Abstract
A large fraction of the genomes of grasses, members of the family Graminae, is composed of retrotransposons. These elements resemble animal retroviruses in their structure and possess a life cycle similar to theirs that includes transcription, translation, and integration of daughter copies. We have investigated if retrotransposons are generally transcribed in the grasses and other plants, and whether the various families of elements are translationally and integrationally active in multiple grass species. A systematic search of 7.8 × 105 publicly available expressed sequence tags from plants revealed widespread retrotransposon transcripts at a frequency of one in 1,000. Monocot retrotransposons found relatively more expressed sequence tags from non-source species than did those of dicots. Antibodies were raised to the capsid protein, GAG, of BARE-1, a transcribed and translated copia-like retrotransposon of barley (Hordeum vulgare). These detected immunoreactive proteins of sizes identical to those of the BARE-1 GAG and polyprotein, respectively, in other species of the tribe Triticeae as well as in oats (Avena sativa) and rice (Oryza sativa). Retrotransposon-based markers showed integrational polymorphisms for BARE-1 in different subfamilies of the Graminae. The results suggest that grasses share families of transcriptionally, translationally, and integrationally active retrotransposons, enabling a comparative and integrative approach to understanding the life cycle of retrotransposons and their impact on the genome.
In most grasses (family Gramineae), genes appear to comprise less than 20% of the genome (Flavell et al., 1977; Barakat et al., 1997), with most of the rest being composed of repetitive DNA. The variation in genome size in eukaryotes, independent of differences in organismal complexity, recognized early on (Thomas, 1971) and referred to as the C-value (genome size) paradox, is particularly apparent in the grasses. Among the diploid grasses, the 1C genome size (DNA content of the unreplicated haploid set of chromosomes) varies from 2.0 × 108 bp in Oropetium thomaeum to 1.3 × 1010 in Lygeum spartum (Bennett et al., 1998). The cereals rice (Oryza sativa, 1C = 4.3 × 108), sorghum (Sorghum bicolor, 1C = 7.2 × 108), maize (Zea mays, 1C = 2.6 × 109), barley (Hordeum vulgare, 1C = 4.5 × 109), and rye (Secale cereale, 1C = 8.0 × 109) are arrayed in between (Kankaanpää et al., 1996; Kurata et al., 1997; Bennett et al., 1998).
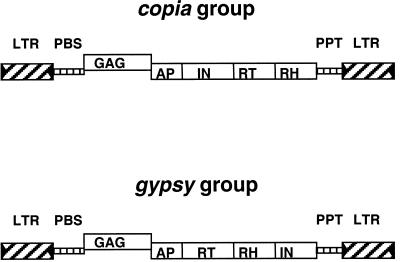
Evidence is accumulating that much of this more than 50-fold variation in genome size is due to variations in the prevalence of one specific class of repetitive DNA, retrotransposons. Retrotransposons are so named because, unlike the DNA transposons such as Ac and En/Spm, they propagate not by cutting and pasting, but by a mechanism of reverse transcription followed by integration of the new cDNA copy back into the genome (Boeke and Corces, 1989; Kumar and Bennetzen, 1999). Their life cycle, encoded products, and structure (Fig. 1) resemble those of the retroviruses; the retroviruses and retrotransposons are thought to be derived from a common ancestor (Xiong and Eickbush, 1990; Doolittle and Feng, 1992; Lazcano et al., 1992). The two classes of retrotransposons, gypsy like and the copia like, differing in the order of their encoded proteins (Fig. 1), are both ubiquitous throughout the plants (Flavell et al., 1992; Voytas et al., 1992; Suoniemi et al., 1998). The replicative nature of retrotransposon mobilization, combined with the large size of the elements (5 to 10 kb), indicates that active retrotransposon families have the potential to be major contributors to variation in genome size.
Figure 1.
Organization of the two major classes of retrotransposons. Both classes are bound by long terminal repeats (LTRs). The LTRs contain inverted repeats (triangles) at their termini. The primer binding site (PBS) and polypurine tract (PPT) are present in most elements and are required for replication by reverse transcriptase (RT). The protein-coding region is frequently separated into two domains by a frame shift (between GAG, the capsid protein, and aspartic proteinase [AP]). The two groups can be distinguished by the placement of integrase (IN), which in copia-like elements precedes the RT and ribonuclease H (RH) but in gypsy-like elements follows these units.
Mapping in the cereals showed that the genes of rice and other cereals are largely syntenic or collinear despite the large differences in genome size (Bennetzen, 2000; Keller and Feuillet, 2000). Detailed comparisons of sequenced regions in the maize and sorghum genomes (Tikhonov et al., 1999) established that their genome size difference was largely (74%) due to accumulation of retrotransposons since the divergence of these species. A major feature of cereal genomes is the localization of genes into “gene islands” interspersed by “repeat seas” (SanMiguel et al., 1996; Ananiev et al., 1998; Panstruga et al., 1998). Sequence analysis of a 66-kb contiguous region of the barley chromosome 2HL (Shirasu et al., 2000) showed the three genes on that stretch to span only 18 kb, yielding a local density only 30% greater than the average for the Arabidopsis genome and within its range (Arabidopsis Genome Initiative, 2000), most of the rest being composed of retrotransposons. If this region is typical, retroelements (intact retrotransposons and their derivatives) account for more than 60% of the barley genome, compared with approximately 5% of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). We showed earlier that a family of copia-like retrotransposons, BARE-1, alone comprises approximately 5% of the barley genome as 13.7 × 103 full-length copies of 8.9 kb and >6 × 104 solo LTRs of 1.8 kb (Vicient et al., 1999), dispersed along the chromosomes (Suoniemi et al., 1996a). Given 2.5 × 104 to 9 × 104 genes (respectively the number for Arabidopsis and the estimated human unigene set) at this density, the genic component of the barley genome would cover 1.5 × 108 to 5.4 × 108 bp, or 3.3% to 12% of the whole.
In barley, the BARE-1 family of retrotransposons (Manninen and Schulman, 1993) is transcribed in somatic tissues from conserved promoters within the LTR (Suoniemi et al., 1996b; Suoniemi et al., 1997). Jääskeläinen et al. (1999) demonstrated that the transcript is also translated, the predicted polyprotein processed, and the cDNA packaged into virus-like particles as seen for retroviruses and active retrotransposons such as Ty1 of yeast (Saccharomyces cerevislae) and copia of Drosophila melanogaster (Miyake et al., 1987; Roth, 2000). Many but not all retrotransposons investigated appear to be quiescent in somatic tissues but activated by stress including protoplast formation (Wessler, 1996; Grandbastien, 1998) and in tissue culture (Grandbastien et al., 1989; Hirochika et al., 1996; Okamoto and Hirochika, 2000).
If retrotranspososons currently have a widespread role as contributors to growth in genome size, one would expect to find evidence for their activity in a broad range of species. Here we have examined whether retrotransposons are generally transcribed in the grasses and other plants, and whether the various families of elements are translationally and integrationally active in multiple grass species. We have systematically searched the expressed sequence tag (EST) databases to find transcripts and used immunoblotting to detect translational products and marker methods to reveal insertion site polymorphisms generated by integration. The results suggest that retrotransposons are widely active in the grasses and that at least some families of retrotransposons are active across multiple species.
RESULTS
Retrotransposon Transcription Is Widespread But Most Prevalent in the Grasses
The many EST sequencing projects currently under way for various plants gave us an opportunity to search for transcripts of known retrotransposons in the public EST databases. A total of 7.8 × 105 ESTs were searched for homologies to the LTRs or internal domains of known retrotransposons, and 934 matches (1.2‰) of the total were found (Table I). The number of public EST sequences varies greatly by species, from almost 1.23 × 105 for soybean (Glycine max) to less than 50 accession numbers each for more than 30 species. Furthermore, it should be emphasized that a taxonomically representative set of ESTs for monocots and dicots is not yet available. For plants with more than 1 × 103 reported ESTs, the monocots, mostly grasses, showed the highest average fraction of retrotransposon-containing accessions, 1.75‰, compared with 1.40‰ for the two conifer species and 0.92‰ for the dicots. The monocots also had both a higher maximum (3.13‰ versus 2.47‰) and a higher minimum (0.98‰ versus 0.26‰) than the dicots. In a recent in silico transcriptional profiling of EST data sets, Bortoluzzi et al. (2000) considered EST frequencies > 3.6‰ to represent abundant, 3.6% to 1.25% moderate, and <1.2‰ rare transcripts. In this framework, retrotransposons are moderately expressed in only three of the nine dicots, but in four of the six monocots analyzed here.
Table I.
Plant EST databases and their share of accessions matching retrotransposons
| Genus | Class/Division | ESTs
|
Genome Size | ||
|---|---|---|---|---|---|
| Total no. | No. matching retrotransposons | ‰ Total | |||
| pg | |||||
| Glycine | Dicot | 122,843 | 153 | 1.25 | 2.25 |
| Arabidopsis | Dicot | 112,467 | 59 | 0.52 | 0.35 |
| Lycopersicon | Dicot | 93,115 | 112 | 1.20 | 2.05 |
| Medicago | Dicot | 82,320 | 21 | 0.26 | 1.75 |
| Zea | Monocot | 73,965 | 184 | 2.49 | 5.45 |
| Oryza | Monocot | 62,420 | 61 | 0.98 | 1.00 |
| Triticum | Monocot | 47,435 | 60 | 1.26 | 11.55 |
| Sorghum | Monocot | 47,098 | 75 | 1.59 | 1.5 |
| Lotus | Dicot | 26,844 | 15 | 0.56 | 0.95 |
| Hordeum | Monocot | 23,947 | 75 | 3.13 | 11.10 |
| Gossypium | Dicot | 23,100 | 57 | 2.47 | 3.23 |
| Pinus | Coniferales | 21,866 | 23 | 1.05 | – |
| Solanum | Dicot | 14,083 | 12 | 0.85 | 1.75 |
| Mesembryanthemum | Dicot | 11,115 | 7 | 0.63 | – |
| Secale | Monocot | 6,574 | 3 | 0.46 | 16.55 |
| Brassica | Dicot | 3,627 | 2 | 0.55 | 1.60 |
| Cryptomeria | Coniferales | 2,293 | 4 | 1.74 | – |
| Suaeda | Dicot | 742 | 2 | 2.70 | – |
| Saccharum | Monocot | 495 | 1 | 2.02 | – |
| Avena | Monocot | 492 | 1 | 2.03 | 8.82 |
| Capsicum | Dicot | 252 | 1 | 3.97 | 8.00 |
| Nicotiana | Dicot | 130 | 3 | 23.08 | 5.85 |
| Allium | Monocot | 79 | 2 | 25.32 | 33.50 |
| Vigna | Dicot | 69 | 1 | 14.49 | – |
Plant ESTs currently represent some 12% of all available ESTs in the EST database (http://www.ncbi.nlm.nih.gov/dbEST/ dbEST_summary.html). The plant ESTs were search for retrotransposon matches.
These data suggest that retrotransposons are generally more transcriptionally active in the grasses than in other groups of plants, although transcription occurs in all groups. An important caveat in these analyses is that the ESTs in the databases are derived from a mixture of cDNA construction and sequencing methods and also display length variations. These factors, together with the inherently partial nature of EST sequences, may cause accessions for retrotransposon transcripts to be missed when searching with particular motifs in the same way it hinders annotation of ESTs generally. In addition, it is not possible to differentiate between retrotransposon transcripts originating from LTRs, from elements transcriptionally active in their own right, and transcripts that are initiated in conventional cellular promoters and read through into solo LTRs or adjacent full-length retrotransposons. However, the tendency of genes in grass genomes to cluster into gene islands, discussed above, would tend to decrease the likelihood of illegitimate transcripts, at least for this group of plants.
Earlier work showed that retrotransposon number and genome size is positively correlated in barley (Vicient et al., 1999; Kalendar et al., 2000). Because transcription is a prerequisite to integration of new retrotransposon copies, one might expect to find a positive association between the fraction of retrotransposon ESTs detected and genome size. The data here (Table I) display a strong and significant correlation between genome size and plant group (Pearson Product Moment, rP= 0.616, P = 0.025), the grasses containing larger genomes than the others, and also a weak but not significant association between genome size and the fraction of ESTs matching retrotransposons (rP = 0.209, P = 0.493). Taking the dicots alone, the correlation between genome size and EST fraction is both strong and significant (rP = 0.895, P = 0.006). However, it should be kept in mind that only transcription and ultimate integration in tissues giving rise to gametes is heritable; the ESTs here are derived from many types of tissues (Table II).
Table II.
Plant retrotransposons and their matching plant ESTs
| Query Retrotransposon
|
Homologous ESTs
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Elementa | Source species | Type | Total ESTs | Matching species | EST source tissueb
|
||||||
| A | B | C | D | E | F | G | |||||
| SIRE-1 | Soybean | Copia | 2 | Soybean | – | – | – | – | 1 | 1 | – |
| Tgmr | Soybean | Copia | 0 | – | – | – | – | – | – | – | – |
| diaspora | Soybean | Gypsy | 3 | Soybean | – | – | 1 | 2 | – | – | – |
| Athila | Arabidopsis | Gypsy | 1 | Arabidopsis | 1 | – | – | – | – | – | – |
| AtRE1 | Arabidopsis | Copia | 0 | – | – | – | – | – | – | – | – |
| AtRE2 | Arabidopsis | Copia | 0 | – | – | – | – | – | – | – | – |
| Ta1 | Arabidopsis | Copia | 0 | – | – | – | – | – | – | – | – |
| Tat1 | Arabidopsis | Gypsy | 0 | – | – | – | – | – | – | – | – |
| Evelknievel | Arabidopsis arenosa | Copia | 0 | – | – | – | – | – | – | – | – |
| RetroLyc | L. peruvianum | Copia | 6 | Cultivated tomato (Lycopersicum esculentum) | – | – | – | – | 3 | 2 | 1 |
| Bs1 | Maize | ? | 1c | Maize | – | – | 1 | – | – | – | – |
| Hopscotch | Maize | Copia | 0 | – | – | – | – | – | – | – | – |
| Opie-2 | Maize | Copia | 21 | Maize | 2 | – | 9 | 3 | 5 | 2 | – |
| PREM-2 | Maize | Copia | 17 | Maize | – | – | 8 | 1 | 7 | 1 | – |
| Stonor | Maize | Copia | 3 | Maize | 1 | – | 2 | – | – | – | – |
| CentA | Maize | Gypsy | 2 | Rice | – | 1 | – | – | – | – | 1 |
| Cinful1 | Maize | Gypsy | 23 | Maize | 3 | – | 7 | 3 | 10 | – | – |
| Cinful2 | Maize | ? | 22 | Maize | 4 | – | 4 | 2 | 12 | – | – |
| Grande1 | Zea diploperennis | Gypsy | 2 | Maize | 2 | – | – | – | – | – | – |
| Reina | Maize | Gypsy | 0 | – | – | – | – | – | – | – | – |
| Tekay | Maize | Gypsy | 2 | Maize | 2 | – | – | – | – | – | – |
| Zeon-1 | Maize | Gypsy | 10 | Maize | 4 | – | 5 | 1 | – | – | – |
| RIRE1 | Oryza australiensis | Copia | 2 | Barley | 1 | – | – | – | – | – | – |
| Bread wheat (Triticum aestivum) | – | – | – | 1 | – | – | – | ||||
| RIRE2 | Rice | Gypsy | 6 | Rice | – | – | – | 1 | 2 | – | 3 |
| RIRE3 | Rice | Gypsy | 8 | Rice | – | – | 7 | – | – | – | – |
| Gossypium arboreum | – | – | 1 | – | – | – | – | ||||
| RIRE7 | Rice | Gypsy | 7 | Rice | – | – | – | – | 1 | – | 1 |
| Maize | – | – | 1 | 1 | 2 | – | – | ||||
| Sorghum | – | – | – | – | – | 1 | – | ||||
| RIRE8 | Rice | Gypsy | 5 | Rice | – | – | 4 | – | – | – | 1 |
| Wis2-1 | Bread wheat | Copia | 19 | Bread wheat | – | – | 1 | 4 | 2 | 2 | – |
| Triticum monococum | – | 1 | – | – | – | – | – | ||||
| Barley | 5 | – | 3 | – | – | – | – | ||||
| Rye | – | – | – | – | – | 1 | – | ||||
| RetroSor1 | Sorghum | Gypsy | 4 | Sorghum | – | – | – | – | 3 | 1 | – |
| Levithan | Sorghum | LTR | 1 | Sorghum | – | – | – | – | 1 | – | – |
| BARE-1 | Barley | Copia | 23 | Barley | 7 | – | 6 | – | – | – | – |
| Bread wheat | – | – | 1 | 5 | 1 | 2 | – | ||||
| Rye | – | – | – | – | – | 1 | – | ||||
| Bagy-1 | Barley | Gypsy | 6 | Barley | 2 | – | 1 | – | – | – | – |
| Bread wheat | – | – | 1 | 1 | – | – | – | ||||
| Rice | – | – | 1 | – | – | – | – | ||||
| Bagy-2 | Barley | Gypsy | 7 | Barley | 4 | – | 1 | – | – | – | – |
| Bread wheat | – | – | – | – | 2 | – | – | ||||
| Cereba | Barley | Gypsy | 5 | Bread wheat | – | – | – | – | 2 | – | – |
| Maize | – | – | 1 | 1 | – | – | – | ||||
| Sorghum | – | – | – | – | – | 1 | – | ||||
| Nikita | Barley | LTR | 1 | Bread wheat | – | – | – | – | 1 | – | – |
| Sabrina | Barley | ? | 29 | Barley | 22 | – | 3 | – | – | – | – |
| Bread wheat | – | – | – | 2 | 1 | – | – | ||||
| Rye | – | – | – | – | – | 1 | – | ||||
| Sukkula | Barley | ? | 34 | Barley | 12 | – | 9 | – | 1 | – | – |
| Bread wheat | 1 | 1 | 2 | 4 | – | 3 | – | ||||
| IFG7 | Pinus radiata | Gypsy | 4 | Pinus taeda | – | 2 | 2 | – | – | – | – |
Published retrotransposon elements were used as query sequences in searches of plant EST accessions in the EST database. The matching ESTs for each retrotransposon are listed according to their source. The databases are not necessarily nonredundant.
The retrotransposon accession nos. are: SIRE-1 (U96295), Tgmr (U96748), diaspora (AF095730), Athila (X81801), AtRE1 (AB021265), AtRE2 (AB021266), Evelknievel (AF039376), Ta1 (X53976), Tat1 (AF056632), RetroLyc (AF228701), Bs1 (X16080), Hopscotch (U12626), Opie-2 (AF090446), PREM-2 (U41000), Stonor (AF082130, AF082134, and AF082133), CentA (AF078917), Cinful1 (AF049110), Cinful2 (AF049111), Grande1 (X97604), Reina (U69258), Tekay (AF050455), Zeon-1 (U11059), RIRE1 (D85597), RIRE2 (AB030283), RIRE3 (AB014738), RIRE7 (AB014740), Wis2-1 (X63184), RetroSor (AF098806), Levithan (U07816), BARE-1 (Z17327), Bagy-1 (Y14573), Bagy-2 (AF254799), Cereba (AF078801), Nikita (AF254799), Sabrina (AF254799), Sukkula (Z17327, Y14573, AFO29897, AF254799), and IFG7 (AJ004945).
Letters represent the tissue from which the EST was obtained: A, Leaf; B, shoot and root; C, flower and other reproductive organs; D, embryo and seed; E, seedling; F, stress-induced plants; and G, cultured cells.
Bs1 has been shown to contain a transduced gene corresponding to a plasma membrane H+-ATPase. For this reason, Bs1 gave homologies with many cDNAs that are transcripts from these genes. Those matches have been removed from the Table.
Grass Species Share Retrotransposon EST Matches
A set of 10 previously described retrotransposons from dicots, 27 from grasses, and one from a pine, including 14 which could be clearly defined as copia like and 18 as gypsy like, were used as query sequences against the EST database and the hits analyzed (Table II). Whereas the analysis for Table I looked for all expressed elements, characterized and uncharacterized, detectable in each plant, Table II reports only those matches for the specific elements listed. For the three dicotyledonous species examined encompassing 3.3 × 105 ESTs, only 12 matches to homologous retrotransposons were found, a fraction of 3.7 × 10−5. The most active of these elements is the copia-like RetroLyc of Lycopersicon peruvianum, which found six matches in the closely related cultivated tomato. Of the six Arabidopsis elements examined, only the gypsy-like Athila matched an EST, a single-leaf accession. In parallel, only 4% of the complete retroelements in the full sequence of the Arabidopsis genome correspond to an Arabidopsis EST (Arabidopsis Genome Initiative, 2000). None of the dicot elements found inter-generic matches. In contrast, the 27 grass retrotransposons identified 259 matches within grasses corresponding to 2.6 × 105 ESTs. Furthermore, the matches from grass retrotransposons show a very different pattern from those of the dicots. Elements of both the gypsy-like and copia-like classes find related and expressed elements in other grass species. Although all of the matches are to cereals, many of the matches cross tribe lines. This is particularly the case for the elements of barley and rice, whereas with the exception of CentA, the maize elements found matches only among maize ESTs.
Expression of BARE-Like Capsid Protein GAG Is Common in the Triticeae
Retroviruses and retrotransposons express their encoded proteins as a single polyprotein or as two (GAG and POL) separated by a frameshift (Fig. 1); these are processed into functional units by the aspartic proteinase present as part of the polyprotein itself (Garfinkel et al., 1991; Katz and Skalka, 1994). Earlier we demonstrated that GAG encoded by BARE-1 is translated and processed to the predicted mature size of 32.0 kD in dry and germinating embryos, leaves, and cell cultures of barley (Jääskeläinen et al., 1999). Oat (Avena sativa), bread wheat, and rye contain elements similar to BARE-1 (Pearce et al., 1997). The only reported full-length retrotransposon from these species is Wis-2 (accession no. X63184), which contains many stop codons interrupting its predicted translation. A reconstructed version of this translation (not shown) reveals only 47% identity and 54% similarity to the BARE-1a (accession no. Z17327) GAG. Here, we have raised antibodies to an expressed, full-length BARE-1 GAG (accession no. AJ295226).
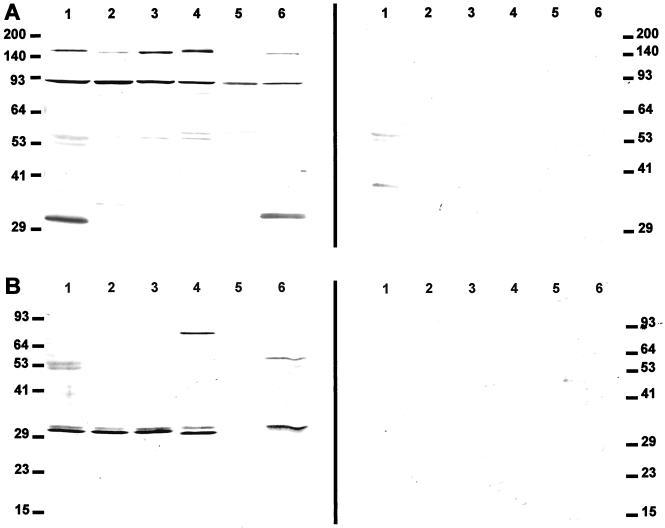
The antibodies recognize proteins of 150 kD and 90 kD on immunoblots of virtually all tested samples, including those of species outside the Triticeae (Fig. 2A). The 150-kD band is weak in lyme grass (Leymus arenarius L. Hochst.) and bread wheat. The 150-kD protein corresponds to the 146.9 kD predicted for the unprocessed polyprotein of BARE-1. The 90-kD band corresponds to the mass predicted for the BARE-1 polyprotein following endoproteolytic cleavage of the reverse transcriptase-ribonuclease H domain. In addition, these antibodies recognized a 31.5-kD protein, matching the predicted size of the mature, proteolytically processed GAG (Jääskeläinen et al., 1999), as well as a 34-kD band from rice which may be its equivalent. The approximately 53- and 54-kD bands and weak 36-kD band in barley on the immunoblots also reacted with the pre-immune serum (shown on the right); otherwise, all reactions seen were specific.
Figure 2.
Immuno-responses of seed proteins separated by SDS-PAGE to anti-GAG antibodies. A, Immunoblot reacted with antibodies made to a full-length BARE-1 GAG (left) and with pre-immune antibodies (right). B, Immunoblot reacted with antibodies made to the N-terminal portion of BARE-1a GAG (left) and with pre-immune antibodies (right). Mr shown on the left axes. Lane 1, Barley var. Himalaya; lane 2, bread wheat var. Tjalve; lane 3, rye var. Riihi; lane 4, oat var. Veli; lane 5, lyme grass; lane 6, rice line IRRI52886.
In other experiments (Fig. 2B), immunoblots were reacted with the antibodies earlier generated to the N-terminal portion of GAG (Jääskeläinen et al., 1999). These antibodies recognized a 31-kD protein, a mass virtually identical to that of BARE-1 GAG, in all species tested; the lyme grass reaction was weak (lane 5) but detectable by eye. The oat protein was slightly smaller, 29 kD. Maize extracts contain a reacting protein of this size as well (not shown). The additional bands seen in the oat and rice lanes may represent intermediates in the proteolytic processing of the polyprotein. As is seen from the negative response to the pre-immune serum on the parallel blot, the recognition of the 29- to 31-kD proteins was specific. The antibody used in Figure 2B, earlier raised to only the amino-terminal half of BARE-1 (Jääskeläinen et al., 1999), visualizes the mature GAG but not the polyprotein. The two antisera were raised to nonidentical BARE-1 variants (85% identity in the overlapping, expressed GAG region) from the genomic population; their differing but specific protein recognition patterns may reflect variations in the processing kinetics of distinct BARE-1 subfamilies. The observed immunoresponse detected for rice is consistent with the RIRE1 retrotransposon of O. australiensis (accession no. D85597) and other Oryza spp. having been reported to be most similar to BARE-1 (Noma et al., 1997). An alignment of the RIRE1 polyprotein (GenPept accession no. BAA22288) to the GAG of BARE-1a shows 48.9% similarity, suggesting that an active RIRE-1-like element may be responsible for the immunoreaction detected.
BARE-1 Appears Integrationally Active in Many Grasses
The creation by LTR retrotransposons of new joints with the flanking genomic DNA upon integration allows molecular fingerprints of the insertion pattern to be detected. Marker bands are generated by PCR using outward facing primers matching retroelement LTRs in combination with primers corresponding to dispersed genomic components, alternatively another retroelement (the inter-retrotransposon amplified polymorphism [IRAP] method; Kalendar et al., 1999), a microsatellite (the retrotransposon-microsatellite amplified polymorphism method; Kalendar et al., 1999), or a restriction site adapter (the sequence-specific amplified polymorphism [SSAP] method; Waugh et al., 1997). Because LTRs do not excise as part of retrotransposition, marker band polymorphisms are generally due to integration of new retrotransposon copies. We established earlier (Gribbon et al., 1999) that the SSAP method displays BARE-1 insertional polymorphisms throughout the Triticeae as well as in oat cultivars. Because SSAP polymorphisms may also be due to variability in the occurrence of restriction sites, we could not attribute them solely to retroelement mobility. Here, we have used a method relying only on LTR primers (IRAP) as well as looked for BARE-1 integrational polymorphisms in grasses distant from the Triticeae.
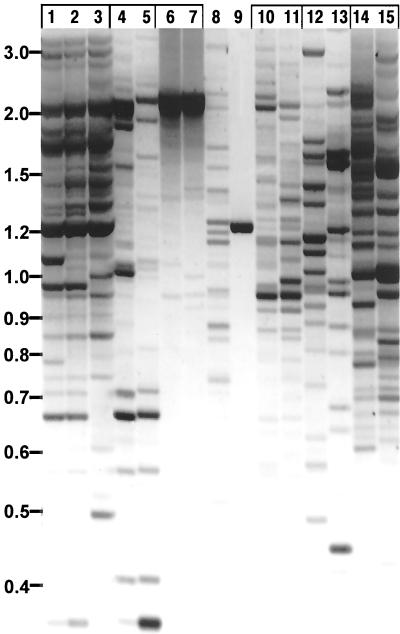
The BARE-1 element appeared among the EST database matches for three species in the tribe Triticeae (Table II), and we performed IRAP for BARE-1 on these and a range of other grass species (Fig. 3). The primers produced bands not only for all the Triticeae accessions and for timothy and oat, like the Triticeae in the subfamily Pooideae, but also for S. maritima and cordgrass of the subfamily Chloridoideae. Polymorphisms were observed between the pairs of, respectively, wheat cultivars, both Nordic spring types, rye lines, timothy lines, and cordgrass species. This suggests that BARE-1 has been integrationally active within these groups of cultivars or accessions since their divergence from their last common ancestor.
Figure 3.
Banding pattern generated by IRAP amplification with primers to retrotransposon BARE-1. The image is a negative of an agarose gel stained with ethidium bromide and visualized under UV light. Lanes of reactions made with accessions of the same genus are grouped. The template DNA was from bread wheat var. Tjalve (lane 1), bread wheat var. Mahti (lane 2), durum wheat (lane 3), Aegilops tauschii line 1691 (lane 4), A. tauschii line 1704 (lane 5), rye line P105 (lane 6), rye line P87 (lane 7), oats var. Veli (lane 8), rice var. IRRI52886 (lane 9), timothy (Phleum pratense) line 22 (lane 10), timothy line 16 (lane 11), Spartina maritima (lane 12), cordgrass (Spartina alterniflora; lane 13), lyme grass (lane 14), Leymus mollis (Lane 15). Marker sizes in kb are indicated on the left axis.
Rice, in the subfamily Erhartoideae, produced a single band, and maize, in the subfamily Panicoideae, produced only very weak bands (data not shown), although other barley retroelements are useful for IRAP in these species (our unpublished results). BARE-1- like elements are translated in rice (Fig. 2), so the data suggest either that few are clustered or that the LTRs are dissimilar from those of BARE-1 of barley. Rye produces many bands with the BARE-1 primers; these are better resolved on sequencing gels than on agarose gels as in Figure 3. A. tauschii represents the D-genome donor to bread wheat. A set of IRAP bands that are both monomorphic and shared between the A. tauschii and bread wheat accessions (Fig. 3) may represent integration events arising in the D genome before the advent of bread wheat.
DISCUSSION
Retrotransposons are well established as ubiquitous and highly prevalent components of plant genomes in general. These elements are nevertheless often referred to as “junk DNA,” implying that they are inert, in contrast to the genes required for cellular function. Their prevalence might be explained by ancient retrotransposition without requiring activity in the present. However, for some individual retroelements, evidence exists for transcription (Suoniemi et al., 1996b; Hirochika, 1997; Vernhettes et al., 1997; Okamoto and Hirochika, 2000), stress activation (Wessler, 1996; Grandbastien, 1998; Kalendar et al., 2000), translation (Hu et al., 1995; Jääskeläinen et al., 1999), and integration at specific loci (Johns et al., 1985; Grandbastien et al., 1989; Hirochika et al., 1996).
Here, we have undertaken to demonstrate that three components of the life cycle of active retrotransposons (transcription, translation, and integration) are widespread. First, we have made a systematic search for transcribed retrotransposon sequences in EST databases. Among the 7.8 × 105 ESTs searched, retrotransposons represent about 1.2‰ of transcripts, the frequency being somewhat higher among monocots than dicots. The frequency would be several-fold higher were only the identifiable ESTs (“hits”) to be considered. Retrotransposons from the grasses tend to match ESTs across multiple genera, whereas elements from the dicots tend to find matches only in their host species. These analyses cannot be regarded as exhaustive because even large EST databases may be 30% to 40% incomplete (Arabidopsis Genome Initiative, 2000; Penn et al., 2000).
Antibodies raised to BARE-1 GAG expressed in Escherichia coli recognized proteins in the seeds of virtually all species tested, not only barley and other grasses of the Triticeae, but also in species in different tribes and subfamilies of the Gramineae. The sizes of the bands detected are almost identical to those predicted for the BARE-1 polyprotein and the proteolytically processed, mature GAG, consistent with expression of polyproteins of size similar to those in the other species. These results are the first evidences for pools of retrotransposon polyproteins in plant cells. They show that BARE-like retrotransposons are translationally active and sufficiently well conserved for immunological cross reaction in a wide range of species in the Graminae. The translation of BARE-like elements in other grasses is consistent with the evidence from the EST database searches that transcriptionally active retrotransposon families are shared among the grasses.
The third line of evidence that grass genomes share families of active retrotransposons is the demonstration that a retrotransposon originally identified in barley can generate polymorphic marker bands in distant species. The IRAP products result from two retrotransposons near enough to each other to permit amplification of a PCR fragment between them. Their prevalence is consistent with the observed retroelement clusters in grass genomes (SanMiguel et al., 1996; Ananiev et al., 1998; Panstruga et al., 1998; Manninen et al., 2000). The IRAP polymorphisms reported here indicate integration events subsequent to the last common ancestors of lines or cultivars outside barley and the tribe Triticeae. Similar findings for other retrotransposons, including gypsy-like Bagy-1, Sukkula, and others (our unpublished observations), together with the EST and translation data here, suggest that the broad activity of retrotransposon families across the grasses may be a general phenomenon.
In recent years, the large-scale syntenic nature of the grass genomes has been recognized and applied (Ahn and Tanksley, 1993; Kilian et al., 1997). Exceptions to the microcolinearity of genes can be due to insertion of transposable elements (Bennetzen, 2000); recombinational loss of sequences intervening between the LTRs of retrotransposons (Shirasu et al., 2000) may help explain other exceptions to microcolinearity caused by deletions. Given synteny and active, shared families of retrotransposons, the grasses may be well suited for a comparative approach to the understanding of the impact of retrotransposons on the genome. Examination of the changes in genome organization among the grasses wrought by specific retrotransposon families may help untangle shared mechanisms of propagation and regulation from the contingent history of a these families in any given species.
MATERIALS AND METHODS
Plant Materials
Barley (Hordeum vulgare var. Himalaya) was obtained from Washington State University (Pullman). Rye (Secale cereale var. Riihi), spring bread wheat (Triticum aestivum vars. Tjalve and Mahti), and durum wheat (Triticum durum) were gifts of the Department of Food Technology, University of Helsinki (Finland). Rye lines P105 and P87 were gifts of Tamara S. Schulko (Academy of Sciences, Minsk, Belarus). Oat (Avena sativa var. Veli) and timothy grass (Phleum pratense) lines 22 and 16 were gifts of Boreal Plant Breeding Ltd. (Jokioinen, Finland). Aegilops tauschii lines 1691 and 1704 were gifts of Bikhram Gill (Kansas State University, Manhattan). Spartina maritima (Curtis) Fernald and cordgrass (Spartina alterniflora Loisel) are from Malika Ainouche (University of Rennes, France). Inbred maize (Zea mays) lines Oh43 and Mo17 were gifts of the U.S. Department of Agriculture, Agricultural Research Service, North Central Regional Plant Introduction Station, Iowa State University, Regional Plant Introduction Station (Ames). Lyme grass (Leymus arenarius L. Hochst.) with a provenance in Eyrarbakkí, Iceland was a gift of Kesara Anamthawat-Jánsson (University of Reykjavík, Iceland). Rice (Oryza sativa) line IRRI 52886 was from the International Rice Research Institute (Los Baños, Laguna, The Philippines).
EST Database Searches
The plant accessions found in the dbEST division of the combined GenBank (release 120.0), EMBL, and DNA Data Bank of Japan databases were searched for similarities to retrotransposons in two ways. First, we selected all entries in the overall nonredundant, combined nucleic acid database containing the words “retrotransposon,” “copia-like,” or “gypsy-like” in the descriptor, as well as all entries of previously published plant retrotransposons. We then selected the parts of these sequences corresponding to retrotransposons. These were used as the query sequences against the dbEST database with the Advanced BLAST program using a cutoff value of 0.0001. Second, for those general database entries having a putative translation, we queried the EST databases using the TBLASTN program applying a cutoff value of 1.0. All searches were done using the online service of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/blast.cgi).
Immunoblotting
Two anti-GAG antisera were used: one (Fig. 2B) was identical to that used earlier (Jääskeläinen et al., 1999), and the other (Fig. 2A) was prepared as follows. The region corresponding to the full-length BARE-1 GAG was isolated from a gag clone (accession no. AJ295226) by amplification with the forward primer 5′ GT TGT AGA CAT ATG GCT CGC GGA GTA GC 3′ (start codon underlined) and the reverse primer 5′ GAC ATG TGG ATC CAA TAT ACC TCA TTT TTC 3′ (stop codon underlined). The forward primer introduced a NdeI site (CATATG) and the reverse primer a BamHI site (GGATCC). The reaction product was digested with BamHI and NheI and cloned into the vector pET14b (Novagen, Madison, WI). This vector tags the N terminus of the protein with a poly-His stretch. The construct was expressed in Escherichia coli and purified on a TALON metal affinity resin (CLONTECH, Palo Alto, CA) column under denaturing conditions according to the manufacturer's instructions. The expressed protein was further purified by SDS-PAGE electrophoresis prior to injection into the rabbit. The antiserum was raised and antibodies purified as previously described (Jääskeläinen et al., 1999). Pre-immune antisera were from the same rabbits and purified as for the anti-GAG antisera.
For immunoblotting, embryo halves of the various seeds were isolated and pulverized under liquid N2, then extracted in 50 mm Tris-HCl (pH 7.5), 10 mm EDTA, 10 mm CHAPS {3-[(3-cholamidoproyl) dimethylammonio]-1-propane-sulfonate), 15 mm KCl, 5 mm MgCl2, 3 mm dithiothreitol, 10 μM trans-epoxysuccinyl-L-leucylamido-(4-guanidino) butane, 4 μM Pepstatin, and 2 μM Leupeptin. The protein contents of the extracts were determined (Bio-Rad Protein Assay, Bio-Rad, Hercules, CA) and an equivalent of 20 μg protein for each sample separated by SDS-PAGE electrophoresis. Electrophoresis, blotting, and immunoreactions were carried out as previously described (Jääskeläinen et al., 1999).
IRAP Polymorphism Detection
The IRAP markers were generated as before (Kalendar et al., 1999; Manninen et al., 2000) in a thermocycler (Master Cycler Gradient, Eppendorf AG, Hamburg, Germany) in 0.2-mL tubes (AB-0337, ABgene, Epsom, Surrey, UK). The 20-μL reactions contained 75 mm Tris-HCl (pH 8.8), 20 mm (NH4)2SO4, 1.5 mm MgCl2, 0.01% (v/v) Tween 20 (polyoxylethylenesorbitan), 20 ng DNA, 200 nm LTR primers, 200 μM dNTPs, and 1.2 units thermostable DNA polymerase (FIREPol, Solis Biodyne, Tartu, Estonia). The BARE-1 LTR primer consisted of 5′ TCC CAT GCG ACG TTC CCC 3′, matching nt 2,116 to 2,133 of accession number Z17327 at 4 nt from the 3′ end of the LTR. The template DNA was isolated as earlier described (Kalendar et al., 1999). The reaction mixture was heated to 94°C for 2 min, then 30 cycles were carried out, which were as follows: 94°C, 20 s; 60°C, 20 s; and 72°C, 2 min. The reaction was terminated by a final extension at 72°C for 10 min followed by maintenance at 4°C. One-fifth of the reaction mixture was analyzed by gel electrophoresis, carried out in 2% (w/v) agarose (RESolute LE agarose, BIOzym, Landgraaf, The Netherlands) at 80 V for 7 h and visualized by staining with ethidium bromide.
ACKNOWLEDGMENTS
The expert technical assistance of Anne-Mari Narvanto (University of Helsinki) is deeply appreciated. The involvement of Alex Baumel and Malika Ainouche (University of Rennes, France) in developing the IRAP markers for Spartina spp. is gratefully acknowledged.
Footnotes
This work was supported by Academy of Finland grants (to C.M.V. and M.J.J.), by an Academy of Finland Senior Fellowship (to A.H.S.), and by the European Union Research Directorate (contract no. QLK5–1999–01499).
LITERATURE CITED
- Ahn S, Tanksley SD. Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev EV, Phillips RL, Rines HW. Complex structure of knob DNA on maize chromosome 9: retrotransposon invasion into heterochromatin. Genetics. 1998;149:2025–2037. doi: 10.1093/genetics/149.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Barakat A, Carels N, Bernardi G. The distribution of genes in the genomes of Gramineae. Proc Natl Acad Sci USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Cox AV, Leitch IJ. Angiosperm DNA C-values database. 1998. http://www.rbgkew.org.uk/cval/database1.html http://www.rbgkew.org.uk/cval/database1.html (February 5, 2001) (February 5, 2001) [Google Scholar]
- Bennetzen J. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Corces VG. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Bortoluzzi S, d'Alessi F, Romualdi C, Danieli GA. The human adult skeletal muscle transcriptional profile reconstructed by a novel computational approach. Genome Res. 2000;10:344–349. doi: 10.1101/gr.10.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF, Feng DF. Tracing the origin of retroviruses. Curr Top Microbiol Immunol. 1992;176:195–211. doi: 10.1007/978-3-642-77011-1_13. [DOI] [PubMed] [Google Scholar]
- Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A. Ty1-copiagroup retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 1992;20:3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RB, Rimpau J, Smith DB. Repeated sequence DNA relationships in four cereal genomes. Chromosoma. 1977;63:205–222. [Google Scholar]
- Grandbastien M-A. Activation of retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. [Google Scholar]
- Garfinkel DJ, Hedge AM, Youngren SD, Copeland TD. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M-A, Spielmann A, Caboche M. Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature. 1989;337:376–380. doi: 10.1038/337376a0. [DOI] [PubMed] [Google Scholar]
- Gribbon BM, Pearce SR, Kalendar R, Schulman AH, Paulin L, Jack P, Kumar A, Flavell AJ. Phylogeny and transpositional activity of Ty1-copiagroup retrotransposons in cereal genomes. Mol Gen Genet. 1999;261:883–891. doi: 10.1007/pl00008635. [DOI] [PubMed] [Google Scholar]
- Hirochika H. Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Das OP, Messing J. Zeon-1, a member of a new retrotransposon family. Mol Gen Genet. 1995;248:471–480. doi: 10.1007/BF02191647. [DOI] [PubMed] [Google Scholar]
- Jääskeläinen M, Mykkänen A-H, Arna T, Vicient C, Suoniemi A, Kalendar R, Savilahti H, Schulman AH. Retrotransposon BARE-1: Expression of encoded proteins and formation of virus-like particles in barley cells. Plant J. 1999;20:413–422. doi: 10.1046/j.1365-313x.1999.00616.x. [DOI] [PubMed] [Google Scholar]
- Johns MA, Mottinger J, Freeling M. A low copy number, copia-like transposon in maize. EMBO J. 1985;4:1093–1102. doi: 10.1002/j.1460-2075.1985.tb03745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar R, Grob T, Regina M, Suoniemi A, Schulman AH. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet. 1999;98:704–711. [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankaanpää J, Mannonen L, Schulman AH. The genome sizes of Hordeumspecies show considerable variation. Genome. 1996;39:730–735. doi: 10.1139/g96-092. [DOI] [PubMed] [Google Scholar]
- Katz RA, Skalka AM. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- Keller B, Feuillet C. Colinearity and gene density in grass genomes. Trends Plant Sci. 2000;5:246–251. doi: 10.1016/s1360-1385(00)01629-0. [DOI] [PubMed] [Google Scholar]
- Kilian A, Chen J, Han F, Steffenson B, Kleinhofs A. Towards map-based cloning of the barley stem rust resistance genes Rpg1 and rpg4using rice as an intergenomic cloning vehicle. Plant Mol Biol. 1997;35:187–195. [PubMed] [Google Scholar]
- Kumar A, Bennetzen J. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Kurata N, Umehara Y, Tanoue H, Sasaki T. Physical mapping of the rice genome with YAC clones. Plant Mol Biol. 1997;35:101–113. [PubMed] [Google Scholar]
- Lazcano A, Valverde V, Hernandez G, P. G, Fox GE, Oro J. On the early emergence of reverse transcription: theoretical basis and experimental evidence. J Mol Evol. 1992;35:524–536. doi: 10.1007/BF00160213. [DOI] [PubMed] [Google Scholar]
- Manninen I, Schulman AH. BARE-1, a copia-like retroelement in barley (Hordeum vulgareL.) Plant Mol Biol. 1993;22:829–846. doi: 10.1007/BF00027369. [DOI] [PubMed] [Google Scholar]
- Manninen O, Kalendar R, Robinson J, Schulman AH. Application of BARE-1 retrotransposon markers to map a major resistance gene for net blotch in barley. Mol Gen Genet. 2000;264:325–334. doi: 10.1007/s004380000326. [DOI] [PubMed] [Google Scholar]
- Miyake T, Mae N, Shiba T, Kondo S. Production of virus-like particles by the transposable genetic element, copia, of Drosophila melanogaster. Mol Gen Genet. 1987;207:29–37. doi: 10.1007/BF00331487. [DOI] [PubMed] [Google Scholar]
- Noma K, Nakajima R, Ohtsubo H, Ohtsubo E. RIRE1, a retrotransposon from wild rice Oryza australiensis. Genes Genet Syst. 1997;72:131–140. doi: 10.1266/ggs.72.131. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Hirochika H. Efficient insertion mutagenesis of Arabidopsis by tissue culture-induced activation of the tobacco retrotransposon Tto1. Plant J. 2000;23:291–304. doi: 10.1046/j.1365-313x.2000.00779.x. [DOI] [PubMed] [Google Scholar]
- Panstruga R, Büschges R, Piffanelli P, Schulze-Lefert P. A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res. 1998;26:1056–1062. doi: 10.1093/nar/26.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SR, Harrison G, Heslop-Harrison JS, Flavell AJ, Kumar A. Characterization and genomic organization of Ty1-copia group retrotransposons in rye (Secale cereale) Genome. 1997;40:1–9. doi: 10.1139/g97-081. [DOI] [PubMed] [Google Scholar]
- Penn SG, Rank DR, Hanzel DK, Barker DL. Mining the human genome using microarrays of open reading frames. Nat Genet. 2000;26:315–318. doi: 10.1038/81613. [DOI] [PubMed] [Google Scholar]
- Roth JF. The yeast Tyvirus-like particles. Yeast. 2000;16:785–795. doi: 10.1002/1097-0061(20000630)16:9<785::AID-YEA550>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. A contiguous 66 kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suoniemi A, Anamthawat-Jónsson K, Arna T, Schulman AH. Retrotransposon BARE-1 is a major, dispersed component of the barley (Hordeum vulgareL.) genome. Plant Mol Biol. 1996a;30:1321–1329. doi: 10.1007/BF00019563. [DOI] [PubMed] [Google Scholar]
- Suoniemi A, Narvanto A, Schulman AH. The BARE-1 retrotransposon is transcribed in barley from an LTR promoter active in transient assays. Plant Mol Biol. 1996b;31:295–306. doi: 10.1007/BF00021791. [DOI] [PubMed] [Google Scholar]
- Suoniemi A, Schmidt D, Schulman AH. BARE-1 insertion site preferences and evolutionary conservation of RNA and cDNA processing sites. Genetica. 1997;100:219–230. [PubMed] [Google Scholar]
- Suoniemi A, Tanskanen J, Schulman AH. Gypsy-like retrotransposons are widespread in the plant kingdom. Plant J. 1998;13:699–705. doi: 10.1046/j.1365-313x.1998.00071.x. [DOI] [PubMed] [Google Scholar]
- Thomas CA. The genetic organization of chromosomes. Annu Rev Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein NM, Bennetzen JL, Avramova Z. Colinearity and its exceptions in orthologous adhregions of maize and sorghum. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernhettes S, Grandbastien MA, Casacuberta JM. In vivo characterization of transcriptional regulatory sequences involved in the defense-associated expression of the tobacco retrotransposon Tnt1. Plant Mol Biol. 1997;35:673–679. doi: 10.1023/a:1005826605598. [DOI] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Anamthawat-Jónsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF, Cummings MP, Konieczny AK, Ausubel FM, Rodermel SR. Copia- like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA. 1992;89:7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W. Genetic distribution of BARE-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP) Mol Gen Genet. 1997;253:687–694. doi: 10.1007/s004380050372. [DOI] [PubMed] [Google Scholar]
- Wessler SR. Turned on by stress. Plant retrotransposons. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]