Abstract
The rice (Oryza sativa) retrotransposon Tos17 is one of a few active retrotransposons in plants and its transposition is activated by tissue culture. Here, we present the characterization of viviparous mutants of rice induced by tissue culture to demonstrate the feasibility of the use of retrotransposon Tos17 as an endogenous insertional mutagen and cloning of the tagged gene for forward genetics in unraveling the gene function. Two mutants were shown to be caused by the insertion of Tos17. Osaba1, a strong viviparous mutant with wilty phenotype, displayed low abscisic acid level and almost no further increase in its levels upon drought. The mutant is shown to be impaired in the epoxidation of zeaxanthin. On the other hand, Ostatc, a mutant with weak phenotype, exhibited the pale green phenotype and slight increase in abscisic acid levels upon drought. Deduced amino acids of the causative genes of Osaba1 and Ostatc manifested a significantly high homology with zeaxanthin epoxidase isolated from other plant species and with bacterial Sec-independent translocase TATC protein, respectively. This is the first example of transposon tagging in rice.
Transposable elements are ubiquitously present in almost all plant genomes and constitute a major portion of the genome (Kumar and Bennetzen, 1999). These elements belong to class I (also called retrotransposons) and class II (DNA-based transposons). Class I elements transpose through reverse transcription of an RNA intermediate, the so-called copy-and-paste mode, and therefore induced mutations are usually stable. On the other hand, class II elements transpose through cut-and-paste mode, thus inducing unstable mutation. These elements comprise well-characterized Ac/Ds, En/Spm, and Mu elements of maize, Tam elements of snapdragon, and dTph1 of petunia. Because of their ability to induce mutations at high frequency and at the same time tag the causative genes molecularly, they have been used as molecular genetic tools for cloning genes (transposon tagging). Among these elements, the maize Ac/Dc element has been successfully used in heterologous plant species such as petunia, tobacco, Arabidopsis, and tomato (Sundaresan, 1996). In Arabidopsis, En/Spm has also been shown to be a powerful tool (Wisman et al., 1998). Besides these elements, T-DNA was also used as an insertional mutagen, which was proved to be very successful in Arabidopsis (Krysan et al., 1999).
Rice (Oryza sativa) is recognized as an important model plant because of its small genome size among cereals and development of efficient transformation system. In contrast to another model plant Arabidopsis, an efficient tagging system, which is one of the most important methods for plant molecular biology, is still not established in rice. Although the exogenous elements such as Ac/Ds and T-DNA are being used in rice as insertional mutagens to establish a tagging system, the use of endogenous insertional mutagens is desirable. That is because mutant lines induced by exogenous insertional mutagens are transgenic and thus a matter for environmental concern. We have succeeded recently in finding active endogenous retrotransposons in rice (Hirochika et al., 1996). The most active one was called Tos17, whose activation by tissue culture resulted in an increase in its copy number by 5 to 30, and these increased Tos17 copies were shown to be inserted throughout the genome (Yamazaki et al., 2001). Important features of Tos17 are summarized as follows (Hirochika, 1997; Hirochika, 1999): (a) transposition can be regulated because Tos17 is activated by tissue culture and becomes silent in regenerated plants; (b) highly mutagenic during tissue culture, Tos17 transposes preferentially into gene-rich, low copy regions and about eight loci on an average are disrupted in each plant regenerated from 5-month-old culture; (c) integration target loci were widely distributed over the chromosomes so that random insertion for saturation mutagenesis is feasible; (d) induced mutations are stable; (e) original copy number is quite low, one to four depending on varieties, so that it is easy to identify the transposed copy responsible for the specific mutation; and (f) endogenous transposon, so that screening and characterization of mutants in the field are possible without considering environmental issues. Considering these features, it is believed that Tos17 can be used as a useful tool for forward and reverse genetic studies of rice. We recently have demonstrated the use of Tos17 for reverse genetic studies (Sato et al., 1999). Therefore, to assess the feasibility of Tos17 in tagging the rice genes and in exploring their biological functions, a forward genetic approach was applied, and approximately 30,000 regenerated rice lines (A. Miyao and H. Hirochika, unpublished data) were screened for viviparous mutants. Although the feasibility of Tos17 based transposon-tagging system was assessed on a part of our viviparous mutant collection, results presented here indicate that Tos17 can be successfully used to tag the rice genes and explore their function.
Abscisic acid (ABA) plays a central role at least in higher plants by regulating plant growth and development, including seed maturation and dormancy as well as adaptation to a variety of environmental stresses (Zeevaart and Creelman, 1988; Bray, 1997). Although considerable progress has been made in understanding and establishing the ABA biosynthetic pathway in plants mostly by characterizing ABA-deficient mutants (Schwartz et al., 1997a, 1997b; Tan et al., 1997; Burbidge et al., 1999; Qin and Zeevaart, 1999; Thompson et al., 2000), only two genes of this pathway have been cloned by transposon-tagging strategy from ABA-deficient mutants: the aba2 gene encoding a zeaxanthin epoxidase (ZEP) was cloned from a wilty mutant of Nicotiana plumbaginifolia (Marin et al., 1996), and the vp14 gene encoding a 9-cis-epoxycarotenoid dioxygenase (Schwartz et al., 1997b) was cloned from a mutant of Zea mays with reduced seed dormancy (Tan et al., 1997). Considering the importance of ABA, and the ease of mutant isolation, viviparous mutants were chosen as one of our targets for tagging experiments.
RESULTS
Screening of the Viviparous Mutant Lines
The regenerated rice lines (R0) were used to identify the viviparous mutants by screening for precocious germination. These lines carry eight newly transposed Tos17 copies per line on average. Of approximately 30,000 regenerated rice lines since 1998, more than 1,400 mutant lines have been found to be of vivipary nature. Precociously germinated seeds were rescued from more than 100 lines and grown in soil at 25°C in green house under 70% to 80% relative humidity to establish/confirm their phenotype and to perform the cosegregation analysis of transposed Tos17 copies. Among the observed phenotypes, wilty, slender, hyper-slender, albino, and pale green were the main phenotypes; many lines showed almost normal growth and wild-type-like phenotype, even though they were from precociously germinated seeds, implying that the causative gene(s) of these mutants might be involved only in dormancy of seeds. Furthermore, the cosegregation analysis of XbaI-digested genomic DNA, isolated from these 100 viviparous mutant lines, was carried out by Southern analyses, and it was found that seven mutant lines cosegregate perfectly with one of the transposed Tos17 copies (data not shown). Characterization of two of seven viviparous mutant lines is presented here.
Tagging of Osaba1 Mutant, Impaired in the Epoxidation of Zeaxanthin, by Tos17
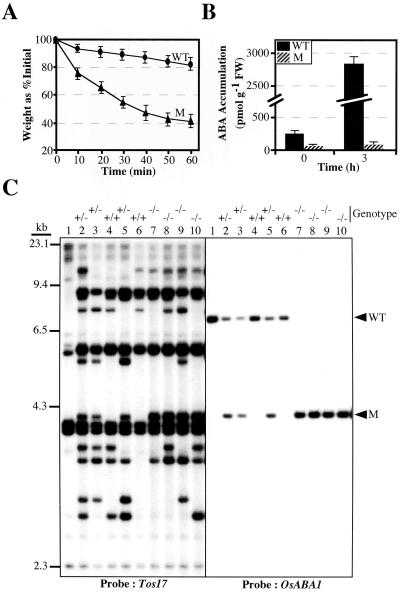
Precociously germinated seeds were rescued, grown, and genomic DNA was isolated from individual seedlings. Genomic DNA digested with XbaI restriction enzyme was subjected to Southern analysis to check the cosegregation of Tos17 with the observed phenotypes. The analysis showed that the phenotype of one of the mutant line, called ND7980, was found cosegregating with one of the transposed Tos17copies, as indicated by lower arrowhead (Fig. 1C; left panel). The cosegregation of Tos17 with the observed phenotype was also confirmed in R2 populations (data not shown). To further confirm the cosegregation, the sequence flanking the cosegregating Tos17 was isolated by inverse PCR and used as a probe to hybridize the same cosegregating membrane used for probing with Tos17 (Fig. 1C; right panel). As expected, only the mutant plants carried homozygous mutations. These results strongly suggest that the mutant phenotype was caused by the insertion of Tos17.
Figure 1.
A, Water loss assay. Detached leaves of wild type (WT) and mutant (M) were weighed every 10 min until 60 min. Mean and sd of three independent experiments are shown. B, ABA levels in WT and M at 0 h and after 3 h of drought treatment. Mean and sd of three independent experiments are shown. C, Tagging of OsABA1 by Tos17. Cosegregation analysis of XbaI-digested genomic DNA (0.5 μg) was carried out by Southern analyses, and probed with a 32P-labeled Tos17 (left) or the cloned OsABA1 gene (right). Numbers on the top of each lane represent individual seedling, and the genotype of each seedling is also given on the top of each lane. Lane 1 carries the genomic DNA of Nipponbare and serves as a control for other seedlings. The size of marker (λDNA digested with HindIII) fragments is indicated on the left hand in kilobase pairs.
Wilty phenotype is one of the characteristic features of ABA-deficient or ABA-insensitive mutants (McCarty, 1995). Therefore, we asked whether the above mutant showing wilty phenotype is an ABA-deficient or an ABA-insensitive mutant. To answer this question, we checked the water loss in the detached leaves and the effect of applied ABA on the mutant seedling. The water loss analysis indicated that the mutant leaves lost more than 50% of their fresh weight (FW) within 50 min at room temperature, whereas in the same period, wild-type leaves lost only 15% FW (Fig. 1A). Furthermore, when young mutant seedlings were treated with alternate day spray of a 40-μm ABA solution for 2 weeks, mutant seedlings were restored to wild-type phenotype; moreover, the detached leaves of ABA-treated mutant seedlings showed water loss almost like wild type (data not shown). Based on above results, it was inferred that ND7980 mutant is most likely to be ABA deficient.
To determine the ABA content in the mutant and its wild type before and after drought treatment, detached leaves gathered from five individual seedlings for three independent assays were collected in the beginning (representing 0 h) and after 3 h of drought treatment (Fig. 1B). ABA levels were measured by using an ELISA kit and an internal (±) ABA as a standard. Wild-type leaves contained 251 ± 30 pmol ABA per gram of FW, whereas the mutant contained only 50 ± 15 pmol ABA. On the other hand, under 3 h of drought condition, ABA level in the leaves of wild type increased tremendously by at least 11-fold to that of the untreated wild-type leaves, whereas the mutant leaves showed only a 1.3-fold increase in ABA level (Fig. 1B). This result further indicates that this mutant is ABA deficient, and the causative gene of this mutant might be involved in ABA biosynthesis.
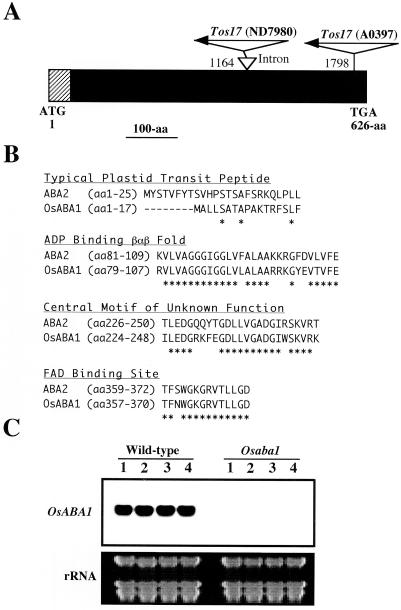
To identify the causative gene of the mutant and to know the Tos17 insertion point, the cDNA library was screened with the cloned Tos17-flanking sequence, and three independent cDNA clones were obtained. The longest cDNA is 2,345 bp long and has a single open reading frame (ORF) of 626 amino acid residues (Fig. 2A). A homology search by BLAST showed its considerable similarity at amino acid level to the gene encoding ZEP isolated from N. plumbaginifolia (66% identity; Marin et al., 1996) and other plant species. Considering the adopted nomenclature in Arabidopsis, where the gene encoding ZEP was called ABA1 (Marin et al., 1996), we call the rice gene as OsABA1 (DDJB Nucleotide Sequence Database accession no. AB050884). As shown in Figure 1C (right), the OsABA1 gene is a single copy gene in the rice genome. Four regions of ZEP are supposed to be of functional importance (Marin et al., 1996). So, we compared these four regions of OsABA1 with the tobacco protein (ABA2), as shown in Figure 2B. One appears to be a typical plastid transit peptide signal, whereas other three regions showing high level of identity are FAD binding site, ADP binding βαβ fold, and the central motif of unknown function.
Figure 2.
Structural features of the OsABA1 and position of Tos17 insertion. A, Diagrammatic representation of the OsABA1 ORF showing the initiation codon (ATG) and the stop codon (TGA), and the positions of Tos17 insertion. Nucleotides of OsABA1 flanking the Tos17 insertion or an intron are also given. Hatched lines indicate the putative chloroplast transit peptide. B, Four regions of the tobacco ZEP (ABA2) protein are compared with the OsABA1 protein. The identical amino acid is marked by asterisk. C, Northern analysis of OsABA1 in wild type and mutant (Osaba1). Total RNA (20 μg) was subjected to northern analysis and probed with the 32P-labeled OsABA1. Equal loading was confirmed by staining with ethidium bromide.
Tos17 was shown to be inserted into one of the introns (Fig. 2A). To examine the effect of the Tos17 insertion on the OsABA1 RNA synthesis, total RNA isolated from four individual seedling leaves of wild-type and of mutant (Osaba1) was subjected to northern analysis. Whereas a single 2.4-kb transcript was detected in wild type, the level of the transcript was decreased dramatically in Osaba1 (Fig. 2C). The existence of the low level of the normal transcript (observed occasionally) may explain the residual level of ABA in the mutant. In the mutant, the large transcript was also occasionally detected. This transcript carries the Tos17 sequence, indicating the normal splicing was inhibited (data not shown).
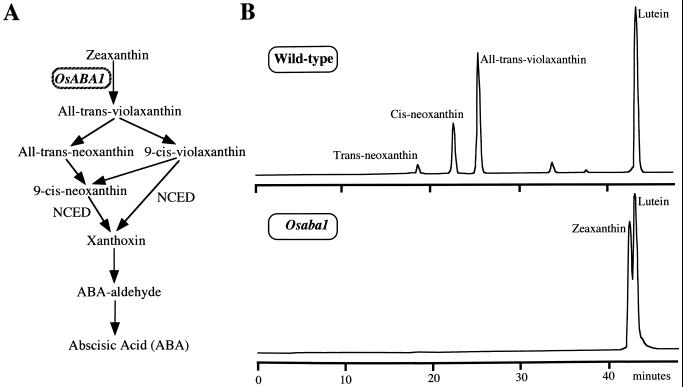
ZEP is shown to converts zeaxanthin to all-trans-violaxanthin by a two steps epoxidation in the ABA biosynthetic pathway (Marin et al., 1996; Fig. 3A).Considering the function of ZEP, the mutation of OsABA1 should cause an accumulation of zeaxanthin and the absence of its derived products of the ABA biosynthetic pathway (Fig. 3A). The study of carotenoid composition carried out in the leaves of both wild type and mutant (Osaba1) revealed significant quantitative differences in zeaxanthin and its derived products, as illustrated in Figure 3B. Zeaxanthin was undetectable in the leaves of wild type but accumulated to a significant level in the mutant leaves. In contrast, cis- and trans-neoxanthin, and all-trans-violaxanthin were undetectable in the mutant leaves. This result along with the above mentioned results clearly demonstrate that the Osaba1 mutant is impaired in the zeaxanthin epoxidation step of ABA biosynthesis.
Figure 3.
Analysis of carotenoid content in the leaves of wild type and Osaba1 mutant. A, The ABA biosynthetic pathway from zeaxanthin. Adopted from the most recently proposed pathway in plants by Qin and Zeevaart (1999). B, Carotenoid profile in the leaves of wild type and mutant (Osaba1). Presented carotenoid profile is one of three independent measurements.
Considering the mutant phenotype and the biochemical function predicted from the mutant gene, we concluded that the OsABA1 is a causative gene for the viviparous mutation. This was further confirmed by the analysis of an allelic mutant. Following the PCR-screening strategy (Hirochika, 1999; Sato et al., 1999; Hirochika et al., 2001), we were able to find an allelic mutant, called A0397, of Osaba1. A0397 was found to carry Tos17 insertion in the OsABA1 coding region in an opposite direction, close to the stop codon (Fig. 2A). As expected from the Tos17 insertion site, A0397 displayed only weak viviparous phenotype, and the homozygous plants can grow normally in the field and set seeds. It is interesting that the leaves of homozygous mutant plants were intermediate in color between green and pale green.
Finding of a Novel Ostatc Mutant Displaying the Pale Green Phenotype Genetically Linked with Tos17
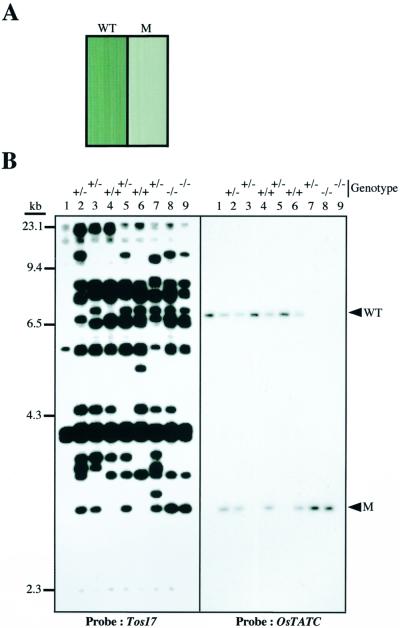
Southern analyses performed using the genomic DNA, isolated from another mutant line (called ND5664) and its wild type, indicated that the pale green phenotype (Fig. 4A) of mutant is tightly cosegregating with one of the transposed Tos17 copies as indicated by lower arrowhead in Figure 4B (left). The cosegregation of Tos17 with the mutant phenotype was also confirmed in R2 population (data not shown). To further confirm the cosegregation, inverse PCR was performed to isolate the sequence flanking the cosegregating Tos17 and used as a probe. As shown in Figure 4B (right), the mutant plants carried homozygous mutation, suggesting that the mutant phenotype is caused by the Tos17 insertion.
Figure 4.
A, The phenotype of Ostatc mutant (M) and wild type (WT) exhibited by 1-week-old seedlings grown on MS2 medium. B, Tagging of OsTATC by Tos17. Cosegregation analysis of XbaI-digested genomic DNA (0.5 μg) was carried out by Southern analyses, and probed with a 32P-labeled Tos17 (left) or the cloned OsTATC gene (right).
To identify the causative gene of this mutant and to know the position of Tos17 insertion, the cDNA library was screened with the isolated sequence flanking the cosegregating Tos17, and four independent cDNA clones were obtained. The longest cDNAs is 1,781-bp long and has a single ORF of 359 amino acid residues (Fig. 5A). Homology search by BLAST manifested its significant similarity to the Arabidopsis genome sequence (67% identity, accession no. AAF18659) annotated as a putative TATC. OsTATC showed a weak but considerable homology with TATC of Eschericia coli (35% identity, 56% similarity). TATC is known to be a Sec-independent translocase protein in bacteria (Berks et al., 2000). The Sec-independent translocase protein in bacteria is one of the components of Sec-independent pathway, which has been termed the TAT (for twin-arginine translocation) system, because precursors are targeted to the pathway by signal peptides (Berks et al., 2000 and references therein). Considering the nomenclature adopted in bacteria for the components of TAT system and as Arabidopsis genome sequence annotated as a putative TATC, we termed the rice gene as OsTATC (DDJB Nucleotide Sequence Database accession nos. AB050885). This OsTATC gene exists as a single copy gene in the rice genome (Fig. 4B; right).
Figure 5.
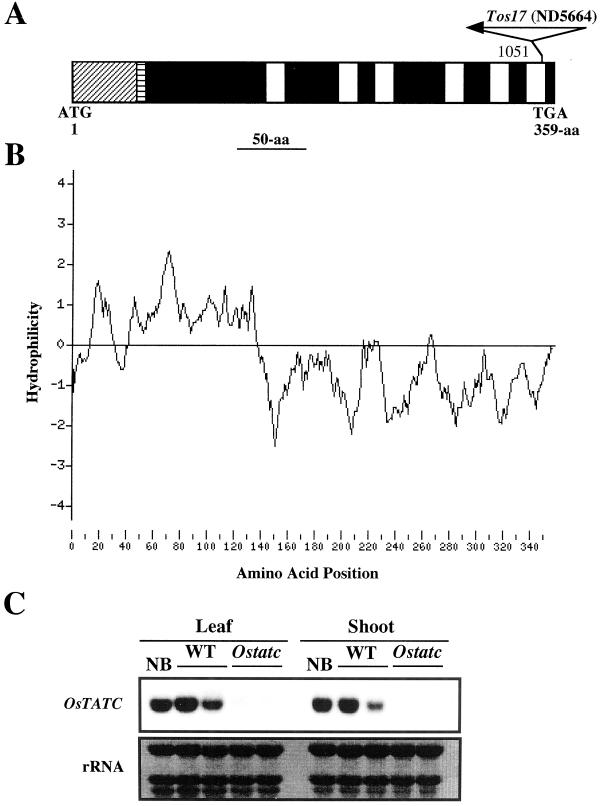
A, Diagrammatic representation of the OsTATC ORF showing the translation start point (ATG) and the stop codon (TGA), and the position of Tos17 insertion. Nucleotide of OsTATC flanking the Tos17 insertion is given. The putative chloroplast transit peptide (hatched lines), mitochondrial targeting peptide (horizontal lines), and six transmembrane helices (clear areas) are also shown. B, Hydrophilicity analysis of OsTATC protein. Hydrophilicity was analyzed using the Tmpred program. C, Northern analysis of OsTATC in wild type and mutant (Ostatc). Total RNA (20 μg), isolated from leaf and shoot of two individual seedlings, was subjected to northern analysis, and probed with the 32P-labeled OsTATC. Equal loading was confirmed by staining with methylene blue.
We performed a computer analysis to know the structural features of this gene. Computer analysis predicted that the OsTATC protein carries the putative chloroplast (Fig. 5A; hatched lines) and mitochondrial (Fig. 5A; horizontal lines) targeting signal peptides, suggesting that OsTATC functions in the organelle. In addition to this, it also carries six transmembrane helices (Fig. 5A; clear areas) of 17 amino acid residues. Furthermore, the hydrophilicity of OsTATC, as shown in Figure 5B, reveals two distinct regions; the first stretch of 140 amino acid residues is almost hydrophilic in nature, whereas the later part of this protein is hydrophobic. In prokaryote, the predicted topological structure of TATC protein has also been shown to possess six transmembrane helices (Berks et al., 2000).
The Tos17 insertion was found to be in the sixth transmembrane helix (Fig. 5A). To examine the effect of Tos17 insertion on the OsTATC RNA synthesis, total RNA isolated from the leaf and shoot of two individual seedlings of both wild type and mutant (Ostatc) was subjected to northern analysis (Fig. 5C). No transcript was detected in both the leaf and shoot of the mutant, whereas a single 1.3-kb transcript was detected in the both leaf and shoot of wild type and Nipponbare, indicating the loss-of-function of OsTATC in the mutant. The Tos17 insertion in the OsTATC ORF and no expression of OsTATC in the Ostatc mutant suggest that Ostatc is a null mutant.
Ostatc Mutant Is Deficient in Chlorophyll, Shows Quick Water Loss, and Fails to Accumulate ABA under Drought
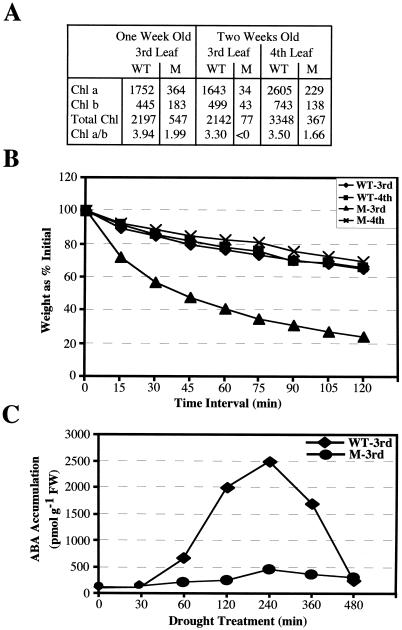
Because the role of TATC in plants, including rice, remains to be elucidated, we studied the ABA-related biochemical and physiological parameters of this mutant. We checked chlorophyll content, water loss, and ABA content in the leaves of wild type and Ostatc mutant (Fig. 6). The chlorophyll content was measured in the third and fourth leaves of 1- and 2-week-old seedlings, respectively (Fig. 6A). As expected from the mutant phenotype, total chlorophyll and chlorophyll a and b were significantly low in the mutant than that in the wild type. The overall chlorophyll in the third leaf of 1- and 2-week-old mutant seedling was only 25% and 4%, respectively, whereas the fourth leaf of 2-week-old mutant seedling was found to have 11% total chlorophyll to that in its wild-type leaf. Furthermore, the detached third leaf showed almost 50% water loss of its FW within 50 min, whereas the fourth leaf of the same 2-week-old mutant seedling showed only 15%, which was also the same in case of wild type (Fig. 6B). Moreover, the third leaf of 2-week-old mutant seedling showed only the basal level of ABA even after drought treatment, whereas the third leaf of wild type showed rapid ABA accumulation just after 30 min of drought treatment, reaching to its maximum near 4 h, followed by decrease in ABA (Fig. 6C). Considering the pleiotropic effect of the mutation, the OsTATC gene might be indirectly involved in the regulation of ABA biosynthesis.
Figure 6.
Biochemical and physiological analyses of Ostatc. A, Total chlorophyll (Chl), chlorophyll a, and chlorophyll b contents in the leaves of M and WT are given as nmol g−1 FW. B, Water loss assay. Detached leaves of M and WT were weighed every 15 min until 120 min. Presented data are the mean of three independent experiments. C, ABA levels in M and WT at 0 min (control) and after the drought treatment. Presented data are the mean of three independent experiments.
DISCUSSION
Because the maize Ac/Ds system was used successfully in several plant species (Sundaresan, 1996), an attempt was also made to develop a transposon-tagging system in rice by using the maize Ac/Ds elements (Izawa et al., 1997); but so far no direct evidence for tagging has come to light. Considering the socioeconomic importance of rice and an international effort to sequence the rice genome (Sasaki and Burr, 2000), it is time to have a well-developed and suitable transposon-tagging system for systematic functional studies of rice genes. Thus, one of the most active retrotransposons, Tos17, was characterized in detail, and it was considered as a tool for forward and reverse genetics of rice (Hirochika, 1999; Yamazaki et al., 2001). Although the feasibility of the use of Tos17 for reverse genetics came to light (Sato et al., 1999), the feasibility of gene tagging by Tos17 using the forward genetic approach remained to be elucidated.
In this study, we have used forward genetic approach to isolate genes involved in ABA biosynthesis by screening for viviparous mutants. Precocious germination is a clear sign of altered ABA levels. By screening for precocious germination, several viviparous mutants of maize have been identified (McCarty, 1995). This approach seems to be suitable for finding both ABA-deficient and -insensitive mutants. More than 1,400 putative mutant lines displaying weak, mild, and strong viviparous phenotypes have so far been identified. The environmental conditions, such as wet soil and relatively high humidity needed for rice growth, might be helpful in triggering even the weak viviparous phenotype, which leads to high frequency of viviparous mutant lines in rice. This implies that rice might have an advantage over other plants for cloning of novel genes. Cloning of a new rice gene, OsTATC, demonstrates that this is the case.
TATC protein, which has only been studied in bacteria, recognizes the twin-Arg signal peptide in its substrates, and is supposed to be encoded in all the genome(s) having a “TAT system” (Berks et al., 2000). In E. coli, at least five components (termed TATA to TATE) of TAT system have been found. TAT system has a fundamental feature of transporting folded proteins across the membranes. Previously, existence of a prokaryotic TATC system with homologs in plastids and mitochondria was considered (Berks et al., 2000). Loss of chlorophyll of Ostatc with its increasing age (Fig. 6A) and which eventually become dead 3 weeks after germination indicates that OsTATC might also be playing an important role at the later stage of leaf development and chloroplast differentiation by means of protein translocation across the membranes. In monocots, an intimate relationship and a precise regulation between leaf development and chloroplast differentiation have been proposed (Kusumi et al., 1997 and references therein). A significant homology and structural similarity between the OsTATC and prokaryotic TATC proteins indicate that the rice genome carries a TAT system for protein translocation of folded proteins, and the OsTATC gene might be one of the potential components of TAT system. Based on typical structural features of OsTATC as a protein translocator and pleiotropic effect of its mutant (Ostatc), the loss-of-function of OsTATC might block the proper development of chloroplast, because most of the proteins/enzymes needed for its development are imported into the chloroplast. So, OsTATC is likely to be indirectly involved in ABA biosynthesis because the initial steps of ABA biosynthetic pathway are catalyzed in the chloroplast. Based on the characterization of bacterial TATC gene, and the presence of its homologs encoded by the plastid genome of the algae and also by the mitochondrial genome of Arabidopsis, it has been proposed previously that TATC-dependent systems operate in chloroplast and mitochondria (Bogsch et al., 1998). Predicted chloroplastic and mitochondrial targeting signal peptides in OsTATC and also in Arabidopsis TATC, encoded by nuclear genomes, support the above proposed idea. Localization of OsTATC in the chloroplast and/or mitochondria and finding of its potential substrate will be needed to throw light on TAT system in rice and in plants, in general.
Preliminary data of cosegregation analysis obtained from 100 viviparous mutant lines, where seven mutant lines were found to cosegregate with one of the transposed Tos17 copies, implies that the tagging frequency of Tos17-based system is 7%. A significant fraction of mutations similarly has also been shown to be not tagged in Ac/Ds and T-DNA-based systems (Bancroft and Dean, 1993; Long et al., 1993; Azpiroz-Leehan and Feldmann, 1997). In these cases, non-tagged mutations are thought to be caused by abortive insertion events, which results in a rearrangement at the target site without integration of the elements (Bancroft and Dean, 1993; Azpiroz-Leehan and Feldmann, 1997). A rearrangement caused by imprecise excision would be an another important factor, as has been discussed in rice, where the Ac/Ds system was used (Izawa et al., 1997). In Tos17-based tagging system, mutations, if not tagged with Tos17, might be due to insertion of unknown transposable elements activated also by tissue culture or due to other types of induced mutations caused by tissue culture (Larkin and Scowcroft, 1981; Dennis et al., 1987). So, to increase the transposon-tagging frequency, it is important to find new transposable elements responsible for tissue culture induced mutations and/or reduced mutation frequency, which is not related with transposable elements. Because transposition of plant retrotransposons including the Tos17 is mainly regulated at the transcriptional level (Hirochika, 1993; Hirochika et al., 1996), it might be possible to alleviate the tissue culture associated problems by modifying the promoter of Tos17. An alternative way is to look for more suitable condition(s) under which Tos17 can be activated.
In conclusion, the evidence provided in the present study demonstrates that Tos17 can be efficiently used for forward genetics of rice. Moreover, isolation of an Osaba1 allelic mutant by PCR screening further confirms that Tos17 is equally applicable for reverse genetic studies.
MATERIALS AND METHODS
Plant Growth
Rice (Oryza sativa) mutant lines were grown in the rice paddy field to identify the viviparous mutants by screening for precocious germination. In green house, seeds were grown at 25°C under a relative humidity of 70% to 80%. In case of Ostatc, seeds were grown in vitro on MS2 medium (Murashige and Skoog basal medium, 1% [w/v] Suc, and 0.5% [w/v] gellan gum, pH 5.8).
Southern Analysis
Rice genomic DNA (0.5 μg), isolated according to the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980), was digested with restriction enzymes over a period of 18 h and separated on a 0.8% (w/v) agarose gel using the Multiblotter MB24 system supplied by Labimap (Plaisir, France). The membrane was prehybridized with Church-Gilbert hybridization buffer (Church and Gilbert, 1984) for 3 h at 65°C. Probe was added directly to the hybridization buffer, and hybridization was continued for 18 h. Probes were labeled by Multiprime DNA labeling system (Amersham, Buckinghamshire, UK) using [α-32P]dCTP (Amersham), and nonincorporated nucleotides were removed by spin chromatography. The hybridized membrane was washed with 2× SSC and 0.1% (w/v) SDS at the same temperature for 1 h, and exposed to an x-ray film (Kodak, Tokyo) for 18 h.
Water-Loss Assay
Plants were grown in soil in a green house (25°C) having relative humidity of 70% to 80%. Detached leaves (300 mg from 10 seedlings) of 2-week-old seedlings were kept on aluminum foil at room temperature (25°C). Weight of leaves was taken at 10-min intervals until 60 min or longer. Each experiment was done in triplicate at the same time.
Determination of ABA Levels
Leaves (100 mg FW) were weighed, freeze dried for 3 h, and extracted for 16 h at 4°C in the dark with MilliQ water (water/tissue ratio 50:1, v/w). Quantitative analyses of ABA were performed using a Phytodetek ABA ELISA kit, supplied by Agdia, and (±) cis-trans ABA (Sigma, St. Louis) as a standard. Data were reported as mean se of three repetitions. For determination of drought induced ABA, detached leaves (100 mg) were kept on aluminum foil under light at 25°C for desired period.
Northern Analysis
Total RNA was isolated from rice seedling leaf or shoot using the Isogen kit as per the protocol provided by the manufacturer (Nippon Gene Co., Tokyo). Total RNA (20 μg) was separated on a 1.2% (w/v) formaldehyde-denaturing agarose gels according to Sambrook et al. (1989) and blotted onto nylon membranes (Hybond N+, Amersham). Hybridization was performed at 65° for 18 h using Church-Gilbert hybridization buffer (Church and Gilbert, 1984). Hybridized membranes were washed with 2× SSC and 0.1% (w/v) SDS at the same temperature for 1 h followed by exposure to x-ray film. Membranes were reprobed after stripping off the previous probe in 0.1% (w/v) hot SDS. Probes were prepared as mentioned above (see “Southern Analysis”).
Inverse PCR
Inverse PCR was basically carried out as described before (Ochman et al., 1988; Triglia et al., 1988) using the Tos17 specific primers, as mentioned elsewhere (Miyao et al., 1998).
DNA Sequencing and Sequence Analysis
Nucleotide sequences were determined using an automated DNA sequencer (models 310 genetic analyzer and 377 DNA sequencer, Applied Biosystems, Foster City, CA) with either the universal or the gene specific primers. All the sequencing data were analyzed using GENETYX software (SDC Software Development, Tokyo). Searches for information and homology of nucleotide and amino acid sequence was analyzed using homology search with BLAST against the sequences in the GenBank and EMBL DNA Database.
Carotenoid Analysis
Plant materials (200 mg FW) were ground in liquid nitrogen and extracted twice with methanol (20 mL each). The extracts were combined, concentrated under vacuum, redissolved in 80% (v/v) methanol (5 mL), and then loaded onto a Bond Elut C18 cartridge column (gel size, 1 g; Varian, Harbor City, CA). Carotenoids were eluted from the column with 15 mL of methanol-water-chloroform (62.5:7.5:30, v/v). Eluate was concentrated under vacuum, redissolved in methanol, and subjected to HPLC analysis.
The HPLC was performed with Waters LC Module I plus equipped with a column of Pegasil-B ODS column (250 mm × 4.6 mm, Senshu Scientific Co., Tokyo). The column was eluted with a mixture of solvent A (85% [v/v] methanol) and solvent B (methanol-chloroform, 1:1, v/v), where the ratio of solvent B was increased from 10% to 30% in 40 min, followed by increase to 100% in 20 min, and continued for further 15 min at 100%. The elute was monitored at 440 nm, and the peak for each carotenoid was collected, concentrated under vacuo, redissolved in ethanol, and analyzed on a Beckman DU-640 spectrophotometer (Fullerton, CA). In each step, samples were protected from light by using aluminum foil as much as possible.
Chlorophyll Determination
Chlorophyll in leaf was determined by extraction with cold 80% (v/v) acetone as described by Arnon (1949).
Construction and Screening of a Rice cDNA Library
Total RNA was isolated from leaves of 2-week-old rice seedlings (Nipponbare) using Isogen kit (Nippon Gene Co.), and poly(A+) RNA was purified using poly(A) mRNA isolation kit (Stratagene, La Jolla, CA). The purified poly(A+) mRNA was used to construct cDNA library using a HybriZAP-2.1 cDNA cloning kit (Stratagene) according to the recommended protocol. Plaques were also screened following the protocol supplied along with the kit and of Sambrook et al. (1989).
Footnotes
This work was supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan; by a grant for the enhancement of Center-of-Excellence; by the special coordination funds for promoting Science and Technology in Japan; and by the Program for Promotion of Basic Research Activities for Innovative Biosciences (to G.K.A.).
LITERATURE CITED
- Arnon DI. Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Bancroft I, Dean C. Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics. 1993;134:1221–1229. doi: 10.1093/genetics/134.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks BC, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Bray EA. Plant responses to water deficit. Trends Plant Sci. 1997;2:48–54. [Google Scholar]
- Bogsch EG, Sargent F, Stanley NR, Ben BC, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 1999;17:427–431. doi: 10.1046/j.1365-313x.1999.00386.x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Brettell RIS, Peacock WJ. A tissue culture induced Adh null mutation of maize results from a single base change. Mol Gen Genet. 1987;210:181–183. [Google Scholar]
- Hirochika H. Activation of tobacco retrotransposons during tissue culture. EMBO J. 1993;12:2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H. Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- Hirochika H. Retrotransposons of rice as a tool for forward and reverse genetics. In: Shimamoto K, editor. Molecular Biology of Rice. Tokyo: Springer-Verlag; 1999. pp. 43–58. [Google Scholar]
- Hirochika H, Miyao A, Yamazaki M, Takeda S, Abe K, Hirochika R, Agrawal GK, Watanabe T, Sugimoto K, Sasaki T. Rice Genetics IV: Proceedings of the Fourth International Rice Genetics Symposium. International Rice Research Institute, Manila, The Philippines (in press) 2001. Retrotransposons of rice as a tool for the functional analysis of genes. [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Ohnishi T, Nakano T, Ishida N, Enoki H, Hashimoto H, Itoh K, Terada R, Wu C, Miyazaki C. Transposon tagging in rice. Plant Mol Biol. 1997;35:219–229. [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Kusumi K, Mizutani A, Nishimura M, Iba K. A virescent gene V1determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J. 1997;12:1241–1250. [Google Scholar]
- Larkin PJ, Scowcroft WR. Somaclonal variation: a novel source of variability from cell culture for plant improvement. Theor Appl Genet. 1981;60:197–214. doi: 10.1007/BF02342540. [DOI] [PubMed] [Google Scholar]
- Long D, Martin M, Sundberg E, Swinburne J, Puangsomlee P, Coupland G. The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Dsinsertion. Proc Natl Acad Sci USA. 1993;90:10370–10374. doi: 10.1073/pnas.90.21.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996;15:2331–2342. [PMC free article] [PubMed] [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- Miyao A, Yamazaki M, Hirochika H. Systematic screening of mutants of rice by sequencing retrotransposon-insertion sites. Plant Biotech. 1998;15:253–256. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA. 1999;96:15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasaki T, Burr B. International rice genome sequencing project: the effort to completely sequence the rice genome. Curr Opin Plant Biol. 2000;3:138–141. doi: 10.1016/s1369-5266(99)00047-3. [DOI] [PubMed] [Google Scholar]
- Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M. Loss-of–function mutations in the rice homeobox gene OSH15affect the architecture of internodes resulting in dwarf plants. EMBO J. 1999;18:992–1002. doi: 10.1093/emboj/18.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Leon-Kloosterziel KM, Koornneef M, Zeevaart JAD. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997a;114:161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997b;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- Sundaresan V. Horizontal spread of transposon mutagenesis: new uses for old elements. Trends Plant Sci. 1996;1:184–190. [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB. Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol. 2000;42:833–845. doi: 10.1023/a:1006448428401. [DOI] [PubMed] [Google Scholar]
- Triglia T, Peterson MG, Kemp DJ. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. Knock-out mutants from an En-1 mutagenized Arabidopsis thalianapopulation generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA. 1998;95:12432–12437. doi: 10.1073/pnas.95.21.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Tsugawa H, Miyao A, Yano M, Wu J, Yamamoto S, Matsumoto T, Sasaki T, Hirochika H (2001) The rice transposon Tos17 prefers low copy sequences as integration targets. Mol Gen Genet (in press) [DOI] [PubMed]
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–473. [Google Scholar]