Abstract
Although the Mutator (Mu) system is well characterized in maize (Zea mays), very little is known about this highly mutagenic system of transposons in other grasses. Mutator is regulated by the MuDR class of elements, which encodes two genes, one of which, mudrA, has similarity to a number of bacterial transposases. Experiments in our laboratory, as well as database searches, demonstrate that mudrA sequences are ubiquitous and diverse in the grasses. In several species it is clear that multiple paralogous elements can be present in a single genome. In some species such as wheat (Triticum aestivum) and rice (Oryza sativa), mudrA-similar sequences are represented in cDNA databases, suggesting the presence of active Mu transposon systems in these species. Further, in rice and in sorghum, mudrA-like genes are flanked by long terminal inverted repeats, as well as the short host sequence direct repeats diagnostic of insertion. Thus, there is ample evidence that systems related to Mu in maize are at least potentially active in a wide variety of grasses. However, the mudrB gene, though important for Mu activity in maize, is not necessarily a component of Mu elements in other grasses.
The Mutator (Mu) system is a highly mutagenic transposable element system that is used extensively for gene identification and mutagenesis in maize (Zea mays). Although a great deal is known about Mu in maize (for review, see Bennetzen, 1996), it is not well characterized in other grass species. However, there is good evidence of Mu-like elements (MuLE) in species as diverse as rice (Oryza sativa; Yoshida et al., 1998; Mao et al., 2000) and Arabidopsis (The Cold Spring Harbor Laboratory, WUGSC, and PE Biosystems Arabidopsis Sequencing Consortium, 2000), suggesting that this family of transposons is widespread in plants. Published reports demonstrate that a Mu transposase ortholog is expressed in rice callus (Yoshida et al., 1998), suggesting potential activity in this species.
In maize the Mu system is composed of a number of different variants, all of which have similar 200-bp terminal inverted repeats (TIRs), but each of which contains unrelated internal sequences (for review, see Chandler and Hardeman, 1992; Bennetzen 1996). All Mu elements are regulated by the MuDR class of elements (Chomet et al., 1991; Hershberger et al., 1991; James et al., 1993). MuDR elements carry two transcribed genes: mudrA and mudrB. The larger of the two, mudrA, encodes a protein, MURA, that has significant similarities to a number of bacterial transposases (Eisen et al., 1994). Hence, this protein is the putative transposase. The smaller gene, mudrB, encodes the protein MURB. This protein functions by an unknown mechanism. However, in our low-copy minimal Mutator line, deletion derivatives that carry and express only mudrA do not appear to cause germinal duplications, although they can still condition somatic excisions of a reporter element (Lisch et al., 1999). Similar experiments using a mudrA-only transgene gave similar results (Raizada and Walbot, 2000), suggesting that, in maize at least, mudrA and mudrB are necessary for a fully active Mu system.
Because grass phylogeny has been the subject of intense scrutiny (Clayton and Renvoize, 1986; Catalan et al., 1997; Kellogg 1998; Mathews et al., 2000 and refs. therein), a great deal is known about the phylogenetic relationships among grass species. Thus, there is an excellent comparative set of data for phylogenetic analysis of Mu evolutionary history. Further, because most of the worlds major crop species are grasses, the prospect of active transposons systems in many of them is an appealing one. For these reasons, an investigation of the Mu system in the grasses represents an excellent opportunity to understand the evolution of a highly complex and active transposon system, and raises the possibility of the development of tagging systems in grasses other than maize.
Our PCR strategies are designed to amplify elements that are most closely related to mudrA in maize. Thus, we would only expect to identify a subset of these sequences, and variations in specific sequences could result in a lack of amplification in any given species. We use DNA-blot hybridization whenever possible to limit this bias. However, even with this control, this work does not provide a comprehensive view of all mudrA-similar sequences. Nevertheless, analysis of our limited data set makes clear that there are multiple families of mudrA-similar sequences in the grasses, each one of which diverges to varying degrees from mudrA in maize.
RESULTS
Potentially Functional mudrA-Homologous Sequences Are Widespread among the Grasses
Our initial survey of the grasses employed DNA blots that were probed with a conserved portion of mudrA from maize and washed at low stringency (see “Materials and Methods”; Fig. 1A). DNA quantities were not generally adjusted for C value, although a relatively low amount of DNA was used for Coix lacryma and sorghum (Sorghum bicolor), both of which have a lower C value than does maize (Kellogg, 1998). These blots revealed the presence of hybridizing fragments in a number of species, including some as distant as two species of bamboo (Fig. 1A, lanes 20 and 21). In general, DNA from species closely related to maize hybridized most efficiently to our mudrA probe. C. lacryma (same subtribe as maize, lane 3) and sorghum (same tribe as maize, lane 4) hybridized less efficiently than expected. In contrast, DNA from three species of the genus Muhlenbergia (members of the Chloridoideae subfamily, lanes 9–11) hybridized more efficiently than expected based on the phylogenetic tree.
Figure 1.
DNA gel blots of various grass species. Abbreviations for grass species are given in Table I. A, DNA was digested with EcoRI + BamHI, blotted, and probed with the HindIII internal fragment of mudrA from maize. The upper portion is the ethidium-stained DNA of each sample before blotting. The tree is derived from (Kellogg, 1998). B, DNA was digested with EcoRI + BamHI, blotted, and probed with the HindIII internal fragment of mudrA from maize. The lower portion is the ethidium-stained DNA of each sample before blotting. C, DNA was digested with EcoRI + HindIII, blotted, and probed with the SalI internal fragment of mudrB.
To confirm these results, DNA samples of Zea luxurians, Tripsacum dactyloides, C. lacryma (at two concentrations), Muhlenbergia macroura, and Sporabolus airoides (same subtribe as Muhlenbergia) were blotted and probed with a portion of mudrA at a medium stringency (Fig. 1B). The results clearly showed that hybridization at this stringency was restricted to Z. luxurians (lane 1), T. dactyloides (lane 2), and M. macroura (lane 5). These results indicate that there is incongruity between the species phylogeny and the distribution of mudrA-similar sequences.
Conserved mudrB Sequences Are Less Widespread than mudrA Sequences
In contrast to mudrA, mudrB-similar sequences do not appear to be widely distributed, and the degree of hybridization matches species phylogeny. At low stringency only the closely related Zeas hybridize well to mudrB (Fig. 1C, lanes 1–3). DNA from a genus in the same subtribe as maize, Tripsacum (lanes 4 and 5), hybridized less efficiently, and DNA from C. lacryma (lane 6) less still. These mudrB hybridization results are as expected for any genomic sequence in these species. In contrast to the results using mudrA as a probe, DNA from species of the Muhlenbergia genus (lanes 10 and 11) does not hybridize to mudrB probe at this stringency.
PCR amplification using primer pairs designed to span the region between mudrA and mudrB were successfully used to produce amplicons that contain sequences of both genes in Z. luxurians and Z. diploperennis, demonstrating that both genes are found together in these species, presumably within MuDR transposons. It is interesting that although the genic regions are quite well conserved, the intergenic region, which contains a great deal of internally repetitive sequence, is poorly conserved (data not shown). Amplification from any species more distantly related to maize than the Zeas, using a variety of mudrB specific primers, has as yet been unsuccessful.
PCR Amplification of mudrA Orthologs
To gain a more detailed understanding of mudrA-similar sequences in various grasses, PCR primers were designed that could amplify a rice mudrA ortholog with a good open reading frame, as well as mudrA from maize. Selection of primer sites was based on the high degree of conservation of certain amino acids motifs present in previously identified rice (Ishikawa et al., 1996; Yoshida et al., 1998; Mao et al., 2000), Arabidopsis (The Cold Spring Harbor Laboratory, WUGSC, and PE Biosystems Arabidopsis Sequencing Consortium, 2000), and maize elements (Hershberger et al., 1991; Qin et al., 1991). It should be emphasized that these primers were not degenerate primers; they were specifically designed to amplify sequences most like mudrA (the only sequence known to be functional) rather than all sequences homologous to mudrA. Thus, a lack of amplification does not necessarily imply an absence of mudrA sequences. As additional sequences became available, amino acid alignments were used to design additional primers using a similar strategy.
Using these primers it was possible to amplify product from all the major subfamilies of grasses and most species examined (Table I). In all cases, an amplicon of the expected size proved to contain sequences similar to mudrA. Additional fragments of other sizes were produced in several of these species, but in all cases those amplicons proved to be completely unrelated to mudrA (data not shown). Because our primers amplified only a portion of the mudrA gene in these species, it is not known whether any particular sequence we obtained lies between TIRs. However, in several species, DNA-blot analysis revealed the presence of multiple fragments, consistent with their identity as transposable elements (Fig. 1A).
Table I.
Species with mudrA-like sequences
| Species | Abbreviation | Primers | No. of Clonesa | Open Reading Frameb |
|---|---|---|---|---|
| Ammophila arenaria | AA | VF1 + VR2 | 5 | No |
| Arundo donax | AD | VF4 + VR2 | 1 | No |
| Aristida purpurea | AP | VF1 + VR2 | 1 | Yes |
| Oat | AS | VF1 + VR2 | 1 | No |
| Bamboo (unknown species) | Bam | RF1 + RR2 | 2 | Yes |
| Briza maxima | BM | VF4 + VR2 | 2 | No |
| Calamagrostis acutifolia | CA | SOF2 + VR2 | 2 | Yes |
| C. lacryma | CL | VF1 + VR2 | 3 | Yes |
| Festuca rubra | FR | SOF2 + VR2 | 1 | No |
| Barley | HV | RF2 + RR2 | 1 | No |
| Muhlenbergia filiformis | MF | SOF2 + VR2 | 5 | Yes |
| M. macroura | MM | RF2 + RR2 | 1 | Yes |
| Muhlenbergia porteri | MP | RF2 + RR2 | 2 | Yes |
| Muhlenbergia rigens | MR | RF2 + RR2 | 2 | Yes |
| Rice | OS | PAc | PA | Yes |
| Panicum virgatum | PV | VF4 + VR2 | 4 | Yes |
| Setaria anceps | SaN | VF1 VR2 | 2 | Yes |
| Setaria sphacelata | SS | VF1 VR2 | 3 | No |
| Sinarundinaria murielae | SM | RF2 + RR2 | 3 | No |
| Sinarundinaria nitida | SN | RF2 + RR2 | 3 | No |
| Sorghum | SB | PA, NAd | 0 | No |
| S. airoides | SA | NA | 0 | – |
| T. dactyloides | TD | RF2 + RR2 | 1 | No |
| Wheat | TA | RF2 + RR2 | 1 | No |
| Veteveria zizanioides | VZ | RF2 + RR2 | 2 | Yes |
| Z. diploperennis | ZD | RF2 + RR2 | 3 | Yes |
| Z. luxurians | ZL | RF2 + RR2 | 3 | Yes |
| Maize | ZM | PA | PA | Yes |
A list of species from which PCR using mudrA-specific primers was attempted. Abbreviations and primers are as listed in “Materials and Methods.” Sequences similar to mudrA in maize were successfully amplified from most species tested.
No. of clones from a given PCR amplification sequenced.
Sequence contains no stop or missense mutations.
Sequence previously available.
No successful amplification.
Selective Constraints on Sequence Variation of mudrA Orthologs
Although many of these nucleotide sequences are quite different from mudrA in maize, ranging from 50% to 60% identity, many of the inferred amino acids are conserved among all or nearly all orthologs (Fig. 2), and the frequency of similar and identical amino acids is considerably higher than that of identical nucleotides in all sequences examined. The nucleotide sequence of the mudrA sequence from M. macroura, for instance, is 65% identical to mudrA in maize, but the protein sequence is 78% similar to MURA in maize. Several of the products we amplified encoded portions of intact open reading frames, consistent with continued function (Table I). It is not surprising that a number of the conserved amino acids are identical or similar to those already identified as being part of a Mutator motif present in mudrA and a large number of bacterial transposases (Eisen et al., 1994). To detect evidence of selection, the method of Nei and Gojobori (1986) was used to compare synonymous with non-synonymous mutations between all possible pairs of sequences. The number of changes for each class of mutation was expressed as a proportion of the number of possible changes, and that figure was subjected to the Jukes Cantor correction for multiple hits. Using only those sequence pairs whose proportion of synonymous mutations was below saturation (<0.75), the average ratio of corrected synonymous to non-synonymous substitutions (dS/dN) was found to be 6.6. Further, in all pairs examined, the uncorrected proportion of synonymous substitution was higher (often at or above saturation) compared with the proportion of non-synonymous substitutions. Taken together, these data clearly suggest that these sequences have been under selective pressure, consistent with the hypothesis that these sequences remained active following divergence from their common ancestor with mudrA in maize.
Figure 2.
Alignment of putative translation products of mudrA-similar sequences from several grass species. Only a subset of the sequences without stop or missense codons are shown. Identical amino acids are shaded dark. Similar amino acids are shaded in gray.
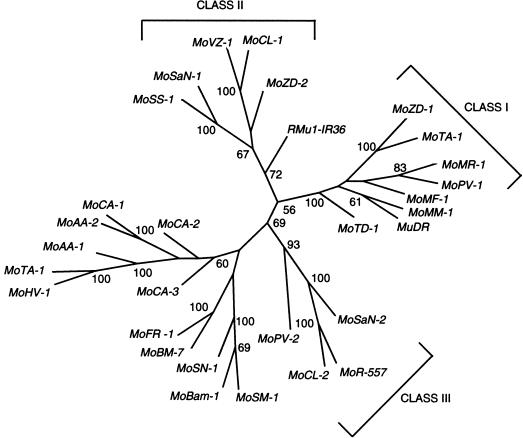
mudrA Sequences Are Organized into Distinct Subfamilies
When these mudrA sequences are subjected to phylogenetic analysis, they fall into several distinct groups or classes (Fig. 3) that are often independent of species phylogenetic relationships. Class I includes mudrA from maize and its close relatives, as well as a sequence (MoPV-1) from P. virgatum (a different subtribe), and all of the sequences from the genus Muhlenbergia (a different subfamily). Class II includes sequences from both species of Setaria we examined (MoSaN-1and MoSS-1), as well as sequences from C. lacryma (MoCL-1), Z. diploperennis (MoZD-2), and V. zizanioides (MoVZ-1). Class III includes one of the sequences from C. lacryma (MoCL-2), one from S. anceps (MoSaN-2), one from P. virgatum (MoPV-2), as well an element from rice (MoOS-557, accession no. BAA89557). Sequences from the Pooidea generally group together, as do those from the three species of Bambusoideae we examined.
Figure 3.
Phylogenetic tree of various mudrA-similar sequences in various grasses. Bootstrap values greater than 50% are indicated at the nodes.
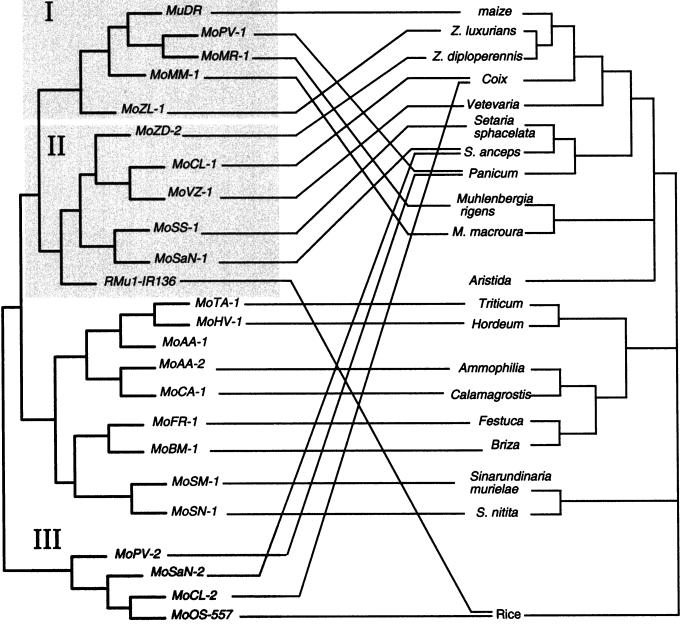
It is quite apparent that phylogenetic trees made from the sequences we amplified do not match that of the grasses in which they are found (Fig. 4). Most striking, the sequences amplified from three Muhlenbergia species (different subfamily than maize), as well as one sequence we have from P. virgatum (different tribe than maize) are consistently more similar to mudrA in maize than are sequences from species as closely related to maize as C. lacryma (same subtribe as maize) and even the sequences from Z. luxurians and Z. diploperennis (same genus as maize).
Figure 4.
Comparison of an unrooted phylogenetic tree of selected mudrA-similar sequences (left) and a tree of the species from which they were derived (right). For clarity, only a subset of the sequences are shown. Shaded boxes indicate class designations shown in Figure 3. Branch lengths of the species tree, which was adapted from Kellogg (1998), are not correct.
Individual genomes can carry more than one class of sequence. For instance, Z. dactyloides and has class I and class II elements, and P. verbatim has class I and class III elements. C. lacryma has representatives of class II and class III elements, but it appears to lack class I elements, the class to which mudrA from maize belongs. The lack of amplification of class I elements in C. lacryma could be due to incomplete sampling due to bias introduced by PCR. However, DNA from the Muhlenbergia species hybridizes more intensely to mudrA than does DNA from C. lacryma (Fig. 1B), suggesting that class I elements are indeed missing in this species. In a similar manner, we were unable to hybridize to sorghum DNA using mudrA from maize, and our PCR from this species using a several primer pairs was unsuccessful. Database searches revealed that there are at least two sorghum elements that have mudrA-similar sequences (MoSB-1 and MoSB-2; Table II), but they are much more distantly related to MuDR than any of the sequences we successfully amplified. It is interesting that one of the two elements, MoSB-1, is closer to a maize element recently identified in the Chandler laboratory, MuSE (M. Stam, D. Selenger, and V.L. Chandler, personal communication) than it is to MuDR in maize (data not shown). The second element, MoSB-2, is quite similar to the group that includes MoOS-557 (Fig. 5B). As in the case of C. lacryma, these data suggest that sorghum may have lost one class of mudrA-containing element, but retained others.
Table II.
Distant relatives of MuDR
| Element | Size | Accession No. | Terminal Inverted Repeatsa | Direct Repeatsb | Expressedc |
|---|---|---|---|---|---|
| MuDR | 4,942 | M76978 | 220 | 9 | Yes |
| RMu1-A23 | 4,374 | AB023047 | 156 | 9 | Yes |
| MoR-557d | 3,420 | BAA89557 | 182 | 9 | Unknown |
| TRAP | 6,823 | CAB51950 | 232 | 15e | Yes |
| MoSB-2 | 16,496f | AF061282 | 185 | 9 | Unknown |
| MoOS-521 | 6,296 | BAA92521 | 218 | 9 | Unknown |
| MoSB-1 | 3,133 | AAD27572 | 114 | 8 | Unknown |
| MuSE | 4,632 | None | 500 | 9 | Unknown |
| Jittery | 3,916 | AF247646 | 185 | 9 | Unknown |
| MoOS-J1 | 4,387 | AC078840 | 140 | 8 | Unknown |
Various MuLE in rice, Arabidopsis, and maize. Elements share long-inverted repeats and sequences similar to mudrA.
Terminal inverted repeat length in base pairs.
Direct host sequence repeat in base pairs.
98%+ homology to a cDNA sequence in the database or direct molecular evidence.
Probable deletion derivative.
Degenerate.
Includes putative insertion of retroelements.
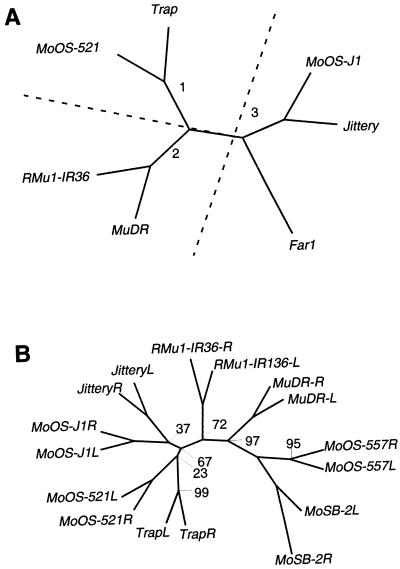
Figure 5.
Phylogenetic trees of several MuLE elements. A, Phylogenetic tree of MuLE elements in rice (MoOS-521, MoOS-J1, and RMu1-IR136), maize (Trap, MuDR, and Jittery), and Arabidopsis (Far1). The tree is derived from parsimony analysis of amino acids from the conserved transposase domain (see Fig. 6) of these sequences. All nodes of this tree had bootstrap values of 100 with the exception of that leading to MoOS-J1 and Jittery, which had a bootstrap value of 85. The numbers refer to the branches. 1, MuDR; 2, Trap; and 3, Jittery. B, A phylogenetic tree of the TIRs of several distinct MuLE elements. Bootstrap values are as indicated at the nodes. Nodes without values ascribed had a value of 100.
DISCUSSION
Our observation that species such as C. lacryma and P. vergatum can carry more than one distinct class of mudrA orthologous sequence is part of a larger pattern of inter- and intraspecific variation. Database searches reveal that there are multiple paralogous mudrA-similar sequences in rice and maize that are even more remotely related to mudrA than our PCR-amplified sequences. We refer to all elements carrying mudrA-similar sequences and long TIRs or their derivatives as MuLEs. Like the sequences we PCR amplified, phylogenetic analysis of these elements does not match that of the species in which they are found (Fig. 5). The presence of these elements suggests that MuLE elements represent a broadly diversified group of transposable elements that can coexist as potentially active elements within a single genome. Thus, the various classes identified in our analysis may represent an earlier stage in a continuous process of element diversification.
In maize, in addition to MuDR, there is a MuLE identified as part of a polycistronic message that includes a Hox1A homolog (Comelli et al., 1999). This element, designated Trap (transposon-associated protein) contains degenerate (imperfect) TIRs and host sequence direct repeats flanking a mudrA-orthologous sequence (accession no. CAB51950). A database search reveals that rice contains an element (MoOS-521, accession no. BAA92521) that is more similar to Trap than it is to MuDR or to other rice mudrA orthologs (Fig. 5), suggesting that the Trap-like branch of elements existed prior to the divergence of rice and maize, and that selection has maintained limited similarity with MuDR in this group. However, the conservation of amino acid sequence between Trap and MoOS-521 suggests that selection has also operated differentially on this group of elements, resulting in a number of branch-specific amino acids (Fig. 6).
Figure 6.
An alignment of the conserved transposase domain of elements from rice, maize, and Arabidopsis. Identical amino acids are shaded dark. Similar amino acids are shaded in gray. “Staff” refers to a transposase from the bacteria Staphylococcus aureus (accession no. AAA88546).
A recently identified transposon, Jittery, in maize has clear similarities to mudrA, and contains characteristic long inverted repeats (H. Dooner; accession no. AF247646). However, it is more similar to sequences in rice and Arabidopsis than to MuDR. In rice, an element with TIRs and host sequence direct repeats (MoOS-J1, accession no. AC078840) clusters with Jittery in a phylogenetic tree (Fig. 5). It is interesting that Jittery is also more closely related to a gene in Arabidopsis, Far1 (accession no. AAD51282), than it is to MuDR in maize. Mutations in Far1 are deficient in the response to far-red light (Hudson et al., 1999). This surprising relationship between a maize transposon and a gene with a clear host function suggests that a MuLE element in Arabidopsis may have been coopted to operate as part of a host-encoded response pathway. This hypothesis is supported by the absence of identifiable TIRs around the Far1 gene, since selection would not favor their retention if the element was no longer a transposable element. In addition to Jittery, Far1, and MoOS-J1, database searches reveal the presence of a number of genomic cDNA sequences from monocots and dicots that appear to be part of this class of sequences (Hudson et al., 1999).
Rice contains an element, RMu1-IR36 (Ishikawa et al., 1996), that is more similar to MuDR than it is to Trap and Jittery class elements (Fig. 5). This element most likely represents the closes relative of MuDR, although it is closer to class II elements than it is to mudrA in maize (Fig. 4).
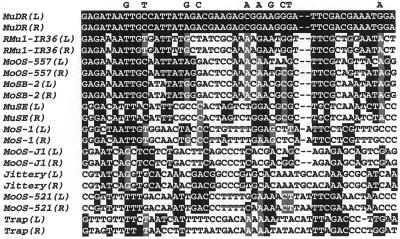
The TIRs of the each of these widely disparate elements have few sequences in common (Fig. 7), although they are all longer on average that the TIRs of other plant transposons (Table II), suggesting a requirement by the mudrA class of transposases for relatively long TIRs. With the exception of Trap whose TIRs are clearly degenerate, the left-right homology of these TIRs range from 85% to 95% over their length. Further, in all cases there is evidence of host sequence direct repeats, consistent with insertion. Phylogenetic analysis of these TIRs reveals that although the tree derived from parsimony analysis is not well supported at all nodes, it is generally consistent with the tree derived from the transposases (Fig. 5B). It is notable that the MoSB-2 (from sorghum) TIRs are 76% identical to those of MoOS-557 (from rice) over the 185-bp length of the MoSB-2 TIR. It is interesting that this TIR sequence in sorghum is identified as a high-copy repeat sequence in the database (“HCSR-2,” accession no. AF061282).
Figure 7.
An alignment of the 5′ portion of the terminal inverted repeats of several MuLE elements. Nucleotides shaded dark are identical to MuDR. Nucleotides shaded gray are identical to one of the active Mu elements in maize.
The database for rice and maize is as yet incomplete, and at least three distinct branches of MuLEs have already been identified in both of them. It is likely that additional subfamilies will be identified as additional sequences for these species are obtained. It is clear from the data already available that MuLE elements represent a broadly diversified and widely distributed group of transposable elements. It is interesting to note that of all the elements identified to date, only MuDR in maize contains the mudrB gene. The mudrB gene, which is required for germinal insertions of Mu elements in maize, matches nothing in the database. Analysis of the natural history of this gene will be the subject of a future report.
Database searches also suggests that mudrA-similar transcripts may be ubiquitous. Our wheat (Triticum aestivum) product is 96% identical over 494 bp to a wheat cDNA (BE497524), suggesting that the element in wheat is expressed. In rice, RMu1-A23 (accession no. AB023047) is 99% identical to a cDNA from rice (accession no. C98506) and a rice cDNA with homology to mudrA has been isolated previously (Yoshida et al., 1998). Further, TBLASTN searches of the cDNA database (National Center for Biotechnology Information, dbest) using MURA protein sequence gives significant hits (e value < e10−7) for additional transcripts in sorghum (accession no. BE600044), and in non-grasses such as Arabidopsis (accession no. AV557094), potato (accession no. BF187650), tomato (accession no. AW979689), and gray mangrove (accession no. AU108521). These results suggest that MuLE sequences are expressed at some level in many species, suggesting that these species may have active transposon systems.
The presence of such a diverse array of Mu-like transposons whose distribution does not always match species phylogenies raises some interesting questions regarding the dynamics of Mu element evolution. Functional transposons are only maintained in a given genome if they can continue to duplicate themselves or if they can provide a benefit to their host. The Far1 gene suggests that some MuLE sequences may provide such a benefit. However, if it is assumed that most Mu transposons are selectively neutral or negative, there are no selective forces maintaining their integrity (Charlesworth, 1988). Only the process of duplication provides the raw material for positive selection on transposases. Any transposon lineage that ceases to transpose will eventually succumb to the random noise of background mutation and become irreversibly inactive (Robertson and Lampe, 1995). However, there are clearly selective pressures to limit the activity of transposable elements (Nuzhdin, 1999). Mutator active lines in maize demonstrate how potentially mutagenic the Mu system can be, and there is an increasing body of evidence that hosts have a variety of mechanisms to prevent transposon activity (for review, see Matzke et al., 1999). To avoid extinction, transposons must stay active, but selection on the negative consequences of transposition leads to repression of transposon activity. The evolutionary history of any transposon reflects a shifting balance between these two forces.
One mechanism that has been proposed for continued activity of transposon lineages is horizontal transfer to transposons from “immunized” hosts in which regulatory mechanisms have evolved to “naive” hosts, which lack such specific mechanisms (Kidwell, 1994). The best-documented case of horizontal transfer is that between Drosophila willistoni and fruit fly (Daniels et al., 1990; Clark et al., 1994). Subsequent to the putative transfer event, P elements spread rapidly through the world-wide population of fruit fly (Kidwell, 1994). There is good evidence that this was not the first such horizontal transfer of P elements (Silva and Kidwell, 2000). In a similar manner, phylogenetic analysis of mariner-like elements suggests many horizontal transfers of these elements may have occurred (Hartl et al., 1997; Robertson, 1997). It has been proposed that such horizontal transfer events are the primary means by which DNA based transposable elements maintain their activity over long periods of time (Robertson and Lampe, 1995).
Given the presence of multiple paralogous lineages of mudrA-containing elements in the grasses, it is difficult to distinguish horizontal transfer from selective retention of particular subfamilies. Based on our phylogenetic analysis of PCR products, as well as our DNA-blot analysis, it appears that class I (the class to which mudrA belongs) sequences are missing in the Panicoids Coix, sorghum, Vetevaria, and Setaria, and in the Chlorinoid S. airoides. The class I sequences are present in the Chlorinoid Muhlenbergia species, and the Panicoid, P. virgatum. These data can be explained by two horizontal transfers (one to P. virgatum and one to Muhlenbergia), or two losses (once in the lineage leading to C. lacryma, sorghum, and V. zizanioides and once in S. airoides). Given the presence of multiple classes of elements that may predate the divergence of all of these species, we find the loss hypothesis to be more parsimonious. Additional sequences using degenerate primers will help to determined whether class I elements are truly missing in all of these species.
Supporting the possibility of horizontal transfer of MuLE elements, some similarities from database searches are strikingly high for a transposable element sequence that would be expected to have a reasonably high mutation rate. For instance, a wheat cDNA (accession no. BE443436) is 86% identical at the nucleotide level and 85% identical and 91% similar at the amino acid level to a portion of MoOS-373 (accession no. BAA82373) from rice. In a similar manner, a class II element from C. lacryma is 96% identical at the nucleotide level to a class II element from V. zizanioides. These results suggests a very strong degree of selection or horizontal transfer between these species or their ancestors.
A second mechanism for continued activity that one could hypothesize is the rapid production of subvarients of transposable elements. If we assume that the amplification of one subclass of elements provokes a response by the host specific in some way to that subclass, then transposon variants may emerge that are different enough to avoid that regulation. We have ample evidence that mudrA-containing elements have produced multiple subvarients that can coexist within the same genome. Despite having been only partially sequenced, rice and maize contain at least three distinct branches of MuLEs. In each case at least one member of each branch appears to have been active recently enough to have retained evidence of insertion in the form of host sequence direct repeats (Table II). The continuing selection for certain key amino acids in all branches (Figs. 2 and 6) suggests continued selection for activity over at least 70 million years. The conservation of amino acid sequence within specific branches such as Trap suggests that more than one active branch of MuLE elements can exist in a given genome. Further, if Mu is any guide, each branch or class of elements may have its own family of nonautonomous elements, as is suggested by the presence of an element in rice that has Mu-like TIRs flanking unrelated sequences (accession no. X16597). It will be interesting to see if and how nonautonomous variants derived from one branch of MuLEs interact with MuLEs from other branches.
The presence of potentially active mudrA-containing transposons in many species, including Arabidopsis and maize, suggests that active Mu transposons have been a component of plant genomes at least since the origin of the monocots. Although we have no direct evidence for active Mu transposons in species other than maize, we do have strong circumstantial evidence for recent, or even current Mu activity in a number of the grasses. The presence of multiple branches and classes of MuLEs within individual genomes suggest that differentiation of elements within individual species lineages may be a common occurrence. A fascinating subject for future investigation will be whether and how and these distinct branches of elements interact with each other and with their hosts.
MATERIALS AND METHODS
DNA Gel-Blot Analysis
DNA extraction and DNA gel-blot analysis was performed according to Dorweiller and coworkers (Dorweiler et al., 2000). Blots were hybridized at 65°C and washed at 60°C in 1× SSPE, 0.2% (w/v) SDS (medium stringency), or at 60°C and washed at 55°C in 1× SSPE, 0.2% (w/v) SDS (low stringency) with three changes of wash solution in each case. The mudrA probe used was the internal HindIII fragment from mudrA from a cDNA from C. hardeman (Hershberger et al., 1995). The mudrB probe was an SalI internal fragment of mudrB (Chomet et al., 1991).
Plant Material
Plant material was collected from the University of California-Berkeley Botanical Garden, grown from seeds supplied by the Royal Botanical Gardens at Kew, and several commercial sources. Accession numbers and collection data are available upon request.
PCR Primers
PCR primers for the initial screen were designed to sequences encoding well-conserved amino acids in rice (Oryza sativa) and maize (Zea mays). One set of primers was specific to mudrA in maize and one set was specific to rice. The rice sequence of RMu1-IR36 was provided by Ryuji Ishikawa, who isolated a MuDR-like element from maize (Ishikawa et al., 1996). This element is quite similar to one deposited in the database by Dr. Ishikawa (RMu1-A23, accession no. AB023047). As expected, based on DNA sequence, both sets of primers could amplify products in both species. However, the rice primers yielded a more distinct amplicon in rice and maize and proved to be more reliable in amplification of discrete bands in a number of species, perhaps due to additional competing sequences present in maize and its close relatives. Therefore, the rice-specific primers were used in most instances. To amplify sequences from which the rice and maize primers did not work, additional primers were employed as additional sequences became available. PCR primers used to amplify the amplicons described in Table I were the following: RF2, CTTAGTGTAAACTCAACTGC; SF2, CTTAATGGTAGGTGGAATGG; VF1, CTTGCTATTTGCACTGATGC; VF4, GCTATGCGACAGTATGCAAT; RR2, GGCTTGCCAGTGTGTTGCCA; and VR2, TGGTTTCCCAGTGTGTTGCC
Letter designations refer to the species from which the primers was designed: R from rice, V from Vetevaria zizanioides, and S from Setaria anceps. PCR conditions were then 94°C for 30 s, 48°C for 45 s, and 72°C for 45 s. The samples were subjected to 35 rounds of amplification, and were then gel isolated, purified using the Qiaquick Gel Extraction kit (Qiagen, Valencia, CA), and subcloned using the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA). Following transformation, when possible, several individual clones from each amplification reaction were sequenced. Sequencing was performed by the University of California, Berkeley sequencing facility, using an ABI sequencer.
DNA Sequence Analysis
Database searches were performed using the BLAST algorithm. Multiple alignments were performed using a web site maintained by the Belozersky Institute of Physico-Chemical Biology (www.genebee.msu.su/services/malign reduced.html) and were corrected by hand based on the correct reading frame of mudrA in maize. The alignments shown in Figures 2, 6, and 7 were displayed using the boxshade web site (www.ch.embnet.org/software/BOX form.html). Phylogenetic analysis was performed using the PHYLIP package (version 3.5) available through the University of Washington. Trees in Figures 3, 4, and 5 were derived from DNA alignments analyzed using DNADIST for distances and NIEGHBOR for tree inference. Distances were bootstrapped 100 times. The maximum likelihood method was employed, using a transition/transversion ratio of 1.4 based on the analysis of the coding sequences from several plant species (Langdon et al., 2000). The phylogenetic tree in Figure 5A was derived using the PROTPARS program from the PHYLIP package, with 100 bootstraps.
Nomenclature
Devising nomenclature for transposable elements is rarely clear-cut, and there are no accepted practices for doing so. However, in the interests of clarity, we have attempted where possible to name these elements systematically. Individual mudrA-like sequences are named with the prefix “Mo” followed by a letters for the genus and species of origin. Thus, a sequence from Zea luxurians is designated “MoZL,” with successive numbers for additional sequences from the same species (e.g. MoZL-1). In those cases where the transposase was previously identified as such in the database, the numerical designation is drawn from the last three letters of the protein accession number of that transposase. Thus, an element from rice carrying a transposase with the accession number BAA89557 is designated ”MoOS-557.“ Previously identified elements such as the Trap (Comelli et al., 1999) and Jittery (H. Dooner: accession no. AF247646) elements from maize are left with their original designation.
We refer to all Mu-like elements as MuLEs. This includes all elements with long TIRs and significant homology to mudrA, as well as derivatives and nonautonomous variants thereof. Major subdivisions, or branches, of this general group of MuLEs include the Jittery branch of elements, the Trap branch of elements, and the MuDR branch of elements. Within the MuDR branch of elements, we designate subclades as classes distinguished by Roman numerals. Thus, the clade that includes MuDR is class I of the MuDR branch of elements. This class includes sequences from maize, and its close relatives, as well as sequences from Muhlenbergia and Panicum.
ACKNOWLEGEMENTS
Thanks to Randall Tyers and Nancy Nelson for a critical reading of the manuscript.
Footnotes
This work was supported entirely by the Novartis Agricultural Discovery Institute Inc. (now Syngenta Agricultural Discovery Institute) University of California-Berkeley Strategic Alliance.
LITERATURE CITED
- Bennetzen JL. The Mutatortransposable element system of maize. Curr Top Microbiol Immunol. 1996;204:195–229. doi: 10.1007/978-3-642-79795-8_9. [DOI] [PubMed] [Google Scholar]
- Catalan P, Kellogg EA, Olmstead RG. Phylogeny of Poaceae subfamily Pooideaebased on chloroplast ndhF gene sequences. Mol Phylogenet Evol. 1997;8:150–166. doi: 10.1006/mpev.1997.0416. [DOI] [PubMed] [Google Scholar]
- Chandler VL, Hardeman KJ. The Mu elements of Zea mays. Adv Genet. 1992;30:77–122. doi: 10.1016/s0065-2660(08)60319-3. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The maintenance of transposable elements in natural populations. Basic Life Sci. 1988;47:189–212. doi: 10.1007/978-1-4684-5550-2_14. [DOI] [PubMed] [Google Scholar]
- Chomet P, Lisch D, Hardeman KJ, Chandler VL, Freeling M. Identification of a regulatory transposon that controls the Mutatortransposable element system in maize. Genetics. 1991;129:261–270. doi: 10.1093/genetics/129.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JB, Maddison WP, Kidwell MG. Phylogenetic analysis supports horizontal transfer of Ptransposable elements. Mol Biol Evol. 1994;11:40–50. doi: 10.1093/oxfordjournals.molbev.a040091. [DOI] [PubMed] [Google Scholar]
- Clayton WD, Renvoize SA. Genera Graminum. London: Her Majesty's Stationary Office; 1986. [Google Scholar]
- Comelli P, Konig J, Werr W. Alternative splicing of two leading exons partitions promoter activity between the coding regions of the maize homeobox gene Zmhox1a and Trap(transposon-associated protein) Plant Mol Biol. 1999;41:615–625. doi: 10.1023/a:1006382725952. [DOI] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A. Evidence for horizontal transmission of the P transposable element between Drosophilaspecies. Genetics. 1990;124:339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL. Mediator of paramutation 1is required for the establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Benito MI, Walbot V. Sequence similarity of putative transposases links the maize Mutatorautonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 1994;22:2634–2636. doi: 10.1093/nar/22.13.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Lohe AR, Lozovskaya ER. Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- Hershberger RJ, Benito MI, Hardeman KJ, Warren C, Chandler VL, Walbot V. Characterization of the major transcripts encoded by the regulatory MuDRtransposable element of maize. Genetics. 1995;140:1087–1098. doi: 10.1093/genetics/140.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger RJ, Warren CA, Walbot V. Mutator activity in maize correlates with the presence and expression of the Mu transposable element Mu9. Proc Natl Acad Sci USA. 1991;88:10198–10202. doi: 10.1073/pnas.88.22.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Lisch D, Freeling M. Screening of Mutator-related sequence in rice. Rice Genet News. 1996;13:146–147. [Google Scholar]
- James MG, Scanlon MJ, Qin M, Robertson DS, Myers AM. DNA sequence and transcript analysis of transposon MuA2, a regulator of Mutatortransposable element activity in maize. Plant Mol Biol. 1993;21:1181–1185. doi: 10.1007/BF00023614. [DOI] [PubMed] [Google Scholar]
- Kellogg EA. Relationships of cereal crops and other grasses. Proc Natl Acad Sci USA. 1998;95:2005–2010. doi: 10.1073/pnas.95.5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. The Wilhelmine E. Key 1991 Invitational Lecture: the evolutionary history of the P family of transposable elements. J Hered. 1994;85:339–346. doi: 10.1093/oxfordjournals.jhered.a111478. [DOI] [PubMed] [Google Scholar]
- Langdon T, Seago C, Mende M, Leggett M, Thomas H, Forster JW, Jones RN, Jenkins G. Retrotransposon evolution in diverse plant genomes. Genetics. 2000;156:313–325. doi: 10.1093/genetics/156.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D, Girard L, Donlin M, Freeling M. Functional analysis of deletion derivatives of the maize transposon MuDRdelineates roles for the MURA and MURB proteins. Genetics. 1999;151:331–341. doi: 10.1093/genetics/151.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Woo S, Sasinowski M, Presting G, Frisch D, Goff S. Rice transposable elements: a survey of 73,000 sequence-tagged- connectors. Genome Res. 2000;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, Tsai RC, Kellogg EA. Phylogenetic structure in the grass family (Poaceae): evidence from the nuclear gene phytochrome B. Am J Bot. 2000;87:96–107. [PubMed] [Google Scholar]
- Matzke MA, Mette MF, Aufsatz W, Jakowitsch J, Matzke AJ. Host defenses to parasitic sequences and the evolution of epigenetic control mechanisms. Genetica. 1999;107:271–287. [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and non-synonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV. Sure facts, speculations, and open questions about the evolution of transposable element copy number. Genetica. 1999;107:129–137. [PubMed] [Google Scholar]
- Qin MM, Robertson DS, Ellingboe AH. Cloning of the Mutator transposable element MuA2, a putative regulator of somatic mutability of the a1-Mum2allele in maize. Genetics. 1991;129:845–854. doi: 10.1093/genetics/129.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada MN, Walbot V. The late developmental pattern of Mutransposon excision is conferred by a cauliflower mosaic virus 35S-driven MURA cDNA in transgenic maize. Plant Cell. 2000;12:5–21. doi: 10.1105/tpc.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. Multiple Marinertransposons in flatworms and hydras are related to those of insects. J Hered. 1997;88:195–201. doi: 10.1093/oxfordjournals.jhered.a023088. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Lampe DJ. Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol Biol Evol. 1995;12:850–862. doi: 10.1093/oxfordjournals.molbev.a040262. [DOI] [PubMed] [Google Scholar]
- Silva JC, Kidwell MG. Horizontal transfer and selection in the evolution of P elements [In Process Citation] Mol Biol Evol. 2000;17:1542–1557. doi: 10.1093/oxfordjournals.molbev.a026253. [DOI] [PubMed] [Google Scholar]
- The Cold Spring Harbor Laboratory WUGSC, Biosystems Arabidopsis Sequencing Consortium PE. The complete sequence of a heterochromatic island from a higher eukaryote. Cell. 2000;100:377–386. [PubMed] [Google Scholar]
- Yoshida S, Tamaki K, Watanabe K, Fujino M, Nakamura C. A maize MuDR-like element expressed in rice callus subcultured with proline. Hereditas. 1998;129:95–99. doi: 10.1111/j.1601-5223.1998.t01-1-00095.x. [DOI] [PubMed] [Google Scholar]