Abstract
Over the last decades, the prevalence of drug-resistance in Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, has increased. These findings have rekindled interest in elucidating the unique adaptive molecular and biochemistry physiology of Mycobacterium. The use of metabolite profiling independently or in combination with other levels of “-omic” analyses has proven an effective approach to elucidate key physiological/biochemical mechanisms associated with Mtb throughout infection. The following review discusses the use of metabolite profiling in the study of tuberculosis, future approaches, and the technical and logistical limitations of the methodology.
Keywords: mycobacteria, metabolomics, anti-TB drugs
1. Introduction
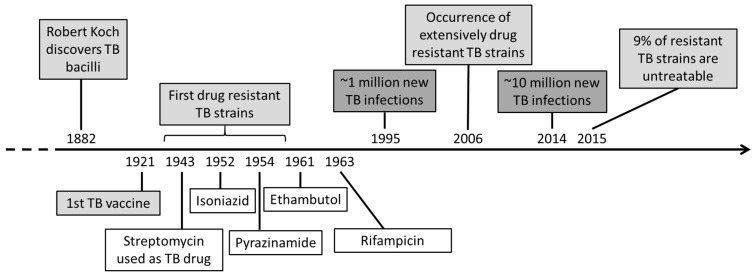
Mycobacterium tuberculosis (Mtb) is the causative agent for tuberculosis (TB), causing millions of new infection cases and deaths every year [1]. Since its discovery in 1882 by Robert Koch, many studies have focused on the understanding of the bacilli as well as the treatment and prevention of TB infection (Figure 1). In 1921, the first TB vaccine (Mycobacterium bovis Bacillus Calmette-Guérin) was discovered, followed by the first anti-TB drugs during WWII [2,3]. Nevertheless, almost a century later, research is still focused on the eradication of the TB epidemic.
Figure 1.
Timeline of tuberculosis (TB) from its discovery to treatments, current case numbers and TB research.
The unique properties of pathogenic mycobacteria provide an advantage over the progression of the infection and against drugs [4,5]. The most prominent properties are the nonreplicating state (NRP), during which the bacilli reduce their metabolic activity to enable long-term viability [6,7], and the mycobacterial cell envelope, which undergoes structural and functional changes under oxygen limiting conditions [8]. The lipid layers of the cell wall form a considerable barrier for the transport of compounds into the cell, preventing drugs from reaching their intracellular targets [7,9,10]. Additionally, the number of mycobacteria developing multidrug-resistance (MDR) to the standard anti-TB drugs increased rapidly over the last few decades [11]. The cause of resistance is known for most of these standard drugs and has resulted in renewed interest for alternative drug target sites [12]. Hence, an important component of studies for new TB therapeutics is the detailed understanding of the metabolism of bacilli across their life cycle [13].
2. Metabolite Profiling a “New” Approach for Drug Discovery
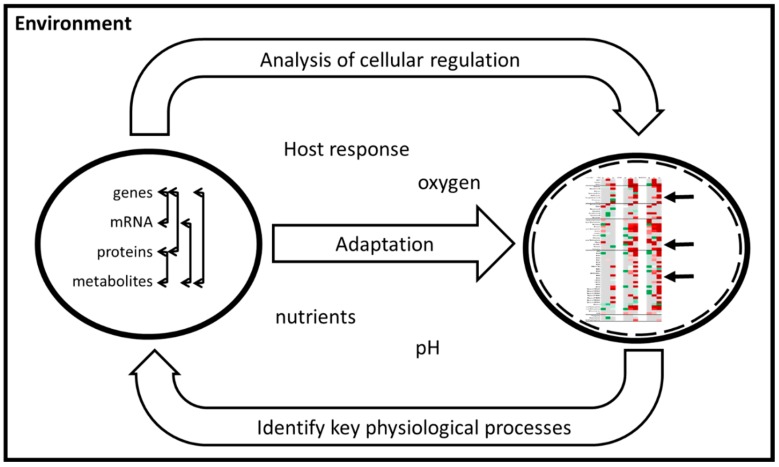
Bacteria are unicellular systems but still have complex cellular regulatory networks that require analysis at different levels (genome, transcriptome, proteome and metabolome) in order to gain a more holistic understanding of the processes involved. Systems Biology as a discipline has evolved and aims to decipher relationships between the different facets of cellular regulation. Underpinning knowledge, from the understanding of the dynamic behaviour of the system as a whole and interactions between the cell/pathogen and its environment/host (Figure 2), can be exploited in the design of new antibiotics [14,15,16]. Many studies, such as the Mtb genome scale model (GSMN), have highlighted that metabolic analysis is needed for a comprehensive analysis and to fill gaps in the reactions predicted from genome annotation [17,18,19].
Figure 2.
Role of systems biology in understanding key physiological processes of the TB bacilli and intracellular regulation under adaptation to the environment. Arrows represent interaction of intracellular regulation molecules (left circle) and changes of metabolites (right circle).
The metabolome comprises small molecular weight molecules (e.g., sugars) as well as components of larger macromolecules (e.g., amino acids for proteins). Metabolic analysis represents a measure of these compounds and components involved in cellular regulation [20] and can be divided into three different types: Chemical fingerprinting (general screen of the metabolome), metabolite profiling (detailed analysis of a defined group of metabolites) and targeted analysis (accurate analysis of specific metabolites) [21]. The analytical platforms for all metabolic analysis include chromatography often coupled to mass spectrometry. To minimise the analytical procedures, the platform applied needs to be able to analyse metabolites varying in mass and polarity. The more advanced the methods become, the easier it is to compare detected features to published metabolite libraries [22,23,24]. Additionally, the aim of the study defines on which metabolite class the analytical focus is based (e.g., end-products such as lipids or metabolites associated with intermediary metabolism) and contributes to the analytical platform used or utilised [22,25,26].
2.1. Understanding Mtb Properties through Metabolite Studies
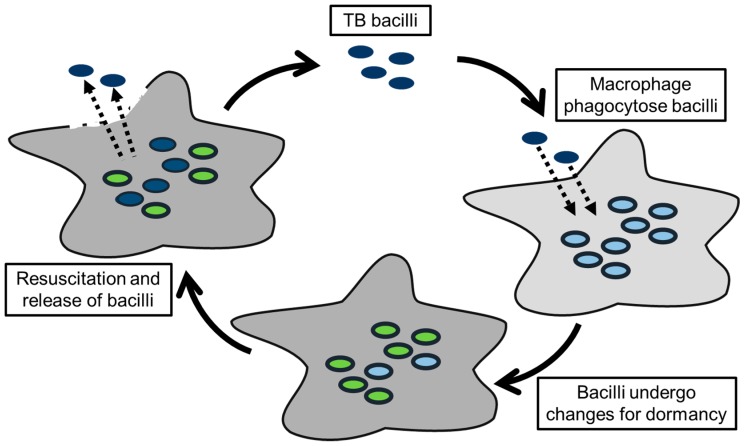
The understanding of pathogens comprises the identification of compounds involved in virulence as well as the elucidation of intracellular changes throughout the infection cycle (Figure 3). The main compounds related to virulence in Mtb are associated with the cell wall and its remodelling/stabilisation during the infection of macrophages. The thickening of the cell wall (higher cross-linking of the peptidoglycan) and modification of cell wall lipids promotes the cell wall rigidity and enables survival in the hostile granuloma environment with a low oxygen content and an acidic pH [27,28]. Many of these lipids (e.g., sulfolipids and trehalose dimycolates) act as virulence factors and induce an immune response in the infected tissue [22,29]. Metabolic analysis facilitated discovery of other cell wall components directly and indirectly involved in virulence. These components are isopentenyl pyrophosphate (IPP)-derived compounds, such as polyprenol carrier lipids, menaquinone, isotuberculosinol and carotenoids, and are involved in the transport of saccharides and mycolic acids for the cell wall, electron transport chain, arrest of phagosomal maturation as well as free radical scavenging [30,31,32,33]. A reduction or even lack of these compounds has been reported to lead to defects of the lipoglycan biosynthesis and a reduction of virulence [34,35,36,37]. In mycobacteria, IPP is produced via a different pathway to that found in human cells, which presents a valuable exploitable drug target [38].
Figure 3.
TB infection cycle indicates intracellular changes of the bacilli during the different stages of the infection. The first adaptations occur during phagocytosis of the bacilli by the macrophages. The inner environment of the macrophages starts changing, which causes further adaptations of the bacilli and can include the switch to dormant bacilli. The last adaptation includes the resuscitation from dormant to active bacilli and the changes during airborne transmission.
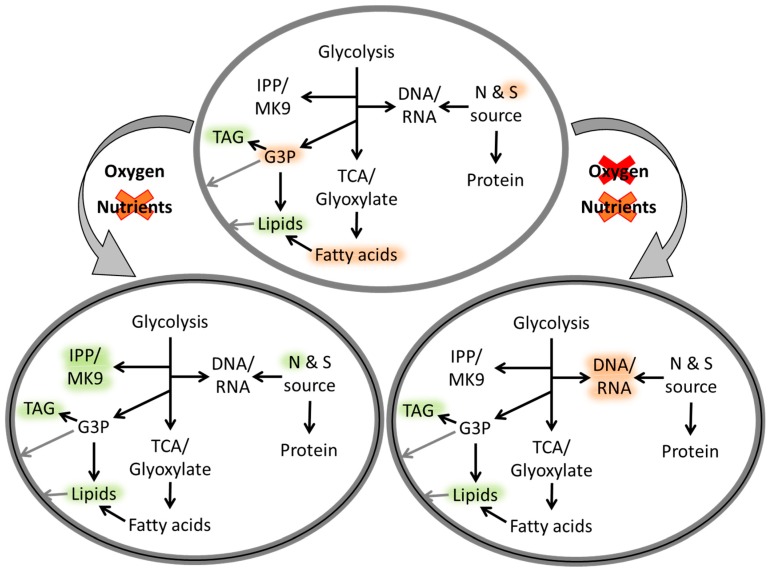
Over the course of the infection, a significant reprogramming of transcription in Mtb can be detected and is primarily influenced by the availability of nutrients and oxygen at each infection stage [39]. The resulting intracellular changes at these stages included the reduction of metabolic activity/transcription, the redirection of the carbon flow and the induction of alternative metabolic pathways, such as the glyoxylate shunt and methyl citrate cycle [7] (Figure 4). The purpose of this reprogramming was attributed to the utilisation of alternative carbon sources (e.g., lipids from the host) and eliminated by-products (e.g., propanoate created during the β-oxidation of odd-number chain fatty acids and production of triacylglycerides as carbon/energy storage) [40,41]. One key enzyme related to most of the processes mentioned was identified as isocitrate lyase (ICL) that converts isocitrate and methyl isocitrate for the alternative TCA cycles [42]. ICL-deficient mycobacteria accumulate precursors associated with these pathways, which leads to bactericidal enzyme inhibition [43].
Figure 4.
Schematic representation of metabolic processes of mycobacterial cells under changes/depletion of oxygen and nutrients levels (grey curved arrows), adapted from Drapal, M.; et al. Microbiology 2016, 162, 1456–1467. Copyright © 2016 Microbiology Society [45]. Intracellular metabolic processes are indicated with black arrows and regulation highlighted as up (green) or down (red).
The culture conditions mimicked by many studies involve a partially aerated or hypoxic NRP state, typically designated on the basis of protocols from Wayne and Hayes [44], which documented an orderly downregulation of the intermediary metabolism, e.g., [27]. Other studies comparing changes of metabolite levels reported a transcriptional mode of regulation under aerated conditions and a more complex metabolic regulation in the presence of hypoxic conditions, e.g., [45]. Despite evident discrepancies about the mode of regulation in previous studies, overall data identified compounds utilised as storage compounds (e.g., succinic acid, triacylglycerides and poly glutamic acid) and arising during the immediate response to resuscitation. Most of these compounds accumulate intracellularly, but some are transported outside the cell, to reduce toxicity and resulting cellular dysfunction [45,46,47]. The process of resuscitation, which can occur quite suddenly [39], is still poorly understood, as it is difficult to establish the exogenous factors influencing this process [7].
2.2. New Drug Targets and Understanding Drug Resistance Based on Metabolic Studies
Much progress has been achieved in identifying new anti-TB drugs and drug targets. However, despite the promising results, many drugs against MDR Mtb strains (e.g., d-cycloserine) tend to lead to serious side effects in the host [48]. In general, research for anti-TB drugs has been focused on intracellular targets both under aerobic and hypoxic conditions and on weakening the mycobacterial cell wall for more drug accessibility. This led to the discovery of metronidazole, which targets DNA in anaerobe bacteria without antagonistic effects in combination with isoniazid or rifampicin [49]. Other studies identified methionine synthesis, important for nucleotide and protein synthesis, as a viable target, as human cells do not synthesize this amino acid [50,51]. The permeability of the cell wall can be influenced by a diverse range of compounds. Examples are trehalose, a saccharide utilised as backbone for mycolic acids, and IPP-derived carrier lipids for cell wall components. The latter can be affected by the inhibition of IPP synthesis through fosmidomycin, a known malaria drug [52,53]. In addition to the permeability, efflux pumps play an important role in decreasing the concentration and therefore efficiency of drugs already within the bacterial cell [54]. Metabolite profiling offers a means of assessing known inhibitor targets, secondary targets and the holistic effects of these drugs/inhibitors on the metabolome under different conditions. In combination with standardised metabolite profiles for known well-characterised drugs, metabolomics can also be used to classify new chemical inhibitors and elucidate their modes of action [55]. Additionally, the metabolic analysis of resistant mycobacteria can help to understand why the resistance occurs and whether/how it can be circumvented as reviewed previously [56,57,58].
3. Advantages and Disadvantages of Metabolite Profiling Studies
3.1. Model Organisms for Mtb
Mtb is logistically difficult to work with due to biosafety risks and slow growth. Hence, suitability of model organisms has led to intense debates [59,60,61]. The most common model species are Mycobacterium smegmatis and M. bovis BCG. M. smegmatis is a fast-growing species and has ~1.7-times larger genome than Mtb [62,63]. M. smegmatis is nonpathogenic, even though it shares 12 out of 19 virulence gene homologues found in Mtb. This challenges the adequacy of M. smegmatis for pathogenicity studies [61]. Furthermore, M. smegmatis develops a spore-like shape under oxygen limitation with a cell morphology different to slow-growing mycobacteria [64]. M. bovis BCG is a slow-grower and closely related to Mtb. They share ~99.9% of the DNA and, for unknown reasons, M. bovis BCG never regained virulence [4,65]. A comparison of five Mycobacterium species (M. bovis BCG, Mycobacterium avium, Mycobacterium intracellulare, M. smegmatis and Mycobacterium phlei) with different growth rates and pathogenicity, showed that: (i) Their only common trait was the metabolic processes related to the cell wall and (ii) the metabolic changes under aerobic and hypoxic conditions emphasise differences on a genetic level between fast- and slow-growing mycobacteria [45]. Conclusions from other studies highlighted that the model Mycobacterium species for Mtb depends on the purpose of the metabolic study, and the use of phenotypically closer-related species (e.g., M. bovis BCG) is recommended for specialised pathways. Nevertheless, a metabolic comparison of nonpathogenic and pathogenic mycobacteria might provide valuable insights into virulence and ‘dormancy’, as the processes involved (e.g., glyoxylate shunt, central carbon metabolism) often also occur in nonpathogenic mycobacteria [42,59,66]. Metabolomic studies facilitate chemotyping different/diverse strains and identifying subtle differences in their steady-state metabolite levels that can eventually be utilised as differentiating biomarkers [42].
3.2. Heterogeneous Cultures
The phenomenon of heterogeneous mycobacterial cultures, comprising active and ‘dormant’ bacilli, bears an obstacle for data interpretation of metabolite profiling [67]. The detected metabolite changes of the culture/tissue samples contain information for both states which cannot be related to one single state and, hence, does not give accurate details about the processes involved in transition between active and ‘dormant’ bacilli. Previous studies implied ‘dormancy’ as a redirection of metabolic activity under unfavourable growth conditions, e.g., [46]. The renewed growth rate and metabolic activity, detected after 28 days under aerated and hypoxic conditions, indicate that the process of ‘dormancy’ does not involve the whole culture [45]. Further analysis of such growth conditions is of importance for a better understanding of the resuscitation process, which could just reside from a delayed log phase of very few active bacilli in the culture instead of the assumed complex and sudden occurrence [7,39].
3.3. In Vitro Versus in Vivo Studies
Metabolite profiling can not only analyse cellular compounds but also the culture composition affecting the metabolism of the strains. In vitro studies represent a regulated environment with set parameters mimicking an in vivo study, which includes a far broader range of parameters and influencing factors, of which many can be potentially unknown. In the case of a Mtb infection, these parameters include all the stages experienced from active replication in the alveolar macrophages to dormant cells within granulomas [7]. Hence, an in vitro study mimicking set conditions might be more helpful, as the first step to elucidate complex processes (e.g., ‘dormant state’ of mycobacteria) compared to an in vivo study. A main disadvantage of an in vitro study is cell concentration, which is usually higher in in vitro studies compared to human sputum with 101–105 cells/ml sputum [68,69].
The introduction of metabolomics for TB research led to major advances in diagnostic tools and the continuous replacement of tradition methods such as sputum smear tests with non-invasive techniques [70,71]. The increasing number of metabolic studies led to the discovery of new TB biomarkers (e.g., volatiles and specific lipids) in plasma and urine samples [24,72]. The advancement of high-resolution metabolomics can distinguish between human and TB metabolites, elucidating the progression of TB in the human host and the host response [73]. Furthermore, the higher resolution of the technique facilitates the identification of biomarkers to a high confidence level with the libraries available [74].
3.4. Limitation to Metabolite Profiling
The metabolome comprises a broad range of compounds that differ in their properties and dynamic range. A complete study of the whole metabolome would require analysis with different platforms and techniques such as steady-state levels or fluxomics [23,26,75]. Studies focused on an overview of the metabolic steady-state levels have shown a strong correlation to the literature and endorsed the importance of metabolic studies [45]. Hence, it is of great value to integrate recently acquired results with published data concerning metabolic biology studies as well as those representing other levels of cellular regulation. This should also include techniques such as electron microscopy to gain a comprehensive understanding of cell biology and how it is influenced by changes in metabolite composition [6].
4. Conclusions
The consistent threat of the TB epidemic and the occurrence of resistance to existing antibiotics challenges the community to better understand Mtb and its infection cycle. Metabolite profiling is a new tool in this research area that can be used to elucidate key physiological properties of Mtb throughout infection. To date, the most prevalent studies relate to cell wall metabolism and the production of storage compounds during ‘dormancy’. The metabolomics platforms now established can be used to (i) generate characteristic biochemical signatures for specific Mycobacterium strains, (ii) identify specific developmental stages in their life cycle and (iii) elucidate metabolic adaptation to their environment. Despite these notable advances, the need for better metabolite annotation and integration with other “omic” technologies to generate exploitable network models remains.
Funding
This work was supported by a Royal Holloway University of London studentship to P.D.F.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.WHO . Global Tuberculosis Report 2018. WHO Press; Geneva, Switzerland: 2018. p. 1. [Google Scholar]
- 2.Luca S., Mihaescu T. History of BCG Vaccine. Mædica. 2013;8:53–55. [PMC free article] [PubMed] [Google Scholar]
- 3.Lugosi L. Theoretical and methodological aspects of BCG vaccine from the discovery of Calmette and Guérin to molecular biology. A review. Tuber. Lung Dis. 1992;73:252–261. doi: 10.1016/0962-8479(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 4.Brosch R., Pym A.S., Gordon S.V., Cole S.T. The evolution of mycobacterial pathogenicity: Clues from comparative genomics. Trends Microbiol. 2001;9:452–458. doi: 10.1016/S0966-842X(01)02131-X. [DOI] [PubMed] [Google Scholar]
- 5.Russell D.G., Cardona P.J., Kim M.J., Allain S., Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anuchin A.M., Mulyukin A.L., Suzina N.E., Duda V.I., El-Registan G.I., Kaprelyants A.S. Dormant forms of Mycobacterium smegmatis with distinct morphology. Microbiology. 2009;155:1071–1079. doi: 10.1099/mic.0.023028-0. [DOI] [PubMed] [Google Scholar]
- 7.Chao M.C., Rubin E.J. Letting sleeping dos lie: Does dormancy play a role in tuberculosis? Annu. Rev. Microbiol. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 8.Gago G., Diacovich L., Arabolaza A., Tsai S.C., Gramajo H. Fatty acid biosynthesis in actinomycetes. FEMS Microbiol. Rev. 2011;35:475–497. doi: 10.1111/j.1574-6976.2010.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakousis P.C., Williams E.P., Bishai W.R. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2008;61:323–331. doi: 10.1093/jac/dkm485. [DOI] [PubMed] [Google Scholar]
- 10.Niederweis M. Nutrient acquisition by mycobacteria. Microbiology. 2008;154:679–692. doi: 10.1099/mic.0.2007/012872-0. [DOI] [PubMed] [Google Scholar]
- 11.WHO . Treatment of Tuberculosis Guidelines. 4th ed. WHO Press; Geneva, Switzerland: 2010. p. 83. [Google Scholar]
- 12.Da Silva P.E.A., Palomino J.C. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: Classical and new drugs. J. Antimicrob. Chemother. 2011;66:1417–1430. doi: 10.1093/jac/dkr173. [DOI] [PubMed] [Google Scholar]
- 13.Jamshidi N., Palsson B. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst. Biol. 2007;1:26. doi: 10.1186/1752-0509-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beste D.J., Bonde B., Hawkins N., Ward J.L., Beale M.H., Noack S., Nöh K., Kruger N.J., Ratcliffe R.G., McFadden J. 13C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 2011;7:e1002091. doi: 10.1371/journal.ppat.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Carvalho L.P.S., Fischer S.M., Marrero J., Nathan C., Ehrt S., Rhee K.Y. Metabolomics of Mycobacterium tuberculosis Reveals Compartmentalized Co-Catabolism of Carbon Substrates. Chem. Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Boshoff H.I., Lun D.S. Systems biology approaches to understanding mycobacterial survival mechanisms. Drug Discov. Today Dis. Mech. 2010;7:e75–e82. doi: 10.1016/j.ddmec.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler P.R., Coldham N.G., Keating L., Gordon S.V., Wooff E.E., Parish T., Hewinson R.G. Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic Mycobacteria. J. Biol. Chem. 2005;280:8069–8078. doi: 10.1074/jbc.M412540200. [DOI] [PubMed] [Google Scholar]
- 18.Sareen D., Newton G.L., Fahey R.C., Buchmeier N.A. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 2003;185:6736–6740. doi: 10.1128/JB.185.22.6736-6740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beste D.J.V., Hooper T., Stewart G., Bonde B., Avignone-Rossa C., Bushell M., Wheeler P., Klamt S., Kierzek A.M., McFadden J. GSMN-TB: A web-based genome scale network model of Mycobacterium tuberculosis metabolism. Genome Biol. 2007;8:R89. doi: 10.1186/gb-2007-8-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrigan G.G., Goodacre R. Introduction. In: Harrigan G.G., Goodacre R., editors. Metabolic Profiling—Its Role in Biomarker Discovery and Gene Function Analysis. Kluwer Academic Publishers; Boston, MA, USA: 2003. pp. 1–9. [Google Scholar]
- 21.Halket J.M., Waterman D., Przyborowska A.M., Patel R.K., Fraser P.D., Bramley P.M. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005;56:219–243. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 22.Layre E., Sweet L., Hong S., Madigan C.A., Desjardins D., Young D.C., Cheng T.Y., Annand J.W., Kim K., Shamputa I.C., et al. A Comparative Lipidomics Platform for Chemotaxonomic Analysis of Mycobacterium tuberculosis. Chem. Biol. 2011;18:1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartain M.J., Dick D.L., Rithner C.D., Crick D.C., Belisle J.T. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J. Lipid Res. 2011;52:861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau S.K.P., Lee K.-C., Curreem S.O.T., Chow W.-N., To K.K.W., Hung I.F.N., Ho D.T.Y., Sridhar S., Li I.W.S., Ding V.S.Y., et al. Metabolomic Profiling of Plasma from Patients with Tuberculosis by Use of Untargeted Mass Spectrometry Reveals Novel Biomarkers for Diagnosis. J. Clin. Microbiol. 2015;53:3750. doi: 10.1128/JCM.01568-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivier I., Loots D.T. A metabolomics approach to characterise and identify various Mycobacterium species. J. Microbiol. Methods. 2012;88:419–426. doi: 10.1016/j.mimet.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y.J., Shui W., Myers S., Feng X., Bertozzi C., Keasling J.D. Central metabolism in Mycobacterium smegmatis during the transition from O(2)-rich to O(2)-poor conditions as studied by isotopomer-assisted metabolite analysis. Biotechnol. Lett. 2009;31:1233–1240. doi: 10.1007/s10529-009-9991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunningham A.F., Spreadbury C.L. Mycobacterial stationary phase induced by low oxygen tension: Cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J. Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sassetti C.M., Rubin E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esmail H., Barry C.E., 3rd, Wilkinson R.J. Understanding latent tuberculosis: The key to improved diagnostic and novel treatment strategies. Drug Discov. Today. 2012;17:514–521. doi: 10.1016/j.drudis.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crick D.C., Schulbach M.C., Zink E.E., Macchia M., Barontini S., Besra G.S., Brennan P.J. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 2000;182:5771–5778. doi: 10.1128/JB.182.20.5771-5778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathew R., Kruthiventi A.K., Prasad J.V., Kumar S.P., Srinu G., Chatterji D. Inhibition of mycobacterial growth by plumbagin derivatives. Chem. Biol. Drug Des. 2010;76:34–42. doi: 10.1111/j.1747-0285.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- 32.Provvedi R., Kocíncová D., Donà V., Euphrasie D., Daffé M., Etienne G., Manganelli R., Reyrat J.M. SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J. Bacteriol. 2008;190:7859–7863. doi: 10.1128/JB.00714-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Besra G.S., Sievert T., Lee R.E., Slayden R.A., Brennan P.J., Takayama K. Identification of the apparent carrier in mycolic acid synthesis. Proc. Natl. Acad. Sci. USA. 1994;91:12735. doi: 10.1073/pnas.91.26.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prach L., Kirby J., Keasling J.D., Alber T. Diterpene production in Mycobacterium tuberculosis. FEBS J. 2010;277:3588–3595. doi: 10.1111/j.1742-4658.2010.07767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann F.M., Xu M., Davenport E.K., Peters R.J. Functional characterization and evolution of the isotuberculosinol operon in Mycobacterium tuberculosis and related Mycobacteria. Front. Microbiol. 2012;3:368. doi: 10.3389/fmicb.2012.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhiman R.K., Mahapatra S., Slayden R.A., Boyne M.E., Lenaerts A., Hinshaw J.C., Angala S.K., Chatterjee D., Biswas K., Narayanasamy P., et al. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol. Microbiol. 2009;72:85–97. doi: 10.1111/j.1365-2958.2009.06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viveiros M., Krubasik P., Sandmann G., Houssaini-Iraqui M. Structural and functional analysis of the gene cluster encoding carotenoid biosynthesis in Mycobacterium aurum A+ FEMS Microbiol. Lett. 2000;187:95–101. doi: 10.1111/j.1574-6968.2000.tb09143.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhao L., Chang W.-C., Xiao Y., Liu H.-W., Liu P. Methylerythritol Phosphate Pathway of Isoprenoid Biosynthesis. Annu. Rev. Biochem. 2013;82:497–530. doi: 10.1146/annurev-biochem-052010-100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wayne L.G., Sohaskey C.D. Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 40.Hampshire T., Soneji S., Bacon J., James B.W., Hinds J., Laing K., Stabler R.A., Marsh P.D., Butcher P.D. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: A model for persistent organisms? Tuberculosis. 2004;84:228–238. doi: 10.1016/j.tube.2003.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirakova T.D., Dubey V.S., Deb C., Daniel J., Korotkova T.A., Abomoelak B., Kolattukudy P.E. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology. 2006;152:2717–2725. doi: 10.1099/mic.0.28993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee K.Y., de Carvalho L.P.S., Bryk R., Ehrt S., Marrero J., Park S.W., Schnappinger D., Venugopal A., Nathan C. Central carbon metabolism in Mycobacterium tuberculosis: An unexpected frontier. Trends Microbiol. 2011;19:307–314. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eoh H., Rhee K.Y. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc. Natl. Acad. Sci. USA. 2014;111:4976–4981. doi: 10.1073/pnas.1400390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wayne L.G., Hayes L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drapal M., Wheeler P.R., Fraser P.D. Metabolite analysis of Mycobacterium species under aerobic and hypoxic conditions reveals common metabolic traits. Microbiology. 2016;162:1456–1467. doi: 10.1099/mic.0.000325. [DOI] [PubMed] [Google Scholar]
- 46.Eoh H., Rhee K.Y. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2013;110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi D., Chandra H., Bhatnagar R. Poly-L-glutamate/glutamine synthesis in the cell wall of Mycobacterium bovis is regulated in response to nitrogen availability. BMC Microbiol. 2013;13:226. doi: 10.1186/1471-2180-13-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halouska S., Chacon O., Fenton R.J., Zinniel D.K., Barletta R.G., Powers R. Use of NMR metabolomics to analyze the targets of D-cycloserine in mycobacteria: Role of D-alanine racemase. J. Proteome Res. 2007;6:4608–4614. doi: 10.1021/pr0704332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wayne L.G., Sramek H.A. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1994;38:2054–2058. doi: 10.1128/AAC.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singhal A., Arora G., Sajid A., Maji A., Bhat A., Virmani R., Upadhyay S., Nandicoori V.K., Sengupta S., Singh Y. Regulation of homocysteine metabolism by Mycobacterium tuberculosis S-adenosylhomocysteine hydrolase. Sci. Rep. 2013;3:2264. doi: 10.1038/srep02264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banks E.C., Doughty S.W., Toms S.M., Wheelhouse R.T., Nicolaou A. Inhibition of cobalamin-dependent methionine synthase by substituted benzo-fused heterocycles. FEBS J. 2007;274:287–299. doi: 10.1111/j.1742-4658.2006.05583.x. [DOI] [PubMed] [Google Scholar]
- 52.Dhiman R.K., Schaeffer M.L., Bailey A.M., Testa C.A., Scherman H., Crick D.C. 1-Deoxy-D-xylulose 5-phosphate reductoisomerase (IspC) from Mycobacterium tuberculosis: Towards understanding mycobacterial resistance to fosmidomycin. J. Bacteriol. 2005;187:8395–8402. doi: 10.1128/JB.187.24.8395-8402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodruff P.J., Carlson B.L., Siridechadilok B., Pratt M.R., Senaratne R.H., Mougous J.D., Riley L.W., Williams S.J., Bertozzi C.R. Trehalose is required for growth of Mycobacterium smegmatis. J. Biol. Chem. 2004;279:28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 54.Balganesh M., Dinesh N., Sharma S., Kuruppath S., Nair A.V., Sharma U. Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob. Agents Chemother. 2012;56:2643–2651. doi: 10.1128/AAC.06003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drapal M., Wheeler P.R., Fraser P.D. The assessment of changes to the nontuberculous mycobacterial metabolome in response to anti-TB drugs. FEMS Microbiol. Lett. 2018;365:fny153. doi: 10.1093/femsle/fny153. [DOI] [PubMed] [Google Scholar]
- 56.Cloete R., Oppon E., Murungi E., Schubert W.-D., Christoffels A. Resistance related metabolic pathways for drug target identification in Mycobacterium tuberculosis. BMC Bioinform. 2016;17:75. doi: 10.1186/s12859-016-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Derewacz D.K., Goodwin C.R., McNees C.R., McLean J.A., Bachmann B.O. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc. Natl. Acad. Sci. USA. 2013;110:2336. doi: 10.1073/pnas.1218524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen L. Antibiotic resistance mechanisms in M. tuberculosis: An update. Arch. Toxicol. 2016;90:1585–1604. doi: 10.1007/s00204-016-1727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyagi J.S., Sharma D. Mycobacterium smegmatis and tuberculosis. Trends Microbiol. 2002;10:68–70. doi: 10.1016/S0966-842X(01)02296-X. [DOI] [PubMed] [Google Scholar]
- 60.Barry I.I.I.C.E. Mycobacterium smegmatis: An absurd model for tuberculosis? Trends Microbiol. 2001;9:473–474. doi: 10.1016/S0966-842X(01)02169-2. [DOI] [PubMed] [Google Scholar]
- 61.Reyrat J.M., Kahn D. Mycobacterium smegmatis: An absurd model for tuberculosis? Trends Microbiol. 2001;9:472–474. doi: 10.1016/S0966-842X(01)02168-0. [DOI] [PubMed] [Google Scholar]
- 62.Gadagkar R., Gopinathan K. Growth of mycobacterium smegmatis in minimal and complete media. J. Biosci. 1980;2:337–348. doi: 10.1007/BF02716867. [DOI] [Google Scholar]
- 63.Scherzinger D., Scheffer E., Bär C., Ernst H., Al-Babili S. The Mycobacterium tuberculosis ORF Rv0654 encodes a carotenoid oxygenase mediating central and excentric cleavage of conventional and aromatic carotenoids. FEBS J. 2010;277:4662–4673. doi: 10.1111/j.1742-4658.2010.07873.x. [DOI] [PubMed] [Google Scholar]
- 64.Smeulders M.J., Keer J., Speight R.A., Williams H.D. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beste D.J., Peters J., Hooper T., Avignone-Rossa C., Bushell M.E., McFadden J. Compiling a molecular inventory for Mycobacterium bovis BCG at two growth rates: Evidence for growth rate-mediated regulation of ribosome biosynthesis and lipid metabolism. J. Bacteriol. 2005;187:1677–1684. doi: 10.1128/JB.187.5.1677-1684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKinney J.D., Honer zu Bentrup K., Munoz-Elias E.J., Miczak A., Chen B., Chan W.T., Swenson D., Sacchettini J.C., Jacobs W.R., Jr., Russell D.G. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 67.Russell D.G., VanderVen B.C., Lee W., Abramovitch R.B., Kim M.J., Homolka S., Niemann S., Rohde K.H. Mycobacterium tuberculosis wears what it eats. Cell Host Microbe. 2010;8:68–76. doi: 10.1016/j.chom.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schoeman J.C., du Preez I., Loots du T. A comparison of four sputum pre-extraction preparation methods for identifying and characterising Mycobacterium tuberculosis using GCxGC-TOFMS metabolomics. J. Microbiol. Methods. 2012;91:301–311. doi: 10.1016/j.mimet.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 69.Lorian V. Differences between in vitro and in vivo studies. Antimicrob. Agents Chemother. 1988;32:1600–1601. doi: 10.1128/AAC.32.10.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pai M., Schito M. Tuberculosis Diagnostics in 2015: Landscape, Priorities, Needs, and Prospects. J. Infect. Dis. 2015;211(suppl_2):S21–S28. doi: 10.1093/infdis/jiu803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang P., Zhang W., Lang Y., Qu Y., Chu F., Chen J., Cui L. Mass spectrometry-based metabolomics for tuberculosis meningitis. Clin. Chim. Acta. 2018;483:57–63. doi: 10.1016/j.cca.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 72.Banday K.M., Pasikanti K.K., Chan E.C.Y., Singla R., Rao K.V.S., Chauhan V.S., Nanda R.K. Use of Urine Volatile Organic Compounds To Discriminate Tuberculosis Patients from Healthy Subjects. Anal. Chem. 2011;83:5526–5534. doi: 10.1021/ac200265g. [DOI] [PubMed] [Google Scholar]
- 73.Weiner J., Maertzdorf J., Sutherland J.S., Duffy F.J., Thompson E., Suliman S., McEwen G., Thiel B., Parida S.K., Zyla J., et al. Metabolite changes in blood predict the onset of tuberculosis. Nat. Commun. 2018;9:5208. doi: 10.1038/s41467-018-07635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collins J.M., Walker D.I., Jones D.P., Tukvadze N., Liu K.H., Tran V.T., Uppal K., Frediani J.K., Easley K.A., Shenvi N., et al. High-resolution plasma metabolomics analysis to detect Mycobacterium tuberculosis-associated metabolites that distinguish active pulmonary tuberculosis in humans. PLoS ONE. 2018;13:e0205398. doi: 10.1371/journal.pone.0205398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zamboni N., Sauer U. Novel biological insights through metabolomics and 13C-flux analysis. Curr. Opin. Microbiol. 2009;12:553–558. doi: 10.1016/j.mib.2009.08.003. [DOI] [PubMed] [Google Scholar]