Abstract
Shiga-toxin producing E. coli (STEC) can cause serious illnesses, including hemorrhagic colitis and hemolytic uremic syndrome. This is the first systematic review of STEC in Brazil, and will report the main serogroups detected in animals, food products and foodborne diseases. Data were obtained from online databases accessed in January 2019. Papers were selected from each database using the Mesh term entries. Although no human disease outbreaks in Brazil related to STEC has been reported, the presence of several serogroups such as O157 and O111 has been verified in animals, food, and humans. Moreover, other serogroups monitored by international federal agencies and involved in outbreak cases worldwide were detected, and other unusual strains were involved in some isolated individual cases of foodborne disease, such as serotype O118:H16 and serogroup O165. The epidemiological data presented herein indicates the presence of several pathogenic serogroups, including O157:H7, O26, O103, and O111, which have been linked to disease outbreaks worldwide. As available data are concentrated in the Sao Paulo state and almost completely lacking in outlying regions, epidemiological monitoring in Brazil for STEC needs to be expanded and food safety standards for this pathogen should be aligned to that of the food safety standards of international bodies.
Keywords: STEC, food microbiology, food-borne diseases, VTEC, EHEC, shiga-toxigenic Escherichia coli, bloody diarrhea
1. Introduction
Brazil is one of the largest producers and exporters of food in the world. Animal products, such as beef, poultry, pork, fish, and crops such as corn, soybeans, and rice represent Brazil’s major exports [1]. However, it is a challenge to maintain both high production efficiency and control physical, chemical and microbiological contamination [2]. In addition, the production of different animals and crops may be carried out in the same geographic region and the use of animal manure as a fertilizer may promote the contamination of fruits and vegetables [3].
The main microorganisms involved in food contamination belong to the Enterobacteriaceae, with Escherichia coli representing a major species. This bacterium represents one of the most extensively studied and was one of the first to be sequenced [4,5]. In addition, Escherichia coli is the main bacterium involved in food contamination in Brazil [6]. It possesses seven pathogenic groups, namely enterotoxigenic (ETEC), enteroinvasive (EIEC), enteropathogenic (EPEC) diffusely adherent (DAEC), invasive adherent (AIEC), enteroaggregative (EAEC), and Shiga-toxin producing (STEC) [7].
These pathogenic groups are involved in outbreaks related to food consumption, and STEC in particular are of importance to public health, potentially causing diarrhea, bloody diarrhea (hemorrhagic colitis), hemolytic uremic syndrome (HUS) and renal injury [8,9,10]. STEC strains may produce two immunologically distinct toxins: Shiga-toxin type 1 and 2. In addition, STEC strains may have a pathogenicity island, LEE (Locus of Enterocyte Effacement), which encodes the proteins that include those responsible for induction of attaching-and-effacing lesions [11,12].
In Brazil, there are no reports of outbreaks involving STEC. The hypothesis is that: (i) Disease outbreaks are not being recorded due to lack of a centralized reporting system or (ii) disease outbreaks are not being recognized as there is no surveillance system for STEC. Farm animals have been shown to be carriers and contamination of food as well as sporadic STEC infections in humans have been reported [13,14]. In addition, STEC infections have been reported in several South American countries, including an endemic issue in Argentina, which borders on Brazil. The possible reasons for this contrast are the differences in cattle breeds, surveillance systems, more livestock confinement system or a combination of these factors. Moreover, Brazil’s large cattle populations pose a direct risk, as STEC infections are mainly linked to beef and milk consumption. Furthermore, understanding the relationships among the STEC serogroups in the different Brazilian regions and sources (livestock, food, humans) in recent years is crucial for the future monitoring and control strategies concerning this relevant pathogen [4]. There are some documents that have compiled global STEC data based on sites of health institutions and overview by continent [15,16]. However, the prevalence and distribution of STEC serogroups in Brazil remains unclear. In this context, the aim of the present study was to conduct the first systematic review of Escherichia coli STEC with a focus on Brazil and to compare the presence of serogroups detected in food products, animals and humans.
2. Materials and Methods
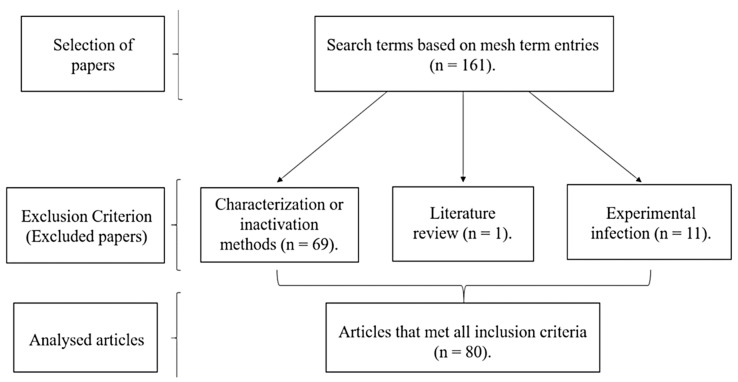
Data were obtained from online databases PubMed, Scielo, Lilacs, Web of Science and Cochrane BVS. The date interval filter was set from January 2000 to December 2018, accessed between 10 September 2018 and 2 January 2019. Papers were selected according to the Prisma guidelines and flow diagram [17]. Therefore, from each database using the Mesh term entries: “Verotoxigenic Escherichia coli” OR “Verotoxigenic” OR “STEC” OR “Shiga Toxigenic E. coli” OR “Shiga Toxigenic Escherichia coli” OR “Shiga Toxin-Producing Escherichia coli” OR “VTEC” OR “Vero Cytotoxin-Producing Escherichia coli” OR “Verotoxigenic E. coli” OR “Verotoxigenic Escherichia coli” OR “Verotoxin-Producing Escherichia coli” AND “Brazil”. Papers in both English and Portuguese were included in this review. Duplicates were traced and excluded. A total of 161 papers were collected, independent of sample size or culture/detection methods. The full text for each paper was obtained and evaluated individually (Figure 1). Papers were excluded when reporting the characterization of strains isolated from another published article, when assessing decreases in intentional contamination (inactivation methods), and when composed of literature reviews or experimental infection. After the application of these criteria, a total of 80 papers were selected.
Figure 1.
Papers selection and exclusion scheme.
3. Results
3.1. Animals
Data on livestock were evaluated in 35 scientific articles, analyzing the presence of STEC through the collection of feces from healthy animals and those with diarrhea (Table 1). The frequency of STEC was heterogeneous and determination of the presence of STEC in herds presents a challenge. STEC contamination rates in cattle ranged from 17.5% to 71.0%, and relevant serogroups O157:H7, O113:H21 and O111 were detected.
Table 1.
Prevalence of Shiga toxin-producing Escherichia coli (STEC) isolated in animals reared in Brazil.
| Host | State | Number of Samples | Prevalence | Serotype or Genes Amplified | Author |
|---|---|---|---|---|---|
| Bovine and Buffaloes | Rio Grande do Sul | 243 feces from dairy cattle | 49% | O157:H-, O91:H-, O125:H-, O119:H-, O112:H-, O29:H- | [19] |
| São Paulo | 182 E. coli isolated from milk samples | 12.08% | stx1 and stx2 | [20] | |
| São Paulo | 153 fecal samples | 25.5% | O113:H21, O157:H7, O111:H-, O22:H8 | [13] | |
| Rio Grande do Sul | 243 feces from dairy cattle | 48.9% | O157, O157:NM, O91:NM, O112:NM | [21] | |
| Minas Gerais | 100 water buffaloes | 37% | O137:H41, O74:H25, O159:H21, O41, O77:H18, O88:H25, O116:H21, O141:H49, O178:H19, O23:H7, O82:H8, O93:H19, O59:H8, O113:H21, O93:H19, O156:H21, O22:H16, O49, O49:H21, O77:H41, O176:H2, O93:H16, O79:H14 | [22] | |
| Minas Gerais | 205 healthy beef and dairy cattle, and 106 reared goats | 57.5% (goats) 39.2% (beef cattle 17.5% in dairy cattle |
O181:H4, O22:H8, O104:H2, O116:H21, O105:H8, O157:H7, O98:H14, O22:H16, O22:H4, O150:H8, O179:H8, O79:H14, O6:H49, O19:H7, O91:H21, O141:H49, O178:H19, O174:H21, O174:H8, O39:H49, O124:H21 | [23] | |
| Paraná | 190 healthy cattle | 36% | O6:H34, O10:H42, O17:H41, O22:H8, O22:H16, O41:H2, O74:H42, O79:H, O82:H8, O98:H41, O110:H2, O113:H21, O117:H8, O124:H11, O159:H21, O174:H21, O175:H21, O178:H19, O179:H8, O181:H4 | [24] | |
| Goiás | 198 rectal swabs | 72.73% | stx1, stx2, ehxA | [25] | |
| Rio Grande do Sul | 108 carcass swabs | 20.37% | O157:H7 | [26] | |
| Birds | São Paulo | 171 fecal samples | 19.4% | stx1 and stx2 | [27] |
| São Paulo | 516 fecal samples | 0.74% | stx 2 | [28] | |
| São Paulo | 446 fecal samples | 9.8% | O6, O48 | [29] | |
| Minas Gerais | 100 fresh fecal samples | 2% | stx1 and stx2 | [30] | |
| Ceará | Case report | - | stx 1 | [31] | |
| São Paulo | 118 pigeons and 38 great egrets | 2.5% (pigeons) | stx 2f | [32] | |
| São Paulo | Case report | - | O137:H6 | [33] | |
| Sheep | São Paulo | 48 sheep | 52.1% | O5:H-, O16:H-, O75:H-, O75:H8, O87:H16, O91:H-, O146:H21, O172:H- OR:H-, ONT:H-, ONT:H16 | [34] |
| São Paulo | 330 feces and 99 carcass samples | 2.72% (Feces), 1.01% (Carcass) | O5, O75, and O91 | [35] | |
| Paraná | Case report | - | O103:H2 | [36] | |
| Paraná | 130 fecal samples | 50% | O76:H19 and O65:H– | [37] | |
| Goiás | 115 E. coli strains | 78.3% | stx1, stx2 | [38] | |
| Calves | Mato Grosso do Sul | 205 E. coli strains | 9.75% (stx1), 6.34% (stx2) | O26:H, O111:H, O118:H16 | [39] |
| São Paulo | 139 diarrheic and 205 non-diarrheic fecal samples | 12.7% | O113:H21, O118:H16, O123:H2, O111:NM, O111:H8 | [40] | |
| São Paulo | 264 diarrheic and 282 healthy fecal samples | 12% | O7:H7, O7:H10, O48:H7, O111:H19, O123:H2, O132:H51, O173:H(-), O175:H49 | [41] | |
| Paraná | 29 diarrheic and 21 healthy fecal samples | 101 strains | O1, O3, O7, O8, O17, O23, O78, O144, O146, ONT, O26, O55, O103, O117, O123, O124, O153, O15, O128, O175, O119, O4, O156 | [42] | |
| Minas Gerais | 850 fecal samples | 20.9% | stx1, eae, iha, toxB, ehxA, efa-1, saa, astA | [43] | |
| Chickens | São Paulo | 110 APEC E. coli samples | 30.90% | stx1, stx2 | [44] |
| Pigs | Mato Grosso | 74 lumen content samples | 2.2% | stx 2 | [45] |
| Dogs | São Paulo | 25 feces from diarrheic dogs | 48% | O157:H7 and stx1, stx2, eaeA | [46] |
| Cats | São Paulo | 40 feces samples and 3 urine infection samples | 0% | - | [47] |
| Dogs and Cats | Paraná | 50 cat feces and 50 dogs | 0% | - | [48] |
| Rabbits | São Paulo | 178 E. coli isolates | 5.05% | O20:H28, O41:H-, O103:H19, O110:H6, O126:H- O126:H20, O128:H2, O132:H2, O153:H7 | [49] |
| Avian Organic Fertilizers | Paraná | 40 fertilizers | 4.7% | stx1, stx2 | [50] |
| Stomoxys calcitrans | Rio de Janeiro | 40 Stomoxys calcitrans flies | 15% | stx1, stx2 and eae | [51] |
In calves, STEC prevalence rates ranged from 12.0% to 20.9%, and serogroups O26, O103 and O111 were detected. These serogroups are monitored in meat by control agencies, such as the EFSA (European Food Safety Authority) and USDA-FSIS (Food Safety and Inspection Service), which require their absence in meat products. In addition, STEC were isolated from 2.7% to 78.3% of sheep and O103:H2 was detected in sheep (Table 1) and has been associated with human disease cases in Brazil [18]. Other animals were also positive for STEC such as pigs (2.2%) and rabbits (5.1%). In chickens, a study evaluated 110 strains of avian pathogenic Escherichia coli (APEC) and noted that stx1 and stx2 were present in 30.9% of the strains, indicating possible dispersion of the stx genes between STEC and APEC. In addition, stx1 and stx2 were detected in 4.7% of poultry litter samples in the south of Brazil (Table 1).
In other species that could possibly act as vectors, STEC rates were 0.7% to 20.4% in wild birds and 15% in stable flies (Stomoxys calcitrans). Prevalence rates in dogs ranged between 0 and 48%, but to date there are no reports of STEC in cats in Brazil.
3.2. Food
The Shiga toxin-producing contamination in food was evaluated in 23 scientific articles (Table 2). In six studies, the presence of STEC in beef was assessed, with prevalence rates ranged from 0 to 27.5%, the serogroups O157 and the “big six” (O26, O45, O103, O111, O121, and O145) were not isolated. In milk, the presence of stx1 and stx2 in E. coli ranged from 0 to 31.1%, but isolates were not serotyped in these studies [52,53,54]. However, STEC were detected in 0 to 14% of cheese samples. In addition, the presence of O111, O55, and O157:H7, serogroups frequently linked to food-borne disease outbreaks worldwide [55] were also found in Brazilian cheese.
Table 2.
Prevalence of Shiga toxin-producing Escherichia coli isolated in food produced in Brazil.
| Matrix | State | Number of Samples | Prevalence | Serotype or Genes Amplified | Author |
|---|---|---|---|---|---|
| Beef | São Paulo | 204 bovine carcass swabs | 27.5% (rainy season), 17.5% (dry season) | stx1 and stx2 | [58] |
| São Paulo | 250 raw ground beef samples | 1.6% | O93:H19, O174:HNT | [59] | |
| São Paulo | 91 beef samples | 2.1% | stx 2 | [60] | |
| São Paulo | 70 raw kibe samples | 2.8% | O125:H19, O149:H8 | [61] | |
| São Paulo | 552 meat products samples | 0% | - | [62] | |
| Rio Grande do Sul | 5 beef jerky samples | 0% | - | [63] | |
| Mato Grosso | 80 samples | 10% | O83:H19, O26:HNT, O73:H45, O8:H21, O79:H7, O113:H21, O22:H16, O117:H7, O21:H19, O132:H21 | [4] | |
| Milk | São Paulo | 30 milk samples | 3.3% | stx1, stx2 | [64] |
| Minas Gerais Rio Grande do Sul |
670 bovine mastitis milk samples | 8.6% | stx1, stx2 | [65] | |
| Rio Grande do Sul | 101 milk samples | 31.1% | stx1, stx2 | [52] | |
| São Paulo | 62 milk samples | 0% | - | [53] | |
| Paraná | 87 milk E. coli strains | 0% | - | [54] | |
| Cheese | Minas Gerais | 50 cheese samples | 14% | O125, O111, O55, O119 | [66] |
| Minas Gerais | 30 cheese samples | 0% | - | [67] | |
| Goiás | 60 cheese samples | 6.7% | O157:H7 | [68] | |
| Minas Gerais | 147 E. coli strains isolated from 38 cheeses | 9.5% | stx 1 | [69] | |
| Water | São Paulo | 133 E. coli isolates | 0.75% | stx 2 | [70] |
| Paraná | 1850 drinking water samples | 0.65% | ehxA, saa, lpfAO113, iha, subAB, cdtV | [71] | |
| Rio de Janeiro | 178 E. coli isolates | 2.8% | stx 1 | [72] | |
| São Paulo | 25 water samples | 19 isolates | stx2, rfbEO157:H7 | [73] | |
| Rio Grande do Sul | 219 water samples | 5.93% | O157:H7 | [74] | |
| Vegetable | Rio Grande do Sul | 260 lettuce samples | 0.76% | O157:H7 | [75] |
| Shrimp meat | São Paulo | 42 chilled shrimp samples | 0% | - | [76] |
Contaminated water is increasingly linked to STEC outbreaks associated with fruits and vegetables in Europe [56]. Although STEC contamination is usually related to products of animal-origin, contamination of plant products occurs as a result of cross-contamination [57]. In Brazil, STEC prevalence rates in water ranged from 0.65 to 5.93% with only a single sample testing positive for O157:H7. Water can also be a source of contamination of plant products. For example, in a study with lettuce, 0.76% of samples were contaminated with O157:H7.
3.3. Human
The presence of STEC contamination in humans was evaluated in 22 scientific articles (Table 3). In five studies, STEC infection was reported in case reports or in samples collected from patients with hemolytic uremic syndrome, and was associated with O26:H11, O103:H2, O165, O157, O157:H7 and O104:H4 (enteroaggregative group with the acquisition of the stx gene). Through whole genome sequencing, the O104:H4 strain in Brazil was found to be similar to a strain isolated from an American citizen diagnosed with hemolytic uremic syndrome (HUS) who had traveled to Germany during the 2011 HUS outbreak [14].
Table 3.
Prevalence of Shiga toxin-producing Escherichia coli isolated in cases of foodborne disease in Brazil.
| State | Number of Samples | Prevalence | Toxin Type | Serotype | Author |
|---|---|---|---|---|---|
| São Paulo | 1010 children feces samples | 0.3% | stx 1 | O111ac | [77] |
| 3 patient strain samples | Case report | stx1 and stx2 | O157:H7 | [78] | |
| 2607 samples from patients with diarrhea | 1.1% | stx1 and stx2 | O55:H19, O93:H19, O118:H16, O157:H7 O111:HNM, O111:H8, O26:H11 | [79] | |
| 1 haemolytic anaemia and 2 faecal with diarrhea samples | Case report | stx1 and stx2 | O103:H2 | [18] | |
| Feces from 19-month-old children | Case report | stx 2 | O165:HNM | [80] | |
| 337 children and 102 HIV adult patients | 1.8% | stx1 and stx2 | O111:HNM, O157:H7, ONT:H2, O103:H2, O118:H16, O77:H18, ONT:H8 | [81] | |
| 13 post-diarrheal HUS cases | Case report | stx2, stx2c | O26:H11, O157:H7, O165:H- | [82] | |
| Stool specimens collected from 115 children | 0.86% | stx1 and stx2 | ONT:H19 | [83] | |
| Stools and ileum biopsy of a 51-year-old woman | Case report | stx 1 | O104:H4 | [14] | |
| 5047 cases of human infection | 4.2% | stx1a, stx1d, stx2a, stx2c, stx2d, and stx2e | O8:H19, O24:H4, O26:H11, O71:H8, O91:H14, O100:HNM, O103:HNM, O111:H11, O111:H8, O111:HNM, O118:H16, O123:H2, O123:HNM, O145:HNM, O153:H21, O153:H7, O157:H7, O177:HNM, O178:H19 | [84] | |
| Bahia | 1233 children feces | 1.6% | stx1, stx2 | O26:H11, O21:H21 | [85] |
| 1233 children feces | 1.6% | Not disclosed | 0% | [86] | |
| 1207 children feces | 0.6% | stx 2 | O157:H7, O26:H11, O111:H- | [87] | |
| Paraná | 306 culture stool samples | 0.65% | stx1, stx2 | O69:H11, O178:H19 | [88] |
| 141 children fecal samples | 2.83% | stx1, stx2 | 0% | [89] | |
| Rio de Janeiro | 307 children samples | 0% | 0% | 0% | [90] |
| Human gastroenteritis caused by E. coli O157:NM | Case report | stx 2 | O157:NM | [91] | |
| Rio Grande do Norte and Santa Catarina | 2 strains | Case report | stx1 and stx2 | O157 | [92] |
| Rio Grande do Sul | 375 children feces samples | 0.26% | stx 2 | O91:H21 | [93] |
| Piauí | 400 children feces samples | 0.4% | Not disclosed | O125 | [94] |
| Espírito Santo | 560 children feces samples | 0.17% | Not disclosed | 0% | [95] |
| Paraíba | 580 children feces samples | 0% | 0% | 0% | [96] |
In other studies of diarrhea or healthy subjects, the serogroups predominantly detected were O26:H11, O103:H2, O111 Not typeable (NT), O118:H16, O165:NT and O157:H7 (Figure 2).
Figure 2.
Main serotypes present in each assessed group reported from 2000 to 2018.
Some of these serogroups were reported in animal and food studies in Brazil. For instance, the O111 and O157 serogroups were verified in all three groups. Our hypothesis is that the dispersion of the strains has contaminated all stages of the food chain (pre and post-processing). Moreover, the O103 strains in animals were linked to those found in humans. In contrast, stains O165 and O118:H16 were only detected in human clinical cases.
4. Discussion
The most frequent serogroups described in peer-reviewed papers in samples collected in Brazil are represented in Figure 2. As with studies in other countries, O157:H7 was the serotype most frequently linked to human cases and also had the highest occurrence rates in animal, food and humans in Brazil. However, due to the large-scale outbreaks related to this serotype in the USA in 1982 and 1993, much effort has been invested in methods to detect this specific serotype. It can be readily isolated as non-sorbitol fermenting colonies (the main O157 characteristic) and identified using PCR primers designed specifically for the O157 antigen or the flagellum H7. Consequently, a higher prevalence of O157:H7 in Brazil might also be related to its ease of detection.
Serogroup O111 was also reported in animals, food and humans and has been linked to clinical human cases in Brazil [13,66,77] and worldwide [55,97]. O111 is one of six non-O157 serogroups classified by the USDA-FSIS (2013) as foodborne pathogens that should be monitored during meat production. Considering the serogroups listed by the USDA-FSIS, three: O157, O111, O26:H11 and O103:H2 have been associated with foodborne disease in Brazil (Figure 2). However, two other non-O157 strains, O165 and O118:H16 are not part of the USDA-FSIS “big six”, but were linked to cases of diarrhea, hemolytic uremic syndrome or hemorrhagic colitis in Brazil.
Serogroup O165 has been reported in cases of hemolytic uremic syndrome in Japan [98], and Germany [99]. This serotype has also been identified in cattle in the United States [100]. In addition, its genome has recently been made available on the GenBank/NCBI platform [101] for comparisons and genetic investigations, emphasizing the increasing importance of this serogroup in food production and human infection. Serotype O118:H16 has been linked to foodborne diseases in Germany [102], the United States [103], and its genome sequence was included in GenBank/NCBI in 2014 [104]. Detection of other new STEC in future cases of human disease is likely.
Studies evaluating different cattle feeds indicate that different diets may increase the acid resistance of Escherichia coli strains, enabling the bacteria to survive during passage through the human stomach [105]. Strain resistance due to diet may be responsible for the selection of some strains of STEC in Brazilian herds. In addition, a recent study by Acquaotta [106] identified a correlation between STEC infections and climatic differences in North America, where more cases of contamination were reported during warm periods as compared to cold periods. This result emphasizes the need for monitoring and control during food production as Brazil’s tropical climate may increase the risk of STEC infections.
Foods showed the greatest diversity among serogroups, followed by animal sources (Figure 2). This diversity may be related to several factors, such as breeding and production heterogeneity, the nature of the animal and food carrier, the innate (animal) or initial (food) microbial load and the applied methodology (antibiotic use for pre-enrichment, colony morphology and analyzed genes). Moreover, serogroups detected in humans display greater homogeneity, perhaps related to toxin subtypes (stx2e) or other genetic factors including virulence [107,108].
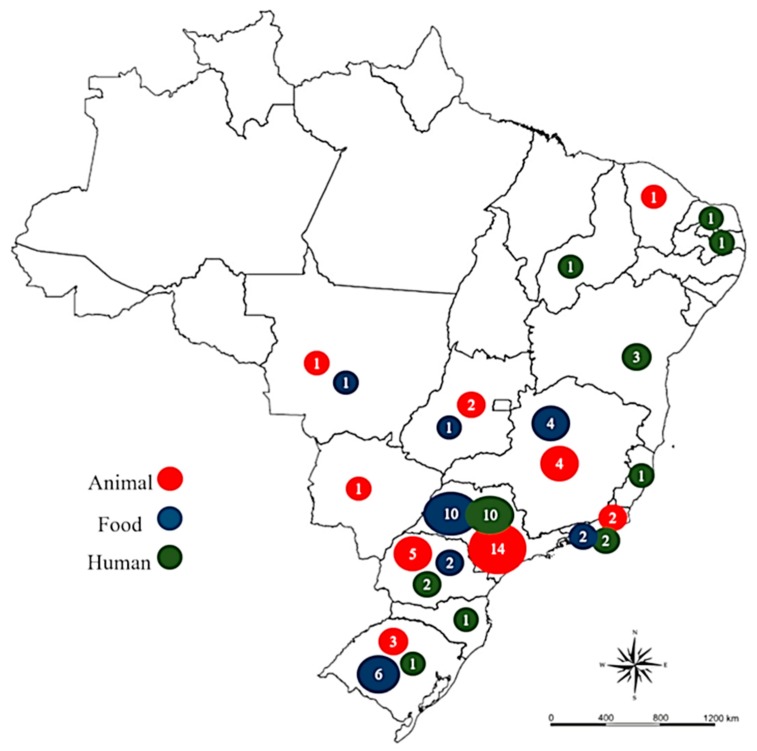
Studies were concentrated mainly in Brazil’s most developed areas and especially in the state of Sao Paulo (Figure 3), with the southern and southeastern regions accounting for 69% of the Brazilian population [6]. However, the majority of animal and grain-production occurs in central Brazil, which is responsible for 34.4% of animal and 42% of grain production [109]. Differences in the number of STEC studies directly reflect national development and demographics. Further studies in the central region would provide a better description of STEC present in livestock and food and would aid in obtaining a more accurate national picture of foodborne STEC risks.
Figure 3.
Distribution of STEC assessment studies carried out in Brazil from 2000 to 2018.
Currently, the main legislation in force in Brazil concerning food production is Resolution 12 of 2001 [110], which applies to coliforms in frozen or fresh meat. This resolution, however, does not call for STEC serogroup analyses in any food products. However, the Ministry of Livestock and Food Supply (MAPA) established an internal standard in 2013, requesting E. coli STEC tests for O157 and the “big six” non-O157 in beef destined for export [111]. However, any data about the frequency of STEC isolation has not been released.
The analysis procedure determined by MAPA is based on the methodology used by the USDA FSIS [112], comprising molecular screening followed by cultivation on Rainbow™ agar O157 (Biolog Inc, Hayward, USA) and serology of the positive isolates. The use of this methodology assists in production control and monitoring of the main STEC, followed by subsequent implementation of measures to combat contamination. However, this measure is only for beef, and it is necessary to extend the standard to other food matrices as the present review demonstrated high STEC contamination rates in milk and cheese, as well as water. The STEC causing infection and for which microbiological monitoring and control during production is required show some geographical variation and new STEC continually evolve. Instead of continuing to add new serogroups to a long list of food adulterants, microbiological monitoring could instead aim for the absence of strains that possess Shiga-toxin gene(s) in their genome.
Moreover, the improvement and cost reduction of molecular tools such as whole genomic sequencing (WGS), metagenomics, and others will facilitate the understanding of serotype epidemiology and the dispersion of these strains in neighboring countries and in other continents. Epigenetics advances will also improve the understanding of gene expression and the impact of good animal management practices, as well as possible genome mutations that may influence virulence and antimicrobial resistance profiles [113].
5. Conclusions
The epidemiological data presented in this review indicate that O157:H7, O26, O103 and O111 strains, classified as foodborne pathogens and monitored by the USDA-FSIS (USA), U.S. FDA, EFSA (European Food Safety Authority - EU) and MAPA (Ministry of Agriculture, Livestock and Supply - Brazil), are currently circulating in different regions of Brazil. Although no STEC outbreak cases in Brazil have been reported, several animal, food and human studies have indicated the presence of STEC in Brazil and it has been related to several foodborne outbreaks around the world. Thus, improved epidemiological monitoring and food production control is necessary. Novel studies should be financed in regions presenting significant agricultural production, such as Brazil’s central region in order to better assess potential threats and prevent human STEC infections.
Author Contributions
V.S.C. and E.E.d.S.F. methodology, investigation and data curation, writing—original draft preparation; K.S., T.M., writing—review and editing; E.E.S.F and C.C.A.-J., visualization, project administration, funding acquisition.
Funding
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior―CAPES/Brazil, Visiting Professor, Process: PVEX-88881.169965/2018-01, Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (Process: 310462/2018-5 and 311422/2016-0) Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro—FAPERJ (grant number E-26/203.049/2017), and Fundação de Amparo a Pesquisa do Estado de Mato Grosso—FAPEMAT (grant number 222388/2015).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.The Observatory of Economic Complexity OEC Brasil (BRA) Exportação, Importação, e Parceiro Comercial. [(accessed on 19 March 2019)];2018 Available online: https://atlas.media.mit.edu/pt/profile/country/bra/
- 2.Stellato G., Storia A.L., Filippis F.D., Borriello G., Villani F., Ercolini D. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl. Environ. Microb. 2016;82:4045–4054. doi: 10.1128/AEM.00793-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atidégla S.C., Huat J., Agbossou E.K., Saint-Macary H., Kakai R.G. Vegetable contamination by the fecal bacteria of poultry manure: Case study of gardening sites in southern Benin. Int. J. Food Sci. Tech. 2016:4767453. doi: 10.1155/2016/4767453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos E.C.C., Castro V.S., Cunha-Neto A., Santos L.F.D., Vallim D.C., Lisbôa R.C., Carvalho R.C.T., Junior C.A.C., Figueiredo E.E.S. Escherichia coli O26 and O113:H21 on carcasses and beef from a slaughterhouse located in Mato Grosso, Brazil. Foodborne Pathog. Dis. 2018;2018:2431. doi: 10.1089/fpd.2018.2431. [DOI] [PubMed] [Google Scholar]
- 5.Gangiredla J., Mammel M.K., Barnaba T.J., Tartera C., Gebru S.T., Patel I.R., Leonard S.R., Kotewicz M.L., Lampel K.A., Elkins C.A., Lacher D.W. Draft genome sequences of Escherichia albertii, Escherichia fergusonii, and strains belonging to six cryptic lineages of Escherichia spp. Genome Announc. 2016;6:e00271-18. doi: 10.1128/genomeA.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brasil. Instituto Brasileiro de Geografia e Estatística―IBGE. População por Região do Brasil. [(accessed on 19 March 2019)];2017 Available online: https://cidades.ibge.gov.br/brasil/panorama.
- 7.Croxen M.A., Law R.J., Scholz R., Keeney K.M., Wlodarska M., Finlay B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013;26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarr P.I., Carre A.G., Chandler W.L. Shiga toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:107–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 9.Feng P.C.H., Delannoy S., Lacher D.W., Bosilevac J.M., Fach P., Beutin L. Shiga Toxin-producing serogroup O91 Escherichia coli strains isolated from food and environmental samples. Appl. Environ. Microb. 2017;83:e01231-17. doi: 10.1128/AEM.01231-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro V.S., Carvalho R.C.T., Conte-Junior C.A., Figueiredo E.E.S. Shiga-Toxin producing Escherichia coli: pathogenicity, supershedding, diagnostic methods, occurrence, and foodborne outbreaks. Compr. Rev. Food Sci. Food Saf. 2017;16:1269–1280. doi: 10.1111/1541-4337.12302. [DOI] [PubMed] [Google Scholar]
- 11.Goncuoglu M., Erol I., Ayaz N.D., Ormanci F.S.B., Kaspar C.W. Isolation and genomic characterization of Escherichia coli O157:H7 in bile of cattle. Ann. Microbiol. 2010;60:293–297. doi: 10.1007/s13213-010-0040-5. [DOI] [Google Scholar]
- 12.Franzin F.M., Sircili M.P. Locus of Enterocyte Effacement: A pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed Res. Int. 2015;2015:534738. doi: 10.1155/2015/534738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irino K., Kato M.A.M.F., Vaz T.M.I., Ramos I.I., Souza M.A., Cruz A.S., Gomes T.A., Vieira M.A., Guth B.E. Serotypes and virulence markers of Shiga toxin-producing Escherichia coli (STEC) isolated from dairy cattle in São Paulo State, Brazil. Vet. Microbiol. 2005;105:29–36. doi: 10.1016/j.vetmic.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Da Silva Santos A., Gomes Romeiro F., Sassaki Y.L., Rodrigues J. Escherichia coli from Crohn’s disease patient displays virulence features of enteroinvasive (EIEC), enterohemorragic (EHEC), and enteroaggregative (EAEC) pathotypes. Gut Pathog. 2015;7:2. doi: 10.1186/s13099-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Food and Agriculture Organization. World Health Organization Shiga Toxin-Producing Escherichia coli (STEC) and Food: Attribution, Characterization and Monitoring. World Health Organization Reports. [(accessed on 25 April 2019)];2019 Available online: https://apps.who.int/iris/handle/10665/272871.
- 16.Majowicz S.E., Scallan E., Jones-Bitton A., Sargeant J.M., Stapleton J., Angulo F.J., Yeung D.H., Kirk M.D. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014;11:447–455. doi: 10.1089/fpd.2013.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moher D., Liberati A., Tetzlaff J., Altman D.G. The PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guth B.E.C., Vaz T.M., Gomes T.A., Chinarelli S.H., Rocha M.M.M., Pestana de Castro A.F., Irino K. Re-emergence of O103:H2 Shiga toxin-producing Escherichia coli infections in Sao Paulo, Brazil. J. Med. Microbiol. 2005;54:805–806. doi: 10.1099/jmm.0.46057-0. [DOI] [PubMed] [Google Scholar]
- 19.Moreira C.N., Pereira M.A., Brod C.S., Rodrigues D.P., Carvalhal J.B., Aleixo J.A. Shiga toxin-producing Escherichia coli (STEC) isolated from healthy dairy cattle in southern Brazil. Vet. Microbiol. 2003;93:179–183. doi: 10.1016/S0378-1135(03)00041-5. [DOI] [PubMed] [Google Scholar]
- 20.Lira W.M., Macedo C., Marin J.M. The incidence of Shiga toxin-producing Escherichia coli in cattle with mastitis in Brazil. J. Appl. Microbiol. 2004;97:861–866. doi: 10.1111/j.1365-2672.2004.02384.x. [DOI] [PubMed] [Google Scholar]
- 21.Sandrini C.N.M., Pereira M., Brod C.S., Carvalhal J.B., Aleixo J.A.G. Escherichia coli verotoxigênica: Isolamento e prevalência em 60 propriedades de bovinos de leite da região de Pelotas, RS, Brasil. Cienc. Rural. 2007;37:175–182. doi: 10.1590/S0103-84782007000100028. [DOI] [Google Scholar]
- 22.Oliveira M.G., Brito J.R.F., Carvalho R.R., Guth B.E.C., Gomes T.A.T., Vieira M.A.M., Kato M.A.M.F., Ramos I.I., Vaz T.M.I., Irino K. Water buffaloes (Bubalus bubalis) identified as an important reservoir of Shiga toxin-producing Escherichia coli in Brazil. Appl. Environ. Microbiol. 2007;73:5945–5948. doi: 10.1128/AEM.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira M.G., Brito J.R.F., Gomes T.A.T., Guth B.E., Vieira M.A., Naves Z.V., Vaz T.M., Irino K. Diversity of virulence profiles of Shiga toxin-producing Escherichia coli serotypes in food-producing animals in Brazil. Int. J. Food Microbiol. 2008;127:139–146. doi: 10.1016/j.ijfoodmicro.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 24.Pigatto C.P., Schocken-Iturrino R.P., Souza E.M., Pedrosa F.O., Comarella L., Irino K., Kato M.A., Farah S.M., Warth J.F., Fadel-Picheth C.M. Virulence properties and antimicrobial susceptibility of Shiga toxin-producing Escherichia coli strains isolated from healthy cattle from Paraná State, Brazil. Can. J. Microbiol. 2008;54:588–593. doi: 10.1139/W08-046. [DOI] [PubMed] [Google Scholar]
- 25.Ferreira M.R.A., Filho E.G.F., Pinto J.F.N., Dias M., Moreira C.N. Isolation, prevalence, and risk factors for infection by shiga toxin-producing Escherichia coli (STEC) in dairy cattle. Trop. Anim. Health Prod. 2014;46:635–639. doi: 10.1007/s11250-014-0541-5. [DOI] [PubMed] [Google Scholar]
- 26.Loiko M.R., de Paula C.M.D., Langone A.C.J., Rodrigues R.Q., Cibulski S., Rodrigues R.D.O., Camargo A.C., Nero L.A., Mayer F.Q., Tondo E.C. Genotypic and antimicrobial characterization of pathogenic bacteria at different stages of cattle slaughtering in southern Brazil. Meat Sci. 2016;116:193–200. doi: 10.1016/j.meatsci.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Gioia-Di Chiacchio R.M.G.D., Cunha M.P.V., Sturn R.M., Moreno L.Z., Moreno A.M., Pereira C.B., Martins F.H., Franzolin M.R., Piazza R.M., Knöbl T. Shiga toxin-producing Escherichia coli (STEC): Zoonotic risks associated with psittacine pet birds in home environments. Vet. Microbiol. 2016;184:27–30. doi: 10.1016/j.vetmic.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Sanches L.A., Gomes M.S., Teixeira R.H.F., Cunha M.P.V., Oliveira M.G.X., Vieira M.A.M., Gomes T.A.T., Knobl T. Captive wild birds as reservoirs of enteropathogenic E. coli (EPEC) and Shiga-toxin producing E. coli (STEC) Braz. J. Microbiol. 2017;48:760–763. doi: 10.1016/j.bjm.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borges C.A., Cardozo M.V., Beraldo L.G., Oliveira E.S., Maluta R.P., Barboza K.B., Werther K., Ávila F.A. Wild birds and urban pigeons as reservoirs for diarrheagenic Escherichia coli with zoonotic potential. J. Microbiol. 2017;55:344–348. doi: 10.1007/s12275-017-6523-3. [DOI] [PubMed] [Google Scholar]
- 30.Silva V.L., Nicoli J.R., Nascimento T.C., Diniz C.G. Diarrheagenic Escherichia coli strains recovered from urban pigeons (Columba livia) in Brazil and their antimicrobial susceptibility patterns. Curr. Microbiol. 2009;59:302–308. doi: 10.1007/s00284-009-9434-7. [DOI] [PubMed] [Google Scholar]
- 31.Bezerra W.G.A., Horn R.V., Silva I.N.G., Siqueira R.A.S., Lucena R.B., Havt A., Medeiros P.H.Q.S., Maciel W.C. Escherichia coli and Salmonella ser. Saintpaul natural co-infection in a free-living ruddy ground dove (Columbina talpacoti): A case report. Arq. Bras. Med. Vet. Zootec. 2017;69:1236–1242. doi: 10.1590/1678-4162-9454. [DOI] [Google Scholar]
- 32.De Oliveira M.C.V., Camargo B.Q., Cunha M.P.V., Saidenberg A.B., Teixeira R.H.F., Matajira C.E.C., Moreno L.Z., Gomes V.T.M., Christ A.P.G., Barbosa M.R.F. Free-ranging synanthropic birds (Ardea alba and Columba livia domestica) as carriers of Salmonella spp. and diarrheagenic Escherichia coli in the vicinity of an urban zoo. Vector-Borne Zoonotic Dis. 2018;18:65–69. doi: 10.1089/vbz.2017.2174. [DOI] [PubMed] [Google Scholar]
- 33.Chiacchio R.M.G., Cunha M.P.V., de Sá L.R.M., Davies Y.M., Pereira C.B.P., Martins F.H., Munhoz D.D., Abe C.M., Franzolin M.R., dos Santos L.F. Novel hybrid of typical enteropathogenic Escherichia coli and Shiga-Toxin-producing E. coli (tEPEC/STEC) emerging from pet birds. Front. Microbiol. 2018;9:2975. doi: 10.3389/fmicb.2018.02975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vettorato M.P., Leomil L., Guth B.E.C., Irino K., Pestana de Castro A.F. Properties of Shiga toxin-producing Escherichia coli (STEC) isolates from sheep in the state of São Paulo, Brazil. Vet. Microbiol. 2003;95:103–109. doi: 10.1016/S0378-1135(03)00153-6. [DOI] [PubMed] [Google Scholar]
- 35.Maluta R.P., Fairbrother J.M., Stella A.E., Rigobelo E.C., Martinez R., de Ávila F.A. Potentially pathogenic Escherichia coli in healthy, pasture-raised sheep on farms and at the abattoir in Brazil. Vet. Microbiol. 2014;169:89–95. doi: 10.1016/j.vetmic.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Martins F.H., Guth B.E.C., Piazza R.M.F., Blanco J., Pelayo J.S. First description of a Shiga toxin-producing Escherichia coli O103:H2 strain isolated from sheep in Brazil. [(accessed on 25 April 2019)];J. Infect. Dev. Ctries. 2014 8:126–128. doi: 10.3855/jidc.3615. Available online: https://jidc.org/index.php/journal/article/view/24423724. [DOI] [PubMed] [Google Scholar]
- 37.Martins F.H., Guth B.E.C., Piazza R.M., Leão S.C., Ludovico A., Ludovico M.S., Dahbi G., Marzoa J., Mora A., Blanco J. Diversity of Shiga Toxin-producing Escherichia coli in sheep flocks of Paraná state, southern Brazil. Vet. Microbiol. 2015;175:150–156. doi: 10.1016/j.vetmic.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Ferreira M.R.A., Silva T.S., Stella A.E., Conceição F.R., dos Reis E.F., Moreira C.N. Detection of virulence factors and antimicrobial resistance patterns in Shiga Toxin-producing Escherichia coli isolates from sheep. Pesqui. Vet. Brasil. 2015;35:775–780. doi: 10.1590/S0100-736X2015000900002. [DOI] [Google Scholar]
- 39.Salvadori M.R., Valadares G.F., Leite D.S., Blanco J., Yano T. Virulence factors of Escherichia coli isolated from calves with diarrhea in Brazil. [(accessed on 25 April 2019)];Braz. J. Microbiol. 2003 34:230–235. doi: 10.1590/S1517-83822003000300009. Available online: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1517-83822003000300009. [DOI] [Google Scholar]
- 40.Leomil L., Aidar-Ugrinovich L., Guth B.E.C., Irino K., Vettorato M.P., Onuma D.L., de Castro A.F. Frequency of Shiga toxin-producing Escherichia coli (STEC) isolates among diarrheic and non-diarrheic calves in Brazil. Vet. Microbiol. 2003;97:103–109. doi: 10.1016/j.vetmic.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Aidar-Ugrinovich L., Blanco J., Blanco M., Blanco J.E., Leomil L., Dahbi G., Mora A., Onuma D.L., Silveira W.D., Pestana de Castro A.F. Serotypes, virulence genes, and intimin types of Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) isolated from calves in São Paulo, Brazil. Int. J. Food Microbiol. 2007;115:297–306. doi: 10.1016/j.ijfoodmicro.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 42.Moura C., Ludovico M., Valadares G.F., Gatti M.S.V., Leite D.S. Detection of virulence genes in Escherichia coli strains isolated from diarrheic and healthy feces of dairy calves in Brazil. Arq. Inst. Biol. 2012;79:273–276. doi: 10.1590/S1808-16572012000200016. [DOI] [Google Scholar]
- 43.Coura F.M., Araújo D.S., Mussi J.M.S., Silva M.X., Lage A.P., Heinemann M.B. Characterization of virulence factors and phylogenetic group determination of Escherichia coli isolated from diarrheic and non-diarrheic calves from Brazil. Folia Microbiol. 2017;62:139–144. doi: 10.1007/s12223-016-0480-9. [DOI] [PubMed] [Google Scholar]
- 44.Bernedo-Navarro R.A., Miyachiro M.M., Conceição R.A., Yano T. Non O157:H7 avian pathogenic Shiga toxin-producing Escherichia coli isolated from lesions on broiler chickens in Brazil. Microbiol Immunol. 2014;58:266. doi: 10.1111/1348-0421.12143. [DOI] [PubMed] [Google Scholar]
- 45.Martins R.P., da Silva M.C., Dutra V., Nakazato L., da Silva Leite D. Preliminary virulence genotyping and phylogeny of Escherichia coli from the gut of pigs at slaughtering stage in Brazil. Meat Sci. 2013;93:437–440. doi: 10.1016/j.meatsci.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Paula C.J.S., Marin J.M. Occurrence of non-O157 Shiga toxin-producing Escherichia coli in dogs with diarrhea. Cienc. Rural. 2008;38:1682–1686. doi: 10.1590/S0103-84782008000600029. [DOI] [Google Scholar]
- 47.Caliman M.W., Marin J.M. Virulence genes in Escherichia coli isolated from feces and urine of cats. Arq. Bras. Med. Vet. Zootec. 2014;66:1287–1290. doi: 10.1590/1678-6694. [DOI] [Google Scholar]
- 48.Puño-Sarmiento J., Medeiros L., Chiconi C., Martins F., Pelayo J., Rocha S., Blanco J., Blanco M., Zanutto M., Kobayashi R. Detection of diarrheagenic Escherichia coli strains isolated from dogs and cats in Brazil. Vet. Microbiol. 2013;166:676–680. doi: 10.1016/j.vetmic.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Penteado A., Ugrinovich L., Blanco J., Blanco M., Blanco J.E., Mora A., Andrade J.R., Corrêa S.S., Pestana de Castro A.F. Serobiotypes and virulence genes of Escherichia coli strains isolated from diarrheic and healthy rabbits in Brazil. Vet. Microbiol. 2002;89:41–51. doi: 10.1016/S0378-1135(02)00148-7. [DOI] [PubMed] [Google Scholar]
- 50.Puño-Sarmiento J., Gazal L., Medeiros L., Nishio E.K., Kobayashi R.K., Nakazato G. Identification of diarrheagenic Escherichia coli strains from avian organic fertilizers. Int. J. Environ. Res. Public Health. 2014;11:8924–8939. doi: 10.3390/ijerph110908924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Castro B.G., de Souza M.M.S., Regua-Mangia A.H., Bittencourt A.J. Occurrence of Shiga-toxigenic Escherichia coli in Stomoxys calcitrans (Diptera: Muscidae) Ver. Bras. Parasitol. Vet. 2013;22:318–321. doi: 10.1590/S1984-29612013000200052. [DOI] [PubMed] [Google Scholar]
- 52.Vendramin T., Kich D.M., Molina R.D., de Souza L.F.V., Salvatori R.U., Pozzobon A., Bustamante-Filho I.C. Molecular screening of bovine raw milk for the presence of Shiga toxin-producing Escherichia coli (STEC) on dairy farms. Food Sci. Technol. 2014;34:604–608. doi: 10.1590/1678-457x.6422. [DOI] [Google Scholar]
- 53.Beraldo L.G., Borges C.A., Maluta R.P., Cardozo M.V., Rigobelo E.C., de Ávila F.A. Detection of Shiga toxigenic (STEC) and enteropathogenic (EPEC) Escherichia coli in dairy buffalo. Vet. Microbiol. 2014;170:162–166. doi: 10.1016/j.vetmic.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Hoffmann S.A., Pieretti G.G., Fiorini A., Patussi E.V., Cardoso R.F., Mikcha J.M. Shiga-toxin genes and genetic diversity of Escherichia coli isolated from pasteurized cow milk in Brazil. J. Food Sci. 2014;79:M1175–M1180. doi: 10.1111/1750-3841.12477. [DOI] [PubMed] [Google Scholar]
- 55.Watahiki M., Isobe J., Kimata K., Shima T., Kanatani J., Shimizu M., Nagata A., Kawakami K., Yamada M., Izumiya H., Iyoda S., Morita-Ishihara T., Mitobe J., Terajima J., Ohnishi M., Sata T. Characterization of enterohemorrhagic Escherichia coli O111 and O157 strains isolated from outbreak patients in Japan. J. Clin. Microbiol. 2014;52:2757–2763. doi: 10.1128/JCM.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gobin M., Hawker J., Cleary P., Inns T., Gardiner D., Mikhail A., McCormick J., Elson R., Ready D., Dallman T., et al. National outbreak of Shiga toxin-producing Escherichia coli O157:H7 linked to mixed salad leaves, United Kingdom, 2016. Euro Surveill. 2018;23:17-00197. doi: 10.2807/1560-7917.ES.2018.23.18.17-00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jongman M., Korsten L. Irrigation water quality and microbial safety of leafy greens in different vegetable production systems: A review. Food Rev. Int. 2018;34:308–328. doi: 10.1080/87559129.2017.1289385. [DOI] [Google Scholar]
- 58.Rigobelo E.C., Stella A.E., Ávila F.A., Macedo C., Marin J.M. Characterization of Escherichia coli isolated from carcasses of beef cattle during their processing at an abattoir in Brazil. Int. J. Food Microbiol. 2006;110:194–198. doi: 10.1016/j.ijfoodmicro.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 59.Bergamini M.A.M., Simões M., Irino K., Gomes T.A.T., Guth B.E.C. Prevalence and characteristics of Shiga toxin-producing Escherichia coli (STEC) strains in ground beef in São Paulo, Brazil. Braz. J. Microbiol. 2007;38:553–556. doi: 10.1590/S1517-83822007000300032. [DOI] [Google Scholar]
- 60.Rodolpho D., Marin J.M. Isolation of Shiga toxigenic Escherichia coli from butcheries in Taquaritinga city, state of São Paulo, Brazil. Braz. J. Microbiol. 2007;38:599–602. doi: 10.1590/S1517-83822007000400004. [DOI] [Google Scholar]
- 61.Peresi J.T.M., Almeida I.A.Z.C., Vaz T.M.I., Hernandes R.T., Teixeira I.S.C., Silva S.I.L., Graciano R.A.S., Pinheiro S.R., dos Santos L.F. Search for diarrheagenic Escherichia coli in raw kibbe samples reveals the presence of Shiga toxin-producing strains. Food Control. 2016;63:165–170. doi: 10.1016/j.foodcont.2015.11.018. [DOI] [Google Scholar]
- 62.Ristori C.A., Rowlands R.E.G., Martins C.G., Barbosa M.L., Dos Santos L.F., Jakabi M., de Melo Franco B.D.G. Assessment of consumer exposure to Salmonella spp., Campylobacter spp., and Shiga Toxin–Producing Escherichia coli in meat products at retail in the city of São Paulo, Brazil. Foodborne Pathog. Dis. 2017;14:447–453. doi: 10.1089/fpd.2016.2270. [DOI] [PubMed] [Google Scholar]
- 63.Fernandes F.P., Voloski F.L.S., Ramires T., Haubert L., Reta G.G., Mondadori R.G., Silva W.P.D., Conceição R.C.D.S.D., Duval E.H. Virulence and antimicrobial resistance of Salmonella spp. and Escherichia coli in the beef jerky production line. [(accessed on 25 April 2019)];FEMS Microbiol. Lett. 2017 364:fnx083. doi: 10.1093/femsle/fnx083. Available online: https://academic.oup.com/femsle/article/364/9/fnx083/3746133. [DOI] [PubMed] [Google Scholar]
- 64.Vicente H.I.G., Amaral L.A., Cerqueira A.M.F. Shigatoxigenic Escherichia coli serogroups O157, O111 and O113 in feces, water and milk samples from dairy farms. Braz. J. Microbiol. 2005;36:217–222. doi: 10.1590/S1517-83822005000300003. [DOI] [Google Scholar]
- 65.Rangel P., Marin J.M. Analysis of Escherichia coli isolated from bovine mastitic milk. Pesqui Vet. Bras. 2009;29:363–368. doi: 10.1590/S0100-736X2009000500001. [DOI] [Google Scholar]
- 66.Paneto B.R., Schocken-Iturrino R.P., Macedo C., Santo E., Marin J.M. Occurrence of toxigenic Escherichia coli in raw milk cheese in Brazil. Arq. Bras. Med. Vet. Zootec. 2007;59:508–512. doi: 10.1590/S0102-09352007000200035. [DOI] [Google Scholar]
- 67.Okura M.H., Marin J.M. Survey of Minas frescal cheese from southwest Minas Gerais for virulence factors and antimicrobial resistance in Escherichia coli isolates. Cienc. Rural. 2014;44:1506–1511. doi: 10.1590/0103-8478cr20131237. [DOI] [Google Scholar]
- 68.Carvalho R.N., Oliveira A.N., Mesquita A.J., Rezende C.S.M., Mesquita A.Q., Romero R.A.M. PCR and ELISA (VIDAS ECO O157®) Escherichia coli O157:H7 identification in Minas Frescal cheese commercialized in Goiânia, GO. Braz. J. Microbiol. 2014;45:7–10. doi: 10.1590/S1517-83822014000100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cardoso P., Marin J.M. Occurrence of non-O157 Shiga toxin-encoding Escherichia coli in artisanal mozzarella cheese in Brazil: risk factor associated with food workers. Food Sci. Technol. 2017;37:41–44. doi: 10.1590/1678-457x.06316. [DOI] [Google Scholar]
- 70.Orsi R.H., Stoppe N.C., Sato M.I.Z., Gomes T.A., Prado P.I., Manfio G.P., Ottoboni L.M. Genetic variability and pathogenicity potential of Escherichia coli isolated from recreational water reservoirs. Res. Microbiol. 2007;158:420–427. doi: 10.1016/j.resmic.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Lascowski K.M.S., Guth B.E.C., Martins F.H., Rocha S.P., Irino K., Pelayo J.S. Shiga toxin-producing Escherichia coli in drinking water supplies of north Paraná State, Brazil. J. Appl. Microbiol. 2013;114:1230–1239. doi: 10.1111/jam.12113. [DOI] [PubMed] [Google Scholar]
- 72.Rebello R.C.L., Regua-Mangia A.H. Potential enterovirulence and antimicrobial resistance in Escherichia coli isolates from aquatic environments in Rio de Janeiro, Brazil. Sci. Total Environ. 2014;490:19–27. doi: 10.1016/j.scitotenv.2014.04.040. [DOI] [PubMed] [Google Scholar]
- 73.Ribeiro L.F., Barbosa M.M.C., Rezende P.F., Guariz C.S., Maluta R.P., Rossi J.R., Rossi G.A., Lemos M.V., do Amaral L.A. Shiga toxigenic and enteropathogenic Escherichia coli in water and fish from pay-to-fish ponds. Lett. Appl. Microbiol. 2016;62:216–220. doi: 10.1111/lam.12536. [DOI] [PubMed] [Google Scholar]
- 74.Decol L.T., Casarin L.S., Hessel C.T., Batista A.C.F., Allende A., Tondo E.C. Microbial quality of irrigation water used in leafy green production in Southern Brazil and its relationship with produce safety. Food Microbiol. 2017;65:105–113. doi: 10.1016/j.fm.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 75.Ceuppens S., Hessel C.T., de Quadros R.R., Bartz S., Tondo E.C., Uyttendaele M. Microbiological quality and safety assessment of lettuce production in Brazil. Int. J. Food Microbiol. 2014;181:67–76. doi: 10.1016/j.ijfoodmicro.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 76.Barbosa L.J., Ribeiro L.F., Lavezzo L.F., Barbosa M.M., Rossi G.A., do Amaral L.A. Detection of pathogenic Escherichia coli and microbiological quality of chilled shrimp sold in street markets. Lett. Appl. Microbiol. 2016;62:372–378. doi: 10.1111/lam.12562. [DOI] [PubMed] [Google Scholar]
- 77.Guth B.E.C., Lopes S.R., Vaz T.M.I. First Shiga toxin-producing Escherichia coli isolate from a patient with hemolytic uremic syndrome, Brazil. Emerg Infect Dis. 2002;8:535–536. doi: 10.3201/eid0805.010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Irino K., Vaz T.M.I., Kato M.A.M.F., Naves Z.V.F., Lara R.R., Marco M.E.C., Rocha M.M.M., Moreira T.P., Gomes T.A.T., Guth B.E.C. O157:H7 Shiga toxin-producing Escherichia coli strains associated with sporadic cases of diarrhea in São Paulo, Brazil. Emerg. Infect. Dis. 2002;8:446–447. doi: 10.3201/eid0804.010490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vaz T.M.I., Irino K., Kato M.A.M.F., Dias Â.M.G., Gomes T.A.T., Medeiros M.I.C., Rocha M.M.M., Guth B.E.C. Virulence properties and characteristics of Shiga toxin-producing Escherichia coli in São Paulo, Brazil, from 1976 through 1999. J. Clin. Microbiol. 2004;42:903–905. doi: 10.1128/JCM.42.2.903-905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Souza R.L. Hemolytic Uremic Syndrome in Pediatric Intensive Care Units in São Paulo, Brazil. Open Microbiol. J. 2011;5:76–82. doi: 10.2174/1874285801105010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Irino K., Vaz T.M.I., Medeiros M.I.C., Kato M.A., Gomes T.A., Vieira M.A., Guth B.E.C. Serotype diversity as a drawback in the surveillance of Shiga toxin-producing Escherichia coli infections in Brazil. J. Med. Microbiol. 2007;5:565–567. doi: 10.1099/jmm.0.46815-0. [DOI] [PubMed] [Google Scholar]
- 82.De Souza R.L., Nishimura L.S., Guth B.E.C. Uncommon Shiga toxin-producing Escherichia coli serotype O165:HNM as cause of hemolytic uremic syndrome in São Paulo, Brazil. Diagn. Microbiol. Infect. Dis. 2007;59:223–225. doi: 10.1016/j.diagmicrobio.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 83.Fernandes M.R., Ignacio A., Martins F.H., Rocha L.B., Piazza R.M., Vaz T.M., Avila-Campos M.J., Nakano V. Presence of Shiga toxin 2e-producing Escherichia coli and atypical enteropathogenic E. coli in an asymptomatic child. JMM Case Rep. 2014;1:e000001. doi: 10.1099/jmmcr.0.000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ori E.L., Takagi E.H., Andrade T.S., Miguel B.T., Cergole-Novella M.C., Guth B.E.C., Hernandes R.T., Dias R.C.B., Pinheiro S.R.S., Camargo C.H. Diarrhoeagenic Escherichia coli and Escherichia albertii in Brazil: Pathotypes and serotypes over a 6-year period of surveillance. Epidemiol. Infect. 2018;147:e10. doi: 10.1017/S0950268818002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Franzolin M.R., Alves R.C.B., Keller R., Gomes T.A., Beutin L., Barreto M.L., Milroy C., Strina A., Ribeiro H., Trabulsi L.R. Prevalence of diarrheagenic Escherichia coli in children with diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz. 2005;10:359–363. doi: 10.1590/S0074-02762005000400004. [DOI] [PubMed] [Google Scholar]
- 86.Barreto M.L., Milroy C.A., Strina A., Prado M.S., Leite J.P., Ramos E.A., Ribeiro H., Alcântara-Neves N.M., Teixeira M.G., Rodrigues L.C. Community-based monitoring of diarrhea in urban Brazilian children: incidence and associated pathogens. Trans. R. Soc. Trop. Med. Hyg. 2006;100:234–242. doi: 10.1016/j.trstmh.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 87.Bueris V., Sircili M.P., Taddei C.R., dos Santos M.F., Franzolin M.R., Martinez M.B., Ferrer S.R., Barreto M.L., Trabulsi L.R. Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz. 2007;102:839–844. doi: 10.1590/S0074-02762007005000116. [DOI] [PubMed] [Google Scholar]
- 88.De Toni F., de Souza E.M., Pedrosa F.O., Klassen G., Irino K., Un Rigo L., Steffens M.B., Fialho O.B., Farah S.M., Fadel-Picheth C.M. A prospective study on Shiga toxin-producing Escherichia coli in children with diarrhea in Paraná State, Brazil. Lett. Appl. Microbiol. 2009;48:645–647. doi: 10.1111/j.1472-765X.2009.02569.x. [DOI] [PubMed] [Google Scholar]
- 89.Garcia P.G., Silva V.L., Diniz C.G. Occurrence and antimicrobial drug susceptibility patterns of commensal and diarrheagenic Escherichia coli in fecal microbiota from children with and without acute diarrhea. J. Microbiol. 2011;49:46–52. doi: 10.1007/s12275-011-0172-8. [DOI] [PubMed] [Google Scholar]
- 90.Regua-Mangia A., Gomes T.A., Vieira M.A., Andrade J.R., Irino K., Teixeira L.M. Frequency and characteristics of diarrhoeagenic Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro, Brazil. J. Infect. 2004;48:161–167. doi: 10.1016/S0163-4453(03)00138-5. [DOI] [PubMed] [Google Scholar]
- 91.Santos R.F.C., Nascimento J.D.S., Geimba M.P., Hessel C.T., Tondo E.C. First report of human gastroenteritis caused by Escherichia coli O157:NM in Brazil. Foodborne Pathog. Dis. 2017;14:665–666. doi: 10.1089/fpd.2017.2296. [DOI] [PubMed] [Google Scholar]
- 92.Blank T.E., Lacher D.W., Scaletsky I.C.A., Zhong H., Whittam T.S., Donnenberg M.S. Enteropathogenic Escherichia coli O157 strains from Brazil. Emerg. Infect. Dis. 2003;9:113–115. doi: 10.3201/eid0901.020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cantarelli V., Nagayama K., Takahashi A., Cauduro P.F., Dias C.A.G., Mezzari A., Brodt T. Isolation of shiga toxin-producing Escherichia coli (STEC) serotype 091:H21 from a child with diarrhea in Porto Alegre city, RS, Brazil. Braz. J. Microbiol. 2000;31:266–270. doi: 10.1590/S1517-83822000000400005. [DOI] [Google Scholar]
- 94.Nunes M.R.C.M., Magalhães P.P., Macêdo A.S., Franco R.T., Penna F.J., Mendes E.N. Attaching and effacing Escherichia coli and Shiga toxin-producing E. coli in children with acute diarrhoea and controls in Teresina/PI, Brazil. Trans. R. Soc. Trop. Med. Hyg. 2012;106:43–47. doi: 10.1016/j.trstmh.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 95.Lozer D.M., Souza T.B., Monfardini M.V., Vicentini F., Kitagawa S.S., Scaletsky I.C., Spano L.C. Genotypic and phenotypic analysis of diarrheagenic Escherichia coli strains isolated from Brazilian children living in low socioeconomic level communities. BMC Infect Dis. 2013;13:418. doi: 10.1186/1471-2334-13-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moreno A.C.R., Filho A.F., Gomes T.A.T., Ramos S.T., Montemor L.P., Tavares V.C., Filho L.S., Irino K., Martinez M.B. Etiology of childhood diarrhea in the northeast of Brazil: significant emergent diarrheal pathogens. Diagn. Microbiol. Infect. Dis. 2010;6:50–57. doi: 10.1016/j.diagmicrobio.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 97.Brooks J.T., Bergmire-Sweat D., Kennedy M., Hendricks K., Garcia M., Marengo L., Wells J., Ying M., Bibb W., Griffin P.M. Outbreak of shiga toxin–producing Escherichia coli O111:H8 infections among attendees of a high school cheerleading camp. Clin. Infect. Dis. 2004;38:190–198. doi: 10.1086/380634. [DOI] [PubMed] [Google Scholar]
- 98.Uchida H., Kanegane H., Yoshiya K., Kitamura K., Ihara T., Kamiya H., Kobayashi Y., Miyazawa H., Takeda T. Four cases of hemolytic uremic syndrome (HUS) associated with serotype O165 verotoxin producing Escherichia coli (VTEC) identified by LPS-Solid phase enzyme-linked immunosorbent assay (ELISA) J. Jpn. Assoc. Infect. Dis. 1995;69:678–683. doi: 10.11150/kansenshogakuzasshi1970.69.678. [DOI] [PubMed] [Google Scholar]
- 99.Bielaszewska M., Prager R., Vandivinit L., Müsken A., Mellmann A., Holt N.J., Tarr P.I., Karch H., Zhang W. Detection and characterization of the fimbrial Sfp cluster in enterohemorrhagic Escherichia coli O165:H25:NM isolates from humans and cattle. Appl. Environ. Microb. 2009;75:64–71. doi: 10.1128/AEM.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moxley R.A., Stromberg Z.R., Lewis G.L., Loy J.D., Brodersen B.W., Patel I.R., Gangiredla J. Haemorrhagic colitis associated with enterohaemorrhagic Escherichia coli O165:H25 infection in a yearling feedlot heifer. JMM Case Rep. 2015;2:1–6. doi: 10.1099/jmmcr.0.005004. [DOI] [Google Scholar]
- 101.Lindsey R.L., Knipe K., Rowe L., Garcia-Toledo L., Loparev V., Juieng P., Trees E., Strockbine N., Stripling D., Gerner-Smidt P. Complete genome sequences of two Shiga Toxin-producing Escherichia coli strains from serotypes O119:H4 and O165:H25. Genome Announc. 2015;3:e01496-15. doi: 10.1128/genomeA.01496-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wieler L.H., Busse B., Steinrück H., Beutin L., Weber A., Karch H., Baljer G. Enterohemorrhagic Escherichia coli (EHEC) strains of serogroup O118 display three distinctive clonal groups of EHEC pathogens. J. Clin. Microbiol. 2000;38:2162–2169. doi: 10.1128/jcm.38.6.2162-2169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klein E.J., Stapp J.R., Clausen C.R., Boster D.R., Wells J.G., Qin X., Swerdlow D.L., Tarr P.I. Shiga toxin-producing Escherichia coli in children with diarrhea: A prospective point-of-care study. J. Pediatr. 2002;14:172–181. doi: 10.1067/mpd.2002.125908. [DOI] [PubMed] [Google Scholar]
- 104.Lindsey R.L., Trees E., Sammons S., Loparev V., Frace M., Strockbine N., Sabol A.L., Sowers E., Stripling D., Martin H., et al. Draft whole-genome sequences of nine non-O157 Shiga toxin-producing Escherichia coli strains. Genome Announc. 2014;2:501–515. doi: 10.1128/genomeA.00501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.[allaway T.R., Carr M.A., Edrington T.S., Anderson R.C., Nisbet D.J. Diet, Escherichia coli O157:H7, and cattle: A review after 10 years. Curr. Issues Mol. Biol. 2009;11:67–79. [PubMed] [Google Scholar]
- 106.Acquaotta F., Ardissino G., Fratianni S., Perrone M. Role of climate in the spread of shiga toxin-producing Escherichia coli infection among children. Int. J. Biometeorol. 2017;61:1647–1655. doi: 10.1007/s00484-017-1344-y. [DOI] [PubMed] [Google Scholar]
- 107.Law D. The history and evolution of Escherichia coli O157 and other Shiga toxin-producing E. coli. World J. Microbiol. Biotechnol. 2000;16:701–709. doi: 10.1023/A:1008927820535. [DOI] [Google Scholar]
- 108.Ranjbar R., Sheikhshahrokh A., Jafari N.J. Shiga (Vero) toxin producing Escherichia coli in various types of food stuffs; virulence factors, O-serogroups and antimicrobial resistance properties. J. Food Saf. 2017;37:e12312. doi: 10.1111/jfs.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brasil. Ministério Da Agricultura, Pecuária e Abastecimento. Centro-Oeste Lidera Produção Agrícola Brasileira. [(accessed on 19 March 2019)];2016 Available online: http://www.agricultura.gov.br/noticias/centro-oeste-lidera-producao-agricola-brasileira.
- 110.Brasil. Agência Nacional de Vigilância Sanitária. Resolução RDC No. 12° de 2001. [(accessed on 19 March 2019)]; Available online: http://portal.anvisa.gov.br/documents/33916/394219/Resolucao_CNNPA_n_12_de_1978.pdf/4f93730f-65b8-4d3c-a362-eae311de5547.
- 111.Brasil. Ministério da Agricultura Pecuária e Abastecimento [Internet]. Norma interna Dipoa/SDA No. 2, de 20 de agosto de 2013. [(accessed on 19 March 2019)]; Available online: http://www.agricultura.gov.br/assuntos/inspecao/produtos-animal/arquivos/escherichia-coli/norma-interna-dipoa-sda-no-2-de-20-de-agosto-de-2013.pdf.
- 112.USDA-FSIS MLG 5B.03 Appendix 4.01, FSIS Laboratory Flow Chart for Detection and Isolation of Non-O157 Shiga Toxin-Producing Escherichia coli (STEC) from Meat Products 2012. [(accessed on 8 March 2013)]; Available online: http://www.fsis.usda.gov/Science/Microbiological_lab_guidebook/index.asp.
- 113.Moudgil P., Bedi J.S., Moudgil A.D., Gill J.P.S., Aulakh R.S. Emerging issue of antibiotic resistance from food producing animals in India: Perspective and legal framework. Food Rev. Int. 2018;34:447–462. doi: 10.1080/87559129.2017.1326934. [DOI] [Google Scholar]