Abstract
Animal microbiomes play an important role in dietary adaptation, yet the extent to which microbiome changes exhibit parallel evolution is unclear. Of particular interest is an adaptation to extreme diets, such as blood, which poses special challenges in its content of proteins and lack of essential nutrients. In this study, we assessed taxonomic signatures (by 16S rRNA amplicon profiling) and potential functional signatures (inferred by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)) of haematophagy in birds and bats. Our goal was to test three alternative hypotheses: no convergence of microbiomes, convergence in taxonomy and convergence in function. We find a statistically significant effect of haematophagy in terms of microbial taxonomic convergence across the blood-feeding bats and birds, although this effect is small compared to the differences found between haematophagous and non-haematophagous species within the two host clades. We also find some evidence of convergence at the predicted functional level, although it is possible that the lack of metagenomic data and the poor representation of microbial lineages adapted to haematophagy in genome databases limit the power of this approach. The results provide a paradigm for exploring convergent microbiome evolution replicated with independent contrasts in different host lineages.
This article is part of the theme issue ‘Convergent evolution in the genomics era: new insights and directions’.
Keywords: haematophagy, microbiome, convergence
1. Background
Diet is recognized as a major factor that shapes the composition and function of gut microbial communities of animals [1,2]. Highly specialized diets often require feeding adaptations as well as physiological adaptations to obtain adequate nutrition from a narrow diet. Apart from host genomic traits that can yield adaptive functions to deal with a specialized diet, the host's gut microbes provide a range of functions that aid in digestion, nutrient release and vitamin synthesis [3]. Thus, we may expect diet specialists to have particularly important gut symbionts that are vital to the diet specialization (e.g. [4–6]). For example, myrmecophagy (ant and termite eating) is a specialized diet that has evolved independently multiple times within mammals such as armadillos, aardvarks, aardwolves, pangolins and anteaters [7]. Delsuc et al. [8] found that the composition of gut bacterial communities of some distantly related myrmecophagous mammals was strikingly similar, indicating a strong convergence of the gut microbiome. This finding suggests that the gut bacteria likely provide essential functions related to the specialized diet and opens new areas of inquiry. More broadly, investigating the role of microbial symbionts in host adaptation can further inform the interconnectedness of host and microbe evolution.
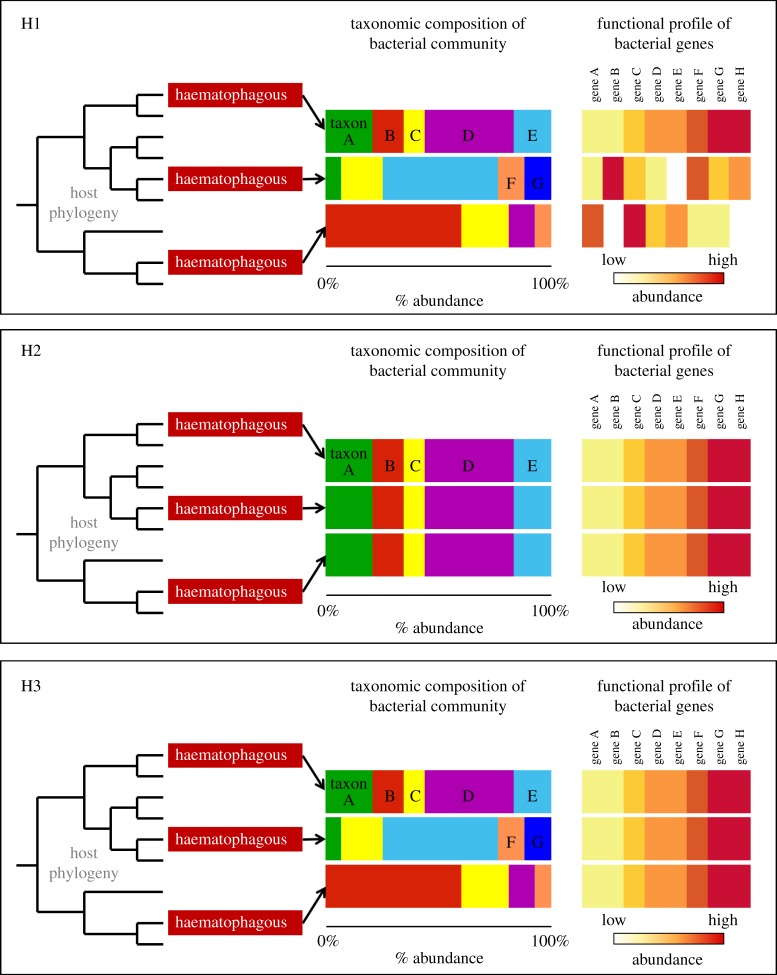
Convergence of the microbiome can be broadly described as either compositional (and therefore also functional) or functional (without taxonomic convergence). Compositional convergence is indicated by shared phylogenetic identity of the gut microbes across distantly related taxa with a convergent trait, and by extension functional convergence is also achieved as closely related taxa tend to carry similar genes (figure 1: H2) (although variable rates of horizontal gene transfer could cause functional profiles among taxa to differ). Functional convergence is indicated by phylogenetically distinct microbes carrying convergent genetic traits that result in convergent functions (figure 1: H3). Alternatively, gut microbes of taxa with a convergent trait may not converge at all (figure 1: H1). Figure 1 provides a conceptual framework for these alternative hypotheses.
Figure 1.
Conceptual diagram of alternative hypotheses of microbiome convergence. Hypothesis 1 (No convergence, H1): no evidence of either compositional or functional convergence of the microbiome. Support for this hypothesis may indicate that the microbiome does not play an important role in the host convergent trait in question. Hypothesis 2 (Taxonomic convergence, H2): the microbiomes of convergent hosts share similar bacterial taxonomic composition. Support for this hypothesis would indicate that the microbes have an important role in the functional trait of the host and that the identity of those microbes is key (or that the trait selects for microbes with specific identities). Hypothesis 3 (Functional convergence): the microbiomes of convergent hosts differ in their taxonomic composition but share key gene functions related to the convergent trait of the host. Support for this hypothesis would indicate that compositionally different microbial communities can converge on the same functional trait to aid the host in achieving a specific function. (Online version in colour.)
Haematophagy is an extremely specialized diet strategy that has evolved in multiple lineages across both vertebrates and invertebrates. Blood, as a source of food, presents some unique challenges nutritionally. While blood is a source of protein and lipids, it lacks many essential nutrients, such as B vitamins, and has high salt content. Moreover, the digestion of blood can release potentially toxic levels of haeme (and thus iron) and urea. We hypothesize that gut microbes may help their hosts deal with these challenges in similar ways across disparate host lineages. We therefore expect to find in their microbiomes, an enrichment in genes or pathways for functions such as B vitamin synthesis, sodium transport and haeme/iron metabolism. The roles of microbial symbionts and some instances of convergence are already well described for haematophagous arthropods and other invertebrates. For example, it has been shown that obligate bacterial symbionts produce B vitamins in a wide range of arthropod hosts [9], and that this function is convergent between some arthropods and the leech Haementeria officinalis [10]. However, equivalent knowledge in haematophagous vertebrate species is lacking.
For this study, we compared the gut bacterial communities of a bat and a bird species alongside closely related non-haematophagous species—in each case, to ask whether there is convergence of the gut bacteria among these haematophagous animals. The common vampire bat (Desmodus rotundus) represents one of the three related haematophagous bats within the family Phyllostomidae. This family includes species with six types of feeding habits, including haematophagy [11]. The clade of haematophagous bats evolved from insectivorous members of the family [11], requiring tremendous physiological evolution in a short evolutionary time frame [11,12]. The vampire finch (Geospiza septentrionalis) is a haematophagous bird endemic to the Galapagos islands (Darwin and Wolf islands, specifically). It feeds on the blood of other birds, such as Nazca and blue-footed boobies [13,14], a trait that is thought to have evolved as a means to supplement their diet, particularly during the dry season when other foods (seeds and fruits) are less available [15,16]. Other species of Geospiza in the Galapagos are not blood-feeders and are famous for their phenotypic diversity of beak forms. Recent evidence suggests the vampire finches are most closely related to ground finches such as G. fuliginosa and G. fortis [17].
2. Material and methods
(a). Sample collection
Representative gut samples were collected from the haematophagous vampire bat (D. rotundus), two non-haematophagous species in the family Phyllostomidae (Jamaican fruit-eating bat (Artibeus jamaicensis) and little yellow-shouldered bat (Sturnira lilium)), the haematophagous vampire finch (G. septentrionalis) and two non-haematophagous congeneric ground finches (sharp-beaked ground finch (G. difficilis) and Genovesa ground finch (G. acutirostris)).
All samples were collected from wild animals (electronic supplementary material, table S1) under approved permits and IACUC protocols. Faecal samples were collected from a single roost of vampire bats in Lamanai, Belize. Colon and anal samples were collected from two more populations of vampire bats and the two species of non-haematophagous bats in the Mexican states of Morelos and Veracruz [18]. Faecal samples from vampire finches were collected from the Galapagos islands of Darwin and Wolf during the blood-feeding season, sharp-beaked ground finch samples were collected from the island of Pinta and Genovesa ground finch samples were collected from the island of Genovesa following standardized methods for birds [19]. Samples were immediately frozen, stored in 95% EtOH or stabilized on Whatman FTA cards until processing (electronic supplementary material, table S1). We should note that although different collection methods can affect the microbial composition, previous studies have shown that the methods used in this study introduce effects that are small relative to inter-individual variation [20,21], and therefore we do not consider this factor in downstream analyses.
(b). Sample and sequence data processing
Protocols for DNA extraction, amplification of the V4 region of the 16S rRNA gene and sequencing followed those outlined for the Earth Microbiome Project [22]. Briefly, DNA was extracted using the Qiagen PowerSoil kit (formerly manufactured by MoBio, Carlsbad, CA, USA) and amplification was targeted using modified versions of the Golay barcoded forward primer 515F and the reverse primer 806R originally designed in [23]. Details of the modifications are described on the Earth Microbiome Project protocol website (http://press.igsb.anl.gov/earthmicrobiome/protocols-and-standards/16s/). Samples were amplified in triplicate and amplicons were pooled in equimolar volumes and cleaned using the Qiagen UltraClean PCR Cleanup Kit (formerly manufactured by MoBio, Carlsbad, CA, USA). DNA sequencing was performed on the Illumina MiSeq and HiSeq platforms at the Biofrontiers Institute Next-Generation Genomics Facility at the University of Colorado Boulder and at the IGM Genomics Center at the University of California San Diego.
Sequence data were uploaded to the public database Qiita (https://qiita.ucsd.edu) and processed using the database's implementation of QIIME2 [24]. Sequence data for additional species of non-haematophagous phyllostomids and tanagers were obtained from public studies 1734 and 2338 in Qiita. The forward sequences were demultiplexed and quality filtered using default settings. They were then trimmed to 100 nt and amplicon sequence variants (ASVs) were identified using Deblur [25]. Taxonomy was assigned to the ASVs by fitting a naive-Bayes classifier trained on the Greengenes v.13_8 99% OTUs reference (515f/806r region) using the sk-learn classifier [26]. A phylogenetic tree was constructed by inserting the sequences into the Greengenes 13_8 reference tree using the QIIME2 plugin q2-fragment-insertion [27], which uses the SEPP insertion method [28]. Sequences taxonomically identified as chloroplast and mitochondrial were removed from the dataset and the data were rarefied to 5000 sequences per sample for downstream analyses.
(c). Analysis of microbial community structure and composition
Pairwise comparisons of community composition (beta diversity) were calculated by computing unweighted UniFrac distances [29] and visualized through ordination using principal coordinates analysis (PCoA) and the visualization software EMPeror [30]. Permutational multivariate analysis of variation (PERMANOVA) from the R package ‘vegan’ (function adonis) [31] was used to statistically partition the sources of variation in community structure. Primary factors known to affect microbiome composition were accounted for before further partitioning variation by listing them first in the models. Because PERMANOVA is sensitive to differences in dispersion between groups, we also tested whether there were significant differences in dispersion among groups using an analysis of multivariate homogeneity (function betadisper).
(d). Metagenome prediction
To predict each metagenome using 16S rRNA amplicon sequences, we used the tool Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt; [32]). ASVs were first clustered into operational taxonomic units (OTUs) at 97% identity against the Greengenes 13_5 representative sequences, as recommended by PICRUSt authors. The data were rarefied to 5000 OTUs per sample and normalized for 16S rRNA gene copy number by dividing the OTU counts by their expected 16S rRNA gene copy number before metagenomes were predicted using the functional Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) database [33]. This database contains information on genes of both known and unknown function, providing groupings of gene orthologues and hierarchical classification of genes into higher-level functions and pathways. We then summarized these gene predictions into pathway predictions by collapsing the gene table of KOs to their higher-level functions.
To identify differences in abundance among microbial taxa, KEGG orthologies and pathways among haematophagous species and their non-haematophagous relatives, we used an analysis of the composition of microbiomes (ANCOM; [34]). Analyses were performed separately for bats and birds and compared for overlapping identified features. Average abundance and average differences in abundance of identified taxa, KOs and pathways were plotted using the R package ggplot2 [35]. For the list of KOs that were identified through ANCOM, we mapped the OTUs contributing to those related to functions of interest (urea, haeme/iron, sodium, amino acids and short-chain fatty acids (SCFAs)) using PICRUSt's metagenome_contributions.py script. The average abundances of these OTUs in the three vertebrate host groups were plotted after removing OTUs with a mean relative abundance lower than 1%.
3. Results
(a). Microbial community structure and composition
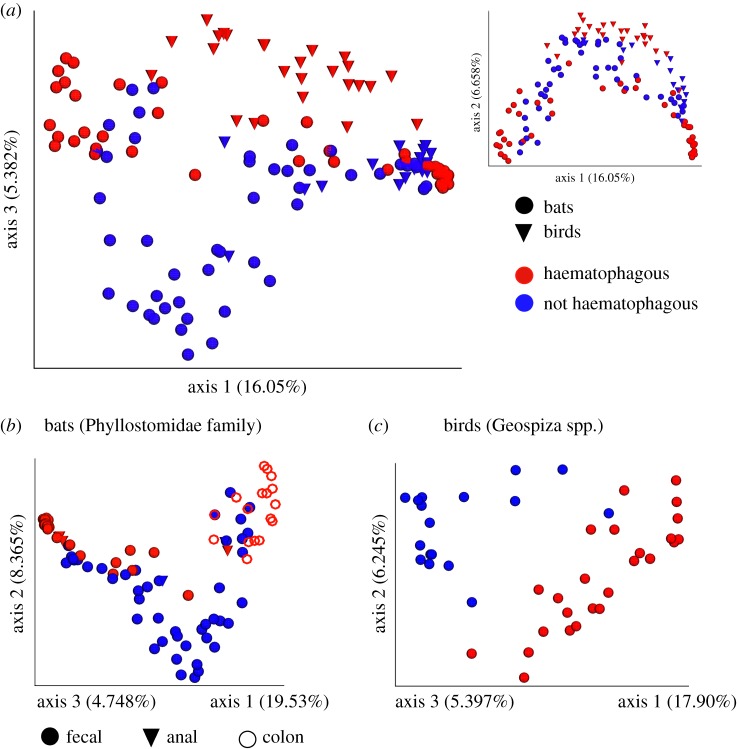
Across birds and bats, haematophagy explains a small but significant amount of the variation in gut microbial community structure (PERMANOVA R2 = 0.02196, p = 0.049; dispersion p = 0.02839). However, after accounting for phylogeny (host Family; R2 = 0.05320, p = 0.001; dispersion p = 1.917e × 10−05), haematophagy explains a larger portion of the remaining variation (R2 = 0.03392, p = 0.001). When considering just bats, vampire bats separate from other phyllostomids (R2 = 0.05402, p = 0.001; dispersion p = 0.9155) after accounting for differences in country of collection (R2 = 0.15160, p = 0.001; dispersion p = 3.696 × 10−09) and effect of sample type (faecal, anal, or colonic) (R2 = 0.04626, p = 0.001; dispersion p = 9.042×10−10) (figure 2b). In birds, there is a clear and strong separation between the vampire finch and other congeners (R2 = 0.34285, p = 0.001; dispersion p = 0.000508) (figure 2c).
Figure 2.
Principal coordinate analysis (PCoA) plots of the unweighted UniFrac distance among samples from haematophagous (red) and non-haematophagous (blue) species. PCoA ordination of distances among all bat and bird samples are shown in (a), with axes 1 and 3 in the larger image showing stratification between haematophagous and non-haematophagous animals, and axes 1 and 2 in the smaller inset. Separate ordinations for bats and birds are shown in (b,c), respectively. In bats, the different sample types collected are also indicated.
(b). Differentially abundant microbial taxa
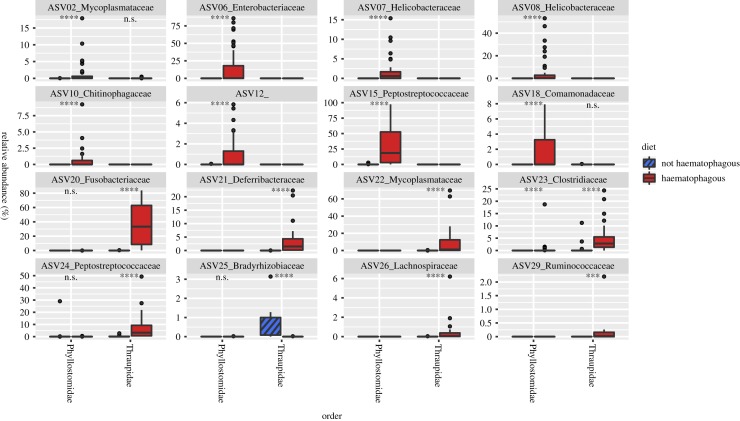
Nineteen ASVs were identified as being differentially abundant between the vampire bat and other phyllostomids, while 29 ASVs were identified between the vampire finch and other Geospiza species (electronic supplementary material, table S2). None of the ASVs identified showed overlap between the two groups, even taxonomically, apart for one exception: ASV15 in bats and ASV24 in birds (figure 3). These ASVs differed by merely one nucleotide in their 100 nt sequence and were taxonomically assigned as belonging to the bacterial family Peptostreptococcaceae. We should note that despite bat faecal, anal and colon samples showing different compositional structure (consistent with what was reported in [36]), all three sample types contained a notable abundance of ASVs identified as Enterobacteriaceae, Helicobacteraceae and Peptostreptococcaceae (ASVs 06–08, and 15), which were virtually absent in samples from non-haematophagous bat species.
Figure 3.
Boxplots show the eight most abundant exact sequence variants (ASVs) identified as being differentially abundant between haematophagous (shown in red) and non-haematophagous (shown in cross-hatched blue) bats (top two rows) and birds (bottom two rows). Stars above the paired boxplots indicate the level of significance when means are compared using a Wilcoxon test (***p < 0.0005, ****p < 0.00005, n.s.: not significant). Except for ASV25, which is lower in the vampire finch, blue bars are not visible because these ASVs were not detected or detected at very low levels in non-haematophagous animals. (Online version in colour.)
(c). Differentially abundant KOs and pathways
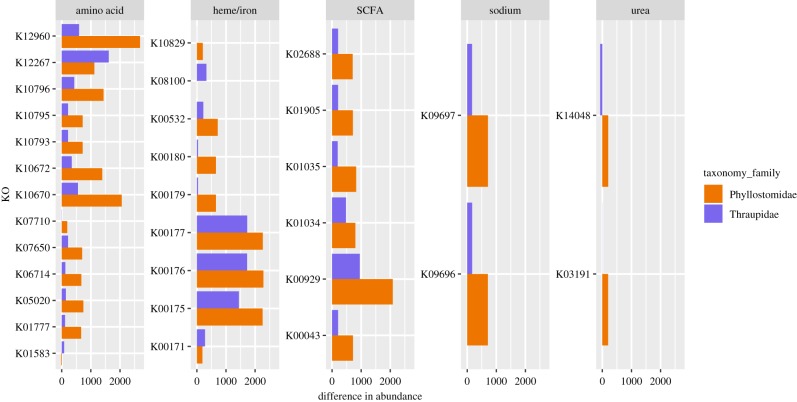
We detected 40 predicted KOs differentially represented in haematophages compared to non-haematophages that overlapped between bats and birds. There were an additional 75 KOs unique to bats and 50 unique to birds, for a total of 125 KOs (electronic supplementary material, table S3). Of these, several were related to the catabolism or metabolism of proteins likely to be important in haematophagous organisms, such as amino acids, SCFAs, urea, sodium and haeme/iron (figure 4). A number of other KOs appeared to contribute to sporulation and movement (flagellar assembly) (electronic supplementary material, table S3). Curiously, nearly all of the KOs detected in bats and birds were relatively higher in the haematophagous species (electronic supplementary material, table S3).
Figure 4.
Predicted orthologous gene families (based on the KEGG database) (KOs) of interest from those detected as differentially abundant between at least one pair of haematophagous versus non-haematophagous species. KOs are separated into broad groups based on whether they are associated with metabolic pathways involving amino acids, haeme/iron, short-chain fatty acids, urea or sodium transport, and bar graphs show the mean difference in abundance between haematophagous and non-haematophagous individuals (all are higher in haematophagous individuals). (Online version in colour.)
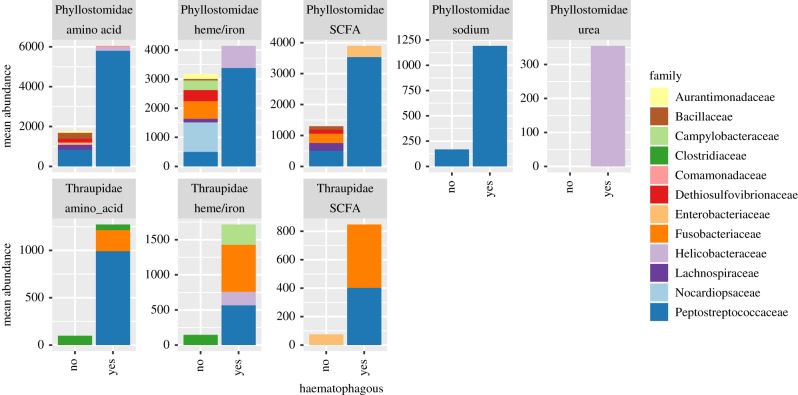
When we summarized which taxa were contributing to some of these predicted KEGG Ortholog groups of interest, we found that OTUs identified as belonging to the family Peptostreptococcaceae were among those most abundant in vampire bats and finches (figure 5). Moreover, Fusobacteria (which contains Cetobacterium somerae), a taxon enriched in vampire finches, were among the taxa contributing to KOs associated with amino acids, haeme/iron and SCFAs.
Figure 5.
Mean relative abundance of OTUs (≥1% mean relative abundance) contributing to KEGG Ortholog groups (KOs) associated with elevated functions of interest. Abundances in bats (Phyllostomidae) are shown in the upper row and birds (Thraupidae) in the lower row. Bar size represents the average relative abundance of OTUs in the samples from a given bacterial group, collapsed to the family taxonomic level. Colours represent different bacterial families.
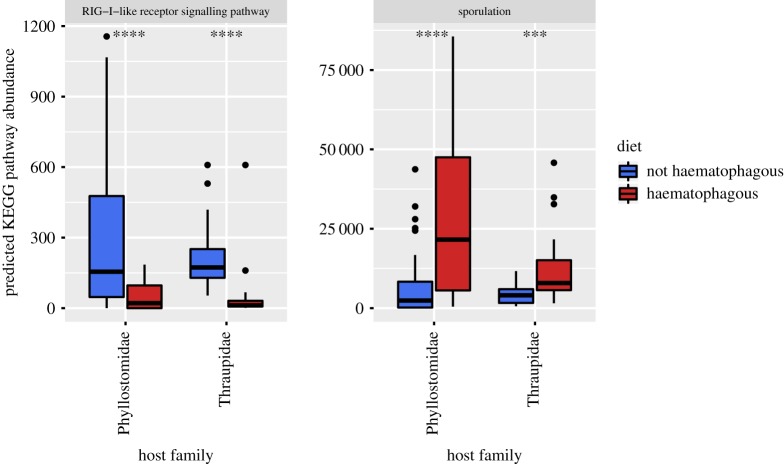
When we collapsed the KOs to higher-level functions (i.e. pathways), we found that only two predicted KEGG pathways overlapped as differentially abundant using ANCOM in both groups (figure 6). Consistent with findings on the individual orthologues, predicted sporulation pathways were enriched in haematophagous bats and birds compared to non-haematophagous relatives. However, one predicted pathway, the retinoic acid-inducible gene-I-like (RIG-I-like) receptor signalling pathway, was depleted in both haematophagous bats and birds.
Figure 6.
Boxplots show the KEGG pathways detected as differentially abundant between both pairs of haematophagous (red) and non-haematophagous (blue) species. Stars above the paired boxplots indicate the level of significance when means are compared using a Wilcoxon test (****p < 0.00005; ***p < 0.0005).
4. Discussion and conclusion
We find some evidence for microbiome convergence among haematophagous birds and bats, although the overall pattern is mixed. For the purposes of discussion, we divide the results into evidence in support of each of the three hypotheses.
(a). Evidence for H1: no convergence
None of the ASVs identified as differentially abundant in the vampire bat, relative to other bat species sampled, overlapped with those identified in vampire finches (electronic supplementary material, table S2). Consequently, we do not find evidence for convergent adaptation to haematophagy at the ASV level. However, we do find evidence that gut taxa may be meeting specific needs related to coping with a blood diet in the two host groups in different ways.
For example, ASVs identified as Helicobacteraceae (e.g. ASVs 07 and 08) were differentially abundant in vampire bats and are often associated with the enteric tracts of animals, including humans, and can be cultured using blood agar [37]. While we are not able to determine whether these and other taxa identified as differentially abundant are merely entering the gut via the blood meal or are resident symbionts capable of providing functions for the host, studies of Helicobacter species suggest that this group may be providing useful functions, at least for the vampire bats. This taxon has been shown to produce high amounts of urease (a function that is also indicated by our PICRUSt results (figure 5)); this enzyme is needed to break down urea, which gets concentrated in vampire bats owing to their high protein intake [38].
As previously observed in haematophagic arthropods [9], we expected an enrichment in either bacteria or functional genes associated with the production of essential B vitamins. However, the only indication of this function that we detected was a higher abundance in vampire finches of ASV20, taxonomically identified as C. somerae (family Fusobacteriaceae, phylum Fusobacteria) (figure 3; electronic supplementary material, table S2). A separate sequence analysis of these vampire finch faecal samples confirmed this finding [39]. C. somerae is known to produce cobalamin (vitamin B12), a vitamin that has recently been found to serve important roles in the microbial metabolism of other essential compounds such as folate, ubiquinone and methionine [40]. Intriguingly, C. somerae has been found to be a dominant resident in the digestive tract of freshwater fish species that do not require dietary vitamin B12 supplementation, such as goldfish, carp and tilapia [41,42]. However, no such elevation of putative B12 (or other B) vitamin-producing taxa was found in the vampire bats.
Functionally, we detected an enrichment in two predicted KOs involved in the sodium transport system in vampire bats (figure 4; electronic supplementary material, table S3). However, the same was not detected in vampire finches (although Peptostreptococcaceae shows functional capacity, the abundance of this taxon in finches is much lower than in the bats), suggesting that vampire finches may not need to deal with salt concentrations as high as those dealt with by the vampire bat. In contrast to the common vampire bat, which feeds primarily on other mammals, the vampire finch feeds on the blood of marine birds, which may have much lower salt concentrations than mammals: in marine birds, the countercurrent blood flow induced by ingestion of seawater causes salt glands to concentrate and remove salt ions from the blood [43]. Future research into the gut metagenomes of the other two vampire bat species, the hairy-legged vampire bat (Diphylla ecaudata) and the white-winged vampire bat (Diaemus youngi), which feed on birds, would provide additional data on whether salt regulation in the diet is a function conferred by gut symbionts.
(b). Evidence for H2: taxonomic and functional convergence
Our PERMANOVA results indicate that there are convergent phylogenetic properties of the overall gut microbiome, particularly between the vampire bat and finch (figure 2). This suggests that high-level properties of the overall community converge, even when ASVs are not shared. However, there is evidence that some lower-level properties are shared among distantly related haematophagous species. Specifically, vampire bats and birds both have comparatively high abundances of a taxon identified as Peptostreptococcaceae, although the sequences of the two ASVs are not identical (figure 3; electronic supplementary material, table S2). Prediction of this bacterial taxon's functional capacity indicates that it can produce proteins involved in iron-sulfur reactions and sodium transport (figure 5). Both of these functions may need to be elevated in haematophagous animals, given the high iron and salt concentrations in blood. This taxon also has predicted functional capacity associated with important molecules such as SCFAs and amino acids (figure 5)
(c). Evidence for H3: functional convergence
We found an enrichment of one predicted KO involved in butyrate synthesis (K00929) but also detected KOs associated with other SCFAs in both haematophagous bats and birds (figure 4; electronic supplementary material, table S3). This is consistent with a previously observed enrichment in enzymes involved in butyrate metabolism in vampire bats when compared to other bats [44].
The predicted enrichment of sporulation pathways in the vampire bat and finch suggests that sporadic blood meals and concomitant pulses of nutrients might select for bacteria with a spore-forming capability that can be dormant for the rest of the time (figure 6). The vampire bat, for example, forages only once nightly, drinking up to 40% of its body weight in blood, but then returns to its roost and stays relatively inactive until the next foraging flight the following night. The vampire finch receives more seasonal pulses of blood in its diet, displaying haematophagy during the dry months of the year ( June–December) when seeds and arthropods are scarce, and resuming their omnivorous diet once the wet season starts ( January–May). Higher abundance of sporulation pathways in these species could, however, also stem from the higher relative abundance of Peptostreptococcaceae (class Clostridia, phylum Firmicutes) present, as a side-effect of the fact that endospore formation is a common trait in Firmicutes.
Intriguingly, RIG-I-like receptors, which are involved in detecting viral RNA and initiating immune responses, were predicted to be found in much lower abundance in both haematophagous bats and birds when compared to their non-haematophagous counterparts (figure 6). This is counterintuitive, because one might have expected that viral RNA detection ability would instead be higher in animals that feed on blood, to guard against blood-borne viruses. Interestingly, Zepeda Mendoza et al. [44] found a low diversity of endogenous retroviral elements in the vampire bat genome compared to other bats, a finding suggestive of there being other mechanisms providing protection from viral infections. Further confirmation of and research into this topic would be valuable.
(d). Overall conclusions
In summary, we do not find evidence of microbiome convergence associated with haematophagy at the ASV level, and weakly at the overall community level, but key taxa and predicted functions converge more strongly between haematophagous bats and birds. Our results indicate that the level of convergence may depend on the phylogenetic scale, with evolutionary history exerting restrictions in terms of how similar host-associated microbiomes can become in response to a similar diet. While exact microbial taxa may be shared less across distantly related host species, functions conferred by different microbial communities may be more likely to converge, although phylogenetic limitations may still exist. In support of this hypothesis, a recent study on the convergence of another specialized diet—folivory—in several lineages of primates found that host phylogeny was a much stronger predictor of gut microbiome structure than the host diet type, but certain functional pathways were similarly depleted in folivorous primates compared to non-folivorous relatives [45]. Yet another possibility is that the host genome provides needed functions rather than the microbiome: for example, Zepeda Mendoza et al. [44] report genes in the vampire bat genome for traits that could be beneficial for feeding on a blood diet. Future comparisons of the genomes and metagenomes of haematophagous organisms may reveal that the adaptation to a blood diet may be contributed differently between the host's and the symbionts’ genomes, depending on the host group.
One important limitation of the present study is that we assess function using PICRUSt rather than directly from metagenomic data. This requires inference from the closest genome, and the observed NSTI (Nearest Sequenced Taxon Index) values are high, especially for bacteria hosted by birds (electronic supplementary material, figure S1). This effect could lead to under-reporting instances of functional convergence. We expect that as genome databases of microbes isolated from blood-feeders improve, and in particular as the annotation of genes involved in blood digestion by microbes becomes clearer (which will also assist greatly in interpretation of shotgun metagenomic data relevant to this problem), our understanding of convergent and non-convergent adaptation to haematophagy will continue to improve.
The approach used in the present study to detect convergence in host-associated microbial symbionts linked to convergent traits in unrelated hosts represents a coarse-level perspective, akin to a satellite view as opposed to a fine-grained, ‘on the ground’ view. The strength of this approach lies in the comparative framework involving host taxa with a trait of interest along with closely related hosts as a reference (e.g. blood-feeding bats and related non-blood feeding bats). Applying analyses that detect specific microbial taxa and predicted microbial gene pathways that are common among the hosts with the convergent trait, but absent or reduced in the respective related hosts without the convergent trait, is key to identifying future areas of deeper study. For example, the present study clearly points to the Peptostreptococcaceae taxon as one of particular interest for future work to further understand the nature of the convergence; however, this could have been missed if the reference hosts were not included. Future work could focus on the taxa of interest with deeper sequencing tools such as shotgun metagenomics or whole genome sequencing (if cultured isolates are obtainable) in order to dive into finer-grain questions. For example, how did haematophagous bats and finches converge on having Peptostreptococcaceae enriched in their respective gut microbiomes? Deeper sequencing of different strains of this bacterial group from different sources could help address whether it fits a model of host–symbiont phylosymbiosis and elucidate the origin of these host–microbe relationships. Lastly, many of the microbial reference databases are still relatively data-poor in their coverage of microbial taxa from exotic animal hosts, thus as these databases improve, so too will our ability to address the finer-grain questions.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The authors thank Gregory Humphrey for molecular and sequencing work, Gail Ackermann for help with data curation and deposition and Kurt Gielow for providing assistance in the field. Mention of trademark does not infer endorsement by the U.S. Federal Government.
Ethics
Finch samples from the Galapagos islands were collected under Contrato Marco de Acceso a los Recursos Genéticos MAE-DNB-CM-2016-0041 and Galapagos National Park permit PC-0615 and were approved by University of Miami IACUC protocol 14–109. Vampire bat samples from Belize were collected under the University of Georgia IACUC protocols AUP A2009-10003-0 and A2014 04-016-Y3-A5. Vampire bat samples from Mexico were collected from animals that were chemically restrained and humanely treated until euthanasia endpoints, in accordance with IACUC Protocol 17164 at the University of California, Davis. Scientific collection permits were issued by the Mexico Ministry of Agriculture, Livestock, Rural Development, Fisheries, and Food (SAGARPA License FAUT—0250) and the Veterinary and Zootechnical School at the National University of Mexico (UNAM Permit B00.02.02/1026 1708).
Data accessibility
Raw sequence data and information about each sample (metadata) are deposited in the public repository Qiita (qiita.ucsd.edu) under study 11 933 and the European Nucleotide Archive (ENA) accession #ERP111040. Jupyter notebooks describing data analyses are available on Github at https://github.com/knightlab-analyses/hematophagy_16S. Existing sequence data from the following datasets were included in this study: Qiita study 1734 (ENA study PRJEB14489): Gut microbiota of phyllostomid bats that span a breadth of diet (published in Thompson et al. [22]); Qiita study 2338 (ENA study PRJEB14818): Microbiome of Seba's short-tailed bats (published in Thompson et al. [22])
Authors' contributions
S.J.S., J.L.M., R.K. and V.J.M. designed the study. S.J.S., D.T.B., M.J.S., B.B.C., A.A.-S., N.S.J., A.J.P., E.N. and J.A.C. performed the fieldwork and collected the samples. S.J.S. analysed the data. S.J.S., R.K. and V.J.M. drafted the manuscript, and all authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This project was funded by the John Templeton Foundation (Grant ID 44000, Convergent Evolution of the Vertebrate Microbiome) and the W.M. Keck Foundation (DT061413) awarded to R.K. and V.J.M., which also supported S.J.S., J.G.S. and J.L.M. Funds for vampire finch work were in part provided by a GAIAS-USFQ Grant to J.A.C. and an NSF-Postdoctoral Grant to D.T.B. Vampire bat work in Mexico by M.J.S., B.B.C. and A.A.-S. was made possible through a grant from U.C. Mexus. Funding for N.S.J. was provided by the Great Lakes Fishery Commission.
References
- 1.Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320, 1647–1651. ( 10.1126/science.1155725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974. ( 10.1126/science.1198719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohl KD, Weiss RB, Cox J, Dale C, Dearing MD. 2014. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 17, 1238–1246. ( 10.1111/ele.12329) [DOI] [PubMed] [Google Scholar]
- 5.Zhu L, Wu Q, Dai J, Zhang S, Wei F. 2011. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl Acad. Sci. USA 108, 17 714–17 719. ( 10.1073/pnas.1017956108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12, 168–180. ( 10.1038/nrmicro3182) [DOI] [PubMed] [Google Scholar]
- 7.McGhee GR. 2011. Convergent evolution: limited forms most beautiful. Cambridge, MA: MIT Press. [Google Scholar]
- 8.Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. 2014. Convergence of gut microbiomes in myrmecophagous mammals. Mol. Ecol. 23, 1301–1317. ( 10.1111/mec.12501) [DOI] [PubMed] [Google Scholar]
- 9.Rio RVM, Attardo GM, Weiss BL. 2016. Grandeur alliances: symbiont metabolic integration and obligate arthropod haematophagy. Trends Parasitol. 32, 739–749. ( 10.1016/j.pt.2016.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manzano-Marín A, Oceguera-Figueroa A, Latorre A, Jiménez-García LF, Moya A. 2015. Solving a bloody mess: B-vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthropods and the leech Haementeria officinalis. Genome Biol. Evol. 7, 2871–2884. ( 10.1093/gbe/evv188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker RJ, Bininda-Emonds ORP, Mantilla-Meluk H, Porter CA, Van Den Bussche RA.. 2012. Molecular time scale of diversification of feeding strategy and morphology in New World Leaf-Nosed Bats (Phyllostomidae): a phylogenetic perspective. In Evolutionary history of bats: fossils, molecules and morphology, Cambridge studies in morphology and molecules: new paradigms in evolutionary biology series, number 2 (eds G Gunnell, N Simmons), pp. 385–409. Cambridge, UK: Cambridge University Press ( 10.1017/CBO9781139045599.012) [DOI]
- 12.Schmidt U, Greenhall A.. 1988. Natural history of vampire bats. Boca Raton, FL: CRC Press ( 10.1201/9781351074919) [DOI]
- 13.Koster F, Koster H. 1983. Twelve days among the ‘vampire finches’ of Wolf Island. Noticias de Galápagos 38, 4–10. [Google Scholar]
- 14.Bowman RI, Billeb SL. 1965. Blood-eating in a Galapagos finch. Living Bird 4, 29–44. [Google Scholar]
- 15.Schluter D, Grant PR. 1984. Ecological correlates of morphological evolution in a Darwin's finch, Geospiza difficilis. Evolution 38, 856–869. ( 10.1111/j.1558-5646.1984.tb00357.x) [DOI] [PubMed] [Google Scholar]
- 16.Schluter D, Grant PR. 1982. The distribution of Geospiza difficilis in relation to G. fuliginosa in the Galápagos Islands: tests of three hypotheses. Evolution 36, 1213–1226. ( 10.1111/j.1558-5646.1982.tb05490.x) [DOI] [PubMed] [Google Scholar]
- 17.Lamichhaney S, et al. 2015. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature 518, 371–375. ( 10.1038/nature14181) [DOI] [PubMed] [Google Scholar]
- 18.Stuckey MJ, Chomel BB, Galvez-Romero G, Olave-Leyva JI, Obregón-Morales C, Moreno-Sandoval H, Aréchiga-Ceballos N, Salas-Rojas M, Aguilar-Setién A. 2017. Bartonella infection in haematophagous, insectivorous, and phytophagous bat populations of Central Mexico and the Yucatan Peninsula. Am. J. Trop. Med. Hyg. 97, 413–422. ( 10.4269/ajtmh.16-0680) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knutie SA, Gotanda KM. 2018. A non-invasive method to collect fecal samples from wild birds for microbiome studies. Microb. Ecol. 76, 851–855. ( 10.1007/s00248-018-1182-4) [DOI] [PubMed] [Google Scholar]
- 20.Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, Knight R. 2016. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 1, e00021-16 ( 10.1128/mSystems.00021-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogtmann E, Chen J, Amir A, Shi J, Abnet CC, Nelson H, Knight R, Chia N, Sinha R. 2017. Comparison of collection methods for fecal samples in microbiome studies. Am. J. Epidemiol. 185, 115–123. ( 10.1093/aje/kww177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson LR, et al. 2017. A communal catalogue reveals Earth's multiscale microbial diversity Nature 551, 457–463. ( 10.1038/nature24621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108, 4516–4522. ( 10.1073/pnas.1000080107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. ( 10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amir A, et al. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191-16 ( 10.1128/mSystems.00191-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedregosa F, et al. 2011. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830. [Google Scholar]
- 27.Janssen S, et al. 2018. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems 3, e00021-18 ( 10.1128/mSystems.00021-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirarab S, Nguyen N, Warnow T. 2012. SEPP: SATé-enabled phylogenetic placement. Pac. Symp. Biocomput. 2012, 247–258. ( 10.1142/9789814366496_0024) [DOI] [PubMed] [Google Scholar]
- 29.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. ( 10.1128/AEM.71.12.8228-8235.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 16 ( 10.1186/2047-217x-2-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oksanen J, et al. 2013. vegan: Community Ecology Package. R package version 2. See https://CRAN.Rproject.org/package=vegan.
- 32.Langille MGI, et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. ( 10.1038/nbt.2676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. ( 10.1093/nar/gkm882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD.. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26, 27663 ( 10.3402/mehd.v26.27663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickham H. 2016. Ggplot2: elegant graphics for data analysis. Berlin, Germany: Springer. [Google Scholar]
- 36.Ingala MR, Simmons NB, Wultsch C, Krampis K, Speer KA, Perkins SL. 2018. Comparing microbiome sampling methods in a wild mammal: fecal and intestinal samples record different signals of host ecology, evolution. Front. Microbiol. 9, 803 ( 10.3389/fmicb.2018.00803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.On SLW, Miller WG, Houf K, Fox JG, Vandamme P. 2017. Minimal standards for describing new species belonging to the families Campylobacteraceae and Helicobacteraceae: Campylobacter, Arcobacter, Helicobacter and Wolinella spp. Int. J. Syst. Evol. Microbiol. 67, 5296–5311. ( 10.1099/ijsem.0.002255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFarland WN, Wimsatt WA. 1969. Renal function and its relation to the ecology of the vampire bat, Desmodus rotundus. Comp. Biochem. Physiol. 28, 985–1006. ( 10.1016/0010-406X(69)90543-X) [DOI] [Google Scholar]
- 39.Michel AJ, et al. 2018. The gut of the finch: uniqueness of the gut microbiome of the Galápagos vampire finch. Microbiome 6, 167 ( 10.1186/s40168-018-0555-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romine MF, et al. 2017. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc. Natl Acad. Sci. USA 114, E1205–E1214. ( 10.1073/pnas.1612360114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugita H, Takahashi J, Miyajima C, Deguchi Y. 1991. Vitamin B12-producing ability of the intestinal microflora of rainbow trout (Oncorhynchus mykiss). Agric. Biol. Chem. 55, 893–894. ( 10.1080/00021369.1991.10870683) [DOI] [Google Scholar]
- 42.Tsuchiya C, Sakata T, Sugita H. 2008. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 46, 43–48. ( 10.1111/j.1472-765x.2007.02258.x) [DOI] [PubMed] [Google Scholar]
- 43.Hughes MR. 2003. Regulation of salt gland, gut and kidney interactions. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 136, 507–524. ( 10.1016/j.cbpb.2003.09.005) [DOI] [PubMed] [Google Scholar]
- 44.Zepeda Mendoza ML, et al. 2018. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat. Ecol. Evol. 2, 659–668. ( 10.1038/s41559-018-0476-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amato KR, et al. 2018. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 13, 576–587. ( 10.1038/s41396-018-0175-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data and information about each sample (metadata) are deposited in the public repository Qiita (qiita.ucsd.edu) under study 11 933 and the European Nucleotide Archive (ENA) accession #ERP111040. Jupyter notebooks describing data analyses are available on Github at https://github.com/knightlab-analyses/hematophagy_16S. Existing sequence data from the following datasets were included in this study: Qiita study 1734 (ENA study PRJEB14489): Gut microbiota of phyllostomid bats that span a breadth of diet (published in Thompson et al. [22]); Qiita study 2338 (ENA study PRJEB14818): Microbiome of Seba's short-tailed bats (published in Thompson et al. [22])