Abstract
An underexplored question in evolutionary genetics concerns the extent to which mutational bias in the production of genetic variation influences outcomes and pathways of adaptive molecular evolution. In the genomes of at least some vertebrate taxa, an important form of mutation bias involves changes at CpG dinucleotides: if the DNA nucleotide cytosine (C) is immediately 5′ to guanine (G) on the same coding strand, then—depending on methylation status—point mutations at both sites occur at an elevated rate relative to mutations at non-CpG sites. Here, we examine experimental data from case studies in which it has been possible to identify the causative substitutions that are responsible for adaptive changes in the functional properties of vertebrate haemoglobin (Hb). Specifically, we examine the molecular basis of convergent increases in Hb–O2 affinity in high-altitude birds. Using a dataset of experimentally verified, affinity-enhancing mutations in the Hbs of highland avian taxa, we tested whether causative changes are enriched for mutations at CpG dinucleotides relative to the frequency of CpG mutations among all possible missense mutations. The tests revealed that a disproportionate number of causative amino acid replacements were attributable to CpG mutations, suggesting that mutation bias can influence outcomes of molecular adaptation.
This article is part of the theme issue ‘Convergent evolution in the genomics era: new insights and directions’.
Keywords: adaptation, CpG, haemoglobin, high altitude, mutation bias, protein evolution
1. Introduction
A question of enduring interest in EvoDevo research concerns the extent to which the biased production of variation during development influences directional trends and patterns of convergence in morphological evolution [1–4]. At the molecular sequence level, an analogous question concerns the extent to which mutational bias in the production of genetic variation influences pathways of molecular evolution [5,6]. Mutation bias is known to exert an important influence on patterns of neutral molecular evolution [7], but our understanding of mutation as an introduction process suggests that it may be an important orienting factor in adaptive evolution as well [8,9]. The modern synthesis originally included a strong position on the importance of recombination, and did not attach much importance to the role of mutation bias [10–13]. When the process of evolution is defined in terms of shifts in the frequencies of pre-existing alleles, recombination serves as the main source of new genetic variation and mutation rates must be of the order of selection coefficients to exert an appreciable influence on the direction of adaptive change [14].
An alternative view is that outcomes and pathways of evolution may be influenced by the rate of mutation even in the presence of selection [5]. For instance, in the simple case of origin-fixation models [8], the substitution rate is given as K = 2Nμλ, where N is the size of a diploid population, μ is the per-copy rate of mutation and λ is the probability of fixation [15]. This model specifies the substitution rate as the product of the rate at which new alleles originate via mutation (2Nμ) and the probability that they become fixed once they arise (λ). In this type of model, mutation bias in the introduction of variation can produce a bias in substitution rates even when the substitutions are beneficial [8]. Results of several experimental evolution studies suggest that mutation bias can influence trajectories of adaptive protein evolution [9,16–20], and Stoltzfus & McCandlish [9] also provide evidence for transition–transversion bias among adaptive substitutions that contributed to natural protein evolution.
One especially powerful means of addressing questions about the role of mutation bias in molecular adaptation is to examine convergent changes in protein function that can be traced to specific amino acid substitutions. The experimental identification of causative substitutions provides a means of testing whether such changes are random with respect to different classes of site or different classes of mutational change. In vertebrate genomes, transition : transversion bias results in especially high rates of change from one pyrimidine to another (C ↔ T) or from one purine to another (G ↔ A). In the genomes of mammals and birds, the dinucleotide CG—often designated ‘CpG’—is a hotspot of nucleotide point mutations due to the effect of methylation on damage and repair. In mammals, the mutation rate at CpG sites is elevated 10-fold for transition mutations and several-fold for transversions [21,22]. A recent mutation-accumulation study in birds confirmed a similar increase in mutation rate at CpG sites [23]. Mammalian and avian genomes both exhibit roughly fivefold depletions of CpG dinucleotides, consistent with elevated rates of mutation at such sites [24].
In studies of haemoglobin (Hb) evolution in high-altitude birds, site-directed mutagenesis experiments have documented three cases in which missense mutations at CpG dinucleotides contributed to derived increases in Hb–O2 affinity: 55Ile → Val (I55V) in the βA-globin gene of Andean house wrens (Troglodytes aedon) [25] and parallel 34Ala → Thr substitutions (A34T) in the αA-globin genes of two different passerine species that are native to the Tibetan Plateau, the ground tit (Parus humilis [=Pseudopodoces humilis]) and the grey-crested tit (Lophophanes dichrous) [26]. The patterns documented in these three high-altitude taxa are consistent with a broader pattern of convergence in Hb function in highland birds [26–30], and the direction of character-state change is consistent with theoretical and experimental results which suggest that it is generally beneficial to have an increased Hb–O2 affinity under conditions of severe environmental hypoxia [30–32]. Moreover, in the case of the high-altitude house wrens, an analysis of genome-wide patterns of nucleotide polymorphism revealed evidence for altitude-related selection on the missense CpG variant [25], providing additional support for the inferred adaptive significance of the affinity-altering amino acid change.
The documented cases in which CpG mutations have contributed to altitude-related changes in Hb function suggest that mutation bias may influence which mutations are most likely to contribute to molecular adaptation. However, a far more comprehensive and systematic analysis is required to draw firm conclusions. Here, we examine existing data from a larger set of case studies in which it has been possible to identify the causative substitutions that are responsible for convergent increases in Hb–O2 affinity in high-altitude birds. Using a dataset of experimentally verified, affinity-enhancing mutations in the Hbs of highland avian taxa, we test whether a disproportionate number involve mutations at CpG sites. The results suggest that mutation bias has influenced outcomes of molecular adaptation.
2. Methods
(a). Sampling design
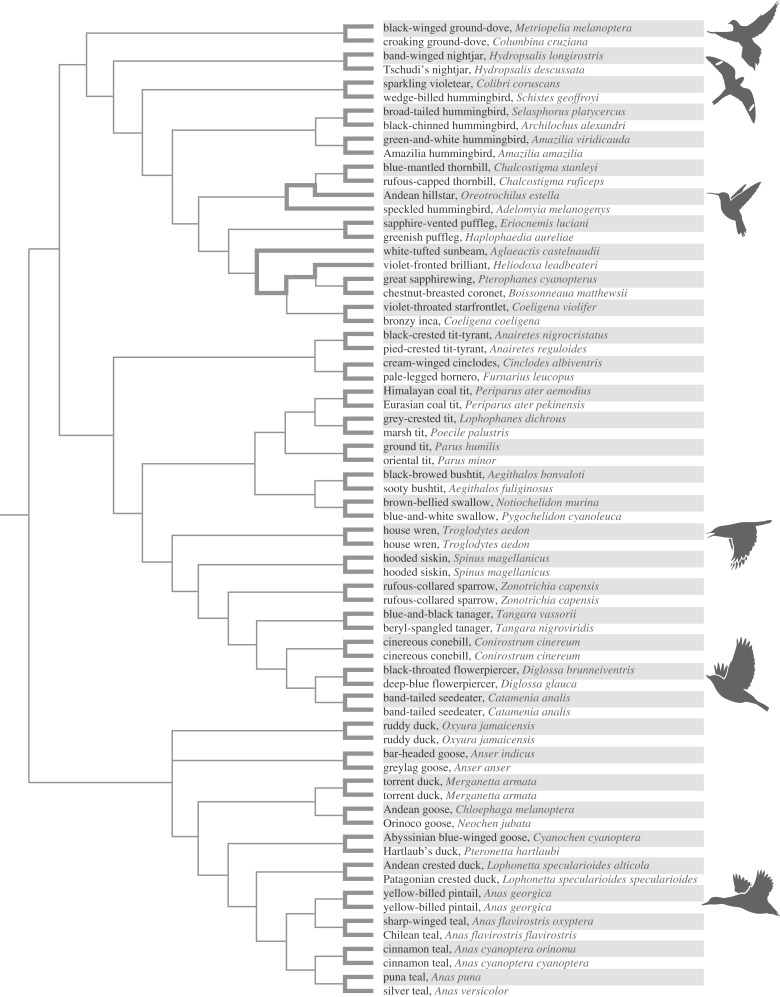
To test for convergent changes in Hb–O2 affinity, we conducted phylogenetically independent comparisons involving 70 avian taxa representing 35 matched pairs of high- versus low-altitude species or subspecies (figure 1). These taxa include ground doves, nightjars, hummingbirds, passerines and waterfowl [25–29,33–36]. All pairwise comparisons involved dramatic elevational contrasts; high-altitude taxa native to very high elevations in the Andes, Ethiopian highlands or Tibetan Plateau (with upper range limits of 3500–5000 m above sea level) were paired with close relatives that typically occur at or near sea level. Thus, the highland members of each taxon pair have upper elevational range limits that likely exceed the threshold at which an increased Hb–O2 affinity becomes physiologically beneficial.
Figure 1.
Phylogenetic relationships of 70 avian taxa used in comparative analyses of Hb function. Rows corresponding to high-altitude taxa are shaded. Branches in bold connect pairs of high- and low-altitude taxa that were used to test for a relationship between Hb–O2 affinity and native elevation. The 35 pairwise comparisons are phylogenetically independent. For information regarding elevational ranges, see [25–29,33].
(b). Experimental measurement of haemoglobin function
For each taxon, we used our previously measured O2 affinities of purified Hbs in the presence of allosteric cofactors: Cl− ions (added as KCl) and inositol hexaphosphate (IHP). IHP is a chemical analogue of inositol pentaphosphate (IPP), which is endogenously produced as a metabolite of oxidative phosphorylation in avian red cells. Using a diffusion-chamber protocol, we measured O2 equilibria of purified Hb solutions at pH 7.40, 37°C in the presence and absence of allosteric cofactors, namely 0.10 M Cl−; 0.1 M HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid); 0.3 mM haeme; and IHP : tetrameric Hb ratio of 2.0 [26,37–39]. The estimated values of P50 (the partial pressure of O2 at which Hb is 50% saturated) provide an inverse measure of Hb–O2 affinity.
Most bird species express two main isoforms of the Hb heterotetramer (α2β2) in adult red blood cells: HbA (the major isoform, with α-chain subunits encoded by the αA-globin gene) and HbD (the minor isoform, with α-chain subunits encoded by the αD-globin gene); both isoforms incorporate the same β-chain subunits [39,40]. In species that expressed both Hb isoforms, we separately measured the O2-binding properties of purified HbA and HbD.
All the experimental data used in the present analysis were published previously [25–29,33–36], and these papers can be consulted for additional details regarding experimental protocols. (For additional methodological details regarding the expression and purification of recombinant Hbs, see [29,41–43].) We restrict the analysis to data based on standardized measurements of purified Hbs, so the observed variation in P50 values is purely genetic, reflecting evolved changes in the amino acid sequences of the α- and β-type subunits of the α2β2 Hb tetramer. This focus on purified Hbs avoids problems associated with the confounding effects of environmentally induced variation.
(c). Testing for convergence in haemoglobin function
In comparative analyses of phenotypic variation, it is important to account for the fact that trait values from different species are not statistically independent because the sampled species did not evolve independently of one another [44]. Accordingly, we used a paired-lineage design [45] to test for a non-random association between Hb–O2 affinity and native elevation in the set of 70 avian taxa. The paired-lineage design restricts comparisons to phylogenetically replicated pairs of high- and low-altitude taxa that were chosen so that there is no overlap in evolutionary paths of descent. Since our comparative analysis includes a phylogenetically diverse range of avian taxa, an advantage of this design is that comparisons can be restricted to closely related species by excluding pairs with long paths between them [30].
(d). Experimental identification of causative mutations
For each pair of high- and low-altitude taxa in which we documented an evolved difference in Hb–O2 affinity, we used a combinatorial protein-engineering approach to identify the causative substitution(s). Specifically, we synthesized recombinant Hbs representing the wild-type Hbs of the high- and low-altitude species, and we used ancestral sequence reconstruction to synthesize the recombinant Hb representing the inferred genotype of the species' most recent common ancestor. By design, our pairs of high- and low-altitude taxa were always very closely related (in many cases representing sister species or nominal subspecies), so the ancestral state estimates were always unambiguous [26,28,29,36]. In cases where the triangulated comparison involving the reconstructed ancestral protein and those from the pair of modern-day descendants confirmed that the highland taxon evolved a derived increase in Hb–O2 affinity, we performed site-directed mutagenesis experiments to measure the additive and non-additive effects of all or nearly all amino acid substitutions that were specific to the highland lineage [25–29,33,36]. In the majority of cases, we synthesized combinatorially complete sets of mutant genotypes in order to test each mutation in all possible multi-site combinations. There were two pairs of bird species for which the site-directed mutagenesis experiments were not combinatorially complete: the comparison between Parus humilis and Parus minor and that between Lophophanes dichrous and Poecile palustris. In both of these pairwise comparisons, we experimentally tested mutations at a single candidate site, so we cannot rule out the possibility that additional, untested substitutions also contributed to the observed differences in Hb–O2 affinity between the major HbA isoforms of the high- and low-altitude species.
(e). Null model for frequency of CpG changes among causative mutations
To assess a role for CpG effects in molecular adaptation, we calculated the expected frequency of adaptive CpG changes for a null model in which CpG status is irrelevant to adaptation, and we compared the observed frequency of adaptive CpG changes to this null expectation. In all cases examined here, outgroup comparisons revealed that the derived, affinity-enhancing amino acid changes were specific to the high-altitude taxa. Using reconstructed ancestral sequences for each pair of taxa, we calculated the null expectation for the frequency of CpG changes for three slightly different models (custom R script, Dryad Digital Repository: https://doi.org/10.5061/dryad.2256f38).
In particular, following Stoltzfus & McCandlish [9], we considered null models in which all mutationally accessible variants are maintained in the population at sufficiently high frequency that selection deterministically chooses the fittest variant independent of its mutation rate or CpG status. Because we do not know a priori which variant will be the fittest, we calculate the expected frequency of CpG-associated changes by assuming that each mutational neighbour has an equal probability of being the most fit. Although the most obvious justification for this kind of calculation is in terms of the population-genetic scenario just described, the same null expectation applies to any scenario in which CpG status (for whatever reason) fails to have any impact on which mutant is chosen from the set of mutationally accessible neighbours.
The three null models differ in whether the mutationally accessible variants are defined at the level of nucleotides, codons or amino acids. In model 1, we define the set of possible mutant alleles as the set of all single-nucleotide mutations, and because each site has the same number of mutational neighbours (i.e. 3), the expected frequency of CpG mutants is simply the frequency of nucleotides found in CpG sites. In model 2, we assume that mutational neighbours that are synonymous or missense mutations are not likely to be chosen as the fittest neighbour, and so we exclude these. Finally, in model 3, we assume that—at any given site—only the amino acid state is relevant to fitness, so each mutationally accessible amino acid state has the same chance of being the most fit, regardless of the number of single-nucleotide changes that would produce that state. Under this model, each amino acid change is assigned to be CpG-associated or not depending on whether or not the underlying nucleotide change in the ancestral codon would have occurred at a CpG site. Because our calculations (for all three models) are based on the reconstructed ancestral sequences, they naturally take into account the observed frequencies of CpG sites, amino acids, synonymous codons and NNC GNN codon pairs in avian globin genes.
We note that for model 3, the structure of the genetic code renders all amino acid substitutions from an ancestral codon unambiguous with respect to whether they are CpG-associated, with exactly one exception. For the sequence TTC GNN, the first codon (which specifies Phe) can reach a neighbour (Leu) by either a CpG mutation (TTC GNN to TTR GNN) or a non-CpG mutation (TTC GNN to CTC GNN). We counted this as a CpG-associated change, which is conservative because it over-estimates the expected frequency of CpG changes, thereby making it more difficult to reject the null hypothesis.
(f). Statistical tests for elevated frequency of CpG-associated changes
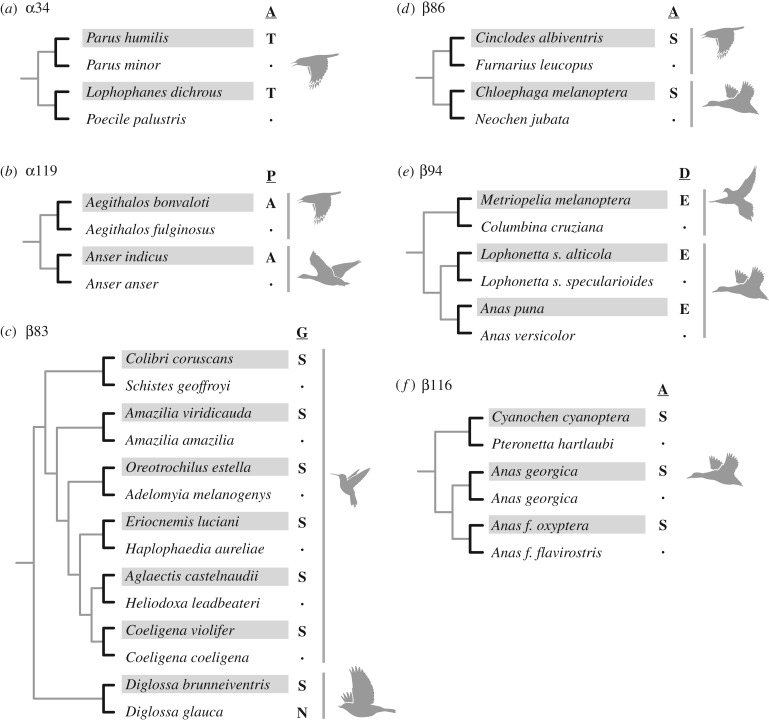
Having calculated the frequencies of CpG-associated changes under our null model, we then calculated the frequency of CpG-associated changes out of all observed adaptive mutations. Following Stoltzfus and McCandlish [9], we note that because identical causative mutations sometimes contributed to adapation in several different high-altitude taxa, it is useful to distinguish between the set of distinct mutations that contribute to adaptation, which we call ‘paths’, and the number of episodes of adaptation each such mutation contributed to, which we call ‘events’. We thus calculated the frequency of CpG-associated causative changes for both paths, where each distinct mutation is weighted equally, and for events, where each mutation is weighted by the number of different adaptive episodes it contributed to. As described below, for the dataset in question, we observe 10 different paths with one to six events per path (see figure 3 for all observed paths with two or more events).
Figure 3.
Phylogenetically replicated substitutions that contributed to derived increases in Hb–O2 affinity in different lineages of high-altitude birds. The select pairs of high- and low-altitude taxa are taken from the set of 70 taxa shown in figure 1. In each case, the high-altitude taxon is denoted by shading and the inferred ancestral amino acid is shown as the reference state. (a) Parallel substitutions at α34 in two high-altitude passerines from the Sino-Himalayan region. Paired comparisons between high- and low-altitude taxa involved ground tit, Parus humilis (high) versus oriental tit, P. minor (low), and grey-crested tit, Lophophanes dichrous (high) versus marsh tit, Poecile palustris (low) [26]. (b) Parallel substitutions at α119 in high-altitude passerine and waterfowl species from the Sino-Himalayan region. Comparisons involved black-browed bushtit, Aegithalos bonvaloti (high) versus sooty bushtit, A. fulginosus (low), and bar-headed goose, Anser indicus (high) versus greylag goose, A. anser (low) [26,35,36]. (c) Parallel and convergent substitutions at β83 in high-altitude hummingbird and passerine species from the Andes. Comparisons involved sparkling violetear, Colibri coruscans (high) versus wedge-billed hummingbird, Schistes geoffroyi (low), green-and-white hummingbird, Amazilia viridicauda (high) versus amazilia hummingbird, A. amazilia (low), Andean hillstar, Oreotrochilus estella (high) versus speckled hummingbird, Adelomyia melanogenys (low), sapphire-vented puffleg, Eriocnemis luciani (high) versus greenish puffleg, Haplophaedia aurelieae (low), white-tufted sunbeam, Aglaeactis castelnaudii (high) versus violet-fronted brilliant, Heliodoxa leadbeateri (low), violet-throated starfrontlet, Coeligena violifer (high) versus bronzy inca, C. coeligena (low), and black-throated flowerpiercer, Diglossa brunneiventris (high) versus deep-blue flowerpiercer, D. glauca (low) [27,29]. Note that the inferred ancestral state for β83 of hummingbirds is glycine (G), but the inferred ancestral state for this same site in Diglossa is asparagine (N). Thus, the βN83S substitution in D. brunneiventris represents a convergent substitution relative to the hummingbird clade. (d) Parallel βA86S substitutions in high-altitude suboscine passerines and waterfowl species from the Andes. Comparisons involved cream-winged cinclodes, Cinclodes albiventris (high) versus pale-legged hornero, Furnarius leucopus (low), and Andean goose, Chloephaga melanoptera (high) versus Orinoco goose, Neochen jubata (low) [28,29]. (e) Parallel βD94E substitutions in high-altitude doves and waterfowl species from the Andes. Comparisons involved black-winged ground dove, Metriopelia melanoptera versus croaking ground dove, Columbina cruziana, Andean crested duck, Lophonetta specularioides alticola (high) versus Patagonian crested duck, L. s. specularioides (low), and Puna teal, Anas puna (high) versus silver teal, A. versicolor (low) [28,29]. (f) Replicated βA116S substitutions in high-altitude waterfowl. Comparisons involve Abyssinian blue-winged goose, Cyanochen cyanoptera (high) versus Hartlaub's duck, Pteronetta hartlaubi (low) and high- versus low-altitude subspecies of yellow-billed pintail, Anas georgica, and sharp-winged teal, A. flavirostris. In the latter two taxon pairs, the derived Ser-β116 variants do not have independent mutational origins in the two highland taxa; the allele-sharing is attributable to a history of introgressive hybridization [28].
After calculating the expected and observed frequencies of CpG-associated causative changes, we tested whether the observed frequency was significantly larger than the expected frequency under our null model. However, these tests were somewhat different depending on whether we calculated the frequency at the level of paths or at the level of events. Under our null hypothesis, each path is either CpG-associated or not CpG-associated independently of all other paths, and each path is CpG-associated with the same probability (as specified by one of the three null models). That is, under our null hypothesis, the number of CpG-associated paths is binomially distributed. We therefore evaluated this null hypothesis against the alternative hypothesis (that the frequency of CpG mutations among paths is greater than the null expectation) using a binomial test (binom.test in the R stats package).
Whereas paths are independent of one another, the causative mutations involved in different events may or may not have distinct mutational origins. Causative alleles that are shared between two or more high-altitude species may be identical by state (distinct mutational origins) or identical by descent (due to retention of ancestral polymorphism or introgressive hybridization). Thus, under the null hypothesis, different events belonging to the same path may not be independently CpG-associated. To account for this potential non-independence, we considered a null model in which the number of CpG-associated events is determined by randomly and independently assigning each observed path to be either CpG-associated or not with probabilities specified by one of the three null models. The frequency of CpG-associated events is then determined by weighting each path by the empirically observed number of events for that path. This choice of null is highly conservative because it increases the variability in the fraction of CpG-associated events relative to a more realistic but harder-to-specify null in which events with distinct mutational origins are treated as independent. To implement this randomization test for whether the number of CpG-associated events exceeds the null expectation, we calculated the exact probability that the number of CpG events is greater than or equal to the observed number (custom R script, Dryad Digital Repository: https://doi.org/10.5061/dryad.2256f38).
(g). Ancestral sequence reconstructions
We used alignments of avian globin sequences in conjunction with previously estimated phylogenetic relationships [40,46] to reconstruct ancestral sequences using baseml, as implemented in PAML 4.7 [47]. We used the substitution model GTR + G in the ancestral sequence reconstructions of both αA- and βA-globin.
3. Results
(a). Convergence in protein function
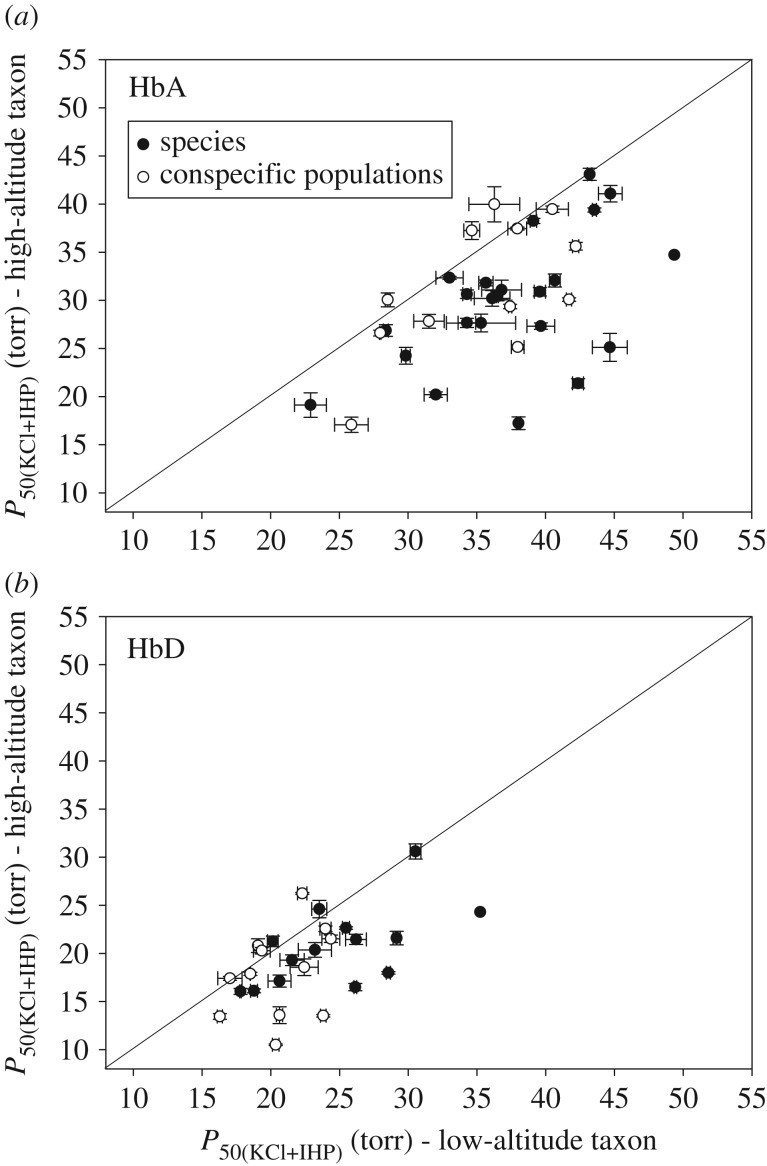
Given that theoretical and experimental results indicate that an increased Hb–O2 affinity can contribute to an enhancement of arterial O2 saturation and, hence, improved tissue O2 delivery under conditions of severe hypoxia [30–32], an obvious prediction is that derived increases in Hb–O2 affinity will have evolved repeatedly in avian taxa that have independently colonized extreme altitudes. We tested this prediction using phylogenetically independent comparisons involving 35 pairs of high- and low-altitude avian taxa (figure 1). The analysis revealed a striking elevational pattern of convergence, as the high-altitude taxon exhibited a higher Hb–O2 affinity in the overwhelming majority of pairwise comparisons. Phylogenetically independent comparisons revealed that highland natives generally have an increased Hb–O2 affinity relative to their lowland counterparts, a pattern consistent for both the major HbA isoform (Wilcoxon's signed-rank test, Z = −4.6844, p < 0.0001, N = 35; figure 2a) and the minor HbD isoform (Z = −3.3144, p = 0.0009, N = 26; figure 2b). In all pairwise comparisons in which the high-altitude taxa exhibited significantly higher Hb–O2 affinities relative to the lowland taxa (N = 35 taxon pairs for HbA, N = 26 for HbD), the measured differences were entirely attributable to differences in intrinsic O2 affinity rather than differences in responsiveness to the inhibitory effects of Cl− ions or IHP [26,28,29,35]. The sample size is smaller for HbD because some species included in our dataset do not express this isoform [29,39,40].
Figure 2.
Convergent increases in Hb–O2 affinity in high-altitude birds. (a) Plot of P50(KCl+IHP) (±s.e.) for HbA in 35 matched pairs of high- and low-altitude taxa. The pairwise comparisons of Hb–O2 affinity involve HbA homologues of closely related species or representative allelic variants from high- and low-altitude populations of the same species. Data points that fall below the diagonal (x = y) denote cases in which the high-altitude member of a given taxon pair possesses a higher Hb–O2 affinity (lower P50). Comparisons involve phylogenetically replicated pairs of taxa, so all data points are statistically independent. (b) Plot of P50(KCl+IHP) (±s.e.) for the minor HbD isoform in a subset of the same taxon pairs in which both members of the pair express HbD. Sample sizes are larger for HbA than for HbD because the two ground dove species (Metriopelia melanoptera and Columbina cruziana) expressed no trace of HbD, and several hummingbird species expressed HbD at exceedingly low levels [27,29]. In such cases, sufficient quantities of HbD could not be purified for measures of O2 equilibria. Black symbols denote comparisons between species, whereas open symbols denote comparisons between high- versus low-altitude populations of the same species. Data from [26,28,29].
(b). Identification of causative mutations that contribute to convergent changes in haemoglobin function
After documenting that high-altitude taxa have convergently evolved derived increases in Hb–O2 affinity, we used results of our previously published site-directed mutagenesis experiments to identify causative amino acid substitutions. Comparative sequence data for the set of 70 taxa revealed phylogenetically replicated replacements at numerous sites in the αA- and αD-globin genes (affecting HbA and HbD, respectively) and in the βA-globin gene (affecting both HbA and HbD) [26–29,33,35,36]. Although we observed numerous parallel substitutions (i.e. independent changes from the same ancestral amino acid to the same derived amino acid) and convergent substitutions (i.e. independent changes from different ancestral amino acids to the same derived amino acid), functional data from native Hb variants and engineered, recombinant Hb mutants revealed that only a subset of replicated replacements actually contributed to convergent increases in Hb–O2 affinity in the different highland taxa. Overall, we identified a total of 22 affinity-enhancing replacements in 20 different high-altitude taxa (table 1). This set of causative changes included two parallel replacements in αA-globin (αA34T and αP119A) and four parallel or convergent replacements in βA-globin (βG83S, βN83S, βA86S, βD94E and βA116S) [26–29,36] (figure 3). Convergent increases in Hb–O2 affinity were largely attributable to non-replicated replacements (divergent substitutions), suggesting that evolutionary increases in Hb–O2 affinity can be produced by amino acid replacements at numerous sites.
Table 1.
Experimentally verified mutations (*, CpG mutations) that contributed to derived increases in Hb–O2 affinity in high-altitude birds.
| high-altitude taxon | affinity-enhancing amino acid change | nucleotide change | reference |
|---|---|---|---|
| bar-headed goose, Anser indicus | α18 G → S | G* to A | [36] |
| grey-crested tit, Lophophanes dichrous | α34 A → T | G* to A | [26] |
| ground tit, Parus humilis | α34 A → T | G* to A | [26] |
| bar-headed goose, Anser indicus | α63 A → V | C* to T | [36] |
| black-browed bushtit, Aegithalos bonvaloti | α119 P → A | C to G | [26] |
| bar-headed goose, Anser indicus | α119 P → A | C to G | [36] |
| house wren, Troglodytes aedon | β55 V → I | G* to A | [25] |
| sparkling violetear, Colubri coruscans | β83 G → S | G to A | [27,29] |
| green-and-white hummingbird, Amazilia viridicauda | β83 G → S | G to A | [27,29] |
| Andean hillstar, Oreotrochilus estella | β83 G → S | G to A | [27,29] |
| sapphire-vented puffleg, Eriocnemis luciani | β83 G → S | G to A | [27,29] |
| white-tufted sunbeam, Aglaeactis castelnaudii | β83 G → S | G to A | [27,29] |
| violet-throated starfrontlet, Coeligena violifer | β83 G → S | G to A | [27,29] |
| black-throated flowerpiercer, Diglossa brunneiventris | β83 N → S | A to G | [29] |
| cream-winged cinclodes, Cinclodes albiventris | β86 A → S | G* to T | [29] |
| Andean goose, Chloephaga melanoptera | β86 A → S | G* to T | [28] |
| black-winged ground dove, Metriopelia melanoptera | β94 D → E | C to G | [29] |
| Andean crested duck, Lophonetta specularioides alticola | β94 D → E | C to G | [28] |
| puna teal, Anas puna | β94 D → E | C to G | [28] |
| yellow-billed pintail, Anas georgica | β116 A → S | G* to T | [28] |
| sharp-winged teal, Anas flavirostris oxyptera | β116 A → S | G* to T | [28] |
| Abyssinian blue-winged goose, Cyanochen cyanoptera | β116 A → S | G* to T | [28] |
(c). Testing for a disproportionate contribution of CpG mutations to the adaptive convergence of haemoglobin function
After experimentally identifying the causative substitutions that are responsible for (putatively adaptive) increases in Hb–O2 affinity in high-altitude birds, we then asked whether mutations at CpG sites made a disproportionate contribution to the observed changes in protein function. The use of site-directed mutagenesis experiments to test functional effects was restricted to the major HbA isoform (table 1), so we focus attention on amino acid replacements in the αA- and βA-globin genes. Note, however, that the βA-globin gene encodes β-type subunits of both HbA and HbD, so the effects of missense mutations in that gene are manifest in both isoforms.
The set of mutational changes in the αA- and βA-globin genes can be described in several ways. There are 22 total substitution events that take place at nine different sites via 10 different mutational paths. There is a difference between sites and paths because there are two different paths of change at site β83: there are parallel Gly → Ser replacements in six different high-altitude hummingbird species and there is a convergent Asn → Ser replacement in one species of high-altitude flowerpiercer (figure 3). The βG83S change, with six events, is the most highly repeated path. The distribution of events for all 10 paths is 6, 3, 3, 2, 2, 2, 1, 1, 1 and 1.
We calculated the expected frequency of CpG-associated changes under a null model in which the CpG status of a mutation is irrelevant to its probability of contributing to adaptation. Because this expectation varies slightly depending on assumptions, we calculated three separate null expectations (see Methods): (i) the frequency that a site in a globin gene is a CpG site, (ii) the frequency that a missense mutation in a globin gene is a CpG mutation, and (iii) the frequency of CpG-associated changes among the set of all one-mutant amino acid neighbours of a given ancestral sequence. The three models are very similar in giving a null expectation of 10 or 11% CpG, as shown in table 2. Thus, for the 10 independent mutational paths, we expect approximately 1 CpG path, and for the 22 events, we expect approximately 2 CpG events. As shown in table 2, the observed number of CpG changes far exceeded the null expectations. Out of 22 events, 10 involve CpG mutations. Out of 10 paths, six involve a CpG mutation. The binomial test revealed that the observed frequency of CpG paths is significantly higher than the null expectation (table 2). Likewise, the observed frequency of CpG mutations among events is significantly higher than the null expectation as determined by the randomization test that accounts for potential non-independence. These results remain qualitatively unchanged if we exclude βV55I from our set of causative mutations on the grounds that our initial discovery of this particular CpG path [25] motivated our investigation of CpG effects in the broader set of taxa. Leaving out βV55I and using model 1 (the most conservative), the probability of observing so many CpG-associated changes is p = 0.001 for paths (binomial test) and p = 0.033 for events (randomization test).
Table 2.
Comparison of observed and expected numbers of CpG mutations that contribute to derived increases in Hb–O2 affinity in high-altitude species. See Methods for an explanation of the difference between mutational paths and events.
| CpG paths (N = 10) |
CpG events (N = 22) |
||||||
|---|---|---|---|---|---|---|---|
| model | expected frequency | expected | observed | p-value | expected | observed | p-value |
| 1 | 0.109 | 1.1 | 6 | 2.4 × 10−4 | 2.4 | 10 | 0.018 |
| 2 | 0.097 | 1.0 | 6 | 1.2 × 10−4 | 2.1 | 10 | 0.013 |
| 3 | 0.096 | 1.0 | 6 | 1.2 × 10−4 | 2.1 | 10 | 0.013 |
4. Discussion
(a). A disproportionate role for CpG mutations in adaptation
Results of our comparative analysis of Hb function reveal that high-altitude birds have generally evolved increased Hb–O2 affinities relative to lowland sister taxa, consistent with earlier reports based on smaller subsets of the data presented here [26,28,29]. The striking pattern of convergence is consistent with the hypothesis that the elevational differences reflect a history of directional selection in high-altitude natives. This pattern of apparently adaptive convergence in protein function allowed us to address a key question: Are some types of amino acid mutation preferentially fixed? If so, to what extent is the observed substitution bias attributable to variation in rates of origin (mutation bias)—that is, variation among sites in rates of mutation to alleles that produce the beneficial change in phenotype? Our analysis of causative amino acid changes revealed that a disproportionate number of affinity-enhancing amino acid replacements were attributable to mutations at CpG dinucleotides, suggesting that mutation bias exerts an influence on patterns of adaptive substitution.
In each of the high-altitude bird species that evolved increased Hb–O2 affinities due to missense CpG mutations, there seems little reason to suppose that the causative amino acid mutations would have had larger selection coefficients (and, hence, higher fixation probabilities) than any number of other possible mutations that could have produced a similar increase in Hb–O2 affinity. However, if CpG mutation occurs at a higher rate than non-CpG mutations, then—in the absence of contributions from standing variation—the bias in mutation rate is expected to influence evolutionary outcomes in the same way as a commensurate bias in fixation probability [5,7,8].
(b). Mutational independence of parallel substitutions
The majority of ‘repeated’ substitutions are authentic parallelisms, where the shared, derived amino acids in two or more high-altitude taxa have independent mutational origins [26,28,29]. However, in the case of the six repeated βG83S changes in high-altitude hummingbirds, we cannot rule out the possibility that some or most of the shared, derived Ser alleles in high-altitude species are identical by descent [27]. Even if the pattern of allele sharing among species is partly attributable to incomplete lineage sorting or introgression, the pattern is still highly non-random with respect to altitude, suggesting a non-random sorting of ancestral polymorphism at β83 (or extensive introgressive hybridization). In other words, the observed pattern indicates that the derived Ser variant was repeatedly driven to fixation in different highland lineages [27]. Similarly, we previously documented that the shared, derived β116Ser variants in the high-altitude subspecies of Anas georgica and Anas flavirostris are identical by descent. In this case, the allele sharing is attributable to introgressive hybridization, and results of population genomic analyses provide strong evidence for both highland taxa that the derived β116Ser variants increased in frequency under the influence of positive directional selection [28].
Thus, in the case of the repeated substitutions at β83 and β116, the derived amino acid variants do not necessarily have distinct mutational origins in different species, but the derived amino acid variants contributed to altitude-related increases in Hb–O2 affinity in each case, and this is what matters for the purposes of our tests. Using both binomial and randomization approaches, we tested the null hypothesis that the contributions of mutations to adaptive changes in Hb function are unrelated to whether they occur at CpG sites or non-CpG sites. Whether fixed alleles in different species are identical by descent is certainly relevant to questions about the prevalence of molecular parallelism and the causes of homoplasy [48,49], but it has no bearing on our tests of whether CpG sites make disproportionate contributions to molecular adaptation. For example, in the case of the six βG83S substitutions in high-altitude hummingbirds, the ‘jackpot’ of six events in the same mutational path does not bias our conclusions because the possibility of such jackpot effects is incorporated into the design of the randomization test, which treats each observed mutational path as if it had a single mutational origin.
(c). Conclusion: mutation bias and substitution bias
We tested whether affinity-enhancing mutations in the Hbs of high-altitude birds are enriched for mutations at CpG dinucleotides relative to the frequency of CpG mutations among all possible missense mutations. The results summarized in tables 1 and 2 indicate that a disproportionate number of causative amino acid replacements were attributable to CpG mutations, suggesting that mutation bias can influence outcomes of molecular adaptation. Moreover, if methylated CpG sites have higher-than-average rates of point mutation, we hypothesize that any given set of adaptive substitutions should be enriched for changes at such sites.
Acknowledgements
The identification of any specific commercial products is for the purpose of specifying a protocol, and does not imply a recommendation or endorsement by the National Institute of Standards and Technology.
Data accessibility
All data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.2256f38 [50].
Authors' contributions
J.F.S., D.M.M. and A.S. conceived the study; J.F.S., D.M.M. and A.S. drafted the manuscript; C.N. and A.V.S. collected experimental data; J.F.S., A.V.S. and C.C.W. analysed experimental data; D.M.M. and A.S. designed and implemented the statistical analysis of CpG effects; A.S. assembled and curated the electronic supplementary material package.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by National Institutes of Health grant no. HL087216 (J.F.S.), National Science Foundation grant nos MCB-1517636 (J.F.S.), RII Track-2 FEC-1736249 (J.F.S.), DEB-1146491 (C.C.W.) and MCB-1516660 (C.C.W.).
References
- 1.Alberch P. 1982. Developmental constraints in evolutionary processes. In Evolution and development (ed. Bonner T.), pp. 313–332. New York, NY: Springer. [Google Scholar]
- 2.Wake DB. 1991. Homoplasy—the result of natural selection, or evidence of design limitations? Am. Nat. 138, 543–567. ( 10.1086/285234) [DOI] [Google Scholar]
- 3.Arthur W. 2000. The concept of developmental reprogramming and the quest for an inclusive theory of evolutionary mechanisms. Evol. Dev. 2, 49–57. ( 10.1046/j.1525-142x.2000.00028.x) [DOI] [PubMed] [Google Scholar]
- 4.Wake DB, Wake MH, Specht CD. 2011. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science 331, 1032–1035. ( 10.1126/science.1188545) [DOI] [PubMed] [Google Scholar]
- 5.Yampolsky LY, Stoltzfus A. 2001. Bias in the introduction of variation as an orienting factor in evolution. Evol. Dev. 3, 73–83. ( 10.1046/j.1525-142x.2001.003002073.x) [DOI] [PubMed] [Google Scholar]
- 6.Stoltzfus A. 2006. Mutationism and the dual causation of evolutionary change. Evol. Dev. 8, 304–317. ( 10.1111/j.1525-142X.2006.00101.x) [DOI] [PubMed] [Google Scholar]
- 7.Stoltzfus A, Yampolsky LY. 2009. Climbing mount probable: mutation as a cause of nonrandomness in evolution. J. Heredity 100, 637–647. ( 10.1093/jhered/esp048) [DOI] [PubMed] [Google Scholar]
- 8.McCandlish DM, Stoltzfus A. 2014. Modeling evolution using the probability of fixation: history and implications. Q. Rev. Biol. 89, 225–252. ( 10.1086/677571) [DOI] [PubMed] [Google Scholar]
- 9.Stoltzfus A, McCandlish DM. 2017. Mutational biases influence parallel adaptation. Mol. Biol. Evol. 34, 2163–2172. ( 10.1093/molbev/msx180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobzhansky T. 1955. Genetics and the origin of species. New York, NY: Wiley and Sons, Inc. [Google Scholar]
- 11.Mayr E. 1959. Where are we? Cold Spring Harb. Symp. Quant. Biol. 24, 1–14. [Google Scholar]
- 12.Stebbins GL. 1959. The synthetic approach to problems of organic evolution. Cold Spring Harb. Symp. Quant. Biol. 24, 305–311. ( 10.1101/SQB.1959.024.01.028) [DOI] [PubMed] [Google Scholar]
- 13.Stoltzfus A. 2017. Why we don't want another ‘Synthesis’. Biol. Direct 12, 23.11 ( 10.1186/s13062-017-0194-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher RA. 1930. The genetical theory of natural selection. Oxford, UK: Oxford University Press. [Google Scholar]
- 15.Kimura M. 1983. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 16.Rokyta DR, et al. 2005. An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nat. Genet. 37, 441–444. ( 10.1038/ng1535) [DOI] [PubMed] [Google Scholar]
- 17.Lozovsky ER, et al. 2009. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl Acad. Sci. USA 106, 12 025–12 030. ( 10.1073/pnas.0905922106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigand MR, Sundin GW. 2012. General and inducible hypermutation facilitate parallel adaptation in Pseudomonas aeruginosa despite divergent mutation spectra. Proc. Natl Acad. Sci. USA 109, 13 680–13 685. ( 10.1073/pnas.1205357109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong A, Rodrigue N, Kassen R. 2012. Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet. 8, e1002928 ( 10.1371/journal.pgen.1002928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couce A, Rodriguez-Rojas A, Blazquez J. 2015. Bypass of genetic constraints during mutator evolution to antibiotic resistance. Proc. R. Soc. B 282, 20142698 ( 10.1098/rspb.2014.2698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepel A, Haussler D. 2004. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 21, 468–488. ( 10.1093/molbev/msh039) [DOI] [PubMed] [Google Scholar]
- 22.Smith T, et al. 2016. Extensive variation in the mutation rate between and within human genes associated with Mendelian disease. Hum. Mutat. 37, 488–494. ( 10.1002/humu.22967) [DOI] [PubMed] [Google Scholar]
- 23.Smeds L, Qvarnstrom A, Ellegren H. 2016. Direct estimate of the rate of germline mutation in a bird. Genome Res. 26, 1211–1218. ( 10.1101/gr.204669.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mugal CF, Arndt PF, Holm L, Ellegren H. 2015. Evolutionary consequences of DNA methylation on the GC content in vertebrate genomes. Genes Genomes Genet. 5, 441–447. ( 10.1534/g3.114.015545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galen SC, et al. 2015. Contribution of a mutational hotspot to adaptive changes in hemoglobin function in high-altitude Andean house wrens. Proc. Natl Acad. Sci. USA 112, 13 958–13 963. ( 10.1073/pnas.1507300112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, et al. 2018. Divergent and parallel routes of biochemical adaptation in high-altitude passerine birds from the Qinghai-Tibet Plateau. Proc. Natl Acad. Sci. USA 115, 1865–1870. ( 10.1073/pnas.1720487115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Projecto-Garcia J, et al. 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc. Natl Acad. Sci. USA 110, 20 669–20 674. ( 10.1073/pnas.1315456110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan C, et al. 2015. Convergent evolution of hemoglobin function in high-altitude Andean waterfowl involves limited parallelism at the molecular sequence level. PLoS Genet. 11, e1005681 ( 10.1371/journal.pgen.1005681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natarajan C, Hoffmann FG, Weber RE, Fago A, Witt CC, Storz JF. 2016. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science 354, 336–340. ( 10.1126/science.aaf9070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storz JF. 2016. Hemoglobin–oxygen affinity in high-altitude vertebrates: is there evidence for an adaptive trend? J. Exp. Biol. 219, 3190–3203. ( 10.1242/jeb.127134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storz JF, Scott GR, Cheviron ZA. 2010. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 213, 4125–4136. ( 10.1242/jeb.048181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storz JF. 2019. Hemoglobin: insights into protein structure, function, and evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 33.Cheviron ZA, et al. 2014. Integrating evolutionary and functional tests of adaptive hypotheses: a case study of altitudinal differentiation in hemoglobin function in an Andean sparrow, Zonotrichia capensis. Mol. Biol. Evol. 31, 2948–2962. ( 10.1093/molbev/msu234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Natarajan C, Moriyama H, Witt CC, Weber RE, Fago A, Storz JF. et al. 2017. Stability-mediated epistasis restricts accessible mutational pathways in the functional evolution of avian hemoglobin. Mol. Biol. Evol. 34, 1240–1251. ( 10.1093/molbev/msx085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jendroszek A, Malte H, Overgaard CB, Beedholm K, Natarajan C, Weber RE, Storz JF, Fago A. et al. 2018. Allosteric mechanisms underlying the adaptive increase in hemoglobin–oxygen affinity of the bar-headed goose. J. Exp. Biol. 221, 185470 ( 10.1242/jeb.185470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natarajan C, Jendroszek A, Kumar A, Weber RE, Tame JRH, Fago A, Storz JF. et al. 2018. Molecular basis of hemoglobin adaptation in the high-flying bar-headed goose. PLoS Genet. 14, e1007331 ( 10.1371/journal.pgen.1007331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber RE, Fago A, Malte H, Storz JF, Gorr TA. et al. 2013. Lack of conventional oxygen-linked proton and anion binding sites does not impair allosteric regulation of oxygen binding in dwarf caiman hemoglobin. Am. J. Physiol. Regul. Integr. Comp. Physiol. 305, R300–R312. ( 10.1152/ajpregu.00014.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storz JF, Natarajan C, Moriyama H, Hoffmann FG, Wang T, Fago A, Malte H, Overgaard J, Weber RE. et al. 2015. Oxygenation properties and isoform diversity of snake hemoglobins. Am. J. Physiol. Regul. Integr. Comp. Physiol. 309, R1178–R1191. ( 10.1152/ajpregu.00327.2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grispo MT, Natarajan C, Projecto-Garcia J, Moriyama H, Weber RE, Storz JF. et al. 2012. Gene duplication and the evolution of hemoglobin isoform differentiation in birds. J. Biol. Chem. 287, 37 647–37 658. ( 10.1074/jbc.M112.375600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opazo JC, Hoffmann FG, Natarajan C, Witt CC, Berenbrink M, Storz JF. et al. 2015. Gene turnover in the avian globin gene family and evolutionary changes in hemoglobin isoform expression. Mol. Biol. Evol. 32, 871–887. ( 10.1093/molbev/msu341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tufts DM, Natarajan C, Revsbech IG, Projecto-Garcia J, Hoffmann FG, Weber RE, Fago A, Moriyama H, Storz JF. et al. 2015. Epistasis constrains mutational pathways of hemoglobin adaptation in high-altitude pikas. Mol. Biol. Evol. 32, 287–298. ( 10.1093/molbev/msu311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natarajan C, Jiang X, Fago A, Weber RE, Moriyama H, Storz JF. et al. 2011. Expression and purification of recombinant hemoglobin in Escherichia coli. PLoS ONE 6, e20176 ( 10.1371/journal.pone.0020176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natarajan C, Inoguchi N, Weber RE, Fago A, Moriyama H, Storz JF. et al. 2013. Epistasis among adaptive mutations in deer mouse hemoglobin. Science 340, 1324–1327. ( 10.1126/science.1236862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garland T Jr, Bennett AF, Rezende EL. 2005. Phylogenetic approaches in comparative physiology. J. Exp. Biol. 208, 3015–3035. ( 10.1242/jeb.01745) [DOI] [PubMed] [Google Scholar]
- 45.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15. ( 10.1086/284325) [DOI] [Google Scholar]
- 46.Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320. ( 10.1126/science.1251385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z. 2007. PAML: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 48.Hahn MW, Nakhleh L. 2016. Irrational exuberance for resolved species trees. Evolution 70, 7–17. ( 10.1111/evo.12832) [DOI] [PubMed] [Google Scholar]
- 49.Storz JF. 2016. Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet. 17, 239–250. ( 10.1038/nrg.2016.11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storz JF, Natarajan C, Signore AV, Witt CC, McCandlish DM, Stoltzfus A. 2019. Data from: The role of mutation bias in adaptive molecular evolution: insights from convergent changes in protein function Dryad Digital Repository. ( 10.5061/dryad.2256f38) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Storz JF, Natarajan C, Signore AV, Witt CC, McCandlish DM, Stoltzfus A. 2019. Data from: The role of mutation bias in adaptive molecular evolution: insights from convergent changes in protein function Dryad Digital Repository. ( 10.5061/dryad.2256f38) [DOI] [PMC free article] [PubMed]
Data Availability Statement
All data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.2256f38 [50].