Abstract
BACKGROUND:
Thalassemia is one of the most common genetic health problems in the world. More than 200 different mutations have been identified in the beta-globin gene and among the 24 β-globin gene mutations in β-thalassemia carriers in the north of Iran IVSII-I G>A mutation has the highest frequency. Using fast, inexpensive, simple and reliable methods for the detection of the mutations in β-thal carriers is very important in prenatal diagnosis, and introduction of alternative methods to the existing ones can help to simplify the detection of mutations. Since its introduction, different methods derived from LAMP have been widely used for SNPs detection.
AIM:
This study was aimed to design a new method for the detection of IVSII-I G>A mutation on β-globin gene based on AS - LAMP technique.
METHODS:
Primer explorer V5 software was used for the design of LAMP primers. Three sets of primers were designed. In the first set, the BIP primers were exactly complementary to the normal and mutant alleles. In the second set, 1 nucleotide (T) was inserted at the 5’ end of BIP primers, and in the last set, one nucleotide at the 5’ end of BIP primer was changed. The other required primers for the LAMP reaction (FIP, B3, and F3) were the same for all 3 sets of primers. The LAMP reaction was applied on three DNA samples (Wild type, Heterozygote and Homozygote for IVSII-I G>A mutation) and synthetic DNA.
RESULTS:
The results of the present study showed that LAMP reaction using three sets of primers could not successfully detect the IVSII-I G > A mutation among subjects DNA sample and synthetic DNA.
CONCLUSION:
Although several studies have successfully used ARMS-LAMP method to detect the SNPs, and other studies use a variety of methods to identify IVSII-I G>A mutation, the present study was unable to differentiate between a normal allele and IVSII-I G>A mutation. Hence further studies are recommended to consider redesigning of primer set, DNA concentration and using commercial LAMP Master Mix to detect the mutation.
Keywords: β-thalassemia, AS-LAMP, IVSII-IG-A mutation
Introduction
Thalassemia is one of the most common genetic health problems in the world resulting from the reduced or absent synthesis of the haemoglobin chains synthesis. Thalassemias are classified into two main types of α- or β based on the absence production of α- or β-globin chains. β-thalassemia is caused by inherited mutations on the β-globin gene that leads to reduced synthesis of the β-globin chain of the haemoglobin. The highest frequency of β-thalassemia mutations is reported from Mediterranean countries, the Middle East, and East Asia. More than 200 different mutations have been identified in the beta-globin gene, causing wide genotypic and phenotypic variability of this blood disorder [1], [2].
Alpha and beta thalassemia mutations are very common in Iran [3], and the highest prevalence of β-Thalassemia (around 10%) has been reported from north of Iran, in Southern coastlines of Caspian Sea, and South of Iran close to the Persian Gulf. The prevalence of β-thalassemia mutations in most parts of the country has been estimated to be 4-8% [4], [5], [6]. Overall, there are 20,000 affected β-thalassemia patients and 3,750,000 carriers of the diseases in Iran [7]. Among the 24 β-globin gene mutations in β-thalassemia carriers in the north of Iran IVSII-I G>A mutation has the highest frequency (61%) [8].
The national program for the prevention of thalassemia in Iran has been started since 1997. This premarital screening program is compulsory in all provinces, and all couples should be screened before they get married. The aim of this program is screening β-thal carriers and preventing the child-birth with β-thal major [9], [10], [11]. Since in lots of the cases in screening program the mutations of the carriers must be identified, using fast, inexpensive, simple and reliable methods for the detection of the mutations in β-thal carriers is very important in prenatal diagnosis, and introduction of alternative methods to the existing ones can help to simplify the detection of mutations.
For the identification of the mutations among β-thal carriers, PCR-based methods are usually used. Although PCR-based methods are cost-effective techniques that are very robust for nucleic acid amplification, they cannot be performed in every lab. Also, the methods require a thermocycling system, which limits their application in the field. There are few genetic laboratories that collaborate with the screening programs to detect the mutations among carriers and patients.
In the past years, numerous nucleic amplification approaches have been introduced that do not need the thermocycling system. Isothermal techniques to amplify nucleic acids include loop-mediated isothermal amplification (LAMP), nucleic acid sequence-based amplification (NASBA), helicase-dependent amplification (HDA), rolling circle amplification (RCA), multiple displacement amplification (MDA), and recombinase polymerase amplification (RPA); these new approaches can easily be used at a constant temperature [12], [13], [14], [15], [16], [17], [18], [19].
The LAMP method was first introduced by Notomi et al., in 2000 [20]. Since then, different methods derived from LAMP have been widely used for pathogen detection [21], [22], [23], [24].
The LAMP method uses four sets of primers that are specially designed to identify six distinct regions on the objective gene. These sets of primers are the outer and inner primers (F3 and B3). The forward inner primer (FIP) consisting of the F2 zone at the 3’ end with the sequence similar to the F1c region at the 5’ end, is complementary to the F2c zone, while the backward inner primer (BIP) consisting of the B2 zone at the 3’ end with the sequence similar to the B1c region at the 5’ end, is complementary to the B2c zone (Figure 1). The two primers are complementary to the downstream zone of the opposite strand in the objective region (F1 and B1) [20].
Figure 1.
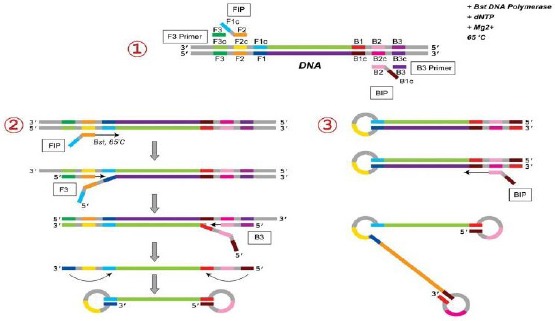
Schematic view of LAMP Primers Using 4 sets of Primers. These sets of primers are the outer and inner primers (F3 and B3). The forward inner primer (FIP) consisting of the F2 zone at the 3’ end with the sequence similar to the F1c region at the 5’ end, is complementary to the F2c zone, while the backward inner primer (BIP) consisting of the B2 zone at the 3’ end with the sequence similar to the B1c region at the 5’ end, is complementary to the B2c zone
The LAMP method yields a substantial amount of amplicons in a short time (around 60 min), which is 103 times higher than the amount produced with PCR [25].
Because the LAMP method uses four primers sets that are specific to the objective regions, it has rapid amplification, a higher amount of amplification products, and smaller detection limits compared with PCR-based methods [23]. Hence, LAMP is more beneficial for point-of-care testing (POCT) than PCR [24].
The LAMP-based methods have rarely been used for genotyping and detecting point mutations. Thus, designing new LAMP methods capable of genotyping could be an alternative method to other genotyping techniques. Moreover, the LAMP technique is a very suitable, easy to use, and cost-effective approach for POCT or on-site testing. This study was aimed to design a new method for the detection of IVSII-I G>A mutation on β-globin gene based on AS (Allele-Specific)-LAMP technique.
Material and Methods
Among the registered patients at the thalassemia research centre, Mazandaran University of medical sciences, the cases that were heterozygote and homozygote for IVSII-I G>A mutation were selected. The mutation in patients was detected using standard ARMS-PCR method and confirmed with PCR-sequencing. The genomic DNA was extracted from peripheral blood applying QIAamp DNA Micro Kit (Qiagen, USA).
Some healthy subjects with no mutations on the β-globin gene were also selected with the help of PCR-sequencing method. Synthetic DNAs complementary to the wild type and mutated region were also designed.
Primer designing for AS-LAMP
For primer designing Primer explorer V5 software, which is a specific tool for the design of LAMP, primers were used (Table 1), and NM_000518.4 reference sequence was considered as a template. To select the primers that could successfully differentiate the IVSII-I G>A mutation (dB SNP rs#:33945777) from normal allele, three sets of primers were designed. In the first set, the BIP primers were exactly complementary to the normal and mutant alleles. In the second set, 1 nucleotide (T) was inserted at the 5’ end of BIP primers, and in the last set, one nucleotide at the 5’ end of BIP primer was changed. The other required primers for the LAMP reaction (FIP, B3, and F3) were the same for all 3 sets of primers (Table 1).
Table 1.
The sequences of primers used for AS-LAMP method for the detection of IVSII-I G>A method
| Set1: BIP-Wild = B1C (5-3) + B2 (5-3) | GTGAGTCTATGGGACGCTTGATTATCCCCTTCCTATGACATGA |
| Set1: BIP-Mutated = B1C M (5-3) + B2 (5-3) | ATGAGTCTATGGGACGCTTGATTATCCCCTTCCTATGACATGA |
| Set 2: BIP-Wild = B1C (5-3) + B2 (5-3) | GTTGAGTCTATGGGACGCTTGATTATCCCCTTCCTATGACATGA |
| Set2: BIP-Mutated = B1C M (5-3) + B2 (5-3) | ATTGAGTCTATGGGACGCTTGATTATCCCCTTCCTATGACATGA |
| Set3: BIP-Wild = B1C (5-3) + B2 (5-3) | GTCAGTCTATGGGACGCTTGATTATCCCCTTCCTATGACATGA |
| Set3: BIP-Mutated = B1C M (5-3) + B2 (5-3) | ATCAGTCTATGGGACGCTTGATTATCCCCTTCCTATGACATGA |
| FIP= F1C (5-3) + F2 (5-3) | CCTGAAGTTCTCAGGATCCACGCCTTTGCCACACTGAGTG |
| F3 | TCACCTGGACAACCTCAA |
| B3 | 5’CCATTCTAAACTGTACCCTG3’ |
Set1: primer designed by http://primerexplorer.jp/lampv5e/index.html, Set2: In this sets of primers 1 nucleotide was inserted at the 5’ end of BIP primers, Set3: The sequence of primers in which one nucleotide at the 5’ end of BIP primer was changed
PCR-Sequencing reaction with B3 and F3 primers
To find that the primers could amplify the target region PCR reaction with the help of F3 and B3 primers were applied, and the PCR product was sequenced using ABI Capillary system.
AS-LAMP reaction
Conventional LAMP mix in total volume of 20µL that contained 2 µL of 10 X Bst DNA polymerase reaction buffer (New England Bio-labs), 8 mM MgSo4, 0.8 M betaine, 1.4 mM of each dNTPs, 1.6 mM FIP, 1.6 mM BIP (wild or Mutated), 0.2 mM B3, 0.2 mM F3 and 8U Bst DNA polymerase (New England Bio-labs). The master mix was dispensed in each microtube, and 50 ng from the DNA template was added to the mixtures. Then the reaction mixes were incubated at 60°C for 60 min. It should be mentioned that for each sample, two LAMP reactions were done, one with wild primers and another with Mutated primer. The LAMP products were run on 1% agarose gel.
Results
PCR sequencing
The results of PCR sequencing with the help of F3 and B3 primer showed that the primers could successfully amplify the target region on the beta-globin gene (Figure 2).
Figure 2.
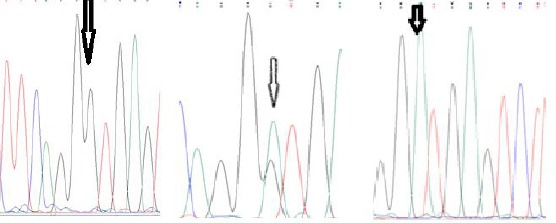
The results of PCR-Sequencing using F3 and B3 primers for the identification of IVSII-I G > A mutation: from left to right: Homozygote wild type, Heterozygote, Homozygote mutated
LAMP reaction with the first primer set
The LAMP reaction using first sets of primers (without any modification) showed that the primers could amplify the target DNA, but this set of primer was unable to differentiate the normal allele from mutated one (Figure 3).
Figure 3.
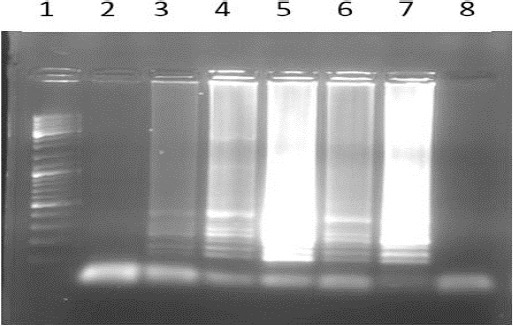
The results of AS LAMP reaction using primers without any modification; 1) Ladder 100bP; 2) Normal DNA with wild type primer; 3) Normal DNA with mutant type primer; 4) Mutant DNA (Homozygote) with wild type primer; 5) Mutant DNA (Homozygote) with mutant type primer; 6) Heterozygote DNA with wild type Primer; 7) Heterozygote DNA with mutant type primer; 8) blank
LAMP reaction with second primer set (with a nucleotide insertion)
Like the LAMP reaction with the modified primers that a nucleotide (T) was inserted one nucleotide before mutation site at the 5 ends of BIP primers indicated that these primers could not differentiate the normal DNA from the mutant one (Figure 4).
Figure 4.
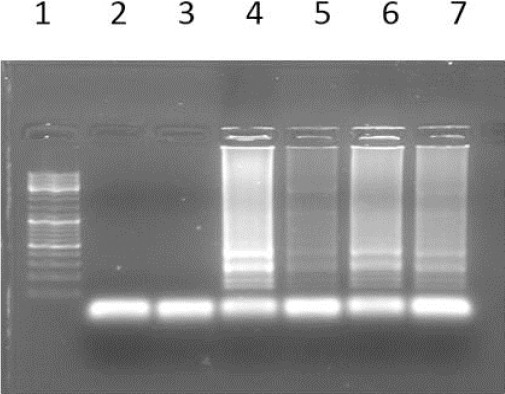
The results of AS LAMP reaction using primers with a T insertion; 1) Ladder 100bP; 2) Normal DNA with wild type primer; 3) Normal DNA with mutant type primer; 4) Mutant DNA (Homozygote) with wild type primer; 5) Mutant DNA (Homozygote) with mutant type primer; 6) Heterozygote DNA with wild type Primer; 7) Heterozygote DNA with mutant type primer; 8) blank
LAMP reaction with third primer set (with a nucleotide substitution at the third nucleotide before mutation site)
The results of LAMP reaction using the third set of primers with a nucleotide substitution at the third nucleotide before mutation site indicated that the LAMP reaction could not successfully distinguish the mutant and normal alleles (Figure 5).
Figure 5.
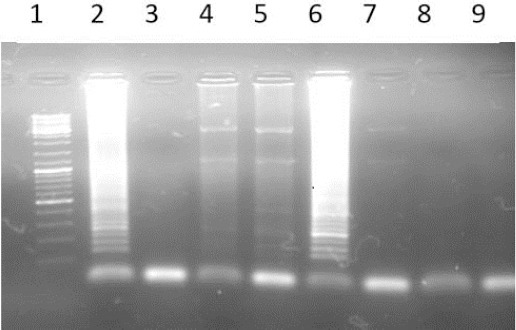
The results of AS LAMP reaction using primers with a nucleotide substitution at the third nucleotide before mutation site; 1) Ladder 100bP; 2) Normal DNA with wild type primer; 3) Normal DNA with mutant type primer; 4) Mutant DNA (Homozygote) with wild type primer; 5) Mutant DNA (Homozygote) with mutant type primer; 6) Heterozygote DNA with wild type Primer; 7) Heterozygote DNA with mutant type primer; 8) blank with wild primer; 9) blank with mutant type primer
The gradient of mgso4 and betain for LAMP reaction
The gradient of mgso4 (8-10-12-16 mM (and betain (0.8-1.6 -2.4-4.8 M) was separately applied to Optimize LAMP reaction, and the results showed that different concentration of these materials was unable to distinguish the mutant and normal alleles.
Discussion
Since the introduction of the LAMP approach, most of the studies have been applying the conventional type of this technique to detect the desired segment of DNA of the pathogens. However, some studies have used this method for genotyping and detection of the SNPs. These studies argued that the AS-LAMP method is a valid method for the detection of the SNPs. the present study aimed to detect the IVSII-I G◊A mutation which is the most frequent mutation among β- thalassemia patients in the north of Iran.
In the ARMS-LAMP method, two sets of LAMP primers are designed to differentiate two different nucleotides at the same position in the target DNA: BIP or FIP primers which are specifically designed complementary to the mutation point at the 3’ end of the B2 primer (5’ end of the BIP primer). The remaining primers, F3, B3, and FIP or BIP, are common for each primer set. Two LAMP reactions should be prepared with each set of primers for each sample.
Several studies have used the AS-LAMP method for SNP detection. For instance, this method was used for the identification of the West African kdr (kdr-w; L1014F) [26]. And G119S ace-1R mutations [27] in field-collected Anopheles gambiae. Yongkiettrakul et al., (2017) designed an SNP-LAMP assay to detect the N51I mutation on the dhfr gene that is associated with pyrimethamine resistance in Plasmodium falciparum with 100% specificity [28].
Ikeda et al. in 2007, designed an ARMS-LAMP approach for the identification of L858R mutation on the epidermal growth factor receptor gene (EGFR) [29]. They used an in situ LAMP in which The FIP and BIP primers were labelled with fluorescein isothiocyanate (FITC) for mutation detection on paraffin-embedded tissues.
In 2017, Tamura et al., Used the ARMS-LAMP method to detect the N526K mutation in the ftsI gene that was involved in H. pylori-resistant Haemophilus influenza with a negative effect on ampicillin beta-lactamase. Using this method, they succeeded in identifying this mutation in the mentioned species [30].
Using the AS-LAMP method, Duan et al., in 2016, identified spot mutations (TTC → TAC, F200Y) in the β2-tubulin gene, which is involved in resistance to benzimidazole in Fusarium asiaticum fungus. The comparison between the results of this method with the PCR test showed that this method could successfully detect this mutation in resistant strains of agricultural products [31].
β-thalassemia is the most common haemoglobin disorder in Iran and IVSII-I G◊A mutation is the most frequent mutation in the north of Iran. Several studies have introduced a variety of methods for the identification of this mutation. Zafari et al., in 2010 developed the real time HRM (High-Resolution Melting) method in which they identified the mutation IVSII-I G◊A on serum samples of pregnant mothers. The purpose of the research was to detect the fetus’s IVSII-I G◊A mutation in the serum of pregnant mothers. The sensitivity and specificity of the introduced method were 92.6% and 82.6%, respectively [32].
Heidari Sharafkhalkali et al., in 2017, using a Ligation Assay method combined with Nanoparticles could successfully identify the IVSII-I G◊A mutation. They used probes that were labelled with magnetic nanoparticles and quantum dots. In the mentioned study, two types of the probe were designed, one complementary to the normal region and another complementary to the mutant area. Fifteen homozygote and 21 heterozygote patients were successfully identified using this method [33].
Hamid et al., in 2012 have also used the HRM (High Resolution Melting) method to identify 4 common mutations in Iran including IVSI-5 (G>C), CD36/37 (-T), IVSII-I (G>A), IVSI-110 (G>A). However, in the IVSII-I G◊A mutation, 2 peaks were observed due to the presence of a polymorphism close to mutation site [34].
Multiplex HRM method to identify common beta-globin gene mutations in Greece was also developed by Chassanidis et al., in 2016. They designed several primers for mutation sites. Several mutations, including IVSII-I, in multiplex conditions, were simultaneously amplified [35].
Haji Hosseini et al., in 2015, using Tetra-Primer ARMS PCR method could detect IVSII-I G◊A and FSC 8/9 InsG mutations in 30 affected beta-thalassemia patients in Isfahan province. This method has been able to differentiate normal, heterozygous, and homozygous samples in patients with 100% specificity and reliability. The frequency of IVSII-I G◊A and FSC 8/9 InsG mutations in Isfahan province were 86.6% and 16.6 % respectively [36].
Honardoost et al., in 2014, compared the results of Tetra Primer ARMS PCR method with conventional ARMS PCR technique to detect IVSII-I mutation in β-thalassemia patients. The PCR-sequencing approach was considered as a standard gold test. Fifty-seven subjects, including 2 homozygous, 49 heterozygotes, and 6 normal samples were analysed using both techniques. They reported that Tetra Primer ARMS-PCR was able to detect IVSII-I mutation with 100% sensitivity and specificity while the conventional ARMS test had a specificity and sensitivity of 100% and 92.55% [37].
In 2015, Alfdali et al., identified common β-globin gene mutations in Egypt using Reverse Dot-Blot method among 40 β-thalassemia patients. Using PCR sequencing as a standard gold method, the results showed that this method had a sensitivity of 92.5% [38].
Although the mentioned studies have successfully used ARMS-LAMP method to detect the SNPs, and other studies use a variety of methods to identify IVSII-I G◊A, the present study was unable to differentiate between a normal allele and IVSII-I G◊A mutation. At the present study, three different primer sets could not detect the mutation in the patient’s DNA and synthetic DNA. Hence further studies are recommended to consider redesigning of primer set, DNA concentration and using commercial LAMP Master Mix to detect the mutation.
Footnotes
Funding: This research did not receive any financial support
Competing Interests: The authors have declared that no competing interests exist
References
- 1.Kosaryan M, Vahidshahi K, Karami H, Forootan MA, Ahangari M. Survival of thalassemic patients referred to the Boo Ali Sina teaching hospital, Sari, Iran. Hemoglobin. 2007;31(4):453–62. doi: 10.1080/03630260701641294. https://doi.org/10.1080/03630260701641294 PMid:17994379. [DOI] [PubMed] [Google Scholar]
- 2.Karimi M, Cohan N, De Sanctis V, Mallat NS, Taher A. Guidelines for diagnosis and management of Beta-thalassemia intermedia. Pediatric hematology and oncology. 2014;31(7):583–96. doi: 10.3109/08880018.2014.937884. https://doi.org/10.3109/08880018.2014.937884 PMid:25247665. [DOI] [PubMed] [Google Scholar]
- 3.Jalali H, Mahdavi MR, Roshan P, Kosaryan M, Karami H, Mahdavi M. Alpha thalassemia gene mutations in neonates from Mazandaran, Iran, 2012. Hematology. 2014;19(4):192–5. doi: 10.1179/1607845413Y.0000000115. https://doi.org/10.1179/1607845413Y.0000000115 PMid:24074530. [DOI] [PubMed] [Google Scholar]
- 4.Rahimi Z, Raygani AV, Merat A, Haghshenass M, Gerard N, Nagel R, et al. Thalassemic mutations in southern Iran. Iranian journal of medical sciences. 2015;31(2) [Google Scholar]
- 5.Kosaryan M, Karami H, Darvishi-Khezri H, Akbarzadeh R, Aliasgharian A, Bromand K. Demographic data of patients with β-thalassemia major recorded in the electronic system in the north of Iran, 2016. Tanzania Journal of Health Research. 2018;20(3) [Google Scholar]
- 6.Valaei A, Bayat F, Kordafshari A, Zeinali S, Karimipoor M. A Novel Polymorphism Causes A Different Restriction Pattern by Rsa I in the β-Globin Gene Cluster:Application in Prenatal Diagnosis. Hemoglobin. 2009;33(6):417–21. doi: 10.3109/03630260903327817. https://doi.org/10.3109/03630260903327817 PMid:19958186. [DOI] [PubMed] [Google Scholar]
- 7.Karimi M, Jamalian N, Yarmohammadi H, Askarnejad A, Afrasiabi A, Hashemi A. Premarital screening for β-thalassaemia in Southern Iran:options for improving the programme. Journal of Medical Screening. 2007;14(2):62–6. doi: 10.1258/096914107781261882. https://doi.org/10.1258/096914107781261882 PMid:17626703. [DOI] [PubMed] [Google Scholar]
- 8.Akhavan-Niaki H, Derakhshandeh-Peykar P, Banihashemi A, Mostafazadeh A, Asghari B, Ahmadifard M-R, et al. A comprehensive molecular characterization of beta thalassemia in a highly heterogeneous population. Blood Cells, Molecules, and Diseases. 2011;47(1):29–32. doi: 10.1016/j.bcmd.2011.03.005. https://doi.org/10.1016/j.bcmd.2011.03.005 PMid:21493114. [DOI] [PubMed] [Google Scholar]
- 9.Samavat A, Modell B. Iranian national thalassaemia screening programme. Bmj. 2004;329(7475):1134–7. doi: 10.1136/bmj.329.7475.1134. https://doi.org/10.1136/bmj.329.7475.1134 PMid:15539666 PMCid:PMC527686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khorasani G, Kosaryan M, Vahidshahi K, Shakeri S, Nasehi MM. Results of the national program for prevention of β-thalassemia major in the Iranian Province of Mazandaran. Hemoglobin. 2008;32(3):263–71. doi: 10.1080/03630260802004269. https://doi.org/10.1080/03630260802004269 PMid:18473242. [DOI] [PubMed] [Google Scholar]
- 11.Fallah MS, Samavat A, Zeinali S. Iranian national program for the prevention of thalassemia and prenatal diagnosis:mandatory premarital screening and legal medical abortion. Prenatal Diagnosis:Published in Affiliation With the International Society for Prenatal Diagnosis. 2009;29(13):1285–6. doi: 10.1002/pd.2373. https://doi.org/10.1002/pd.2373 PMid:19953581. [DOI] [PubMed] [Google Scholar]
- 12.Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies-a review. Nucleosides, Nucleotides, and Nucleic Acids. 2008;27(3):224–43. doi: 10.1080/15257770701845204. https://doi.org/10.1080/15257770701845204 PMid:18260008. [DOI] [PubMed] [Google Scholar]
- 13.Karami A, Gill P, Motamedi MHK, Saghafinia M. A review of the current isothermal amplification techniques:Applications, advantages and disadvantages. J Global Infect Dis. 2011;3(3):293–302. https://doi.org/10.4103/0974-777X.83538. [Google Scholar]
- 14.Guichón A, Chiparelli H, Martınez A, Rodrıguez C, Trento A, Russi JC, et al. Evaluation of a new NASBA assay for the qualitative detection of hepatitis C virus based on the NucliSens®Basic Kit reagents. Journal of clinical virology. 2004;29(2):84–91. doi: 10.1016/s1386-6532(03)00091-x. https://doi.org/10.1016/S1386-6532(03)00091-X. [DOI] [PubMed] [Google Scholar]
- 15.Bremer J, Nowicki M, Beckner S, Brambilla D, Cronin M, Herman S, et al. Comparison of two amplification technologies for detection and quantitation of human immunodeficiency virus type 1 RNA in the female genital tract. Journal of clinical microbiology. 2000;38(7):2665–9. doi: 10.1128/jcm.38.7.2665-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent M, Xu Y, Kong H. Helicase?dependent isothermal DNA amplification. EMBO reports. 2004;5(8):795–800. doi: 10.1038/sj.embor.7400200. https://doi.org/10.1038/sj.embor.7400200 PMid:15247927 PMCid:PMC1249482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fire A, Xu S-Q. Rolling replication of short DNA circles. Proceedings of the National Academy of Sciences. 1995;92(10):4641–5. doi: 10.1073/pnas.92.10.4641. https://doi.org/10.1073/pnas.92.10.4641 PMid:7753856 PMCid:PMC42000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature genetics. 1998;19(3):225. doi: 10.1038/898. https://doi.org/10.1038/898 PMid:9662393. [DOI] [PubMed] [Google Scholar]
- 19.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al. Comprehensive human genome amplification using multiple displacement amplification. Proceedings of the National Academy of Sciences. 2002;99(8):5261–6. doi: 10.1073/pnas.082089499. https://doi.org/10.1073/pnas.082089499 PMid:11959976 PMCid:PMC122757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic acids research. 2000;28(12):e63–e. doi: 10.1093/nar/28.12.e63. https://doi.org/10.1093/nar/28.12e63 PMid:10871386 PMCid:PMC102748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafati A, Gill P. Microfluidic method for rapid turbidimetric detection of the DNA of Mycobacterium tuberculosis using loop-mediated isothermal amplification in capillary tubes. Microchimica Acta. 2015;182(3-4):523–30. https://doi.org/10.1007/s00604-014-1354-y. [Google Scholar]
- 22.Li Y, Fan P, Zhou S, Zhang L. Loop-mediated isothermal amplification (LAMP):a novel rapid detection platform for pathogens. Microbial pathogenesis. 2017;107:54–61. doi: 10.1016/j.micpath.2017.03.016. https://doi.org/10.1016/j.micpath.2017.03.016 PMid:28323152. [DOI] [PubMed] [Google Scholar]
- 23.Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP):recent progress in research and development. Journal of infection and chemotherapy. 2013;19(3):404–11. doi: 10.1007/s10156-013-0590-0. https://doi.org/10.1007/s10156-013-0590-0 PMid:23539453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP):a rapid, accurate, and cost-effective diagnostic method for infectious diseases. Journal of infection and chemotherapy. 2009;15(2):62–9. doi: 10.1007/s10156-009-0669-9. https://doi.org/10.1007/s10156-009-0669-9 PMid:19396514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law JW-F, Ab Mutalib N-S, Chan K-G, Lee L-H. Rapid methods for the detection of foodborne bacterial pathogens:principles, applications, advantages and limitations. Frontiers in microbiology. 2015;5:770. doi: 10.3389/fmicb.2014.00770. https://doi.org/10.3389/fmicb.2014.00770 PMid:25628612 PMCid:PMC4290631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Namountougou M, Diabaté A, Etang J, Bass C, Sawadogo SP, Gnankinié O, et al. First report of the L1014S kdr mutation in wild populations of Anopheles gambiae M and S molecular forms in Burkina Faso (West Africa) Acta tropica. 2013;125(2):123–7. doi: 10.1016/j.actatropica.2012.10.012. https://doi.org/10.1016/j.actatropica.2012.10.012 PMid:23128044. [DOI] [PubMed] [Google Scholar]
- 27.Badolo A, Bando H, Traoré A, Ko-Ketsu M, Guelbeogo WM, Kanuka H, et al. Detection of G119S ace-1 R mutation in field-collected Anopheles gambiae mosquitoes using allele-specific loop-mediated isothermal amplification (AS-LAMP) method. Malaria journal. 2015;14(1):477. doi: 10.1186/s12936-015-0968-9. https://doi.org/10.1186/s12936-015-0968-9 PMid:26620269 PMCid:PMC4665897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yongkiettrakul S, Kampeera J, Chareanchim W, Rattanajak R, Pornthanakasem W, Kiatpathomchai W, et al. Simple detection of single nucleotide polymorphism in Plasmodium falciparum by SNP-LAMP assay combined with lateral flow dipstick. Parasitology international. 2017;66(1):964–71. doi: 10.1016/j.parint.2016.10.024. https://doi.org/10.1016/j.parint.2016.10.024 PMid:27816495. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda S, Takabe K, Inagaki M, Funakoshi N, Suzuki K. Detection of gene point mutation in paraffin sections using in situ loop?mediated isothermal amplification. Pathology international. 2007;57(9):594–9. doi: 10.1111/j.1440-1827.2007.02144.x. https://doi.org/10.1111/j.1440-1827.2007.02144.x PMid:17685931. [DOI] [PubMed] [Google Scholar]
- 30.Tamura S, Maeda T, Misawa K, Osa M, Hamamoto T, Yuki A, et al. Development of a highly resolved loop-mediated isothermal amplification method to detect the N526K ftsI mutation of β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Journal of microbiological methods. 2017;141:108–14. doi: 10.1016/j.mimet.2017.08.008. https://doi.org/10.1016/j.mimet.2017.08.008 PMid:28807759. [DOI] [PubMed] [Google Scholar]
- 31.Duan Y, Yang Y, Li T, Zhao D, Cao J, Shi Y, et al. Development of a rapid and high?throughput molecular method for detecting the F200Y mutant genotype in benzimidazole?resistant isolates of Fusarium asiaticum. Pest management science. 2016;72(11):2128–35. doi: 10.1002/ps.4243. https://doi.org/10.1002/ps.4243 PMid:26823005. [DOI] [PubMed] [Google Scholar]
- 32.Zafari M, Gill P, Kowsaryan M, Alipour A, Banihashemi A. High-resolution melting analysis for noninvasive prenatal diagnosis of IVS-II-I (GA) fetal DNA in minor beta-thalassemia mothers. The Journal of Maternal-Fetal &Neonatal Medicine. 2016;29(20):3323–8. doi: 10.3109/14767058.2015.1124263. [DOI] [PubMed] [Google Scholar]
- 33.Motovali-Bashi M, Gill P. The sensitive detection of IVSII-1 (G? A) mutation in beta globin gene using a Nano-based ligation genotyping system. Gene. 2018 doi: 10.1016/j.gene.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Hamid M. Designing a Rapid Screening Method for Detection of Four? Common Beta Globin Gene Mutations in Iranian Population by? High-Resolution Melting Analysis. Pathobiology Research. 2018;21(2):101–6. [Google Scholar]
- 35.Chassanidis C, Boutou E, Voskaridou E, Balassopoulou A. Development of a high-resolution melting approach for scanning beta globin gene point mutations in the Greek and other Mediterranean populations. PloS one. 2016;11(6):e0157393. doi: 10.1371/journal.pone.0157393. https://doi.org/10.1371/journal.pone.0157393 PMid:27351925 PMCid:PMC4924799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hajihoseini S, Motovali-Bashi M, Honardoost MA, Alerasool N. Tetra-primer ARMS PCR optimization for detection of IVS-II-I (GA) and FSC 8/9 InsG mutations in β-thalassemia major patients in Isfahan population. Iranian journal of public health. 2015;44(3):380. [PMC free article] [PubMed] [Google Scholar]
- 37.Honardoost MA, Tabatabaeian H, Akbari M, Salehi M. Investigation of sensitivity, specificity and accuracy of Tetra primer ARMS PCR method in comparison with conventional ARMS PCR, based on sequencing technique outcomes in IVS-II-I genotyping of beta thalassemia patients. Gene. 2014;549(1):1–6. doi: 10.1016/j.gene.2014.05.071. https://doi.org/10.1016/j.gene.2014.05.071 PMid:24946023. [DOI] [PubMed] [Google Scholar]
- 38.El-Fadaly N, Abd-Elhameed A, Abd-Elbar E, El-Shanshory M. Accuracy of reverse Dot-Blot PCR in detection of different β-Globin gene mutations. Indian Journal of Hematology and Blood Transfusion. 2016;32(2):239–43. doi: 10.1007/s12288-015-0553-y. https://doi.org/10.1007/s12288-015-0553-y PMid:27065589 PMCid:PMC4788995. [DOI] [PMC free article] [PubMed] [Google Scholar]


