Summary
The statistical complexity of heredity has long been evident, but its molecular origins remain elusive. To investigate, we charted 90 comprehensive genotype-to-phenotype maps in a large population of wild diploid yeast. In contrast to longstanding assumptions, all types of genetic variation contributed similarly to phenotype. Causal synonymous and regulatory variants exhibited distinct molecular signatures, as did nonlinearities in heterozygote fitness that likely contribute to hybrid vigor. Highly pleiotropic variants altered disordered sequences within signaling hubs, and their effects correlated across environments – even when antagonistic – suggesting that large fitness gains bring concomitant costs. Natural genetic networks defined by the causal loci differed from those determined by precise gene deletions or protein-protein interactions. Finally, we found that traits that would appear omnigenic in less powered studies do in fact have finite genetic determinants. Integrating these molecular principles will be crucial as genome reading and writing become routine in research, industry, and medicine.
eTOC Blurb
The heritability of quantitative traits is intrinsically complex, but identifying its molecular origins is crucial for understanding how phenotypes emerge from genomes. Using a powerful genetic mapping approach, we discovered the molecular signatures of natural genetic variants that are important for phenotype. Many variants impact multiple traits, and their effects often switch between environments. Although they can be extremely complex, quantitative traits have finite linear contributors that can be comprehensively charted.
Graphical Abstract
INTRODUCTION
The intrinsic complexity of heritable traits has long been appreciated. Following the rediscovery of the work of Mendel more than a century ago, geneticists developed theories of heredity encompassing polygenicity, heterosis, and pleiotropy even before the molecular nature of the gene was understood (Fisher, 1919). Despite the evident statistical impact of these phenomena, we still lack a detailed molecular understanding of their origins. The idea that quantitative traits are driven by very large numbers of underlying loci has returned to prominence since the advent of practical whole-genome sequencing, but this ‘omnigenic’ model remains largely untested (Boyle et al., 2017; Wray et al., 2018).
The budding yeast Saccharomyces cerevisiae has been a workhorse for establishing the architecture of heredity (Bloom et al., 2013; Costanzo et al., 2010) because targeted deletions and mapping studies using inbred crosses have much greater power to detect small effects and second- and third-order interactions than genome-wide association studies in humans (Wu et al., 2017). However, studies in yeast and other models have been limited in important respects. First, the effect of natural genetic variation (e.g. missense variants) is seldom as dramatic as the deletion of an entire open reading frame (Roy et al., 2018). Second, the haplotype blocks identified as causal often encompass many candidate variants. Lastly, most large-scale genetic mapping and deletion screening studies in yeast have been conducted in haploid strains, precluding the exploration of hybrid vigor and other diploid-specific phenomena.
To investigate the complexity of natural genotype-to-phenotype maps, we constructed a panel of 18,126 fully genotyped F6 diploid S. cerevisiae segregants derived from wild yeast isolates. The scale of our experiments provided statistical power to discover quantitative trait loci (QTLs) of small effect and the high meiotic crossover density in the segregants allowed us to resolve a substantial fraction of these QTLs to single causal nucleotides (quantitative trait nucleotides or QTNs). In contrast to deletion studies, our approach is highly sensitive to the wide range of effects on phenotype caused by the natural genetic variants. In all, we discovered 18,007 QTLs and 3,394 QTNs for 90 quantitative traits, implicating 1,644 of the 6,604 protein-coding genes in S. cerevisiae. This high-resolution atlas of heredity allowed us to define molecular mechanisms of polygenicity, heterosis, pleiotropy, and gene × environment interactions and to estimate the distribution of fitness effects of extant genetic variants.
RESULTS
A powerful, high-resolution genetic mapping panel
Typical genetic mapping panels in model organisms contain many more segregating markers than genotyped individuals (Bloom et al., 2013), with few exceptions (Ehrenreich et al., 2010); the same is true of genome-wide association studies (GWAS) of humans (Visscher et al., 2017). Moreover, marker variants are usually in strong linkage disequilibrium with other nearby variants, and when microarrays are used for genotyping these haplotype blocks may also contain additional, unidentified polymorphisms (Schaid et al., 2018). Furthermore, causal variants may be rare, reducing their statistical effect on the mapping population as a whole (Gibson, 2012).
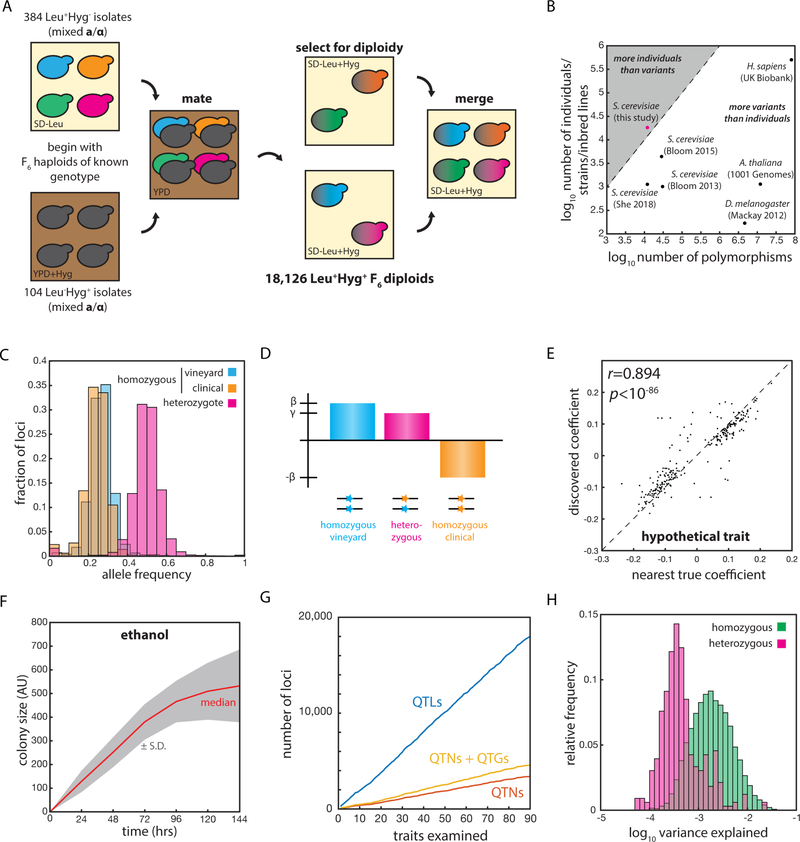
To address these shortcomings, we generated a panel of 18,126 fully genotyped F6 diploid progeny of a cross between a pathogenic S. cerevisiae isolate, YJM975, from an immunocompromised patient in Italy (McCullough et al., 1998), and a Zinfandel grape isolate, RM11–1a, from a California vineyard (Török et al., 1996) [Fig. 1A]. In contrast to most previous mapping panels in model organisms, our population contains more genotyped individuals than segregating genetic variants [Fig. 1B]. The use of inbred S. cerevisiae strains also results in highly uniform minor allele frequencies, allowing equivalent sensitivity to the effects of all segregating polymorphisms (Li et al., 2017) [Fig. 1C]. Importantly, the segregating variants are in very low linkage disequilibrium, allowing in silico allele swaps to identify causal variants with single-nucleotide resolution (She and Jarosz, 2018). We first mapped the linear contributions of homozygous loci, and then considered nonlinearities attributable to partial or overdominance by allowing heterozygous loci to adopt coefficients that deviated from the homozygous midpoints [Fig. 1D].
Fig. 1: An extremely large panel of fully genotyped diploid yeast to inventory complex heredity.
(A) Mating scheme used to construct the diploid segregant collection. (B) Number of segregating genetic variants and number of genotyped individuals in various mapping panels. (C) Relative frequencies of RM11 homozygotes (blue), YJM975 homozygotes (orange), and heterozygotes (magenta) across the 12,054 polymorphic loci in the panel. (D) Scheme illustrating the linear mixed model used to describe phenotype. β represents the homozygous locus effect and γ the heterozygous locus effect, if any. (E) Correlation (Pearson’s r) between effect size in the model and nearest true locus effect for an example simulated trait. (F) Growth of segregant panel on S-CSM + 2% ethanol solid medium. (G) Total number of QTLs, QTGs, and QTNs discovered with increasing number of quantitative traits examined. (H) Histogram of variance explained per QTL for linear (homozygous, green) and nonlinear (heterozygous, magenta) contributions across all 90 quantitative traits. See also Figure S1.
To characterize the performance of our mapping panel and analysis procedure, we performed rigorous simulations using 50 highly complex ground-truth genetic architectures. Each hypothetical trait comprised N = 275 underlying causal loci of random effect and sign, with realistic levels of Gaussian noise based on the broad-sense heritability of the traits we mapped. Comparison of in silico mapping of the simulated phenotypes to the known architectures revealed that our regression approach recalled 94 ± 2.8% of true underlying QTL and identified true underlying QTN with 92 ± 7.1% precision (N = 50 hypothetical traits; mean ± S.D.) [Fig. S1A]. Moreover, our mapping procedure accurately captured the effect size of the true causal loci for a range of underlying effect sizes (Pearson’s r = 0.93 ± 0.03 for N = 50 traits; mean ± S.D.) [Fig. S1A, e.g. Fig. 1E]. These comprehensive tests confirmed that our panel was powered to dissect highly complex traits into their constituent loci, accurately identify effect sizes, and identify the genes or causal variants associated with each QTL. We were able to not only identify nearly all linear contributors to phenotype, but also to resolve them to single genes, and, in many cases, single nucleotides.
Diverse contributors to complex traits
We next phenotyped the segregant panel in fifteen environmental conditions (including various carbon sources and toxins, an FDA-approved drug, and other stresses) across six time points [e.g. Fig. 1F]. We considered each time point in each environment as a separate quantitative trait, and the ~ 1,600,000 growth measurements allowed us to identify 18,007 QTLs at an empirical false discovery rate of 1.5 ± 2.1% (by permutation test; mean ± S.D.), with 200 ± 52.2 QTLs identified per trait (mean ± S.D.) [Fig. S1B]. Our model explained 72.8 ± 18.5% of the broad sense heritability across the 90 traits examined (mean ± S.D.) [Fig. S1C] and we readily discovered loci explaining as little as 0.01% of phenotypic variance [Fig. S1DE]. The remaining ‘missing heritability’ is likely due to second- or higher-order genetic interactions (Bloom et al., 2015a; Poelwijk et al., 2017). Most phenotypic variance was explained by linear homozygous contributions (N = 3165), but numerous heterozygous contributions (of N = 229 total) had effect sizes comparable to homozygous terms [Fig. 1H].
Of the QTLs identified, we unambiguously mapped 3,394 with single-nucleotide resolution, corresponding to 1,608 unique causal variants. An additional 1,166 QTLs could be resolved to a single quantitative trait gene (QTG) [Fig. 1G]. Strikingly, fully 24.9% of the 6,604 protein-coding genes (and 13.3% of the individual segregating polymorphisms) were implicated in determining growth across this comparatively small number of environments. Thus, it is possible that most segregating variants have the potential to significantly contribute to phenotype in the highly complex, varied environments faced by S. cerevisiae in nature (Jakobson et al., 2019).
Molecular mechanisms of coding, non-coding, and extragenic causal variants
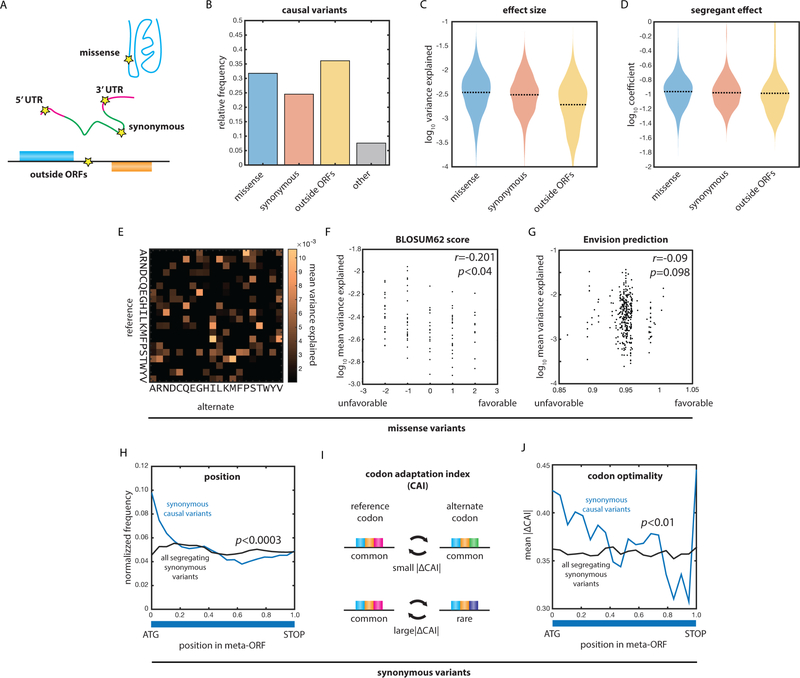
The large number of QTNs we identified allowed us to examine diverse molecular contributions to heredity [Fig. 2AB]. Missense variants exhibited the greatest variance explained, followed closely by synonymous and extragenic variants [Fig. 2C]. These same trends were reflected in the regression coefficients, which better represent the impact of the alleles in each segregant [Fig. 2D; Fig. S1F]. Many different amino acid substitutions were represented in the pool of causal missense variants [Fig. 2E], and their effect sizes correlated with molecular expectation: substitutions with lower BLOSUM62 scores (i.e. more perturbative amino acid substitutions) were of larger effect (Pearson’s r = −0.201, p < 0.04) [Fig. 2F]. Despite this, the effect of each variant remained context-dependent, and BLOSUM62 scores were only modestly predictive of the effect sizes, underscoring the importance of explicitly assessing the effects of coding variants (Diss and Lehner, 2018; Fowler and Fields, 2014). A recently developed method based on deep learning of saturating mutagenesis data (Gray et al., 2018) was less predictive than BLOSUM62, perhaps because natural missense variants are conservative relative to the broad spectrum of variants explored in saturation mutagenesis studies [Fig. 2G].
Fig. 2: Diverse molecular mechanisms underlie genetic complexity.
(A) Classes of molecular variation responsible for phenotoype. (B) Relative frequencies of all types of causal variants. (C) Variance explained by missense, synonymous, and extragenic causal variants. (D) Effect size of missense, synonymous, and extragenic causal variants. (E) Mean variance explained by missense variants (ordinate: reference residue; abscissa: alternate residue). (F) Mean variance explained as a function of BLOSUM62 score for missense variants. Correlation by Pearson’s r. (G) Mean variance explained as a function of Envision score for missense variants. Correlation by Pearson’s r. (H) Position of synonymous causal variants (blue) and all segregating synonymous variants (black) as a function of position within the meta-ORF (ATG: start codon; TAA: stop codon). P value by Kolmogorov-Smirnov test. (I) Schematic of large and small changes in codon optimality. (J) Mean absolute change in codon adaptation index (CAI) as a function of position within the meta-ORF. P value by Kolmogorov-Smirnov test. See also Figure S1.
Although missense variants had the largest effect on phenotype (p < 0.001 by two-sample Kolmogorov-Smirnov test), the effect-size distributions of all variant classes overlapped [Fig. 2C]. Synonymous natural variants, often regarded as unlikely to significantly affect phenotype (Kumar et al., 2009), had median effect sizes that were comparable to those of missense variants and larger than those of extragenic variants (p < 10−6 by two-sample Kolmogorov-Smirnov test). To probe the molecular origin of this relationship, we assessed the relative positions of causal synonymous variants within a genome-wide meta-open reading frame (ORF) relative to all synonymous variants segregating in the cross [Fig. 2H]. Causal variants were strongly enriched at the 5’ end of the meta-ORF relative to all synonymous variants (p < 0.0003 by two-sample Kolmogorov-Smirnov test). Moreover, synonymous causal variants at the 5’ end of the meta-ORF exhibited larger changes in codon adaptation index compared to all segregating synonymous variants (CAI; p < 0.01 by two-sample Kolmogorov-Smirnov test) [Fig. 2IJ] (Drummond et al., 2006). There was no correlation between the sign of the change in CAI and the sign of the effect on phenotype, likely because both increased and decreased translation could improve growth, depending on the gene product. Our data are consistent with a model in which effects due to synonymous codons are most pronounced in the early stages of translation and folding, possibly as part of an adaptive ‘translation ramp’ that favors slow translation at the beginning of genes (Tuller et al., 2010).
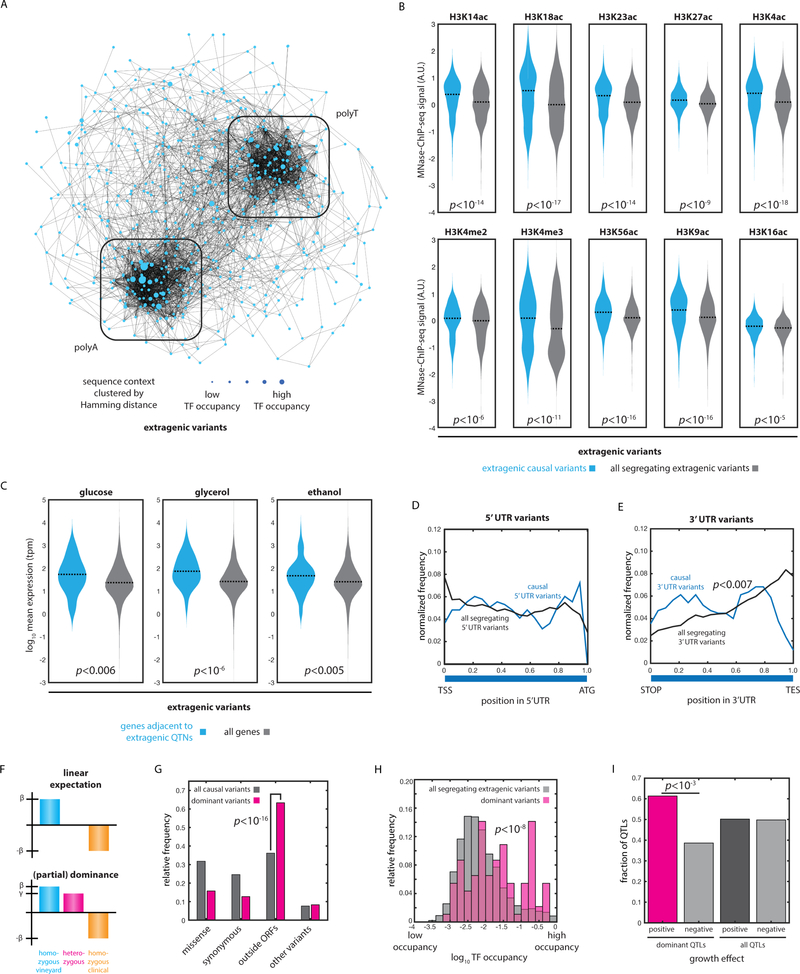
Next, we examined causal variants lying outside annotated ORFs. The positions of these causal variants were not distinct from all segregating extragenic variants relative to the positions of annotated transcriptional start (TSS) and end (TES) sites [Fig. S2AB]. However, when we examined predicted transcription-factor occupancy (Pachkov et al., 2007), we found that extragenic causal variants were enriched at sites of both high and low transcription-factor binding [Fig. S2C]. That is, these variants may act either by changing transcription-factor affinity or by influencing genome structure and accessibility independent of transcription factor binding sites. The former could directly precipitate changes in transcriptional regulation, consistent with the observation of abundant cis-eQTLs in S. cerevisiae (Kita et al., 2017). The latter may be attributable to perturbations in poly(dA:dT) tracts that are involved in nucleosome organization (Segal and Widom, 2009). Indeed, our analysis of the local sequence context (± 5 nt) of these causal variants revealed striking clusters of polyA and polyT motifs [Fig. 3A]. More than 77% of extragenic causal variants were in contexts with greater than 50% A/T content, and 23.6% were in contexts with greater than 75% A/T content.
Fig. 3: Molecular signatures of causal regulatory variation.
(A) Spring-embedded network representation of local sequence context of extragenic causal variants, weighted by Hamming distance. Boxes indicate polyA- and polyT-enriched clusters. Nodes are sized by predicted transcription factor occupancy. (B) Mean MNase-ChIP-seq signal for the indicated histone marks for nucleosomes within 200 nt of causal extragenic variants (blue) and all segregating extragenic variants (grey). Bonferroni-corrected p values by Kolmogorov-Smirnov test. (C) Mean expression (TPM) across both parental diploids (N = 3 biological replicates per strain) for genes adjacent to extragenic QTN and all genes, measured during growth in media containing 2% glucose, 2% glycerol, and 2% ethanol, as indicated. p values by Kolmogorov-Smirnov test. (D) Position of all causal variants in the 5’ UTR (blue) and all segregating variants in the 5’ UTR (black) as a function of position within the pseudo-UTR (TSS: transcription start site; ATG: start codon). p value by Kolmogorov-Smirnov test. (E) Position of all causal variants in the 3’ UTR (blue) and all segregating variants in the 3’ UTR (black) as a function of position within the pseudo-UTR (TAA: stop codon; TES: transcription end site). p value by Kolmogorov-Smirnov test. (F) Impact of dominance in our linear mixed model of phenotype; shown is an example of partial dominance. (G) Variant types of all causal variants (grey) and dominant causal variants (magenta). p value by Fisher’s exact test. (H) Transcription factor occupancy at positions of all extragenic causal variants (grey) and dominant variants (magenta). p value by Kolmogorov-Smirnov test. (I) Number of dominant QTLs with a positive (magenta) or negative (grey) effect as compared to all QTLs. p value by Fisher’s exact test. See also Figure S2.
Reasoning that biophysical perturbations to the genome might impact chromatin structure and thus transcriptional regulation, we next examined the histone marks near extragenic causal variants (Weiner et al., 2015). We found enrichments for many histone marks associated with open, active chromatin in the vicinity of causal variants, including H3K14ac, H3K18ac, H3K4me3, and H3K9ac [Fig. 3B], and the prevalence of many of these marks near causal variants was significantly correlated with effect size [Fig. S2D]. Together, these data indicated that extragenic variants in open chromatin adjacent to actively transcribed genes were more likely to impact phenotype.
To test this hypothesis, we measured genome-wide mRNA levels by RNA-seq in the diploid parental strains (RM11a/α and YJM975a/α) during exponential-phase growth in several conditions (2% glucose, 2% glycerol, and 2% ethanol; Supplementary File 2). The mean expression (averaged across both parental genetic backgrounds) was significantly higher for genes adjacent to identified extragenic QTNs in that environment as compared to all genes (p < 0.006; p < 10−6; p < 0.005 respectively by two-sample Kolmogorov-Smirnov test) [Fig. 3C]. Extragenic QTNs were also adjacent to genes with higher variability in expression, although this effect was driven primarily by the correlation between mean expression and expression variability [Fig. S2E]. In contrast, no enrichment for increased expression was observed for genes containing missense QTNs [Fig. S2F], indicating that extragenic QTNs were indeed playing a preferential role in controlling the expression of actively transcribed genes. The fold change in expression of a gene between the homozygotes was not predictive of QTG status, perhaps because the expression levels in the parents represent a complex integration of both cis and trans regulation (Wittkopp et al., 2004).
Many causal variants arose in the 5’ and 3’ UTRs of ORFs. Causal variants in the 5’ UTRs exhibited no significant spatial enrichment [Fig. 3D], whereas those in 3’ UTRs were markedly enriched at the 5’ (ORF-proximal) end of the 3’ UTR and depleted near the end of the transcript relative to all segregating 3’ UTR variants, suggesting a role for these variants in translation termination (p < 0.007 by two-sample Kolmogorov-Smirnov test) [Fig. 3E]. The nucleotides immediately following the STOP codon are known to impact the efficiency of translation termination (Namy et al., 2001), and other 3’ UTR variants may impact the stability of the mRNA transcript as a whole (Shalgi et al., 2005).
Finally, we used a sign test (Fraser et al., 2010) to search for a signature of lineage-specific selection across the many linear contributors to phenotype that we observed. All of the causal variants we identified likely have a selection coefficient greater than ~1/Ne, the threshold for the action of selection (which is very small, ~10−5–10−6, for organisms such as fungi). Therefore, one would expect to observe a coherent signature of adaptation as evidenced by spatial clusters of variants from one parent with the same effect on phenotype, as was recently observed in S. cerevisiae for other causal variants (Sharon et al., 2018). Indeed, we observed that nearby pairs of variants from the same parent were significantly more likely than would be expected by chance to have the same effect on phenotype [Fig. S2G], even over long genomic distances. This observation suggests that selection has acted coherently on variants of widely varying effect size (Jakobson et al., 2019), perhaps as a consequence of the recent adaptation of RM11 and YJM975 to their fermentation and human host-associated niches, respectively.
Dominance loci disrupt regulation
Heterosis, also called hybrid vigor, is the tendency for hybrids to outperform their parents. The phenomenon is widespread in organisms from yeast to agricultural crops, yet our understanding of its molecular origins is limited to a few individual cases (Chen, 2013). In addition to diverse linear contributors to phenotype, we identified extensive nonlinearities in the behavior of heterozygotes [Fig. 3F]. We refer to these loci collectively as dominance QTNs, encompassing partial, under-, and over-dominance. The dominance QTNs we discovered (N = 229) were strongly enriched for extragenic (presumably regulatory) variants relative to all causal variants we identified (p < 10−16; Fisher’s exact test) [Fig. 3G]. Moreover, dominance QTNs were enriched in regions of high transcription-factor occupancy relative to all segregating extragenic variants (p < 10−8 by two-sample Kolmogorov-Smirnov test) [Fig. 3H]. Lastly, the coefficients for dominance loci were predominantly positive, i.e., heterozygotes typically exhibited greater growth than would be expected from a linear model (178 positive coefficients of 290 total; 61.4%; p < 10−3 by Fisher’s exact test) [Fig. 3I]. Theory predicts that this skew in heterozygote fitness should naturally result from adaptation in diploid populations (Sellis et al., 2011). Together, these observations suggest a molecular model of dominance in which changes in regulatory interactions disrupt processes that would ordinarily limit growth under stress (Bar-Zvi et al., 2017), driving hybrid vigor.
Heterozygotes of the Rds1 transcription factor (RDS1Gln695/Lys695), for instance, exhibited improved growth in 2% galactose relative to the expectation based on a linear model. The polymorphic residue in this protein, Gln695, is located near the C-terminus, distal to the N-terminal DNA-binding region. A neighboring homozygous variant, Rds1Ser352Asn, affected growth in 2% ethanol, 2% glycerol, 2% raffinose, 2% maltose, and at 37 °C, suggesting that the Rds1 regulatory hub may play a role in adaptation to many environments despite the low copy number of the Rds1 protein when cells are grown in rich medium (Kulak et al., 2014). Indeed, genes observed to be upregulated by Rds1 form a highly connected network (protein-protein interaction p < 10−16; STRING database) that is enriched for genes involved in ‘starch and sucrose metabolism’ (p < 0.001, STRING database). The reported targets include YGP1, SPI1, and GLK1, all of which are differentially regulated in response to stress- and metabolism-related reprogramming (Stanley et al., 2010).
Two other non-coding variants exhibiting dominance also seemed likely to affect gene regulation: a TTTTTT deletion at position 134,112 of chromosome X, lying between INO1 and VPS35, and a T insertion at position 409,806 of chromosome VII, lying between TIF463 (eIF4G) and RPT6. Both variants are in poly(dA:dT) tracts, which are associated with transcriptional regulation and, as noted above, are enriched for extragenic causal variants (Yagil, 2006). These observations are consistent with a model of heterosis in which a single copy of a polymorphism, whether in a diffusible factor or a regulatory region, can be sufficient to meaningfully alter a gene control program.
Natural causal variants form coherent biological networks distinct from those defined by whole-gene deletions
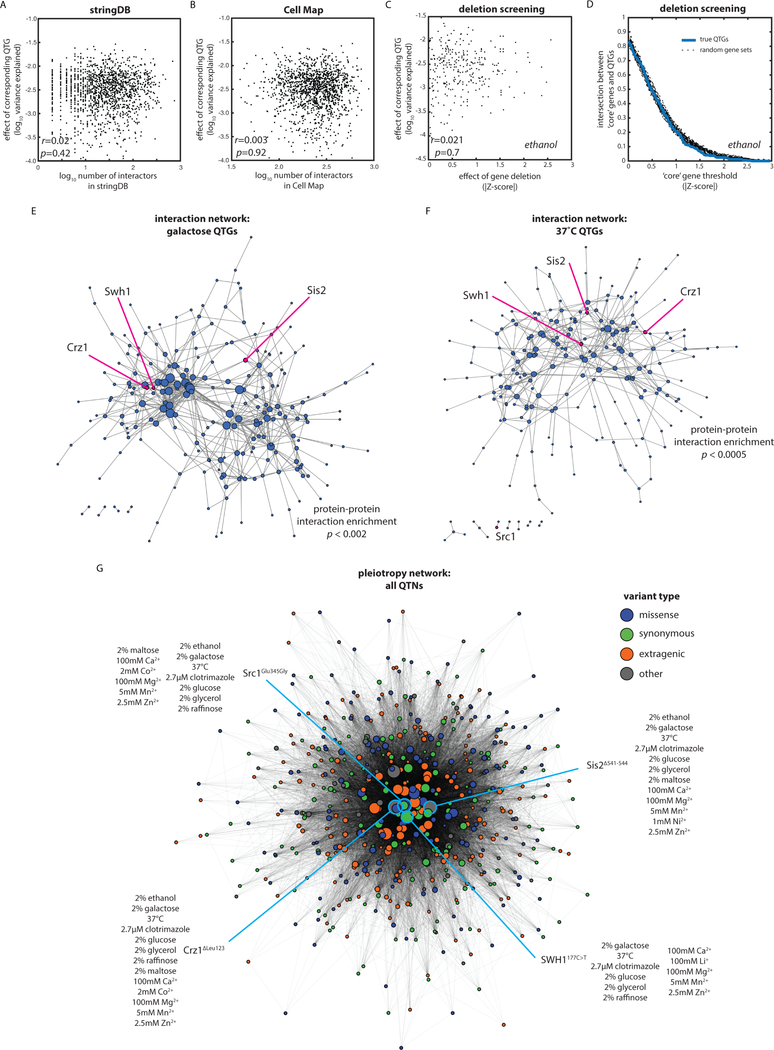
In yeast, most previous efforts to identify causal genes and the interactions between them have focused on precise ORF deletions (Costanzo et al., 2010, 2016; Giaever et al., 2002). We tested whether the topology of the variant-to-phenotype mapping we discovered was similar to those determined previously. We first compared the effect size of each identified QTG to the number of total genetic and physical interactors with that gene in the STRING database (derived from known and predicted protein-protein and genetic interactions) [Fig. 4A] (Szklarczyk et al., 2017). We also compared the effect size of each QTG to the number of significant genetic interactors of the gene in the Cell Map database (derived from genetic interaction scores in precise double-deletion backgrounds) [Fig. 4B] (Costanzo et al., 2016). The connectivity of the gene was not predictive of effect size in either case. Finally, because these databases are based primarily on data collected in rich medium, and to control for growth assay format, we phenotyped the S. cerevisiae deletion and DAmP (hypomorphic allele) collections (Breslow et al., 2008; Giaever et al., 2002) in 2% ethanol under the same growth conditions used in our mapping experiment. Even in this case, the effect size of QTGs in 2% ethanol was not significantly correlated with the strength of its corresponding deletion or DAmP allele phenotype [Fig. 4C]. Although we and others have previously found that some genes are identified as important in both deletion and mapping studies (She and Jarosz, 2018), our observations suggest that the genotype-to-phenotype map for natural genetic variation is fundamentally topologically distinct from that derived from gene deletions.
Fig. 4: The variant-to-phenotype map is distinct from other maps of cellular connectivity.
(A) QTG effect size as a function of (A) connectivity in STRING database, (B) connectivity in the Cell Map, and (C) gene deletion or DAmP allele effect. Correlations by Pearson’s r. (D) Enrichment of QTGs in core gene sets as defined by a sliding window of absolute Z-score of gene deletion or DAmP allele effect. Dashed lines show the same enrichment calculated for random gene sets of the same size. (E and F) Spring-embedded protein-protein interaction maps from STRING database for QTG in (E) galactose and (F) 37°C with edges weighted by interaction strength. Nodes are sized by interaction degree. Highlighted are key, highly pleiotropic genes. (G) Spring-embedded network representation of QTNs (nodes) connected by edges weighted by the number of traits in which the variants are jointly causal and colored by the type of molecular variation as indicated. Nodes sized by extent of pleiotropy of the QTN. Four key pleiotropic QTNs are highlighted in blue.
One method to define the ‘core’ genes important for a trait in yeast is to measure the phenotype of all precise ORF deletions. We defined core genes for growth on 2% ethanol using a sliding window based on the effect size we measured for the precise gene deletions [Supplementary File 3]. At no effect size threshold were core genes defined in this manner enriched for QTGs relative to randomly selected sets of genes [Fig. 4D], although the networks of QTGs we identified were in some cases enriched for protein-protein interactions [Fig. 4EF]. Notably, however, causal variants that impacted large numbers of quantitative traits (highlighted and discussed in more detail below) were not central to these networks, again suggesting that the network of phenotypic connectivity between genetic variants [Fig. 4G] is distinct from the molecular networks defined by genetic or physical interactions. Together, these results suggest that gene hits from deletion screens should not be the only loci at which to search for consequential natural genetic variation.
Abundant synergy and antagonism in pleiotropy and gene × environment interactions
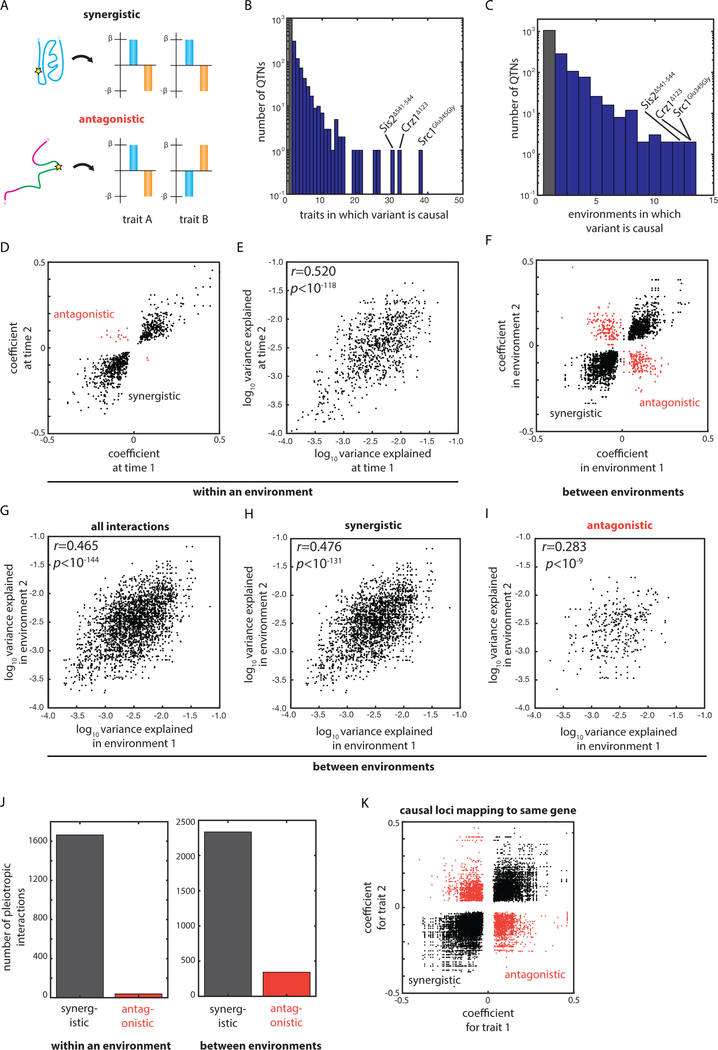
Pleiotropy and gene × environment interactions impact the topology of an organism’s fitness landscape by embedding synergies and tradeoffs in the effects of genetic changes on fitness (Wagner and Zhang, 2011). Many detrimental alleles in humans are thought to have been maintained due to antagonistic pleiotropy (Corbett et al., 2018) and theory predicts that the fate of newly arising variants is strongly influenced by their varying effects across environments (Pavličev and Cheverud, 2015). Yet these effects have remained difficult to assess because mapping intervals typically contain multiple candidate causal variants (Solovieff et al., 2013). Our discovery of large numbers of unambiguous causal variants for numerous quantitative traits allowed us to systematically assess the prevalence of synergistic and antagonistic interactions in the genotype-to-phenotype map of our segregant panel [Fig. 5A].
Fig. 5: Abundant synergistic and antagonistic interactions resolved to single nucleotides.
(A) Schematic of the phenotypic effects of synergistic and antagonistically pleotropic variants. (B) Histogram of number of traits for which each QTN was identified as causal. (C) Histogram of number of environments for which each QTN was identified as causal. (D) Plot of discovered coefficients at each pair of time points for which a given QTN was identified as causal within a given environment. (E) Plot of variance explained at each pair of time points for which a given QTN was identified as causal within a given environment. (F) Plot of discovered coefficients in each pair of quantitative traits for which a given QTN was identified as causal between environments. (G) Plot of variance explained in each pair of quantitative traits for which a given QTN was identified as causal between environments. Plot of variance explained in each pair of quantitative traits for which a given QTN was identified as (H) synergistically pleiotropic or (I) antagonistically pleiotropic between environments. (J) Number of pleiotropic interactions identified as synergistic or antagonistic (left) within an environment or (right) between environments. (K) Plot of discovered coefficients for each pair of quantitative traits in which a QTL could be attributed to a given QTG. Correlations by Pearson’s r. See also Figure S3.
The growth of a segregant was generally correlated across traits, and this growth correlation (in terms of Pearson’s r) was itself correlated with the extent of QTN overlap (a proxy for pleiotropy and gene × environment interactions) [Fig. S3AB]. This suggested that such interactions were common and predominantly synergistic, resulting in macroscopic phenotypic correlation. This was indeed the case at the molecular level: 39.5% of QTNs influenced more than one of the quantitative traits we examined [Fig. 5B]. Moreover, 34% of QTNs influenced growth in more than one distinct environment [Fig. 5C].
The vast majority (97.7%) of pleiotropic interactions within a given environmental condition were synergistic across time points, suggesting that pronounced tradeoffs between growth in the lag and exponential growth phases, for instance, were uncommon [Fig. 5DJ]. Since the genetic architectures of traits measured at successive time points are not independent, we separately assessed gene × environment interactions occurring between traits measured independently. In this case, by contrast, a substantial fraction (12.8%) of gene × environment interactions were antagonistic [Fig. 5FJ]. These antagonistic interactions imply significant intrinsic phenotypic tradeoffs, even amongst the relatively small array of conditions we tested. The instances we identified involved diverse biological processes, including glycosylation and ergosterol biosynthesis, and diverse types of variation, including missense, regulatory, and synonymous variants [Fig. S4ABC].
It is challenging to assign ‘true’ pleiotropy on the basis of mapping studies that lack nucleotide resolution because it remains formally possible that distinct causal variants within the mapping window result in spurious apparent pleiotropy (Solovieff et al., 2013). Moreover, other variants within a gene could have opposing effects on a different phenotype, convoluting interpretation of the gene product’s biological role. Our results suggest that these concerns are well founded: many QTLs mapping to the same gene had distinct (often opposing) effects [Fig. 5K].
Although variants often exhibited opposing effects across environments, the effect sizes of common QTNs across traits were significantly correlated, even for antagonistic gene × environment interactions [Fig. 5EGHI]. This implicates certain variants as key nodes in the mapping of genotype to phenotype, regardless of whether the allele is beneficial or detrimental in a given condition, and suggests that large fitness gains may be intrinsically tied to substantial fitness losses in other environments.
Highly pleiotropic variants alter disordered regions of signaling hubs
The three most synergistically pleiotropic variants we identified were associated with key hubs in cellular information flow and exhibited coherent effects on phenotype. The most pleiotropic causal variant was a missense mutation (Glu345Gly) in the inner nuclear membrane protein Src1 [Fig. S4D]; the effect on growth was in the same direction across all 38 traits for which the locus affected phenotype. Src1 binds chromosomes at the telomere and sub-telomere and mediates transcription of sub-telomeric genes, including many associated with alternative carbon source and phosphate metabolism (Grund et al., 2008). Residue 345 of the protein is nuclear-facing in both splice variants of Src1, implicating SRC1Glu345Gly in telomere and sub-telomere binding and thus the transcriptional regulation of a wide array of genes.
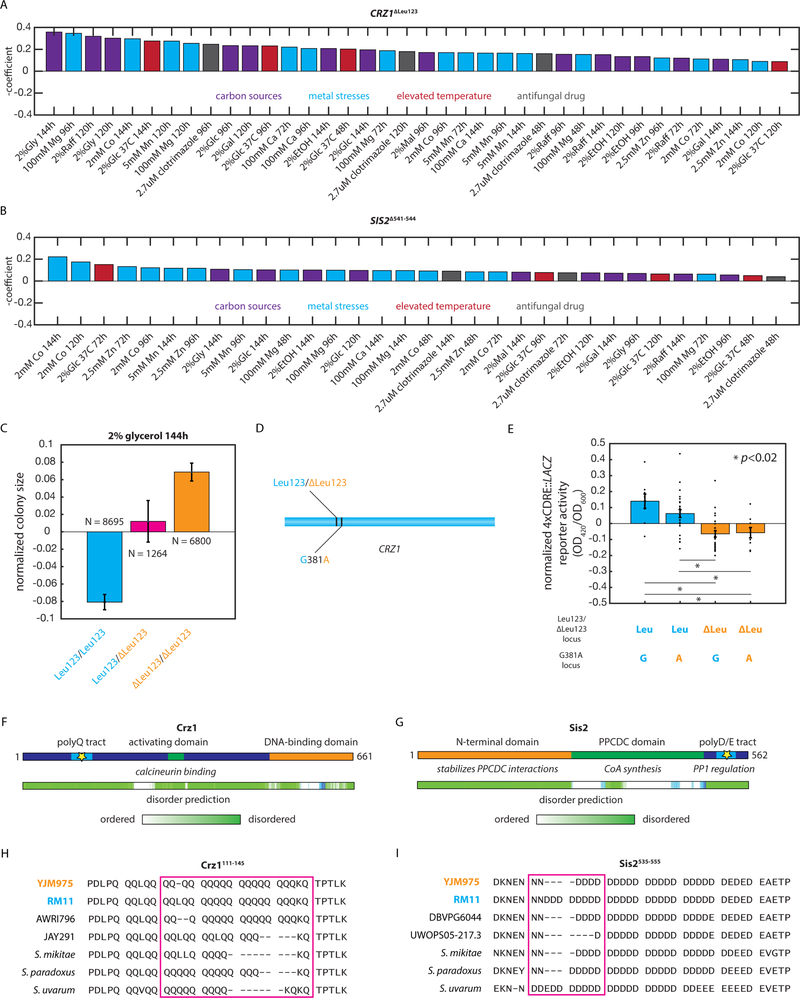
Two other highly pleiotropic variants employed a common mechanism: contractions in disordered protein regions. The first, CRZ1ΔLeu123, removed Leu123 from a poly-Q tract in the disordered N-terminal transcriptional activation domain of the transcription factor Crz1, an archetypal calcineurin target (from the same family as human NFAT) that mediates the transcriptional response to a broad range of insults (Matheos et al., 1997; Stathopoulos and Cyert, 1997). The CRZ1ΔLeu123 deletion exhibited a coherent phenotypic effect, with the same effect direction in all 32 traits [Fig. 6A]. We confirmed the widespread phenotypic importance of the CRZ1 gene product by measuring the growth of a strain with a targeted deletion of the gene in the same battery of growth conditions used for mapping: the absence of CRZ1 exerted a significant effect in 11 of the 15 environments tested (p < 0.05; Student’s two-tailed T test) [Fig. S4]. The CRZ1ΔLeu123 QTN was associated with substantial phenotypic changes in many traits [e.g. Fig. 6C] but was in very close proximity to another segregating variant (CRZ1381G>A) [Fig. 6D]. To confirm that the statistically identified QTN was responsible for changes in calcineurin-dependent signaling, we measured the activity of a 4xCDRE::LACZ reporter (Stathopoulos and Cyert, 1997) in representative haploid F6 progeny of each ditype of these two loci. Signaling was impaired in segregants bearing the CRZ1ΔLeu123 QTN and not the neighboring synonymous variant [Fig. 7E].
Fig. 6: Highly pleiotropic variants affect key cellular signaling hubs.
(A) Effect of the CRZ1ΔLeu123 QTN in the indicated conditions. (B) Effect of the SIS2Δ541−544 QTN in the indicated conditions. (C) Normalized colony sizes of F6 diploid progeny with the indicated genotypes after 96h of growth on media containing 2% glycerol. Error bars show s.e.m. (D) Diagram indicating the locations of the CRZ1ΔLeu123 QTN and the neighboring synonymous CRZ1381G>A variant in the CRZ1 gene. (E) Normalized 4xCDRE::LACZ reporter activity for at least N = 3 F6 haploid segregants of the indicated genotypes measured in biological triplicate. Bars show mean across the genotype; error bars show s.e.m. (F) Diagram of the Crz1 protein and predicted disorder from the Database of Disordered Protein Predictions (D2P2); the identified causal variant is indicated with a star. (G) Diagram of the Sis2 protein and predicted disorder from D2P2; the identified causal variant is indicated with a star. (H) Multiple sequence alignment of the Crz1111−145 region in the indicated S. cerevisiae strains and other yeast species. (I) Multiple sequence alignment of the Sis2516−555 region in the indicated S. cerevisiae strains and other yeast species. See also Figure S4.
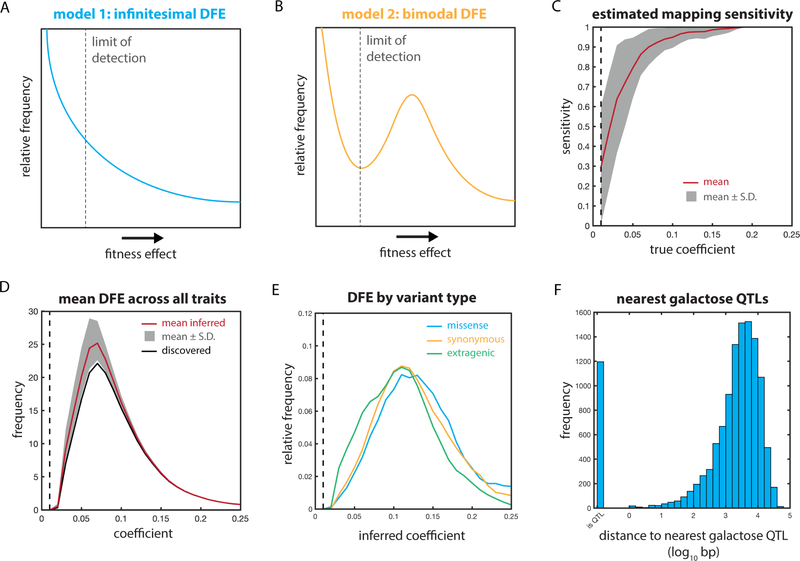
Fig. 7: Inference of the distribution of fitness effects of extant mutations.
Hypothetical true underlying fitness effect distributions under (A) infinitesimal and (B) bimodal models. (C) Sensitivity of the mapping procedure to QTLs as a function of the true effect size, for N = 10 simulations with 250 underlying causal variants. (D) Inferred true underlying fitness effect distribution (per trait). (E) Inferred average underlying fitness effect distributions based on discovered coefficients for missense (blue), synonymous (orange), and extragenic (green) variants. (F) Distance of each segregating polymorphism in the mapping panel to the nearest identified QTL for growth on 2% galactose. See also Figure S5.
The other highly pleiotropic variant, SIS2Δ541−544, removed four Asp residues from a long, unstructured acidic tract at the protein’s C-terminus. Sis2 is a bifunctional protein that plays an enzymatic role in Coenzyme A biosynthesis, conferred by its N-terminal and central domains, and engages in a regulatory interaction with protein phosphatase 1 (PP1), conferred by the disordered C-terminal domain in which the causal amino acid contraction was located (Nadal et al., 1998). This case is emblematic of the power of our mapping approach: resolving the causal variant to the C-terminal domain clarified the function of the variant and the mechanism of pleiotropy. Once again, the phenotypic effects of the variant were uniform, exhibiting a coherent effect in all 30 traits [Fig. 6B]. PP1 inhibition modulates the phosphorylation of Crz1, in turn controlling its nuclear localization and transcriptional activity (Ruiz et al., 2003). Indeed, deletion of SIS2 was even more pleiotropic than the loss of CRZ1: a strain lacking SIS2 exhibited a phenotype in all 15 mapped environments (p < 0.05; Student’s two-tailed T test) [Fig. S4E]. Our data suggest that the SIS2Δ541−544 variant impacts stress-responsive transcription by altering PP1 inhibition and in turn altered Crz1 activity. Full transcriptional activation of a 4xCDRE::LACZ reporter of Crz1 activity (Stathopoulos and Cyert, 1997) required SIS2, further supporting this model [Fig. S4F].
The mechanistic convergence between these highly pleiotropic variants was two-fold, and suggestive of general principles for pleiotropic molecular variation. First, both polymorphisms occurred in disordered regions [Fig. 6FG]; these regions are increasingly recognized as key facilitators of regulatory interactions in cells (Uversky, 2014). Strong positive selection has recently been identified in disordered regions in H. sapiens (Afanasyeva et al., 2018), suggesting that this kind of variant may be of broad importance. Second, both variants were in proteins related to a key regulatory process: signaling through the calcineurin and PP1 hubs, transduced by the transcription factor Crz1 (Ruiz et al., 2003; Thewes, 2014). Interestingly, both the polyglutamine tract of Crz1 and the acidic tract of Sis2, in which the two causal variants occurred, are highly polymorphic across S. cerevisiae strains and among related budding yeast species [Fig. 6HI] (Bergström et al., 2014). This, combined with the observation of extensive synergistic pleiotropy, suggests that these loci may be important sites for the generation of phenotypic heterogeneity.
Distribution of fitness effects of extant genetic variation
The distributions of fitness effects (DFEs) of new and existing mutations are key in understanding the process, history, and potential of natural selection and evolution (Loewe and Hill, 2010; Orr, 2010). Diverse experimental approaches have measured the DFE of new mutations in bacteria and eukarya (Frenkel et al., 2014; Koufopanou et al., 2015; Robert et al., 2018). These experiments can be expected to sample, on average, mutations of larger effect than exist in the wild, since the distributions at hand have not been subject to substantial purifying selection (other than to avoid lethality). Conversely, computational sequence-based approaches are subject to the limitation that we cannot accurately predict the effects of variants from sequence alone. Our mapping approach bridges this divide by combining direct measurement of the fitness effects of causal variants with uniform sampling of a real, naturally occurring distribution of fitness effects.
The DFE for a monogenic trait would have very few causal variants; at the other extreme of complexity, a trait that was truly infinitesimal (in Fisher’s sense of the limit of all segregating Mendelian factors (Barton et al., 2017)) would present as many causal variants as there are extant variants, likely with a modal effect approaching zero [Fig. 7A]. Alternatively, a polygenic trait might instead exhibit a bimodal distribution with two distinct classes of variants: ‘causal’ variants with a measurable effect on phenotype and ‘nearly neutral’ variants whose effects are essentially negligible [Fig. 7B]. The shape of the DFE and the number of distinct classes of variants it contains have been the subject of much theoretical and experimental attention (Boucher et al., 2016; Orr, 2003; Rice et al., 2015).
We estimated the sensitivity of our mapping panel to causal loci of varying effect by performing in silico mapping of simulated hypothetical traits with underlying causal variants of known effect size [Fig. 7C]. We subsequently estimated the true underlying distribution of fitness effects across the real quantitative traits we examined [Fig. 7D]. The apparent distribution we detected suggested that the true underlying distribution of fitness effects was bimodal: a discrete set of variants impacted each quantitative trait in a manner categorically distinct from the other, nearly neutral polymorphisms.
The shape of this inferred underlying distribution was similar for missense, synonymous, and extragenic variants [Fig. 7E]. It was also robust to the underlying distribution of effect sizes used to estimate sensitivity: we could accurately recover the distribution for normal, uniform, and monotonically decreasing effect-size distributions [Fig. S5A–I]. We observed no evidence of the Beavis effect, wherein genetic mapping can overestimate the effect of discovered QTLs due to closely linked causal variants (Beavis et al., 1991; King and Long, 2017); in fact, the coefficients were mildly deflated rather than overestimated. Our estimates of sensitivity were also robust to very large numbers of causal variants underlying the hypothetical traits used for calibration, ranging from 250 to 1000 causal loci (of 12,054 segregating polymorphisms) per trait [Fig. S5J–M].
The omnigenic model as recently articulated by Pritchard and colleagues is distinct from the limit of Fisher’s model (Boyle et al., 2017; Liu et al., 2018). It does not posit that all variants are causal per se, but rather that sufficiently many ‘peripheral’ genes contribute to phenotype that the majority of (or perhaps all) polymorphisms are in linkage disequilibrium with a causal variant. This results in apparent universal causality from the perspective of GWAS, as nearly all genomic regions make a meaningful predictive contribution. Our observations are broadly consistent with this model: 30.0% of segregating polymorphisms in our mapping panel are within 1 kb of a QTL marker for growth in 2% galactose, for example, [Fig. 7F] and 91.6% of segregating variants are within 1 kb of a QTL marker for at least one quantitative trait. The genome-wide distribution of QTLs we observe thus reconciles the statistical and theoretical architecture of the omnigenic model (Boyle et al., 2017; Liu et al., 2018) with our observation that not all segregating variants impact every quantitative trait.
DISCUSSION
Resolving large numbers of QTLs to individual causal variants allowed us to quantitatively address long-standing questions regarding the architecture of complex traits. Although coding variants had on average the greatest effect on phenotype, non-coding variants, including synonymous codons, had remarkably similar effect sizes. Indeed, the importance of such genetic variation has been noted in organisms from Salmonella to humans (Kristofich et al., 2018; Supek et al., 2014). These observations suggest that the conventional wisdom regarding the prioritization of putative causal variants demands revision, as does the bioinformatic practice of assuming the neutrality, or near-neutrality, of ‘silent’ variants relative to missense mutations. The importance of codon choice is evident from empirical studies in both bacteria and eukarya (Frumkin et al., 2018; Goodman et al., 2013; Tuller et al., 2010). Although the effects we observe can in principle be explained by many mechanisms, the pronounced biases in position and codon optimality suggest that the effects of changes in codon choice are primarily manifested early in translation. Nature, it seems, uses synonymous codons as a means to tune translation rate, and presumably protein level, in a manner orthogonal to amino acid identity.
Variants outside of ORFs exhibited two likely modes of action that are not mutually exclusive, impacting the binding of transcription factors directly and changing the local structural genomic context by altering the biophysical properties of the surrounding DNA. This idea was reinforced by our observation that causal variants were associated with open chromatin, in concordance with a privileged role for polymorphisms in transcriptionally active regions of the genome in determining phenotype (Roytman et al., 2018; Schaub et al., 2012; Trynka et al., 2013). Our results provide rigorous evidence in favor of the pragmatic assumptions underlying the integration of tissue-specific epigenomic and expression data with lower-resolution genetic mapping results (GTEx Consortium, 2017).
Antagonistic pleiotropy and antagonistic gene × environment interactions, phenomena of fundamental importance in understanding the patterns of emergence and fixation of novel molecular variation (Qian et al., 2012) as well as in designing safe and effective interventions for genetic medicine (Carter and Nguyen, 2011; Rodríguez et al., 2017), were strikingly common. Indeed, our observations likely represent a lower bound on the true extent of pleiotropy and gene × environment interactions, as unresolved QTLs for one trait may be attributable to mapped QTNs for another. Two of the three most pleiotropic variants were coding variants in disordered regions of important cell-signaling proteins, Crz1 and Sis2, suggesting that the alteration of the interactions (Babu et al., 2011) mediated by disordered regions, long regarded as inert linkers, may be a general mechanism for the generation of phenotypic heterogeneity by minimal genetic change (Jakobson and Jarosz, 2018). Our findings offer fertile ground for the future study of specific examples of pleiotropy in a variety of genes, pathways, and environments and describe convolutions to the genotype-to-phenotype relationship that likely abound in the wild (Manuck and McCaffery, 2014; Via and Lande, 1987). It will also be fruitful in future to examine other phenotypes; the predominance of synergistic interactions observed here may not extend to traits beyond growth and proliferation.
Our results help to reconcile a vigorous debate (Cox, 2017; Liu, 2017; McMahon, 2017) regarding the omnigenic model: while seemingly equivalent to Fisher’s ‘infinitesimal’ model (which assumes infinitely many segregating causal alleles), an apparently omnigenic relationship between genotype and phenotype can often arise under realistic linkage disequilibrium without all segregating variants impacting a trait. We find that most quantitative traits likely comprise sufficiently many underlying causal loci as to appear omnigenic from the perspective of a typically (under)powered GWAS; nonetheless, enumerating these many contributors may be feasible.
Substantial opportunities remain for improvement in the construction of inbred mapping populations. Most notably, although our mapping panel has advantages in the detection and resolution of causal variants, it is not currently feasible to detect all epistatic interactions. Indeed, once cis- and trans-effects are considered, there exist in principle nearly one billion possible second-order terms, a problem of underdetermination exacerbated by our use of diploids. These higher-order interactions are known to be significant in many molecular contexts (Heck et al., 2006; Olson et al., 2014; Poelwijk et al., 2017) and in complex traits (Bloom et al., 2015b; Forsberg et al., 2017), and are therefore important for a full understanding of heredity. The residual unexplained broad-sense heritability in our experiments was likely due primarily to these second- and higher-order effects. Much larger mapping panels will be required to address these questions while maintaining sufficiently low linkage disequilibrium to enable in silico fine-mapping.
Despite astounding advances in our ability to edit genes and even synthesize genomes, a conundrum persists: if one were to design an entire genome from scratch, what sequence should be chosen? Gene-level information is insufficient to answer this question if the goal is to optimize one or more quantitative traits. Despite the evident complexity of the problem, our results indicate that it will be possible to elucidate not only linear and dominant contributions to the genotype-to-phenotype relationship at the molecular level, as we have done, but also to thoroughly define higher-order contributions at nucleotide resolution. The use of appropriate experimental and statistical approaches in model organisms such as budding yeast will be critical in advancing our fundamental understanding of the highly complex relationships between genotypes and expressed traits. Indeed, fully defining such relationships in sufficient detail may well prove intractable in humans and other metazoans without establishing their underlying architecture in models amenable to maximum-resolution quantitative genetics. Understanding the nature of quantitative traits will, in turn, become crucial as genome reading and writing become routine elements of not only scientific research, but also medicine and industry.
STAR Methods
Contact for Reagent and Resource Sharing
Address requests to Daniel F. Jarosz (jarosz@stanford.edu).
Experimental Model and Subject Details
The founding parental strains of the inbred cross are RM11a (MATa ho::kanMX ura3Δ0 leu2Δ0) and YJM975α (MATα ho::hygMX uraΔ3::KanMX his3Δ::NatMX), both part of the Saccharomyces Genome Resequencing project and available from the NCYC. Also used were BY4741a (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), BY4741a ΔCRZ1 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 crz1Δ::kanMX), BY4741a ΔSIS2 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sis2Δ::kanMX), and the complete BY4741a gene deletion and DAmP allele collection (Dharmacon/Thermo). The derivation and genotypes of the F6 haploid progeny are described in detail elsewhere (She and Jarosz, 2018). The phased genotypes of the F6 diploid progeny used here for genetic mapping are available at github.com/cjakobson/mapping.
Method Details
Yeast propagation and phenotyping
Cells were revived from frozen stocks by pinning first to appropriate selective liquid medium (SD-Leu; SD+Hyg; SD-Leu+Hyg) using a Singer ROTOR robotic pinning instrument and thence to selective solid medium. Cells were transferred to various solid media (synthetic medium with yeast nitrogen base without amino acids, complete supplement mixture, and carbon source and drug/stressor as indicated; carbon source is 2% glucose if not indicated otherwise; see Table S2) for phenotyping using the same instrument and propagated for 144 hours. Growth was measured every 24 hours in 384-spot format by scanning the plates; colony size was quantified using the SGAtools suite (Wagih et al., 2013). Custom MATLAB code was used to normalize and Z-score colony sizes. Broad-sense heritability was estimated from biological replicates using a linear mixed-effects model with random effects (Bloom et al., 2013).
Cross construction
We previously genotyped 1,125 F6 haploid progeny of a cross between RM11–1a and YJM975 (She and Jarosz, 2018). Of these, we selected 384 Leu+Hyg− and 104 Leu−Hyg+ F6 progeny (mixed Mat a/α) to generate the diploid panel used here. Each Leu−Hyg+ haploid was mated to all 384 Leu+Hyg− progeny on solid YPD medium for 24 hours, then transferred to solid diploid-selective medium (SD-Leu+Hyg), grown for 48 hours, then transferred to selective solid medium once more for 48 hours. The 384 Leu+Hyg− progeny are approximately one-half MATa, so each mating yielded approximately 192 diploid progeny. Complementary mating type plates were merged so that each final plate of the collection contains two combined collections of diploids, with each collection sharing one parent. This is important to avoid conflating plate-to-plate variability effects, which are included in the regression model, with the effect of sharing a parent. Diploid selection plates were imaged before merging and positions that aberrantly contained cells in both source plates were excluded from the subsequent regression. The final collection contains 18,126 unique F6 diploid strains stored in 52 384-well plates. Please contact jarosz@stanford.edu for information regarding obtaining the segregant panel.
Genetic mapping
Diploid genotypes were constructed based on the haploid genotypes determined previously (She and Jarosz, 2018); phasing information was retained (although not used here) and loss of heterozygosity was neglected, as the diploids were propagated minimally before phenotyping. Variants present in the haploids had been called previously using SnpEff (Cingolani et al., 2012), and the coordinates shown are with reference to the strain S288C. The .vcf file associated with the segregating variants is available along with the full genotype matrix and code for the mapping procedure at github.com/cjakobson/mapping. 5’ and 3’ UTR variants were assigned on the basis of transcription start and end sites as reported in (Nagalakshmi et al., 2008). Extragenic QTNs were assigned to the nearest TSS.
The full 18,126-strain homozygous genotype matrix, comprising 12,054 markers, was regressed against normalized, Z-scored phenotype vectors for each quantitative trait using a forward stepwise selection routine. Strains lacking genotype or phenotype information were excluded from the regression. For the first iteration of the regression, only homozygous loci were included, with homozygous RM/RM loci taking a coefficient of +1 and homozygous YJM/YJM loci taking a coefficient of −1. Pseudogenotypes for plates and plate edges were included to account for positional effects. Markers were included in the model at a threshold of p < 10−3 by F-test and removed at a threshold of p < 10−2 by the same test. Following regression on the homozygous loci, the genotype matrix was expanded to 24,108 markers, with heterozygous loci taking coefficient +1 and homozygous loci taking coefficient 0 in the additional columns. Pseudogenotypes for plates and plate edges were again included. Forward stepwise selection was repeated using the same cutoffs with the final terms from the homozygous regression constituting the initial model.
Following marker selection by forward selection, causal variants with p < 10−5 were fine-mapped by in silico allele swaps, as described previously (She and Jarosz, 2018). Briefly, all pairs of candidate loci within 10 markers of the putative marker from forward selection were compared by ANOVA, and a true causal variant (QTN) was declared when the null hypothesis that the marker in question was not causal could be rejected with respect to all other variants in the window. When multiple candidate QTNs could not be unambiguously distinguished but were all associated with a single gene, a QTG was declared instead. Otherwise, we recorded the marker as a QTL and did not include it in subsequent gene- or variant-specific analyses. Runtime for the entire mapping procedure (coarse and fine mapping) using MATLAB varied between 60 min and 120 min using hardware as described below, depending on the complexity of the trait. All mapped loci are annotated in Supplementary File 1.
False discovery was controlled first at the level of QTLs by forward selection of the true genotype matrix against randomly permuted real data. An inclusion criterion of p < 10−3 for stepwise selection and a final p-value cutoff of 10−5 yielded acceptable false discovery rates as shown in Table S3. However, this approach does not give information on the accuracy of the fine-mapping procedure, nor on the recovery of the true underlying effect size of the causal variants. We therefore also conducted extensive simulations using 50 known ground-truth genetic architectures with 250 underlying linear causal variants and 25 underlying dominant causal variants, each with random, normally distributed effects, and using the true genotype structure of the panel [Fig. S1A]. Simulations were conducted with broad-sense heritability H2 ~ 0.85, commensurate with that observed for the majority of traits we examined [Fig. S1C]. Our multivariate regression approach, followed by fine-mapping, performed favorably relative to a univariate method using Pearson’s r to independently correlate each marker with phenotype [Fig. S1G], which lacked the power to detect QTLs of small effect even in our very large mapping population. Hypothetical traits were used to tune the regression procedure, but the same training traits were not used to generate the validation results presented.
The causal genes identified across all traits formed a densely connected protein-protein interaction network (enrichment p < 10−11 by STRING) and were enriched for 112 GO terms, including cellular metabolic process (FDR < 0.004), response to stress (FDR < 0.006), ion binding (FDR < 0.003), and response to nutrient levels (FDR < 0.03).
Gene expression analysis
Samples of exponentially growing (OD600 ~ 0.5–1.0) 5 mL liquid cultures of homozygous diploid RM11a/α and YJM975a/α strains (in the media conditions indicated) were harvested in biological triplicate, snap-frozen, and stored at −80°C. RNA extraction, mRNA isolation by polyA enrichment, cDNA library preparation, and DNA sequencing were performed at the Beijing Genomics Institute, which returned ~20M clean, trimmed reads per sample deposited as GSE123702 at the NIH GEO. Reads were mapped and transcript abundances estimated using Kallisto (Bray et al., 2016) on the basis of the Ensemble S. cerevisiae cDNA reference. Estimated transcripts per million abundances for each ORF can be found in Supplementary File 2.
Inference of distribution of fitness effects
In order to estimate the sensitivity of the mapping panel and regression procedure to QTL of varying effect size, we generated hypothetical traits with H2 ~ 0.85 and 250, 500, or 1000 underlying causal variants, with effect sizes drawn from normal, uniform, or quadratically decreasing distributions. The mapping procedure was conducted on N = 10 simulated traits for each underlying architecture using the true genotype structure of the panel, and sensitivity for each effect-size bin was estimated as the number of detected QTLs with effect sizes within the given range divided by the number of true underlying variants with those effect sizes. The true underlying effect size distribution was estimated as the number of discovered QTLs within a given effect size range for all the actual traits we considered divided by the estimated sensitivity for that bin based on the simulations.
Phenotyping of the S. cerevisiae ORF deletion collection
Strains bearing precise ORF deletions or hypomorphic DAmP alleles (Thermo Sci.) were revived from frozen stocks in liquid YPD medium, spotted to YPD solid medium, and grown on solid medium in 384-spot format (synthetic medium with yeast nitrogen base without amino acids, complete supplement mixture, and 2% ethanol) in biological duplicate for 48 hours. After 48 hours, growth was quantified as described above. Effect size of each deletion or hypomorphic allele was estimated as the Z-score of the mean colony size for that strain. The 100 core genes with the largest effect sizes were enriched for protein-protein interactions (p < 0.03; STRING database) and enriched for logically connected annotations (organelle organization, p < 10−4; mitochondrial translation, p < 10−3; mitochondrial matrix, p < 0.05; STRING database).
Bioinformatic analyses
Chromosome and nucleotide positions of the extragenic variants were used to retrieve the surrounding nucleotide sequence of the S288C reference genome. Histone marks were averaged across all histones centered within 200 nt of the variant (Weiner et al., 2015). Similarly, TF occupancy from SwissRegulon was retrieved based on the chromosome and nucleotide positions of the extragenic variants relative to the S288C reference genome. Codon adaptation index was calculated as fcodon i/max(fcodons for that residue) across the S288C reference genome, without adjustment for expression level. All statistical comparisons were conducted relative to all segregating variants of a given class within the mapping panel, not relative to simulated or uniform distributions.
Reporter of calcineurin-dependent signaling
Calcineurin-dependent transcriptional activation was measured using the 4xCDRE::lacZ reporter plasmid pAMS366 (Stathopoulos and Cyert, 1997). The LacZ enzyme activity assay was conducted using standard methods. Briefly, cells with genotypes as indicated and bearing the episomal reporter were subcultured to OD600 ~ 0.1 in SD-CSM liquid medium with or without 100 mM Ca2+, as indicated. After 4 hours of growth at ~21°C, cell density at 600 nm was recorded and the cells were lysed for 90 min at 37°C using 10 g/L SB3–14 detergent in Z buffer (Miller, 1972). Following lysis, one-half of the reaction volume of 2 g/L chromogenic o-NPG substrate in prewarmed Z buffer was added and the mixture incubated at 37°C for 120 min. Samples were briefly centrifuged at ~2,000 × g to pellet insoluble cell debris and the supernatant transferred to a new plate. The LacZ hydrolysis product o-nitrophenol was monitored at 420 nm, and CDRE activity was calculated as A420/A600, adjusted for background scattering and nonspecific hydrolysis in lysate of cells lacking the reporter plasmid.
Computational methods and resources
Most computation was performed using MATLAB (MathWorks) on a MacBook Pro computer (2.7 GHz Intel Core i7; 16 GB RAM). Mapping was conducted using MATLAB on Sherlock nodes with 64 GB RAM. Also used were the SGATools (Wagih et al., 2013), clustalOmega (Sievers et al., 2011), D2P2 (Oates et al., 2013); Panther/GO (The Gene Ontology Consortium, 2017), SwissRegulon (Pachkov et al., 2007), Phyre2 (Kelley et al., 2015), SGRP (Bergström et al., 2014; Liti et al., 2009), and CellMap (Costanzo et al., 2016) webservers. The genotype and phenotype data are too large to include as supplementary files; all code required for mapping and validation, including segregant genotypes and all actual and simulated growth data, is deposited at github.com/cjakobson/mapping. Code used to generate the figures is available upon request to Daniel F. Jarosz (jarosz@stanford.edu).
Quantification and Statistical Analysis
Student’s two-tailed T-test, F-test, binomial test, two-sample Kolmogorov–Smirnov test, and Fisher’s exact test were performed in MATLAB. The Bonferroni correction was applied in the case of multiple testing. In mapping analyses, strains missing either genotype or phenotype information were excluded. Mean, median, standard deviation, and standard error of the mean are variously shown as indicated in the legends.
Data and Software Availability
All genetic mapping code is deposited at github.com/cjakobson/mapping. Other dependencies, including genotype and phenotype data too large to host on GitHub, can be downloaded from the link in the GitHub readme. mRNA-seq data are available at GSE123702 at the NIH GEO.
Supplementary Material
Supplementary File 1: Annotated list of QTLs, QTGs, and QTNs identified in this study; to accompany Figure 1
Supplementary File 2: Estimated abundances of mRNAs measured in this study; to accompany Figure 3
Supplementary File 3: Results of the 2% ethanol deletion and DAmP screen; to accompany Figure 4
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| SB3–14 | Sigma | 40772–50G |
| o-NPG | Bio Basic | ND0382 |
| Deposited Data | ||
| mRNA-seq data | This paper | GSE123702 |
| F6 diploid genotype data | This paper | github.com/cjakobson/mapping |
| F6 diploid growth data | This paper | github.com/cjakobson/mapping |
| Experimental Models: Organisms/Strains | ||
| RM11 | NCYC | (Liti et al., 2009) |
| YJM975 | NCYC | (Liti et al., 2009) |
| F6 haploid progeny | Jarosz Laboratory | (She and Jarosz, 2018) |
| F6 diploid progeny | This paper | N/A |
| BY4741 gene deletion/DAmP collection | Dharmacon/Thermo | N/A |
| Recombinant DNA | ||
| 4xCDRE::LACZ reporter (pAMS366) | Cyert Laboratory | (Stathopoulos and Cyert, 1997) |
| Software and Algorithms | ||
| Genetic mapping code | This paper | github.com/cjakobson/mapping |
| MATLAB R2016B | MathWorks | N/A |
| Kallisto | Pachter Lab | https://pachterlab.github.io/kallisto/download |
Acknowledgments
We thank Hunter Fraser, Joanna Wysocka, Jonathan Pritchard, and the members of the Jarosz Lab for critical review of the manuscript. Some of the computing for this project was performed on the Sherlock cluster. We would like to thank Stanford University and the Stanford Research Computing Center for providing computational resources and support that contributed to these research results. This work was supported by the National Institutes of Health (NIH-1F32-GM125162 to CMJ & NIH-DP2-GM119140 to DFJ), the National Science Foundation (NSF-MCB116762 to DFJ), a Kimmel Scholar award (to DFJ), a Searle Scholar Award (14-SSP-210 to DFJ), a Vallee Scholar award (to DFJ) and a Science and Engineering Fellowship from the David and Lucile Packard Foundation (to DFJ).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afanasyeva A, Bockwoldt M, Cooney C, Heiland I, and Gossmann TI (2018). Human long intrinsically disordered protein regions are frequent targets of positive selection. Genome Res. gr.232645.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, van der Lee R, de Groot NS, and Gsponer J (2011). Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol 21, 432–440. [DOI] [PubMed] [Google Scholar]
- Barton NH, Etheridge AM, and Véber A (2017). The infinitesimal model: Definition, derivation, and implications. Theor. Popul. Biol 118, 50–73. [DOI] [PubMed] [Google Scholar]
- Bar-Zvi D, Lupo O, Levy AA, and Barkai N (2017). Hybrid vigor: The best of both parents, or a genomic clash? Curr. Opin. Syst. Biol 6, 22–27. [Google Scholar]
- Beavis WD, Grant D, Albertsen M, and Fincher R (1991). Quantitative trait loci for plant height in four maize populations and their associations with qualitative genetic loci. Theor. Appl. Genet 83, 141–145. [DOI] [PubMed] [Google Scholar]
- Bergström A, Simpson JT, Salinas F, Barré B, Parts L, Zia A, Ba N, N A, Moses AM, Louis EJ, et al. (2014). A High-Definition View of Functional Genetic Variation from Natural Yeast Genomes. Mol. Biol. Evol 31, 872–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite T-LV, and Kruglyak L (2013). Finding the sources of missing heritability in a yeast cross. Nature 494, 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Kotenko I, Sadhu MJ, Treusch S, Albert FW, and Kruglyak L (2015a). Genetic interactions contribute less than additive effects to quantitative trait variation in yeast. Nat. Commun 6, ncomms9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Kotenko I, Sadhu MJ, Treusch S, Albert FW, and Kruglyak L (2015b). Genetic interactions contribute less than additive effects to quantitative trait variation in yeast. Nat. Commun 6, ncomms9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JI, Bolon DNA, and Tawfik DS (2016). Quantifying and understanding the fitness effects of protein mutations: Laboratory versus nature. Protein Sci. 25, 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, and Pritchard JK (2017). An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, and Weissman JS (2008). A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5, 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AJ, and Nguyen AQ (2011). Antagonistic pleiotropy as a widespread mechanism for the maintenance of polymorphic disease alleles. BMC Med. Genet 12, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2013). Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet 14, 471–482. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, and Ruden DM (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett S, Courtiol A, Lummaa V, Moorad J, and Stearns S (2018). The transition to modernity and chronic disease: mismatch and natural selection. Nat. Rev. Genet 19, 419–430. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. (2010). The Genetic Landscape of a Cell. Science 327, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox N (2017). Comments on Pritchard Paper. J. Psychiatry Brain Sci [Google Scholar]
- Diss G, and Lehner B (2018). The genetic landscape of a physical interaction. ELife 7, e32472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Raval A, and Wilke CO (2006). A Single Determinant Dominates the Rate of Yeast Protein Evolution. Mol. Biol. Evol 23, 327–337. [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, and Kruglyak L (2010). Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464, 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1919). XV.—The Correlation between Relatives on the Supposition of Mendelian Inheritance. Earth Environ. Sci. Trans. R. Soc. Edinb 52, 399–433. [Google Scholar]
- Forsberg SKG, Bloom JS, Sadhu MJ, Kruglyak L, and Carlborg Ö (2017). Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nat. Genet 49, 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, and Fields S (2014). Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Moses AM, and Schadt EE (2010). Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc. Natl. Acad. Sci 107, 2977–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel EM, Good BH, and Desai MM (2014). The Fates of Mutant Lineages and the Distribution of Fitness Effects of Beneficial Mutations in Laboratory Budding Yeast Populations. Genetics 196, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumkin I, Lajoie MJ, Gregg CJ, Hornung G, Church GM, and Pilpel Y (2018). Codon usage of highly expressed genes affects proteome-wide translation efficiency. Proc. Natl. Acad. Sci 201719375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- Gibson G (2012). Rare and common variants: twenty arguments. Nat. Rev. Genet 13, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DB, Church GM, and Kosuri S (2013). Causes and Effects of N-Terminal Codon Bias in Bacterial Genes. Science. [DOI] [PubMed] [Google Scholar]
- Gray VE, Hause RJ, Luebeck J, Shendure J, and Fowler DM (2018). Quantitative Missense Variant Effect Prediction Using Large-Scale Mutagenesis Data. Cell Syst. 6, 116–124.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund SE, Fischer T, Cabal GG, Antúnez O, Pérez-Ortín JE, and Hurt E (2008). The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J. Cell Biol 182, 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GTEx (2017). Genetic effects on gene expression across human tissues. Nature 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JA, Argueso JL, Gemici Z, Reeves RG, Bernard A, Aquadro CF, and Alani E (2006). Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. U. S. A 103, 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobson CM, and Jarosz DF (2018). Organizing biochemistry in space and time using prion-like self-assembly. Curr. Opin. Syst. Biol 8, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobson CM, She R, and Jarosz DF (2019). Pervasive function and evidence for selection across standing genetic variation in S. cerevisiae. Nat. Commun 10, 1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EG, and Long AD (2017). The Beavis Effect in Next-Generation Mapping Panels in Drosophila melanogaster. G3 Genes Genomes Genet. 7, 1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita R, Venkataram S, Zhou Y, and Fraser HB (2017). High-resolution mapping of cis-regulatory variation in budding yeast. Proc. Natl. Acad. Sci 114, E10736–E10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufopanou V, Lomas S, Tsai IJ, and Burt A (2015). Estimating the Fitness Effects of New Mutations in the Wild Yeast Saccharomyces paradoxus. Genome Biol. Evol 7, 1887–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristofich J, Morgenthaler AB, Kinney WR, Ebmeier CC, Snyder DJ, Old WM, Cooper VS, and Copley SD (2018). Synonymous mutations make dramatic contributions to fitness when growth is limited by a weak-link enzyme. PLOS Genet. 14, e1007615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, and Mann M (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, and Ng PC (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Li X, Kim Y, Tsang EK, Davis JR, Damani FN, Chiang C, Hess GT, Zappala Z, Strober BJ, Scott AJ, et al. (2017). The impact of rare variation on gene expression across tissues. Nature 550, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C (2017). A Case for Core Genes. J. Psychiatry Brain Sci [Google Scholar]
- Liu X, Li YI, and Pritchard JK (2018). Trans effects on gene expression can drive omnigenic inheritance. BioRxiv 425108. [DOI] [PMC free article] [PubMed]
- Loewe L, and Hill WG (2010). The population genetics of mutations: good, bad and indifferent. Philos. Trans. R. Soc. B Biol. Sci 365, 1153–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuck SB, and McCaffery JM (2014). Gene-environment interaction. Annu. Rev. Psychol 65, 41–70. [DOI] [PubMed] [Google Scholar]
- Matheos DP, Kingsbury TJ, Ahsan US, and Cunningham KW (1997). Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11, 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough MJ, Clemons KV, Farina C, McCusker JH, and Stevens DA (1998). Epidemiological investigation of vaginal Saccharomyces cerevisiae isolates by a genotypic method. J. Clin. Microbiol 36, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon F (2017). Casting a Shadow of Doubt over the GWAS Parade. J. Psychiatry Brain Sci [Google Scholar]
- Miller JH (1972). Miller JH.. Experiments in Molecular Genetics (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: ). [Google Scholar]
- Nadal E. de, Clotet J, Posas F, Serrano R, Gomez N, and Ariño J (1998). The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc. Natl. Acad. Sci 95, 7357–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, and Snyder M (2008). The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Hatin I, and Rousset J-P (2001). Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, et al. (2013). D2P2: database of disordered protein predictions. Nucleic Acids Res. 41, D508–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CA, Wu NC, and Sun R (2014). A Comprehensive Biophysical Description of Pairwise Epistasis throughout an Entire Protein Domain. Curr. Biol 24, 2643–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA (2003). The Distribution of Fitness Effects Among Beneficial Mutations. Genetics 163, 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA (2010). The population genetics of beneficial mutations. Philos. Trans. R. Soc. B Biol. Sci 365, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachkov M, Erb I, Molina N, and van Nimwegen E (2007). SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 35, D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavličev M, and Cheverud JM (2015). Constraints Evolve: Context Dependency of Gene Effects Allows Evolution of Pleiotropy. Annu. Rev. Ecol. Evol. Syst 46, 413–434. [Google Scholar]
- Poelwijk FJ, Socolich M, and Ranganathan R (2017). Learning the pattern of epistasis linking genotype and phenotype in a protein. BioRxiv 213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Ma D, Xiao C, Wang Z, and Zhang J (2012). The Genomic Landscape and Evolutionary Resolution of Antagonistic Pleiotropy in Yeast. Cell Rep. 2, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DP, Good BH, and Desai MM (2015). The Evolutionarily Stable Distribution of Fitness Effects. Genetics 200, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert L, Ollion J, Robert J, Song X, Matic I, and Elez M (2018). Mutation dynamics and fitness effects followed in single cells. Science 359, 1283–1286. [DOI] [PubMed] [Google Scholar]
- Rodríguez JA, Marigorta UM, Hughes DA, Spataro N, Bosch E, and Navarro A (2017). Antagonistic pleiotropy and mutation accumulation influence human senescence and disease. Nat. Ecol. Evol 1, 0055. [DOI] [PubMed] [Google Scholar]
- Roy KR, Smith JD, Vonesch SC, Lin G, Tu CS, Lederer AR, Chu A, Suresh S, Nguyen M, Horecka J, et al. (2018). Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat. Biotechnol 36, 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roytman M, Kichaev G, Gusev A, and Pasaniuc B (2018). Methods for fine-mapping with chromatin and expression data. PLOS Genet. 14, e1007240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Yenush L, and Ariño J (2003). Regulation of ENA1 Na+-ATPase Gene Expression by the Ppz1 Protein Phosphatase Is Mediated by the Calcineurin Pathway. Eukaryot. Cell 2, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Chen W, and Larson NB (2018). From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, and Snyder M (2012). Linking disease associations with regulatory information in the human genome. Genome Res. 22, 1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, and Widom J (2009). Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol 19, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellis D, Callahan BJ, Petrov DA, and Messer PW (2011). Heterozygote advantage as a natural consequence of adaptation in diploids. Proc. Natl. Acad. Sci 108, 20666–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Lapidot M, Shamir R, and Pilpel Y (2005). A catalog of stability-associated sequence elements in 3’ UTRs of yeast mRNAs. Genome Biol. 6, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Chen S-AA, Khosla NM, Smith JD, Pritchard JK, and Fraser HB (2018). Functional Genetic Variants Revealed by Massively Parallel Precise Genome Editing. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She R, and Jarosz DF (2018). Mapping Causal Variants with Single-Nucleotide Resolution Reveals Biochemical Drivers of Phenotypic Change. Cell 172, 478–490.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, and Smoller JW (2013). Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet 14, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Bandara A, Fraser S, Chambers P. j., and Stanley G. a. (2010). The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol 109, 13–24. [DOI] [PubMed] [Google Scholar]
- Stathopoulos AM, and Cyert MS (1997). Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11, 3432–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Miñana B, Valcárcel J, Gabaldón T, and Lehner B (2014). Synonymous Mutations Frequently Act as Driver Mutations in Human Cancers. Cell 156, 1324–1335. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. (2017). The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes S (2014). Calcineurin-Crz1 Signaling in Lower Eukaryotes. Eukaryot. Cell 13, 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török T, Mortimer RK, Romano P, Suzzi G, and Polsinelli M (1996). Quest for wine yeasts—An old story revisited. J. Ind. Microbiol 17, 303–313. [Google Scholar]
- Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, and Raychaudhuri S (2013). Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet 45, 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, and Pilpel Y (2010). An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation. Cell 141, 344–354. [DOI] [PubMed] [Google Scholar]
- Uversky VN (2014). Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front. Mol. Biosci 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, and Lande R (1987). Evolution of genetic variability in a spatially heterogeneous environment: effects of genotype–environment interaction. Genet. Res 49, 147–156. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, and Yang J (2017). 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet 101, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O, Usaj M, Baryshnikova A, VanderSluis B, Kuzmin E, Costanzo M, Myers CL, Andrews BJ, Boone CM, and Parts L (2013). SGAtools: one-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 41, W591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, and Zhang J (2011). The pleiotropic structure of the genotype–phenotype map: the evolvability of complex organisms. Nat. Rev. Genet 12, 204–213. [DOI] [PubMed] [Google Scholar]
- Weiner A, Hsieh T-HS, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, and Friedman N (2015). High-Resolution Chromatin Dynamics during a Yeast Stress Response. Mol. Cell 58, 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, and Clark AG (2004). Evolutionary changes in cis and trans gene regulation. Nature 430, 85–88. [DOI] [PubMed] [Google Scholar]
- Wray NR, Wijmenga C, Sullivan PF, Yang J, and Visscher PM (2018). Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell 173, 1573–1580. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zheng Z, Visscher PM, and Yang J (2017). Quantifying the mapping precision of genome-wide association studies using whole-genome sequencing data. Genome Biol. 18, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagil G (2006). DNA tracts composed of only two bases concentrate in gene promoters. Genomics 87, 591–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Annotated list of QTLs, QTGs, and QTNs identified in this study; to accompany Figure 1
Supplementary File 2: Estimated abundances of mRNAs measured in this study; to accompany Figure 3
Supplementary File 3: Results of the 2% ethanol deletion and DAmP screen; to accompany Figure 4
Data Availability Statement
All genetic mapping code is deposited at github.com/cjakobson/mapping. Other dependencies, including genotype and phenotype data too large to host on GitHub, can be downloaded from the link in the GitHub readme. mRNA-seq data are available at GSE123702 at the NIH GEO.