Abstract
Identification of genes with variants causing non-syndromic hearing loss (NSHL) is challenging due to genetic heterogeneity. The difficulty is compounded by technical limitations that in the past prevented comprehensive gene identification. Recent advances in technology, using targeted capture and next-generation sequencing (NGS), is changing the face of gene identification and making it possible to rapidly and cost-effectively sequence the whole human exome. Here, we characterize a five-generation Chinese family with progressive, postlingual autosomal dominant nonsyndromic hearing loss (ADNSHL). By combining population specific mutation arrays, targeted deafness genes panel, whole exome sequencing (WES), we identified PDE1C (Phosphodiesterase 1C) c.958G>T (p.A320S) as the disease-associated variant. Structural modeling insights into p.A320S strongly suggest that the sequence alteration will likely affect the substrate-binding pocket of PDE1C. By whole-mount immunofluorescence on postnatal day 3 mouse cochlea, we show its expression in outer (OHC) and inner (IHC) hair cells cytosol co-localizing with Lamp-1 in lysosomes. Furthermore, we provide evidence that the variant alters the PDE1C hydrolytic activity for both cAMP and cGMP. Collectively, our findings indicate that the c.958G>T variant in PDE1C may disrupt the cross talk between cGMP-signaling and cAMP pathways in Ca2+ homeostasis.
Keywords: Hearing loss, Whole exome sequencing, Mutation, Cyclic nucleotides, Phosphodiesterases
Hearing loss (HL) is an economically and socially important cause of human morbidity, affecting over 5% of the world’s population, 360 million people. About 2 to 3 out of every 1,000 children in the United States are born with a detectable level of HL in one or both ears (Vohr 2003). Approximately 37.5 million American adults (15%) aged 18 and over report some trouble hearing and this number is expected to gradually increase from 44.11 million in 2020 to 73.50 million by 2060 (Blackwell et al. 2014; Goman et al. 2017). A minimum of 50% of HL is predicted to be hereditary and NSHL accounts for more than 70% of this. The inheritance pattern is autosomal dominant (AD) in nearly all families affected by post-lingual, progressive NSHL with an onset often spanning the second through fifth decades of life (Petersen 2002). To date, there are 59 unique ADNSHL loci mapped; however, only 35 causative genes implicated in a wide variety of biological functions have been cloned since 1997 (http://hereditaryhearingloss.org/).
Although there has been much success using traditional strategies, such as linkage mapping and positional candidate approaches, NGS technology is changing the field of genetics by increasing the speed and efficacy of detecting variants for all Mendelian disorders, including NSHL. HL arises due to variants in broad subsets of genes and the diverse genetic underlying defects often make the initial development and progression of HL indistinguishable. Understanding the molecular basis of the auditory system, one of the human body’s most complex and delicate sensory systems, requires a fundamental knowledge of how the participating genes function. Elucidation of the molecular outcomes of pathogenic variants has translational implications to improve diagnostic, prognostic, and therapeutic medical care.
Here we report the identification of a variant (c.958G>T; p.A320S; rs775633137; ClinVar: 433528) in the PDE1C (Phosphodiesterase 1C) gene that is associated with progressive, postlingual ADNSHL in a large multi-generational Chinese family. Cyclic nucleotide phosphodiesterases (PDEs) catalyze the hydrolysis of intracellular second messengers, cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP), to produce 5’-AMP and 5’-GMP, respectively. Since these second messengers play a critical role in many cellular homeostatic processes, dysregulation of their signals and crosstalk of cellular signaling pathways promotes maladaptive cellular function and underpins human diseases. Based on their molecular sequence, kinetic properties (substrate specificity), regulation and pharmacological characteristics, mammalian PDEs can be classified into 11 families (Thompson and Appleman 1971). Deregulation of PDE isozymes have been widely implicated in many pathologies including erectile dysfunction (PDE5) (Tsertsvadze et al. 2009), cardiovascular diseases (PDE3, 4, 5) (Van Wagoner and Lindsay 2012; Giannetta et al. 2012; Movsesian et al. 2011), pulmonary inflammatory conditions (PDE4) (Antoniu 2006), auto-immune diseases (PDE3, 4, 5, 7) (Bjorgo et al. 2011; Hertz and Beavo 2011) and cognition and memory disorders (PDE1, 2, 3, 4, 7, 9, 10A) (Xu et al. 2011).
Materials and Methods
Subjects and clinical evaluation
We ascertained a five-generation family from central China with ADNSHL. The clinical history was taken and a physical examination carried out on the family members, with special emphasis on identifying potential environmental causes of HL such as infections, trauma, and exposure to ototoxic drugs including aminoglycoside use and noise, and for evidence of syndromic forms of deafness. Otoscopic examination was done, and air conduction threshold measurements made at 250 Hz, 500 Hz, 1 kHz, 2 kHz, 4 kHz, 6 kHz, and 8 kHz. Bone conduction thresholds were determined to exclude a conductive component in patients with HL. Oto‐immittance measurements were carried out in all subjects. A history of motor development was obtained and Romberg testing was performed in all family members for assessment of the integrity of the entire proprioceptive pathway, and vestibular function was further evaluated by ice water caloric tests using Frenzel's glasses in selected patients. The study was approved by the ethics committee of Henan Province People’s Hospital. The study was performed in accordance with relevant guidelines and regulations. The written informed consent was obtained from all adult subjects and guardians on behalf of the children prior to the clinical evaluation and blood sample collection.
Preparation of DNA
Peripheral blood samples were collected from patients and their lineal relatives. Genomic DNA was extracted from whole blood using the Gentra Puregene DNA isolation kit (QIAGEN, Hilden, Germany). Agarose gel and Qubit (Life technologies, CA, USA) were used for DNA quantity and quality assessment.
Mutation screening and gene identification
Screening of common deafness gene mutations using a DNA microarray
Hereditary hearing loss allele-specific PCR based universal array (ASPUA) (CapitalBio, Beijing, China) was used to simultaneously screen 9 common deafness causing variants in all affected family members (GJB2: c.35delG, c.176del16, c.235delC, c.299–300delAT; GJB3: c.538C>T (R180X); SLC26A4: c.IVS7–2A>G, c.2168A>G (p.H723R); mtDNA 12S rRNA: 1555A>G, 1494C>T). Multiplex allele-specific PCR was performed and chips were imaged with a LuxScan TMHT 24 Microarray Scanner (CapitalBio, Beijing, China) (Li et al. 2008).
Comprehensive targeted gene panel testing
A custom capture panel (MiamiOtoGenes), with a target size of approximately 1.158 MB encompassing 3494 regions, was designed to include all exons, 5' UTRs and 3' UTRs of 180 known and candidate deafness causing genes using the Agilent SureDesign online tool (https://earray.chem.agilent.com/suredesign/). The Agilent's SureSelect Target Enrichment (Agilent, Santa Clara, CA) of coding exons and flanking intronic sequences in-solution hybridization capture system was used following the manufacturer's standard protocol. Adapter sequences for the Illumina HiSeq2000 were ligated and the enriched DNA samples were prepared using the standard methods for the HiSeq2000 instrument (Illumina) (Yan et al. 2016).
Whole exome sequencing and data analysis
Enrichment of coding exons and flanking intronic regions was performed using a solution hybrid selection method with the SureSelectXT human all exon 60 Mb v6 kit (Agilent Technologies) following the manufacturer’s standard protocol. The Illumina CASAVA v1.8 pipeline was used to assemble 99 bp sequence reads. Burrows-Wheeler Aligner (BWA) was applied for alignment of sequence reads to the human reference genome (hg19) (Li and Durbin 2010) and variants were called using FreeBayes (Garrison and Marth 2012). Genesis 2.0 (https://www.genesis-app.com/) was then used for variant filtering based on quality/score read depth and minor allele frequency (MAF thresholds of 0.0005 for ADNSHL variants) as reported in dbSNP141, the National Heart, Lung, and Blood Institute Exome Sequencing Project Exome Variant Server, Seattle, WA Project, (Exome Variant Server, 2012). Exome Aggregation Consortium (ExAC) browser (http://exac.broadinstitute.org/), the 1000 Genome Project Database and our internal database of > 3,000 samples from European, Asian, and American ancestries. Variants meeting these criteria were further annotated based on their presence and pathogenicity information in Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk), the Deafness Variation Database (DVD) (deafnessvariationdatabase.org), and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/). Copy number variations (CNV) calling was performed using an R-based tool (Nord at al. 2011). This method normalizes read-depth data by sample batch and compares median read-depth ratios using a sliding-window approach.
Confirmation and Segregation analysis
Samples from all available members from the family were subjected to Sanger sequencing to determine whether the potential mutations in identified genes co-segregated with the disease phenotype. Specific exons containing candidate variations were amplified by polymerase chain reaction (PCR) using primer sets designed via Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer /primer3_www.cgi). The PCR products were sequenced using BigDye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA, USA) and analyzed using an ABI 3700XL Genetic Analyzer. Finally, the genotype of the variants were confirmed by sequence analysis with the Sequencing Analysis software v5.2 (Applied Biosystems Corps., Foster City, CA, USA).
Antibodies and whole mount immunofluorescence staining
The primary antibodies used were as follows: PDE1C rabbit polyclonal antibody raised against amino acids 448–709 of human PDE1C (H-262; catalogue number sc-67323, Santa Cruz Biotechnology). Anti-LAMP1 mouse monoclonal antibody (H4A3; catalogue number ab25630, Abcam). Secondary antibodies were goat anti-rabbit IgG conjugated to Alexa Fluor 488 (A-11001; Molecular Probes) and anti-mouse conjugated to AlexaFluor-568 (A-11011; Molecular Probes). Phalloidin-CF405M (Biotium; catalogue number 00034) was used for counterstaining. For immunofluorescence, posnatal day 3 (P3) mice were killed and inner ear sensory epithelia were then microdissected and fixed with 4% paraformaldehyde in phosphate buffered saline (PBS; pH 7.4) for 20 min at room temperature, then permeabilized with 0.5% Triton X-100 for 15 min, and blocked overnight at 4 °C in filtered 4% bovine serum albumin (BSA) in PBS. The tissue was first incubated with primary antibodies for 2 h, rinsed with PBS three times, then stained with goat anti-rabbit for pde1c and anti-mouse for Lamp-1 conjugated antibodies and subsequently counterstained with phalloidin-CF405M (Biotium; catalogue number 00034). Confocal imaging was accomplished using a TiE inverted fluorescence microscope (Nikon Instruments) equipped with a spinning disk confocal head (Yokogawa), DU-888 EMCCD (Andor) and Apo TIRF 1.45 NA objective. Image acquisition was managed through NIS-Elements software (Nikon Instruments).
PDE1C purified proteins and assay of phosphodiesterase activity
The wild type E. coli expressed and purified recombinant human PDE1C protein (NP 005011) with N-terminal HIS tag was obtained from OriGene (TP760557). For p.A320S (c.954G>T; NM_005020) mutant, site-directed mutagenesis was performed on the full-length RG205291 (OriGene) and was then subcloned into pEX-N-His vector and expressed in E. coli and purified. The Abcam colorimetric PDE activity assay kit (ab139460) that combines a dual enzyme system with green assay reagent for phosphate detection, was used to detect phosphodiesterase (PDE) activity.
Results
Whole exome sequencing (WES) of family members
The ascertained family was a five-generation pedigree with 62 members, 28 of whom suffered HL (Fig.1A). DNA was available for 20 family members for analysis. The affected family members showed post-lingual, progressive, bilateral and symmetric NSHL. The onset of HL in most cases was in the third decade of life with subsequent gradual progression from mild to profound HL involving the high frequencies in the majority of patients. Representative audiograms of 4 affected individuals (IV: 6, IV: 13, IV: 19, IV: 22) are shown in Figure 1 B. HL was variable, not only between generations but also within generations. The earliest onset of HL in the family was reported in a fifth generation individual at the age of 14 years.
Fig. 1.
Identification of a variant in the PDE1C gene underlying ADNSHL in a Chinese family. A: The family pedigree with genotypes at PDE1C c.958G > T (p.A320S). Filled symbols refer to subjects with progressive sensorineural hearing loss. B: Pure-tone audiometry of 4 affected family members showing high frequency hearing loss. C: Partial PDE1C sequence chromatograms are shown for one affected individual (left) and one normal control (right). The position of the c.958G > T (p.A320S) mutation is indicated with a red arrow. D: The variant site (box) is perfectly preserved across the species. E: And this variant is also highly conserved among human PDEs (blue arrow).
We first excluded the involvement of common Chinese deafness-causing variants in GJB2, SLC26A4, and mtDNA 12SrRNA in this family using a population specific microarray chip. We have also ruled out the involvement of 180 known/suspected deafness genes by a comprehensive targeted gene panel testing (Yan et al. 2016). We then subjected 2 affected family members (IV: 3, IV: 6) to WES. On average, each exome in the family produced 83,000,000 reads. Approximately, 90,000 single nucleotide variants and 9,500 INDELS per sample were obtained before variant filtering. Coverage of targeted exons for 2X, 10X, 20X and 30X were 99%, 89%, 78% and 68%, respectively. We filtered the variants according to the autosomal dominant inheritance model, functional significance (i.e., nonsense, missense, or splice site), single-nucleotide variants shared by the affected individuals and minor allele frequency (MAF) thresholds of 0.0005 as reported in public databases. This simple filtering strategy rapidly extracted only 1 heterozygous variant shared by the 2 affected siblings subjected to WES. This sequence change is located in the PDE1C (hg19) gene on chromosome 7p14.3 at 31,890,328, c.958G>T (NM_001191058) leading to p.A320S (NP_001177987.2). Its co-segregation with the phenotype in the family as an autosomal dominant trait was confirmed via Sanger sequencing that revealed that all of the 15 available affected family members were heterozygous for this variant, while it was not observed in the 5 family members with normal hearing (Fig. 1A, C). The missense variant was reported in only in East Asians in the Exome Aggregation Consortium (ExAC) browser (http://exac.broadinstitute.org/) and in the Genome Aggregation Database (gnomAD, Broad Institute) in 6/8596 and in 14/18820 alleles, respectively, with a global minor allele frequency (MAF) ~ 0.00005. But it was absent in 430 ethnic-matched control chromosomes. Nevertheless, population-based databases should be interpreted with caution, because individual-level phenotype data are not available. Thus, as such, presence or absence of statistical significance cannot always be used as evidence against or for pathogenicity.
We screened copy number variations (CNVs) from whole exome sequencing data by investigating whether there were combined effects of the identified variants with CNVs in the same locus for the studied probands. There were no CNVs encompassing two or more exons gains nor losses found, thus, excluding their involvement in the etiology of the hearing loss in the family.
The p.A320S is highly conserved in different species (Fig. 1D) and the evolutionary conservation of the involved amino acid residue is further corroborated by genomic evolutionary rate profiling conservation score (GERP) and PhastCons scores of 5.91 and 1, respectively. Furthermore, multiple sequence alignment revealed that the residue at this position is highly conserved among PDEs (Fig. 1E). Moreover, the observed Ala to Ser amino acid change at position 320 is predicted to be deleterious by the Sorting intolerant from tolerant (SIFT) score of 0.02 and damaging according to PolyPhen (score=1).
Structural modeling of A320S in PDE1C that causes ADNSHL
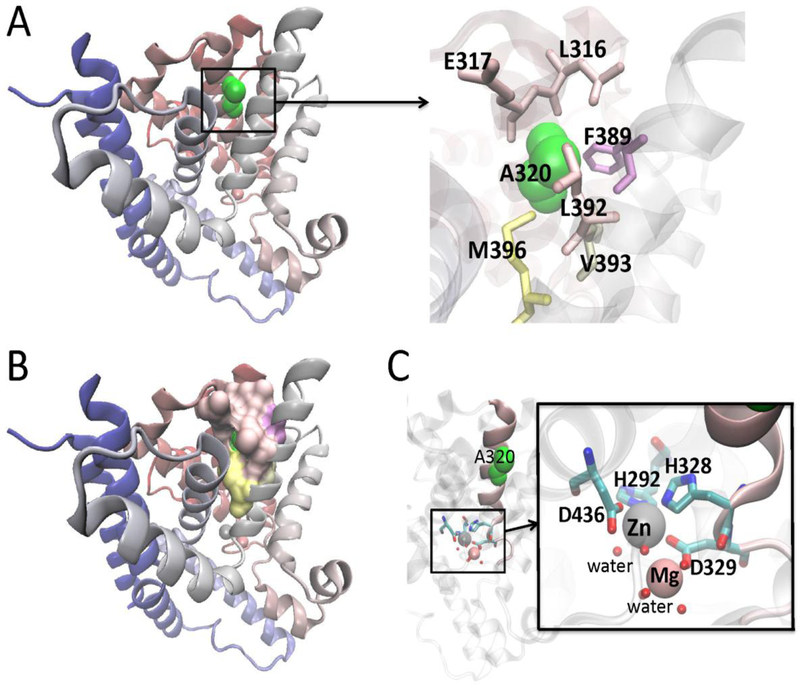
PDEs are modular proteins that exhibit a common structural organization, with a conserved carboxy-terminal catalytic core (250–300 amino acids) and highly divergent N-terminal regulatory regions that target individual PDEs to different subcellular locations and signalosomes (Houslay et al. 2007; Kritzer et al. 2012; Lee et al. 2013). The PDE1 isoforms are the only mammalian PDEs that are activated by intracellular Ca2+ levels and the Ca2+ binding protein, calmodulin (Teo and Wang 1973). X-ray crystal structures of isolated catalytic domains of PDE families have demonstrated that the catalytic domains of PDEs fold into a novel compact structure composed of 17 α helices. These form three subdomains with active site of PDEs buried in a deep pocket located at the junction of these three subdomains. This pocket contains 12 of the 21 residues absolutely conserved across all PDE families (Xu et al. 2000; Conti and Beavo 2007). In the site, there are two metal ions that are coordinated by residues from the three subdomains, which help to hold the subdomains together and are required for the catalytic activity of PDE (Hardman et al. 1971). The first metal, identified as a zinc (Zn2+) ion, is deeply buried in the binding pocket and has been found to play a structural role and essential for catalysis (Xu et al. 2000; Conti and Beavo 2007). The second metal ion, thought to be Mg2+ is less deeply located in the substrate binding pocket. Thus the catalytic center of PDEs is formed by a binuclear motif comprised of a “tightly gripped’ Zn2+ ion and a loosely held Mg2+.
We constructed a molecular model of PDE1C based on the template pdb 1TZA using SwissModel (Biasini et al. 2014). As shown in Fig. 2A-B, A320 is predicted to form a hydrophobic core and in the PDE catalytic pocket, H328 binds to Zn2+ and D329 to Mg2+. The replacement of alanine by serine at 320 may have a direct influence on the wall of the substrate-binding pocket through the two ions binding residues (Fig. 2C). It might create a relaxation of the connecting helices. Subtle structural changes resulting from such looseness could alter the enzyme binding capacity and affect the energy difference between the rotameric complexes of the glutamine switch Q427 with cAMP and cGMP sufficiently to change substrate preference.
Fig. 2.
3-D structure modeling of the A320S mutation, based on the template pdb 1TZA. A: A320 is shown in green. The residues around A320 are also highlighted. B: Ala320 forms a hydrophobic core as in most helix-bundles. C: In the catalytic pocket, H328 binds to Zn2+ and D329 to Mg2+. An alanine to serine amino acid substitution at codon 320 may have a direct impact on the substrate binding pocket through the two ions binding residues. Figures were made with Visual Molecular Dynamics (VMD 1.9.2) (Humphrey et al., 1996). (http://www.ks.uiuc.edu/Research/vmd/).
Expression analysis of PDE1C in the cochlea.
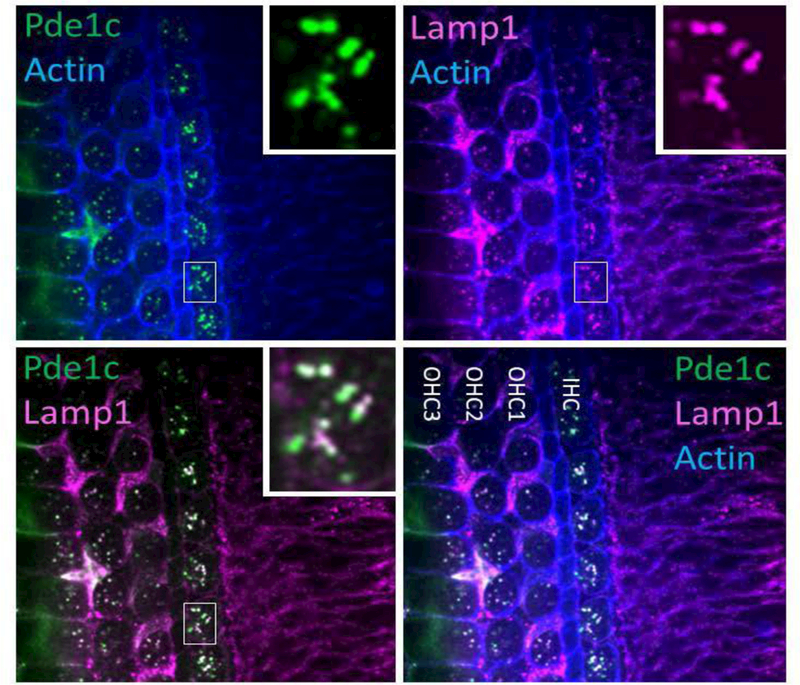
Since the location of a protein in a cell is key to its function, we investigated expression of Pde1c in the cochlea. Transcriptome analysis showed that Pde1c is expressed in cochlear and vestibular hair cells as well as support cells and spiral ganglion neurons in the inner ear (SHIELD; https://shield.hms.harvard.edu) (Scheffer et al. 2015; Shen et al. 2015). We examine its expression using a rabbit polyclonal antibody against amino acids 448–709 of the human PDE1C protein. We show in wholemount immunofluorescence staining of P3 mouse cochlea using this antibody that Pde1c is expressed in outer (OHC) and inner (IHC) hair cells cytosol co-localizing with Lamp-1 in lysosomes. A diffuse and barely detectable cytosolic immunofluorescence can also be seen in hair cells and supporting cells (Fig. 3). Our results thus suggest that PDE1C may play important roles in the regulation of activation of lysosomes, degradation of proteins and internalizations of receptors.
Fig. 3.
Wholemount immunofluorescence on P3 mouse cochlea. The panel reveals a strong Pde1c immunofluorescence in outer and inner hair cells cytosol colocalizing with Lamp-1 immunofluorescence in Lysosomes. A diffuse and barely detectable cytosolic immunofluorescence can also be seen in hair cells and supporting cells.
The presence and functional importance of PDE1C in olfactory sensory tissues are well established. It has been shown that this isozyme modulates the amplitude and duration of the cAMP signal in sensory cilia in response to odorant stimulation (Yan et al. 1995). The cAMP directly opens olfactory cyclic nucleotide-gated (CNG) ion channels, resulting in influx of Ca2+ and Na+ and membrane depolarization (Firestein 2000; Ma 2007), which results in efflux of Cl− and further depolarization, contributing to the generation of action potentials (Kurahashi and Yau 1993; Kleene 1993; Lowe and Gold 1993; Reisert et al. 2003). PDE1C has also been demonstrated to be a major regulator of smooth muscle cells proliferation. Mechanistic studies revealed that PDE1C plays a critical role in regulating the stability of growth factor receptors, including the platelet-derived growth factor receptor beta (PDGFRβ) known to be important in pathological vascular remodeling and that a transmembrane adenylate cyclase (tmAC)-derived cAMP dependent protein kinase (PKA) signaling is critical in promoting PDGFRβ internalization and endocytosis. The upregulation of PDE1C has been found to suppress PDGFRβ degradation through an endosome/lysosome and a low density lipoprotein receptor-related protein-1-(LRP1) dependent mechanism. This PDE1 regulation of PDGFRβ protein is thought to be mediated via modulation of PKA-dependent phosphorylation of LRP1 (Cai et al. 2015).
Catalytic potential of wild type and mutant PDE1C proteins for hydrolysis of cyclic nucleotides
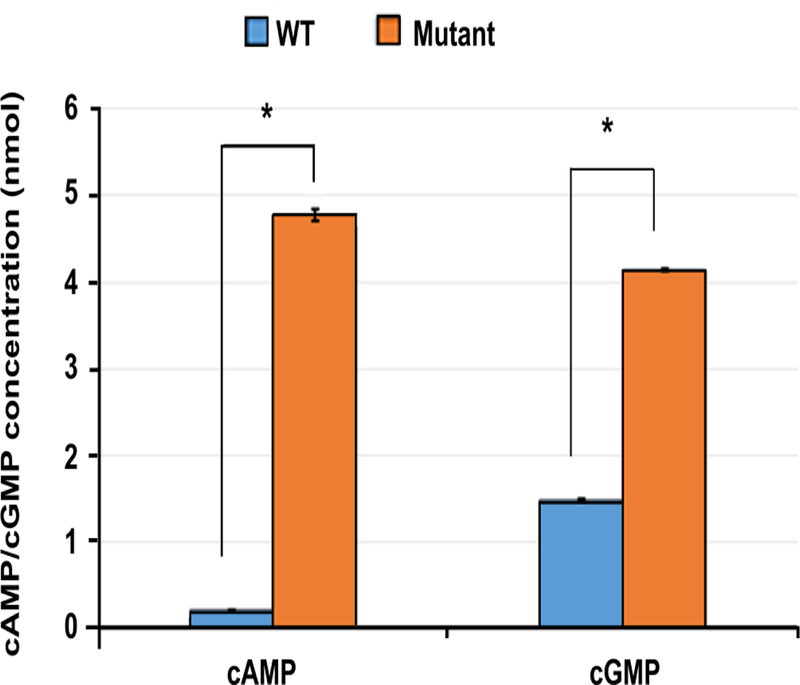
To investigate the functional consequence of the PDE1C missense c.958G>T; p.A320S) mutation, recombinant proteins expressed in E. coli were produced with DNA sequences of PDE1C TrueORF clone RC205291, encoding human full-length PDE1C Protein (NP_005011). Purified wild type and mutant proteins were analyzed for catalytic activity (cAMP and cGMP hydrolysis) using the Abcam colorimetric PDE activity assay kit (ab139460). We show in Fig. 4 that the variant alters the PDE hydrolytic activity for both cAMP and cGMP with an approximately 10 fold increase in cAMP and 3 fold for cGMP compared to the wild type (p<0.01). These data suggest that the missense variant enhance phosphodiesterase activity and thus decrease cAMPs and cGMPs level.
Fig. 4.
The p.A320S variant alters the PDE hydrolytic activity for both cAMP and cGMP with an approximately 10-fold increase in cAMP and three-fold for cGMP compared to the wild-type.
Discussion
PDEs are enzymes that have the unique function of regulating the cellular levels of the second messengers, cAMP and cGMP, by controlling their rates of hydrolysis. Current data suggest that individual isozymes modulate specific cell signaling pathways. Here, we show that a dominant variant in the PDE1C gene is associated with NSHL in a Chinese family. The amino acid Alanine 320 in the catalytic domain affects a residue that is fully conserved among vertebrate PDE1C proteins and PDE gene family. Using purified proteins, we observe that mutant protein exhibited a highly significant PDE hydrolytic activity of both cAMP and cGMP, resulting in a decrease in cAMPs and cGMPs level. These changes in the rate and capacity for cyclic nucleotides hydrolysis suggest a decrease in the amplitude and/or duration of their signals within the cells.
The cAMP and cGMP signaling pathways have been described in the auditory system. In a series of studies, evidence was found for the physiological interactions between arginine vasopressin (AVP) and type 2 receptor (V2R) in the endolymphatic sac via cAMP-dependent signaling for regulation of membranous fluid turnover in the inner ear (Kitahara et al. 2008). The clinical implication is that elevated plasma concentrations of endogenous vasopressin may stimulate the V2R-aquaporin-2 (AQP2) system and hypersensitize the cAMP-linked signaling, causing overflow of endoplasmic fluid, leading to endolymphatic hydrops and subsequent attacks of vertigo, with HL and tinnitus in Meniere’s disease (Maekawa et al. 2010). Furthermore, it is well-established that in the inner ear Ca2+ can influence mechanotransduction via cAMP, which has been shown to affect the response-displacement curve of the transducer (Ricci and Fettiplace 1997). This signaling pathway may involve cAMP production by Ca2+-calmodulin activated type I adenyl cyclase (Drescher et al. 1997), cAMP-induced activation of protein kinase A and phosphorylation of the mechanotransduction channel or the myosin motor (Fettiplace and Ricci 2003).
On the other hand, cGMP-Prkg1 pathway has been found to facilitate protective processes in response to traumatic events and Prkg1-null mice have been shown to have a normal hearing threshold, but were more susceptible to noise-induced hearing loss (NIHL) and showed markedly less recovery than wild type mice following acoustic trauma. The expression of Prkg1 in sensory cells and neurons of the inner ear partly overlapped that of cGMP-dependent phosphodiesterase Pde5 and pharmacologic inhibition of Pde5 in wildtype mice and rats has been reported to protect from noise-induced cochlear damage. This otoprotective effect of Pde5 inhibition is believed to be mediated through the cGMP-Prkg1 signaling pathway, characterized by an increase in cGMP and the subsequent activation of Prkg1 in OHCs (Jaumann et al. 2012). We can thus speculate that decrease in cGMP level due to enhancement of hydrolytic activity of mutated PDE1C and subsequent changes in its downstream effectors could be one of the mechanisms by which the A320S variant in the PDE1C gene cause HL in the Chinese family. Furthermore, there is evidence that the effects of nitric oxide (NO) in cellular signaling are related to its ability to regulate Ca2+ homeostasis through the activation of the NO-cGMP-PKG pathway (Clementi 1998) and studies also showed that NO affects the ATP-induced Ca2+ response via this NO pathway in OHCs, IHCs, spiral ganglion neurons (SGNs) (Shen et al. 2003; Shen et al. 2005; Shen et al. 2006; Yukawa et al. 2005). NO is synthesized by three isoforms of NO synthase (NOS). The neuronal isoform (nNOS) and the endothelial isoform (eNOS) of NOS are constitutive and Ca2+/calmodulin -dependent, whereas the inducible NOS is Ca2+-independent (Griffith and Stuehr 1995). Both nNOS and eNOS are present in several cell types in the cochlea, including IHCs and OHCs and thought to contribute to the NOS signal in inner ear (Gosepath et al 1997; Riemann and Reuss 1999). The ATP-induced NO production is believed to be mainly due to the Ca2+ influx through the activation of P2X receptor (Yukawa et al 2005). The role of intrinsic feedback pathways of purinergic signaling through ATP activation provides endogenous cochlear tissue protection against noise damage. This purinergic regulation of hearing sensitivity is characterized by the lack of temporary shift of the auditory threshold (TTS) in P2X2 receptor knockout mice. The P2rx2-null mice were also more vulnerable than wild-type littermates to permanent hearing loss, supporting the protective role of P2X2 receptor signaling pathway in NIHL. The P2x2-null mice were also more vulnerable than wild-type littermates to permanent HL (Housley et al. 2013). We have also shown that in human, a defect in P2X2 causes DFNA41 and mutation carriers in this gene with history of occupational noise exposure have increased TTS of 10–20 dB compared to carriers with no previous noise exposure (Yan et al. 2013). Collectively, these findings suggest that the cochlear P2X2 receptor/ ATP-gated ion channel signaling pathway confers protection from NIHL and the absence or mutation of P2X2 receptor increases susceptibility to NIHL and presbycusis.
Elevated Ca2+ levels in the cochlea may link to NOS production as well as triggering apoptotic and necrotic cell death pathways by oxidative stress and excitotoxicity. NOS/cGMP/cGMP-dependent protein kinase I signaling is a very complex system. There are myriad possibilities for physiological or pathophysiological events to affect the signaling process that may vary among tissues. These include localization/activity of cGMP-hydrolyzing PDEs. The cGMP-hydrolyzing activity of PDE1C, a Ca2+/calmodulin –dependent isozyme, is largely determined by intracellular calcium level and also more likely by specific pools of calcium that enter the cell in response to different stimuli (Miller et al. 2009). Elevated Ca2+ levels in the cochlea may link to NOS production as well as triggering apoptotic and necrotic cell death pathways by oxidative stress and excitotoxicity. We propose thus that PDE1C is a locus for cross-talk between cGMP-signaling and cAMP pathways in Ca2+ homeostasis through the activation of the nitrite oxide (NO)-cGMP-PKG- signaling processes and Ca2+ - mechanotransduction – cAMP adaptation in auditory hair cells pathways.
In summary we show that in cochlea, PDE1C is located in the lysosomes where it may function as a “housekeeping” PDE to maintain cellular cGMP level. Given that PDE1 dependent cGMP hydrolyzing is predominant during elevation in intracellular calcium [Ca2+]i (Miller et al. 2009), we hypothesize that mutation in PDE1C results in disruption of calcium homeostasis and cause deafness and that the cochlear PDE1C/cGMP signaling pathway may confer under stress conditions.
Acknowledgments
Immunofluorescence confocal imaging was performed in Dr. Bechara Kachar’s lab (NIDCD-NIH; Bethesda, MD). This study was supported by the National Natural Science Foundation No. 81771024, 81771023, and medical science and technology project of Henan Province No. 87 from China; R01 DC05575, R01 DC01246, and R01 DC012115 from the National Institutes of Health/National Institute on Deafness and Other Communication Disorders, and Action on hearing loss grant.
Footnotes
Present address: Laboratory of Cell Structure & Dynamics, NIDCD, NIH, Bethesda, MD 20892.
Author Contributions
L.W., F.Y., D.Y., L.Q., M.G., R.M., T.L., A.S.K., Y.L., P.C., S.H.B., S.L., and X.L planned and performed the study and participated in writing the manuscript. All authors read and approved the final version of the manuscript.
Conflict of interest The authors declare no conflict of interest.
References
- Vohr B. Overview: infants and children with hearing loss—part I. Ment Retard Dev Disabil Res Rev. 2003;9:62–64. doi: 10.1002/mrdd.10070. [DOI] [PubMed] [Google Scholar]
- Blackwell DL, Lucas JW, Clarke TC. Summary health statistics for U.S. adults: National Health Interview Survey, 2012. Vital Health Stat. 2014;10:1–161. [PubMed] [Google Scholar]
- Goman AM, Reed NS, Lin FR. Addressing estimated hearing loss in adults in 2060. JAMA Otolaryngol Head Neck Surg. 2017;143:733–734. doi: 10.1001/jamaoto.2016.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MB. Non-syndromic autosomal-dominant deafness. Clin Genet. 2002;62:1–13. doi: 10.1034/j.1399-0004.2002.620101.x. [DOI] [PubMed] [Google Scholar]
- Thompson WJ, Appleman MM. Cyclic nucleotide phosphodiesterase and cyclic AMP. Ann N Y Acad Sci. 1971;185:36–41. doi: 10.1111/j.1749-6632.1971.tb45233.x. [DOI] [PubMed] [Google Scholar]
- Tsertsvadze A, Fink HA, Yazdi F, MacDonald R, Bella AJ, Ansari MT, Garritty C, Soares-Weiser K, Daniel R, Sampson M, Fox S, Moher D, Wilt TJ. Oral phosphodiesterase- 5 inhibitors and hormonal treatments for erectile dysfunction: A systematic review and meta-analysis. Ann Intern Med. 2009;151:650–661. doi: 10.7326/0003-4819-151-9-200911030-00150. [DOI] [PubMed] [Google Scholar]
- Van Wagoner DR, Lindsay BD. Phosphodiesterase-4 activity: A critical modulator of atrial contractility and arrhythmogenesis. J Am Coll Cardiol. 2012;59:2191–2192. doi: 10.1016/j.jacc.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannetta E, Isidori AM, Galea N, Carbone I, Mandosi E, Vizza CD, Naro F, Morano S, Fedele F, Lenzi A. Chronic Inhibition of cGMP phosphodiesterase 5A improves diabetic cardiomyopathy: A randomized, controlled clinical trial using magnetic resonance imaging with myocardial tagging. Circulation. 2012;125:2323–2333. doi: 10.1161/CIRCULATIONAHA.111.063412. [DOI] [PubMed] [Google Scholar]
- Movsesian M, Wever-Pinzon O, Vandeput F. PDE3 inhibition in dilated cardiomyopathy. Curr Opin Pharmacol. 2011;11:707–713. doi: 10.1016/j.coph.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniu SA. Roflumilast for the treatment of chronic obstructive pulmonary disease. Curr Opin Investig Drugs. 2006;7:412–417. [PubMed] [Google Scholar]
- Bjorgo E, Moltu K, Tasken K. Phosphodiesterases as targets for modulating T-cell responses. Handb Exp Pharmacol. 2011;204:345–363. doi: 10.1007/978-3-642-17969-3_15. [DOI] [PubMed] [Google Scholar]
- Hertz AL, Beavo JA. Cyclic nucleotides and phosphodiesterases in monocytic differentiation. Handb Exp Pharmacol. 2011;204:365–390. doi: 10.1007/978-3-642-17969-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang HT, O'Donnell JM. Phosphodiesterases in the central nervous system: Implications in mood and cognitive disorders. Handb Exp Pharmacol. 2011;204:447–485. doi: 10.1007/978-3-642-17969-3_19. [DOI] [PubMed] [Google Scholar]
- Houslay MD, Baillie GS, Maurice DH. cAMP-Specific phosphodiesterase-4 enzymes in the cardiovascular system: a molecular toolbox for generating compartmentalized cAMP signaling. Circulation Res. 2007;100:950–966. doi: 10.1161/01.RES.0000261934.56938.38. [DOI] [PubMed] [Google Scholar]
- Kritzer MD, Li J, Dodge-Kafka K, Kapiloff MS. AKAPs: the architectural underpinnings of local cAMP signaling. J Mol Cell Cardiol. 2012;52:351–358. doi: 10.1016/j.yjmcc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LCY, Maurice DH, Baillie GS. Targeting protein-protein interactions within the cyclic AMP signaling system as a therapeutic strategy for cardiovascular disease. Future Med Chem. 2013;5:451–464. doi: 10.4155/fmc.12.216. [DOI] [PubMed] [Google Scholar]
- Teo TS, Wang JH. Mechanism of activation of a cyclic adenosine 3':5'- monophosphate phosphodiesterase from bovine heart by calcium ions. Identification of the protein activator as a Ca2+ binding protein. J Biol Chem. 1973;248:5950–5955. [PubMed] [Google Scholar]
- Xu RX, Hassell AM, Vanderwall D, Lambert MH, Holmes WD, Luther MA, Rocque WJ, Milburn MV, Zhao Y, Ke H, Nolte RT. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Ann Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- Hardman JG, Beavo JA, Gray JP, Chrisman TD, Patterson WD, Sutherland EW. The formation and metabolism of cyclic GMP. Ann N Y Acad Sci. 1971;185:27–35. doi: 10.1111/j.1749-6632.1971.tb45232.x. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer DI, Shen J, Corey DP, Chen ZY. Gene expression by mouse inner ear hair cells during development. J Neurosci. 2015;35:6366–6380. doi: 10.1523/JNEUROSCI.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Scheffer DI, Kwan KY, Corey DP. SHIELD: an integrative gene expression database for inner ear research. Database (Oxford) 2015 doi: 10.1093/database/bav071. bav071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Zhao AZ, Bentley JK, Loughney K, Ferguson K, Beavo JA. Molecular cloning and characterization of a calmodulin-dependent phosphodiesterase enriched in olfactory sensory neurons. Proc Natl Acad Sci U S A. 1995;92:9677–9681. doi: 10.1073/pnas.92.21.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein S. How the olfactory system makes sense of scents. Nature. 2000;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- Ma M. Encoding olfactory signals via multiple chemosensory systems. Crit Rev Biochem Mol Biol. 2007;42:463–480. doi: 10.1080/10409230701693359. [DOI] [PubMed] [Google Scholar]
- Kurahashi T, Yau KW. Co-existence of cationic and chloride components in odorant-induced current of vertebrate olfactory receptor cells. Nature. 1993;363:71–74. doi: 10.1038/363071a0. [DOI] [PubMed] [Google Scholar]
- Kleene SJ. Origin of the chloride current in olfactory transduction. Neuron. 1993;11:123–132. doi: 10.1016/0896-6273(93)90276-w. [DOI] [PubMed] [Google Scholar]
- Lowe G, Gold GH. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature. 1993;366:283–286. doi: 10.1038/366283a0. [DOI] [PubMed] [Google Scholar]
- Reisert J, Bauer PJ, Yau KW, Frings S. The Ca-activated Cl channel and its control in rat olfactory receptor neurons. J Gen Physiol. 2003;122:349–363. doi: 10.1085/jgp.200308888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Nagel DJ, Zhou Q, Cygnar KD, Zhao H, Li F, Pi X, Knight PA, Yan C. Role of cAMP-phosphodiesterase 1C signaling in regulating growth factor receptor stability, vascular smooth muscle cell growth, migration, and neointimal hyperplasia. Circ Res. 2015;116:1120–1132. doi: 10.1161/CIRCRESAHA.116.304408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara T, Kubo T, Okumura S, Kitahara M. Effects of endolymphatic sac drainage with steroids for intractable Meniere’s disease: a long-term follow-up and randomized controlled study. Laryngoscope. 2008;118:854–861. doi: 10.1097/MLG.0b013e3181651c4a. [DOI] [PubMed] [Google Scholar]
- Maekawa C, Kitahara T, Kizawa K, Okazaki S, Kamakura T, Horii A, Imai T, Doi K, Inohara H, Kiyama H. Expression and translocation of aquaporin-2 in the endolymphatic sac in patients with Meniere's disease. J Neuroendocrinol. 2010;22:1157–1164. doi: 10.1111/j.1365-2826.2010.02060.x. [DOI] [PubMed] [Google Scholar]
- Ricci AJ, Fettiplace R. The effects of calcium buffering and cyclic AMP on mechano-electrical transduction in turtle auditory hair cells. J Physiol. 1997;501:111–124. doi: 10.1111/j.1469-7793.1997.111bo.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher MJ, Barretto RL, Chaturvedi D, Beisel KW, Hatfield JS, Khan KM, Drescher DG. Expression of adenylyl cyclase type I in cochlear inner hair cells. Brain Res Mol Brain Res. 1997;45:325–330. doi: 10.1016/s0169-328x(97)00007-7. [DOI] [PubMed] [Google Scholar]
- Fettiplace R, Ricci AJ. Adaptation in auditory hair cells. Curr Opin Neurobiol. 2003;13:446–451. doi: 10.1016/s0959-4388(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Jaumann M, Dettling J, Gubelt M, Zimmermann U, Gerling A, Paquet-Durand F, Feil S, Wolpert S, Franz C, Varakina K, Xiong H, Brandt N, Kuhn S, Geisler HS, Rohbock K, Ruth P, Schlossmann J, Hütter J, Sandner P, Feil R, Engel J, Knipper M, Rüttiger L. cGMP-Prkg1 signaling and Pde5 inhibition shelter cochlear hair cells and hearing function. Nat Med. 2012;18:252–259. doi: 10.1038/nm.2634. [DOI] [PubMed] [Google Scholar]
- Clementi E. Role of nitric oxide and its intracellular signaling pathways in the control of Ca2+ homeostasis. Biochem Pharmacol. 1998;55:713–718. doi: 10.1016/s0006-2952(97)00375-4. [DOI] [PubMed] [Google Scholar]
- Shen J, Harada N, Yamashita T. Nitric oxide inhibits adenosine 5′-triphophate- induced Ca2+ response in inner hair cells of the guinea pig cochlea. Neurosci Lett. 2003;337:135–138. doi: 10.1016/s0304-3940(02)01320-4. [DOI] [PubMed] [Google Scholar]
- Shen J, Harada N, Nakazawa H, Yamashita T. Involvement of the nitric oxide/cyclic GMP pathway and neuronal nitric oxide synthase in ATP-induced Ca2+ signalling in cochlear inner hair cells. Eur J Neurosci. 2005;21:2912–2922. doi: 10.1111/j.1460-9568.2005.04135.x. [DOI] [PubMed] [Google Scholar]
- Shen J, Harada N, Nakazawa H, Kaneko T, Izumikawa M, Yamashita T. Role of nitric oxide on ATP-induced Ca2+ signaling in outer hair cells of the guinea pig cochlea. Brain Res. 2006;1081:101–112. doi: 10.1016/j.brainres.2005.12.129. [DOI] [PubMed] [Google Scholar]
- Yukawa H, Shen J, Harada N, Cho-Tamaoka H, Yamashita T. Acute effects of glucocorticoids on ATP-induced Ca2+ mobilization and nitric oxide production in cochlear spiral ganglion neurons. Neuroscience. 2005;130:485–496. doi: 10.1016/j.neuroscience.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Stuehr DJ. Nitric oxide synthase: properties and catalytic mechanism. Ann Rev Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- Gosepath K, Gath I, Maurer J, Pollock JS, Amedee R, Förstermann U, Mann W. Characterization of nitric oxide synthase isoforms expressed in different structures of the guinea pig cochlea. Brain Res. 1997;747:26–33. doi: 10.1016/s0006-8993(96)01149-3. [DOI] [PubMed] [Google Scholar]
- Riemann R, Reuss S. Nitric oxide synthase in identified olivocochlear projection neurons in rat and guinea pig. Hear Res. 1999;135:181–189. doi: 10.1016/s0378-5955(99)00113-6. [DOI] [PubMed] [Google Scholar]
- Housley GD, Morton-Jones R, Vlajkovic SM, Telang RS, Paramananthasivam V, Tadros SF, Wong AC, Froud KE, Cederholm JM, Sivakumaran Y, Snguanwongchai P, Khakh BS, Cockayne DA, Thorne PR, Ryan AF. ATP-gated ion channels mediate adaptation to elevated sound levels. Proc Natl Acad Sci U S A. 2013;110:7494–7499. doi: 10.1073/pnas.1222295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Zhu Y, Walsh T, Xie D, Yuan H, Sirmaci A, Fujikawa T, Wong AC, Loh TL, Du L, Grati M, Vlajkovic SM, Blanton S, Ryan AF, Chen ZY, Thorne PR, Kachar B, Tekin M, Zhao HB, Housley GD, King MC, Liu XZ. Mutation of the ATP-gated P2X(2) receptor leads to progressive hearing loss and increased susceptibility to noise. Proc Natl Acad Sci U S A. 2013;110:2228–2233. doi: 10.1073/pnas.1222285110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Oikawa M, Cai Y, Wojtovich AP, Nagel DJ, Xu X, Xu H, Florio V, Rybalkin SD, Beavo JA, Chen YF, Li JD, Blaxall BC, Abe J, Yan C. Role of Ca2+/calmodulin-stimulated cyclic nucleotide phosphodiesterase 1 in mediating cardiomyocyte hypertrophy. Circ Res. 2009;105:956–964. doi: 10.1161/CIRCRESAHA.109.198515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Pan Q, Guo YG, Li Y, Gao HF, Zhang D, Hu H, Xing WL, Mitchelson K, Xia K, Dai P, Cheng J. Construction of a multiplex allele-specific PCR-based universal array (ASPUA) and its application to hearing loss screening. Hum Mutat. 2008;29:306–314. doi: 10.1002/humu.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Tekin D, Bademci G, Foster J, Cengiz FB, Kannan-Sundhari A, Guo SS, Mittal R, Zou B, Grati M, Kabahuma RI, Kameswaran M, Lasisi TJ, Adedeji WA, Lasisi AO, Menendez I, Herrera M, Carranza C, Maroofian R, Crosby AH, Bensaid M, Masmoudi S, Behnam M, Mojarrad M, Feng Y, Duman D, Mawla AM, Nord AS, Blanton SH, Liu XZ, Tekin M. Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum Genet. 2016;135:953–961. doi: 10.1007/s00439-016-1697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 arXiv preprint arXiv:1207.3907. [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]