Abstract
To enhance our understanding of the genetic basis of nitrogen use efficiency in maize (Zea mays), we have developed a quantitative genetic approach by associating metabolic functions and agronomic traits to DNA markers. In this study, leaves of vegetative recombinant inbred lines of maize, already assessed for their agronomic performance, were analyzed for physiological traits such as nitrate content, nitrate reductase (NR), and glutamine synthetase (GS) activities. A significant genotypic variation was found for these traits and a positive correlation was observed between nitrate content, GS activity and yield, and its components. NR activity, on the other hand, was negatively correlated. These results suggest that increased productivity in maize genotypes was due to their ability to accumulate nitrate in their leaves during vegetative growth and to efficiently remobilize this stored nitrogen during grain filling. Quantitative trait loci (QTL) for various agronomic and physiological traits were searched for and located on the genetic map of maize. Coincidences of QTL for yield and its components with genes encoding cytosolic GS and the corresponding enzyme activity were detected. In particular, it appears that the GS locus on chromosome 5 is a good candidate gene that can, at least partially, explain variations in yield or kernel weight. Because at this locus coincidences of QTLs for grain yield, GS, NR activity, and nitrate content were also observed, we hypothesize that leaf nitrate accumulation and the reactions catalyzed by NR and GS are coregulated and represent key elements controlling nitrogen use efficiency in maize.
In the last three decades, nitrogen fertilization has been a powerful tool in increasing grain yield (GY), especially for cereals such as maize (Zea mays) and wheat. However, the current agricultural and economic environment means that farmers must optimize the application of nitrogen fertilizers to avoid pollution by nitrates and to preserve their economic margin. Therefore, it has become of major importance to select for cereal cultivars that absorb and metabolize nitrogen in the most efficient way for grain or silage production.
In the majority of crop species, including grasses, the plant life cycle can be roughly divided into two main phases. During the vegetative growth phase young developing roots and leaves behave as sink organs that efficiently absorb and assimilate minerals such as inorganic nitrogen for amino acid and protein synthesis. During the remobilization phase leaves start to behave as source organs translocating carbon and organic molecules to ensure the formation of new developing tissues and/or storage tissues involved in plant survival such as seeds, tubers, bulbs, or trunks (Masclaux et al., 2000b). A better understanding of the metabolic and genetic control of acquisition and recycling during these two phases of plant growth and development is therefore of particular importance not only to improve crop quality and productivity, but also to avoid excessive use of fertilizers.
Until now, a number of studies have been undertaken by plant molecular physiologists to decipher the regulatory control mechanisms involved during the transition from sink to source organs (Harrison et al., 2000; Hellman et al., 2000; Lewis et al., 2000; Masclaux et al., 2000b). However, these approaches that involve whole plant physiology and/or transgenic plants are limited in that they only allow the role of a single or limited number of enzymes or regulatory elements to be identified and do not account for the variation of complex traits such as nitrogen use efficiency (NUE) often found in agronomic applications.
Conventional breeding procedures have been performed empirically over the last two decades by selecting the most appropriate traits in terms of yield or technological characteristics to improve plant productivity (Masclaux et al., 2000b; Richards, 2000). Although these approaches have been successful in terms of yield enhancement, there have so far been no real attempts to understand in a more integrated manner the physiological and genetic basis of these improvements, especially in relation to NUE.
At present, the use of quantitative genetic studies associated with the use of molecular markers may be a way to identify genes involved in the genetic variation of a complex character (Causse et al., 1995; Prioul, 1995). The combination of agronomic and physiological studies with quantitative genetic approaches will allow the use of molecular markers to identify key structural or regulatory loci involved in the expression of a quantitative trait (Causse et al., 1995; Prioul et al., 1997) and the selection of genotypes more efficient in terms of nitrogen use. Furthermore, the recent development of genome sequencing and mapping projects in a number of model crop species will be a valuable tool, allowing the precise location of key genes influencing the expression of desired traits. In turn, this new strategy will be of great potential for plant breeders to carrying out of marker assisted selection for improved NUE in relation to yield (Ribaud and Hoisington, 1998).
Because NUE is defined as the ratio of GY to nitrogen supplied (by soil and fertilizer) for a given level of fertilization differences in GY match differences in NUE. Thus, in selecting improved cultivars, breeders empirically select those that are more efficient in terms of nitrogen absorption and utilization. As modern maize genotypes were selected in the presence of high fertilization, they were consequently selected for their adaptation to high input (Castleberry et al., 1984). However, expression of genetic variability for GY is largely dependent on the level of nitrogen fertilization. The existence of an interaction of genotype x level of fertilization was shown in maize by various investigators (Moll et al., 1987; Landbeck, 1995; Bertin and Gallais, 2000a). In addition, it was found that correlations among various agronomic traits were very different depending upon the level of nitrogen fertilization (Di Fonzo et al., 1982; Bertin and Gallais, 2000a, 2000b). At high nitrogen input, variation in NUE was explained by variation in nitrogen uptake capabilities, whereas at low nitrogen input, variation in NUE was mainly due to differences in nitrogen utilization efficiency defined as the ratio GY/nitrogen uptake. These differences in the expression of genetic variability were further confirmed following the detection of specific quantitative trait loci (QTL) for a given level of fertilization (Agrama et al., 1999; Bertin and Gallais, 2000b). This suggests that several sets of genes are differentially expressed according to the amount of nitrogen provided to the plant (Bertin and Gallais, 2000a, 2000b).
In parallel with these agronomic studies, several investigators found that it is possible to detect genetic variation and select new genotypes that show increased or decreased activities of several enzymes involved in the nitrogen assimilatory pathway (Groat et al., 1984; Sherrard et al., 1986; Degenhart et al., 1992; Harrison et al., 2000). In particular, in maize hybrids a few attempts have been made to correlate the efficiency of primary nitrogen assimilation and nitrogen remobilization with yield and its components (Reed et al., 1980; Purcino et al., 1998). As a result of these studies it was concluded that increases in GY observed during the two last decades were not due to additional enhancement in inorganic nitrogen assimilation, but rather due to a better NUE as a result of a more efficient nitrogen remobilization. In particular, leaf longevity was shown to be one of the main factors responsible for yield increase in modern maize hybrids (Tollenaar, 1991; Ma and Dwyer, 1998). Extension of leaf metabolic activity improved the ratio between the assimilate supply from source leaves and demand in sink leaves during grain filling and was independent of the level of fertilization in the soil (Racjan and Tollenaar, 1999a, 1999b). During this metabolic process the putative role of enzymes involved in inorganic nitrogen assimilation and recycling such as nitrate reductase (NR), cytosolic Gln synthetase (GS1), and Glu dehydrogenase (GDH) was suggested (Lea and Ireland, 1999).
These physiological-agronomic studies prompted us to develop a quantitative genetic approach using molecular markers to obtain more information on the genetic basis of NUE in relation to yield using maize as a model crop. This species was chosen for study because of its world-wide economic importance and because of its high level of genetic polymorphism for molecular markers (Mann, 1999). This latter characteristic allowed the construction of a saturated restriction fragments length polymorphism (RFLP) map based on a population of recombinant inbred lines (RILs) using markers of known or unknown function. These RFLP markers included probes for genes involved in various regulatory and metabolic functions including carbon assimilation (Causse et al., 1995). In the present work a particular effort was devoted to mapping genes encoding proteins and enzymes involved in nitrogen assimilation and recycling for further QTL detection. Another advantage of using maize is its high capacity to absorb and metabolize organic and remobilize inorganic nitrogen (Cliquet et al., 1990). Since yield and its components depend largely on these three metabolic processes, maize is one of the best model plants to combine physiological and agronomic studies. In addition, maize is a C4 grass that compared with the majority of other plant species, has developed during its evolution a specific cellular compartmentation that allows a better metabolic efficiency in terms of carbon and nitrogen assimilation (Oaks, 1994).
The novelty of our approach was to study, in parallel, agronomic and physiological traits for the detection of QTLs and to interpret their causal relationships in an integrated manner. Coincidences between QTLs for agronomic traits and QTLs for physiological traits related to NUE will therefore give a physiological meaning to the QTLs for the agronomic traits. In addition, comapping of agronomic and physiological QTLs with genes encoding enzymes involved in nitrogen metabolism will give a genetic meaning to these QTLs. Several colocations of QTLs for yield and its components with the genes encoding enzymes involved in nitrogen assimilation were detected. Moreover, in many cases QTLs for the corresponding enzyme activity were detected on the same chromosomal fragments, suggesting that these genes may be considered as candidate genes influencing NUE and thus the expression of the corresponding agronomic and/or physiological traits. This study, therefore, represents the first attempt to dissect the genetic variability of a complex trait such as NUE and to identify some of its key physiological components that may influence the productivity of a crop plant. The physiological role of these components is further discussed in relation to nitrogen assimilation and management, leading to the conclusion that they could constitute good markers for selection to optimize plant performance and rationalize the use of nitrogen fertilizers in the future.
RESULTS AND DISCUSSION
Correlations between Physiological Traits in Young Vegetative Plants and Agronomic Traits for Yield at Adult Stage
To study correlations between agronomic and physiological traits we first established a database for agronomic traits related to yield and its components from field experiments performed over a 2-year period (Bertin, 1997; Bertin and Gallais, 2000a). As described in the “Material and Methods” section, RILs (selected from those used to build the genetic map) were crossed to an unrelated inbred line used as a tester. The resultant crosses reflect more accurately the performance of hybrids, which are always used for grain or silage maize production and as such, all measurements of agronomic traits were performed on these plants. For the determination of physiological traits related to nitrogen metabolism in young vegetative plants the RILs were not crossed to the tester. As a consequence, genetic variation for the physiological traits and detection of the corresponding QTL was expected to be greater. However, this type of analysis was at the expense of the correlation between agronomic and physiological traits. In addition, correlations for yield were calculated from adult plants grown under both low (N−) and high nitrogen (N+) input. This is justified because it was previously shown in field-grown maize plants that a shortage in mineral nitrogen does not significantly affect the growth of young vegetative plants (nitrogen from the soil being sufficient) and actually has a relatively low effect until the flowering period (Bertin and Gallais, 2000a).
For the agronomic traits the means and heritabilities of the 2-year experiment are presented in Table I (Bertin and Gallais, 2000b). thousand kernel weight (TKW) was the trait exhibiting the greatest heritability (0.81), whereas the lowest (0.13) was for grain nitrogen yield (GNY). As a consequence, genetic correlation with such a trait will be poorly estimated. For the physiological traits, the means and heritabilities of the 2-year experiments are presented in Table II. Heritability was about 0.69 for leaf NO3− content and 0.75 for glutamine synthetase (GS) activity. Because leaf NR activity was measured only in 1998, its heritability (0.87) was inflated. The phenotypic and genotypic correlations between agronomic and physiological traits are presented in Table III.
Table I.
Means and heritabilities for agronomic traits
| Trait | N−
|
N+
|
||
|---|---|---|---|---|
| Mean | Heritability | Mean | Heritability | |
| GY | 49.9 | 0.52 | 81.3 | 0.69 |
| KN | 229 | 0.54 | 339 | 0.74 |
| TKW | 229 | 0.81 | 252 | 0.89 |
| GNY | 0.92 | 0.13 | 1.56 | 0.34 |
| GMEa | 48.5 | 0.75 | 38.0 | 0.71 |
Results are the mean of a 2-year field experiment (1994 and 1995). Plants were grown at high (N+) and low (N−) nitrogen input as described in “Materials and Methods.” GY, Grain yield expressed (10−1 t ha−1); KN, kernel no. per plant; TKW, thousand kernels wt (g); GNY, grain nitrogen yield (g); GME, grain metabolic efficiency.
GME is the ratio grain yield/N uptake in aerial biomass.
Table II.
Means and heritabilities for physiological traits
| Trait | Mean | Heritability |
|---|---|---|
| Leaf NO3− content | 32.03 | 0.69 |
| Leaf NR activity | 204.95 | 0.87 |
| Leaf GS activity | 525.20 | 0.75 |
Results are the mean of a 2-year greenhouse experiment (1998 and 1999). Plants were grown hydroponically on a nutrient solution containing 1 mM NO3− as described in “Materials and Methods.” GS activity is expressed as nmol mn−1 mg protein−1 and NR activity as μmol h−1 mg−1 protein. NO3− concentration is expressed in mg dry wt−1.
Table III.
Phenotypic and genotypic correlations between agronomic and physiological traits
| Trait | N+
|
N−
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GY | KN | TKW | GNY | GME | GY | KN | TKW | GNY | GME | |
| Leaf NO3− content | 0.33a | 0.17 | 0.19b | 0.24c | 0.19b | 0.26c | 0.18b | 0.25c | 0.34a | 0.09 |
| (0.54) | (0.23) | (0.26) | (0.82)d | (0.30) | (0.44) | (0.29) | (0.29) | (1.00)d | (0.14) | |
| Leaf NR activity | −0.19a | −0.02 | −0.25b | 0.01 | −0.17 | −0.22b | −0.17 | −0.12 | −0.04 | −0.02 |
| (−0.28) | (−0.03) | (−0.30) | (0.03) | (−0.20) | (−0.32) | (−0.25) | (−0.15) | (−0.10) | (−0.10) | |
| Leaf GS activity | 0.15 | 0.07 | 0.15 | 0.28a | 0.08 | 0.25c | 0.21b | 0.13 | 0.09 | 0.28b |
| (0.24) | (0.12) | (0.19) | (0.93)d | (0.13) | (0.41) | (0.32) | (0.16) | (0.26) | (0.43) | |
N+ corresponds to plants grown in the field under high nitrogen input (N+ = 175 kg N/ha) and N− to plants grown in the field with no nitrogen fertilization (soil N content = 60 kg N/ha). The physiological traits measured in young vegetative plants grown under greenhouse-controlled conditions are: Leaf NO3− content (mg dry wt−1), leaf NR activity (μmol h−1 mg−1 protein), and leaf GS activity (nmol mn−1 mg−1 protein). The agronomic characters are GY (10−1 t ha−1), KN, TKW (g), GNY (g), and GME (ratio grain yield/N uptake in aerial biomass). Results are the mean of a 2-year experiment except for NR activity.
Phenotypic correlations significant respectively at 0.001, 0.05, and 0.01 probability levels. Genotypic correlations are indicated below in parentheses.
Poor estimation of genotypic correlation due to a large environmental effect.
A significant and positive correlation between leaf NO3− content of young vegetative plants and GY and GNY was always observed regardless of the level of fertilization of adult plants (Table III). As far as yield components are concerned, it appears that a higher correlation was observed between leaf NO3− content and TKW than with kernel number per plant (KN; Table III). Because we observed a highly significant correlation between leaf NO3− content and total plant NO3− content (r = 0.70) and no correlation with fresh plant biomass (data not shown), it was concluded that the leaf NO3− content mirrors the capacity of the plant to absorb and store mineral nitrogen. This NO3− pool is usually stored in the cell vacuole and serves as an osmoticum and as a source of mineral nitrogen when the soil supply becomes depleted (McIntyre, 1997; Crawford and Glass, 1998). Our results suggest that this pool of NO3− constitutes a major source of nitrogen that can be further metabolized and translocated to the grain and that can subsequently participate in the grain filling process. Grain filling appears to be largely under the control of nitrogen availability, since recent findings have demonstrated that nitrogen translocation facilitates kernels utilization of sugars (Below et al., 2000). All together these results suggests that the capacity for grain production is predetermined by the plants ability to absorb and store mineral nitrogen in its early phases of development. Similar conclusions were drawn by Teyker et al. (1989) and Plénet and Lemaire (1999) following the observation that in addition to what is required for its vegetative growth, the plant must absorb and store an excess of mineral nitrogen, which is then further metabolized and translocated to the kernels. It is proposed that leaf nitrate content at the early stages of plant development may be a good marker to select genotypes with enhanced GY and grain nitrogen content. This idea is in agreement with previous studies performed on maize hybrids in which the efficiency of primary nitrogen assimilation and nitrogen remobilization was correlated with yield and its components (Reed et al., 1980; Purcino et al., 1998). This prompted us to further investigate if within our RIL population genetic variability for metabolic processes involved in nitrogen assimilation could be detected.
One of the main enzymes involved in the assimilation and recycling of mineral nitrogen is GS (EC 6.3.1.2), which catalyzes the ATP-dependent conversion of Gln into Glu, utilizing ammonia as a substrate (Cren and Hirel, 1999; Lea and Ireland, 1999). As a consequence, our working hypothesis was that the rate of ammonium assimilation derived from NO3− reduction and/or organic nitrogen recycling was of major importance for plant NUE. Therefore, GS activity was used as a marker in the analysis of the correlation between physiological and agronomic traits. The finding that total leaf GS activity was positively correlated to GY, KN, and grain metabolic efficiency (GME) under low nitrogen input and to GNY at high nitrogen input (Table III) is in agreement with our hypothesis. This result is not surprising considering on the one hand the role of the plastidic isoenzyme (GS2) in the process of primary nitrogen assimilation and on the other hand the role of the cytosolic GS isoenzyme (GS1) during the recycling of organic nitrogen (Masclaux et al., 2000a). The role of GS1 during nitrogen remobilization has been already highlighted in maize hybrids containing lower amounts of nitrate, suggesting an active contribution of cytosolic GS during proteic nitrogen recycling (Purcino et al., 1998). Under nitrogen-limiting conditions the positive correlation found between GS activity and kernel number suggests that a high GS activity is required to avoid embryo abortion just after fertilization (Below, 1995).
It can be argued that the rate of NO3− reduction and the rate of ammonia assimilation contributeto the reduced nitrogen, which is further metabolized and translocated during grain filling. However, we observed a negative correlation between leaf NADH-NR (EC 1.6.6.1) activity in young vegetative plants (although measured only in 1998) and GY of mature plants. Such a negative correlation was also found for TKW under high nitrogen input (Table III). These data therefore suggest that when grain filling is high, the capacity of the plant to reduce NO3− is low. Similar conclusions were drawn by Reed et al. (1980) who showed that higher yields were obtained in genotypes exhibiting low NR activity.
QTLs Detection for Leaf NO3− Content, GS and NR Activity, and Coincidence with Agronomic Traits
QTLs for agronomic traits were identified in previous studies (Bertin, 1997; Bertin and Gallais, 2000a). QTLs for the traits considered in this study they are presented in Table IV. QTLs detected for physiological traits are presented in Table V. The position of the different QTLs on the maize RFLP map is shown in Figure 1.
Table IV.
QTLs detected for grain yield, its components and some related traits by simple interval mapping
| Trait | N Level | R2pa | R2gb | Chrc | Location
|
Confidence Intervald | LOD | Additive Effecte | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | +cM | Distance cM | ||||||||
| GY (10−1 t ha−1) | N+ | 0.40 | 0.59 | 1 | gsy282a | +7 | 160 | 146–176 | 2.4 | 2.66 |
| 1 | umc161 | +10 | 234 | 228–240 | 3.7 | 2.35 | ||||
| 3 | gsy224a | +21 | 78 | 66–90 | 3.4 | 2.53 | ||||
| 4 | gsy82r | +3 | 178 | 168–216 | 2.1 | −1.90 | ||||
| 5 | gsy258a | +13 | 186 | 170–196 | 2.5 | 2.69 | ||||
| N− | 0.23 | 0.44 | 3 | umc60 | +5 | 86 | 74–96 | 3.0 | 2.70 | |
| 5 | umc39b | +12 | 162 | 146–196 | 2.2 | 2.70 | ||||
| KN | N+ | 0.40 | 0.54 | 1 | gsy282a | +11 | 164 | 130–178 | 3.0 | 10.0 |
| 1 | gsy145 | +15 | 102 | 92–124 | 3.9 | 12.9 | ||||
| 1 | umc161 | +10 | 234 | 230–240 | 4.4 | 11.3 | ||||
| 3 | gsy298c | +25 | 120 | 108–120 | 2.5 | 9.2 | ||||
| 6 | umc39d | +28 | 88 | 72–94 | 1.9 | 8.4 | ||||
| 8 | gsy224b | +16 | 142 | 118–168 | 2.5 | 14.0 | ||||
| N− | 0.07 | 0.16 | 3 | umc60 | +7 | 88 | 72–112 | 2.0 | 8.9 | |
| TKW (g) | N+ | 0.48 | 0.54 | 1 | umc67 | +2 | 82 | 76–88 | 3.8 | −6.9 |
| 2 | gsy348c | +14 | 44 | 32–64 | 2.6 | −5.9 | ||||
| 4 | gsy156 | +6 | 120 | 106–128 | 4.7 | −8.3 | ||||
| 4 | umc66 | +20 | 154 | 134–174 | 2.6 | −9.1 | ||||
| 5 | gsy258a | +8 | 176 | 162–192 | 2.9 | 7.9 | ||||
| N− | 0.39 | 0.46 | 1 | umc67 | +2 | 82 | 74–114 | 2.9 | −5.8 | |
| 4 | gsy431 | +13 | 72 | 62–102 | 3.4 | −7.4 | ||||
| 4 | gsy156 | +16 | 120 | 104–130 | 3.2 | −6.6 | ||||
| 5 | gsy258a | +10 | 178 | 154–194 | 2.7 | 7.4 | ||||
| GNY (g) | N+ | 0.13 | 0.38 | 1 | adh1i | +1 | 204 | 198–210 | 3.1 | 0.047 |
| 1 | umc161 | +6 | 230 | 216–236 | 3.2 | 0.051 | ||||
| GME | N+ | 0.23 | 0.32 | 3 | gsy224a | +19 | 76 | 62–92 | 3.1 | 1.02 |
| N− | 0.25 | 0.33 | 1 | bnl829 | +16 | 250 | 238–258 | 2.3 | 1.06 | |
| 9 | umc113 | +4 | 6 | 2–26 | 3.6 | 1.37 | ||||
| 9 | bnl510 | +3 | 48 | 38–56 | 3.0 | 1.21 | ||||
Results are the mean of a 2-year field experiment (1994 and 1995). Plants were grown at high (N+) and low (N−) nitrogen input as described in “Materials and Methods.”
Percentage of phenotypic variance explained by the markers.
Percentage of genotypic variance explained by the markers R2g = R2p/h2.
Chromosome no.
Approximate confidence interval (LOD − 1).
Additive effect in testcross value with positive value for parent Io.
Table V.
QTLs for physiological traits
| Trait | R2pa | R2gb | Chrc | Location
|
Confidence Intervald | LOD | Additive Effecte | ||
|---|---|---|---|---|---|---|---|---|---|
| Marker | + cM | Distance cM | |||||||
| Leaf NO3− content | 0.28 | 0.40 | 2 | gsy1b1 | +3 | 24 | 20–80 | 2.82 | −3.66 |
| 2 | gsy108 | +10 | 58 | 68–80 | 2.92 | −4.034 | |||
| 5 | gsy154 | +6 | 74 | 96–136 | 3.36 | 4.123 | |||
| 5 | gsy343b | +9 | 114 | 150–184 | 3.50 | 4.452 | |||
| 5 | umc39b | +10 | 160 | 14–30 | 2.86 | 4.229 | |||
| Leaf NR activity | 0.36 | 0.42 | 5 | gsy343b | +5 | 110 | 98–116 | 8.31 | −79.29 |
| 5 | gsy258a | +6 | 174 | 146–190 | 2.19 | 49.30 | |||
| Leaf GS activity | 0.52 | 0.69 | 1 | gsy143b | +6 | 14 | 4–56 | 3.50 | 32.95 |
| 1 | gsy304 | +2 | 42 | 8–56 | 3.76 | 32.42 | |||
| 1 | gsy52r | +2 | 190 | 156–198 | 1.96 | 23.10 | |||
| 5 | gsy343b | +10 | 106 | 94–120 | 3.56 | 19.24 | |||
| 5 | gsy258a | +0 | 168 | 160–176 | 5.85 | 27.31 | |||
| 9 | gsy330 | +4 | 164 | 146–164 | 5.58 | 22.53 | |||
Results are the mean of a 2-year greenhouse experiment (1998 and 1999). Plants were grown hydroponically on a nutrient solution containing 1 mm NO3− as described in “Materials and Methods.”
Percentage of phenotypic variance explained by the markers.
Percentage of genotypic variance explained by the markers R2g = R2p/h2.
Chromosome no.
Approximate confidence interval (LOD − 1).
Additive effect in testcross with positive value for parent Io.
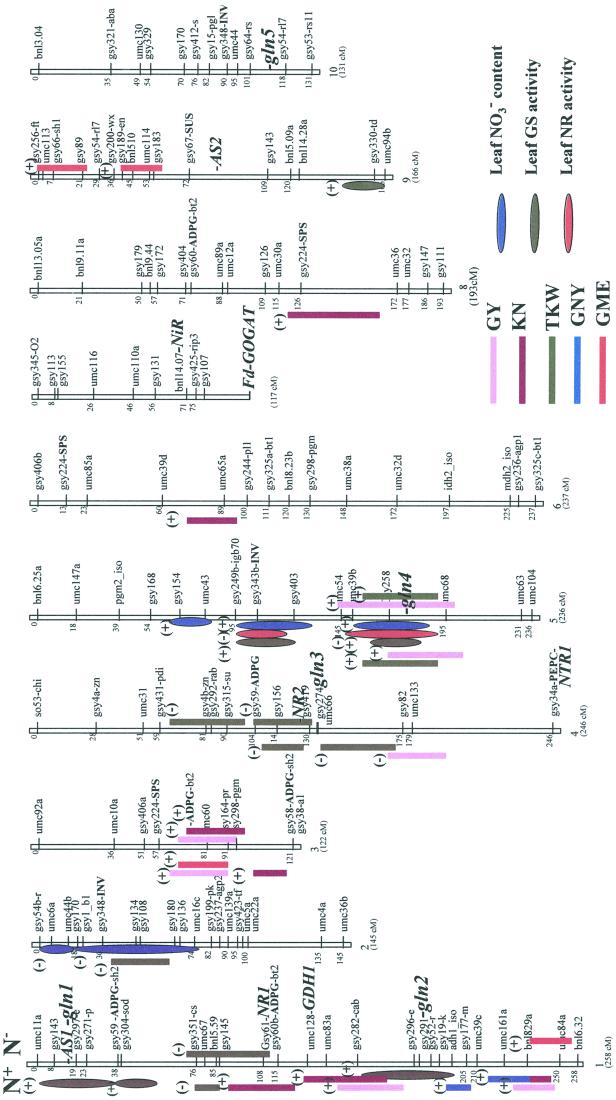
Figure 1.
Coincidences between QTLs for physiological traits and traits related to GY and its components. Location of the QTLs for physiological traits on the maize RFLP genetic map are indicated by oval symbols: blue for leaf NO3− content, green for leaf GS activity, and red for leaf NR activity. Locations of the QTLs for agronomic traits are indicated by vertical bars. Bars on the left side of the chromosomes are for plant grown under high nitrogen input (N+) and bars on the right side of the chromosomes are for plants grown under low nitrogen input (N−). Favorable allele from the parental line Io is indicated by (+) and unfavorable allele by (−). The position of the loci for genes encoding enzymes involved in nitrogen assimilation is indicated in bold italics: AS1 and AS2 (Asn synthetase 1 and 2); Fd-GOGAT (Ferredoxin-dependent Glu synthase); GDH1 (GDH 1); gnl1 to 5 (Gln synthetase 1 to 5); NR (nitrate reductase); NiR (nitrite reductase); and NTR1 (high-affinity nitrate transporter). The position of the loci for genes encoding enzymes involved in carbon assimilation is indicated in small bold characters; see Causse et al. (1995). ADPG, ADP Glc pyrophosphorylase; INV, invertase; SPS, Suc phosphate synthase.
Five QTLs for leaf NO3− content explaining 28% of the phenotypic variation were detected (Table V): two were located on chromosome 2 (near markersgsy1b1 and gsy 108), both with the favorable allele from the parental line F2, and the other three were located on chromosome 5 (markers gsy 154, gsy 343b, and gsy258) with the favorable allele from the parental line Io. One of the QTLs for leaf NO3− content (marker gsy108), located on chromosome 2, coincided positively with a QTL for TKW when plants were grown under high nitrogen input. One of the QTLs for leaf NO3− content located on chromosome 5 (marker gsy 258) was also positively coincident with a QTL for GY and TKW, regardless of the nitrogen fertilization level.
These results are in agreement with our previous analysis showing that there was a positive correlation between leaf NO3− content of young developing plants, GY, and TKW in field-grown mature plants independent of the level of fertilization (Table III). In addition, the identification of two QTLs for leaf NO3− content (common to QTLs for yield and its components) indicates that at least on chromosome 2 and chromosome 5 such QTLs are putatively involved in controlling NO3− accumulation. Therefore, the formation of a NO3− storage pool appears to be genetically controlled and is likely to be of major importance in the subsequent steps leading to its assimilation into organic matter that is used during grain filling.
Although QTLs for maximal leaf NADH-NR activity were detected using measurements performed on plants from only one of the 2-year experiments, we found on chromosome 5 two main regions influencing the enzyme activity (markers gsy 343b and gsy 258). They explained 36% of the observed phenotypic variation, or 42% of the genetic variation, which is high if we consider that only two QTLs for this trait were detected. One of the QTLs for NADH-NR activity located on chromosome 5 (marker gsy 258) with the favorable allele from the parental line F2, was negatively coincident with a QTL for yield both detected under low or high nitrogen fertilization conditions. These results are consistent with the observed negative correlation between GY or TKW and leaf NR activity (Table III). As shown in Figure 1, NADH-NR activity did not colocalize with either of the two structural genes (NTR1 and NTR2) located on chromosome 1 and 4, respectively. This result is not surprising if we consider that gene and protein expression are subjected to multiple regulations at the transcriptional and post-translational level, respectively (Campbell, 1999). This leads to the suggestion that genes encoding some of these regulatory elements are present in the two DNA regions identified on chromosome 5.
Six QTLs for total leaf GS activity were detected explaining 52% of the phenotypic variation, all six with the favorable alleles from the parental line Io. Three were located on chromosome 1 (marker gsy 143, gsy 304, and gsy52r). Two were localized on chromosome 5 (markers gsy 343b and gsy 258). The other one was located on chromosome 9 (marker gsy 330). It is interesting that out of these six QTLs we found three colocalized with genes encoding cytosolic GS quoted as gln1, gln2, and gln4 on the genetic map. This result suggests that for these three genes the final leaf cytosolic enzyme activity is mostly regulated at the transcriptional level. In contrast, for the other cytosolic GS gene gln3 located on chromosome 4 and the gene encoding plastidic GS (gln5) located on chromosome 10, other regulatory mechanisms acting at the posttranscriptional and/or -translational levels are likely to be involved in controlling the corresponding enzyme activity (Cren and Hirel, 1999). The detection of three additional QTLs (out of six) for leaf GS activity that did not colocalize with GS structural genes indicates that some loci located on different chromosome segments may be partly involved in the regulation of cytosolic and plastidic GS activity.
One of the most striking findings of this study was a positive coincidence of two QTLs for GS activity and QTLs for yield and its components (TKW and KN). One was located on chromosome 1 (coincidence with gln2 locus), which is coincident with a QTL for yield and kernel number at high nitrogen input, and one on chromosome 5 (coincidence with gln4 locus), which is coincident with QTLs for yield and TKW and independent of the nitrogen fertilization level. Such positive coincidences are consistent with the positive correlation observed between GY and GS activity, particularly at low nitrogen input. However, QTLs for yield on chromosome 5 can be considered as common to both nitrogen levels, because the favorable allele was detected under low and high levels of nitrogen fertilization. In contrast, only in N+ conditions did the QTL for yield colocalize with leaf GS activity on chromosome 1.
Although both QTLs for leaf GS activity coincided with QTLs for GY, the QTL identified on chromosome 1 seems to be related to the number of kernels whereas, that localized on chromosome 5 seems to influence kernel weight. This observation may be explained by the nonoverlapping function of the different GS genes in different organs or tissues and according to the plant developmental stage (Sakakibara et al.1992a; Li et al., 1993; Rastogi et al., 1998). It can therefore be hypothesized that the relative contribution of the corresponding GS isoenzyme activity in synthesizing or recycling organic nitrogen necessary for grain filling is finely balanced, not only depending on the plant developmental stage, but also on soil nitrogen availability. However, due to the complexity of the different GS isoenzyme distribution in the chloroplasts (GS2) and in the cytosol (GS1) of mesophyll and bundle sheath cells (Becker et al., 2000) and the number of analysis to be performed, it was not possible at this stage in our investigation to determine which GS isoenzyme was involved in each organ or cell type.
Nevertheless, the GS gene present at the gln4 locus appears to be a good candidate gene controlling NUE and influencing yield. It encodes a mRNA constitutively expressed in the different organs of maize, pGS112 (Sakakibara et al., 1992b). Since its transcriptional activity was not modified by NO3− or when the plants were nitrogen starved (Sakakibara et al., 1992a), it can be considered as a housekeeping gene responsible for ammonia assimilation and or recycling during plant growth and development.
One of the most interesting results was the significant number of QTLs coincident on chromosome 5 with the gln4 locus corresponding to the gene encoding cytosolic GS (Sakakibara et al., 1992b; Li et al., 1993) and which were always detected on the same genomic region over the 2-year experiment (Fig. 1). They included QTLs for GY, TKW, leaf GS activity, NR activity, and leaf NO3− content, leading to the suggestion that NO3− availability and the reactions catalyzed by NR and GS are key steps in the NUE for seed production. However, the negative effect of the QTL for NR activity was revealed by the negative additive effect of the allele from Io. This result is consistent with the negative impact of a capacity of NO3− reduction on yield and its components as already discussed in the previous section. In contrast, the QTLs for leaf NO3− content and leaf GS activity, both coincident with a favorable allele originating from the parental line Io, confirm the positive effect of these two traits on yield found in the correlation studies.
It is surprising that we did not find any coincidences between QTLs for GNY and GME and the three measured physiological traits related to nitrogen assimilation even though coincidences were observed with QTLs for GY and its components. This observation suggests that regulatory mechanisms others than those directly linked to primary nitrogen assimilation and recycling are involved in controlling the amount of nitrogen allocated to the grain. Therefore, the influence of GS and NR activity on GY and its components appears to be physiologically more relevant compared with GNY and GME, which represent only theoretical agronomic parameters. However, it cannot be excluded that the accuracy of the QTL detection for GNY and GME was not sufficient due to the relatively low number (77) of RILs examined, particularly if QTL effects for these two agronomic traits were relatively low.
CONCLUSION
NUE in plants is a complex trait that in addition to soil nitrogen availability can also depend on a number of internal and external factors such as photosynthetic carbon fixation to provide precursors required for amino acid biosynthesis or respiration to provide energy (Lewis et al., 2000). Our study confirmed that although this trait is controlled by a large number of loci acting individually or together, depending on carbon and nitrogen availability (Scheible et al., 1997) or being differentially expressed according to plant developmental stage (Masclaux et al., 2000a), it is possible to find enough phenotypic and genotypic variability to partially understand the genetic basis of NUE and thus identify some of the key components of yield in maize.
Several previous investigators have shown that in crops there is a significant genetic variability in several steps of nitrogen metabolism including nitrogen absorption, assimilation, and recycling (Masclaux et al., 2000b). It was suggested that these steps were of major importance in controlling yield and its components. However, these studies were mainly based on the correlations made between metabolite content or enzyme activity and yield or biomass production in contrasting genotypes (Reed et al., 1980; Purcino et al., 1998). In this investigation we confirmed that most of the traits of performing genotypes in terms of nitrogen metabolite content and activity of enzymes involved in nitrogen reduction and assimilation can also be found in a population of RILs in which the expression of these physiological traits is extended. Our results strengthened the concept that yield improvement in maize can be achieved by selecting genotypes with a high capacity to store NO3− in the leaves and a low capacity to reduce inorganic nitrogen during the vegetative phase of plant development (Plénet and Lemaire, 1999). In addition, the organic nitrogen supply from source leaves during grain filling seems to be of major importance in selecting genotypes with enhanced yield. In particular, the process of nitrogen remobilization was shown to be dependent on leaf longevity rather than the level of fertilization in the soil (Racjan and Tollenaar, 1999a, 1999b). Although the physiological and molecular mechanisms controlling the ratio of assimilate supply from source leaves to the demand in sink organs such as grains have still not been fully elucidated (Masclaux et al., 2000b), there are strong lines of evidence that suggest that enzymes such as cytosolic GS and possibly GDH play a central role in recycling organic nitrogen released from protein hydrolysis during leaf senescence (Reed et al., 1980). In this study, particularly when nitrogen is limiting, the finding that within the population of RILs a positive correlation was observed between leaf GS activity and several agronomic traits related to yield reinforces this hypothesis. In addition, the colocalization of two QTLs for yield with two loci for GS1 structural genes and two QTLs for leaf GS activity strongly support the current consensus that the GS enzymatic activity in the leaf cytosol is one of the major steps controlling organic matter reallocation from source to sink organs. Previous studies have already demonstrated that when GS1 is overexpressed in Lotus, nitrogen remobilization was prematurely induced leading to early senescence of the plant (Vincent et al., 1997). In rice (Yamaya, 1999) and wheat (D. Habash, personal communication) preliminary investigations made on plants with enhanced or decreased GS1 activity indicates that GY and grain nitrogen content is modified. In other species such as tobacco (Migge et al., 2000) or poplar (Gallardo et al., 1999), overexpression of GS2 or GS1 significantly increased plant biomass production at the early stage of plant development.
In addition, the triple colocalization of leaf GS activity, leaf NR activity, and leaf NO3− content found in two loci on chromosome 5 is in favor of the hypothesis that signals derived from the ammonia assimilatory pathway interact with NO3− uptake and reduction (Scheible et al., 1997). One of these two loci comapping with the gln4 locus and two QTLs for yield merits further analysis to identify whether common regulatory gene(s) or element(s) may be involved in the concerted regulation of these three metabolic processes. This will be achieved by increasing the number of RILs analyzed and by the use of highly RIL lines to refine the chromosomal zone involved and to decrease the distance between the flanking markers. Validation of the putative effect of the QTL for GS1 activity will be performed in parallel by overexpressing the corresponding gene or by introducing the favorable allele in an unfavorable genetic background.
We have also to bear in mind that coincidence between QTLs for yield and its components and leaf NO3− content, NR, and GS activity were found following the physiological analysis of young vegetative plants. It is well known that depending on whether young or senescing tissues are being examined the relative amounts of the various nitrogen metabolites can be variable (Masclaux et al., 2000a). In addition, the different genes encoding GS can be differentially expressed according to the physiological status and the developmental stage of the plant (Cren and Hirel, 1999). It is therefore possible that other QTLs related to yield could be identified when measuring the same physiological parameters at different stages of plant development. This may also be true for the other proteins (nitrate transporters) or enzymes involved in nitrogen assimilation (NR), reassimilation (ferredoxin-dependent Glu synthase), and translocation (GDH and Asn synthetase) for which we did not find any QTLs colocalizing with those relating to yield or its components. As such, the aim of our future studies will be to investigate the genetic basis of NUE in a spatial and temporal manner.
We have shown in the present study that genetic variability for NUE can be studied in a more targeted and integrated manner by the means of a quantitative genetic approach using molecular markers and combining agronomic and physiological studies. This approach will certainly be increasingly used in the future to identify new genes or loci involved in the regulation of these metabolic pathways and their interconnection with carbon assimilation and recycling and to select genotypes that assimilate or re-mobilize nitrogen more efficiently. The recent development of genome sequencing and mapping projects in a number of model crop species such as maize (Running et al., 2000) will be a valuable tool allowing the precise location of QTLs associated with the desired traits. In addition, genetic characterization of the identified QTLs through sequence analysis will certainly allow the identification of possible structural or regulatory genes controlling NUE during plant development and according to different environmental conditions.
MATERIALS AND METHODS
Plant Material for Agronomic Studies
Data obtained by Bertin and Gallais (2000a) served as an agronomic reference for the studies performed on young developing plants. A total of 99 RILs crossed to a common tester (F252) were grown in the field on two levels of nitrogen fertilization (N+ = 175kg nitrogen/ha and N−= no nitrogen fertilization) over 2 consecutive years (1994 and 1995) as described by Bertin and Gallais (2000a, 2000b). In N−, all nitrogen was provided by the soil (estimated at about 60 kg/ha). The RILs were an F6 generation derived from a cross between a French flint and early line of maize (Zea mays; F2) and an iodent late line (Io). Such a population of RILs was chosen since the two parental lines are highly complementary in terms of heterotic grain productivity. Furthermore, the agronomic study of the parents revealed differences in their NUE (Bertin and Gallais, 2000a, 2000b). Several traits were measured at flowering and grain harvest. In the present study traits used for correlation studies and QTL detection were GY and its components: KN and TKW, and GME corresponding to the ratio GY/total nitrogen absorbed by the aerial biomass. For more details about the procedure used to measure the agronomic traits, see Bertin and Gallais (2000a, 2000b).
Plant Material for Physiological Studies
Due the limited capacity of our plant growth hydroponic system (240 plants), physiological studies in the young stages were developed only on 77 RILs randomly chosen from the 99 lines used to perform the agronomic studies. In addition, the two parental lines (F2 and Io) and the tester (F252) were used as internal controls in all the subsequent measurements. To avoid heterogeneity in the germination time, imbibition of the seeds was performed at 6°C in the dark for 3 d. Seedlings were then transferred onto sand and watered daily on a nutrient solution containing 5.6 mm K+, 3.4 mm Ca2+, 0.9 mm Mg2+, 0.9 mm H2PO4−, and 21.5 μm Fe (Sequestrene Ciba-Geigy, Basel, 23 μm B, 9 μm Mn, 0.30 μm Mo, 0.95 μm Cu, and 3.50 μm Zn. Nitrogen was supplied as 1 mm NO3−. After 1 week when two to three leaves had emerged, the 77 RIL and the three control lines were randomly placed on a 130-L aerated hydroponic culture unit containing the same nutrient solution used for seedling growth. The nutrient solution was replaced daily. The experiment was performed in triplicate for each RIL (three hydroponic units with 80 lines per hydroponic unit) and the three hydroponic culture units were kept 18 d in a greenhouse in 1998 (May 18–June 5) and 17 d in 1999 (June 18–July 5). Plants were harvested at the 6 to 7 leaf stage between 9 to 12 am and separated into young leaves (three youngest leaves), stems, and roots. The samples were immediately placed in liquid N2 and then stored at −80°C until further analysis. Leaf NO3− content, leaf NADH-NR activity, and leaf GS activity were selected as representative marker metabolites and enzyme activities of primary nitrogen assimilation in young developing plants (Masclaux et al., 2000b). Therefore, these parameters were measured on pooled frozen young leaf samples collected from each experiment, except for leaf NR activity, which was measured only on plants grown in 1998. Measurements were performed twice on two different extractions of the three replicates. For correlation studies and QTL detection, leaf NO3− concentration and enzyme activities were calculated from the average value of the different measurements.
Protein Extraction, Enzymatic Assay, Metabolite Extraction, and Analyses
Protein extraction was carried out on 250 mg of frozen leaf material as described earlier (McNally et al., 1983). GS activity was assayed using the biosynthetic activity as described by O'Neal and Joy (1973). GS activity was expressed as nmol mn−1 mg−1 protein. NR was extracted and the maximal extractable activity measured as described by Ferrario-Méry et al. (1998). NR activity was expressed as μmol h−1 mg −1 protein and proteins were quantified using the Bradford method (Bradford, 1976). For NO3− determination, a 20-mg aliquot of lyophilized young leaf tissue was extracted successively with 80% ethanol, with 50% ethanol, and finally with water (Rochat and Boutin, 1989). NO3− concentration expressed in milligrams of dry weight−1 was determined according to the method of Cataldo et al. (1975).
Gene Mapping and QTL Detection
For the mapping of genes encoding enzymes and proteins involved in nitrogen metabolism we used the RFLP genetic map published by Causse et al. (1996) containing 152 marker loci corresponding to a total map length of 1,813 cM. The mean interval between two markers, depending on the chromosome, varies from 8 to 18 cM. The cDNA probes used for mapping were as follows: a high affinity NO3− transporter, NTR1, (B. Hirel, unpublished data) isolated from a NO3− induced maize root seedlings cDNA library using a barley cDNA clone as a probe (Trueman et al., 1996); two NADH-NR, NR1 and NR2 (Long et al., 1992); nitrite reductase, NiR; (Lahners et al., 1988); GDH, GDH1, (Sakakibara et al., 1995); four cytosolic GS, gln1, gln2, gln3, and gnl4, plastidic GS, gln5, (Sakakibara et al., 1992b), ferredoxin-dependent Glu synthase, Fd-GOGAT, (Sakakibara et al., 1992b), and Asn synthetase (AS1 and AS2; Chevalier et al., 1996), which was located on two loci. The loci corresponding to gnl1, 2, 3, 4, and 5 corresponds to the GS genes named pGS122, pGS134, pGS107, pGS112, and pGS202 by Sakakibara et al. (1992b) and GS1–1, GS1–2, GS1–4, GS1–3, and GS2 by Li et al. (1993).
QTLs were detected using the QTL software (Utz and Melchinger, 1995) following simple interval mapping. Only QTLs with an LOD score greater than 2 were considered (Lander and Botstein, 1989). To represent a QTL on the map taking into account error in the location, we give the chromosome region corresponding to a LOD greater than the maximum LOD minus 1, which is not a true confidence interval. It is called an LOD-1 interval. It generally overestimates the confidence interval. Two QTLs of different traits will be declared as coincident when their LOD-1 intervals overlap. A coincidence will be said to be positive when there is coincidence of favorable (or unfavorable) alleles for both traits. The coincidence will be said to be negative when there is coincidence of a favorable allele for one trait with an unfavorable allele for the other trait. For each trait we have calculated the percentage of phenotypic (R2p) and genotypic variation (R2g) explained by the markers. In addition, for each QTL detected, the estimated additive effect (one-half of the difference between the estimated value of the two homozygous genotypes at the QTL) is presented.
Statistical Analysis
To determine whether any of the main physiological traits measured in young vegetative plants could explain the genetic variability of yield and its components of field-grown plants, correlations were made between the two sets of traits using, in each case, the average of the values obtained from the 2-year experiments. However, such correlations can be affected by variation due to environmental effects. We therefore calculated genetic correlations following removal of variability due to environmental effects. Since the two experiments were performed independently, the phenotypic covariance between the two developmental stages was equal to the genotypic covariance. As a consequence, the genetic correlation was equal to the phenotypic correlation divided by the product of the square root heritabilities of the two traits (Becker, 1984). Heritabilities for physiological traits were deduced from the analysis of variance of each trait. Heritabilities for agronomic traits have been calculated previously (Bertin and Gallais, 2000a). Significance (difference from 0) for the correlations is given only for phenotypic correlations. It is also an approximate test for the genotypic correlations. Accuracy on the genotypic correlations depend mainly on the accuracy of measurements on each trait (when the heritability is low, the accuracy on genotypic correlation is low). However, it is not possible to calculate an accurate confidence interval.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Jakson Hoarau (Université de Paris Sud, Orsay, France) for providing the maize root cDNA library. We are also grateful to Dr. Judith Harrison for helpful discussion and for proofreading the manuscript. Thanks to Bertrand and Christiane Auclair, Valerie Combes, François Gosse, Brigitte Mauze, and Jean-Marc Monties for technical assistance.
LITERATURE CITED
- Agrama HAS, Zacharia AG, Said M, Tuinstra M. Identification of quantitative trait loci for nitrogen use efficiency in maize. Mol Breed. 1999;5:187–195. [Google Scholar]
- Becker TW, Carrayol E, Hirel B. Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta. 2000;211:800–806. doi: 10.1007/s004250000355. [DOI] [PubMed] [Google Scholar]
- Becker WA. Manual of quantitative genetics. Ed 4. WA: Academic Enterprises, Pullman; 1984. [Google Scholar]
- Below FE. Nitrogen metabolism and crop productivity. In: Pessarakli M, editor. Handbook of Plant and Crop Physiology. New York: Marcel Dekker; 1995. pp. 275–301. [Google Scholar]
- Below FE, Cazetta JO, Seebauer JR. Physiology and Modeling Kernel Set in Maize. Crop Science Society of America and American Society of Agronomy special publication number 29. 2000. Carbon/nitrogen interactions during ear and kernel development of maize. [Google Scholar]
- Bertin P. Bases génétiques et physiologiques de la valorisation de la fumure azotée chez le maïs. PhD thesis. Université de Paris XI; 1997. [Google Scholar]
- Bertin P, Gallais A. Physiological and genetic basis of nitrogen use efficiency in maize: I. Agrophysiological results. Maydica. 2000a;45:53–66. [Google Scholar]
- Bertin P, Gallais A (2000b) Physiological and genetic basis of nitrogen use efficiency in maize: II. QTL detection and coincidences. Maydica (in press)
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Campbell WH. Nitrate reductase structure, function, and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Physiol. 1999;50:277–303. doi: 10.1146/annurev.arplant.50.1.277. [DOI] [PubMed] [Google Scholar]
- Castleberry RM, Crum CW, Krull CF. Genetic yield improvement of US maize cultivars under varying fertility and climatic environments. Crop Sci. 1984;24:33–36. [Google Scholar]
- Cataldo DA, Haroon M, Schrader TE, Youngs VL. Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Ann. 1975;6:71–80. [Google Scholar]
- Causse M, Rocher JP, Henry AM, Charcosset A, Prioul JL, de Vienne D. Genetic dissection of the relationship between carbon metabolism and early growth in maize, with emphasis on key-enzyme loci. Mol Breed. 1995;1:259–272. [Google Scholar]
- Causse M, Santoni S, Damerval C, Maurice A, Charcosset A, Deatrick J, de Vienne D. A composite map of expressed sequences in maize. Genome. 1996;39:418–432. doi: 10.1139/g96-053. [DOI] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Just D, Raymond P. Metabolic regulation of asparagine synthetase gene expression in maize (Zea mays L.) root tips. Plant J. 1996;9:1–11. doi: 10.1046/j.1365-313x.1996.09010001.x. [DOI] [PubMed] [Google Scholar]
- Cliquet JB, Deléens E, Bousser A, Martin M, Lescure JC, Prioul JL, Mariotti A, Morot-Gaudry JF. Estimation of C and N allocation during stalk elongation by 13C and 15N tracing in Zea mays L. Plant Physiol. 1990;92:79–87. doi: 10.1104/pp.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. [Google Scholar]
- Cren M, Hirel B. Glutamine synthetase in higher plants: regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol. 1999;40:1187–1193. [Google Scholar]
- Degenhart NR, Barnes DK, Vance CP. Divergent selection for nodule aspartate aminotransferase and asparagine synthetase activities in alfalfa. Crop Sci. 1992;32:313–317. [Google Scholar]
- Di Fonzo N, Motto M, Maggiore T, Sabatino R, Salamini F. N uptake, translocation and relationships among N related traits in maize as affected by genotype. Agronomie. 1982;2:789–796. [Google Scholar]
- Ferrario-Méry S, Valadier M-H, Foyer C. Overexpression of nitrate reductase in tobacco delays drought-induced decreases in nitrate reductase activity and mRNA. Plant Physiol. 1998;117:293–302. doi: 10.1104/pp.117.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Fu J, Canton FR, Garcia-Gutierez A, Canovas F, Kirby E. Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta. 1999;210:19–26. doi: 10.1007/s004250050649. [DOI] [PubMed] [Google Scholar]
- Groat RC, Vance CP, Barnes DK. Host plant nodule enzymes associated with selection for increased N2 fixation in alfalfa. Crop Sci. 1984;24:895–898. [Google Scholar]
- Harrison J, Brugière N, Phillipson B, Ferrario-Méry S, Becker T, Limami A, Hirel B. Manipulating the pathway of ammonia assimilation through genetic manipulation and breeding: consequences on plant physiology and development. Plant Soil. 2000;221:81–93. [Google Scholar]
- Hellman H, Barker L, Funk D, Frommer W. The regulation of assimilate allocation and transport. Aust J Plant Physiol. 2000;27:583–594. [Google Scholar]
- Lahners C, Kramer V, Back E, Privalle L, Rothstein S. Molecular cloning of a complementary DNA encoding maize nitrite reductase. Plant Physiol. 1988;88:741–746. doi: 10.1104/pp.88.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landbeck MV. Untersuchungen zur genetischen verbesserung der anbaueigung von körnermais unter produktionsbedingungen mit verringerter sticksoffversorgung. PhD thesis. Universität Hohenheim; 1995. [Google Scholar]
- Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Ireland RJ. Plant amino acids. In: Singh BK, editor. Nitrogen Metabolism in Higher Plants. New York: Marcel Dekker; 1999. pp. 1–47. [Google Scholar]
- Lewis CE, Noctor G, Causton D, Foyer C. Regulation of assimilate partitioning in leaves. Aust J Plant Physiol. 2000;27:507–519. [Google Scholar]
- Li MG, Villemur R, Hussey PJ, Silflow CD, Gantt JS, Snustad DP. Differential expression of six glutamine synthetase genes in Zea mays. Plant Mol Biol. 1993;23:401–407. doi: 10.1007/BF00029015. [DOI] [PubMed] [Google Scholar]
- Long DM, Oaks A, Rothstein SJ. Regulation of maize root nitrate reductase mRNA levels. Physiol Planta. 1992;85:561–566. [Google Scholar]
- Ma BL, Dwyer ML. Nitrogen uptake and use in two contrasting maize hybrids differing in leaf senescence. Plant Soil. 1998;199:283–291. [Google Scholar]
- Mann CC. Crop scientists seek a new revolution. Science. 1999;283:310–314. [Google Scholar]
- Masclaux C, Quilleré I, Gallais A, Hirel B (2000b) The challenge of remobilization in plant nitrogen economy: a survey of physio-agronomic and molecular approaches. Ann Appl Biol (in press)
- Masclaux C, Valadier MH, Brugiere N, Morot-Gaudry JF, Hirel B. Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management. Planta. 2000a;211:510–518. doi: 10.1007/s004250000310. [DOI] [PubMed] [Google Scholar]
- McIntyre GI. The role of nitrate in the osmotic and nutritional control of plant development. Aust J Plant Physiol. 1997;24:103–118. [Google Scholar]
- McNally SF, Hirel B, Gadal P, Mann AF, Stewart GR. Glutamine synthetase in higher plants: evidence for a specific isoform content related to their possible physiological role and their compartmentation within the leaf. Plant Physiol. 1983;72:22–25. doi: 10.1104/pp.72.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migge A, Carrayol E, Hirel B, Becker TW. Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta. 2000;2:252–260. doi: 10.1007/PL00008132. [DOI] [PubMed] [Google Scholar]
- Moll RH, Kamprath EJ, Jackson WA. Development of nitrogen efficient prolific hybrids of maize. Crop Sci. 1987;27:181–186. [Google Scholar]
- Oaks A. Efficiency of nitrogen utilization in C3 and C4 cereals. Plant Physiol. 1994;106:407–414. doi: 10.1104/pp.106.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal D, Joy KD. Glutamine synthetase of pea leaves: I. Purification, stabilization and pH optima. Arch Biochem Biophys. 1973;159:113–122. doi: 10.1016/0003-9861(73)90435-9. [DOI] [PubMed] [Google Scholar]
- Plénet D, Lemaire G. Relationships between dynamics of nitrogen uptake and dry matter accumulation in maize crops: determination of critical N concentration. Plant Soil. 1999;216:65–82. [Google Scholar]
- Prioul JL. Corn distribution of photoassimilates and source-sink relationship. In: Zamski E, Shaffer AA, editors. Photoassimilates Distribution in Plants and Crops. New York: Marcel Dekker; 1995. [Google Scholar]
- Prioul JL, Quarrie S, Causse M, de Vienne D. Dissecting complex physiological functions into elementary components through the use of molecular quantitative genetics. J Exp Bot. 1997;48:1151–1163. [Google Scholar]
- Purcino AAC, Arellano C, Athwal GS, Huber SC. Nitrate effect on carbon and nitrogen assimilating enzymes of maize hybrids representing seven eras of breeding. Maydica. 1998;43:83–94. [Google Scholar]
- Racjan I, Tollenaar M. Source:sink ratio and leaf senescence in maize: I. Dry matter accumulation and partitioning during grain filling. Field Crop Res. 1999a;60:245–253. [Google Scholar]
- Racjan I, Tollenaar M. Source:sink ratio and leaf senescence in maize: II. Nitrogen metabolism during grain filling. Field Crop Res. 1999b;60:255–265. [Google Scholar]
- Rastogi R, Chourey PS, Muhitch MJ. The maize glutamine synthetase gene is preferentially expressed in kernel pedicels and is developmentally regulated. Plant Cell Physiol. 1998;39:443–4446. doi: 10.1093/oxfordjournals.pcp.a029388. [DOI] [PubMed] [Google Scholar]
- Reed AJ, Below FE, Hageman RH. Grain protein accumulation and the relationship between leaf nitrate reductase and protease activities during grain development in maize: I. Variation between genotypes. Plant Physiol. 1980;66:164–170. doi: 10.1104/pp.66.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaud JM, Hoisington D. Marker assisted selection: new tools and strategies. Trends Plant Sci. 1998;3:236–239. [Google Scholar]
- Richards RA. Stable traits to increase crop photosynthesis and yield of grain crops. J Exp Bot. 2000;51:447–458. doi: 10.1093/jexbot/51.suppl_1.447. [DOI] [PubMed] [Google Scholar]
- Rochat C, Boutin JP. Carbohydrates and nitrogenous compounds changes in the hull and in the seed during the pod development of pea. Plant Physiol Biochem. 1989;67:10–15. [Google Scholar]
- Running M, Scanlon M, Sinha N. Maize genetics: 2000 and beyond. Plant Cell. 2000;12:830–835. doi: 10.1105/tpc.12.6.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Fujii K, Sugiyama T. Isolation and characterization of a cDNA encoding maize glutamate dehydrogenase. Plant Cell Physiol. 1995;36:789–797. doi: 10.1093/oxfordjournals.pcp.a078823. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kawabata S, Hase T, Sugiyama T. Differential effects of nitrate and light on the expression of glutamine synthetase and ferredoxin-dependent glutamate synthase in maize. Plant Cell Physiol. 1992a;33:1193–1198. [Google Scholar]
- Sakakibara H, Kawabata S, Takahashi H, Hase T, Sugiyama T. Molecular cloning of the family of glutamine synthetase genes from maize: expression of genes for glutamine synthetase and ferredoxin-dependent glutamate synthase in photosynthetic and non-photosynthetic tissues. Plant Cell Physiol. 1992b;33:49–58. [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Müller-Röber B, Caboche M, Stitt M. Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell. 1997;9:783–798. doi: 10.1105/tpc.9.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard JH, Lambert RJ, Below FE, Durand RT, Messmer MJ, Willman MR, Winkels CS, Hageman RH. Use of physiological traits, especially those of nitrogen metabolism for selection in maize. In: Neyra CA, editor. Biochemical Basis of Plant Breeding. 2, Nitrogen Metabolism. Boca Raton, FL: CRC Press; 1986. pp. 109–130. [Google Scholar]
- Teyker RH, Moll NA, Jackson NA. Divergent selection among maize seedlings for nitrate uptake. Crop Sci. 1989;29:879–884. [Google Scholar]
- Tollenaar M. Physiological basis of genetic improvement of maize hybrids in Ontario from 1959 to 1988. Crop Sci. 1991;31:119–124. [Google Scholar]
- Trueman LJ, Richardson A, Forde BJ. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinardtii and Aspergillus nidulans. Gene. 1996;175:223–231. doi: 10.1016/0378-1119(96)00154-0. [DOI] [PubMed] [Google Scholar]
- Utz HF, Melchinger AE. PLABQTL a computer program to map QTL. JQTL. 1995;1:2. [Google Scholar]
- Vincent R, Fraisier V, Chaillou S, Limami MA, Deléens E, Phillipson B, Douat C, Boutin JP, Hirel B. Overexpression of a soybean gene encoding cytosolic glutamine synthetase in shoots of transgenic Lotus corniculatus L. plants triggers changes in ammonium assimilation and plant development. Planta. 1997;201:424–433. doi: 10.1007/s004250050085. [DOI] [PubMed] [Google Scholar]
- Yamaya T. Proceedings of the International Symposium on Nitrogen and Carbon Utilization in Plants: Molecular Approaches for the Improvement of Productivity. Sendai, Japan: Gonryo-Kailan; 1999. Nitrogen recycling and utilization in rice: molecular aspects and application of QTL analysis; pp. 11–12. [Google Scholar]