Abstract
Escherichia coli are opportunistic pathogens with the potential to cause a variety of infections in both humans and animals and in many cases have developed antimicrobial resistance. In this study, we characterized extended-spectrum cephalosporin resistant (ESCR) E. coli isolates from diseased companion animals (dogs, cats, and horses) and related the results to clinical findings. ESCR E. coli clinical isolates obtained over a 6-year period were screened for extended-spectrum β-lactamase (ESBL) and/or plasmid mediated AmpC (pAmpC) and virulence markers likely to be associated with extraintestinal pathogenic E. coli (ExPEC). ESBL and/or pAmpC genetic determinants were identified in 79.9% of the ESCR E. coli isolates with blaCTX-M genes being the most common ESBL genotype of which blaCTX-M-15, blaCTX-M-14, and blaCTX-M-55 were the most prevalent. In addition, blaCMY -2 was the most common genotype identified amongst pAmpC producing isolates. Phylogenetic group typing showed that B2 was the most prevalent phylogroup among the ESCR E. coli isolates, followed by the closely related phylogroups D and F which are also associated with extra-intestinal infections. ESCR was also identified in phylogroups commonly regarded as commensals (B1, A, and C). Virulence factor (VF) scores >2 were mostly present amongst isolates in phylogroup B2. Higher virulence scores were found in isolates lacking ESBL/pAmpC resistance genes compared with those carrying both genes (p < 0.05). Five of phylogroup B2 isolates, were typed to the pandemic virulent O25b-ST131 clone and three ST131 isolates carrying blaCTX-M-15 belonged to the subclade C2/H30Rx whilst one isolate carrying blaCTX-M-27 typed to the recently described sub-clade C1-M27. MLST typing also identified other sequence types commonly associated with infections in humans (ST410, ST10, and ST648). Most ESCR E. coli isolates obtained in pure growth were cultured from normally sterile body sites (mostly from urinary tract infections, UTIs) whilst only a small proportion were obtained from body sites populated with commensal flora (p < 0.0001). Our study has shown that ExPEC ESBL/pAmpC producing E. coli isolates are common amongst companion animal isolates and are associated with colonization and infection. In addition, their isolation from a normally sterile site is likely to be clinically significant and warrants antimicrobial treatment.
Keywords: ESBL, Escherichia coli, virulence, antimicrobial resistance, companion animals, United Kingdom
Introduction
Escherichia coli are Gram-negative bacteria belonging to the Enterobacteriaceae family, which are ubiquitous colonizers of the gastro-intestinal (GI) tract in humans and companion animals. The Enterobacteriaceae, most notably E. coli and Klebsiella pneumoniae, are among the most important agents of serious hospital acquired and community-onset bacterial infections in humans and resistance to antimicrobial agents in these bacteria has become an increasingly serious problem (Paterson, 2006). Over the last two decades, extended-spectrum β-lactamase (ESBL)-producing E. coli have been isolated with increasing frequency from human and animal samples (Coque et al., 2008; Dolejska et al., 2013). ESBLs confer resistance to extended-spectrum cephalosporins (ESCs, e.g., ceftiofur, cefpodoxime, ceftazidime, ceftriaxone) and monobactams, and often express resistance to non-β-lactam antimicrobials leaving only limited therapeutic options (Pitout and Laupland, 2008). In addition, isolates carrying plasmid-mediated cephalosporinases (pAmpC) have a broader spectrum of resistance, including the cephamycins and are not inhibited by β-lactamase inhibitors, resulting in resistance to almost all therapeutically available β-lactam agents (Pitout, 2012). E. coli is commonly identified from clinical infections in companion animals, including urinary, respiratory, skin and soft tissues, GI tract, joint and wound infections (Guardabassi et al., 2004; Zogg et al., 2018). In veterinary medicine, the β-lactam agents are arguably the most important and widely used antimicrobial class for treating bacterial infections. Recent United Kingdom data shows that β-lactam agents are prescribed in 43.6% of canine and 70.8% of feline antimicrobial agent prescriptions (Singleton et al., 2017). Their widespread usage may be due to their safety, antimicrobial spectrum, availability of orally bioavailable formulations, favorable pharmacokinetic properties for treating commonly encountered infections and the low cost of many products. First generation cephalosporins and amoxicillin + clavulanic acid are among the most commonly prescribed drugs for dogs (Murphy et al., 2012), whilst penicillin (with or without an aminoglycoside) is commonly prescribed for horses (Hughes et al., 2013). Worryingly, recent data has shown that cefovecin, a third generation cephalosporin, has become the most frequently prescribed antimicrobial agent in the United Kingdom for cats (Singleton et al., 2017).
Similar to the epidemiology of ESBL associated infections in people, the CTX-M type ESBLs (CTX-M-1, -14, and -15) are common amongst companion animal isolates and SHV-12 and TEM-52 have also been reported (Ewers et al., 2012). In addition, there seems to be a match in the geographic distribution of the human and animal CTX-M enzymes, with CTX-M-1 predominating in Africa and Europe, CTX-M-14 in Asia and North America and CTX-M-15 β-lactamase in North America, Europe, and Africa (Rubin and Pitout, 2014).
The occurrence of ESBL/AmpC producing E. coli has been reported in both healthy and diseased companion animals (Gandolfi-Decristophoris et al., 2013; Bogaerts et al., 2015), with some reports indicating high fecal carriage rates amongst healthy animals (Maddox et al., 2012; Baede et al., 2015). However, the clinical significance of these isolates and their involvement in animal disease occurring as either opportunistic pathogens or simple colonizers, has been very rarely investigated. Additionally, the lack of clinical and laboratory guidelines for the management of ESBL/pAmpC-producing animal infections, leads to important dilemmas for veterinary microbiologists in deciding when is appropriate to provide susceptibility results to support antimicrobial therapy for these infections. Additional clinical or laboratory information, which could support the distinction between colonization and true infection associated with ESBL/AmpC producing Enterobacteriaceae is critical in order to avoid unnecessary use of antimicrobials in companion animals.
Thus, this study aimed to characterize ESBL/pAmpC producing E. coli isolates from diseased companion animals (including dogs, cats and horses), with a focus on relating their genetic background to clinical and paraclinical findings in order to determine the role they play in companion animal infections.
Materials and Methods
Bacterial Isolates
Escherichia coli isolates were obtained from companion animal clinical specimens submitted to the Microbiology Diagnostic Laboratory, Liverpool Institute of Veterinary Science between January 2010 and August 2016. Clinical specimens, mainly originated from animals admitted to the University of Liverpool Small Animal and Equine Referral Hospitals, included urine samples collected by cystocentesis, body fluids, tissue biopsy samples, bile and/or liver biopsy, swabs from skin infections and otitis externa cases, fecal samples from diarrheic patients or animals suspected of GI disease. These were plated out aerobically on culture media following site-specific culture protocols, including 5% sheep blood agar (Oxoid, Basingstoke, United Kingdom) and were incubated at 37°C for 1–7 days. Clinical isolates presumptively identified as E. coli based on a positive reaction on Eosin Methylene Blue Agar (EMBA; Oxoid, Basingstoke, United Kingdom) and which showed reduced susceptibility to cefpodoxime (10 μg) and/or cefoxitin, were selected for this study. Species identification of clinical isolates was performed using API 20E identification kits (Biomerieux, France) and also by PCR detection of the uidA gene for confirmation of E. coli (McDaniels et al., 1996). Bacterial culture data, including specimen site, the type of bacterial growth obtained (pure or mixed, repeat E. coli isolation from the same case) and the number of bacterial species isolated from each specimen were recorded in the Veterinary Diagnostic Laboratory database.
Phenotypic Testing for ESBL/pAmpC Production
Cefpodoxime was used as a marker for extended-spectrum cephalosporin resistance (ESCR) and for ESBL screening. All cefpodoxime-resistant E. coli isolates were tested for ESBL production by combination disc testing (CDT) using third generation cephalosporins alone (cefpodoxime, ceftazidime, and cefotaxime) and in combination with clavulanic-acid (MAST Group, United Kingdom) on Mueller-Hinton Agar (Oxoid, Basingstoke, United Kingdom). Results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) interpretative criteria (CLSI, 2012). In addition, isolates displaying resistance to cefoxitin (30 μg) and cefotaxime (30 μg) or ceftazidime (30 μg), were considered presumptive AmpC producers (CLSI, 2012). To demonstrate the co-production of ESBL and AmpC β-lactamase, cefoxitin resistant isolates were further tested with cefepime (30 μg) and cefepime/clavulanic acid (30/10 μg). E. coli ATCC 25922 was used as control strain for susceptibility testing, whilst ESBL and pAmpC producing E. coli isolates, previously confirmed by PCR and sequencing, were used as positive controls for the phenotypic ESBL/AmpC testing. Ertapenem (10 μg) and meropenem (10 μg) were also included routinely for carbapenem-resistance screening whilst ciprofloxacin (5 μg) and enrofloxacin (5 μg) were used for confirming fluoroquinolone-resistance in isolates typed to the ST131 H30Rx lineage.
Characterization of ESBL and pAmpC Resistance Genes
Cell lysates obtained from all investigated isolates were screened by PCR product sequencing for the presence of genes encoding for β-lactamases (blaCTX-M, blaSHV, blaTEM, blaOXA) and plasmid-mediated blaAmpC variants (ACC, FOX, MOX, DHA, CIT, and EBC) as previously described (Essack et al., 2001; Pérez-Pérez and Hanson, 2002; Dallenne et al., 2010). DNA sequencing was performed on amplicons for all the CTX-M and pAmpC positive isolates. For this, PCR amplicons were purified with the Macherey-Nagel NucleoSpin Gel and PCR Clean-up (Thermo Fisher Scientific, United Kingdom). The nucleotide sequences were compared with those present in GenBank1 to identify the specific gene variants involved.
Molecular Typing
An extended PCR scheme described by Clermont et al. (2013) was used to assign the ESBL/AmpC E. coli isolates to one of the seven phylogenetic groups (A, B1, B2, C, D, E, F). Isolates not matching any of the profiles of the initial quadruplex scheme were recorded as Unknown phylogroup (U). Selected CTX-M-positive isolates [at least one from each site of infection and carrying unique resistance genes combinations (i.e., blaCTX-M and blaTEM, or blaCTX-M and blaSHV)] were further typed by multi-locus sequence typing (MLST) which was performed as previously described (Wirth et al., 2006). All isolates which typed to phylogroup B2 were screened for the presence of the O25b-ST131 clone by allele-specific PCR (Clermont et al., 2008). Identification of E. coli ST131 clade, including the subclade C/H30, C/H30-R with its H30Rx and non-Rx sublineages was performed by PCR-based multiplex ST131 clade assay (Matsumura et al., 2017).
Virulence Gene and Extraintestinal Pathogenic E. coli (ExPEC) Characterization
All ESCR isolates were screened by a multiplex PCR assay designed to detect virulence markers likely to be associated with extraintestinal pathogenic E. coli (ExPEC) (Johnson et al., 2015). ExPEC status was assigned according to the definition proposed by Johnson and Russo (2005) which defined ExPEC isolates on the basis of the presence of ≥2 virulence associated genes (VAGs) or gene sets, including papAH and/or papC, sfa/focDE [central region of sfa (S fimbriae) and foc (F1C fimbriae) operons], afa/draBC (Dr antigen-binding adhesins), kpsMII (group II capsule Polysaccharides), and iutA (ferric aerobactin receptor) (Johnson et al., 2015). A virulence score was assigned to each isolate based on the number of virulence genes detected, with pap elements counting collectively as a single trait.
Clinical and Paraclinical Data
To evaluate the likelihood of ESCR E. coli isolates being associated with clinical infections, overall bacterial culture results (including results of repeated sampling from the same case), hematological results (where available) and clinical data were collated. From these data, and after reviewing referral letters, patient’s clinical finding and ancillary tests, which were available in 146/164 cases, a decision of evidence of infection was made for each case. Confirmation of association with infection was provided by the presence of at least one of the following: rods/coccobacilli in cytology smears (either Diff-quick or Gram-stained), raised peripheral white blood cell count, raised inflammatory markers (C-reactive protein when tested) and/or the final clinical diagnosis of infection. Where none of these criteria were met and the isolate was obtained from a mixed bacterial culture, the isolation of an ESBL/AmpC-producing E. coli was considered to be due to contamination.
Statistical Analysis
GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, United States) was used for data analysis. We used the chi-squared test to assess the correlation between ExPEC status, phylogroup and presence of antimicrobial resistance genes. Mann–Whitney U test was used to assess the virulence factor score correlation with phylogroup and presence of antimicrobial resistance genes. P-value < 0.05 was considered statistically significant.
Results
Prevalence of ESCR, ESBL, and pAmpC Producing E. coli Isolates
During the study period (January 2010–August 2016), 876 E. coli isolates (dogs n = 748, cats n = 76 and horses n = 52) were obtained from the clinical specimens submitted from companion animals. Of these, ESCR was identified in 164 (18.7%) non-repetitive E. coli isolates obtained from dogs (n = 138), cats (n = 8) and horses (n = 18) which displayed inhibition zones (≤25 mm) for cefpodoxime (CPD) with or without cefoxitin resistance.
The ESCR E. coli isolates were cultured from urine (n = 49), wound swabs (n = 41), GI tract samples, mainly feces from diarrheic animals (n = 30), skin infections or otitis cases (n = 23), body fluids or tissue biopsies (n = 9), bile or liver samples (n = 12). Combination disc tests for phenotypic ESBL testing of the 164 ESCR isolates resulted in 53 isolates displaying an ESBL phenotype (53/164, 32.3%), 67 isolates showed a presumptive AmpC phenotype (67/164, 40.9%) and 11 isolates (11/164, 6.7%) demonstrated a phenotype suggesting mixed ESBL/AmpC co-production (resistance to cefoxitin, cefotaxime or ceftazidime and demonstrating synergy when tested with cefepime, a fourth generation cephalosporin and cefepime/clavulanic acid). Thirty-three ESCR isolates (33/164, 20.0%) produced inconclusive phenotypic ESBL/pAmpC results (Table 1). No carbapenem-resistance was detected in the ESCR isolates.
Table 1.
Phenotypic and genotypic characterization of extended-spectrum cephalosporin resistant Escherichia coli isolates obtained from companion animals clinical specimens.
| Phenotype | β-lactamases enzyme | Number of isolates | Phylogroup | ST | Year of isolation | Source | Body site |
|---|---|---|---|---|---|---|---|
| ESBL (n = 53) | CTX-M-15 | 7 | A, B1 (n = 4) B2 (n = 2) U (n = 1) |
86 (1) 23 (1) 623 (1) 3880 (1) 4184 (1) |
2010 (n = 1), 2013 (n = 1), 2014 (n = 3) |
Dog (n = 5), Horse (n = 2) |
GI tract (n = 3), SI/O (n = 1), Urine (n = 1), Wound (n = 2) |
| CTX-M-15, OXA | 10 | A, B1 (n = 2) B2 (n = 3) C (n = 4) D (n = 1) |
131 (3) 167 (1) 1284 (1) 2348 (1) |
2010 (n = 4), 2011 (n = 3), 2012 (n = 1) |
Dog (n = 9), Horse (n = 1) |
GI tract (n = 4), Hepato-biliary (n = 1), Urine (n = 1), SI/O (n = 3), Wound (n = 1) |
|
| CTX-M-15, TEM, OXA | 2 | D (n = 1) F (n = 1) |
648 (1) | 2011 | Dog | BF/BP (n = 2), SI/O (n = 3) |
|
| CTX-M-15, TEM | 3 | A | 10 (1) 167 (1) |
2011 | Dog | Wound (n = 3) | |
| CTX-M-14 | 6 | A (n = 3) B2 (n = 2) C (n = 1) |
10 (2) 95 (1) 209 (1) |
2012 (n = 2) 2014 (n = 1), 2015 (n = 3) |
Dog (n = 2), Cat (n = 1), Horse (n = 1) |
GI tract (n = 1), SI/O (n = 1), Urine (n = 4) |
|
| CTX-M-14, TEM | 8 | A (n = 7), D (n = 1) |
10 (1) 209 (1) 617 (2) |
2010 (n = 5), 2015 (n = 2), 2016 (n = 1) |
Dog (n = 8) | BF/BP (n = 2), Hepato-biliary (n = 2) GI tract (n = 1), Urine (n = 3) |
|
| CTX-M-55 | 2 | B2 (n = 2) | 1340 (2) | 2015 (n = 2) | Dog (n = 2) | GI tract (n = 2) | |
| CTX-M-55, TEM | 3 | A (n = 1) B2 (n = 2) |
167 (1) 1340 (2) |
2015 (n = 2), 2016 (n = 1) |
Dog (n = 3) | Urine (n = 2), GI tract (n = 1), |
|
| CTX-M-55, OXA | 1 | B2 | 131 | 2015 | Dog | Urine | |
| CTX-M-55, TEM, SHV-12 | 1 | B1 | 5036 | 2013 | Horse | Wound | |
| CTX-M-1 | 3 | B1 (n = 2) D |
47923509 | 2013 (n = 2) 2014 (n = 1) |
Dog | GI tract | |
| CTX-M-1, TEM | 1 | B1 | 641 | 2013 | Horse | Wound | |
| CTX-M-27 | 1 | B2 | 131 | 2010 | Dog | Hepato-biliary | |
| CTX-M-28, TEM | 3 | B1 (n = 3) | 3743448 (2) | 2015 | Dog (n = 2), Horse (n = 1) |
SI/O (n = 2), Wound (n = 1) |
|
| CTX-M-24, TEM | 1 | A | – | 2014 | Dog | Wound (n = 1) | |
| CTX-M-24, OXA | 1 | C | 3163 | 2012 | Dog | Wound (n = 1) | |
| AmpC (n = 67) | CMY-2 | 23 | B1 (n = 4) B2 (n = 9) C/F (n = 3) D (n = 7) |
– | 2010 (n = 2), 2011 (n = 1), 2012 (n = 6) 2013 (n = 4), 2014 (n = 5), 2015 (n = 4), 2016 (n = 1) |
Dog (n = 23) | Wound (n = 7), Urine (n = 6), GI tract (n = 5), SI/O (n = 4), Hepato-biliary (n = 1) |
| CMY-2, TEM | 30 | A/B1 (n = 12) B2 (n = 12) C (n = 3) F (n = 3) |
2348 (1) | 2010 (n = 1), 2011 (n = 2), 2012 (n = 12) 2013 (n = 4), 2014 (n = 4), 2015 (n = 6) |
Dog (n = 28), Cat (n = 1), Horse (n = 1) |
Urine (n = 15), GI tract (n = 5), Wound (n = 4), BF/BP (n = 3), Hepato-biliary (n = 2), SI/O (n = 1) |
|
| CMY-2, TEM, OXA | 3 | B2 (n = 1) C (n = 1) D (n = 1) |
410 (1) | 2012 (n = 1), 2013 (n = 2) |
Dog | Urine (n = 1), SI/O (n = 1), GI tract (n = 1) |
|
| CMY-2, OXA | 2 | A (n = 1) C (n = 1) |
– | 2010 (n = 1), 2012 (n = 1) |
Dog (n = 2) | Wound (n = 2) | |
| CMY-2, TEM, DHA | 1 | B2 | – | 2015 | Dog | SI/O | |
| DHA | 1 | B1 | – | 2015 | Dog | Wound | |
| DHA, TEM | 3 | A/B1 (n = 2) F (n = 1) |
– | 2013 (n = 1), 2015 (n = 1), 2016 (n = 1) |
Dog | Hepato-biliary (n = 1), Urine (n = 1), Wound (n = 1) |
|
| DHA, TEM, OXA | 1 | B1 | – | 2015 | Dog | Wound | |
| EBC | 1 | B2 | – | 2014 | Horse | Wound | |
| EBC, TEM | 2 | B2 | – | 2014 | Horse | Wound (n = 2) | |
| ESBL/AmpC(n = 11) | CTX-M-15, OXA, CMY-2 | 4 | B2 (n = 1) C (n = 3) |
1485 (1) | 2012 (n = 3), 2013 (n = 1) |
Dog | BF/BP (n = 1), SI/O (n = 1), Urine (n = 1), Wound (n = 1) |
| CTX-M-15, TEM, OXA, CMY-2 | 3 | B2 | 410 (1) 3163 (1) |
2011 (n = 1), 2012 (n = 2) |
Dog | GI tract | |
| CTX-M-15, TEM, OXA, DHA | 1 | D | – | 2013 | Dog | SI/O | |
| CTX-M-24, CMY-2, TEM, OXA | 1 | U | – | 2013 | Dog | Hepato-biliary | |
| CTX-M-24, CMY-2, TEM, SHV-12 | 1 | C | 410 | 2013 | Horse | BF/BP | |
| CTX-M-55, TEM, CMY- 2 | 1 | A | 167 | 2013 | Dog | GI tract (n = 1) | |
| Inconclusive phenotype (n = 33) | TEM, OXA | 1 | B1 | – | 2014 | Dog | Bile |
| TEM | 13 | A/B1 (n = 10) B2 (n = 2) U (n = 1) |
– | 2010 (n = 2), 2011 (n = 1), 2012 (n = 1), 2013 (n = 1), 2014 (n = 3), 2015 (n = 2), 2016 (n = 3) |
Dog (n = 8), Cat (n = 2), Horse (n = 3) |
BF/BP (n = 1), GI tract (n = 2), SI/O (n = 2), Urine (n = 5), Wound (n = 3) |
|
| OXA | 4 | B1 (n = 1), F (n = 2), U (n = 1) |
– | 2012 (n = 3), 2014 (n = 1) |
Dog (n = 3), Horse (n = 1) |
Wound (n = 2), Urine (n = 1), SI/O (n = 1) |
|
| – | 15 | B1 (n = 2) B2 (n = 12) C (n = 1) |
– | 2010 (n = 1), 2011 (n = 1), 2012 (n = 1), 2013 (n = 2), 2014 (n = 3), 2015 (n = 5), 2016 (n = 2) |
Dog (n = 11), Cat (n = 4) |
Urine (n = 7), Wound (n = 3), SI/O (n = 3) Hepato-biliary (n = 2) |
|
ST, sequence type by multi-locus sequence typing; BF/BP, body fluids/biopsies; SI/O, skin infection/otitis.
Molecular Characterization of ESBL/pAmpC Genes
Overall, ESBL and/or pAmpC encoding genes were found in 79.9% (131/164) of the ESCR E. coli isolates. blaCTX-M genes were the most common ESBL genotype (48.9%, 64/131) of which the most prevalent were blaCTX-M-15 (46.9%, 30/64), blaCTX-M-14 (21.9%, 14/64), and blaCTX-M-55 (12.5%, 8/64). blaCTX-M-1 and blaCTX-M-24 were each found in four isolates; blaCTX-M-28 was found in three isolates whilst only one blaCTX-M-27 E. coli isolate was identified in a liver tissue biopsy from a dog. In addition, blaSHV -12 (n = 2) was detected in two isolates, which both co-harbored CTX-M genes (blaCTX-M-55 and blaCTX-M-24, respectively). The vast majority of CTX-M-type ESBL producing isolates also carried blaTEM (n = 29/64) and/or blaOXA (n = 23/64). A number of ESCR isolates (n = 18/164) only produced TEM or OXA β-lactamases whilst in 15/164 isolates, most of which belonged to phylogroup B2 (n = 12/15), none of the investigated bla genes were detected. Resistance to ESC via plasmid mediated AmpC genes was detected in 51.2% (67/131) of the ESBL and/or pAmpC producing isolates, where blaCMY -2 was the most common pAmpC gene identified (59/67), whilst in some isolates pAmpC genes were co-harbored with blaCTX-M-15 (n = 8), blaCTX-M-24 (n = 2) or blaCTX-M-55 (n = 1). Additionally, blaDHA and blaEBC were detected in seven canine isolates and three equine isolates, respectively. No blaACC, blaMOX or blaFOX plasmid mediated AmpC resistance genes were detected in the investigated isolates.
Molecular Typing of E. coli Isolates
Phylogenetic group B2, which is associated with extra-intestinal infections in humans, was the most commonly identified phylogroup among the ESCR E. coli isolates (33.5%, 55/164); phylogroups D and F, which are closely related, and can also be associated with extra-intestinal infections, represented 12.20% (20/164) of the isolates. The phylogroups generally regarded as commensal strains (A, B1) were also associated with ESCR where phylogroup B1 was the most predominant (n = 35, 21.3%), closely followed by phylogroup A (n = 31, 18.9%), C (n = 19, 11.5%), and U (n = 4, 2.4%).
MLST typing identified ST410, recently reported as a new successful pandemic clone (Schønning et al., 2018), as the most prevalent amongst ESBL/pAmpC producing and MLST tested isolates (6/131, all phylogroup C), three of which were both CTX-M- and pAmpC –producers. ST410 was identified in isolates obtained from five dog clinical specimens [urine (n = 2), abdominal fluid (n = 2), tissue biopsy] and one from a horse abdominal fluid isolate. In addition, other important human E. coli clones involved in ESBL dissemination were identified amongst CTX-M-producers such as ST10 in four dog isolates, three CTX-M-14 and one CTX-M-15 producers, all phylogroup A, obtained from wound, urine (n = 2) and tissue biopsy; ST167 in one CTX-M-15 and two CTX-M-55 dog isolates from wound, feces and urine, and also one equine CTX-M-15 producing horse isolate obtained from a wound swab; ST648 (phylogroup D) was identified in one isolate obtained from an interdigital abscess from a dog.
Five E. coli isolates were typed to ST131-B2 (5/164, 3.1%) by both allele specific PCR and MLST and were CTX-M-15, -14, -55, or -27 producers. All ST131-B2 isolates were from dogs with three isolates from sterile sites (two from bile, one from urine) whilst two isolates were from a colon biopsy and a skin swab. ST131-clade specific PCR showed that 4/5 ST131 isolates belonged to Clade C/H30; by using the same assay, one isolate carrying blaCTX-M-27 was typed to the recently described sub-clade C1-M27; three isolates carrying blaCTX-M-15 and showing fluoroquinolone resistance were typed to the subclade C2 known as H30Rx (C2/H30Rx). The remaining ST131 isolates carried blaCTX-M-55 and demonstrated fluoroquinolone resistance but remained unclassified by this assay.
Virulence Score (VS) and ExPEC Status of ESCR Isolates
The majority of ESCR E. coli isolates included in this study were obtained from non-GI tract sites (103/131, 81.7%). As expected, most of the isolates with virulence factor (VF) scores >2 belonged to phylogroup B2 (32/55 isolates, 58%) and phylogroup D and F (in 5/20 isolates, 25%), followed by phylogroup A (5/31 isolates 16%), phylogroup C 19 isolates (2/19 isolates 10.5%) and phylogroup B1 (3/35 isolates8.6%).
P fimbrial structural subunits encoded by papA, papC or papE (75/164, 45.7%) were the most commonly identified virulence genes, followed by iutA (61/164, 37.2%), which encodes for a ferric aerobactin receptor involved in the iron uptake and kpsII gene (42/164, 25.6%) which is involved in capsular polysaccharide production. The B2 phylogroup showed a significantly higher virulence score compared to all the other groups (p < 0.01) and more than half of B2 strains (32/55) where defined as ExPEC. Phylogroups D and F, which can also be associated with ExPEC, demonstrated a higher virulence score compared to isolates belonging to group A, B1, C, and U but this difference was not statistically significant (Table 2).
Table 2.
Distribution of average virulence scores and ExPEC status in relation to their phylogroups and resistance genes identified.
| Phylogroup n (%) | ESBL/AmpC type n (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A/B1 (n = 66) |
B2 (n = 55) |
D and F (n = 20) |
C and U (n = 23) |
CTX-M group 1 (n = 36) |
CTX-M- group 9 (n = 18) |
AmpC (n = 67) |
ESBL/AmpC (n = 11) |
Additional β-lactamase producers (n = 18) |
Inconclusive phenotype∗ (n = 15) |
|
| Average virulence | ||||||||||
| score | 0.76 | 2.70 | 1.00 | 0.91 | 1.11 | 1.82 | 1.28 | 0.7 | 1.47 | 3.4 |
| hlyD | 1 | 14 | 1 | 0 | 2 | 1 | 6 | 0 | 2 | 5 |
| papA | 4 | 17 | 1 | 2 | 3 | 6 | 8 | 0 | 2 | 5 |
| kpsII | 3 | 29 | 8 | 2 | 9 | 2 | 20 | 1 | 4 | 6 |
| iutA | 26 | 18 | 6 | 11 | 17 | 9 | 20 | 6 | 8 | 1 |
| papC | 6 | 16 | 0 | 3 | 2 | 6 | 7 | 0 | 3 | 7 |
| sfa1 | 3 | 16 | 1 | 1 | 1 | 1 | 8 | 0 | 3 | 8 |
| afa1 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| papE | 6 | 18 | 2 | 2 | 2 | 6 | 6 | 0 | 5 | 9 |
| cnf1 | 3 | 17 | 0 | 0 | 3 | 1 | 6 | 0 | 1 | 9 |
| ExPEC | 8 (12.12%) | 32 (58.18%) | 5 (25.00%) | 3 (13.05%) | 8 (22.22%) | 8 (44.44%) | 13 (19.40%) | 0 (0%) | 6 (33.33%) | 8 (53.33%) |
ExPEC, extraintestinal pathogenic E. coli, defined operationally as the presence of >2 of the virulence genes investigated (papA and/or papC, counted as one). ∗Isolates lacked all beta-lactam resistance genes, which they were screened for.
Analysis of virulence factor scores, in relation to carriage of resistance genes, also showed differences in the frequency of virulence genes amongst isolates carrying or lacking ESBL/pAmpC genes. In particular, the ExPEC status and virulence score average were higher for isolates lacking the screened ESBL/pAmpC resistance genes (here defined as isolates with an inconclusive phenotype) and lower for isolates possessing both ESBL and AmpC genes (p < 0.05) (Table 2).
Body Site Distribution
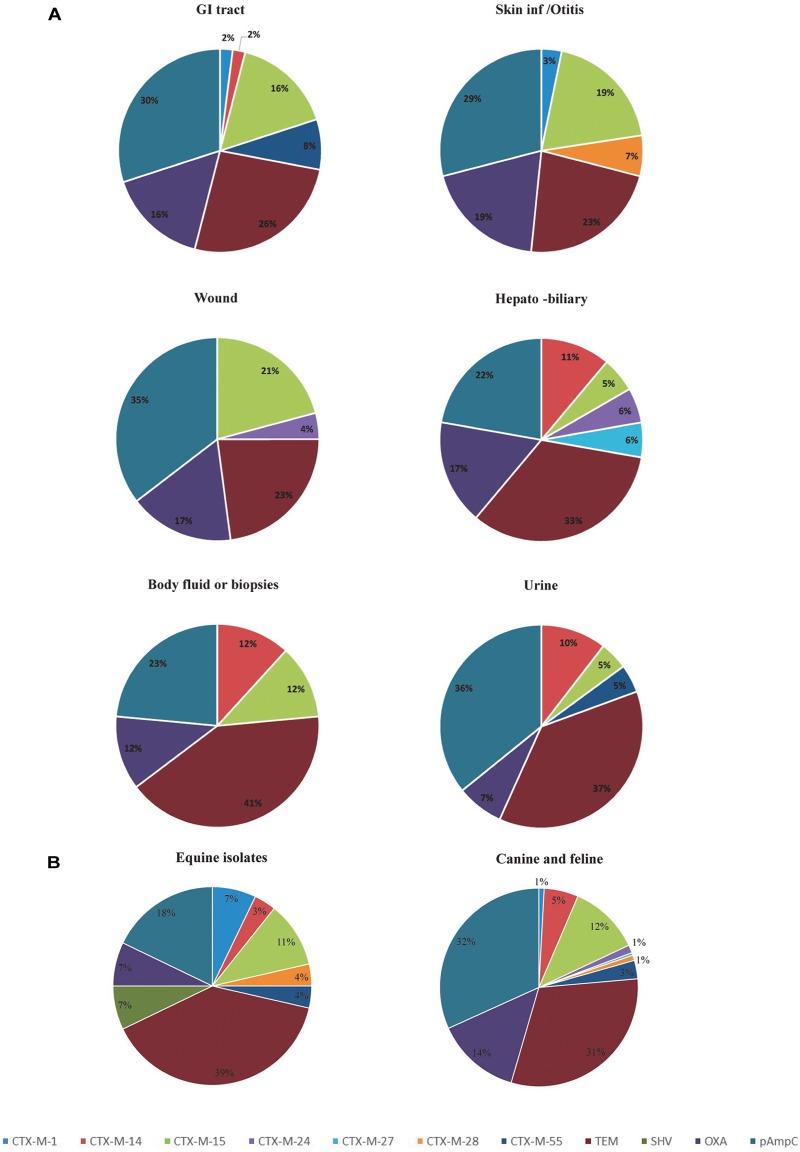
Analysis of the frequency of ESBL/pAmpC genotypes according to the site of isolation, shows that isolates from the GI tract, hepatobiliary infections and urine carry a greater variety of ESBL/pAmpC resistance genes compared to isolates from the skin/soft tissue infections, body fluids and wound infections (Figure 1A). With regard to the host species involved, horse isolates showed a wider variety of bla genes despite the low numbers of isolates compared to canine isolates (Figure 1B). However, blaCTX-M-15 was the predominant ESBL genotype identified in both canine and equine isolates (19.9 and 16.6% of isolates, respectively), as well as in GI tract (35.7%), skin/otitis isolates (31.6%) and wound isolates (19.2%) from cats and dogs. CTX-M-14 β-lactamase producers, although rare in wound infections and GI tract isolates (0 and 2%, respectively), were a relatively common ESBL type found in isolates from body fluids/biopsy samples (25.0%), hepatobiliary infections (18.2%), and urine (15.9%). In addition, around half of the ESCR isolates obtained from dogs harbored pAmpC genes (72/138, 52.2%), either alone or in combination with other β-lactamase encoding genes, with frequencies ranging from 36% for hepatobiliary infections to 55% in the case of urine samples. Moreover, blaCMY -2 alone or in combination with blaTEM was the most common genotype associated with ESCR E. coli isolates from UTI from dogs (23/46, 50.0%).
FIGURE 1.
Diversity and distribution of ESBL and pAmpC producing Escherichia coli according to the clinical specimens (A) and species (B). GI, Gastrointestinal tract.
Clinical/Paraclinical Data Analysis
Clinical or laboratory data used to evaluate evidence of infection was available in 111 of 146 cases (76.0%). Overall, 70 ESCR E. coli isolates were obtained from normally sterile body sites (body fluids/biopsy, urine collected by cystocentesis and hepatobiliary samples) and 54 of these were pure cultures (54/70, 77.1%). Whereas from 94 ESCR isolates, obtained from body sites populated with commensal flora (skin, ear canal, GI tract), only 14 E. coli isolates were obtained in pure growth (14/94, 14.8%) (p < 0.0001).
Bacterial culture results of ESCR E. coli isolates (Table 3), also shows that the vast majority of the isolates originating from urinary tract infections (UTIs) were obtained in pure culture (45/49, 91.8%) and in almost half of the cases, the clinical records showed repeated isolation of an ESBL/AmpC producing isolate (20/49, 40.8%) indicating persistence or re-infection; this was significantly higher than the frequency of repeated isolation from skin/ear infections (p = 0.0029) and GI tract (p < 0.00001). In addition, in the majority of UTI cases where an ESCR E. coli isolate was obtained (30/49, 61.2%), there was at least one clinical or paraclinical test result supportive of infection (either rods on cytology, hematuria or pollakiuria, neutrophilia). However, in 11 UTI cases (11/49, 22.4%) isolation of ESBL/pAmpC producing bacteria was not accompanied by clinical signs, suggesting transient colonization of the bladder or urinary tract or possibly subclinical infection; clinical/paraclinical data was missing in 8 UTI cases. In contrast, the majority of ESCR isolates from skin infections/otitis, GI tract and wounds were obtained in mixed cultures (80/94, 85.1%). The role played by the ESCR or specifically the ESBL/AmpC producing bacteria is difficult to ascertain in samples yielding a mixed bacterial growth (polymicrobial cultures often with other Gram-negative bacteria also involved); however, in most of these cases (64/94, 69.1%) there was clinical evidence of infection (mainly inflammation and purulent discharge).
Table 3.
Clinical and laboratory data associated with extended-spectrum cephalosporin resistant Escherichia coli clinical isolates.
| Phylogroup(s) (n) | VF | ST (n- if >1 isolates) | ESBL/AmpCBeta-lactamase | Narrow-spectrum beta-lactamase | Type of growth | Repeat isolation | Evidence of clinical infection | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≥2(n) | ≤2(n) | Pure (n) | Mixed (n) | Yes (n) | No (n) | Yes (n) | No (n) | |||||
| Urine (n = 49) | B2 (16) | 1 | CTX-M-14 | 15 | 1 | 3 | 12 | 10 | 5 | |||
| 1 | 1 | ST1340; ST131 | CTX-M-55 | TEM (1)OXA (1) | ||||||||
| 5 | 1 | CMY-2 | TEM (4) | |||||||||
| 2 | TEM | |||||||||||
| 5 | – | |||||||||||
| D and F (6) | 1 | DHA | TEM | 6 | 0 | 3 | 2 | 4 | 0 | |||
| 4 | CMY-2 | TEM (1) | ||||||||||
| 1 | CTX-M-14 | TEM | ||||||||||
| A (6) | 1 | ST167; | CTX-M-55 | 5 | 1 | 4 | 2 | 5 | 1 | |||
| 4 | 1 | ST10 (2); ST209 (2); ST617 | CTX-M-14 | TEM (3) | ||||||||
| B1 (15) | 1 | ST23 | CTX-M-15 | 14 | 1 | 8 | 7 | 7 | 4 | |||
| 1 | 8 | CMY-2 | TEM (8) | |||||||||
| 1 | 2 | TEM (2),OXA (1) | ||||||||||
| 2 | ||||||||||||
| C (5) | 1 | CTX-M-15, CMY-2 | OXA | 4 | 1 | 2 | 3 | 4 | 1 | |||
| 1 | ST410 | CTX-M-15 | OXA | |||||||||
| 1 | 2 | ST410 (1) | CMY-2 | TEM (3),OXA (1) | ||||||||
| U | 1 | TEM | 1 | 0 | – | – | – | – | ||||
| Total | 22 | 27 | 45 | 4 | 20 | 26 | 30 | 11 | ||||
| Body fluid/biopsies (n = 9) | B2 (1) | 1 | CMY-2 | TEM | 0 | 1 | 1 | 0 | 1 | 0 | ||
| F (2) | 1 | CTX-M-15 | TEM, OXA | 1 | 1 | 0 | 2 | 1 | 0 | |||
| 1 | ST2348 | CMY-2 | TEM | |||||||||
| A (4) | 1 | TEM | 1 | 3 | 2 | 1 | 3 | 0 | ||||
| 1 | 1 | ST10, ST617 | CTX-M-14 | TEM (2) | ||||||||
| 1 | CMY-2 | TEM | ||||||||||
| C (2) | 1 | ST410 | CTX-M-15, CMY-2 | OXA | 2 | 0 | 1 | 1 | 2 | 0 | ||
| 1 | ST410 | CMY-2, SHV | TEM | |||||||||
| Total | 2 | 7 | 4 | 5 | 4 | 4 | 7 | 0 | ||||
| Hepato-biliary (n = 12) | B2 (4) | 1 | ST131 | CTX-M-27 | 3 | 1 | 1 | 3 | 4 | 0 | ||
| 1 | ST131 | CTX-M-15 | OXA | |||||||||
| 1 | CMY-2 | |||||||||||
| 1 | ||||||||||||
| F (1) | 1 | CMY-2 | TEM | 1 | 0 | 0 | 1 | 0 | 1 | |||
| A (3) | 2 | CTX-M-14 | TEM (2) | 0 | 3 | 0 | 3 | 3 | 0 | |||
| 1 | DHA | TEM | ||||||||||
| B1 (1) | 1 | TEM, OXA | 0 | 1 | 1 | 0 | 1 | 0 | ||||
| C (2) | 1 | CMY-2 | TEM | 1 | 1 | 1 | 1 | 1 | 1 | |||
| 1 | ||||||||||||
| U (1) | 1 | CTX-M-24, CMY-2 | TEM, OXA | 0 | 1 | 1 | 0 | 1 | 0 | |||
| Total | 3 | 9 | 5 | 7 | 4 | 8 | 10 | 2 | ||||
| Skin infection/otitis (n = 23) | B2 (11) | 2 | ST131, ST3880 | CTX-M-15 | OXA (1) | 1 | 10 | 1 | 9 | 10 | 0 | |
| 1 | ST1485 | CTX-M-15, CMY-2 | OXA | |||||||||
| 1 | ST95 | CTX-M-14 | ||||||||||
| 2 | 2 | CMY-2 | TEM | |||||||||
| 3 | ||||||||||||
| D and F (3) | 1 | 1 | ST2348 | CTX-M-15 | TEM (1), OXA (2) | 0 | 3 | 0 | 2 | 2 | 0 | |
| 1 | CMY-2 | TEM, OXA | ||||||||||
| A (2) | 2 | TEM (2) | 0 | 2 | 1 | 0 | 1 | 0 | ||||
| B1 (5) | 1 | ST4792 | CTX-M-1 | 0 | 5 | 0 | 4 | 3 | 1 | |||
| 2 | ST448 (2) | CTX-M-28 | TEM (2) | |||||||||
| 2 | CMY-2 | TEM (1) | ||||||||||
| C (1) | 1 | CTX-M-15 | OXA | 0 | 1 | 1 | 0 | 1 | 0 | |||
| U (1) | 1 | OXA | 0 | 1 | 0 | 1 | 1 | 0 | ||||
| Total | 9 | 14 | 1 | 22 | 3 | 16 | 18 | 1 | ||||
| GI tract (n = 30) | B2 (15) | 1 | 1 | ST131; ST623 | CTX-M-15 | OXA (1) | 3 | 12 | 3 | 12 | 8 | 7 |
| 2 | ST3163 (2) | CTX-M-15, CMY-2 | TEM (2), OXA (2) | |||||||||
| 2 | 1 | ST1340 (2) | CTX-M-55 | TEM (1) | ||||||||
| 2 | 6 | CMY-2 | TEM (5), OXA (1) | |||||||||
| D (1) | 1 | ST3509 | CTX-M-1 | 05 | 1 | 0 | 1 | 0 | 1 | |||
| A (6) | 1 | CMY-2 | TEM | 0 | 5 | 0 | 5 | 1 | 4 | |||
| 2 | ST1284; ST4184 | CTX-M-15 | OXA (1) | |||||||||
| 1 | ST617 | CTX-M-14 | TEM | |||||||||
| 1 | ST167 | CTX-M-55, CMY-2 | TEM | |||||||||
| 1 | TEM | |||||||||||
| B1 (3) | 2 | CMY-2 | 1 | 2 | 0 | 2 | 2 | 0 | ||||
| 1 | TEM | |||||||||||
| C (4) | 1 | ST410 | CTX-M-15, CMY-2 | TEM, OXA | 0 | 4 | 0 | 4 | 2 | 2 | ||
| 2 | CTX-M-15 | OXA (2) | ||||||||||
| 1 | CTX-M-14 | |||||||||||
| U (1) | 1 | CTX-M-15 | 0 | 1 | 0 | 1 | 1 | 0 | ||||
| Total | 5 | 25 | 4 | 25 | 3 | 25 | 14 | 14 | ||||
| Wound (n = 41) | B2 (8) | 1 | 1 | CMY-2 | TEM (2) | 0 | 8 | 1 | 7 | 7 | 1 | |
| 3 | EBC | TEM (2) | ||||||||||
| 2 | 1 | |||||||||||
| D and F (7) | 2 | OXA | 1 | 6 | 1 | 6 | 6 | 1 | ||||
| 5 | CMY-2 | |||||||||||
| A (10) | 1 | CTX-M-24 | TEM | 3 | 7 | 3 | 7 | 7 | 3 | |||
| 2 | CTX-M-15, DHA or CMY | TEM, OXA | ||||||||||
| 3 | ST167, ST10 | CTX-M-15 | OXA (1), TEM (2) | |||||||||
| 2 | CMY-2 | OXA (1), TEM (1) | ||||||||||
| 2 | TEM | |||||||||||
| C (5) | 1 | ST410 | CTX-M-15, CMY-2 | OXA | 1 | 4 | 2 | 3 | 4 | 1 | ||
| 1 | ST3163 | CTX-M 24 | OXA | |||||||||
| 3 | CMY-2 | OXA (1) | ||||||||||
| B1 (11) | 1 | 1 | ST641 | CTX-M-1 | TEM (1) | 4 | 7 | 2 | 6 | 6 | 2 | |
| 2 | ST86 | CTX-M-15 | ||||||||||
| 1 | ST3743 | CTX-M-28 | TEM | |||||||||
| 1 | ST5036 | CTX-M-55, SHV | TEM | |||||||||
| 3 | DHA | OXA (1), TEM (2) | ||||||||||
| 1 | CMY-2 | TEM | ||||||||||
| 1 | ||||||||||||
| Total | 7 | 34 | 9 | 32 | 9 | 29 | 30 | 8 | ||||
ST, sequence type by multi-locus sequence typing; VF, virulence factor score.
Analysis of E. coli phylogroups in relation to the infection site and carriage of resistance genes, shows that the phylogroups usually associated with ExPEC (B2, D, and F) and those regarded as commensals (A, B1, C), were both associated with ESCR and ESBL/AmpC production and were isolated in largely similar proportions from UTIs (22 versus 27), hepatobiliary infections (5 versus 7), GI tract specimens (16 versus 14); however, higher numbers of the ExPEC associated phylogroups (B2, D, and F) were found amongst skin infections and otitis isolates (14 versus 9) and lower numbers amongst body fluids/biopsy isolates (3 versus 6) and wound infections (15 versus 26) (Table 3). Analysis of VF distribution among these two groups [likely ExPEC (B2, D, and F) versus likely commensal (A, B1, and C)] shows that, with the exception of UTI isolates, where there were a similar number of isolates from the two groups with a VF ≥ 2, in general isolates of phylogroups B2, D, and F carried more virulence genes (≥2); this was observed especially in hepatobiliary, skin infections and GI tract isolates. In addition, clinical data analysis also showed that in 70.6% of the infections associated with ESCR E. coli of phylogroup B2, D, and F there was clinical/paraclinical evidence of an active infection.
Discussion
Despite the increasing prevalence of ESBL/pAmpC producing Enterobacteriaceae reported in animals and the increasing rate of isolation by veterinary laboratories, there is still little information regarding the role that ESBL/AmpC producing E. coli isolates play as infectious agents in diseased companion animals. Many studies have investigated the zoonotic risks and the prevalence of fecal carriage or have characterized the ESBL/AmpC producing isolates from clinical specimens in animals (Dierikx et al., 2012; Hordijk et al., 2013; Maeyama et al., 2018; Zogg et al., 2018). However, very few studies (mainly human) have linked the molecular characteristics of ESBL/pAmpC producing isolates with clinical data in order to determine the clinical significance of ESCR E. coli (Sidjabat et al., 2009; Schoevaerdts et al., 2011; Rodríguez-Baño et al., 2012). Thus, the current study investigated the molecular characteristics of ESBL/pAmpC producing E. coli isolates from various companion animal infections in relation to clinical and paraclinical information, which could provide an indication of their role as infecting agents or occasional contaminants in clinical specimens. Comparison with our previous work shows an important increase of ESCR E. coli prevalence in clinical isolates investigated in our laboratory over time, with a rise from 7% (2010/2011) (Timofte et al., 2016) to 21% in the current study (samples from 2010/2016), highlighting the need for greater understanding of their association with infection in order to improve disease management and infection control.
Molecular characterization has shown that ST410 was the most common ST identified in clinical isolates from dogs and horses in this study, both in CTX-M- and pAmpC producers which is concerning given that this clone was recently described as a high-risk multi-drug resistant (MDR) clone with increased potential for inter-species transmission (Schaufler et al., 2016). Also, recently attention has been drawn to the emergence of E. coli D-ST648 in companion animals, a clone which combines characteristics of multidrug resistance and virulence, similar to those observed in E. coli ST131-B2 (Ewers et al., 2014). In our study, we describe the isolation of a D-ST648-CTX-M-15 E. coli from an interdigital abscess from a dog, and to the best of our knowledge, this has not been reported previously in United Kingdom animals. The pandemic virulent E. coli ST131-B2 clone was rare among the ESCR isolates analyzed (five isolates); nonetheless, its isolation occurred in three different years (2010, 2011, and 2015) which suggests persistence in animal populations and raises the potential of zoonotic risks represented by ESBL-producing E. coli in companion animals. Previous studies have shown that ST131-B2 has driven the dissemination of the CTX-M β-lactamase (particularly CTX-M-15) worldwide (Petty et al., 2014) and recent epidemiologic and whole genome phylogeny studies have shown that the clonal structure of E. coli ST131 is highly variable including subclones with particular resistance traits (Price et al., 2013). These studies have also shown that specific sublineages are particularly responsible for ST131/CTX-M-15 dissemination, especially those recently named as clone C or C/H30, which encompasses two sublineages [C/H30-R (non-Rx) and C2/H30Rx] both characterized by fluoroquinolone resistance (Matsumura et al., 2017). In our study, one blaCTX-M-27 positive isolate was typed to the C1/H30-R (recently described as the C1-M27 sublineage) and three blaCTX-M-15 and fluoroquinolone resistant isolates were typed to the C2/H30Rx sublineage. The C1-M27 clone is responsible for the epidemic spread of ST131-CTX-M-27 in Japan (Matsumura et al., 2015) where a high prevalence of E. coli ST131 H30R/non-Rx lineage (likely the C1-M27 sublineage) was also found in isolates from dogs (Maeyama et al., 2018). A recent study looking at the prevalence and characteristics of ST131 clone among ESBL-producing E. coli colonized patients from four European hospitals, also showed a changing epidemiology of ST131 in Europe and the emergence of C1-M27 clade (Merino et al., 2018). Ghosh et al. (2017) examined the presence of ST131-CTX-M-27 producing E. coli amongst isolates from the environment, food, livestock, humans, and companion animals in Germany and found that this lineage was exclusively found in human isolates and showed an increasing prevalence from 0 to 45% over a period of 7 years. E. coli ST131 H30/H30-Rx subclones causing UTI in companion animals in Europe have been previously reported in companion animals from Portugal (Belas et al., 2018) but to the best of our knowledge, this is the first report of ST131 C1-M27 sublineage in companion animals outside Japan.
Overall, the molecular characterization of resistance genes and genetic background of the isolates analyzed this study has identified a diversity of blaCTX-M genes in E. coli isolates. The blaCTX-M variants, CTX-M-14 and CTX-M-15 generally predominate amongst human and animal E. coli isolates; however, our study shows a decreasing occurrence of CTX-M-15 β-lactamase producers over the last few years in favor of new variants, in particular CTX-M-55. In the United Kingdom, this latter blaCTX-M variant has been isolated from turkey meat (Randall et al., 2014; Parker et al., 2016), but it was not previously identified from companion animals in the United Kingdom or Europe. However, CTX-M-55 is among the predominant gene type in E. coli isolated from companion animals in Asian countries (Kawamura et al., 2017).
Phylogroup typing of ESCR E. coli isolates showed a high occurrence of B2 strains (33.5%) which is markedly higher than the frequency reported in a similar study conducted in Germany where 2.4% of the strains were assigned to this phylogroup (Schmiedel et al., 2014; Wagner et al., 2014). B2 strains are frequently associated with a higher number of virulence determinants (Valentin et al., 2014) and as expected, the virulence score of B2 strains (and to a lesser extent D and F) in this study was markedly higher than of other phylogroups; therefore, isolates typed to phylogroup B2, D, or F were more frequently classified as ExPEC. Furthermore, in the majority of the infections associated with phylogroups B2, D, and F there was clinical/paraclinical evidence of infection that supports their association with true infection. The average virulence score in the ESCR isolates was lower in isolates carrying both ESBL/pAmpC genes (VS = 0.7) and higher in isolates lacking these genes and categorized as inconclusive phenotype (VS = 3.4). This is in accordance with the general concept that in most cases, increased resistance in E. coli isolates can be associated with decreased virulence due to survival fitness costs (Beceiro et al., 2013). However, the existence of already established (ST131) or emerging (ST648) clones combining virulence and multidrug-resistance via production of CTX-M β-lactamase (Ewers et al., 2014; Petty et al., 2014), provides the evidence that the opposite is also true and occurrence of such clones in companion animals clinical specimens is worrying both due to therapeutic limitations and public health concerns.
With regards to the body site distribution of isolates carrying different ESBL/pAmpC genes, it is interesting to notice that blaCTX-M-15 and blaCTX-M-1 have been found more frequently in samples of the GI tract but also in wound and skin infection isolates, where contamination of the wound due to the licking in dogs is very common. Carriage of ESBL/pAmpC producing E. coli of phylogroup B2, D, and F in the GI tract suggests ExPEC intestinal carriage and a potential reservoir of multi-drug resistant ExPEC producing bacteria in animal patients. Previous studies have shown that colonization with E. coli of phylogroup B2 in healthy individuals is commonly identified in fecal E. coli in healthy humans and animals (Dale and Woodford, 2015; Alonso et al., 2017) and our findings shows that in diseased companion animals, fecal ExPEC isolates can also be ESBL/pAmpC producers. Moreover, we have identified ExPEC ESBL/pAmpC producers amongst clinical isolates from all specimens including UTIs, body fluids/biopsies, skin, and wound infections. This is an important finding for both patient management and infection control in veterinary settings; ExPEC are considered opportunistic colonizing pathogens (Price et al., 2017) and intestinal carriage or skin/wound colonization in immunocompromised patients can lead to infection. Equally important, colonized patients can be a source for hospital dissemination of ExPEC ESBL/pAmpC producers as well as posing a risk for interspecies transmission and a risk for owner and family members. Therefore, infection control measures need to be implemented upon detection of such bacteria to limit their dissemination within veterinary premises, which needs to be coupled by providing owners with information on the significance of isolation of ESBL/pAmpC producers from their pets and provide, for example, hand hygiene advice to reduce transmission risks.
Accurate and rapid laboratory detection of ESBL/AmpC phenotypes is critical for supporting management of patients and infection control in hospitals and clinics. Whilst detection of ESBL/pAmpC E. coli colonized patients will be always useful for epidemiology and infection control, distinction between colonization and true infection is critical for patient therapeutic management and reducing unnecessary antibiotic use. In our study, more ESCR isolates were obtained from body sites populated with commensal flora where the role played by the ESCR/ESBL/AmpC producing bacteria in the infection is more difficult to establish as cultures generally result in polymicrobial growth. Conversely, the vast majority of ESCR isolates obtained from normally sterile sites were obtained as pure cultures (77%). Urine samples collected by cystocentesis (where the risk of contamination during collection is the lowest) yielded pure cultures in 91% cases and in most of these (61%) at least one criteria supportive of infection was present. These findings suggest that obtaining an ESCR E. coli isolate in pure culture, from a normally sterile site is likely to be clinically significant and warrants antimicrobial treatment. Whenever possible, cytology and Gram-staining can be employed to aid decisions regarding the likely infecting pathogen by revealing the phagocytozed organisms with matching cellular morphology of those obtained from bacterial cultures. Nevertheless, careful assessment of each case is critical for appropriate antimicrobial selection as agents selected to treat the infecting pathogen (other than an ESBL/AmpC producer) may include β-lactam agents and which in turn may select for ESBL/AmpC producers. Repeating bacterial cultures following application of a stringent aseptic technique for specimen collection can also help and should be recommended to distinguish between contamination, transient colonization and true infection.
Our study has shown that a variety of ESBL/pAmpC producing E. coli isolates, including ExPEC, can be associated with both colonization and infection in companion animals. However, this work was performed on a bacterial population originating from cases seen in the Small Animal and Equine Referral Hospitals and may not be directly comparable to the general companion animal population. Future prospective studies are necessary to monitor the duration of ESBL/pAmpC producing E. coli colonization and identify factors that may trigger the transition from colonization to infection. Better understanding and differentiation of ESBL/pAmpC producing E. coli isolates obtained from clinical specimens as true infecting agents or simply transient colonizers will reduce antimicrobial use and therefore preserve the efficacy of currently available antimicrobials and promote antimicrobial stewardship in veterinary practice.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
AB performed molecular testing, analyzed the data, and wrote the manuscript. AB, GP, and VS reviewed clinical data and performed statistical analysis. AB, AW, CE, FZ, and EM performed phenotypic and molecular characterization of isolates. AB, FZ, EM, and DT analyzed and interpreted the data. DT planned and coordinated the study, analyzed the data, and wrote the manuscript. All authors revised and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank clinicians from the Small Animal and Equine Referral Hospitals for their support of this project.
Funding. This work was funded by the Institute of Veterinary Science, University of Liverpool.
References
- Alonso C. A., González-Barrio D., Ruiz-Fons F., Ruiz-Ripa L., Torres C. (2017). High frequency of B2 phylogroup among non-clonally related fecal Escherichia coli isolates from wild boars, including the lineage ST131. FEMS Microbiol. Ecol. 93:fix016. 10.1093/femsec/fix016 [DOI] [PubMed] [Google Scholar]
- Baede V. O., Wagenaar J. A., Broens E. M., Duim B., Dohmen W., Nijsse R., et al. (2015). Longitudinal study of extended-spectrum-β-lactamase- and AmpC-producing Enterobacteriaceae in household dogs. Antimicrob. Agents Chemother. 59 3117–3124. 10.1128/AAC.04576-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Tomás M., Bou G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26 185–230. 10.1128/CMR.00059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas A., Marques C., Aboim C., Pomba C. (2018). Emergence of Escherichia coli ST131 H30/H30-Rx subclones in companion animals. J. Antimicrob. Chemother. 74 266–269. 10.1093/jac/dky381 [DOI] [PubMed] [Google Scholar]
- Bogaerts P., Huang T.-D., Bouchahrouf W., Bauraing C., Berhin C., El Garch F., et al. (2015). Characterization of ESBL- and AmpC-producing Enterobacteriaceae from diseased companion animals in Europe. Microb. Drug Resist. 21 643–650. 10.1089/mdr.2014.0284 [DOI] [PubMed] [Google Scholar]
- Clermont O., Christenson J. K., Denamur E., Gordon D. M. (2013). The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5 58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- Clermont O., Lavollay M., Vimont S., Deschamps C., Forestier C., Branger C., et al. (2008). The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61 1024–1028. 10.1093/jac/dkn084 [DOI] [PubMed] [Google Scholar]
- CLSI (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI document M100-S22. Wayne: CLSI, 10.1038/nprot.2008.226 [DOI] [Google Scholar]
- Coque T. M., Baquero F., Canton R. (2008). Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eur. Commun. Dis. Bull. 13 1–11. 10.1128/AAC.49.7.2693-2700.2005 [DOI] [PubMed] [Google Scholar]
- Dale A. P., Woodford N. (2015). Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J. Infect. 71 615–626. 10.1016/J.JINF.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Dallenne C., da Costa A., Decré D., Favier C., Arlet G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Dierikx C. M., van Duijkeren E., Schoormans A. H. W., van Essen-Zandbergen A., veldman K., Kant A., et al. (2012). Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67 1368–1374. 10.1093/jac/dks049 [DOI] [PubMed] [Google Scholar]
- Dolejska M., Villa L., Hasman H., Hansen L., Carattoli A. (2013). Characterization of IncN plasmids carrying blaCTX-M-1and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J. Antimicrob. Chemother. 68 333–339. 10.1093/jac/dks387 [DOI] [PubMed] [Google Scholar]
- Essack S. Y., Hall L. M. C., Pillay D. G., McFadyen M. L., Livermore D. M. (2001). Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45 88–95. 10.1128/AAC.45.1.88-95.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C., Bethe A., Semmler T., Guenther S., Wieler L. H. (2012). Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18 646–655. 10.1111/j.1469-0691.2012.03850.x [DOI] [PubMed] [Google Scholar]
- Ewers C., Stamm I., Pfeifer Y., Wieler L. H., Kopp P. A., Schønning K., et al. (2014). Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J. Antimicrob. Chemother. 69 2676–2680. 10.1093/jac/dku217 [DOI] [PubMed] [Google Scholar]
- Gandolfi-Decristophoris P., Petrini O., Ruggeri-Bernardi N., Schelling E. (2013). Extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy companion animals living in nursing homes and in the community. Am. J. Infect. Control 41 831–835. 10.1016/j.ajic.2012.11.013 [DOI] [PubMed] [Google Scholar]
- Ghosh H., Doijad S., Falgenhauer L., Fritzenwanker M., Imirzalioglu C., Chakraborty T. (2017). blaCTX-M-27-encoding Escherichia coli sequence type 131 lineage C1-M27 clone in clinical isolates, Germany. Emerg. Infect. Dis. 23 1754–1756. 10.3201/eid2310.170938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardabassi L., Schwarz S., Lloyd D. H. (2004). Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54 321–332. 10.1093/jac/dkh332 [DOI] [PubMed] [Google Scholar]
- Hordijk J., Schoormans A., Kwakernaak M., Duim B., Broens E., Dierikx C., et al. (2013). High prevalence of fecal carriage of extended spectrum β-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 4:242 10.3389/fmicb.2013.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L. A., Pinchbeck G., Callaby R., Dawson S., Clegg P., Williams N. (2013). Antimicrobial prescribing practice in UK equine veterinary practice. Equine Vet. J. 45 141–147. 10.1111/j.2042-3306.2012.00602.x [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Porter S., Johnston B., Kuskowski M. A., Spurbeck R. R., Mobley H. L. T., et al. (2015). Host characteristics and bacterial traits predict experimental virulence for Escherichia coli bloodstream isolates from patients with urosepsis. Open Forum Infect. Dis. 2:ofv083. 10.1093/ofid/ofv083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Russo T. A. (2005). Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 295 383–404. 10.1016/j.ijmm.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Kawamura K., Sugawara T., Matsuo N., Hayashi K., Norizuki C., Tamai K., et al. (2017). Spread of CTX-Type extended-spectrum β-lactamase-producing Escherichia coli isolates of epidemic clone B2-O25-ST131 among dogs and cats in Japan. Microb. Drug Resist. 23 1059–1066. 10.1089/mdr.2016.0246 [DOI] [PubMed] [Google Scholar]
- Maddox T. W., Clegg P. D., Diggle P. J., Wedley A. L., Dawson S., Pinchbeck G. L., et al. (2012). Cross-sectional study of antimicrobial-resistant bacteria in horses. Part 1: prevalence of antimicrobial-resistant Escherichia coli and methicillin-resistant Staphylococcus aureus. Equine Vet. J. 44 289–296. 10.1111/j.2042-3306.2011.00441.x [DOI] [PubMed] [Google Scholar]
- Maeyama Y., Taniguchi Y., Hayashi W., Ohsaki Y., Osaka S., Koide S., et al. (2018). Prevalence of ESBL/AmpC genes and specific clones among the third-generation cephalosporin-resistant Enterobacteriaceae from canine and feline clinical specimens in Japan. Vet. Microbiol. 216 183–189. 10.1016/J.VETMIC.2018.02.020 [DOI] [PubMed] [Google Scholar]
- Matsumura Y., Johnson J. R., Yamamoto M., Nagao M., Tanaka M., Takakura S., et al. (2015). CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J. Antimicrob. Chemother. 70 1639–1649. 10.1093/jac/dkv017 [DOI] [PubMed] [Google Scholar]
- Matsumura Y., Pitout J. D. D., Peirano G., DeVinney R., Noguchi T., Yamamoto M., et al. (2017). Rapid identification of different Escherichia coli sequence type 131 clades. Antimicrob. Agents Chemother. 61:e179-17. 10.1128/AAC.00179-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniels A. E., Rice E. W., Reyes A. L., Johnson C. H., Haugland R. A., Stelma G. N. (1996). Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and β-D- glucuronidase. Appl. Environ. Microbiol. 62 3350–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino I., Hernández-García M., Turrientes M.-C., Pérez-Viso B., López-Fresneña N., Diaz-Agero C., et al. (2018). Emergence of ESBL-producing Escherichia coli ST131-C1-M27 clade colonizing patients in Europe. J. Antimicrob. Chemother. 73 2973–2980. 10.1093/jac/dky296 [DOI] [PubMed] [Google Scholar]
- Murphy C. P., Reid-Smith R. J., Boerlin P., Weese J. S., Prescott J. F., Janecko N., et al. (2012). Out-patient antimicrobial drug use in dogs and cats for new disease events from community companion animal practices in Ontario. Can. Vet. J. 53 291–298. 10.1016/j.aquaculture.2003.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D., Lemma F., Randall L. (2016). Industry survey of Great Britain-sourced broilers for extended-spectrum β-lactamase and Amp-C β-lactamase-producing Escherichia coli. Vet. Rec. 178:474. 10.1136/vr.103579 [DOI] [PubMed] [Google Scholar]
- Paterson D. L. (2006). Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 34 S20–S28. 10.1016/J.AJIC.2006.05.238 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez F. J., Hanson N. D. (2002). Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40 2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty N. K., Ben Zakour N. L., Stanton-Cook M., Skippington E., Totsika M., Forde B. M., et al. (2014). Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. U.S.A. 111 5694–5699. 10.1007/s00769-008-0365-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout J. D., Laupland K. B. (2008). Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8 159–166. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Pitout J. D. D. (2012). Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev. Anti. Infect. Ther. 10 1165–1176. 10.1586/eri.12.110 [DOI] [PubMed] [Google Scholar]
- Price L. B., Hungate B. A., Koch B. J., Davis G. S., Liu C. M. (2017). Colonizing opportunistic pathogens (COPs): the beasts in all of us. PLoS Pathog. 13:e1006369. 10.1371/journal.ppat.1006369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L. B., Johnson J. R., Aziz M., Clabots C., Johnston B., Tchesnokova V., et al. (2013). The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e377-13. 10.1128/mBio.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. P., Lemma F., Rogers J. P., Cheney T. E. A., Powell L. F., Teale C. J. (2014). Prevalence of extended-spectrum-β-lactamase-producing Escherichia coli from pigs at slaughter in the UK in 2013. J. Antimicrob. Chemother. 69 2947–2950. 10.1093/jac/dku258 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Baño J., Mingorance J., Fernández-Romero N., Serrano L., López-Cerero L., Pascual A., et al. (2012). Virulence profiles of bacteremic extended-spectrum β-Lactamase-Producing Escherichia coli: association with epidemiological and clinical features. PLoS One 7:e44238. 10.1371/journal.pone.0044238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J. E., Pitout J. D. D. (2014). Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet. Microbiol. 170 10–18. 10.1016/j.vetmic.2014.01.017 [DOI] [PubMed] [Google Scholar]
- Schaufler K., Semmler T., Wieler L. H., Wöhrmann M., Baddam R., Ahmed N., et al. (2016). Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410-another successful pandemic clone? FEMS Microbiol. Ecol. 92:fiv155. 10.1093/femsec/fiv155 [DOI] [PubMed] [Google Scholar]
- Schmiedel J., Falgenhauer L., Domann E., Bauerfeind R., Prenger-Berninghoff E., Imirzalioglu C., et al. (2014). Multiresistant extended-spectrum β-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol. 14:187. 10.1186/1471-2180-14-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoevaerdts D., Bogaerts P., Grimmelprez A., de Saint-Hubert M., Delaere B., Jamart J., et al. (2011). Clinical profiles of patients colonized or infected with extended-spectrum beta-lactamase producing Enterobacteriaceae isolates: a 20 month retrospective study at a Belgian University Hospital. BMC Infect. Dis. 11:12. 10.1186/1471-2334-11-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schønning K., Stegger M., Sjöström K., Nielsen J., Wang M., Hansen D. S., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e337-18. 10.1128/msphere.00337-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidjabat H. E., Paterson D. L., Qureshi Z. A., Adams-Haduch J. M., O’Keefe A., Pascual A., et al. (2009). Clinical features and molecular epidemiology of CMY-type β-lactamase–producing Escherichia coli. Clin. Infect. Dis. 48 739–744. 10.1086/597037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton D. A., Sánchez-Vizcaíno F., Dawson S., Jones P. H., Noble P. J. M., Pinchbeck G. L., et al. (2017). Patterns of antimicrobial agent prescription in a sentinel population of canine and feline veterinary practices in the United Kingdom. Vet. J. 224 18–24. 10.1016/j.tvjl.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofte D., Maciuca I. E., Williams N. J., Wattret A., Schmidt V. (2016). Veterinary hospital dissemination of CTX-M-15 extended-spectrum beta-lactamase–producing Escherichia coli ST410 in the United Kingdom. Microb. Drug Resist. 22 609–615. 10.1089/mdr.2016.0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin L., Sharp H., Hille K., Seibt U., Fischer J., Pfeifer Y., et al. (2014). Subgrouping of ESBL-producing Escherichia coli from animal and human sources: an approach to quantify the distribution of ESBL types between different reservoirs. Int. J. Med. Microbiol. 304 805–816. 10.1016/j.ijmm.2014.07.015 [DOI] [PubMed] [Google Scholar]
- Wagner S., Gally D. L., Argyle S. A. (2014). Multidrug-resistant Escherichia coli from canine urinary tract infections tend to have commensal phylotypes, lower prevalence of virulence determinants and ampC-replicons. Vet. Microbiol. 169 171–178. 10.1016/j.vetmic.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60 1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogg A. L., Simmen S., Zurfluh K., Stephan R., Schmitt S. N., Nüesch-Inderbinen M. (2018). High prevalence of extended-spectrum β-Lactamase Producing Enterobacteriaceae among clinical isolates from cats and dogs admitted to a Veterinary Hospital in Switzerland. Front. Vet. Sci. 5:62 10.3389/fvets.2018.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.