish population. Unbiased risk estimates for Intellectual Disability and epilepsy were obtained from within the population cohort only, since these diagnoses were not part of case selection. We followed these analyses with IPS-weighted multistate survival models to determine the effect of 22q11.2 rearrangements on diagnostic trajectories over the age span of the study. As with the univariate models, the multistate model was IPS-weighted, stratified on gender, and included 22q11 rearrangement (none, deletion, or duplication) as a predictor. The outcomes in this model are age of onset for each possible transition state from the following list: ID alone, Autism alone, ADHD alone, ID and Autism, ID and ADHD, Autism and ADHD. The baseline hazard was allowed to differ for all state transitions. T-tests were used to assess whether prevalence of commonly-associated congenital abnormalities differed between clinically undiagnosed (no reference in the Danish Cytogenetic Central Register (DCCR)) and diagnosed (reported with 22q11.2DS in DCCR) deletion carriers.
The National Ethical Committee of Denmark (project id: 1-10-72-287-12), the Danish Data Protection Agency (project id: 2012-41-0110), and the steering committees of DNSB and DCCR approved the study.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, writing of the report or decision to submit the paper for publication. The corresponding author, Thomas Werge, had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
After quality control, a total of 76,128 individuals (n=25,704 random population and n=51,493 cases, n=1,069 overlap) were included in the study. We identified 34 with a 22q11.2 deletion and 104 with the reciprocal duplication. The majority had a 3Mb deletion (n=31) or duplication (n=97) whereas three carried the nested 1·5Mb deletion, and seven had a 1·5Mb duplication. Overall mean age was 17·4 and 19·2 years for the deletion and duplication carriers at the end of 2012, respectively (eTable 1). The false positive and negative rates were comparable and low for deletions and duplications indicating acceptable validity of the CNV calls (Supplementary Results), although the false negative rate for duplications might be slightly higher in samples with poorer data quality (eFigure 1).
Within the representative population sample (n=25,704), the prevalence of 22q11.2 deletions and duplications was 1:3,672 (proportion=0·00027, 95% CI: [0·00012, 0·00057]) and 1:1,606 (proportion=0·00062, 95% CI: [0·00040, 0·00107]), respectively (Table 1). This implies that there are 403 deletion (CI = [179, 851]) and 1016 duplication (CI = [597, 1,598]) carriers for all of Denmark in the iPSYCH age cohort. The prevalence of both deletions and duplications was higher in those with a neuropsychiatric or developmental disorder (Table 1). We observed no mortality after age one year among the carriers of 22q11 rearrangements.
Table 1.
Prevalence of 22q11.2 Deletions and Duplications in Population Sample and Neuropsychiatric Disorders
| Diagnostic Groupsa | 22q11.2 deletion | 22q11.2 duplication | |||
|---|---|---|---|---|---|
| n | (%) | n | (%) | ||
| Population-based sample | (n=25,704) | 7 | (0.027) | 17 | (0.066) |
| Attention-Deficit/Hyperactivity Disorder | (n=17,048) | 11 | (0.065) | 46 | (0.27) |
| Autism spectrum disorder | (n=14,618) | 14 | (0.10) | 31 | (0.2) |
| Bipolar Disorder | (n=l,588) | 0 | (0.0) | <3 | (−) |
| Depression | (n=20,579) | 5 | (0.024) | 21 | (0.10) |
| Schizophrenia | (n=2,878) | <3 | (−) | 4 | (0.14) |
| Intellectual Disability | (n=4,108) | 20 | (0.48) | 10 | (0.24) |
| Epilepsy | (n=2,529) | 9 | (0.35) | <3 | (−) |
| Any diagnosis above | (n=45,288) | 30 | (0.066) | 86 | (0.19) |
Sample sizes are calculated based on post quality control (See Supplementary Methods). Attention-Deficit/Hyperactivity Disorder (F90.0); Autism spectrum disorder (F84.0-F84.9); Bipolar Disorder (F30-F31); Depression: single and recurrent depression (F32-F33); Schizophrenia (F20); Intellectual Disability (F70-F79); Epilepsy (G40).%:
The frequency of congenital abnormalities among duplication carriers was only marginally above the general population level, but was significantly lower than observed within the 22q11.2DS group (eTable2). In addition, deletion carriers had lower average gestational age and birthweight, whereas duplication carriers were at population levels (eTable2). Moreover, the deletion, but not the duplication, was strongly associated with epilepsy (Table 2). Cross referencing to the DCCR showed that 70% of deletion carriers, born between 1996–2005, had a clinical genetic diagnosis and higher prevalence of congenital abnormalities compared to genetically undiagnosed carriers (eTable3).
Table 2:
Univariate Cox Regression Models for the Association of Neuropsychiatric and Developmental Disorders among Individuals with a 22q11.2 Deletion or Duplication
| 22qll.2 deletion | 22qll.2 duplication | Mantel-Hanzel p-valueb | Concordance index (95% Cl)c | |||||
|---|---|---|---|---|---|---|---|---|
| HR | (95% Cl) | P | HR | (95% Cl) | P | |||
| Attention- | ||||||||
| Deficit/Hyperactivity | 2·60 | (0·87, 7·73) | 0·09 | 4·01 | (2·15, 7·45) | 1·17 × l0‒5 | 6·04E-06 | 0·77 (0·68, 0·87) |
| Disorder | ||||||||
| Autism spectrum disorder | 2·95 | (112, 7·80) | 0·03 | 3·81 | (1·93, 7·50) | 0·0001 | 1·27E-06 | 0·78 (0·68, 0·88) |
| Depression | 1·01 | (0·28,3·71) | 0·99 | 2·16 | (0·99,5·06) | 0·08 | 0·44 | 0·63 (0·47, 0·79) |
| Schizophrenia | 2·24 | (0·45,12·15) | 0·35 | 1·90 | (0·53, 7·37) | 0·36 | 0·27 | 0·71 (0·49, 0·93) |
| Intellectual Disability | 71·56 | (25·34, 202·10) | 7·77 × 10‒16 | 20·71 | (5·19,82·65) | 1·77 × l0‒5 | <2E-16 | 0·98 (0·97, 1·00) |
| Moderate to severe Intellectual disability | 82·44 | (21·82, 311.50) | 7·77 × 10‒11 | 32·16 | (8·08, 128·00) | 8·47 × l0‒7 | <2E-16 | 0·98 (0·95, 1·00) |
| Epilepsya | 28·41 | (10·82, 74·58) | 1·06 × 10‒11 | - | - | - | <2E-16 | 0·87 (0·73, 1·00) |
Association analysis was left out since no duplication carriers were epileptic.
P-value for Mantel-Cox non-parametric test of differences in survival curves, using survdiff function from R package survival
Concordance index computed for deletions/duplications simultaneously using concordance.index function from R package survcomp, stratifying on gender and with inverse probability of sampling weights. All analyses are performed as univariate Cox proportional hazard regression stratifying by gender and adjusting for the population prevalence for each disease (except for Epilepsy, and Intellectual Disability (and Mod. to Sev. Int. Disability) – here the actual population-based cohort has been applied. See method section). Robust standard errors are used to produce 95% CI and p-values.
Note: HR; hazard ratio, CI; confidence interval; Attention-Deficit/Hyperactivity Disorder (F90.0); Autism spectrum disorder (F84.0–F84.9); Depression: single and recurrent depression (F32-F33); ---Schizophrenia (F20); Intellectual Disability (F70-F79); Moderate to Severe Intellectual Disability (F71-F79); Epilepsy (G40, ICD-10 code).Bipolar Disorder was left out of the analysis since the number of affected subjects was too small.
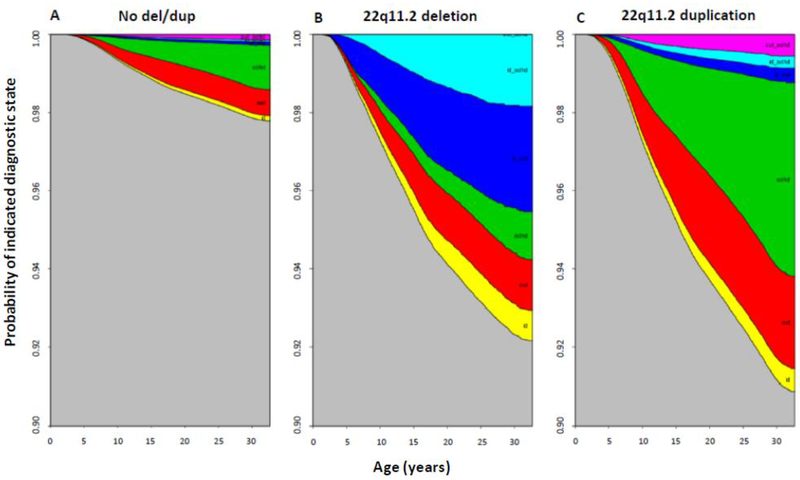
Hazard ratios for neuropsychiatric and developmental disorders were significantly elevated for both deletions and duplications except for Major Depressive Disorder and Schizophrenia (Table 2). To evaluate disease trajectories for individuals with and without rearrangements, we calculated state transition probabilities as a function of age using a multistate survival model (eTable4).25 The probability of being healthy was considerably higher for individuals without 22q11.2 rearrangements (Figure 1). At age 32 only 2·5% of non-carriers were diagnosed with one or more of these disorders, whereas approximately 10% of all deletion or duplication carriers had developed Autism, Attention-Deficit/Hyperactivity Disorder, and/or ID (Figure 1). The probability of having Autism or Attention-Deficit/Hyperactivity Disorder alone was higher among duplication carriers compared to individuals with the deletion who were more often comorbid with Intellectual Disability. While approximately 6% of all deletion carriers had a medical history of Intellectual Disability and Autism or Attention-Deficit/Hyperactivity Disorder, only 1·8 % of those with the duplication had Intellectual Disability with ASD or Attention-Deficit/Hyperactivity Disorder by 32 years of age (Figure 1).
Figure 1: Stacked Transition Probabilities from IPS-weighted Multistate Survival Models for Individuals with and without the 22q11.2 Rearrangement.
The distance between two adjacent lines represents the probability of being in a given state (e.g. AttentionDeficit/Hyperactivity Disorder or Autism) in A) random population sample (no deletion or duplication), B) the identified 22q11.2 deletion carriers, and C) the identified 22q11.2 duplication carriers. Only those diagnoses which showed significant deletion/duplication effects were included in the trajectory analysis. Corresponding coefficient estimates given in Supplementary eTable4. Note: ASD; Autism spectrum disorder; ADHD; AttentionDeficit/Hyperactivity Disorder, ID; Intellectual Disability.
Color code shown from top down: pink: ASD and ADHD, turquoise: ID and ADHD; dark blue: ID and ASD, green: ADHD only, red: ASD only, yellow: ID only.
Discussion
We provide the first population-based estimate of the prevalence and age-dependent mortality of both 22q11.2 duplications and deletions along with population-based risk estimates for a spectrum of neuropsychiatric and developmental and neurological disorders. In addition, we present populationunbiased developmental trajectories of psychiatric disorders of both rearrangement types from one to thirty-two years of age.
Prevalence of 22q11.2 Rearrangements
Our prevalence estimate of the duplication at age one is considerably lower than previous estimates of 1:320 and 1:700 derived from clinical cohorts (unpublished data),11,12 but within the range found among controls in prospective studies of Schizophrenia.9,18 Moreover, the prevalence for the deletion is congruent with the generally accepted estimate of 1:4000 (range: 1:2000 −1:9700).14,15
Contrary to the expectation that there should be a higher ratio of deletions to duplications, we observed a prevalence of 22q11.2 duplications that was over twice that of deletions (p = 0·0007) which supports that the deletion segregates less than the duplication. While we observe a mortality rate of zero in our cohort selected as individuals alive at age one, the lower prevalence of deletions than duplications may occur due to embryonic death or early postnatal death. Three additional lines of evidence support this interpretation: first, 22q11.2 deletions segregate less often in families than duplications;17 second, deletions have been linked to spontaneous abortions;26 and third, infant mortality is high among deletion carriers.16 Hence, the incidence of deletions at birth is likely to exceed 1:3672 as previously reported.14
Risk for Psychopathology
This study illustrates the major impact rearrangements within the 22q11.2 locus have on trajectories of psychiatric illness in the general population. The most notable difference between rearrangements was that duplication carriers were more often diagnosed with Autism or Attention-Deficit/Hyperactivity Disorder alone, whereas deletion carriers more often had comorbid Intellectual Disability. While the strong association between Attention-Deficit/Hyperactivity Disorder and the duplication is novel, our results corroborate previous findings and illustrate that Intellectual Disability12 and Autism27 are relatively common phenotypes in duplication carriers. However, duplication carriers with Intellectual Disability have less severe intellectual constraints (i.e. low-average to mild Intellectual Disability) relative to deletion carriers who have higher prevalence of moderate to severe Intellectual Disability (Table 2). Interestingly, we found a larger proportion of duplication carriers within the Schizophrenia cohort (0.15%) compared to previous studies (0·014 %),1,18 however the sample, with low mean age, is underpowered to draw definitive conclusions regarding associations with Schizophrenia
Given marginal differences in psychiatric disorders between deletion and duplication carriers, our findings imply that type of dysregulation of the affected genes might not be as important for the disease outcome as it is for disease severity. Taken together, our results support that for many of the psychiatric disorders, 22q11.2 duplication carriers need the same careful and persistent clinical attention as individuals with the deletion.
Disease prevalence estimates were considerably lower than in previous clinical studies.28 For the early onset disorders where we expect complete ascertainment in samples of this mean age, this suggests that previous non-population based estimates are highly biased due to mode of ascertainment. However, for later onset disorders such as Schizophrenia and Major Depressive Disorder, the population prevalence in our sample is an underestimate of prevalence by older ages. This almost certainly accounts for the observation that the risk estimate for Schizophrenia in deletion carriers was lower than previous odds ratios derived from large-scale cross sectional studies1 and the recent ageand sex-adjusted Schizophrenia incidence rate ratios (IRR: 6·6–8) in clinically ascertained 22q11.2 deletion carriers reported by our group.19,29
The diagnostic standards for e.g. Autism may differ across studies, which may lead to discrepancies in prevalence estimates. For example, Autism prevalence may be over-reported in studies that use community-rated symptoms rather than gold standards of diagnosing Autism, as in the egalitarian Danish hospital setting. Conversely, we cannot exclude that less severe presentations of Autism may not have met the threshold for referral to child and adolescent psychiatric examination. Second, Attention Deficit/Hyperactivity Disorder may often present as the inattentive type without hyperactivity in 22q11.2DS, which is not included in the diagnosis examined here. Third, while our Schizophrenia sample included the entire spectrum of patients (i.e. from first admitted to chronic) it was limited in size and represents a young cohort (eTable3). Moreover, previous cross-sectional studies in Schizophrenia have included more severely affected cohorts comprised of treatment-resistant patients18 or those with multiple hospitalizations.9 Given these limitations we postulate that the lack of association between 22q11.2 rearrangements and Schizophrenia in our study is an indication that the true population-based risk ratio is somewhat lower than those reported in traditional studies.1 This is also supported by our recent studies of Schizophrenia spectrum disorders in 22q11.2 deletion carriers.19,29 Our study also reveals the risk of mental illness in non-clinically ascertained (i.e. non-congenital) individuals who harbor 22q11.2 rearrangements, and supports the notion that Schizophrenia case-control studies might be biased by comorbidity with Autism, epilepsy and Intellectual Disability among cases leading to inflated estimate of Schizophrenia risk in individuals with the 22q11 deletion. Using logistic regression controlling for age and gender, individuals with schizophrenia diagnoses had significantly higher levels of autism (p<2E-16), intellectual disability (p=6.82E-7) and epilepsy (p=0.004).
Clinical Identification of 22q11.2DS and 22q11.2DupS
Access to the DCCR enabled us to assess clinical detection rate and clinical indicators of 22q11.2 deletions. A large proportion of deletion carriers missed in clinical ascertainment (~60%), and our data indicate that clinical awareness depends on presence of congenital abnormalities such as cleft palate and heart malformations. Importantly, the occurrence of psychosis, Autism and Attention-Deficit/Hyperactivity Disorder is independent of malformation and dysmorphism (data not shown), and emphasizes that clinical awareness is warranted to ensure proper treatment of adjoining psychiatric and neurological conditions. Although 22q11.2 duplications are more prevalent than deletions, most duplication carriers escape clinical ascertainment,12 which may reflect low prevalence of congenital abnormalities. Thus, obvious clues, which direct clinical suspicion towards the presence of a genetic syndrome, are lacking. Altogether, our results suggest that clinical awareness should extend beyond somatic traits and emphasize psychiatric features, further emphasizing the role of genetic testing in the routine diagnosis of psychiatric disorders, especially as the cost of such testing decreases.
Perspectives
We demonstrate that even for well-known rearrangements at 22q11.2 our findings have clinical implications for both genetic counseling and health care. Despite relatively lower psychiatric risk than anticipated, many carriers with psychiatric disorders, but without obvious somatic comorbidities, escape clinical detection and hence have reduced attention to adjoining morbidities like the 22q11.2 deletion associated hypocalcemia, which in combination with antipsychotics and antidepressants lowers the threshold for seizures.30 Since we provide the first population-based estimate showing a high risk for epilepsy, this underscores the importance of proper clinical identification. Our study exemplifies that survival analysis at the level of populations is necessary for genomics to reliably inform clinical practice.
Strengths and Limitations
The strength of this study is the case-cohort design, with utilization of the comprehensive nation-wide register information on all inpatient and outpatient contacts to the free, egalitarian, and national health care system. Due to the age of the biobank, the study was based on cohorts with age below 33 and consequently small sample sizes for later onset disorders reduced the power to detect associations. Register diagnoses have high validity31 and enable accurate risk estimation for neuropsychiatric disorders among people who live long enough to potentially meet the diagnostic criteria. However, the pleiotropic nature of CNVs challenges the diagnostic ICD-10 system that was not developed to capture the clinical manifestations of these syndromes. The study leverages hospital contacts, and does not include milder symptomatology not referred for hospital evaluation or overlooked due to severe psychopathology. Estimates may be affected by observer bias as CNV carriers are more likely than the general population to receive medical attention, although the relatively low risk estimates imply that this bias is less relevant. Estimates are resistant to false negative discoveries since the ability to call CNVs is independent of phenotype. We cannot exclude that the duplication prevalence is slightly underestimated at the lower end of the quality scale, while the high true positive discovery rate of deletions supports the validity of the risk estimates.
Conclusions
In summary, our study provides population prevalence estimates and reports increased risk of neuropsychiatric and developmental disorders at the population level for both 22q11.2 duplications and deletions. Trajectories highlight that the diverging pattern between duplications and deletions are primarily caused by comorbid Intellectual Disability in deletion carriers, suggesting that modelling duplications might be more appropriate for studies of single diagnoses. Current clinical awareness may be driven by somatic clues but high risk for several neuropsychiatric and developmental disorders advocates for more extended use of genetic testing in child and adolescent psychiatry.
Supplementary Material
Research in context
Evidence before this study
The pathogenic nature of neuropsychiatric copy number variants (CNVs) has been acknowledged for decades. However, the lack of unbiased estimates of population prevalence, mortality, disease risks, and diagnostic trajectories (e.g., temporal ordering of comorbidities) has been equally recognized, hindering epidemiologically informed guidance and optimization of healthcare. There is an even more severe shortage of insight into the clinical consequences and utility of other gene disrupting variants identified through sequencing.
Chromosomal rearrangements at the chr22q11.2 locus exemplify the challenge of clinical disease genomics. Prevalence estimates of the chr22q11.2 deletion vary considerably from 1:2,000–1:9,700 presumably due to epidemiological pitfalls such as ascertainment bias or unrecognized early mortality due to somatic complications. Similarly, unbiased estimates of disease risk that would otherwise inform genetic counselling, family planning, and guide clinical surveillance programs are lacking. While our previous report on a clinically ascertained cohort of Danish patients with 22q11.2 rearrangements was the first to provide neuropsychiatric and developmental disorder risk estimates, this register-based study was still, by definition, unable to assess individuals that escape clinical ascertainment and thus prevented estimation of population frequencies, mortality, causes of death as well as disease risk and trajectories.
Added value of this study
This study adds to the existing literature as it is the first to provide the true population prevalence, trajectory of disease risk, and mortality of 22q11.2 CNVs utilizing the unbiased, representative Danish iPSYCH population case-cohort. Importantly, this study also reveals the risk of mental illness in nonclinically ascertained (i.e. non-congenital) individuals who harbor 22q11.2 rearrangements, and allows us to address the notion that Schizophrenia in CNV carriers is often comorbid with Autism, epilepsy and Intellectual Disability.
Implications of all the available evidence
The study illustrates the potential of population-based approaches for translational genomics, resolves long-standing clinical questions about chromosome 22q11.2 aberrations, and advocates for more extensive use of genetic testing in pediatrics and psychiatry.
Acknowledgement
We are very thankful for the kind support provided by Jan Hansen, Karen Brøndum-Nielsen, the scientific board of the Danish Cytogenetic Central Register, and Niels Felsted for helping out with data management. This research has been conducted using the Danish National Biobank resource supported by the Novo Nordisk Foundation and is funded by grants from the Capital Region’s Research Foundation for Mental Health Research, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (R102-A9118 and R155-2014-1724). Thomas Sparsø is funded by the Lundbeck Foundation (R171-2014-1194). Wesley Thompson is partially funded by NIH R01GM104400. The funding bodies have had no impact on the study design, data collection, analysis, or interpretation and have not been involved in writing up the manuscript.
Funding
Capital Region’s Research Foundation for Mental Health Research, The Lundbeck Foundation and National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
All authors have no conflicts of interest.
References
- 1.CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium, Psychosis Endophenotypes International Consortium. Contribution of copy number variants to schizophrenia frm a genome-wide study of 41,321 subjects. Nat Genet 2017; 49: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nurnberger JI Jr, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet 2017; 18: 362–76. [DOI] [PubMed] [Google Scholar]
- 3.McDonald-McGinn DM, Sullivan KE, Marino B, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primers 2015; 1: 15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driscoll DA, Salvin J, Sellinger B, et al. Prevalence of 22q11 microdeletions in DiGeorge and velocardiofacial syndromes: implications for genetic counselling and prenatal diagnosis. J Med Genet 1993; 30: 813–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucan M, Abrahams BS, Wang K, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet 2009; 5: e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders SJ, He X, Willsey AJ, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015; 87: 1215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shishido E, Aleksic B, Ozaki N. Copy-number variation in the pathogenesis of autism spectrum disorder. Psychiatry Clin Neurosci 2014; 68: 85–95. [DOI] [PubMed] [Google Scholar]
- 8.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet 2011; 43: 838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szatkiewicz JP, O’Dushlaine C, Chen G, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry 2014; 19: 762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strehlow V, Swinkels MEM, Thomas RH, et al. Generalized Epilepsy and Myoclonic Seizures in 22q11.2 Deletion Syndrome. Mol Syndromol 2016; 7: 239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Campenhout S, Devriendt K, Breckpot J, et al. Microduplication 22q11.2: a description of the clinical, developmental and behavioral characteristics during childhood. Genet Couns 2012; 23: 135–48. [PubMed] [Google Scholar]
- 12.Ou Z, Berg JS, Yonath H, et al. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med 2008; 10: 267–77. [DOI] [PubMed] [Google Scholar]
- 13.Kirov G, Rees E, Walters JTR, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry 2014; 75: 378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shprintzen RJ. Velo-cardio-facial syndrome In: Cassidy SB, Allason J, eds. Management of Genetic Syndromes. New York: Wiley-Liss, 2005: 615–32. [Google Scholar]
- 15.Tézenas Du Montcel S, Mendizabai H, Aymé S, Lévy A, Philip N. Prevalence of 22q11 microdeletion. J Med Genet 1996; 33: 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan AK, Goodship JA, Wilson DI, et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet 1997; 34: 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahoo T, Theisen A, Rosenfeld JA, et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med 2011; 13: 868–80. [DOI] [PubMed] [Google Scholar]
- 18.Rees E, Kirov G, Sanders A, et al. Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry 2014; 19: 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoeffding LK, Trabjerg BB, Olsen L, et al. Risk of Psychiatric Disorders Among Individuals With the 22q11.2 Deletion or Duplication: A Danish Nationwide, Register-Based Study. JAMA Psychiatry 2017; 74: 282–90. [DOI] [PubMed] [Google Scholar]
- 20.iPSYCH Initiative. You are here: iPSYCH THE LUNDBECK FOUNDATION INITIATIVE FOR INTEGRATIVE PSYCHIATRIC RESEARCH. http://ipsych.au.dk/.
- 21.Pedersen CB, Bybjerg-Grauholm J, Pedersen MG, et al. The iPSYCH2012 case-cohort sample: new directions for unravelling the genetic and environmental architecture of severe mental disorders. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Therneau Terry M. and Grambsch Patricia M.. Modeling Survival Data: Extending the Cox Model. New York: Springer, 2000. [Google Scholar]
- 23.de Wreede LC, Fiocco M, Putter H. mstate: An R Package for the Analysis of Competing Risks and Multi-State Models. J Stat Softw 2011; 38: 1–30. [Google Scholar]
- 24.Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. J Clin Epidemiol 1999; 52: 1165–72. [DOI] [PubMed] [Google Scholar]
- 25.Putter H, Fiocco M, Geskus RB. Tutorial in biostatistics: competing risks and multi-state models. Stat Med 2007. http://onlinelibrary.wiley.com/doi/10.1002/sim.2712/full. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Cheng Q, Meng L, et al. Clinical application of SNP array analysis in first-trimester pregnancy loss: a prospective study. Clin Genet 2017; 91: 849–58. [DOI] [PubMed] [Google Scholar]
- 27.Wenger TL, Miller JS, DePolo LM, et al. 22q11.2 duplication syndrome: elevated rate of autism spectrum disorder and need for medical screening. Mol Autism 2016; 7: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider M, Debbané M, Bassett AS, et al. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 2014; 171: 627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vangkilde A, Olsen L, Hoeffding LK, et al. Schizophrenia Spectrum Disorders in a Danish 22q11.2 Deletion Syndrome Cohort Compared to the Total Danish Population--A Nationwide Register Study. Schizophr Bull 2016; 42: 824–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wither RG, Borlot F, MacDonald A, et al. 22q11.2 deletion syndrome lowers seizure threshold in adult patients without epilepsy. Epilepsia 2017; 58: 1095–101. [DOI] [PubMed] [Google Scholar]
- 31.Uggerby P, Østergaard SD, Røge R, Correll CU, Nielsen J. The validity of the schizophrenia diagnosis in the Danish Psychiatric Central Research Register is good. Dan Med J 2013; 60: A4578. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.