Abstract
Klebsiella pneumoniae is a clinically relevant pathogen and a frequent cause of hospital-acquired (HA) and community-acquired (CA) urinary tract infections (UTI). The increased resistance of this pathogen is leading to limited therapeutic options. To investigate the epidemiology, virulence, and antibiotic resistance profile of K. pneumoniae in urinary tract infections, we conducted a multicenter retrospective study for a total of 81 isolates (50 CA-UTI and 31 HA-UTI) in Portugal. The detection and characterization of resistance and virulence determinants were performed by molecular methods (PCR, PCR-based replicon typing, and multilocus sequence typing (MLST)). Out of 50 CA-UTI isolates, six (12.0%) carried β-lactamase enzymes, namely blaTEM-156 (n = 2), blaTEM-24 (n = 1), blaSHV-11 (n = 1), blaSHV-33 (n = 1), and blaCTX-M-15 (n = 1). All HA-UTI were extended-spectrum β-lactamase (ESBL) producers and had a multidrug resistant profile as compared to the CA-UTI isolates, which were mainly resistant to ciprofloxacin, levofloxacin, tigecycline, and fosfomycin. In conclusion, in contrast to community-acquired isolates, there is an overlap between virulence and multidrug resistance for hospital-acquired UTI K. pneumoniae pathogens. The study is the first to report different virulence characteristics for hospital and community K. pneumoniae pathogens, despite the production of β-lactamase and even with the presence of CTX-M-15 ESBL, a successful international ST15 clone, which were identified in both settings. This highlights that a focus on genomic surveillance should remain a priority in the hospital environment.
Keywords: Klebsiella pneumoniae, multidrug resistance, virulence genes, urinary tract infections
1. Introduction
The Gram-negative Klebsiella pneumoniae is a clinically relevant pathogen that has the propensity to acquire multidrug resistance (MDR), thus limiting the therapeutic options for treating related infections such as pneumonia, liver abscess, meningitis, bloodstream infections, and urinary tract infections (UTIs) [1].
K. pneumoniae is the second most frequent etiological agent involved in community-acquired (CA) UTIs [2,3], and it is one of the top three pathogens of international concern documented in the 2017 World Health Organization’s (WHO) Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics [4].
Extended-spectrum β-lactamases (ESBL) are bacterial enzymes that confer resistance against a number of commonly used classes of β-lactam antibiotics [5]. The emergence of antimicrobial resistance, as Klebsiella pneumoniae carbapenemases (KPC) and cefotaximases (CTX-M) enzymes production, additional to the rapid worldwide spread of K. pneumoniae ESBL producers is posing a serious threat to global health [6], despite the implementation of local and national guidelines [7] as well as the development innovative approaches that are being considered for therapeutics [8].
K. pneumoniae utilizes a variety of virulence factors, especially capsule polysaccharides, adhesins, and determinants for iron acquisition, which are used for survival and immune evasion during infection [9]. Typically, K. pneumoniae is an opportunistic pathogen, which mostly affects those with weakened immune systems and tends to cause hospital-acquired (HA) infections [10]. However, virulent K. pneumoniae serotypes can lead to neonatal sepsis in immunocompromised patients [11], hospital intensive care unit patients [12], or previously healthy persons, and in all cases, it can cause life-threatening infections [13]. We previously reported a high prevalence of virulence determinants on multidrug-resistant sequence type (ST)14 KPC-3 carbapenemase K. pneumoniae [14] and detailed the possibility of this microorganism preceding difficult-to-treat and fatal infections, including those caused by other Gram-negative pathogens, such as A. baumannii [15]. However, despite the recent interest in this relationship, the interplay between resistance and virulence in K. pneumoniae clinical isolates remains poorly understood [9], and to our knowledge, the pathogenic potential of K. pneumoniae in urinary tract infections, especially in the community setting, and its resistance profile have not yet been characterized.
Therefore, we conducted a multicenter retrospective study for a total of 81 isolates (50 CA-UTI and 31 HA-UTI) in Portugal with the main goal of investigating the virulence and antibiotic resistance of K. pneumoniae in urinary tract infections recovered from hospital and community clinic environments.
2. Materials and Methods
2.1. Bacterial Isolates
A total of 81 non-duplicated K. pneumoniae clinical isolates from urinary tract infections were studied. Only one isolate was considered per patient. The CA-UTI K. pneumoniae isolates (n = 50) were recovered from 10 community laboratories located in Portugal. Only K. pneumoniae isolates were selected for the study. The specimens were consecutively collected in the period from January to March 2010. The HA-UTI K. pneumoniae isolates (n = 31) were collected from hospitalized patients at a tertiary care university hospital center located in Lisbon between 1980 and 2013 and were selected from the Faculty of Pharmacy, University of Lisboa (FFUL) collection based on the source of isolation (urine only), by beta-lactamase type, and within this, by random selection. All uropathogens were obtained as part of routine care and were recovered under standard operating procedures. The identification of bacteria and β-lactamase production was primarily performed by microbiology laboratories using conventional methods or automated systems such as Vitek2® (BioMérieux, Marcy, l’Étoile, France) or MicroScan® (Snap-on, Kenosha, WI, USA). Thereafter, the bacteria were sent to the Faculty of Pharmacy, Department of Microbiology and Immunology Laboratory for specific molecular studies. The isolates were frozen in brain heart infusion (BHI) broth (VWR Prolabo, Lisboa, Portugal) with 15% glycerol at −80 °C. Bacterial growth was done using BHI broth (18 h, 37 °C) and the bacteria were later seeded in Luria–Bertani agar (VWR Prolabo®, Lisboa, Portugal).
2.2. Antimicrobial Susceptibility Testing and Phenotypic Detection of Extended-Spectrum β-lactamase (ESBL) Production
Antimicrobial susceptibility testing was performed using the standardized Kirby–Bauer disk diffusion technique, in accordance with the European Committee on Antimicrobial Susceptibility Testing (EUCAST) for Antimicrobial Susceptibility Testing guidelines; the detailed methodology is available at http://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/. Detailed instructions for Mueller–Hinton agar medium (VWR Prolabo, Lisboa, Portugal), including preparation and storage, are also available in the same EUCAST guidelines document. Quality control strains were included to monitor the performance of the test. The K. pneumoniae clinical isolates was tested for their susceptibility to the following antimicrobial agents: amoxicillin/clavulanic acid (20 µg/10 µg), ceftazidime (10 µg), cefotaxime (5 µg), cefoxitin (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), gentamicin (10 µg), imipenem (10 µg), meropenem (10 µg), ertapenem (10 µg), tigecycline (15 µg), and fosfomycin (200 µg). The inhibition zones were interpreted in accordance with EUCAST, except for fosfomycin, which was interpreted using breakpoints proposed by the Clinical and Laboratory Standard Institute guidelines [16]. Zone diameters of susceptibility categories were formed according to the proposed definitions of the 2018 EUCAST Steering Committee [17], which are available at http://www.eucast.org; namely, categories included (1) susceptible, standard dosing regimen (S); (2) susceptible, increased exposure (I); (3) and resistant (R). Multidrug resistance (MDR) was defined as non-susceptibility to at least one agent in three or more antimicrobial categories [18].
The phenotypic double-disk synergy test (DDST), which involves the application of disks containing ceftazidime next to a disk with amoxicillin–clavulanic acid on Mueller–Hinton agar plates, was performed on the K. pneumoniae uropathogens that showed resistance to at least one of the tested third-generation cephalosporins (cefotaxime, ceftazidime). The methodology and interpretation of results was done according to the EUCAST guidelines for the detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance, which are available at http://www.eucast.org/resistance_mechanisms/.
2.3. Detection of Resistance and Virulence Genes
Isolates were screened by the polymerase chain reaction (PCR) technique using specific primers for the detection of β-lactamase-associated genes (blaDHA, blaCMY, blaCTX-M, blaSHV), including carbapenemase genes (blaVIM, blaIMP, blaOXA, and blaKPC). Moreover, the presence of the following K2 capsule serotypes (K2A) were also investigated; type 1 and type 3 fimbrial adhesins (fimH and mrkD, namely the subtypes mrkDV1 and mrkDV2-4); siderophore aerobactin (iucC); haemolysin (khe); regulator of mucoid phenotype A (rmpA); and hypermucoviscosity phenotype (magA) virulence genes. Among the 79 known capsular serotypes of K. pneumoniae, K1 and K2 have been shown to be the most prevalent [19] and clinically relevant, despite their association with invasive infections [20,21,22]. The search for only the K2 capsular type was justified considering that K1 is mainly related to hypervirulent K. pneumoniae variants (hvKP), which cause pyogenic liver abscesses and other infections [23] and are mainly found in Asian countries [21], while K2 is usually related to non-hvKP isolates, which cause severe and difficult-to-treat infections [21,24,25]. Additionally, hypervirulent K. pneumoniae isolates are defined by either capsular type K1 or K2 (which confers different virulence characteristics such as serum resistance) or the presence of one of the following virulence genes: aerobactin (iuc), rmpA/rmpA2, and/or salmochelin (iro) [23].
The PCR methodology, including primer sequences, lengths of expected PCR products, quality control strains, as well the purification technique and nucleotide sequences analysis were previously described in detail [14].
2.4. Multilocus Sequence Typing
A total of twenty-four isolates, namely the eleven KPC-3 carbapenemase producers and thirteen CTX-M-15 cefotaximase extended-spectrum beta-lactamase-producing K. pneumoniae isolates (12 HA-UTI and 1 CA-UTI) were selected for multilocus sequence typing (MLST) after sequencing (Macrogen, Inc., Korea). Primers, PCR reaction conditions, and detailed methodology were in accordance with those previously described by Diancourt, L. et al. [26]. The allele attribution and sequence type (ST) identification was done with the K. pneumoniae MLST database platform from Institute Pasteur (http://www.pasteur.fr/mlst/; Last accessed on 2 May 2018).
2.5. PCR-based Replicon Typing
The molecular identification and classification of plasmids, namely the identification of origins of plasmid replicates belonging to different incompatibility groups, was performed by the PCR-based replicon typing (PBRT) technique described by Carattoli, A. et al. [27].
2.6. Statistical Analysis
The statistical analysis used the Fisher’s exact test, using the computer program available at http://www.graphpad.com/quickcalcs/index. A probability value of p ≤ 0.05 was considered to indicate statistical significance.
2.7. Ethical Approval
The Ethics Committee of the Faculty of Medicine, Universidade de Lisboa approved this study proposal. All isolates were recovered as part of routine testing, studied anonymously, and the epidemiological data were obtained retrospectively from clinical records.
3. Results
3.1. Antimicrobial Susceptibility
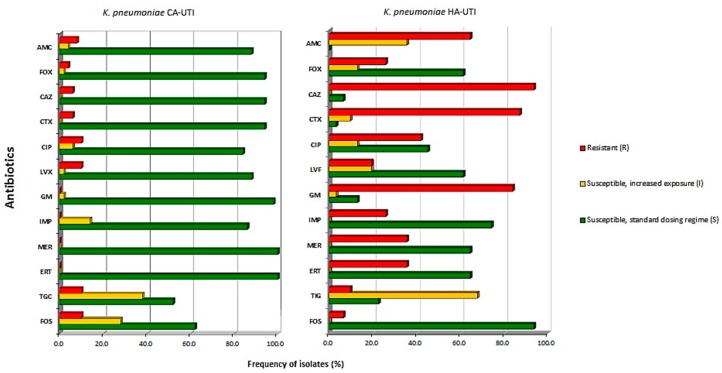
The susceptibility of the CA-UTI K. pneumoniae isolates was found to be 100% (50/50) to meropenem and ertapenem, 98% (49/50) to gentamicin, 94% (47/50) to ceftazidime, cefoxitin, and cefotaxime, 88% (44/50) to amoxicillin/clavulanic acid and levofloxacin, 86% (43/50) to imipenem, and 84% (42/50) to ciprofloxacin. Higher resistance rates were found for ciprofloxacin, levofloxacin, tigeciclin, and fosfomycin with 10% (5/50) resistant isolates for each. Fourteen percent (7/50), 28% (14/50), and 38% (19/50) of the isolates belonged to category I (susceptible, increased exposure) for imipenem, fosfomycin, and tigeciclin, respectively. The results are shown in Figure 1. On the other hand, the HA-UTI K. pneumoniae isolates showed a full antimicrobial resistance profile, namely, 93.5% (29/31), 87.1% (27/31), and 64.5% (20/31) of the isolates were resistant to ceftazidime, cefotaxime, and amoxicilin/clavulanic acid, respectively. Additionally, 83.9% (26/31) and 41.9% (13/31) of the isolates were resistant to gentamicin and ciprofloxacin, respectively. The most active antibiotics were shown to be fosfomycin (93.5%, 20/31), imipenem (74.2%, 23/31), and meropenem, and ertapenem (64.5%, 20/31 each). Imipenem and fosfomycin were found to be non-susceptible, increased exposure (I) isolates.
Figure 1.
Antimicrobial susceptibilities of the K. pneumoniae clinical isolates (n = 81). The isolates were recovered from community-acquired (CA)-urinary tract infection (UTI) and hospital-acquired (HA)-UTI patients. The CA isolates were consecutively collected while the HA isolates were selected from the FFUL collection based on the source of isolation (urine only), beta-lactamase type produced, and within this, by random selection. Legend: AMC, amoxicillin/clavulanic acid; FOX, cefoxitin; CAZ, ceftazidime; CTX, cefotaxime; CIP, ciprofloxacin; LVF, levofloxacin, GM, gentamicin; IMP, imipenem; MER, meropenem; ERT, ertapenem; TIG, tigecycline; FOS, fosfomycin; FFUL, Faculty of Pharmacy University of Lisboa.
3.2. Identification of the β-Lactamases
PCR amplification of β-lactamase genes was performed in CA-UTI and HA-UTI K. pneumoniae isolates. ESBL production was detected in six (12.0%) CA-UTI K. pneumoniae isolates, namely, blaTEM-156 (n = 2), blaTEM-24 (n = 1), blaSHV-11 (n = 1), blaSHV-33 (n = 1), and blaCTX-M-15 (n = 1). The results are presented in Table 1. The multidrug resistant HA-UTI K. pneumoniae isolates carried the ESBLs blaTEM-10 (19.3%, 6/31), blaTEM-24 (9.6%, 3/31), blaCTX-M-15 (38.7%, 12/31), blaKPC-3 (19.3%, 6/31), blaKPC-3 and blaSHV-11 (3.2%, 1/31), and blaKPC-3 and blaCTX-M-15 (12.9%, 4/31). The genes blaDHA, blaCMY, blaIMP, blaVIM, blaNDM, and blaOXA were not detected in our study. The replicon typing plasmids found are also presented in Table 1. The conjugative plasmid IncFIA was the most commonly found (66.7%, 4/6) CA-UTI K. pneumoniae β-lactamase producer, while HI1 (CTX-M-15 producers), A/C, and F (KPC-3 producers) were mainly present (38.7% 12/31, 22.6% 7/31, and 25.8% 8/31, respectively) on the HA-UTI K. pneumoniae isolates The plasmids were non-typeable by the former scheme among eight isolates.
Table 1.
Antibiotic resistance profile, distribution of virulence genes, and plasmid content of β-lactamases-producing K. pneumoniae community-acquired UTI isolates.
| Isolate ID | Date of Isolation | β-Lactamase Produced | Antibiotics Resistance Profile | Virulence Profile | PBRT | MLST | |
|---|---|---|---|---|---|---|---|
| CA-UTI isolates | 201-010 | 2010 | TEM-156 | CIP, LVF | khe, mrkDV1 | FIA | NP |
| 201-075 | 2010 | SHV-11 | CIP, FOS | mrkDV2-4 | FIA, X, Y | NP | |
| 201-076 | 2010 | TEM-24 | LVF, FOS | No VG | X, A/C | NP | |
| 201-094 | 2010 | TEM-156 | AMC, FOS | mrkDV1 | W | NP | |
| 208-309 | 2010 | SHV-33 | NR | fimH, mrkDV2-4, iucC | FIA, Y | NP | |
| 212-193 | 2010 | CTX-M-15 | AMC, CAZ, CTX, CIP, LVF | khe, iucC | FIA, HI2 | ST15 | |
| HA-UTI isolates | 625 | 1980 | TEM-10 | GM | K2, fimH, khe, mrkDV1 | NT | NP |
| 683 | 1999 | TEM-10 | AMC, CTX, CAZ, GM | K2, fimH, khe, mrkDV1 | NT | NP | |
| 684 | 1999 | TEM-10 | AMC, CAZ, GM | K2, fimH, khe, mrkDV1 | NT | NP | |
| 712 | 1999 | TEM-24 | AMC, CTX, CAZ | fimH, khe, mrkDV1 | NT | NP | |
| 721 | 1999 | TEM-10 | AMC, CTX, CAZ, GM | fimH, khe, mrkDV1 | NT | NP | |
| 732 | 2000 | TEM-10 | AMC, CTX, CAZ, GM | fimH, khe, mrkDV1 | NT | NP | |
| 749 | 2001 | TEM-24 | AMC, FOX, CTX, CAZ, CIP | fimH, khe | NT | NP | |
| 770 | 2001 | TEM-24 | AMC, FOX, CTX, CAZ, CIP | fimH, khe | NT | NP | |
| 775 | 2001 | TEM-10, CTX-M-15 | CAZ, GM | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 847 | 2003 | CTX-M-15 | AMC, CTX, CAZ, GM, CIP | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 931 | 2004 | CTX-M-15 | CTX, CAZ, GM, CIP | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 1119 | 2005 | CTX-M-15 | CTX, CAZ, GM, CIP | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 1263 | 2007 | CTX-M-15 | GM, CIP | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 2323 | 2008 | CTX-M-15 | AMC, CTX, CAZ, GM | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 2325 | 2008 | CTX-M-15 | AMC, CTX, CAZ, GM | fimH, mrkDV1 | HI1 | ST15 | |
| 2386 | 2008 | CTX-M-15 | CTX, CAZ, GM | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 2394 | 2008 | CTX-M-15 | CTX, CAZ, GM | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 2398 | 2008 | CTX-M-15 | CTX, CAZ, GM, CIP | fimH, mrkDV1 | HI1 | ST15 | |
| 2400 | 2008 | CTX-M-15 | CTX, CAZ, GM | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 2414 | 2008 | CTX-M-15 | CTX, CAZ, GM, CIP | fimH, khe, mrkDV1 | HI1 | ST15 | |
| 2909 | 2009 | KPC-3, SHV-11 | AMC, CTX, CAZ, IMP, GM, MEM, ERT | K2, fimH, khe, mrkDV1 | F | ST14 | |
| 2954 | 2010 | KPC-3 | AMC, FOX CTX, CAZ, IMP, GM, LVF, TIG, MEM, ERT | K2, fimH, khe, mrkDV1 | A/C, F | ST14 | |
| 3108 | 2010 | KPC-3 | AMC, CTX, CAZ, TIG, MEM, ERT | khe, mrkDV1, iucC | FIA, F | ST14 | |
| 29078 | 2010 | KPC-3, CTX-M-15 | AMC, FOX, CTX, CAZ, IMP, GM, MEM, ERT, FOS | fimH, khe, mrkDV1,iucC | FIA, F | ST14 | |
| 43201 | 2010 | KPC-3, CTX-M-15 | AMC, FOX, CTX, CAZ, IMP, GM, CIP, LVF, MEM, ERT | K2, fimH, khe, mrkDV1,iucC | A/C | ST14 | |
| 61095 | 2011 | KPC-3, CTX-M-15 | AMC, CTX, CAZ, IMP, GM, CIP, LVF, MEM, ERT, TIG | K2, fimH, mrkDV1 | A/C, F | ST14 | |
| 72562 | 2011 | KPC-3 | AMC, FOX, CTX, CAZ, MEM, ERT | fimH, mrkDV1,iucC | A/C | ST14 | |
| 87582 | 2012 | KPC-3 | AMC, FOX, CTX, CAZ, IMP, GM, CIP, LVF, MEM, ERT, FOS | fimH, khe, mrkDV1,iucC | F | ST14 | |
| 91488 | 2012 | KPC-3, CTX-M-15 | CTX, CAZ, GM, CIP, LVF, MEM, ERT | K2, fimH, khe, mrkDV1 | A/C, F | ST14 | |
| 22073 | 2013 | KPC-3 | AMC, FOX, CTX, CAZ, IMP, GM, CIP, LVF, MEM, ERT | K2, fimH, khe, mrkDV1 | A/C | ST14 | |
| 31149 | 2013 | KPC-3 | AMC, CTX, CAZ, IMP, GM, MEM, ERT | K2, fimH, mrkDV1,iucC | A/C, F | ST14 |
Legend: CA-UTI, community-acquired urinary tract infection; HA-UTI, hospital-acquired urinary tract infection; ID, identification; Nr., number; No VG; no virulence genes found; NR, no resistance found; CIP, ciprofloxacin; LVF, levofloxacin; AMC, amoxicillin/clavulanic acid; FOS, fosfomycin; CAZ, ceftazidime; CTX, cefotaxime; FOX, cefoxitin; GM, gentamicin; IMP, imipenem; MER, meropenem; ERT, ertapenem; TIG, tigecycline; PBRT, PCR-based replicon typing; NT, non-typeable; NP, not performed; MLST, multilocus sequence typing; ST, sequence type; KPC, Klebsiella pneumoniae carbapenemase; CTX, cefotaximase.
3.3. Virulence Genes
Of all the K. pneumoniae UTI clinical isolates (n = 81), whose results are shown in Table 2, the major virulence genes identified were fimH (63%, 51/81), mrkDV1 (49.3%, 40/81), and khe (60.5%, 49/81). A total of 38.2% (31/81) of the isolates showed the presence of mrkDV2-4, while 18.5% (15/81) and 16.0% (13/81) showed the iucC and K2A genes, respectively. No magA and rmpA genes were amplified. The type 3 fimbrial adhesin mrkDV1 was predominant in HA-UTIs (96.8%, 30/31) but not in CA-UTIs (20.0%, 10/50), while mrkDV2-4 was the most common type 3 fimbrial adhesin (62.0%, 31/50 vs. 20.0% 10/50) in CA-UTIs, and it was only found in these isolates. Additionally, higher virulence potential was found in the HA-UTI isolates with an average of 3.29 virulence factors per isolate in HA-UTI vs. 1.94 virulence factors per isolate in CA-UTI.
Table 2.
Distribution of virulence genes in K. pneumoniae isolates from community-acquired urinary tract infections (CA-UTI) and hospital-acquired urinary tract infections (HA-UTI).
| Virulence Factor | Target Gene | Nr. of CA-UTI Isolates (%) | Nr. of HA-UTI Isolates (%) | Total (%) | |
|---|---|---|---|---|---|
| (n = 50) | (n = 31) | (n = 81) | |||
| Fimbrial adhesins | Type 1 | fimH | 20 (40.0) | 31 (100.0) * | 51 (63.0) |
| Type 3, variant 1 | mrkDV1 | 10 (20.0) | 30 (96.8) * | 40 (49.3) | |
| Type 3, variant 2-4 | mrkDV2-4 | 31 (62.0) | 0 * | 31 (38.2) | |
| Toxin | Haemolysin | khe | 23 (46.0) | 26 (83.9) * | 49 (60.5) |
| Capsular type | K2 serotype | K2A | 3 (6.0) | 10 (32.2) * | 13 (16.0) |
| Siderophore | Aerobactin | iucC | 10 (20.0) | 5 (16.1) | 15 (18.5) |
| Protectins or invasins | Mucoviscosity phenotype | magA | 0 | 0 | 0 |
| Regulator of mucoid phenotype | rmpA | 0 | 0 | 0 | |
Legend: CA-UTI, community-acquired urinary tract infection; HA-UTI, hospital-acquired urinary tract infection; Nr., number; * p-values less than 0.05 were considered statistically significant.
In order to identify how the virulence genes were simultaneously present on the same isolate, we characterized the virulence profiles (VP) of both CA- and HA-UTI K. pneumoniae uropathogens. The results are presented in Table 3.
Table 3.
Characterization of virulence profiles of community- and hospital-acquired UTI K. pneumoniae isolates.
| Nr. of Virulence Genes | Virulence Profile (VP) | Virulence Genes | Nr. Isolates (%) | ||
|---|---|---|---|---|---|
| CA-UTI n = 50 (100) |
HA-UTI n = 31 (100) |
Total n = 81 (100) |
|||
| 0 VF | 1 | No VF | 8 (16.0) | 0 | 8 (9.9) |
| 1 VF | 2 | khe | 5 (10.0) | 0 | 5 (6.2) |
| 3 | fimH | 1 (2.0) | 0 | 1 (1.2) | |
| 4 | mrkDV1 | 2 (4.0) | 0 | 2 (2.5) | |
| 5 | mrkDV2-4 | 3 (6.0) | 0 | 3 (3.7) | |
| 2 VF | 6 | fimH, khe | 2 (4.0) | 2 (6.3) | 4 (4.9) |
| 7 | fimH, mrkDV1 | 0 | 3 (9.4) | 3 (3.7) | |
| 8 | khe, iucC | 1 (2.0) | 0 | 1 (1.2) | |
| 9 | iucC, mrkDV2-4 | 2 (4.0) | 0 | 2 (2.5) | |
| 10 | khe, mrkDV1 | 2 (4.0) | 0 | 2 (2.5) | |
| 11 | khe, mrkDV2-4 | 5 (10.0) | 0 | 5 (6.2) | |
| 12 | fimH, mrkDV2-4 | 5 (10.0) | 0 | 5 (6.2) | |
| 13 | fimH, khe, mrkDV1 | 2 (4.0) | 13 (40.6) | 15 (18.5) | |
| 14 | fimH, khe, iucC | 1 (2.0) | 0 | 1 (1.2) | |
| 15 | fimH, khe, mrkDV2-4 | 2 (4.0) | 0 | 2 (2.5) | |
| 3 VF | 16 | fimH, iucC, mrkDV1 | 2 (4.0) | 1 (3.1) | 3 (3.7) |
| 17 | fimH, iucC, mrkDV2-4 | 4 (8.0) | 0 | 4 (4.9) | |
| 18 | khe, mrkDV1, iucC | 0 | 1 (3.1) | 1 (1.2) | |
| 19 | K2, khe, mrkDV1 | 2 (4.0) | 0 | 2 (2.5) | |
| 20 | K2, fimH, mrkDV1 | 0 | 1 (3.1) | 1 (1.2) | |
| 21 | K2, fimH, khe, mrkDV2-4 | 1 (2.0) | 0 | 1 (1.2) | |
| 4 VF | 22 | K2, fimH, khe, mrkDV1 | 0 | 7 (21.9) | 7 (8.6) |
| 23 | fimH, khe, mrkDV1, iucC | 0 | 2 (6.3) | 2 (2.5) | |
| 5 VF | 24 | K2, fimH, khe, mrkDV1, iucC | 0 | 1 (3.1) | 1 (1.2) |
Legend: VF, virulence factor; VP, virulence profile; CA-UTI, community-acquired urinary tract infection; HA-UTI, hospital-acquired urinary tract infection.
A total of 24 different virulence profiles was identified in our study (Table 3). The most frequent virulence profile found was VP13 (fimH, khe, mrkDV1), which was observed in 18.5% (15/81) of the isolates, but mainly in HA-UTI isolates (40.6% HA-UTI vs. 4.0% CA-UTI, p < 0.05). A similar finding was obtained for VP22 (K2, fimH, khe, mrkDV1) which showed a prevalence of 8.6% (7/81) but was only found in HA-UTI isolates (21.9%, 7/31). Similarly, virulence profiles with no virulence genes (9.9%, 8/81) and one virulence factor, namely VP2 with the virulence factor khe, accounted for 6.2% (5/81) of the isolates, and this type of profile was only found in CA-UTI K. pneumoniae isolates. All HA-UTI isolates harbored at least one virulence gene. Moreover, the HA-UTIs presented only nine virulence profiles (VP 13 and VP 22 with two and four virulence genes, respectively, accounted for 62.5% of the isolates) while CA-UTI virulence profiles were distributed among 18 virulence profiles, and the most frequent profiles, VP 1 and VP2, presented 0 and 1 virulence factors, respectively, and accounted for only 26.0% of the CA-UTI isolates.
3.4. MLST Results
The clonal relatedness of the MDR CTX-M-15 ESBL K. pneumoniae producers, including one isolate identified in the community setting, was investigated by multilocus sequence typing (MLST). CTX-M-15 was included due to its clinical relevance to human health and because it is the only extended-spectrum-β-lactamase shared between hospital and community settings. The ST15 clone was identified in all CTX-M-15 isolates studied (100.0%, 13/13).
4. Discussion
Multidrug resistance and virulence are typically observed in separate K. pneumoniae populations [28] and their interplay remains poorly understood [9]. Recent reports have shown that K. pneumoniae strains can accumulate, increasing their pathogenicity and causing severe and difficult-to-treat infections. However, the available reports mainly cover the hypervirulent phenotypes of K. pneumoniae (mainly related to pyogenic liver abscesses and meningitis [28,29]), focus on carbapenemase producers [30], and were performed in the hospital setting [14,25]. Our study characterizes the virulence and antibiotic resistance traits of K. pneumoniae in urinary tract infections recovered from both community and hospital settings. Additionally, this is the first report of a community-acquired urinary tract infection related to the CTX-M-15 ESBL K. pneumoniae producer, which belongs to the successful international ST15 clone, in Portugal.
The majority of CA-UTI K. pneumoniae pathogens characterized in our study were susceptible isolates; the antimicrobial susceptibility was >80% in eight out of ten tested antibiotics. However, the number of isolates belonging to category I (susceptible, increased exposure) found for imipenem (14%) deserves particular attention considering that the isolates were inhibited in vitro by a concentration of antibiotics that is associated with an uncertain therapeutic effect. In Portugal, carbapenems consumption is increasing [31]. Portugal is now one of the top consumers in Europe, and resistance to antibiotics of this class has changed from 5.2% in 2016 [32] to 8.6% in 2017 [33] according to the resistance surveillance network (EARS-Net) of the European Centre for Disease Prevention and Control (ECDC). Strict monitoring of imipenem consumption and continuous monitoring of evolutionary trends in the susceptibility patterns of K. pneumoniae, especially regarding community-acquired infections, is mandatory.
Limited data are available on the susceptibility to recently accessible antimicrobial agents, such as tigecycline, along with some “older” antibiotics, such as fosfomycin, particularly on multidrug and ESBL-producing Gram-negative microorganisms [34]. The in vitro activities of these antibiotics were determined against K. pneumoniae isolates recovered from community- and hospital-acquired urinary tract infections. Surprisingly, a significantly lower susceptibility (62% CA-UTI vs. 94% MDR HA-UTI) to the fosfomycin antibiotic was found for CA-UTI K. pneumoniae isolates when compared with HA-UTIs. Similarly, susceptibility rates of 96% for MDR Enterobacteriaceae [35] and 89% for K. pneumoniae KPC producers were found [14], including a report showing that extremely drug-resistant K. pneumoniae KPC producers, including tigecycline and colistin resistant types, were susceptible to fosfomycin [36]. However, lower fosfomycin activity has been previously reported, perhaps due to differences in local epidemiology [29]. The present study demonstrates that fosfomycin, an older antimicrobial agent, should be considered to be an emerging treatment option for difficult-to-treat hospital-acquired urinary tract infections caused by K. pneumoniae uropathogens, including MDR isolate multidrugs, but caution should be applied for its use as an alternative agent for outpatient therapy of UTIs.
The presence of virulence factors has an important contribution to the pathogenesis of K. pneumoniae [13] and to the development of severe and invasive forms of infection, not only in immunocompromised individuals but also in previously healthy adults [11,25,37]. Bandeira et al. support the hypothesis that biofilms formed on medical devices can promote the onset and spread of healthcare-associated infections, and they reported that biofilm-forming bacteria are generally more resistant to antibiotics [37]. In our study, the most frequent virulence factors found were fimH and the mrkD, which encode type 1 and type 3 fimbrial adhesins, respectively, which mediate binding to epithelial cells of the urinary tract and promote biofilm development [38,39,40]. In fact, despite the findings of the present study being similar to those of other studies that have reported the ubiquitous nature of these fimbriae in K. pneumoniae [40,41], a significantly higher prevalence of fimbriae on HA uropathogens when compared with CA isolates was found in our study. This could lead to the infection of medical devices and may explain their persistence and difficulty of eradication in the hospital setting.
The genes involved in the synthesis of siderophores have dual roles as they can also inhibit T cell proliferation, promoting host immunosuppression, in addition to their role in enhancing iron uptake [42]. The iron siderophore aerobactin synthase gene (iucC) was detected in 19% of the isolates and showed similar prevalence between CA and HA uropathogens. Despite the prevalence found in our study being lower than that previously reported (32%) [25], it is unusual to have high identification of community-provenance isolates. The rmpA and magA genes were not found in our uropathogens. These genes are frequently identified in K. pneumoniae causing liver abscesses, particularly in Asian countries [43,44]. The capsule polysaccharide K2 serotype has been previously reported as a major contributor to the virulence of K. pneumoniae isolates [45]. It confers resistance to phagocytosis [46] and is related to severe infections [47]. In our study, 16.0% (13/81) of the isolates showed the K2 capsular serotype, mainly MDR HA uropathogens. Despite the results being in accordance with previous reports [45], unusual capsular types have been described in carbapenem-resistant K. pneumoniae strains, such as K64 and K62. [48]. Therefore, future studies on virulence should consider the characterization of capsular types for Klebsiella spp. strains [47].
Differences between uropathogens from the hospital and community settings were also found in terms of the accumulation of virulence genes by each isolate. The K. pneumoniae recovered from the hospital setting showed higher pathogenic potential, with higher genomic complexity and adaptation to two specific profiles. These findings have important clinical relevance, considering that the presence of virulence factors in K. pneumoniae has been previously described as the most prominent cause of death in patients before starting antibiotic therapy [49]. The virulence profiles VP13 (fimH, khe, mrkDV1) and VP22 (K2, fimH, khe, mrkDV1) were predominant in the hospital-acquired collection. The accumulation of type 1 and type 3 fimbrial adhesins with haemolysin, a toxin that has cytolytic and cytotoxic activity against a wide range of mammalian cell types [50], seems to be an effective virulence combination. Additionally, it is notable that both major virulence profiles identified in HA-UTIs had the same virulence genes, with or without the presence of capsular serotype K2.
High diversity and low accumulation of virulence genes in the same isolate was identified in K. pneumoniae community uropathogens, reinforcing the low pathogenicity of these isolates reported in our study.
A proficient pathogen should be virulent, resistant to antibiotics, and epidemic [9]. Genotyping of the multidrug-resistant CTX-M-15-producing K. pneumoniae isolates by MLST recovered the ST15 clone in isolates from both hospital and community settings. Of relevance, no associations between the ST15 MLST type, antimicrobial resistance, and virulence profiles, or between the ESBLs produced and the virulence profiles, were found in our study. However, there was a clear accumulation of virulence factors in highly multidrug resistant K. pneumoniae clinical isolates recovered from hospital-acquired urinary tract infections when compared with those recovered from the community setting, which showed low resistance and a low virulent potential profile. These results indicate a direct relationship and relevant clinical interplay between resistance and virulence in K. pneumoniae clinical isolates, and the supremacy of antimicrobial resistance appears to be the leading factor.
In 2018, we reported that the ST11 that belongs to the CC258 group, one of the most threating MDR Gram-negative bacterias circulating in nosocomial settings worldwide [51], has been replaced by the ST14 clone KPC-3 carbapenemase-producing K. pneumoniae isolates [14]. In the present study we report the identification of the ST15 clone of CTX-M-15-producing K. pneumoniae in both the hospital and community settings and demonstrate that this clone can accumulate virulence and MDR. ST15 is described as a successful international clone that is present worldwide and has recently been associated with colistin-resistant infections [52]. It is highly virulent and resistant [53] and was one of the first K. pneumoniae isolates in the United States to be reported to the Centers for Disease Control and Prevention (CDC) as non-susceptible to all drugs tested, including all beta-lactams, colistin, and tigecycline [54]. A limitation of the present study is that we showed only a small part of the real epidemiological situation considering the collection studied. Also, despite being a multicenter study, not all districts of Portugal were included, and the period of study can be considered too large for a short number of isolates. Additionally, only CTX-M-15 and KPC-3 producers were characterized by MLST. Future studies should expand the collection of pathogens tested and the number of centers involved in order to provide a more complete picture of the dissemination of K. pneumoniae in Portugal. However, a complete microbiological and molecular characterization of K. pneumoniae strains collected from urinary tract infections was performed, including antimicrobial resistance profiling, ESBL characterization, and detection of virulence determinants. Thus, the results found in this study have great importance from both the clinical and research points of view.
5. Conclusions
In conclusion, there is an overlap between virulence and multidrug resistance in MDR hospital-acquired UTI K. pneumoniae pathogens but not in community-acquired isolates. Different virulence characteristics were reported, despite the production of β-lactamase and even in the presence of the successful international CTX-M-15 ESBL ST15 clone in both settings. These results highlight that the genomic surveillance focus should remain a priority in the hospital environment.
Acknowledgments
The authors would like to thank all the members of the community and hospital microbiology laboratories for the collaboration, isolation, and identification of the clinical isolates.
Author Contributions
Conceptualization, C.C. and A.D.; Methodology, C.C.; Investigation, C.C.; Formal Analysis, C.C., A.D.; Resources, L.L, A.D., and J.M.-C.; Data Curation, C.C., A.D., L.L., and J.M.-C.; Writing—Original Draft Preparation, C.C.; Writing—Review and Editing, A.D.; Supervision, A.D.
Funding
This research was funded by the Research Institute for Medicines (iMed.ULisboa), Faculty of Pharmacy, Universidade de Lisboa (ULisboa).
Conflicts of Interest
J.M.-C. received research grants administered through his university and honoraria for serving on the speaker’s bureaus of Pfizer, Gilead, and Novartis. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Podschun R., Ullmann U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998;11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keynan Y., Rubinstein E. The changing face of Klebsiella pneumoniae infections in the community. Int. J. Antimicrob. Agents. 2007;30:385–389. doi: 10.1016/j.ijantimicag.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Lin W.H., Wang M.C., Tseng C.C., Ko W.C., Wu A.B., Zheng P.X., Wu J.J. Clinical and microbiological characteristics of Klebsiella pneumoniae isolates causing community-acquired urinary tract infections. Infection. 2010;38:459–464. doi: 10.1007/s15010-010-0049-5. [DOI] [PubMed] [Google Scholar]
- 4.WHO Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. [(accessed on 27 February 2017)]; Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/
- 5.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 6.Butler C.C. Antibiotics: Responding to a Global Challenge. Antibiotics. 2012;1:14–16. doi: 10.3390/antibiotics1010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kock R., Siemer P., Esser J., Kampmeier S., Berends M.S., Glasner C., Arends J.P., Becker K., Friedrich A.W. Defining Multidrug Resistance of Gram-Negative Bacteria in the Dutch-German Border Region-Impact of National Guidelines. Microorganisms. 2018;6:11. doi: 10.3390/microorganisms6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo R., Kolesnikova I., Kim T., Gupta S., Pericleous A., Kadouri D.E., Connell N.D. Susceptibility of Virulent Yersinia pestis Bacteria to Predator Bacteria in the Lungs of Mice. Microorganisms. 2018;7:2. doi: 10.3390/microorganisms7010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennequin C., Robin F. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 10.Russo T.A., Shon A.S., Beanan J.M., Olson R., MacDonald U., Pomakov A.O., Visitacion M.P. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical“ K. pneumoniae thereby enhancing its virulence. PLoS ONE. 2011;6:e26734. doi: 10.1371/journal.pone.0026734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaertynov K.S., Anokhin V.A., Rizvanov A.A., Davidyuk Y.N., Semyenova D.R., Lubin S.A., Skvortsova N.N. Virulence Factors and Antibiotic Resistance of Klebsiella pneumoniae Strains Isolated From Neonates With Sepsis. Front. Med. 2018;5:225. doi: 10.3389/fmed.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira R.L., da Silva B.C.M., Rezende G.S., Nakamura-Silva R., Pitondo-Silva A., Campanini E.B., Brito M.C.A., da Silva E.M.L., Freire C.C.M., da Cunha A.F., et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and beta-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front. Microbiol. 2018;9:3198. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B., Zhao Y., Liu C., Chen Z., Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9:1071–1081. doi: 10.2217/fmb.14.48. [DOI] [PubMed] [Google Scholar]
- 14.Caneiras C., Lito L., Mayoralas-Alises S., Diaz-Lobato S., Melo-Cristino J., Duarte A. Virulence and resistance determinants of Klebsiella pneumoniae isolated from a Portuguese tertiary university hospital centre over a 31-year period. Enferm. Infec. Microbiol. Clin. 2018 doi: 10.1016/j.eimc.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Caneiras C., Calisto F., Jorge da Silva G., Lito L., Melo-Cristino J., Duarte A. First Description of Colistin and Tigecycline-Resistant Acinetobacter baumannii Producing KPC-3 Carbapenemase in Portugal. Antibiotics. 2018;7:96. doi: 10.3390/antibiotics7040096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute . Document M2-A11. 11th ed. CLSI; Wayne, PA, USA: 2014. Performance standards for antimicrobial disk susceptibility tests; approved standard. [Google Scholar]
- 17.European Committee on Antimicrobial Susceptibility Testing (EUCAST); 2018. [(accessed on 28 February 2019)]. EUCAST Proposes to Retain Susceptibility Categories “S, I, and R” but to Change the Definitions to “Susceptible, Standard Dosing Regimen”, “Susceptible, Increased Exposure”, and “Resistant”. Available online: http://www.eucast.org/ [Google Scholar]
- 18.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 19.Lin J.C., Koh T.H., Lee N., Fung C.P., Chang F.Y., Tsai Y.K., Ip M., Siu L.K. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 2014;6:21. doi: 10.1186/1757-4749-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Compain F., Babosan A., Brisse S., Genel N., Audo J., Ailloud F., Kassis-Chikhani N., Arlet G., Decre D. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J. Clin. Microbiol. 2014;52:4377–4380. doi: 10.1128/JCM.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siu L.K., Yeh K.M., Lin J.C., Fung C.P., Chang F.Y. Klebsiella pneumoniae liver abscess: A new invasive syndrome. Lancet Infect. Dis. 2012;12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Xie Y., Li G., Liu J., Li X., Tian L., Sun J., Ou H.Y., Qu H. Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence. 2018;9:510–521. doi: 10.1080/21505594.2017.1421894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyres K.L., Wick R.R., Judd L.M., Froumine R., Tokolyi A., Gorrie C.L., Lam M.M.C., Duchene S., Jenney A., Holt K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:e1008114. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struve C., Roe C.C., Stegger M., Stahlhut S.G., Hansen D.S., Engelthaler D.M., Andersen P.S., Driebe E.M., Keim P., Krogfelt K.A. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. MBio. 2015;6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasfi R., Elkhatib W.F., Ashour H.M. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci. Rep. 2016;6:38929. doi: 10.1038/srep38929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diancourt L., Passet V., Verhoef J., Grimont P.A., Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Bialek-Davenet S., Criscuolo A., Ailloud F., Passet V., Jones L., Delannoy-Vieillard A.S., Garin B., Le Hello S., Arlet G., Nicolas-Chanoine M.H., et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014;20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ku Y.H., Chuang Y.C., Chen C.C., Lee M.F., Yang Y.C., Tang H.J., Yu W.L. Klebsiella pneumoniae Isolates from Meningitis: Epidemiology, Virulence and Antibiotic Resistance. Sci. Rep. 2017;7:6634. doi: 10.1038/s41598-017-06878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melo R.D.C.A., de Barros E.M., Loureiro N.G., de Melo H.R., Maciel M.A., Souza Lopes A.C. Presence of fimH, mrkD, and irp2 virulence genes in KPC-2-producing Klebsiella pneumoniae isolates in Recife-PE, Brazil. Curr. Microbiol. 2014;69:824–831. doi: 10.1007/s00284-014-0662-0. [DOI] [PubMed] [Google Scholar]
- 31.Antimicrobial Consumption Database (ESAC-Net) [(accessed on 7 September 2018)]; Available online: https://ecdc.europa.eu/en/antimicrobial-consumption/database/country-overview.
- 32.Data from the ECDC Surveillance Atlas-Antimicrobial Resistance. [(accessed on 7 September 2018)]; Available online: https://ecdc.europa.eu/en/antimicrobial-resistance/surveillance-and-disease-data/data-ecdc.
- 33.Surveillance of Antimicrobial Resistance in Europe 2017. [(accessed on 2 February 2019)]; Available online: https://ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017.
- 34.Alfouzan W., Dhar R., Nicolau D.P. In Vitro Activity of Newer and Conventional Antimicrobial Agents, Including Fosfomycin and Colistin, against Selected Gram-Negative Bacilli in Kuwait. Pathogens. 2018;7:75. doi: 10.3390/pathogens7030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee S., Sengupta M., Sarker T.K. Fosfomycin susceptibility among multidrug-resistant, extended-spectrum beta-lactamase-producing, carbapenem-resistant uropathogens. Indian J. Urol. 2017;33:149–154. doi: 10.4103/iju.IJU_285_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endimiani A., Patel G., Hujer K.M., Swaminathan M., Perez F., Rice L.B., Jacobs M.R., Bonomo R.A. In vitro activity of fosfomycin against blaKPC-containing Klebsiella pneumoniae isolates, including those nonsusceptible to tigecycline and/or colistin. Antimicrob. Agents Chemother. 2010;54:526–529. doi: 10.1128/AAC.01235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bandeira M., Carvalho P.A., Duarte A., Jordao L. Exploring Dangerous Connections between Klebsiella pneumoniae Biofilms and Healthcare-Associated Infections. Pathogens. 2014;3:720–731. doi: 10.3390/pathogens3030720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson J.G., Murphy C.N., Sippy J., Johnson T.J., Clegg S. Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 2011;193:3453–3460. doi: 10.1128/JB.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy C.N., Clegg S. Klebsiella pneumoniae and type 3 fimbriae: Nosocomial infection, regulation and biofilm formation. Future Microbiol. 2012;7:991–1002. doi: 10.2217/fmb.12.74. [DOI] [PubMed] [Google Scholar]
- 40.Stahlhut S.G., Tchesnokova V., Struve C., Weissman S.J., Chattopadhyay S., Yakovenko O., Aprikian P., Sokurenko E.V., Krogfelt K.A. Comparative structure-function analysis of mannose-specific FimH adhesins from Klebsiella pneumoniae and Escherichia coli. J. Bacteriol. 2009;191:6592–6601. doi: 10.1128/JB.00786-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahlhut S.G., Chattopadhyay S., Struve C., Weissman S.J., Aprikian P., Libby S.J., Fang F.C., Krogfelt K.A., Sokurenko E.V. Population variability of the FimH type 1 fimbrial adhesin in Klebsiella pneumoniae. J. Bacteriol. 2009;191:1941–1950. doi: 10.1128/JB.00601-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Autenrieth I., Hantke K., Heesemann J. Immunosuppression of the host and delivery of iron to the pathogen: A possible dual role of siderophores in the pathogenesis of microbial infections? Med. Microbiol. Immunol. 1991;180:135–141. doi: 10.1007/BF00206117. [DOI] [PubMed] [Google Scholar]
- 43.Mei Y.F., Liu P.P., Wan L.G., Liu Y., Wang L.H., Wei D.D., Deng Q., Cao X.W. Virulence and Genomic Feature of a Virulent Klebsiella pneumoniae Sequence Type 14 Strain of Serotype K2 Harboring blaNDM-5 in China. Front. Microbiol. 2017;8:335. doi: 10.3389/fmicb.2017.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pomakova D.K., Hsiao C.B., Beanan J.M., Olson R., MacDonald U., Keynan Y., Russo T.A. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumoniae: An emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 45.Lin Y.C., Lu M.C., Tang H.L., Liu H.C., Chen C.H., Liu K.S., Lin C., Chiou C.S., Chiang M.K., Chen C.M., et al. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: Comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol. 2011;11:50. doi: 10.1186/1471-2180-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turton J.F., Perry C., Elgohari S., Hampton C.V. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 2010;59:541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 47.Brisse S., Passet V., Haugaard A.B., Babosan A., Kassis-Chikhani N., Struve C., Decre D. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 2013;51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Y.J., Lin T.L., Lin Y.T., Su P.A., Chen C.T., Hsieh P.F., Hsu C.R., Chen C.C., Hsieh Y.C., Wang J.T. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by WZC sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother. 2015;59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawlor M.S., O’Connor C., Miller V.L. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 2007;75:1463–1472. doi: 10.1128/IAI.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koczura R., Kaznowski A. Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog. 2003;35:197–202. doi: 10.1016/S0882-4010(03)00125-6. [DOI] [PubMed] [Google Scholar]
- 51.Bowers J.R., Kitchel B., Driebe E.M., MacCannell D.R., Roe C., Lemmer D., de Man T., Rasheed J.K., Engelthaler D.M., Keim P., et al. Genomic Analysis of the Emergence and Rapid Global Dissemination of the Clonal Group 258 Klebsiella pneumoniae Pandemic. PLoS ONE. 2015;10:e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matheeussen V., Xavier B.B., Mermans I., De Weerdt A., Lammens C., Goossens H., Jansens H., Malhotra-Kumar S. Emergence of colistin resistance during treatment of recurrent pneumonia caused by carbapenemase producing Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 2019 doi: 10.1016/j.diagmicrobio.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 53.Lam M.M.C., Wyres K.L., Wick R.R., Judd L.M., Fostervold A., Holt K.E., Lohr I.H. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 2019 doi: 10.1093/jac/dkz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Man T.J.B., Lutgring J.D., Lonsway D.R., Anderson K.F., Kiehlbauch J.A., Chen L., Walters M.S., Sjolund-Karlsson M., Rasheed J.K., Kallen A., et al. Genomic Analysis of a Pan-Resistant Isolate of Klebsiella pneumoniae, United States 2016. MBio. 2018;9 doi: 10.1128/mBio.00440-18. [DOI] [PMC free article] [PubMed] [Google Scholar]