Abstract
Background
The growing importance of the ubiquitous fungal genus Trichoderma (Hypocreales, Ascomycota) requires understanding of its biology and evolution. Many Trichoderma species are used as biofertilizers and biofungicides and T. reesei is the model organism for industrial production of cellulolytic enzymes. In addition, some highly opportunistic species devastate mushroom farms and can become pathogens of humans. A comparative analysis of the first three whole genomes revealed mycoparasitism as the innate feature of Trichoderma. However, the evolution of these traits is not yet understood.
Results
We selected 12 most commonly occurring Trichoderma species and studied the evolution of their genome sequences. Trichoderma evolved in the time of the Cretaceous-Palaeogene extinction event 66 (±15) mya, but the formation of extant sections (Longibrachiatum, Trichoderma) or clades (Harzianum/Virens) happened in Oligocene. The evolution of the Harzianum clade and section Trichoderma was accompanied by significant gene gain, but the ancestor of section Longibrachiatum experienced rapid gene loss. The highest number of genes gained encoded ankyrins, HET domain proteins and transcription factors. We also identified the Trichoderma core genome, completely curated its annotation, investigated several gene families in detail and compared the results to those of other fungi. Eighty percent of those genes for which a function could be predicted were also found in other fungi, but only 67% of those without a predictable function.
Conclusions
Our study presents a time scaled pattern of genome evolution in 12 Trichoderma species from three phylogenetically distant clades/sections and a comprehensive analysis of their genes. The data offer insights in the evolution of a mycoparasite towards a generalist.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5680-7) contains supplementary material, which is available to authorized users.
Keywords: Ankyrin domains, CAZymes, Core genome, Environmental opportunism, Gene gain, Gene loss, SSCPs, Orphans
Background
The Sordariomycetes, one of the largest classes in the Division Ascomycota, display a wide range of nutritional strategies including saprotrophy and biotrophic interactions with bacteria, plants, animals, fungi or other organisms [1]. Within them, the highest number of known genera is found in the order Hypocreales [2] that comprises half of the whole-genome sequenced species of Sordariomycetes (Nov. 2017, NCBI Taxonomy Browser). Molecular data suggest that the ancestors of the Hypocreales evolved some 170–200 Mya as fungi associated with plants either as parasites or saprotrophs [3]. The diversification into extant taxa was accompanied by several intra- and interkingdom host shifts involving fungi, higher plants, and animals [4]. Among them, parasites of animals likely appeared first in the Jurassic period, and specialized entomoparasitic families developed during the Cretaceous period, thereby following the diversification of herbivory insects and angiosperms [3].
Mycoparasitic fungi can be found in species from several fungal taxa [5], but only the Hypocreales contain exclusively fungicolous genera, i.e. Hypomyces, Escovopsis, and Trichoderma. The ancestor of these mycoparasitic fungi likely evolved at the same time as the entomoparasites, but the time and events of Trichoderma evolution are not known.
Among these fungicolous fungal genera, Trichoderma is the largest taxon with many ubiquitously distributed species. Detailed ecological and biogeographic surveys of Trichoderma [6–9] revealed that species of this genus are most frequently found on the fruiting bodies of other fungi and the dead wood colonized by them. While mycoparasitism in Hypomyces is frequently species-specific and restricted to fruiting body forming Basidiomycota [10], the genus Trichoderma is unique as many of its species can parasite also on Ascomycetes and even on phylogenetically close species [11].
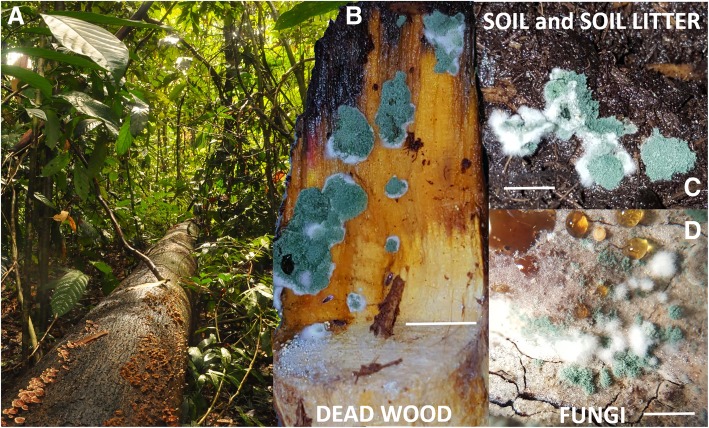
An analysis of the genomes from three species of Trichoderma (T. reesei, T. virens, T. atroviride) suggested that mycoparasitism as an innate property of Trichoderma [12], but these species are also characterized by considerable nutritional versatility [13]: in addition to acting as mycoparasite, which promoted its use as a biocontrol agent against plant pathogenic fungi [12, 14], Trichoderma has become an opportunistic infectant of humans [12]. To date, Trichoderma is rarely reported as a parasite on plants and invertebrates but it can colonize plants as a symptomless endosymbiont [12]. Finally, many species of the genus grow efficiently on dead plant biomass and one of its species - T. reesei - is a major industrial source of cellulases and hemicellulases. Interestingly, the most opportunistic Trichoderma species may also grow in soil where they can either establish in a bulk soil or colonize rhizosphere. As plants usually positively respond to the presence of Trichoderma, this property attracts raising attention for the use of these fungi in biofertilizers. The fact that some Trichoderma species can feed on plant, fungal and animal bodies characterizes them as generalists. (Fig. 1).
Fig. 1.
Trichoderma spp. in nature. a The fallen log of the dead wood colonized by the other fungi represents the major ecological niche for Trichoderma spp. b Trichoderma atroviride on dead wood. c T. harzianum on soil. d T. simonsii on the sporocarps of Stereum sp. Some species may also colonize soil and become endophytes. Scale bar on B and C corresponds to 1 cm
It is not known how generalism evolved from the phytosaprotrophic background of the Hypocreales. Chaverri and Samuels [15] compared a phylogenetic tree of the genus Trichoderma with the habitats from which the individual species had been isolated and concluded that the evolution of the genus involved several interkingdom host jumps and that preference for a special habitat was gained or lost multiple time. It has been argued that the versatility of Trichoderma’s nutritional strategies can be described by the expansions of the spectrum of hosts and substrates due to enrichment of its genome by the laterally transferrered genes required for the feeding on the plant biomass [11].
The hypothesis of this work was that a comparative genomics of those species of Trichoderma which are most frequently sampled (and therefore must be most successful generalists) and an analysis of their pattern of gene evolution would reveal the evolutionary events that shaped the nutritional expansions and environmental generalism. In addition, identification of the gene inventory of the Trichoderma core genome (i.e. the genes that are present in all species) and its intersection with genomes of other fungi would reveal the specific genomic features of these industrially-relevant fungi.
Although the sequences of several Trichoderma genomes have already been published [11, 16–24], detailed genome wide analyses have been published for only three of them (T. reesei, T. virens and T. atroviride [11, 16, 25–27]). To test the hypothesis raised above, we have analysed the evolution and the gene inventory of the genomes from 13 Trichoderma isolates that represent 12 species with a worldwide distribution and are members of three major infrageneric groups [7].
Results
Selection of the most common Trichoderma species
To reveal the most frequently sampled species in the genus Trichoderma, we have first calculated the number of nucleotide sequences deposited for Trichoderma spp. in NCBI GenBank (see Methods). There is today general agreement that the new Trichoderma spp. can only be defined by at least three or more gDNA sequences while the analysis of usually two DNA barcode fragments is required for the species identification [7–9]. The number of gene sequences in NCBI per each species may therefore roughly correspond to the number of isolates detected for this species and thus approximate the frequency of the general species occurrence. This analysis revealed (Additional file 1) that most species (80%) were relatively rare as they were represented by < 50 gene sequences each, whereas 35 species (12% of the total number of species) were represented by more than 100 nucleotide sequences each. Of these, 84% of nucleotide sequences were attributed to a small group of common species: T. harzianum sensu lato (also deposited as T. lixii or Hypocrea lixii) was responsible for 32% (9532 sequences) of total sequences. This was followed by T. asperellum, T. atroviride, T. longibrachiatum, T. gamsii, and T. virens that were represented by > 1000 sequences each and therefore also frequent.
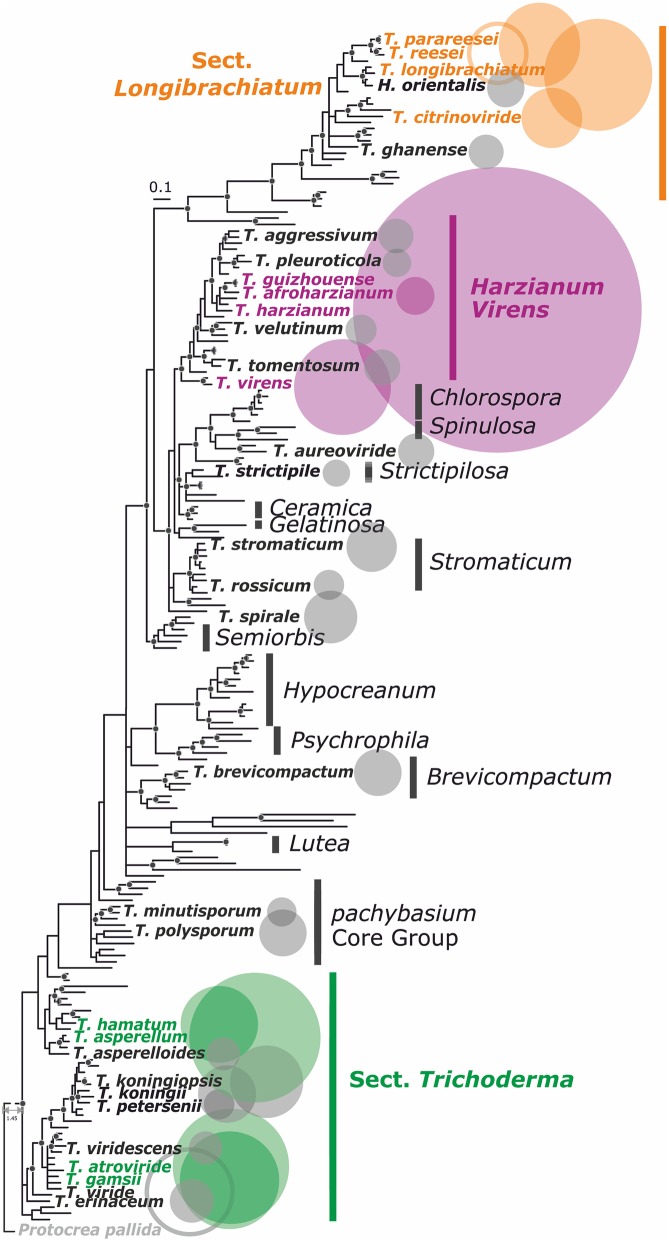
The mapping of the nucleotide sequence abundance (Additional file 1) on the phylogenetic tree of the genus consisting of > 200 species showed that the most frequent species are not present in one or a few infraneric groups but are distributed among different clades (Fig. 2). The most frequent and putatively environmentally successful species are found in the Harzianum/Virens clades (HV), section Longibrachiatum (SL) and section Trichoderma (ST). As all of the common species are profound generalists with cosmopolitan distribution, this property could either be an evolutionary old adaptation lost by other (rare) Trichoderma species or the nutritional versatility independenty evolved in each phylogenetic group of the genus.
Fig. 2.
Phylogeny of the genus Trichoderma and occurrence of the most common species. Phylogeny of Trichoderma based on Bayesian analysis of the rpb2 gene (see Methods for details). Only species with major abundance (> 100 nucleotide sequences deposited in NCBI GenBank, April 2018,) are shown. The number of core nucleotide sequences deposited in GeneBank is indicated by the size of the filled circles with T. pleuroticola, N = 103 being the smalles shown. Sections Longibrachiatum and Trichoderma and the Harzianum/Virens clade are indicated by colored vertical bars. Rare Trichoderma spp. (< 100 nucleotide sequences known in public databases) are not shown. Circles for T. reesei and T. viride likely represent false positive values as T. reesei is most studied species, while T. viride is the oldest Trichoderma species name that was assigned to all strains before DNA barcoding became available
General comparison of the genomes of twelve Trichoderma species
Based on the above analysis, we compared the genomes from 12 Trichoderma species: T. reesei, T. longibrachiatum, T. citrinoviride, T. parareesei from section Longibrachiatum (SL), T. harzianum (the the ex-type strain CBS226.95 marked with “T” throughout the manuscript, and strain TR274), T. guizouense, T. afroharzianum, T. virens from Harzianum and Virens clades (HV), and T. atroviride, T. gamsii, T. asperellum, and T. hamatum from section Trichoderma (ST). The relation between the species is shown on Fig. 2. The species concept of T. harzianum has recently been revised [28] and it is not known what percentage of the newly defined species would account for “T. harzianum” entries in GeneBank. We therefore included T. guizhouense and T. afroharzianum, two species with worldwide distribution [28, 29], and two strains of T. harzianum (one from Northern Europe and one from Brazil [30]) in this study. T. parareesei and T. gamsii were included because they are sibling species of T. reesei and T. atroviride, respectively (Fig. 2).
As already mentioned, the genome sequencing for some of these species has been reported before. For the sequencing of the new strains and annotation improvement of previously published genomes of T. reesei and T. hamatum see Methods and Additional files 2, 3, and 4. The Trichoderma genomes vary in size (33 - 41 Mb), species from SL having the smallest ones (Table 1). Consequently, the number of predicted genes in Trichoderma varies between 9292 and 14,095, which is in the range of that in other Sordariomycetes genomes (https://genome.jgi.doe.gov/fungi/fungi.info.html) [31]). In correlation with the genome size, species from SL also contain the smallest gene inventory (9292–10,938 genes). As shown in Table 1, most of the genomes were 94 to 97% complete as predicted by BUSCO [32], only T. longibrachiatum displaying a lower value (86%). The absence of a gene in the latter species was therefore treated with caution if it was found in all other species from SL.
Table 1.
Properties of the Trichoderma genomes and gene distribution
| Clade | Species | Strain | Genome size (Mbp) | Total genes | Complete-ness (%) | Fragmen-ted (%) | Missing (%) | Orthologs and paralogs |
|---|---|---|---|---|---|---|---|---|
| Longibrachiatum | T. reesei a | QM6a | 32.7 | 9877 | 96.9 | 2.4 | 0.7 | 8090 |
| RUT C30 | 34.2 | 10877 | ||||||
| T. longibrachiatum | ATCC18648 | 31.74 | 10938 | 86.3 | 7.9 | 5.8 | 8229 | |
| T. citrinoviride | TUCIM 6016 | 33.2 | 9737 | 94.1 | 3.1 | 2.8 | 7834 | |
| T. parareesei | CBS125925 | 32.07 | 9292 | 95.3 | 3.7 | 1 | 8328 | |
| Harzianum/Virens Clades | T. harzianum | CBS 226.95 | 40.9 | 14095 | 98.1 | 1.4 | 0.5 | 9921 |
| TR257 | 39.4 | 13932 | 97.2 | 2 | 0.8 | 9870 | ||
| T. afroharzianum | T6776 | 39.7 | 11297 | 95.1 | 1.8 | 3.1 | 9541 | |
| T. guizhouense | NJAU4742 | 38.8 | 11297 | 98.3 | 1.2 | 0.5 | 9246 | |
| T. virens | Gv29–8 | 40.52 | 12427 | 97.8 | 1.9 | 0.3 | 9795 | |
| Trichoderma | T. atroviride | IMI 206040 | 36.4 | 11863 | 97.5 | 2.1 | 0.4 | 9301 |
| T. gamsii | T6085 | 37.9 | 10709 | 94.1 | 2.1 | 3.8 | 8825 | |
| T. asperellum | CBS433.97 | 37.66 | 12586 | 97.9 | 1.4 | 0.7 | 9143 | |
| T. hamatum | GD12 | 38.43 | 10520 | 98.6 | 0.7 | 0.7 | 9030 |
a numbers show data (from left to right) obtained in this paper and by Li et al. [27] (if available)
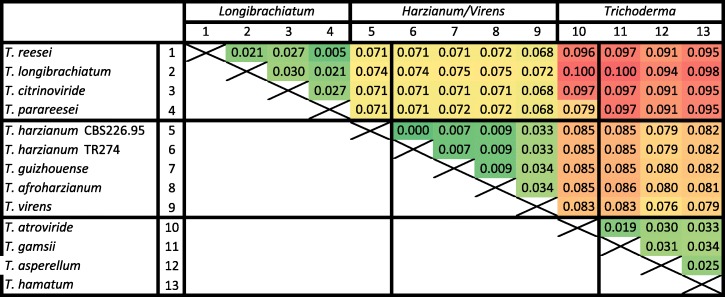
The average protein sequence similarity within Trichoderma orthologues ranges from 90% (SL vs ST) and 92.5% (SL vs HV) to 97–99% within species from the same sections/clades (Table 2). Amino acid similarity with other Hypocreales was still high (75–78.4%), but considerably lower with Neurospora crassa and Chaetomium globosum (58–60%) (Additional file 5).
Table 2.
Pairwise genetic distance between orthologous proteins from 13 Trichoderma strainsa
acolors show relative high (red), intemediate (yelow) and low (green) values
Evolution of the twelve Trichoderma species
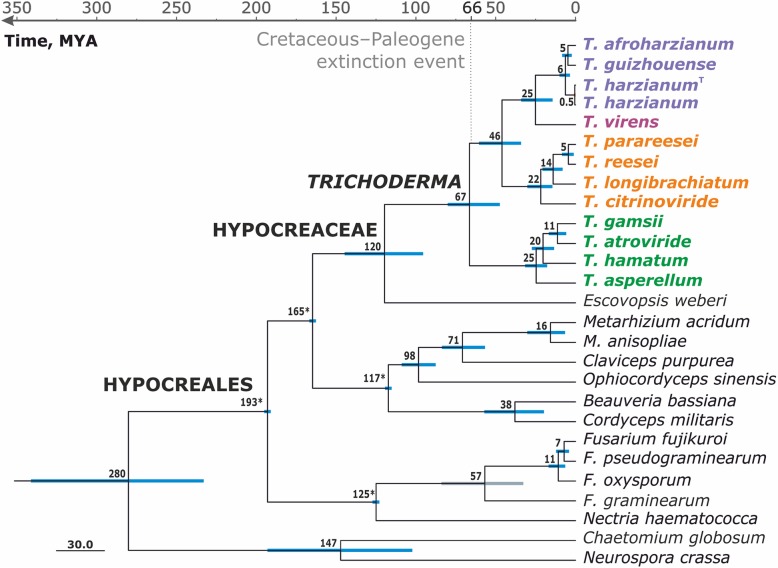
To learn the evolution of Trichoderma, we subjected the 13 strains (twelve species), twelve other fungi of the Hypocreales, and two phylogenetically more distant Sordariomycetes - N. crassa, C. globosum (outgroup) - to a time-scaled phylogenetic analysis using 638 orthogous genes (see Methods). The resulting tree (Fig. 3) shows that Trichoderma evolved 66.5 (± 15) mya, next to the Cretaceous-Paleogene (K-Pg) extinction event characterized by massive extinction of plants and large animals [33]. The two sections (SL, ST) and the HV clade of extant Trichoderma species appear to have arisen 25–21 mya what corresponds to the late Paleogene/early Miocene. The tree thereby confirms the ancient nature of section ST [12], and documents that SL and HV are monophyletic and evolved later. The divergency between the two strains of T. harzianum (i.e. one from Europe, UK, and one from South America, Brazil) was calculated to have occurred 460,000 years ago. Speciation of T. afroharzianum and T. guihouense can be dated around 5–6 Mya, which is comparable to that between T. reesei and T. parareesei, what justifies their recognition as separate species [28].
Fig. 3.
Bayesian chronogram obtained based on the concatenated alignment of 638 core orthologous proteins of Hypocreales and the two other Sordariomycetes. All nodes were supported with PP = 1. Chronological estimations are given in a geological time scale in Mya, and the numbers represent the corresponding node age. Numbers with asterisks at nodes indicate calibration points against the origin of Hypocreales (see Methods for details). Bars correspond to 95% confidence interval in time estimation based on the lognormal relaxed clock
Trichoderma gene inventory
To analyze and compare the gene inventory of Trichoderma, all putative proteins from the thirteen Trichoderma genomes and the fourteen other Sordariomycete species shown in Fig. 3 were analysed by the Markov Cluster Algorithm (MCL). We detected 19,332 clusters (Additional file 6), of which 7923 clusters contained genes from at least two Trichoderma species from each section. Further, 2095 clusters were shared by species from one or two sections/clades. HV exhibited the highest number of unique genes and also shared the highest number of genes with ST (Table 3). No PFAM domain could be assessed to 1980 of the above 19,332 clusters of which 1485 were present in single copies in each strain (or absent from only one species) and therefore putative orthologs.
Table 3.
Distribution of Trichoderma genes in sections, clades and species
| Clade | Species | Present in | Absent from | Total: | ||||
|---|---|---|---|---|---|---|---|---|
| All clusters with at least one gene from Trichoderma | At least two species from each clade | the clade only | SL | HV | ST | |||
| 13,089 | 7923 | 80/745/286 | 1083 | 68 | 335 | |||
| SL | T. reesei | 8775 | 8176 | 80 | 38 | 232 | 8526 | |
| T. longibrachiatum | 8636 | 7951 | 80 | 46 | 249 | 8326 | ||
| T. citrinoviride | 9205 | 8436 | 81 | 48 | 275 | 8840 | ||
| T. parareesei | 8757 | 8105 | 81 | 37 | 234 | 8457 | ||
| HV | T. harzianum T | 12737 | 9419 | 778 | 1038 | 300 | 11535 | |
| T. harzianum | 12698 | 9392 | 763 | 1016 | 296 | 11467 | ||
| T. guizhouense | 10996 | 9036 | 533 | 826 | 230 | 10625 | ||
| T. afroharzianum | 10811 | 8802 | 500 | 867 | 232 | 10401 | ||
| T. virens | 11474 | 9341 | 410 | 875 | 276 | 10902 | ||
| ST | T. atroviride | 10738 | 9009 | 267 | 841 | 54 | 10171 | |
| T. gamsii | 10039 | 8501 | 267 | 780 | 31 | 9579 | ||
| T. asperellum | 10595 | 8886 | 266 | 839 | 47 | 10038 | ||
| T. hamatum | 10164 | 8694 | 241 | 833 | 29 | 9797 | ||
MCL clusters with the highest number of genes (> 2000 in all 13 Trichoderma strains) comprised those encoding the fungal-specific Zn2Cys6 transcriptional regulators, solute transporters of the major facilitator superfamily (MSF) and short-chain dehydrogenases/reductases (SCDR) (Table 4). In addition, the 13 Trichoderma genomes had more than thousand genes that encoded proteins with ankyrin repeats, alpha/beta type hydrolases, protein kinases, zinc-dependent alcohol dehydrogenases, FAD-binding oxidases, methyltransferases and AAA + -ATPases. With respect to clade-specific distribution, the highest number of genes for individual protein families were present in species of HV and ST and were in 1.5–2-fold excess over those in SL. Proteins with a NmrA domain (regulators of GATA-type transcriptional regulators; [34]) were present in HV and ST even in threefold numbers (Table 4). Eighty percent of the genes encoding ankyrins, heteroincompatibility (HET) proteins, zinc-dependent alcohol dehydrogenases, cytochrome P450 monooxygenases and NmrA-like proteins were present in multigene clusters, suggesting that their evolution involved gene duplications. On the other hand, gene clusters encoding protein kinases, AAA + -ATPases, amino acid transporters, DEAE-box helicases or proteins wth an RMM_1 (ribonucleotide reductase M1) domain occurred in the same number in all species (Table 4).
Table 4.
PFAM group members with more than 500 genes in the 13 Trichoderma isolates
| clusters | genes per cluster | total genes | genes/ species | HV/SLa | HV/STa | ST/SLa | singletonsb | multiplesc | C/Sd (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zn2Cys6 transcriptional regulators | PF04082 | 238 | 12.3 | 2929 | 225.3 | 1.69 | 1.21 | 1.40 | 84 | 154 | 64.7 |
| MFS permeases | PF07690 | 172 | 17.3 | 2972 | 228.6 | 1.55 | 1.06 | 1.46 | 67 | 105 | 61.0 |
| short-chain dehydrogenases/reductases | PF00106 | 121 | 17.6 | 2129 | 163.8 | 1.68 | 1.18 | 1.42 | 46 | 75 | 62.0 |
| ankyrin-containing proteins | PF00023 | 106 | 14.4 | 1524 | 117.2 | 1.84 | 1.05 | 1.75 | 20 | 86 | 81.1 |
| alpha-beta-hydrolases | PF00561, 07859, 02230 | 96 | 14.4 | 1382 | 106.3 | 2.02 | 1.27 | 1.59 | 29 | 67 | 69.8 |
| protein kinases | PF00069 | 98 | 12.4 | 1218 | 93.7 | 1.15 | 1.11 | 1.03 | 60 | 38 | 38.8 |
| zinc-dependent alcohol dehydrogenases | PF00107 | 66 | 17.6 | 1159 | 89.2 | 1.89 | 1.17 | 1.61 | 15 | 51 | 77.3 |
| FAD-binding oxidases | PF01494, 01565 | 84 | 13.8 | 1158 | 89.1 | 1.56 | 1.21 | 1.29 | 25 | 59 | 70.2 |
| methyltransferases | PF00891 | 91 | 12.7 | 1157 | 89.0 | 1.24 | 1.09 | 1.14 | 46 | 45 | 49.5 |
| AAA + -ATPasesAAA+ − ATPases | PF00004 | 85 | 13.3 | 1130 | 86.9 | 1.13 | 1.06 | 1.07 | 54 | 31 | 36.5 |
| cytochrome P450 monooxygenases | PF00067 | 59 | 15.9 | 940 | 72.3 | 1.65 | 1.69 | 0.97 | 14 | 45 | 76.3 |
| sugar transporters | PF00083 | 65 | 14.2 | 923 | 71.0 | 1.53 | 1.17 | 1.31 | 33 | 32 | 49.2 |
| ABC-transporters | PF00005 | 48 | 16.3 | 780 | 60.0 | 1.22 | 1.13 | 1.07 | 21 | 27 | 56.3 |
| vegetative heteroincompatibility (HET) proteins | PF06985, 07217, 17,108 | 72 | 10.5 | 753 | 57.9 | 1.94 | 1.24 | 1.56 | 13 | 59 | 81.9 |
| aminotransferases | PF01490 | 49 | 13.8 | 675 | 51.9 | 1.47 | 1.18 | 1.25 | 21 | 28 | 57.1 |
| amino acid permeases | PF00324 | 48 | 13.1 | 630 | 48.5 | 1.17 | 1.10 | 1.06 | 30 | 18 | 37.5 |
| amidases | PF01979, 04909 | 37 | 17.0 | 629 | 48.4 | 1.93 | 1.20 | 1.62 | 12 | 25 | 67.6 |
| acetytransferase | PF00583, 00797, 13,302, 13,523 | 49 | 12.2 | 600 | 46.2 | 1.26 | 1.10 | 1.15 | 23 | 26 | 53.1 |
| DEAD-box helicases | PF00270 | 42 | 13.5 | 567 | 43.6 | 0.97 | 0.96 | 1.01 | 33 | 9 | 21.4 |
| NmrA-like proteins, NAD-binding negative regulators of GATA-binding proteins | PF05368 | 41 | 13.5 | 552 | 42.5 | 3.01 | 1.11 | 2.70 | 10 | 31 | 75.6 |
| DnaJ molecular chaperone | PF00226 | 42 | 12.8 | 537 | 41.3 | 1.02 | 0.99 | 1.04 | 19 | 23 | 54.8 |
| RRM_1 RNA binding proteins | PF00076 | 42 | 12.8 | 537 | 41.3 | 1.00 | 0.99 | 1.01 | 40 | 2 | 4.8 |
a -ratio of the number of genes in all species belonging to one of the Trichoderma sections or clades
b - genes which are present in one a single copy per cluster
c - genes that occur in more than one copy per cluster in at least one species
d - percentage of clusters containing multiple genes
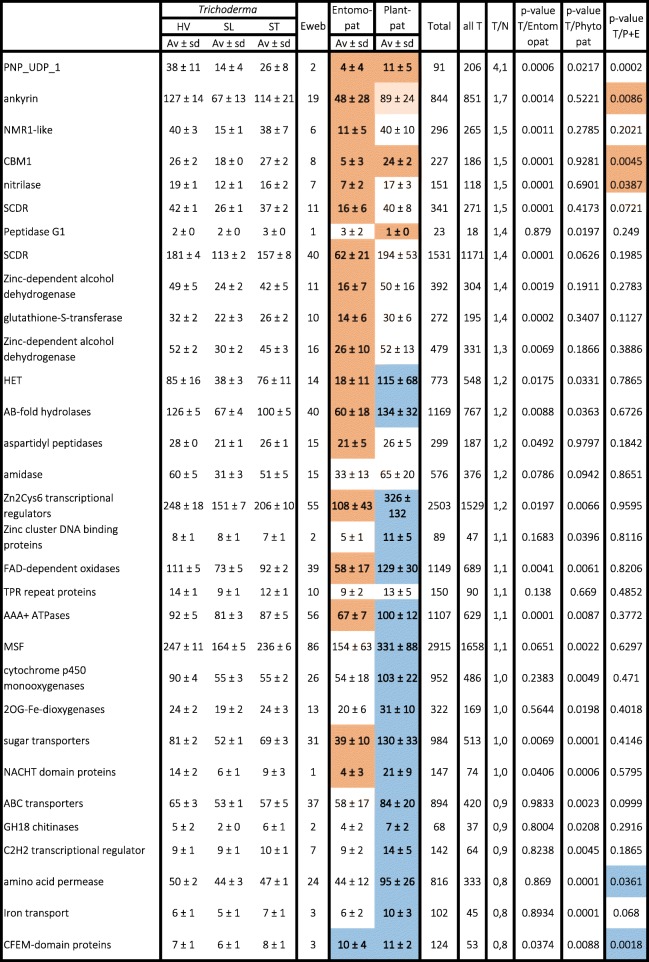
A comparison of the clusters found in Trichoderma to the other 14 Sordariomycetes showed that 2584 gene clusters were present in all Trichoderma spp. and at least one of the other 14 Sordariomycetes (Table 5; Additional file 6). Of them, eighty-six clusters were shared exclusively between Trichoderma and the entomoparasitic Hypocreales, and 700 clusters shared only between Trichoderma and the Nectriaceae. Genes encoding purine/uridine nucleoside phosphorylases, and CBM1-type cellulose binding domains were enriched in Trichoderma, and genes containing ankyrin-containing proteins were also significantly more abundant although they did not gain scientific support with respect to the phytopathogens. The majority of the other gene families were significantly more abundant in Trichoderma than in the entomopathogenes, but many of them less abundant than in the phytopathogens (Table 5; Additional file 7). Some gene families (HET- and NACHT domain proteins, AB-hydrolases, Zn2Cys6 transcriptional regulators, FAD dependent oxidases, AAA+ ATPases, transporter of the major facilitator superfamily, sugar transporters, cytochrome P450 monooxygenases and 2-oxoglutarate-dependent (FeII)-dioxygenases) were significantly more abundant in Trichoderma than in the entomopathogenes but clearly less abundant than in the phytopathogenes (Table 5). Genes encoding proteins with CFEM-domains were present in a lower number in Trichoderma than the entomo- and phytopathogenes.
Table 5.
OrthoMCL clusters shared between Trichoderma and other Sordariomycetes Fungi
Entomopat. - six species of Entomopathogenic Hypocreales, Plant pat. - five species of plant pathogenic Hypocreales, Eweb - Escovopsis weberi, Av ± sd – average ± standard deviation; For strain abbreviations, see Methods. T - Trichoderma, N, Nectriaceae. PFAM categories printed in bold specify those that are significantly (ANOVA coupled with Dunnett’s post-test, P < 0.05) different compared with Trichoderma species
Brown background: enriched in Trichoderma; blue background: less abundant in Trichoderma
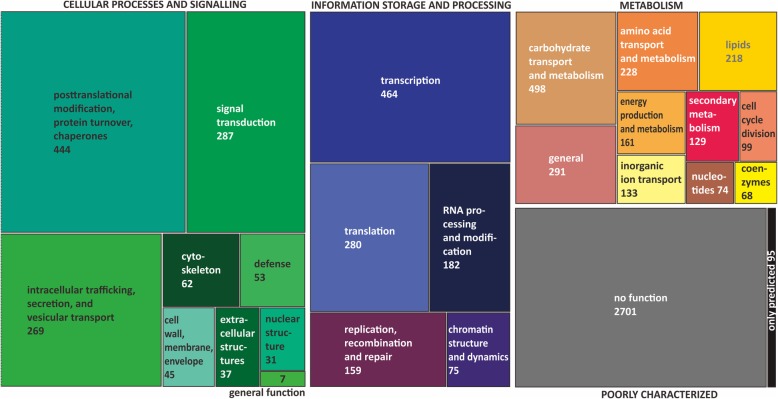
The Trichoderma core genome
Exactly seven thousand genes had orthologs in all twelve Trichoderma species, and therefore represent the Trichoderma core genome. The automatically predicted encoded proteins were manually curated (Additional file 8) and at least a putative function based on a conserved protein domain could be attributed to 4413 of them. Using the KOG (eukaryotic orthologous groups) classification scheme [35], the genes classified as “metabolism” (1809) and “poorly characterized” (2587) constituted the two largest groups (Fig. 4). At the level of individual KOG families, “posttranslational modification, protein turnover and chaperones”, “transcription” and “carbohydrate transport and metabolism” contained more than 400 genes (Fig. 4). Genes encoding glycoside hydrolases (191) and fungal specific Zn2Cys6 transcription factors (173) were the most abundant protein families in the Trichoderma core genome (Table 5), followed by glycoside transferases [36], and C2H2-type transcription factors (45). Among the group of proteins for which only a general function could be predicted, solute transporters of the major facilitator superfamily (151), and short-chain dehydrogenases/reductases [37] contributed to a major part of the KOG group “Metabolism”.
Fig. 4.
The structure of Trichoderma core genomes as revealed based on 13 strains. The number of genes of the core genome for which a KOG classification was obtained. The total number of genes in the core genome is 7000. The size of the boxes represents the abundance of the genes within the main KOG classifications (Cellular processes and signaling – green shades; Information storage and processing – violet shades; Metabolism – reddish shades; Poorly characterized -Grey shade. Predicted ORFs are shown in black). The numbers specify the numbers of core genome genes that belong to the respective functional groups
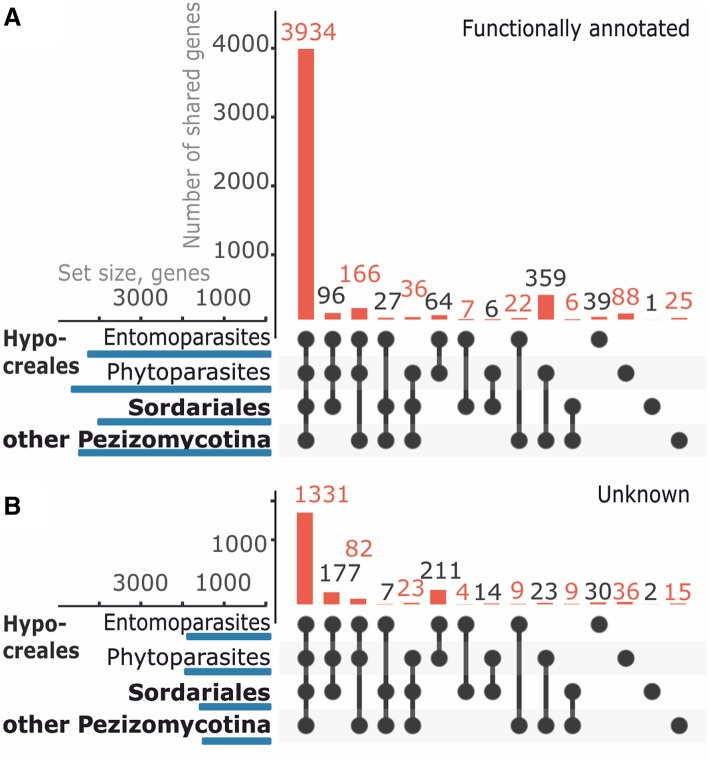
We compared the Trichoderma core genome to the complete genome of other fungi (nine of the Sordariomycetes investigated above, two species from the Eurotiomycetes, one of the Dothidiomycetes and one of the Leotiomycetes; see Methods) using a reciprocal BLAST approach and Intervene [38] (Fig. 5). This showed that 3934 of those 4413 genes of the core genome, for which a putative function could be identified during manual annotation, had orthologs in all of them (Fig. 5). In addition, 642 were shared between a subset of the fungal orders and families tested. The largest number (359) was shared between Trichoderma and all other fungi except for the entomoparasites, and 166 genes were only present in the Sordariomycetes but not in fungi from other classes.
Fig. 5.
The share of Trichoderma core genome with genomes of other fungi. Genes of the Trichoderma core genome which have orthologs in other fungi (a, functionally annotated genes; b, unknown genes). The analysis was performed with Intervene [39]. The vertical bars and numbers indicate the number of genes that are shared by the fungal groups as indicated by the circles below the graph. The horizontal bar over the fungal groups indicates the total number of genes with orthologs in Trichoderma. Hypocreales entomoparasites were estimated based on the genomes of Beauveria bassiana, Cordyceps militaris, Metarhizium acridum and M. robertsii; Hypocreales phytoparasites were estimated based on the genomes of Fusarium graminearum, Nectria haematococca, and F. verticillioides; for Sordariales the genomes of Neurospora crassa and Chaetomium globosum were used. Other Pezizomycotina were assessed based on the analysis of the genomes of Chochliobolus heterostrophus, Exophiala xenobiotica, Aspergillus fumigatus, A. oryzae and Oidiodendron maius (see methods for details)
A similar search for the functionally uncharacterized proteins revealed that 1331 of them were shared with all other fungi. The number of those shared only between some orders or families suggests a phylogenetic relationship: 211 of them were present only in the Hypocreaceae, and 177 in all Sordariomycetes (Fig. 5).
We conclude from these data that 80.7% of the genes encoding functionally predictable proteins and 67.4% of the genes encoding functionally not predictable proteins in the Trichoderma are already been present in the ancestor of Eurotiomycetes and Sordariomycetes and are therefore at least 250 million years old.
Comparing the intraspecific genome differences between the two isolates of T. harzianum showed that 1699 genes of T. harzianumT (12%) were absent from the other strain, and 1419 genes present in the latter (10.1%) absent from the type strain. Most of these genes encoded orphan proteins for the species, and a function could only be predicted for 158 and 160 genes in T. harzianumT and T. harzianum TR247, respectively. Their properties are described in Additional file 9.
We also compared the genomes of T. longibrachiatum and T. citrinoviride - the two species that are more frequently encountered as opportunistic pathogens of immunocomproized humans [40] - and identified 94 genes that were only present in these two species but absent from all others and could therefore belinked for their pathogenicity (Additional file 10).
105 genes of the core genome were present in all 12 Trichoderma species but not found in any other fungus. They thus fulfil the criterium of “genus specific orphans” and we will use the term “orphans” for them further throughout the manuscript. No function could be predicted for any of these genes.
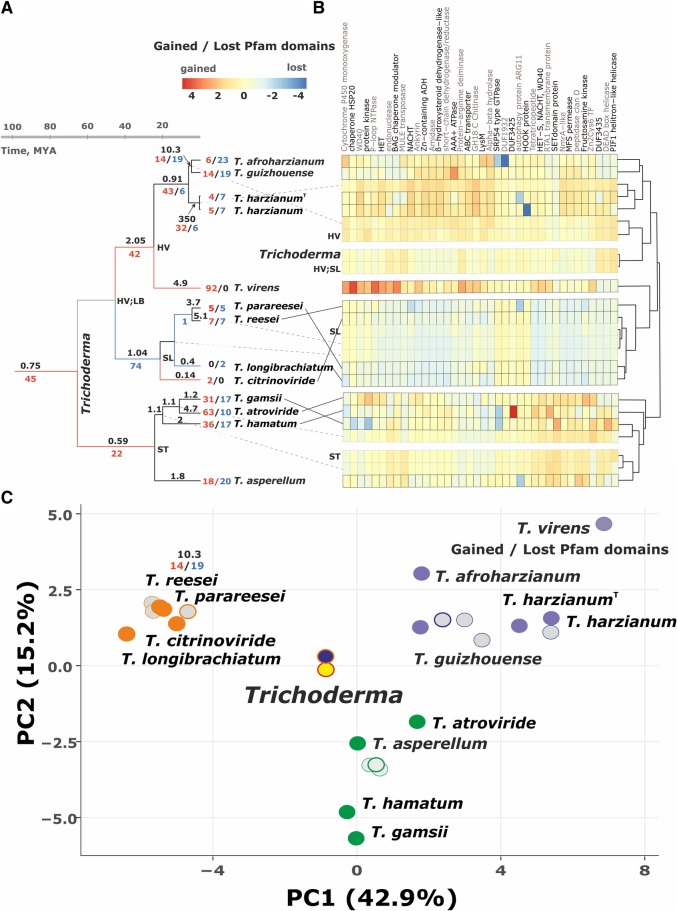
Gene expansion and contraction during evolution of Trichoderma species
We used the likelihood approach implemented in CAFÉ to identify individual gene families that evolved at rates of gain or loss that were significantly higher than the genome-wide averages in Trichoderma [41]. As shown in Fig. 6a, the origin of Trichoderma from its ancestor started with significant gene expansion (13 gene clusters comprising 45 genes gained, and none lost), and this expansion continued in the ancestors of ST (22 genes) and HV (42 genes) (Fig. 6). The rate of gene changes per mya increased significantly during the evolution of HV. The highest number of gene families gained by the Trichoderma ancestor were those encoding HET/NACHT and ankyrin proteins, and these two groups became further enriched during the evolution of HV and ST (Fig. 6b). The ancestor of SL, on the other hand, lost 74 genes and did not gain any. Interestingly, they also comprised a high number of genes for ankyrin and HET/NACHT domain proteins.
Fig. 6.
Genome evolution in Trichoderma. a time scaled evolutionary tree: red branches indicate only gains; blue branches only losses; black branches both gains and losses. Numbers over the branches indicate the number of gene changes per Mya; numbers below the branches indicate the number of gains (red) and losses (blue). b Heat map representing Pfam domains identifiend for OrthoMCL clusters that we gained or lost in the course of Trichoderma evolution. Framed rectangules correspond to extant species. Pale color used for hypothetical taxonomic units (HTUs, ancestral states). c. Principal component analysis based on the number of genes per each Pfam group that have been influenced by gene gain and loss in Trichoderma. Filled cicles correspond to extant Trichoderma species as shown on A. Grey circles correspond to HTUs per each infrageneric group (see a and b). Bold lined circles show the ancestral node for the respective section or clade. Circles with red/yellow and orangy/blue color show the ancestral node for the genis Trichoderma and SL/HV groups
While these data show that the origin the genus Trichoderma and two of its clades/sections (HV, ST) underwent strong gene expansion whereas SL exhibits significant gene contraction, a deeper look into the gene evolution at the level of individual species revealed a mosaic of gain and loss events (Fig. 6a and b). Exceptions were T. longibrachiatum which shows only gene losses (but these data must be viewed with caution because of the higher incompleteness of its genome; see above), and T. citrinoviride which displays only gains. These data suggest that the extant taxa of Trichoderma are reforming their genomes at an increased rate, which is particularly reflected in T. harzianum because the two isolates of this species differed remarkably in their gene loss and gain.
The principal component analysis revealed that the tree different strategies in gene gain and loss that are characteristic for each section or clade (Fig. 6c). As all the tested species are nutritionally versatile, common and cosmopolitan, this pattern of group-specific evolution points to the importance of the core genome is the basis for the generalism.
Since the evolution of the Trichoderma genomes from their ancestor from 120 (±21) to 66 (±15) mya occured entirely by gene expansion (no gene losses revealed by the CAFÉ analysis, Figure 6a,b), we wondered whether this was due to a small genome in its putative ancestor. We therefore extended the CAFE analysis to all available Hypocreales genomes. Unfortunately, at the 99% probability used for Trichoderma, this analysis yielded no data which is probably due to the insufficient number of genomes that are currently available for the predictions over such long evolutionary interval. Reducing the probability level to 95%, however, revealed that the evolution after the split from the entomoparasite branch (184.6 ± 8 mya; see Fig. 3) and the obligate mycoparasite Escovopsis weberi (119.8 ± 21 mya) was accompanied by a total of 23 gain losses and only a single gain (Additional file 11). The ancestors of the genus Trichoderma may therefore have indeed been subject to a significant genome contraction.
The Trichoderma genomes reveals the potential for heterothallic sexual reproduction
Most species of Trichoderma are found in nature in their sexual form (teleomorph) [8, 9], although the most generalist species are frequently and some even exclusively been isolated as anamorphs. Among the strains investigated in this paper, teleomorphs were only known for T. reesei (most frequent), T. citrinoviride (frequent), T. virens (very rarely) and T. atroviride (rarely) [42–44]. Population genetic evidence for the absence of sexual recombination has been shown for T. longibrachiatum, T. parareesei, T. harzianum and T. afroharzianum [29, 45]. The structure of the population of other species is not known. We therefore looked for the presence of mating type genes in the thirteen strains. As can be seen in Table 6, we found either MAT1–1 or MAT1–2 idiomorphs in all of them, consistent with the view that Trichoderma is heterothallic. The distribution of the known sexually recombining and non-recombining species investigated in this study on the phylogenetic tree suggests that the gain or loss of this trait occurred several times during the evolution of Trichoderma.
Table 6.
Mating type genes in Trichoderma
| mating protein | mating protein | mating protein | mating protein | |
|---|---|---|---|---|
| MAT1-2-1 | MAT1-1-1 | MAT 1-1-2 | MAT 1-1-3 | |
| T. reesei | 124341 | |||
| T. longibrachiatum | 1427955 | 1467528 | 1388533 | |
| T. citrinoviride | 1107806 | |||
| T. parareesei | -b | -b | OTA08401 | |
| T. harzianum T | 104176 | |||
| T. harzianum | 434806 | 863060 | 863056 | |
| T. guizhouense | OPB38549 | |||
| T. afroharzianum | KKO 5631 | |||
| T. virens | 60622 | |||
| T. atroviride | 33998 | |||
| T. gamsii | TGAM01v2_08385 | |||
| T. asperellum | 64910 | 158842 | 451243 | |
| T. hamatum | 12232 | 12231a | 12231a |
a annotated as one protein
b no ortholog detected by Blastp against NCBI database
Sensing of a potential mating partner is a prerequisite for sexual reproduction and fulfilled by the pheromone system [46]. The genes involved in this process were found in all Trichoderma spp. and are given in Additional file 12.
Major aspects of Trichoderma metabolism
Carbon metabolism
Carbon metabolism of Trichoderma has so far mainly been studied in T. reesei only and with respect to the catabolism of hemicellulose and pectin monomers [39, 47, 48]. We have therefore manually annotated all genes of the core genome that are putatively involved in carbon metabolism. The majority of these genes has already been described in detail for T. reesei, T. atroviride and T. virens, and we refrain from repeating these data here [25]. Yet we detected some novel features, such as the presence of an extracellular glucose oxidase, D-xylulose-5-phosphate/D-fructose-6-phosphate ketolases, enzymes for D-erythroascorbic acid biosynthesis, and a glutathione-linked methanol degradation pathway. These findings are described in some detail in Additional file 13.
Extracellular polymer hydrolysis
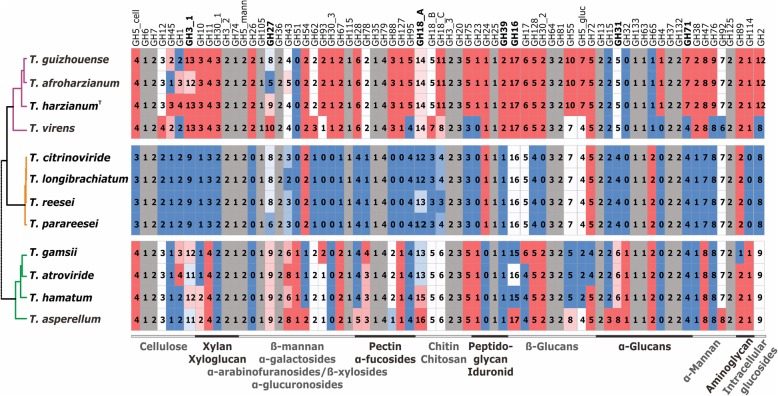
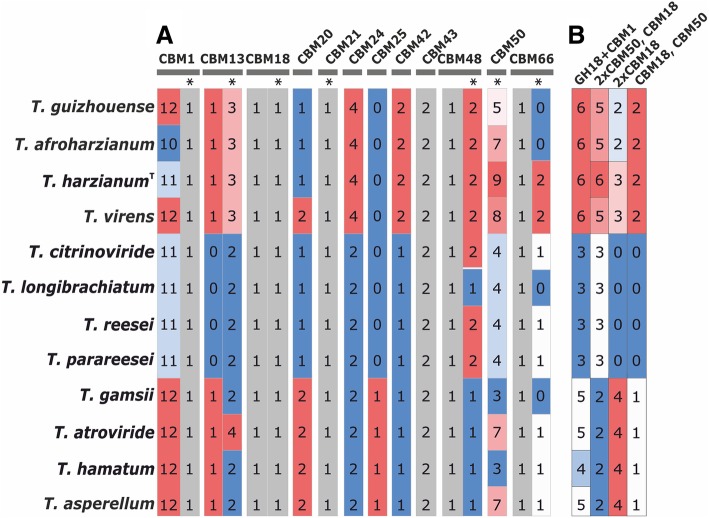
A unique feature of filamentous fungi is that they live in their mostly macromolecular substrates, and therefore must have efficient extracellular systems for the hydrolysis of the respective polymers (mainly polysaccharides and proteins). In Trichoderma, this is nicely reflected in the glycoside hydrolases (GHs) which comprise one the most abundant groups of its genome. In addition, accessory enzymes for the GHs (polysaccharide lyases, polysaccharide binding proteins, carbohydrate esterases, and auxiliary oxidative enzymes), which are classified in the CAZy (carbohydrate active enzymes) database [49] are required to aid in the hydrolysis of the respective polymeric substrates. The distribution of these genes in individual Trichoderma taxa is shown in Figs. 7 and 8, Additional file 14:.as can be seen, Trichoderma comprises between 269 and 361 CAZyme genes (glycoside transferases not counted), of which 195 were found in the core genome. GHs accounted for more than 50% of the CAZymes, GH18 chitinases, GH16 ß-1,3/1,4-glucanases and GH3 ß-glycosidases being present in the highest numbers. When the GHs are sorted according to the type of their polysaccharide substrates, GHs acting on chitin and ß-glucan again comprised the highest numbers, followed by enzymes acting on α-mannan (Fig. 7). Many of the latter are probably involved in cell wall and glycoprotein modification. The function of these genes and their distribution in the 12 Trichoderma species is described in detail in Additional file 14. Interestingly, the composition of CAZomes (estimated based on the respecive GH families) is almost invariable in SL, although the phylogenetic analysis revealed genetic distances between species in this section to be similar to those between species in other groups (compare the CAZyme-based cladogram on Fig. 7 and the phylogram on Fig. 3). The CAZymes of HV is significantly enriched compared to ST and SL. This enrichment is not biased towards a special substrate.
Fig. 7.
The glycosyl hydrolase (GH) inventory of Trichoderma. GHs are ordered according to their substrate (in broader sense), which is given on the right side. GHs which contain enzymes that act on different substrates (GH3, GH5, GH30) are indicated by extended numbers. GH18 chitinases are grouped into A (no binding domain), B (attached CBM1) and C (attached GH18 and/or GH50; see Fig. 8). Numbers mean the number of genes belonging to the respective GH family in their genomes. The cladogram on the left is based on the total number of genes per each GH family, complete linkage, Eucleadian distance
Fig. 8.
Type and presence of carbohydrate binding domains in Trichoderma. a The summary of domains: those columns marked with an asterisk indicate individual domains, i.e. domains which occur as separate proteins and are not attached to another enzyme. b Patterns of CBMs in GH18 chitinases
Apart of polysaccharides, proteins hydrolyzed by various proteases provide a major nutritional source for fungi. Some of protease families are also important for the digestion of proteins secreted by competing organisms [50–52] or hosts. We screened the secretome of the 12 Trichoderma genomes for proteases using the MEROPS database (see Methods for details). This demonstrated the presence of A1 aspartyl proteases, G1 eqolisins (previously termed “pepstatin-insensitive aspartyl proteases”), C13 legumain-type cysteine proteases, eight metalloprotease families (InhA-like peptidases, M6; carboxypeptidases, M14; glutamate carboxypeptidases, M20; methionine aminopeptidases, M24; aminopeptidase Y, M28; deuterolysin, M35 and fungalysin, M36), and six families of serine proteases (S1 chymotrypsins, S8 subtilisins, S10 carboxypeptidases, S28 an S51 dipeptidases, and S53 sedolisins) in Trichoderma (Additional file 15). Aspartyl proteases, subtilisins, sedolisins, and aminopeptidase Y were present in the highest numbers of isoenzymes. Family S10 was particularly abundant in HV, and S53 in HV and ST. In summary, however, the number of Trichoderma proteases is comparable to that of many other fungi [50–52], and we found no protease family that was specifically expanded or contracted in Trichoderma. Proteases have been speculated to be a component allowing niche differentiation between the ascomycetes and the basidiomycetes, particularly towards adaptation to pathogenicity by the former [52]. However, our data suggest that the primeval proteolytic arsenal of Trichoderma was sufficient for the acquisition of the mycoparasitic lifestyle and its more recent expansion towards generalism.
Secondary metabolism
Secondary metabolites (SM) are an intrinsic feature of most Pezizomycotina, because they participate in cellular signalling, competition, pathogenicity, and metal ion uptake [53]. Trichoderma too has been shown to be a proliferic producer of SMs [54, 55]. Unfortunately, the genes encoding these SMs and even the species identity of the SM producing isolates are in most cases unknown. We identified 10–25 polyketide synthase (PKS), 12–34 non-ribosomal polypeptide synthetase (NRPS)-, and 6–14 terpenoid synthase (TS) encoding genes in the 12 species (see Additional files 16 and 17), of which 6 PKS, 10 NRPS and 3 TS genes were present in the core genome.
In contrast to PKS, NRPS and TS, Trichoderma seems not to synthesize alkaloids, as we could not find the genes encoding the precursor dimethylallyl tryptophan synthases (DMATS; [56]) in any of studied genomes.
Small cysteine-rich secreted proteins
Fungi have developed several families of small, secreted proteins that are characterized by an enhanced content of cysteines, and which are believed to function in various ways in the communication between the fungi and other organisms [57]. Three different protein families can be distinguished in this group: small (< 300 amino acids) secreted and cysteine rich proteins (SSCPs); hydrophobins; and cerato-platanins (Table 7). The number of so detected SSCPs was surprisingly diverse (Additional file 18), but the variation was species- and not section-specific. Trichoderma is rich in hydrophobins, ranging from 7 in all species of section SL to 16 in T. atroviride. Six of them are conserved across all twelve Trichoderma species (see Additional file 18). As for cerato-platanin proteins, which represent fungal-specific, small and secreted proteins that are believed to be important for interaction with other organisms and eliciting defense reactions in plants [58–60], three genes (epl1, epl2, epl3) are present in the core genome. A detailed description of the small cysteine rich proteins is presented in Additional file 18.
Table 7.
Number and types of small secreted cystein-rich proteins in Trichoderma
| SSCPs | HFBs | Cerato-platanins | |||
|---|---|---|---|---|---|
| class II | pseudo-class I | ||||
| SL | T. reesei | 39 | 7 | 3 | |
| T. longibrachiatum | 89 | 7 | 3 | ||
| T. citrinoviride | 50 | 7 | 3 | ||
| T. parareesei | 27 | 7 | 3 | ||
| HV | T. harzianum | 113 | 12 | 3 | 3 |
| T. afroharzianum | 66 | 9 | 3 | 3 | |
| T. guizhouense | 44 | 10 | 2 | 3 | |
| T. virens | 65 | 12 | 3 | 3 | |
| ST | T. atroviride | 75 | 13 | 3 | 3 |
| T. gamsii | 42 | 12 | 3 | 3 | |
| T. asperellum | 125 | 11 | 3 | 3 | |
| T. hamatum | 62 | 11 | 3 | 3 | |
Trichoderma orphan genes
The Trichoderma core genome contained 105 orphan genes (vide supra; Additional file 19). While they comprised only 1.5% of the genes in the core genome, orphan genes restricted to sections/clades or evensingle species were much more abundant (on the average 17.4, 13.0 and 10.1% in SL, ST and HV, respectively; and even higher within the pool of species-specific genes (see also Additional file 3).
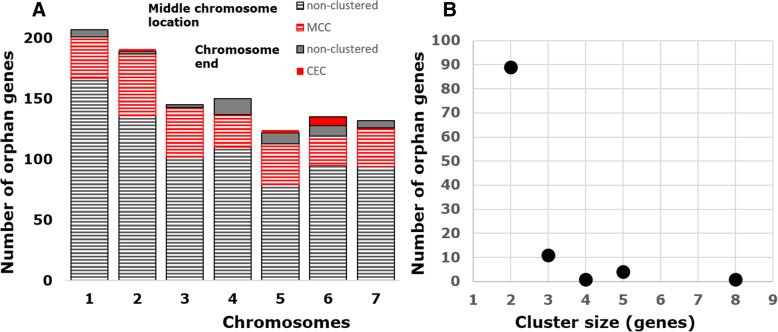
Subjecting the orphans to analysis in the conserved domain database [61] failed to detect any known domain. Orphans have been shown to occur in gene clusters and to be enriched in subtelomeric regions in Plasmodium, yeast, Aspergillus and Neurospora [62–64]. To test whether this is also the case for Trichoderma, we made use of the complete annotated chromosomes of T. reesei [26, 27], and mapped its 1126 orphans on them (Additional file 19). Most of them occurred as single genes, and less than 10% of them were located within 100 kb from the chromosome ends (Additional file 19). Twentyfive to 30% of the orphan genes indeed occurred in gene pairs or clusters which in majority comprised two, but sometimes more and in a single case even eight genes. The proportion of the clusters that occurred in subtelomeric regions was again lower than 10% (Fig. 9; Additional file 19), suggesting that – similarly to the single orphan genes - the clustered orphans of T. reesei are not enriched near the chromosome ends. Interestingly, none of the T. reesei orphan genes that were present in the core genome was located near the chromosome ends, suggesting that the latter location is a species- or clade-specific feature. We however note that in several cases single genes in the core genome occurred as doublettes or triplets in T. reesei.
Fig. 9.
Numbers and clusters of orphan genes in the chromosomes of T. reesei. a Numbers and clusters; “chromosome end” specifies those orphans that are located with 100 kb from either chromosome end; “middle” specifies those that are located in the remaining area. b Number of orphan genes in clusters of varying size. The T. reesei chromosomes, published by Li et al. [27] were used for these investigations
To analyse the evolution of the orphans, we measured the selection pressure acting on them by calculating the ratio of non-synonymous and synonymous amino acid substitution (dN/dS) for 53 orphan genes that are present in the Trichoderma core genome and whose nucleotide sequence could be unambiguously aligned. A dN/dS = 1 would indicate neutral evolution and dN/dS < 1 can be interpreted as evidence for purifying selection [65]. The data obtained (Additional file 20) show that most of these genes is under strong purifying selection. An assessment of dN/dS for the clade-specific orphans was not possible, because their nucleotide sequences were too polymorphic to be aligned.
Discussion
This study is based on the genome sequences of 12 of the most common Trichoderma species. Although this number represents only a few percent of Trichoderma spp. described today, the selected species are members of three phylogenetically distant sections and clades, and the results therefore enable a broader insight of the genus. Also, these species were most frequent in our own studies of soil or rhizosphere sampled in different geographic regions such as the Canary Islands, Sardinia, Columbia, Egypt, China, Israel, South-East Asia, Siberia and many others [6–8, 13] and may therefore be called cosmopolitan. Because several of the twelve species that were selected by this study are used as bioeffectors in biocontrol products against plant pathogenic fungi, stimulate plant growth and immunity, are opportunistic pathogens of immunocompromised humans and are causative agents of the green mold disease on mushroom farms [6, 13], they can be considered as environmental opportunists in a broad sense. Although species in the each of the sections and clades have unique morphological features, their overall ecological features are similar: they are mycoparasites, can feed on cellulolytic material, and can establish themselves in soil and colonize the rhizosphere. This may suggest that these species maintained the “opportunistic” features from a common ancestor what may be reflected in the core genome.
We have therefore investigated the evolution and the therefrom arosen changes in the gene inventory of the selected 12 species. Although all the genomes were still incomplete, the small predicted percentage of missing genes (2–5% for all species except T. longibrachiatum) makes it probable that we have identified all gene families that are relevant for the interpretations and conclusions in this paper. We particularly emphasize that the differences in gene numbers that we considered relevant were in most cases several folds higher than the number of putatively missed genes.
Our results reveal that the the mycoparasitic Hypocreales deversified between 100 and 140 mya, the ancestor of Trichoderma evolved around the time of the K-Pg, and the formation of the three infrageneric groups studied (ST, SL and HV) occured 40–45 mya after the K-Pg event. The uncertainty in chronological dating makes it impossible to decide whether the genus Trichoderma arose before or after K-Pg. However, we have recently proposed that the genus Trichoderma has obtained most of the genes encoding plant cell wall degrading CAZymes required for phytosaprotrophic growth, by the lateral gene transfer [11] that likely took place before the diversification into infrageneric groups. The most likely interpretation of these data is therefore that Trichoderma was one of the fungal genera that participated in the strong burst in fungal populations that fed on the decaying biomass of the plants killed by the K-Pg [66]. Whether or not this increase in the number of fungi stimulated mycoparasitism can only be speculated, but clearly a successful antagonism and the ability to kill its competitor may have aided Trichoderma in establishing a high population density on decaying plant biomass. Moreover, the ability to endoparasitise closely related species (up to adelphoparasitism) could favor host/parasite DNA exchanges and further contribute to the formation of the unique core genome of Trichoderma [11].
Despite the standard deviation in the dating of fungal phylogenies, our data strongly suggest that the evolution of the three Trichoderma sections/clades investigated in this study occured after the K-Pg event. The origin of extant species in the three sections/clades occured in the early oligocene (20–30 mya), a phase characterized by cooler seasons and a significant extinction of the invertebrate marine fauna [67]. It is intriguing that this split led to an increased rate of gene gain and genome expansion in HV, whereas the formation of SL was accompanied by a significant gene loss. Kelkar and Ochman [68] reported that Pezizomycotina genomes in the size of 25- to 75 Mb (which includes all Trichoderma spp. investigated in this study) exhibit a positive correlation between decreased genome size and increased genetic drift, and vice versa. On a first glance, this observation may not be applicable to genome contraction in SL, because it concerned genes from nearly all functional categories and thus was not specifically directed to support a certain trait. Alternatively, our results could be explained by the streamlining hypothesis [69], which considers selection for a more economical lifestyle as the major driving force for genome reduction. According to this model, the presence or absence of multiple genes for the same function may produce only a small effect on the performance of the organism and thus have only little benefits for the cell. Sun and Blanchard [70] considered that this scenario would most likely occur in relative stable environments where competition for nutrients is severe, and where a smaller genome has the ecological advantage of spending less energy for growth and development. We speculate that HV and ST – but not SL - used this alternative for further ecological success.
One of the hypotheses for this work was that gene families that were gained during Trichoderma evolution and are more abundant in Trichoderma than in other related fungi could give further insights about how this genus became an environmental opportunist. Gene families that were gained in highest number by Trichoderma were those encoding proteins with an ankyrin-repeat, proteins with a HET domain and MSF transporters. In addition, protein families that were present in higher numbers than in other Sordariomycetes were PNP_UDP_1 nucleotide phosphorylases, and NmrA-like transcriptional regulators.
The ankyrin repeats - tandemly repeated modules of about 33 amino acid that form two α-helices separated by a loop – are among the most common protein-protein interaction motifs known. They occur in a high number of proteins mainly from eukaryotes and have functions in cell cycle regulation, mitochondrial enzymes, cytoskeleton interactions, signal transduction and stress resistance [71, 72]. So far, proteins with ankyrin repeats have not been systematically characterized from Pezizomycotina, but an expansion of proteins containing ankyrin repeat proteins has been reported for the insect endosymbiotic bacterium Wolbachia [73]. Ankyrins have therefore been suggested to play an important role in endosymbiosis of this bacterium [73]. The higher number of proteins with this protein-protein interaction module in Trichoderma than in other fungi (with Nectria being the only exception) may suggest that its signalling and metabolic processes are more tightly coordinated than in other fungi which could ultimately result in enhanced fitness in its habitat.
Another group of proteins that made up for a significant portion of the genes gained by HV ans ST are the fungal HET (heterokaryon incompatibility) proteins. They have already received considerable attention of fungal genetisists because of their role as key players in recognition and response to non-self during cell fusion, which allows different individuals of the same species to maintain intergity and individuality [74–76]. HET proteins that contain an N-terminal HET effector domain, a central GTP binding site and a C-terminus consisting of highly conserved WD40 tandem repeats have been defined as HNWD protein family [77]. Lamacchia et al. [78] recognized that proteins of this family have similarity to pathogen-recognition receptors in plant and animals and proposed that these genes might also have a function in the recognition and response to other pathogenic species [78, 79]. With respect to Trichoderma, we extend this hypothesis by speculating that they could also play a role in recognition of mycoparasitic hosts, which is a challenging objective for further studies.
Apart of these two striking examples, the expansion of genes encoding NmrA-like proteins (which function as repressors of GATA-type transcription factors; [34]) and Zn2Cys6-transcriptional regulators in HV is also of interest, because we did not notice an expansion of protein kinase families. We therefore assume that speciation in this clade is accompanied by a diversification and fine tuning of transcriptional regulation, whereas regulation at the posttranscriptional level occurs mainly by the canonical signalling pathways in a similar way as in other fungi.
Based on the analysis of T. reesei, T. virens and T. atroviride it was previously concluded that the genus has only a small arsenal of secondary metabolite synthases [12]. The present comparison shows that this only true for the PKS and NRPS in species of SL. Compared with the Aspergillus spp., which are considered as being particularly rich in secondary metabolites [80], T. harzianum has a higher number of NRPS (twenty-nine). The number of PKS in T. virens is in the average [25] of those present in Aspergilli. In the case of terpenoid synthases, Trichoderma contains 12 to 17 genes and therefore clearly outnumbers the Aspergillus spp. that have only 2 to 10. However most of these secondary metabolite synthases – especially those for terpenoids - have not yet been characterized. The relation between the many secondary metabolites reported in Trichoderma and the genes responsible for their synthesis is therefore not known, which defines yet one more intriguing field for further studies.
We have also annotated the complete CAZome of all 12 Trichoderma species, which revealed the presence of some proteins like the GH4 α-glucosidases or the AA11 chitin monooxygenases that have not yet been described to occur Trichoderma. In addition, we detected that Trichoderma possesses a rich arsenal of carbohydrate-binding domains, which occur as fusions to GHs, CEs or AAs, but also as individual secreted proteins. The CBM1 cellulose-binding domain and the CBM50/LysM chitin/peptidoglycan binding domains have been already described in detail [81, 82], but we also found a high number of additional CBMs that putatively bind to starch, fructans and hemicelluloses. It therefore appears that Trichoderma makes significant use of these domains, and this could result in faster and more competitive degradation of the respective polymers. In these regards it is also of interest that HV possesses GH18 group C chitinases that contain both CBM18 as well as CBM50/LysM chitin binding domains, which have not yet been reported elsewhere. The possible differences in binding of CBM18 and CBM50/LysM to chitin are not known, however, which makes a speculation about the advantage of their arrangement in GH18 group C chitinases of HV Trichoderma difficult.
Finally, a striking feature in all Trichoderma genomes was the high number of orphan genes, of which only a very small number is also present in the core genome. The origin of orphan genes has been postulated to be either the consequence of gene duplication events and rearrangement processes followed by fast divergence, or of de novo evolution out of non-coding genomic regions [83]. Our data showed that - in the case of T. reesei - only a fifth of the orphan genes occured in clusters that could be indicative of gene duplications, and only a very small portion of orphans (clustered and non-clustered) occured near the telomeres, a frequent area for gene duplications. Our data therefore do not support gene duplication as the major mechanism for the emergence of orphan genes. The question whether the Trichoderma orphans originate de novo (see above) cannot be answered by our data. Published transcriptome data from T. reesei and T. virens [84, 85] show that about 40% of the orphan genes are indeed expressed, and therefore represent protogenes [86]. Our data suggest that the Trichoderma species-specific orphan genes evolve so fast that their sequences diverge beyond recognition, as already discussed for insects [87]. The biological merit, if any, needs further investigations to become understood, however.
Conclusions
This paper highlights the evolution of twelve Trichoderma species that are most frequently observed in nature and which belong to three different Trichoderma sections/clades and documents the gene inventory of the core genome and the individual species. The data reveal a high genomic diversity both at the section and clade level and on the species-level, which is reflected by the fact that only 50–75% of the genes are conserved in all twelve species. The high polymorphism in ankyrin and HET genes, but also of such encoding transcription factors, enzymes for carbohydrate and secondary metabolism illustrates that Trichoderma belongs to those genera of fungi which constantly re-shape their genome for fast responses and successful competition in potentially novel habitats. These properties are exactly what one would also expect from an environmental opportunist and generalist.
The data presented in this paper will likely become a starting point for mining Trichoderma genomes for enzymes or secondary metabolites, and for selection of candidate genes for manipulating strains towards desired behaviour in their application. Sequencing and annotation of genomes of species outside the currently investigated clades will be facilitated by the curated protein identification encoded by the core Trichoderma genome. This may likely lead to the detection of still new features not seen in species from sections Longibrachiatum, and Trichoderma and in Harzianum/Virens clades.
Finally, our data raise the genus Trichoderma to the level of the few fungal taxa for which genome sequences of several different species are available, such as Aspergillus and Fusarium, and which strongly facilitated studies on various aspects of the molecular physiology of these fungi. Our data for Trichoderma now offers such a basis as well.
Methods
In silico screening for most common Trichoderma species
An in-silico screening for most common Trichoderma species whose nucleotide sequences are deposited in GenBank yielded 29,911 sequences for 292 species (April 2018). Sequences collected for undefined species (“cf. Trichoderma” or “Trichoderma sp.”), poorly characterized species (i.e. that are represented by less than 3 nucleotide sequences), or sequences arising from whole genome sequencing projects were excluded. T. reesei is a special case, because most of its sequences represented genes of only a single isolate (QM6a and its mutants), what is related to its industrial application. The total number of sequences from individual T. reesei isolates is estimated to be 30, which is rather small. T. reesei was nevertheless included in this study becuase its genome sequence and annotation were already available [16] and considered to be a good basis for comparison to the more abundant species of section SL.
Trichoderma genomes
All but one (T. harzianum TR274) Trichoderma genome sequences were taken from JGI and NCBI databases (see “data access” below for numbers), and have been published [11, 12, 16, 17, 19, 22, 23].
T. harzianum TR274 has been isolated from soil in southeast of Brazil [29]. The genome was sequenced paired-end 2 × 250 bp using Miseq technology (Illumina™) and assembled with AllpathsLG [88] using maximum coverage of 120X. The genome was annotated using the Mycocosm annotation pipeline [31], and all data generated are available at the Mycocosm portal (https://genome.jgi.doe.gov/mycocosm/home).
Re-annotation of the T. hamatum GD12 genome
The T. hamatum genome is available in the public domain only in the form of assembled nucleotide scaffolds (accession number ANCB00000000.2). We performed structural annotation using the MAKER genome annotation pipeline v2.31.8 [89] with the gene predictor Augustus (http://bioinf.uni-greifswald.de/augustus/) trained with gene models from Fusarium graminearum. All proteins and transcripts from the Trichoderma ssp. analyzed in this study were used as gene model support. For functional annotation of translated proteins in the T. hamatum GD12, we performed InterProScan5 (http://www.ebi.ac.uk/interpro/) annotation, using stand-alone version 55 with the following embedded programs: SignalP4.1 [90], PFAM v.29 [91], Interpro [92] and GeneOntology (http://www.geneontology.org/).
Other fungal genomes analysed
The Ascomycota that were used in this study in comparison to Trichoderma, their habitats, taxonomic position and published genome sequences are given in Additional file 21.
Annotation of the Trichoderma proteomes
We first searched all 13 Trichoderma genome databases for orthologs in the T. reesei QM6a and RUT C-30 genome by reciprocal blastp, using a treshhold of < E-35 (this value turned out to retrieve the highest percentage of hits that were confirmed by reciprocal blastp in a series of trials with different E treshhold values). Data obtained for T. reesei QM6a and RUT C-30 were combined and pruned to contain individual genes only once. The BLAST servers of the Joint Genome Institute were used for most Trichoderma spp. A local blastp for T. parareesei and T. guizhouense was established at the server of the Institute of Chemical, Environmental and Bioscience Engineering, TU Wien. For T. gamsii, T. afroharzianum, and T. hamatum no individual BLAST server was available, and their predicted proteome therefore re-assessed by blastp in the NCBI Blast server. The so predicted proteins were cross-checked by Pfam v. 29 [91] using a TimeLogic Decypher machine and an <E-35 treshold, and Interpro [92].
Conserved protein domains in proteins were further veryfied by Blastp against NCBI’s conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; [61], using a treshhold of < E-05. Putative localization of proteins was analyzed using SignalP (for secreted proteins; http://www.cbs.dtu.dk/services/SignalP/), TargetP (for possible mitochondrial location; http://www.cbs.dtu.dk/services/TargetP/) and TMHMM (for preduction of transmembrane helixes in proteins; http://www.cbs.dtu.dk/services/TMHMM/). In all three methods, only hits with p < 0.05 were used.
In addition, we performed Ortho MCL clustering [93] with an inflation parameter of 1.5 on protein sequences from 26 predicted full proteomes (thirteen Trichoderma spp. and 13 from Hypocreales and Sordariomycete outgroups). A protein was considered specific to an organism subset if it was found at least in all but one of the organisms of the subset, but not in any organisms outside the subset.
Identification of specific protein families
Annotation of the genes encoding carbohydrate active enzymes (CAZymes) in the 13 Trichoderma genomes was performed using the Carbohydrate-Active Enzyme database and CAZy nomenclature (http://www.cazy.org/), by comparing each protein model from the genome by the sequence similarity search tool (BLAST) to a collection of protein modules corresponding to catalytic and carbohydrate-binding modules derived from CAZy. Individual hits were then compared by HMMer to models corresponding to each CAZy family to allow an assignment of each identified protein.
Proteases were identified by analysis of the proteomes of the 13 strains in the MEROPs database (https://www.ebi.ac.uk/merops/) and the corresponding nomenclature used to specify them.
Identification of PKS, NRPS and terpenoid synthases was performed with Antismash [94] and SMURF (http://www.jcvi.org/smurf). Potential orthologs of PKS genes in different Trichoderma spp. were determined by phylogenetic analysis, using the KS domain (PKS) and adenylation domain (NRPS). The Maximum Likelihood method, based on the Poisson correction model [88], was used to infer the evolutionary history. Branches corresponding to partitions with a boostrap coefficient of < 50% (1000 replicates) are collapsed.
To identify SSCPs, the proteomes of the 13 Trichoderma strains were first filtered with Microsoft Excel for those that have a protein size less then 300 amino acids and contain ≥5% cysteines and the detected candidates then subjected to SignalP analysis [90]. Among this subset of proteins, hydrophobins were visually identified by the presence of 8 cysteines, of which C2/C3 and C6/C7 occured as pairs. Ceratoplatanins were identified by the presence of 4 cysteines and blastp against the NCBI database. The remaining proteins were considered as uncharacterized SSCPs.
Analysis of genome completeness
To access the completeness of the genomes, we conducted a BUSCO v2 (Benchmarking Universal Single-Copy Orthologs) search our genomes for orthologues to each of 3725 Sordariomycete orthologous genes [32].
Generation of a time-scaled phylogeny of the Hypocreaceae
We estimated the phylogeny of the 27 Ascomycota species in our analysis using the protein sequences of 638 orthologs present in single copy in all species, identified using Proteinortho5 [95]. For each set of orthologous proteins, we produced multiple alignments using MAFFT [96] with the auto settings and identified conserved alignment blocks using Gblocks v0.19b [97]. The final concatenated alignment used for phylogenetic reconstruction analysis consisted of 259,738 amino acid positions. Clade ages were estimated using the tool CladeAge [98] described in Matschiner et al. [99]. Four ancestral nodes were used for the time calibration: a common ancestral node of the order Hypocreales was calibrated for a central 95% range of 190–196 Mya [3], a common ancestral node between families Hypocreaceae, Ophiocordycipitaceae and Clavicipitaceae was calibrated for a central 95% range of 162–168 Mya [100], a common ancestral node of Clavicipitaceae crown group for a central 95% range of 114–120 Mya [100] and a common ancestral node of Nectriaceae crown group for a central 95% range of 122–128 Mya [3]. Species within these clades were forced to form a monophyletic group to constrain the tree topology. The selection of best amino acid substitution model was done using ProtTest 3 [171} based on BIC criterion. A MCMC analyses were carried out with a chain length of 20,000,000 sampling on every 1000 generation in BEAST V2.4.0 [98], using JTT I + G + F and the lognormal relaxed clock was used for determination of the clade ages. Their combined logs for the analyses for each dataset were diagnosed using Tracer v1.6 to confirm that the effective sample size is above 200 for the estimated parameters. In TreeAannotator v2.4.0 (in the BEAST package [98]), 25% of the first total trees were discarded, 0.9 was used as posterior probability limit and node heights were estimated using mean heights in order to obtain the maximum clade credibility tree. The final tree with node ages and an automatic reverse scale axis was visualized and obtained using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Approximate 95% confidence interval was obtained by selecting “Height Highest Probable Density of 95%” for node bars in FigTree to show the age in the chronogram.
Analysis of Trichoderma phylogeny
The nucleotide sequence of a fragment of the rpb2 (RNA-polymerase II encoding gene) was retrieved from NCBI GenBank for 196 species of Trichoderma, and aligned. 808 nucleotides were then used for Bayesian analysis. Two independent MCMC runs were performed with 10 million generations and sampling frequency after each 100 generations; the first 800 trees have been removed. An earlier version of this tree, which does not make reference to the abundancy of species, has been published [7].
Analysis of protein family evolution
The evolution of protein family size variation (expansion or contraction) was analyzed by CAFÉ [41] (using the orthoMCL table with an e-value ≤1e-20, and an inflation parameter of 1.5) with a p-value of 0.01 and applying a stochastic model of gene death and birth.
Analysis of dN and dS
We estimated non-synonymous nucleotide substitutions (dN) and synonymous substitutions (dS) using PAML [101] with model M0 in pairwise mode implemented with custom shell scripts and calculated average dN/dS.
Estimates of evolutionary divergence between protein sequences
Analyses were conducted using the JTT matrix-based model and the rate variation among sites was modeled with a gamma distribution (shape parameter = 4). The analysis involved 27 species, same used to build time-scaled phylogenetic tree. All positions containing gaps and missing data were eliminated. There was a total of 380,905 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [101].
Data access
Genome assembly and annotations are available at at the JGI fungal genome portal MycoCosm [31] and are available at DDBJ/EMBL/GenBank under the following accessions: T. reesei, PRJNA225530; T. parareesei, LFMI00000000, T. longibrachiatum, MBDJ00000000; T. citrinoviride, MBDI00000000; T. harzianum CBS226.95, MBGI00000000; T. harzianum TR274, NQLC00000000; T. guizhouense, LVVK00000000; T. afroharzianum, JOKZ00000000; T. virens, PRJNA264113; T. atroviride, PRJNA164112; T. gamsii, JPDN00000000; T. asperellum, MBGH00000000; T. hamatum, ANCB00000000. The revised protein sequences and annotations of the T. reesei and T. hamatum genomes are included in the paper (Additional files 3 and 4).
Additional files
Abundancy of genes for Trichoderma species in GenBank. (XLSX 17 kb)
Improvement of published genome annotations and verification of strains. (PDF 185 kb)
Manually annotated genomes of T. reesei and T. hamatum. (XLSX 982 kb)
Protein sequences of T. hamatum, fasta format. (TXT 5440 kb)
Pairwise amino acid distances between Trichoderma and other Sordariomycetes fungi. (XLSX 16 kb)
OrthoMCL clusters of proteins encoded by Trichoderma and other fungi. (XLSX 6578 kb)
OrthoMCL clusters shared between Trichoderma and other Sordariomycete fungi. (XLSX 35 kb)
The Trichoderma core genome. (XLSX 1010 kb)
Intraspecific variation in Trichoderma harzianum as estimated based on the analysis of the two strains. (PDF 232 kb)
Unique genes shared only between facultative pathogenic Trichoderma species. (PDF 165 kb)
Gene clusters gained or lost in the evolution of Hypocreaceae before the origin of Trichoderma. (XLSX 24 kb)
Trichoderma genes involved in sensing of the mating partner. (PDF 285 kb)
Carbon metabolism in Trichoderma. (PDF 224 kb)
Polysaccharide decomposition by Trichoderma. (PDF 201 kb)
Extracellular proteolytic enzymes in Trichoderma. (XLSX 19 kb)
Secondary metabolism genes of Trichoderma. (PDF 318 kb)
PKS, NRPS, PKS-NRPS hybrids and terpenoid synthases in Trichoderma. (XLSX 45 kb)
Small cysteine-rich secreted effector proteins in Trichoderma. (PDF 210 kb)
Location of orphan genes and orphan gene clusters on the chromosomes of T. reesei. (XLSX 40 kb)
Evolution of the orphan genes present in the Trichoderma core genome. (XLSX 13 kb)
Ascomycete genomes that were used in this study as a comparison to Trichoderma. (XLSX 14 kb)
Acknowledgements
The authors are grateful to Dr. David Studholme, University of Exeter, for providing the full MAKER annotation of the T. hamatum genome.
Funding
The work performed by the Nanjing Agricultural University, China, was supported by the National Key Research and Development Program of China (2017YFD 0200806). This study was supported by grants of the Austrian Science Foundation (FWF) to ISD (P25613-B20) and CPK (I-1249). AS was supported by scholarships provided by CAPES foundation (99999.001745/2014–00) and CNPq (141180/2012–9). AS and EFN were supported by CNPq grant (482697/2011–3). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. B.H. gratefully acknowledges funding from IDEX Aix Marseille (Grant Microbio-E, 2015–2017).
Availability of data and materials
The raw data for this analysis were the 13 Trichoderma genome databases, which can be downloaded from the sources cited above. The reference genomes of other fungi that we used were downloaded from Mycocosm [24] and the NCBI database (http://www.ncbi.nlm.nih.gov/).
Authors’ contributions
AS, GM, KC, BH, JZ, FC, EMK, AGK, AK, RB, SS and IVG carried out in silico experiments and analysed data. CPK, AS, IVG and ISD conceived and designed the study, carried out the data analysis, interpretation, and discussion, and wrote the manuscript with comments from BH, GM, GV, EFN, and QS. CPK, AS, KC and ISD completed the supplements. CPK and ISD prepared the figures. All authors read and approved the final manuscript.
Ethics approval and consent to participate
No ethical approval was needed for this study.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qirong Shen, Email: qirongshen@njau.edu.cn.
Igor V. Grigoriev, Email: ivgrigoriev@lbl.gov
Irina S. Druzhinina, Email: Irina.druzhinina@njau.edu.cn
References
- 1.Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia. 2006;98(6):1076–1087. doi: 10.1080/15572536.2006.11832635. [DOI] [PubMed] [Google Scholar]
- 2.Kirk PM, Cannon PF, Minter DW, Stalpers JA. Dictionary of the Fungi. 10. Wallingford: CAB International; 2008. p. 332. [Google Scholar]
- 3.Sung G-H, Poinar GO, Spatafora JW. The oldest fossil evidence of animal parasitism by fungi supports a cretaceous diversification of fungal-arthropod symbioses. Mol Phylogenet Evol. 2008;49:495–502. doi: 10.1016/j.ympev.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 4.Spatafora JW, Sung G-H, Sung J-M, Hywel-Jones NL, White JF. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol Ecol. 2007;16:1701–1711. doi: 10.1111/j.1365-294X.2007.03225.x. [DOI] [PubMed] [Google Scholar]
- 5.Chenthamara K, Druzhinina IS. Ecological genomics of mycotrophic fungi. In: Druzhinina IS, Kubicek CP, editors. The Mycota: environmental and microbial relationships. 3. 2013. pp. 215–245. [Google Scholar]
- 6.Druzhinina Irina S., Kubicek Christian P. The Ecological Genomics of Fungi. Hoboken, NJ: John Wiley & Sons, Inc; 2013. Ecological Genomics ofTrichoderma; pp. 89–116. [Google Scholar]
- 7.Atanasova L, Druzhinina IS, Jaklitsch WM. Twohundred Trichoderma species recognized on the basis of molecular phylogeny. In: Mukherjee P, Horwitz BA, Singh US, Mukherjee M, Schmoll M, editors. Trichoderma: biology and applications. UK: CABI International; 2013. p. 10–42.
- 8.Jaklitsch WM, Voglmayr H. Biodiversity of Trichoderma (Hypocreaceae) in southern Europe and Macaronesia. Stud Mycol. 2015;80:1–87. doi: 10.1016/j.simyco.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaklitsch WM. European species of Hypocrea part I. the green-spored species. Stud Mycol. 2009;63:1–91. doi: 10.3114/sim.2009.63.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poldmaa K. Three species of Hypomyces growing on basidiomata of Stereaceae. Mycologia. 2003;95:921–933. doi: 10.1080/15572536.2004.11833051. [DOI] [PubMed] [Google Scholar]
- 11.Druzhinina IS, Chenthamara K, Zhang J, Atanasova L, Yang D, Miao Y, et al. Massive lateral transfer of genes for lignocellulolytic enzymes to the mycoparasitic fungal genus Trichoderma from its herbivore hosts. PLoS Genet. 2018; ms accepted for publication. [DOI] [PMC free article] [PubMed]
- 12.Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, et al. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol. 2011;9(10):749–759. doi: 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 14.Hermosa R, Rubio MB, Cardoza RE, Nicolás C, Monte E, Gutiérrez S. The contribution of Trichoderma to balancing the costs of plant growth and defense. Int Microbiol. 2013;16(2):69–80. doi: 10.2436/20.1501.01.181. [DOI] [PubMed] [Google Scholar]
- 15.Chaverri P, Samuels GJ. Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifts in ecology. Evolution. 2013;67(10):2823–2837. doi: 10.1111/evo.12169. [DOI] [PubMed] [Google Scholar]
- 16.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 17.Studholme DJ, Harris B, Le Cocq K, Winsbury R, Perera V, Ryder L, et al. Investigating the beneficial traits of Trichoderma hamatum GD12 for sustainable agriculture-insights from genomics. Front Plant Sci. 2013;4:258. doi: 10.3389/fpls.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie BB, Qin QL, Shi M, Chen LL, Shu YL, Luo Y, et al. Comparative genomics provide insights into evolution of Trichoderma nutrition style. Genome Biol Evol. 2014;6(2):379–390. doi: 10.1093/gbe/evu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang D, Pomraning K, Kopchinskiy A, Karimi Aghcheh R, Atanasova L, Chenthamara K, et al. Genome Sequence and Annotation of Trichoderma parareesei, the Ancestor of the Cellulase Producer Trichoderma reesei. Genome Announc. 2015;3(4). [DOI] [PMC free article] [PubMed]
- 20.Shi-Kunne X, Seidl MF, Faino L, Thomma BP. Draft Genome Sequence of a Strain of Cosmopolitan Fungus Trichoderma atroviride. Genome Announc. 2015;3(3):e00287–e00215. doi: 10.1128/genomeA.00287-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo HC, Wang TY, Chen PP, Chen RS, Chen TY. Genome sequence of Trichoderma virens FT-333 from tropical marine climate. FEMS Microbiol Lett. 2015;362(7). [DOI] [PubMed]
- 22.Baroncelli R, Piaggeschi G, Fiorini L, Bertolini E, Zapparata A, Pè ME, et al. Draft Whole-Genome Sequence of the Biocontrol Agent Trichoderma harzianum T6776. Genome Announc. 2015;3(3). [DOI] [PMC free article] [PubMed]
- 23.Baroncelli R, Zapparata A, Piaggeschi G, Sarrocco S, Vannacci G. Draft Whole-Genome Sequence of Trichoderma gamsii T6085, a Promising Biocontrol Agent of Fusarium Head Blight on Wheat. Genome Announc. 2016;4(1). [DOI] [PMC free article] [PubMed]
- 24.Compant S, Gerbore J, Antonielli L, Brutel A, Schmoll M. Draft genome sequence of the root-colonizing fungus Trichoderma harzianum B97. Genome Announc. 2017;5(23). [DOI] [PMC free article] [PubMed]
- 25.Schmoll M, Dattenböck C, Carreras-Villaseñor N, Mendoza-Mendoza A, Tisch D, Alemán MI, et al. The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol Mol Biol Rev. 2016;80(1):205–327. doi: 10.1128/MMBR.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Druzhinina IS, Kopchinskiy AG, Kubicek EM, Kubicek CP. A complete annotation of the chromosomes of the cellulase producer Trichoderma reesei provides insights in gene clusters, their expression and reveals genes required for fitness. Biotechnol Biofuels. 2016;9:75. doi: 10.1186/s13068-016-0488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li WC, Huang CH, Chen CL, Chuang YC, Tung SY, Wang TF. Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol Biofuels. 2017;10:170. doi: 10.1186/s13068-017-0825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaverri P, Branco-Rocha F, Jaklitsch W, Gazis R, DegenkoSL T, Samuels GJ. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia. 2015;107(3):558–590. doi: 10.3852/14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Druzhinina IS, Kubicek CP, Komoń-Zelazowska M, Mulaw TB, Bissett J. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol. 2010;10:94. doi: 10.1186/1471-2148-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steindorff AS, Ramada MH, Coelho AS, Miller RN, Pappas GJ, Jr, Ulhoa CJ, et al. Identification of mycoparasitism-related genes against the phytopathogen Sclerotinia sclerotiorum through transcriptome and expression profile analysis in Trichoderma harzianum. BMC Genomics. 2014;15:204. doi: 10.1186/1471-2164-15-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42(1):D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 33.Keller G, Sahni A, Bajpai S. Deccan volcanism, the KT mass extinction and dinosaurs. J Biosci. 2009;34:709–728. doi: 10.1007/s12038-009-0059-6. [DOI] [PubMed] [Google Scholar]
- 34.Stammers DK, Ren J, Leslie K, Nichols CE, Lamb HK, Cocklin S, et al. The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 2001;20(23):6619–6626. doi: 10.1093/emboj/20.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland IW. Polysaccharide lyases. FEMS Microbiol Rev. 1995;16(4):323–347. doi: 10.1111/j.1574-6976.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 37.Aranda-Martinez A, Lenfant N, Escudero N, Zavala-Gonzalez EA, Henrissat B, Lopez-Llorca LV. CAZyme content of Pochonia chlamydosporia reflects that chitin and chitosan modification are involved in nematode parasitism. Environ Microbiol. 2016;18(11):4200–4215. doi: 10.1111/1462-2920.13544. [DOI] [PubMed] [Google Scholar]
- 38.Khan A, Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics. 2017;18:287. doi: 10.1186/s12859-017-1708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seiboth B, Metz B. Fungal arabinan and L-arabinose metabolism. Appl Microbiol Biotechnol. 2011;89(6):1665–1673. doi: 10.1007/s00253-010-3071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhls K, Lieckfeldt E, Börner T, Guého E. Molecular reidentification of human pathogenic Trichoderma isolates as Trichoderma longibrachiatum and Trichoderma citrinoviride. Med Mycol. 1999;37:25–33. doi: 10.1080/02681219980000041. [DOI] [PubMed] [Google Scholar]
- 41.de Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 42.Kuhls K, Lieckfeldt E, Samuels GJ, Kovacs W, Meyer W, Petrini O, et al. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc Natl Acad Sci U S A. 1996;93(15):7755–7760. doi: 10.1073/pnas.93.15.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaverri P, Samuels GJ, Stewart EL. Hypocrea virens sp. nov., the teleomorph of Trichoderma virens. Mycologia. 2001;93:1113–1124. doi: 10.1080/00275514.2001.12063245. [DOI] [Google Scholar]
- 44.Dodd SL, Lieckfeldt E, Samuels GJ. Hypocrea atroviridis sp. nov., the teleomorph of Trichoderma atroviride. Mycologia. 2003;95(1):27–40. doi: 10.1080/15572536.2004.11833129. [DOI] [PubMed] [Google Scholar]
- 45.Druzhinina IS, Komoń-Zelazowska M, Atanasova L, Seidl V, Kubicek CP. Evolution and ecophysiology of the industrial producer Hypocrea jecorina (anamorph Trichoderma reesei) and a new sympatric agamospecies related to it. PLoS One. 2010;5(2):e9191. doi: 10.1371/journal.pone.0009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieuwenhuis BP, Aanen DK. Sexual selection in fungi. J Evol Biol. 2012;25(12):2397–2411. doi: 10.1111/jeb.12017. [DOI] [PubMed] [Google Scholar]
- 47.Mojzita D, Herold S, Metz B, Seiboth B, Richard P. L-xylo-3-hexulose reductase is the missing link in the oxidoreductive pathway for D-galactose catabolism in filamentous fungi. J Biol Chem. 2012;287(31):26010–26018. doi: 10.1074/jbc.M112.372755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Druzhinina IS, Kubicek CP. Familiar stranger: ecologicagenomics of the model saprotroph and industrial enzyme producer Trichoderma reesei breaks the stereotypes. Adv Appl Microbiol. 2016;95:69–147. doi: 10.1016/bs.aambs.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Gu F, Wu R, Yang J, Zhang KQ. Phylogenomic evolutionary surveys of subtilase superfamily genes in fungi. Sci Rep. 2017;7:45456. doi: 10.1038/srep45456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Zhang KQ. Independent expansion of zincin metalloproteinases in Onygenales fungi may be associated with their pathogenicity. PLoS One. 2014;9:e90225. doi: 10.1371/journal.pone.0090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muszewska A, Stepniewska-Dziubinska MM, Steczkiewicz K, Pawlowska J, Dziedzic A, Ginalski K. Fungal lifestyle reflected in serine protease repertoire. Sci Rep. 2017;7:9147. doi: 10.1038/s41598-017-09644-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macheleidt J, Mattern DJ, Fischer J, Netzker T, Weber J, Schroeckh V, et al. Regulation and role of fungal secondary metabolites. Annu Rev Genet. 2016;50:371–392. doi: 10.1146/annurev-genet-120215-035203. [DOI] [PubMed] [Google Scholar]
- 54.Mukherjee PK, Horwitz BA, Kenerley CM. Secondary metabolism in Trichoderma--a genomic perspective. Microbiology. 2012;158(1):35–45. doi: 10.1099/mic.0.053629-0. [DOI] [PubMed] [Google Scholar]
- 55.Sivasithamparam K, Ghisalberti EL. Secondary Metabolism in Trichoderma and Gliocladium. In: Kubicek CP, Harman GE, editors. Trichoderma and Gliocladium. Vol. 1. Basic Biology, Taxonomy and Genetics. London: Taylor and Francis Ltd; 1998. pp. 139–191. [Google Scholar]
- 56.Yu X, Li S-M. Prenyltransferases of the dimethylallyltryptophan synthase superfamily. Methods Enzymol. 2012;516:259–278. doi: 10.1016/B978-0-12-394291-3.00005-8. [DOI] [PubMed] [Google Scholar]
- 57.Stergiopoulos I, de Wit PJ. Fungal effector proteins. Annu Rev Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 58.Djonović S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM. Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant-Microbe Interact. 2006;19(8):838–853. doi: 10.1094/MPMI-19-0838. [DOI] [PubMed] [Google Scholar]
- 59.Gaderer R, Bonazza K, Seidl-Seiboth V. Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl Microbiol Biotechnol. 2014;98(11):4795–4803. doi: 10.1007/s00253-014-5690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaderer R, Lamdan NL, Frischmann A, Sulyok M, Krska R, Horwitz BA, Seidl-Seiboth V. Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol. 2015;15:2. doi: 10.1186/s12866-014-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. NCBI's conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasuga T, Mannhaupt G, Glass NL. Relationship between phylogenetic distribution and genomic features in Neurospora crassa. PLoS One. 2009;4(4):e5286. doi: 10.1371/journal.pone.0005286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scherf A, Figueiredo LM, Freitas-Junior LH. Plasmodium telomeres: a pathogen's perspective. Curr Opin Microbiol. 2001;4:409–414. doi: 10.1016/S1369-5274(00)00227-7. [DOI] [PubMed] [Google Scholar]
- 64.Wortman JR, Fedorova N, Crabtree J, Joardar V, Maiti R, et al. Whole genome comparison of the A. fumigatus family. Med Mycol. 2006;44:S3–S7. doi: 10.1080/13693780600835799. [DOI] [PubMed] [Google Scholar]
- 65.Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- 66.Vajda V, McLoughlin S. Fungal proliferation at the cretaceous-tertiary boundary. Science. 2004;303(5663):1489. doi: 10.1126/science.1093807. [DOI] [PubMed] [Google Scholar]
- 67.Ivany LC, Patterson WP, Lohmann KC. Cooler winters as a possible cause of mass extinctions at the Eocene/Oligocene boundary. Nature. 2000;407(6806):887–890. doi: 10.1038/35038044. [DOI] [PubMed] [Google Scholar]
- 68.Kelkar YD, Ochman H. Causes and consequences of genome expansion in fungi. Genome Bio Evol. 2012;4(1):13–23. doi: 10.1093/gbe/evr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun Z, Blanchard JL. Strong genome-wide selection early in the evolution of Prochlorococcus resulted in a reduced genome through the loss of a large number of small effect genes. PLoS One. 2014;9(3):e88837. doi: 10.1371/journal.pone.0088837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13(6):1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45(51):15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 73.Fenn K, Blaxter M. Wolbachia genomes: revealing the biology of parasitism and mutualism. Trends Parasitol. 2006;22(2):60–65. doi: 10.1016/j.pt.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 74.Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 2000;64:489–502. doi: 10.1128/MMBR.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saupe SJ, Clavé C, Sabourin M, Bégueret J. Characterization of hch, the Podospora anserina homolog of the het-c heterokaryon incompatibility gene of Neurospora crassa. Curr Genet. 2000;38(1):39–47. doi: 10.1007/s002940000130. [DOI] [PubMed] [Google Scholar]
- 76.Wu J, Glass NL. Identification of specificity determinants and generation of alleles with novel specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa. Mol Cell Biol. 2001;21(4):1045–1057. doi: 10.1128/MCB.21.4.1045-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chevanne D, Bastiaans E, Debets A, Saupe SJ, Clavé C, Paoletti M. Identification of the het-r vegetative incompatibility gene of Podospora anserina as a member of the fast evolving HNWD gene family. Curr Genet. 2009;55:93–102. doi: 10.1007/s00294-008-0227-5. [DOI] [PubMed] [Google Scholar]
- 78.Lamacchia M, Dyrka W, Breton A, Saupe SJ, Paoletti M. Overlapping Podospora anserina transcriptional responses to bacterial and fungal non self indicate a multilayered innate immune response. Front Microbiol. 2016;7:471. doi: 10.3389/fmicb.2016.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paoletti M, Saupe SP. Fungal incompatibility: evolutionary origin in pathogen defense? Bioessays. 2009;31:1201–1210. doi: 10.1002/bies.200900085. [DOI] [PubMed] [Google Scholar]
- 80.de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017;18(1):28. doi: 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akcapinar GB, Kappel L, Sezerman OU, Seidl-Seiboth V. Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr Genet. 2015;61(2):103–113. doi: 10.1007/s00294-014-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shoseyov O, Shani Z, Levy I. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev. 2006;70(2):283–295. doi: 10.1128/MMBR.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12(10):692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 84.Bischof R, Fourtis L, Limbeck A, Gamauf C, Seiboth B, Kubicek CP. Comparative analysis of the Trichoderma reesei transcriptome during growth on the cellulase inducing substrates wheat straw and lactose. Biotechnol Biofuels. 2013;6(1):127. doi: 10.1186/1754-6834-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morán-Diez ME, Trushina N, Lamdan NL, Rosenfelder L, Mukherjee PK, Kenerley CM, et al. Host-specific transcriptomic pattern of Trichoderma virens during interaction with maize or tomato roots. BMC Genomics. 2015;16:8. doi: 10.1186/s12864-014-1208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, et al. Proto-genes and de novo gene birth. Nature. 2012;487(7407):370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 2013;5(2):439–455. doi: 10.1093/gbe/evt009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goldman N. Variance to mean ratio, R(t), for poisson processes on phylogenetic trees. Mol Phylogenet Evol. 1994;3:230–239. doi: 10.1006/mpev.1994.1025. [DOI] [PubMed] [Google Scholar]
- 89.Campbell MS, Holt C, Moore B, Yandell M. Genome Annotation and Curation Using MAKER and MAKER-P. Curr Protoc Bioinformatics. 2014;48:4.11.1–39. [DOI] [PMC free article] [PubMed]
- 90.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 91.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, et al. InterPro in 2017 — beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of Ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, et al. AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Web Server issue):W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 98.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matschiner M, Musilová Z, Barth JM, Starostová Z, Salzburger W, Steel M, Bouckaert R. Bayesian phylogenetic estimation of clade ages supports trans-Atlantic dispersal of cichlid fishes. Syst Biol. 2017;66(1):3–22. doi: 10.1093/sysbio/syx005. [DOI] [PubMed] [Google Scholar]
- 100.Yang E, Lingling X, Ying Y, Xinyu Z, Meichun X, Chengshu W, Zhiqiang A, Xingzhong L. Origin and evolution of carnivorism in the Ascomycota (fungi) Proc Natl Acad Sci. 2012;109(27):10960–10965. doi: 10.1073/pnas.1120915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Abundancy of genes for Trichoderma species in GenBank. (XLSX 17 kb)
Improvement of published genome annotations and verification of strains. (PDF 185 kb)
Manually annotated genomes of T. reesei and T. hamatum. (XLSX 982 kb)
Protein sequences of T. hamatum, fasta format. (TXT 5440 kb)
Pairwise amino acid distances between Trichoderma and other Sordariomycetes fungi. (XLSX 16 kb)
OrthoMCL clusters of proteins encoded by Trichoderma and other fungi. (XLSX 6578 kb)
OrthoMCL clusters shared between Trichoderma and other Sordariomycete fungi. (XLSX 35 kb)
The Trichoderma core genome. (XLSX 1010 kb)
Intraspecific variation in Trichoderma harzianum as estimated based on the analysis of the two strains. (PDF 232 kb)
Unique genes shared only between facultative pathogenic Trichoderma species. (PDF 165 kb)
Gene clusters gained or lost in the evolution of Hypocreaceae before the origin of Trichoderma. (XLSX 24 kb)
Trichoderma genes involved in sensing of the mating partner. (PDF 285 kb)
Carbon metabolism in Trichoderma. (PDF 224 kb)
Polysaccharide decomposition by Trichoderma. (PDF 201 kb)
Extracellular proteolytic enzymes in Trichoderma. (XLSX 19 kb)
Secondary metabolism genes of Trichoderma. (PDF 318 kb)
PKS, NRPS, PKS-NRPS hybrids and terpenoid synthases in Trichoderma. (XLSX 45 kb)
Small cysteine-rich secreted effector proteins in Trichoderma. (PDF 210 kb)
Location of orphan genes and orphan gene clusters on the chromosomes of T. reesei. (XLSX 40 kb)
Evolution of the orphan genes present in the Trichoderma core genome. (XLSX 13 kb)
Ascomycete genomes that were used in this study as a comparison to Trichoderma. (XLSX 14 kb)
Data Availability Statement
The raw data for this analysis were the 13 Trichoderma genome databases, which can be downloaded from the sources cited above. The reference genomes of other fungi that we used were downloaded from Mycocosm [24] and the NCBI database (http://www.ncbi.nlm.nih.gov/).