Significance
We report on a zebrafish model of hearing loss in Usher syndrome type IIIA caused by pathogenic variant c.144T>G (p.N48K) in the clarin1 gene (CLRN1). We expressed CLRN1c.144T>G in wild-type or clarin1 knockout zebrafish and then probed these animals with genetic or pharmaceutical small-molecular agents. Our findings revealed that the CLRN1N48K protein is transported to the hair cell bundle via an unconventional secretory pathway, and activation of this pathway using artemisinin, an antimalarial drug, enhanced CLRN1N48K levels in hair cell bundles and rescued their functions. Our report demonstrates a paradigm to evaluate the functional consequence of human pathogenic variants in hair cells in vivo and reveals artemisinin’s therapeutic potential to mitigate sensory deficits in CLRN1c.144T>G and other endoplasmic reticulum aggregation disorders.
Keywords: USH3A, hearing loss, mitigation
Abstract
The pathogenic variant c.144T>G (p.N48K) in the clarin1 gene (CLRN1) results in progressive loss of vision and hearing in Usher syndrome IIIA (USH3A) patients. CLRN1 is predicted to be an essential protein in hair bundles, the mechanosensory structure of hair cells critical for hearing and balance. When expressed in animal models, CLRN1 localizes to the hair bundle, whereas glycosylation-deficient CLRN1N48K aggregates in the endoplasmic reticulum, with only a fraction reaching the bundle. We hypothesized that the small amount of CLRN1N48K that reaches the hair bundle does so via an unconventional secretory pathway and that activation of this pathway could be therapeutic. Using genetic and pharmacological approaches, we find that clarin1 knockout (clrn1KO/KO) zebrafish that express the CLRN1c.144T>G pathogenic variant display progressive hair cell dysfunction, and that CLRN1N48K is trafficked to the hair bundle via the GRASP55 cargo-dependent unconventional secretory pathway (GCUSP). On expression of GRASP55 mRNA, or on exposure to the drug artemisinin (which activates GCUSP), the localization of CLRN1N48K to the hair bundles was enhanced. Artemisinin treatment also effectively restored hair cell mechanotransduction and attenuated progressive hair cell dysfunction in clrn1KO/KO larvae that express CLRN1c.144T>G, highlighting the potential of artemisinin to prevent sensory loss in CLRN1c.144T>G patients.
Hereditary hearing loss (HHL) is a common sensory disorder associated with various gene mutations and is often linked to defects in the mechanosensory “hair” cells of the inner ear (1–3). To gain insight into the role of the human gene in hair cell function and hearing loss, orthologs of HHL-associated genes and their pathogenic counterparts are investigated in animal models (4–10). Research into the etiology and treatment of HHL would be expedited if we can directly investigate HHL-associated human genes and their pathogenic variants in hair cells in vivo.
A paradigm for this scenario is the pathogenic variant c.144T>G in the human clarin1 (CLRN1) gene, which causes Usher syndrome IIIA (USH3A), a rare genetic disorder that results in progressive loss of vision and hearing (11, 12). The pathogenic variant c.144T>G in CLRN1 causes the amino acid asparagine (N) at position 48, a conserved N-glycosylation site, to be replaced by lysine (K) in human clarin1 protein (CLRN1N48K). In vitro studies have demonstrated that translation of the CLRN1c.144T>G pathogenic variant produces glycosylation-deficient CLRN1, which is retained in the endoplasmic reticulum (ER) and is prone to degradation by the proteasome (13–15). Furthermore, when expressed in wild-type (WT) mouse or zebrafish hair cells, CLRN1 localizes to the hair bundle, whereas CLRN1N48K is largely retained intracellularly, with only a fraction reaching the bundle (16). Here, we investigated the pathway that enabled glycosylation-deficient CLRN1N48K to reach the hair bundle and whether pharmaceutical activation of this pathway could be therapeutic. We chose the zebrafish for this investigation for the following reasons. First, zebrafish larvae are transparent with accessible sensory cells, permitting direct observation of hair bundle morphology, localization of fluorescent fusion proteins in vivo, and quantification of hair cell function via electrophysiological recordings. Second, considerable evidence indicates conservation of gene function across species, from zebrafish to humans (8–10).
In this report, we first determined whether the expression of human CLRN1 could functionally substitute for a zebrafish host’s clarin1 to establish conservation of function across species and suitability of a fish as a host for this study. This laid the groundwork that allowed us to develop a zebrafish model for an inner ear disorder associated with the pathogenic variant CLRN1c.144T>G. Our investigation revealed that the small amount of CLRN1N48K that reaches the hair bundle is assisted by the GRASP55 cargo-dependent unconventional secretory pathway (GCUSP). Furthermore, we demonstrated that the molecular sequelae associated with the retention of CLRN1N48K in the ER are the likely cause of progressive loss of hair cells and hearing. Most significantly, we demonstrated that the pharmaceutical agents that induce the GCUSP, such as the antimalarial drug artemisinin (ART), enhance the localization of CLRN1N48K to hair bundles and restore hair cell mechanotransduction function similar to WT levels, effectively attenuating the progressive loss of hair cell function. These results reveal the potential of GCUSP activators as therapeutics to prevent sensory loss in CLRN1c.144T>G USH3A patients and in other disorders that involve ER aggregation of mutant protein.
Results
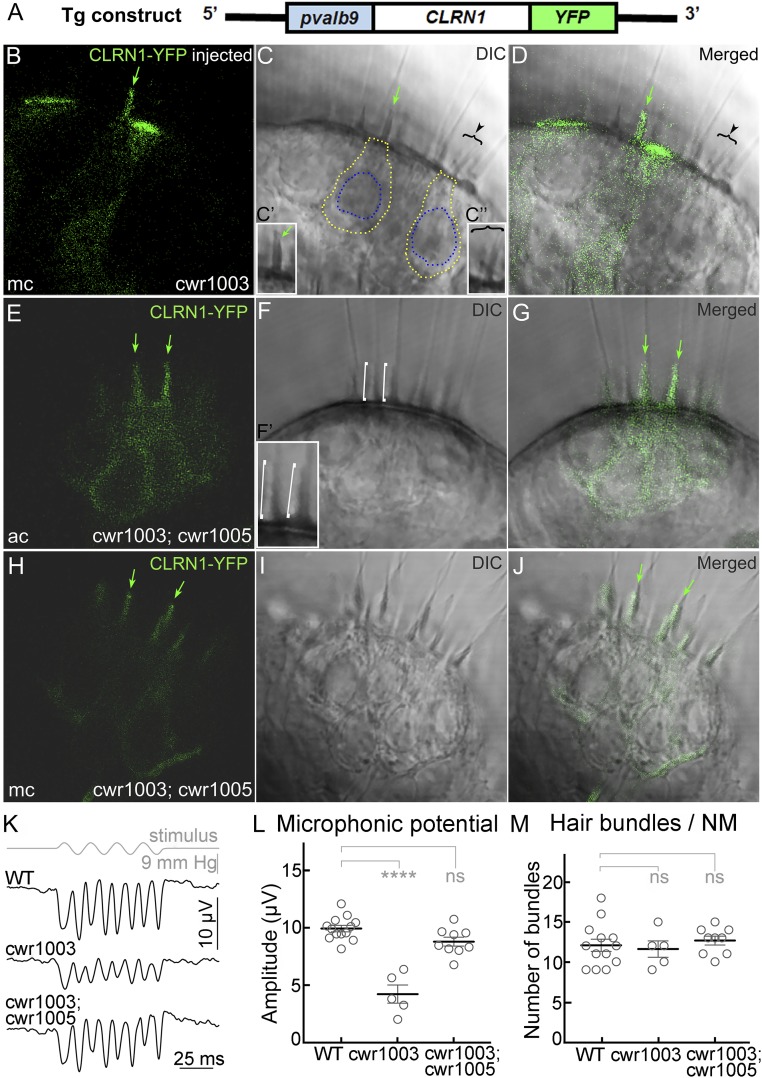
The Human Clarin1 Protein Completely Rescues the Hair Cell Phenotype of clrn1KO/KO Zebrafish.
Based on the functional analysis of clarin1 orthologous proteins in mice and zebrafish, and the localization of CLRN1 expressed in the hair cells of WT zebrafish, it was hypothesized that CLRN1 plays a role in preserving hair bundle structure and function (14, 16, 17). To test this hypothesis, we first used somatic-cell expression of a specific transgene in zebrafish. This involved the injection of the Tg(pvalb9:Hsa.CLRN1-YFP) transgene construct (Fig. 1A) into single-cell–stage clrn1KO/KO zebrafish (cwr1003) embryos (F0 generation) and fluorescence imaging at 6 days post fertilization (dpf). The pvalb9 promoter sequence directs transgene expression primarily in hair cells (18). In somatic-cell expression (in contrast to germline expression), only a few cells in each of the F0 generation larvae are expected to express the transgene. This allows for a side-by-side comparison of the phenotype of cells expressing the transgene to adjacent cells negative for transgene expression. Representative images from the inner ear of the injected larvae (n = 30) showed that only YFP-positive hair cells display WT bundle morphology (cone shaped; green arrow in Fig. 1 B–D and single cone-shaped bundle enlarged in the Inset, Fig. 1C′), whereas the hair bundles of the surrounding YFP-negative hair cells were splayed or disrupted (brackets with the arrowhead, Fig. 1 B–D, and single splayed bundle enlarged in the Inset, Fig. 1C″), similar to the cwr1003 hair cells reported earlier (16). The ability of CLRN1 to restore normal hair bundle structure in all hair cells in the cwr1003 larvae was confirmed by the analysis of the transgenic cwr1003 larva (cwr1003; cwr1005) (SI Appendix, Table S1) that stably express CLRN1-YFP in hair cells (n = 50). Representative images of hair cells from the inner ear of cwr1003; cwr1005 larvae are shown in Fig. 1 E–J; all of the hair cells shown here express YFP and have cone-shaped bundles, demonstrating uniform rescue (Fig. 1 E–J). Representative bundles are marked in Fig. 1F, and in the enlarged Inset (Fig. 1F′). To determine whether rescue of structure translates to rescue of function, the mechanosensory function of the bundles in the cwr1003; cwr1005 line was evaluated. Microphonic potentials were measured in response to fluid jet stimulation of hair cells in the neuromasts of larvae from WT (9.977 ± 0.283 µV; mean ± SEM), cwr1003 (4.22 ± 0.801 µV; mean ± SEM), and cwr1003; cwr1005 (8.828 ± 0.392 µV; mean ± SEM) genotypes at 6 dpf (Fig. 1 K–M). The amplitude of the microphonic potentials showed the complete rescue of mechanotransduction function in the hair cells of cwr1003; cwr1005 larvae compared with WT larvae (one-way ANOVA, P = 0.1 is not significant; Fig. 1L). In comparison, microphonic potentials were three times lower in cwr1003 than in WT larvae (one-way ANOVA, P < 0.0001; Fig. 1L), even though no significant difference was found between the numbers of hair cells in the neuromasts of the above-mentioned three groups (Fig. 1M). The cwr1003; cwr1005 larvae were phenotypically indistinguishable from the WT larvae. Specifically, startle response (depends on hearing), swimming behavior, and survival were similar to those of the age-matched WT larvae (Fig. 2 K and L; and SI Appendix, Table S2). These results confirm our hypothesis and support several important conclusions, including (i) CLRN1 function is conserved across species, (ii) CLRN1 expression in hair cells is sufficient to prevent inner ear dysfunction associated with loss-of-function mutations in CLRN1, (iii) the fusion of YFP to the C terminus of CLRN1 does not affect the protein’s localization or function, and (iv) the zebrafish is a suitable in vivo model for the functional analysis of WT and pathogenic missense variants, such as CLRN1c.144T>G.
Fig. 1.
CLRN1 restores hair bundle structure and function in cwr1003 hair cells. (A) Schematic representation of the DNA construct used to generate the cwr1003; cwr1005 transgenic line; pvalb9, promoter sequence from the zebrafish parvalbumin 9 gene. (B–D) Cone-shaped hair bundle morphology (green arrow) observed in hair cells of the medial crista (mc) expressing CLRN1-YFP, and split or splayed hair bundle morphology observed in adjacent YFP-negative hair cells (arrowhead and bracket), as revealed by the DIC (C) and merged (D) images. The marked bundles are enlarged in the Insets (C′ and C″). (E–J) The hair bundle morphology is uniformly cone-shaped (similar to WT) in all hair cells in the anterior crista (ac) and medial crista (mc) of cwr1003; cwr1005 larvae, which express CLRN1-YFP. DIC images reveal cone-shaped bundles across both cristae (F and I), and merged images (G and J) confirm colocalization of the YFP signal and cone-shaped bundles. The green arrows in B–E, G, H, and J point to cone-shaped or rescued bundle morphology in CLRN1-YFP–expressing hair cells; white brackets in the DIC image (F) point to two representative cone-shaped bundles enlarged in the Inset (F′). (Magnification: 63×.) (B–J) The 1-µm middle (optical) section images of the crista captured from live larvae at 6 dpf. (K–M) Microphonic potentials recorded from the neuromast (NM) hair cells of cwr1003; cwr1005, and control siblings. (K) Representative traces of microphonic potentials recorded from different genotypes, and stimulus are shown. The top trace shows pressure applied to the stimulating puff pipette. (L) Summary of the microphonic potential peak-to-peak amplitudes at twice the stimulus frequency. Average values of microphonic potentials obtained from the lateral line neuromasts of WT, cwr1003, and cwr1003; cwr1005 larvae at 6 dpf. (M) Number of hair bundles per neuromast from WT, cwr1003, and cwr1003; cwr1005 larvae. Analysis of the same larvae is shown in L and M (n = 13 in WT; n = 5 in cwr1003; and n = 9 in cwr1003; cwr1005 groups). Data shown in L and M represent the mean ± SEM. Asterisks indicate statistical significance, and “ns” indicates nonsignificance, one-way ANOVA with Tukey’s multiple-comparisons test: ns, P > 0.05; ****P < 0.0001.
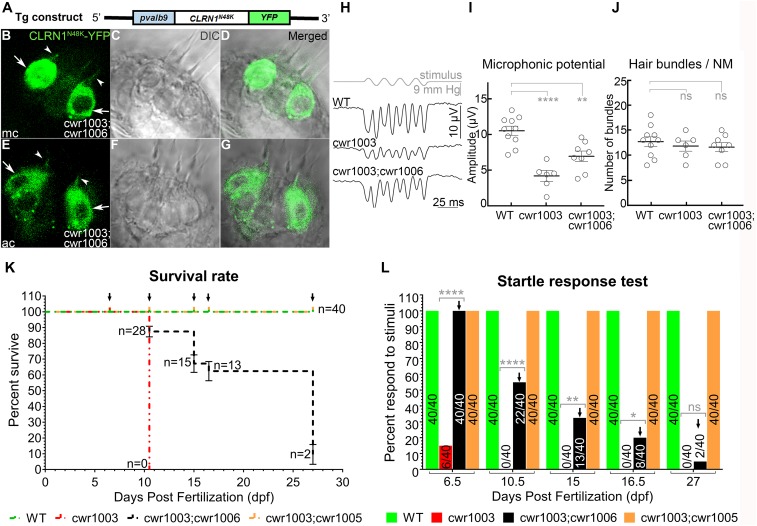
Fig. 2.
Progressive loss of hair cell function in cwr1003 larvae expressing CLRN1N48K. (A) Schematic representation of the DNA construct used to generate the cwr1003; cwr1006 transgenic line. (B–G) Representative images of hair cells from the medial crista (mc) (B) and anterior crista (ac) (E). The majority of CLRN1N48K expressed in the cwr1003 hair cells was mistargeted (arrows, B and E), and relatively small amounts reached the hair bundle (arrowhead, B and E) The bundle morphology in cwr1003; cwr1006 larvae was cone shaped. Merged images of YFP and DIC panels reveal that the YFP signal coincides with the cone-shaped bundle morphology. The DIC image of the corresponding crista in C, D, F, and G indicates the presence of CLRN1N48K in the bundle rescue cwr1003 larvae hair bundle phenotype. Images captured from live larvae at 6 dpf. Images shown are single plane (optical section) images and are 1-µm middle section of the crista. (Magnification: 63×.) (H–J) Microphonic potentials from hair cells of the neuromast from cwr1003; cwr1006 larvae reveals that CLRN1N48K expression improves mechanosensory function of cwr1003 larvae bundles but fails to reach WT levels. (H) Representative traces of microphonic potentials recorded from WT, cwr1003, and cwr1003; cwr1006 larvae are shown here. The top trace shows pressure applied to the stimulating puff pipette. (I) Summary of microphonic potential peak-to-peak amplitudes at twice the stimulus frequency. Average values of microphonic potentials obtained from lateral line neuromasts of zebrafish larvae from the following genotypes, WT (n = 10), cwr1003 (n = 6), and cwr1003; cwr1006 (n = 8), at 6–7 dpf. (J) The number of hair cells per neuromast were quantified in the same WT, cwr1003, and cwr1003; cwr1006 larvae used to analyze microphonic potential amplitudes. (K) Quantification of the cwr1003; cwr1006 zebrafish survival rate in comparison with cwr1003 zebrafish using the Kaplan–Meier survival analysis. (L) Number of larvae that displayed startle response diminished over time, and it coincided with the number of larvae that survived, indicative of a close relationship between hair cell function and survival, and variable-onset progressive loss of hair cell function in cwr1003; cwr1006 larvae. Data in I and J represent the mean ± SEM. Asterisks indicate statistical significance, and “ns” indicates nonsignificance, one-way ANOVA with Tukey’s multiple-comparisons test: ns, P > 0.05; *P ≤ 0.05; **P < 0.01; ****P < 0.0001.
The cwr1003 Zebrafish Expressing CLRN1c.144T>G Display Variable, Progressive Loss of Hair Cell Function.
To understand how CLRN1c.144T>G-expressing hair cells acquire function and whether that pathway could be manipulated to sustain hair cell function, we generated cwr1003 zebrafish that stably express CLRN1N48K-YFP from the transgene Tg(pvalb9:Hsa.CLRN1_c.144T>G-YFP) (Fig. 2A) in hair cells (cwr1003; cwr1006) (SI Appendix, Table S1). In contrast to the localization of CLRN1-YFP (Fig. 1), CLRN1N48K-YFP remained largely intracellular (arrow, Fig. 2 B and E), with a relatively small amount reaching the bundle (arrowhead, Fig. 2 B and E). Importantly, all of the CLRN1N48K-YFP–positive hair cells showed WT-like cone-shaped bundle morphology (Fig. 2 D and G). Moreover, microphonic potentials recorded from neuromasts of cwr1003; cwr1006 larvae (6.906 ± 0.725 µV; mean ± SEM) were considerably larger than those of cwr1003 mutants (4.058 ± 0.724 µV; mean ± SEM) (one-way ANOVA, P = 0.037; Fig. 2I). The observed preservation of WT-like cone-shaped hair bundle structure in cwr1003; cwr1006 larvae (Fig. 2 C and F), and the relative improvement in the function of cwr1003; cwr1006 hair cells in comparison with cwr1003 hair cells (Fig. 2 H–J), strongly suggested that CLRN1N48K-YFP can deliver CLRN1-mediated function in the bundle. Although the expression of CLRN1N48K-YFP in the cwr1003 background larvae (cwr1003; cwr1006) improved the microphonic potentials of the hair cells by nearly twofold compared with the cwr1003 control, these extracellular potentials are not equivalent to those recorded from the hair cells of WT zebrafish larvae (10.48 ± 0.643 µV; mean ± SEM) (one-way ANOVA, P = 0.003; Fig. 2I). These findings suggest that the reduced hair cell mechanotransduction in CLRN1c.144T>G-expressing hair cells is caused by a reduced function or amount of the mutant protein reaching the bundle.
At 6.5 dpf, WT larvae often swim toward the middle or closer to the surface of the E3 medium in the Petri dish and escape swiftly from the field of view when startled by tapping the dish with a metal probe as described previously (16, 19). Unlike WT larvae, at 6.5 dpf, about 40% (16 of 40) of cwr1003; cwr1006 larvae exhibited subdued swimming behavior and an attenuated startle response; they tended to settle at the bottom of the dish and move in “short spurts,” displaying an incomplete escape response to tapping. Some larvae required several taps to stimulate the animals to move out of the field of view (SI Appendix, Fig. S1, Movies S1 and S2). To monitor the preservation of hair cell function over time, larvae from several clutches were observed longitudinally from 6.5 to 27 dpf for survival, swimming behavior, and startle response (Fig. 2 K and L and SI Appendix, Table S2). Although many of the cwr1003; cwr1006 larvae survived beyond 12 dpf, which was significantly improved viability relative to the cwr1003 larvae (95% of cwr1003 larvae did not survive beyond 12 dpf), only a small percentage of cwr1003; cwr1006 larvae (5%; n = 2) survived beyond 25 dpf. During this period (6.5–25 dpf), the startle response of the cwr1003; cwr1006 larvae declined, indicating that the sensory function of hair cells deteriorated in most of these larvae (Fig. 2 K and L and SI Appendix, Table S2). However, ∼5% of the cwr1003; cwr1006 zebrafish did survive to adulthood, which was long enough for them to breed. Overall, these results show that the cwr1003; cwr1006 zebrafish larvae display progressive loss of hair cell function with a variable onset, and that while their hair cell function is superior to that of cwr1003 larvae, it fails to reach WT levels.
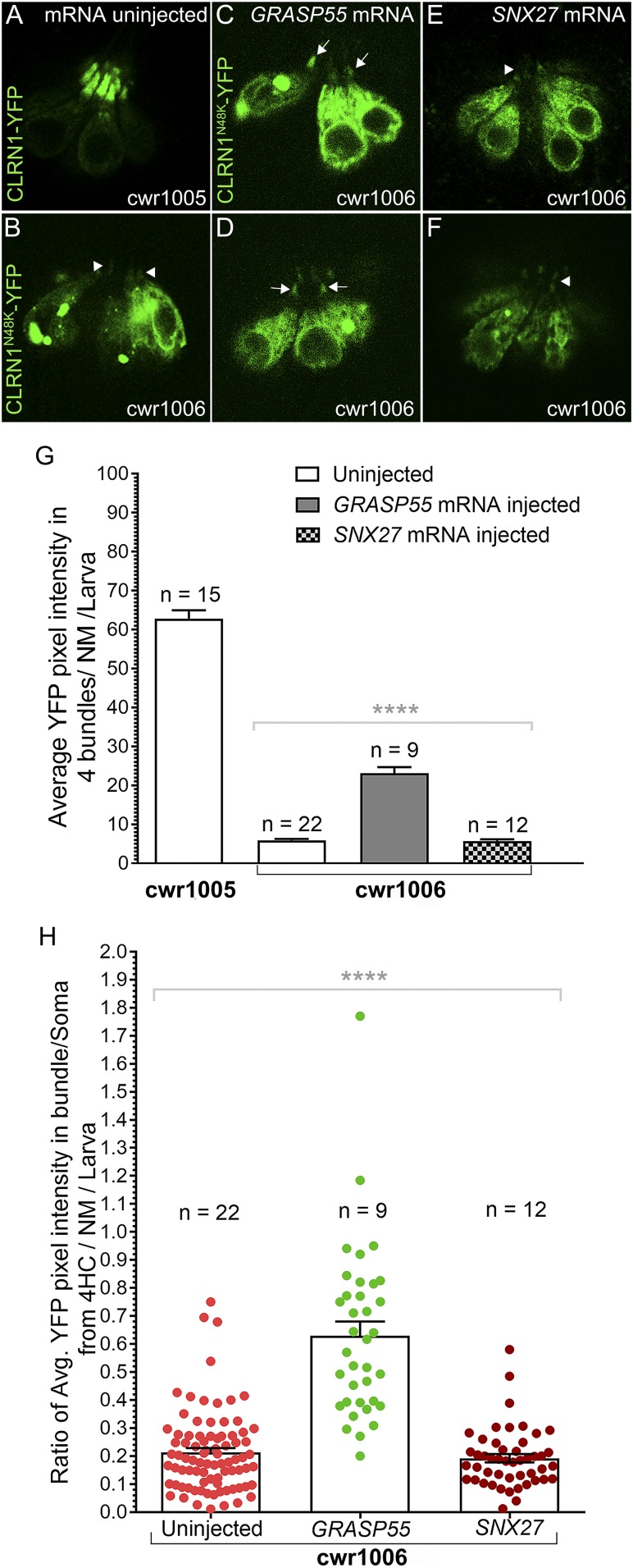
CLRN1N48K Depends on an Unconventional Secretory Pathway Cargo Protein, GRASP55, to Localize to the Hair Bundles.
Our results showed that, although CLRN1N48K-YFP remained largely intracellular, a relatively small amount reached the bundle contributing to a normal hair bundle structure and a partial restoration of the hair cell function of cwr1003; cwr1006 larvae (Fig. 2 B–H). Therefore, next we investigated the mechanism mediating the localization of glycosylation-deficient CLRN1N48K to the bundle of hair cells. The cargo proteins GRASP55 and SNX27 are known to assist ER-sequestered proteins in trafficking to the plasma membrane via a Golgi-independent, unconventional protein secretory pathway (20, 21). Furthermore, GRASP55 and SNX27 play a role in sorting internalized, misfolded or unfolded transmembrane proteins that contain the PDZ type I domain binding site (PDB type I) to the plasma membrane via different unconventional protein secretory pathways under certain physiological conditions, such as during the unfolded protein response (UPR)-triggered up-regulation of ER stress signaling (20, 22). Since CLRN1 contains a type I PDB site, we tested whether trafficking of CLRN1N48K-YFP to the hair bundle is associated with GRASP55 or SNX27. We injected GRASP55 or SNX27 mRNA into single-cell–stage cwr1006 embryos (WT zebrafish line stably expressing CLRN1N48K-YFP) (SI Appendix, Table S1) and quantified the amount of CLRN1N48K-YFP reaching the bundle at 4 dpf. Uninjected cwr1005 and cwr1006 larvae served as the control (Fig. 3 A and B). For this experiment, we chose the WT background to avoid potential confounding factors (changes that culminate in hair cell degeneration) in the cwr1003 background that may affect the interpretation of results.
Fig. 3.
GRASP55 expression enhances hair bundle localization of CLRN1N48K. (A–F) Hair cells in the neuromast of cwr1006 larvae were observed for changes in the localization pattern of CLRN1N48K-YFP in the GRASP55 or SNX27 mRNA-injected groups (C–F) and in the uninjected controls (A and B). Compared with the control (arrowheads, B), hair cells from GRASP55 mRNA injected larvae showed an increased amount of CLRN1N48K-YFP in the bundle as reflected by increased intensity of the YFP signal in the bundle (arrows, C and D), but no such difference was noticed in SNX27 mRNA-injected larvae (arrowheads, E and F). Images in A–F were captured from live larvae, and these images represent a single plane (optical sections) of 1-µm thickness. (Magnification: 40×.) (G) Quantification of YFP intensity in the bundle of cwr1006 larvae injected with GRASP55 or SNX27 mRNA compared with YFP intensity in the bundle of uninjected cwr1006 larvae. (H) The ratio of average YFP intensity in the bundle versus the soma quantified from four hair cells (4HC) per neuromast per larva from either of the GRASP55 or SNX27 mRNA-injected and uninjected control groups. Data in G and H represent the mean ± SEM. Asterisks indicate statistical significance; one-way ANOVA with Tukey’s multiple-comparisons test: ns, P > 0.05; ****P < 0.0001.
In the GRASP55 mRNA injected cwr1006 group, 9 of 25 larvae showed an increase in the intensity of YFP in the hair bundles (Fig. 3 C and D) compared with the hair bundles from the uninjected control cwr1006 larvae (Fig. 3B). By contrast, none of the 25 (0 of 25) cwr1006 larvae injected with SNX27 mRNA (Fig. 3 E and F) showed any appreciable difference in the intensity of YFP in the hair bundles relative to the uninjected control cwr1006 larvae (n = 22; Fig. 3B). Quantification of the amount of CLRN1N48K-YFP that localized to the hair bundle by counting YFP pixel intensities revealed that compared with the uninjected control cwr1006 larvae (YFP pixels, 5.927 ± 0.32; mean ± SEM) and SNX27 mRNA-injected cwr1006 group (YFP pixels, 5.745 ± 0.42; mean ± SEM), the GRASP55 mRNA-injected cwr1006 hair cells showed a significantly increased amount of CLRN1N48K-YFP present in the hair bundles (YFP pixels, 23.12 ± 1.56; mean ± SEM) (Fig. 3G). To show that the increased intensity of the YFP-tagged protein in the bundle is due to increased trafficking and not an artifact of variable expression of the injected transcript and/or the YFP-tagged protein in the hair cells, we calculated the ratio of changes in the YFP pixels in the bundle versus its soma for four hair cells per neuromast per larva. The YFP pixels ratio for GRASP55 mRNA-injected larvae was higher (0.63 ± 0.05; mean ± SEM) compared with the SNX27 mRNA-injected (0.193 ± 0.015; mean ± SEM) or uninjected cwr1006 (0.213 ± 0.016; mean ± SEM) controls (Fig. 3H). The differences in the ratio were statistically significant (P < 0.0001) (Fig. 3H). We also carried out similar analysis for the cwr1005 larvae injected with GRASP55 mRNA, to show that the GRASP55 mRNA injection did not increase or decrease the level of CLRN1-YFP in the bundle or soma (SI Appendix, Fig. S2). These results show that increased trafficking of CLRN1N48K to the bundle is associated with GRASP55 mRNA injection, and it supports the hypothesis that CLRN1N48K is transported to the hair bundle by the GRASP55 cargo-dependent unconventional secretory pathway (GCUSP) and not the SNX27-dependent unconventional secretory pathway.
Sarco/Endoplasmic Reticulum Ca2+-ATPase Inhibitors Rectify the CLRN1c.144T>G Pathogenic Variant Phenotype.
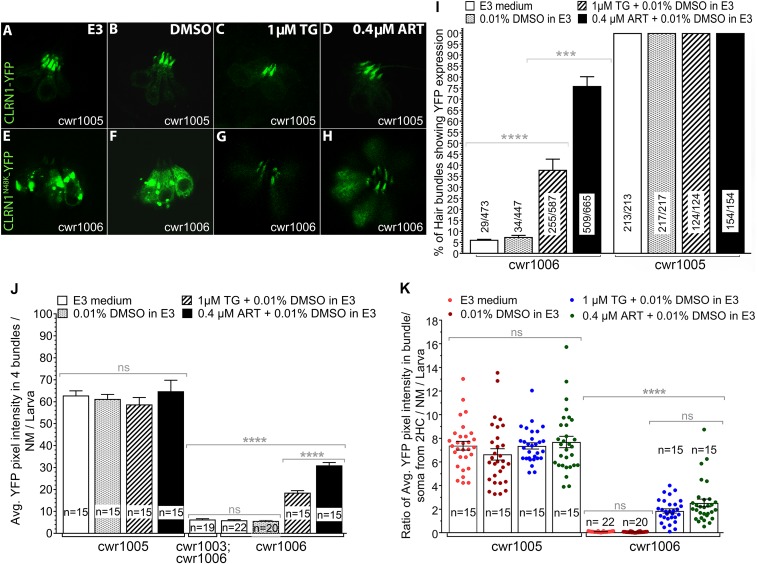
Next, we tested whether activation of the GCUSP would liberate the ER-retained CLRN1N48K protein and enhance its trafficking to the hair cell plasma membrane and hair bundle. We used two inhibitors of the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA): thapsigargin (TG), an anticancer drug, and ART, an antimalarial drug. SERCA controls the levels of ER luminal calcium stores, with its inhibition resulting in reduced calcium levels (23), which in turn triggers the UPR and the GCUSP in mammalian cells (20).
Zebrafish larvae from the cwr1005 and cwr1006 zebrafish lines at 3 dpf were treated with five different TG and ART doses and faced five different drug exposure durations, selected based on earlier reported nonlethal doses of these drugs that perturb ER Ca2+ levels and trigger the unconventional protein secretory pathway (SI Appendix, Table S3 and S4) (24). Live confocal images of neuromast hair cells were captured from the treated and untreated groups (Fig. 4 A–H), and the amounts of CLRN1N48K-YFP and CLRN-YFP that localized to the hair bundles of cwr1006 and cwr1005, respectively, were quantified at various time points after treatment (SI Appendix, Tables S3 and S4). The results were most effective in the 1 µM TG dosage group at 4 h of treatment and the 0.4 µM ART-treated group at 4 h (Fig. 4). At these concentrations, ART was more potent than TG at restoring CLRN1N48K-YFP levels in the hair bundles (Fig. 4 I–K). Specifically, the number of hair bundles showing strong expression of CLRN1N48K-YFP protein was significantly higher (P < 0.0001) in the ART-treated group (76%), compared with the TG-treated group (38%) and to hair bundles from untreated CLRN1N48K-YFP hair cells (<5%) (Fig. 4I). Although the levels achieved after treatment were still lower than those observed in WT CLRN1, the average amount of CLRN1N48K-YFP per bundle (quantified as YFP pixels) in cwr1006 larvae treated with TG or ART was threefold and fivefold higher, respectively, than the untreated control or the control treated with the vehicle cwr1006 (Fig. 4J). To determine whether the increased level of CLRN1N48K-YFP in the bundle is due to increased trafficking and not an artifact of variable CLRN1N48K-YFP expression in the transgenic cwr1006 line, we determined the ratio of fluorescence in the bundle versus its soma for two hair cells per neuromast per larva following exposure to TG and ART (Fig. 4K). The data showed an increased ratio of CLRN1N48K-YFP in the cwr1006 larvae upon exposure to TG or ART compared with the unexposed cwr1006 larvae. Compared with its unexposed counterpart, no significant change in the ratio was observed in the cwr1005 larvae exposed to TG or ART (Fig. 4K), indicating that exposure to these drugs did not alter the level or localization of WT CLRN1 in hair cells (Fig. 4 C and D). The results showed that modulating the SERCA pump activity increases the levels of CLRN1N48K-YFP in the hair bundle and supports the role of the GCUSP in liberating the ER-sequestered CLRN1N48K protein and trafficking it to the hair cell plasma membrane.
Fig. 4.
SERCA inhibitors TG and ART enhance trafficking of CLRN1N48K to the hair bundle. SERCA inhibitors do not affect localization or expression levels of CLRN1 in hair cells. SERCA inhibitors induced release of CLRN1N48K retained in the ER and enhanced trafficking of the mutant protein to the hair bundle. (A–D) Neuromast hair cells from cwr1005 larvae at 4 dpf, and (E–H) neuromast hair cells from cwr1006 larvae at 4 dpf. (A and E) E3 medium. (B and F) Treated with 0.01% DMSO in E3 medium. (C and G) Treated with 1 µM TG. (D and H) Treated with 0.4 µM ART. Images (A–H) were captured from live larvae at 40× magnification. (I) Hair bundle localization of CLRN1 or CLRN1N48K in control and TG- or ART-treated larvae were quantified as a function of YFP intensity in the bundle. In all larvae analyzed, hair cells of the otic and middle neuromasts were considered for statistical analysis. Data represented as a percentage of the number of hair cells expressing CLRN1-YFP or CLRN1N48K-YFP in the hair bundle divided by the total number of hair cells considered for analysis. (J) Quantification of YFP intensity in the bundle from TG- or ART-treated and control groups. (K) Ratio of average YFP intensity in the bundle versus soma quantified from hair cells of the neuromast from TG- or ART-treated and control groups. Data in J and K represent the mean ± SEM. Asterisks indicate statistical significance, and “ns” indicates nonsignificance, one-way ANOVA with Tukey’s multiple-comparisons test: ****P < 0.0001; ***P < 0.001; ns, P > 0.05.
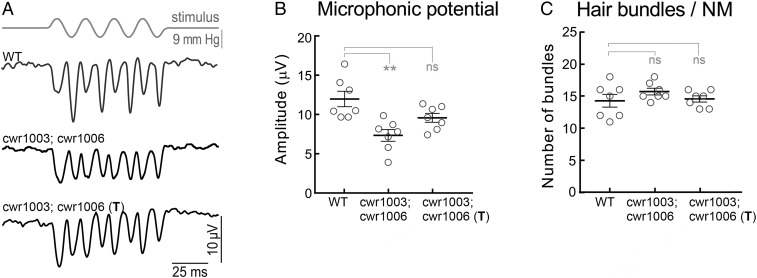
Next, we tested whether increased trafficking of CLRN1N48K to the hair bundle induced by the SERCA inhibitor ART resulted in improved hair cell function. We treated cwr1003; cwr1006 larvae with 0.4 µM ART for 4 h/d over 4 consecutive days, from 3 to 6 dpf, and then we recorded microphonic potentials from the hair cells of the L1 and L2 neuromasts at 6 dpf. The microphonic potential values of the treated cwr1003; cwr1006 larvae were comparable to the traces and values recorded from the WT larvae (Fig. 5 A and B) and the cwr1003; cwr1005 larvae (Fig. 1 K and L), and the difference was not statistically significant. The microphonic potentials data demonstrate that ART treatment restored hair cell function in cwr1003; cwr1006 larvae. Furthermore, these results support the idea that the reduced hair bundle function observed in the untreated cwr1003; cwr1006 larvae (Fig. 5 A and B) is less likely due to attenuated function of the mutant protein and more likely due to reduced amounts of CLRN1N48K reaching the hair bundles.
Fig. 5.
ART treatment rescued hair cell mechanotransduction in cwr1003; cwr1006 larvae. (A) Representative microphonic potential traces recorded from lateral line neuromasts of WT, cwr1003; cwr1006, and ART-treated cwr1003; cwr1006 (T) zebrafish larvae at 6 dpf. The top trace shows pressure applied to the stimulating puff pipette. (B) Summary of microphonic potential peak-to-peak amplitudes at twice the stimulus frequency. Average values of microphonic potentials were obtained from WT, cwr1003; cwr1006, and cwr1003; cwr1006 (T) larvae. (C) Number of hair cells per neuromast from WT, cwr1003; cwr1006, and cwr1003; cwr1006 (T) larvae. Analysis of the same larvae (n = 7 in all groups) is shown in B and C. Data in B and C represent the mean ± SEM. Asterisks indicate statistical significance, and “ns” indicates nonsignificance, one-way ANOVA with Tukey’s multiple-comparisons test: ns, P > 0.05; **P < 0.01.
Next, to evaluate the efficacy of ART at the organism level, we exposed cwr1003; cwr1006 larvae to a defined regimen of ART and determined the degree of rescue in swimming behavior and prolongation of life span as it correlates to the durable rescue of hair cell function in mutant larvae. For this experiment, cwr1003; cwr1006 larvae were exposed to one of two regimens of ART in a 12-well sterile culture plate: regimen I, 0.4 µM ART from 3 to 7 dpf (during inner ear development and hair cell maturation); and regimen II, 0.4 µM ART from 3 to 12 dpf (extended to postdevelopment and maturation period), and these larvae were observed longitudinally from 6.5 to 27 dpf (SI Appendix, Table S2). The percentage of larvae that survived after ART treatment improved at each time point from 10.5 to 27 dpf, with survival at 27 dpf significantly enhanced from 5% in untreated cwr1003; cwr1006 larvae to 40% and 45% in regimens I and II groups, respectively. In addition, 40% and 45% of the survivor larvae in regimen groups I and II, respectively, showed WT swimming behavior (SI Appendix, Table S2), indicating a tight correlation between normal swimming behavior and improved life span in cwr1003; cwr1006 larvae. These data also revealed that increasing the duration of treatment increases the percentage of larvae with normal swimming behavior at 27 dpf (SI Appendix, Table S2). Overall, these results demonstrate proof of concept that ART treatment effectively preserved hair cell function and delayed the hair cell deterioration associated with the USH3A causative CLRN1N48K protein, thus highlighting the potential use of the antimalarial drug ART to preserve hearing in CLRN1c.144T>G USH3A patients.
Discussion
Using zebrafish as an in vivo model, our study set out to investigate the functional consequence of expressing CLRN1c.144T>G in hair cells, and to decipher the mechanism that enabled glycosylation-deficient CLRN1N48K to reach the hair bundle, and whether pharmaceutical activation of this mechanism or pathway was therapeutic. To this end, we developed a zebrafish model of hearing loss associated with the USH3A pathogenic variant CLRN1c.144T>G.
Previously, coding sequences of either CLRN1 or CLRN1c.144T>G fused to that of the YFP reporter gene were each expressed in the hair cells of WT zebrafish to assess localization of CLRN1 and CLRN1N48K proteins in hair cells (16). Since the native clrn1 gene was present in the host cells, these transgenic zebrafish behaved like WT clutch mates (without the transgene); we could not access the functional consequence of expressing CLRN1 or CLRN1c.144T>G in hair cells or pursue other aims of our study outlined above. Here, we present improved zebrafish models that enabled functional, pathologic, and therapeutic investigation of the USH3A causative pathogenic variant CLRN1c.144T>G in hair cells in vivo. By expressing CLRN1c.144T>G in clrn1-null zebrafish, we were able to investigate (i) its role in hair cell function and hearing loss, (ii) the pathway that assists CLRN1N48K translocation to the hair bundle, and (iii) the drugs that activate this pathway. In contrast to the complete and stable rescue of hair cell function, hearing, and balance in clrn1-null zebrafish expressing CLRN1, the progressive phenotype observed in clrn1-null zebrafish expressing CLRN1c.144T>G mimics progressive inner ear dysfunction in CLRN1c.144T>G USH3A patients. Our report demonstrates a paradigm to model human deafness by using a human ortholog in a null background. This paradigm may be generally applicable and could be used to model other monogenic forms of deafness.
Our results demonstrate that the CLRN1c.144T>G pathogenic variant does not compromise the ability of the mutant protein to mediate clarin1 function; if the retention of the mutant protein in the ER can be alleviated, its localization to mutant hair cells can be rescued, along with CLRN1 function. Our genetic and pharmacological investigation of the CLRN1N48K model revealed that a human deafness-associated protein is trafficked via the GCUSP. In support of this observation, we discovered that treating larvae expressing CLRN1N48K-YFP with the GCUSP activators, TG or ART, resulted in the liberation of ER-sequestered CLRN1N48K to enhance the mutant protein’s localization to the hair bundle. Additionally, ART treatment restored mechanotransduction to CLRN1N48K-expressing hair cells, to a level that was comparable to WT cells, thereby restoring hearing and balance to the zebrafish model.
The molecular sequelae associated with the retention of CLRN1N48K in the ER of hair cells is the likely cause of the progressive hearing loss that characterizes USH3A. The N48K missense variant abolishes a consensus glycosylation site on CLRN1 (25), a change that is proven to impair its glycosylation and ER exit (13). These observations led us to investigate the UPR-induced ER stress response as a possible mechanism to explain the CLRN1N48K phenotype. It is known that if ER stress cannot be reversed due to the constitutive accumulation of the misfolded protein, cellular functions deteriorate and cell death ensues (20, 26–29). The UPR-induced ER stress signals are reported to trigger the phosphorylation of GRASP55, a PDZ type I domain-containing protein, and its localization to the ER, where it mediates the selective, Golgi-independent, unconventional secretion of ER-retained cargos of misfolded N-glycoproteins with the PDB type I sequence (20, 30–32). An alternative mechanism for secretion of ER-retained cargos is the SNX27-dependent, PDZ domain type I-directed trafficking of PDB type I cargo proteins from the endosomes to the plasma membrane (22). Given that CLRN1 is an N-glycoprotein with a PDB type I sequence, we hypothesized that under UPR-induced ER stress, either GRASP55 or SNX27 might unconventionally traffic CLRN1N48K to the plasma membrane and hair bundle, thereby delivering clarin1-mediated function to the hair bundle. Here, we reveal the GRASP55-assisted trafficking of CLRN1N48K to the hair bundle (Fig. 3). Although the unconventional trafficking of misfolded proteins reduces their load in the ER, the constitutive expression and accumulation of misfolded proteins in the ER is believed to lead to chronic stress of this organelle, leading to ER stress-induced cellular dysfunction and cell death. We propose that this mechanism could account for the attenuation of hair cell function and the progressive loss of hearing associated with the pathogenic variant CLRN1c.144T>G. Since CLRN1c.144T>G is also associated with the attenuation of retinal cell function and progressive loss of vision in USH3A subjects, we speculate that the molecular sequelae associated with the retention of CLRN1N48K in the ER of hair cells is mirrored by the sensory cells of the retina.
YFP and its variants are versatile reporters used in various in vivo applications (33–35). However, one concern here is whether fusion of YFP to CLRN1N48K could somehow affect CLRN1N48K targeting in hair cells and may be beneficial in the context of GCUSP pathway activation. Available evidence argues otherwise. First, if YFP impaired targeting of CLRN1N48K in hair cells, then we would expect to see mistargeting of CLRN1-YFP as well. CLRN1-YFP expressed in mouse (14) or zebrafish hair cells (Fig. 1E) reach the membrane, consistent with the prediction that clarin-1 is a membrane protein (25). Furthermore, CLRN1-YFP expression restored hair bundle structure and function in clrn1KO/KO (cwr1003; cwr1005) larvae comparable to WT larvae (Fig. 1 and SI Appendix, Table S2). These results show that YFP fusion preserved CLRN1’s function and localization in hair cells. Second, irrespective of the size or amino acid sequence of the reporter used, CLRN1N48K was mistargeted. The 9-aa (∼1-kDa) HA “tag” is derived from human influenza hemagglutinin sequence, whereas YFP, a variant of GFP derived from the jellyfish Aequorea victoria (33), is ∼25 kDa. CLRN1N48K-HA in HEK293 cells (13, 14, 15) or CLRN1N48K-YFP in mouse (14) or zebrafish hair cells (Fig. 2) was mistargeted. Third, CLRN1 is predicted to be an N-linked glycoprotein. N-linked glycosylation is often used for recognition by the chaperone proteins, calnexin and calreticulin, necessary for proper posttranslational processing and targeting. When CLRN1-HA was expressed in HEK293 cells, CLRN1 was N-glycosylated, and it localized to the membrane; however, CLRN1-HA remained intracellular when these cells were treated with an inhibitor of glycosylation (tunicamycin) (13). This shows mistargeting of CLRN1N48K is linked to the glycosylation deficiency caused by the N48K mutation. Fourth, GCUSP-activated rescue of the mutant protein retained in the ER is not YFP-dependent; it has been shown to work with other mutant proteins fused to smaller, non-YFP tags. For example, similar to CLRN1N48K, ∆F508 mutation in CFTR affects its targeting to the plasma membrane, and it is largely retained in the ER. Activation of GCUSP in HEK293 cells expressing HA-∆F508-CFTR via GRASP55 expression or exposure to TG enhanced surface expression of the ∆F508-CFTR and chloride transport function; GCUSP activation in HEK cells expressing CFTR showed no significant difference compared with control (20).
Although both ART and TG resulted in the release of CLRN1N48K from the ER and enhanced hair bundle localization, the pharmacological properties of ART were more desirable at treating the sensory deficits in cwr1003; cwr1006 larvae. For example, at a 60% lower concentration, ART (at a dose of 0.4 µM) was twofold more effective than TG (at a dose of 1 µM) at restoring CLRN1N48K to the hair bundle (Fig. 4). Consistent with this higher efficacy, 0.4 µM ART also restored hair cell function to a level comparable with WT hair cells (Fig. 5). In addition, ART was less deleterious to CLRN1N48K larvae at doses used to treat malaria (36). In contrast, TG was found to be less tolerated by larvae at higher concentrations, which is in agreement with detrimental effects observed for this antitumoral drug on normal endothelial cells, fibroblasts, and osteoblasts, as well as on cancer cells (37). The water-soluble derivative of ART, artesunate (38), is more potent than ART in the treatment of malaria and has an acceptable safety profile (39). We predict that artesunate will be more effective than ART at inducing GCUSP, and it will show an equal or greater efficacy at a lower dose compared with ART in rescuing CLRN1N48K-associated phenotypes. Since the same pathogenic variant causes both retinal and hair cell defects, these antimalarial drugs could be used, in principle, to mitigate both eye and ear disorder in CLRN1c.144T>G USH3A patients. Furthermore, ART-induced GUSCP activation could potentially be beneficial for other pathogenic variants of CLRN1 that fail to exit the ER, such as CLRN1A123D, CLRN1L150P, and CLRN1M120K (40), expanding the scope of ART as a repurposable drug among USH3A subjects.
We reported on the development of a small molecule, BF844, that reduced degradation of the CLRN1N48K protein expressed in HEK293 cells in culture, and preserved hearing in a mouse model of progressive hearing loss in USH3A (15). The BF844 study demonstrated proof of concept that small-molecule therapy mitigates hearing loss associated with the CLRN1c.144T>G pathogenic variant in the clarin1 gene in a mammalian model. However, BF844’s precise mechanism of action in the hair cells remains to be elucidated. While efforts are underway to further characterize BF844 and advance it for regulatory approval, repurposing an approved drug has several advantages (41, 42). The small-molecule ART is an ideal drug for repurposing since its mechanism of action as a SERCA inhibitor (24, 43) and safety profile in humans (39) are well established. Furthermore, this report reveals that, at the dose deemed effective in the fish model, ART is not toxic to the organism or hair cells, and it does not have any discernible impact on hair cells expressing normal human CLRN1 protein.
Here, we report a zebrafish model of hearing loss in USH3A associated with the CLRN1c.144T>G pathogenic variant. By expressing this human pathogenic variant in clrn1-null zebrafish, we investigate its role in hair cell function and hearing loss. Our findings show that mutant CLRN1N48K becomes sequestered in the ER but that small amounts reach hair cells via the GCUSP. ART, an antimalarial drug that activates this pathway, can enhance CLRN1N48K levels in hair cell bundles and rescue their functions, highlighting its therapeutic potential. Furthermore, our study shows proof of concept that zebrafish could be used to directly investigate the functional consequence of a missense variant in a human gene in hair cells in vivo, and the “humanized” zebrafish model could be used to screen for drugs that can be repurposed. Since HHL is associated with pathogenic variants in various genes (3) and can affect the hair cells (2, 6), the reported strategy could have broad implications. Our report also reveals that the disease-modifying potential of these antimalarial drugs could be harnessed to treat other monogenic disorders associated with accumulation of the glycosylation-deficient mutant protein in the ER.
Materials and Methods
Zebrafish.
All experiments were conducted using the Tübingen (Tü) strain of zebrafish of either sex. Previously generated zebrafish lines and those generated as part of this study, along with their nomenclature and abbreviated symbols used, are listed in SI Appendix, Table S1. The animal protocols used in this report were approved by the Case Western Reserve University Institutional Animal Care and Use Committee (protocol approval number 2013-136). All zebrafish were maintained and bred using standard procedures (44).
Generation and Characterization of cwr1003 Zebrafish Line that Expresses CLRN1-YFP in Few Somatic (Hair) Cells.
To examine whether CLRN1-YFP expression in cwr1003 hair cells will rescue the bundle phenotype, somatic-cell expression of a transgene containing the CLRN1-YFP coding sequence under the control of the pvalb9 promoter [Tg(pvalb9:Hsa.CLRN1-YFP) transgene plasmid construct] was tested. This involved injection of the Tg(pvalb9:Has.CLRN1-YFP) transgene construct into single-cell–stage cwr1003 zebrafish embryos (F0 generation), and fluorescence imaging was done at 6 days post fertilization (dpf). The pvalb9 promoter sequence used in the transgene construct directs transgene expression primarily to hair cells in zebrafish (18). In somatic-cell expression (in contrast to germline expression), only a few cells in each of the F0 generation larvae are expected to express the transgene. The cwr1003 zebrafish larvae that express the CLRN1-YFP transgene in the hair cells were generated as follows. The Tg(pvalb9:Hsa.CLRN1-YFP) plasmid [the new nomenclature for parvalbumin-3b (pvalb3b) is pvalb9] (16), along with Tol2 RNA, was injected into cwr1003 embryos at the single-cell stage. Transgene expression was analyzed in live larvae at 6 dpf using an inverted Leica SP8 confocal microscope equipped with a 63× oil-immersion objective lens. All of the cwr1003 larvae injected with the Tg(pvalb9:Hsa.CLRN1-YFP) construct were anesthetized using MS-222 (Sigma-Aldrich). Larvae expressing CLRN1-YFP in their hair cells were identified by screening for YFP expression in hair cells of the injected larvae, using a fluorescence stereomicroscope (Leica MZFLIII). Then, to compare the difference in the hair bundle structure of CLRN1-YFP–expressing cwr1003 hair cells to the adjacent CLRN1-YFP–negative cwr1003 hair cells, we captured z-stacks of single-plane images of the inner ear organs in the YFP channel (for CLRN1-YFP expression) and in the differential interference contrast (DIC) channel (to check the hair cell bundle structure) using confocal microscopy, and the YFP and DIC images were compared parallel for the CLRN1-YFP expression pattern and the change in the hair bundle morphology.
Generation and Characterization of cwr1003 Zebrafish Lines That Stably Express CLRN1-YFP or CLRN1N48K-YFP in Hair Cells.
To generate cwr1003 zebrafish that stably express either CLRN1-YFP (cwr1003; cwr1005) or CLRN1N48K-YFP (cwr1003; cwr1006) in the hair cells, we outcrossed the cwr1003 zebrafish with the cwr1005 and cwr1006 zebrafish lines reported earlier (16). The larvae from the generated cwr1003; cwr1005, and cwr1003; cwr1006 stable lines were used to study the changes in the cwr1003 hair bundle phenotype associated with the presence of CLRN1-YFP or CLRN1N48K-YFP. Confocal microscopy images were captured in live larvae at 6 dpf as described in the previous experiment.
The startle response and swimming behavior of the cwr1003; cwr1005 and cwr1003; cwr1006 larvae, as well as that of the WT and cwr1003 larvae, were recorded at 6.5, 10.5, 16.5, and 27 dpf, as described earlier (SI Appendix, Table S2) (16). Microphonic potentials were recorded at 6 dpf to confirm neuromast hair cell function in cwr1003; cwr1005 and cwr1003; cwr1006 larvae, as described earlier (16).
Transient Expression of Unconventional Secretory Pathway Cargos in the Hair Cells of cwr1006 Larvae.
The cDNAs that encode human Golgi reassembly stacking protein 55 (GRASP55) [National Center for Biotechnology Information (NCBI) accession number NM_015530.3] and human sorting nexin family member 27 (SNX27; NCBI accession number NM_030918.5) were amplified from human retina RNA (Clontech Laboratories) using infusion primer pairs: F1, 5′-GGCAGATCCACCATGatgggctcctcgcaaagcgtc-3′, and R1, 5′-CTAGTCAGTCACTAGttaaggtgactcagaagcat-3′ for GRASP55, and F2, 5′-GGCAGATCCACCATgatggcggacgaggacgggga-3′, and R2, 5′-CTAGTCAGTCACTAgctaatattcctcttttctccac-3′, for SNX27, respectively. Uppercase and lowercase letters denote vector and gene of interest sequences, respectively. The amplicon was subcloned into the pT3TS vector digested with NcoI and SpeI (In-Fusion Cloning Kit; Clontech Laboratories) to generate pT3TS/GRASP55 and pT3TS/SNX27 constructs. Both GRASP55 and SNX27 mRNA were synthesized from the XbaI linearized pT3TS/GRASP55 and pT3TS/SNX27 constructs using the mMESSAGE mMACHINE T3 kit (Ambion). The quality of the synthesized GRASP55 and SNX27 mRNA was analyzed using an Agilent 2100 Bioanalyzer. Either GRASP55 or SNX27 mRNA was injected into cwr1005 and cwr1006 embryos at the single-cell stage. At 4 dpf, GRASP55 mRNA or SNX27 mRNA-injected cwr1005 and cwr1006 larvae were examined for a change in the localization pattern of CLRN1-YFP or CLRN1N48K-YFP in the hair cells and bundles (Fig. 3 and SI Appendix, Fig. S2).
Using ImageJ software (version 1.48) (45), we quantified the amount of CLRN1-YFP and CLRN1N48K-YFP that localized to the hair bundle in the injected and uninjected groups from the genotypes cwr1005 and cwr1006, respectively. To quantify the YFP fluorescence intensity (YFP pixels), the unadjusted 1-µm middle section of neuromast images from the above-mentioned groups were analyzed. The YFP fluorescence intensity was reported as the background-subtracted integrated density value (46). The representative images chosen for the figures (Fig. 3 and SI Appendix, Fig. S2) were near the mean value of the groups. Z-stack images of neuromast hair cells were captured using a Leica SP2 confocal microscope, and YFP fluorescence measurements in the hair bundles and their somas were obtained using ImageJ from the following: (i) control, uninjected larvae; and (ii) larvae injected with transgenes expressing unconventional secretory pathway cargo proteins, GRASP55 or SNX27. A triangle-shaped region of interest covering the area of the hair bundle and the area covering its soma, in a 1-µm-thick middle section of a z-stack image of neuromast hair cells were used to obtain YFP fluorescence pixel intensity measurements from the hair bundle (Ihair bundle) and its soma (Isoma); and to nullify the background intensity, pixels from an area without hair cells (Ibackground) in the same image were measured. Fluorescence intensity of YFP for each hair bundle (Iload in hair bundle) and its soma (Iload in soma) in a given neuromast was normalized (Iload in hair bundle = Ihair bundle − Ibackground, and Iload in soma = Isoma − Ibackground). We analyzed four hair cells per neuromast in a larva, and the relative ratio of YFP fluorescence pixel intensity per neuromast was represented as an average of ratio of the YFP pixels in four hair bundles and its soma (Irelative average load = Mean of ratio of Iload from four bundles and soma in a neuromast/larva) (Figs. 3H and 4K and SI Appendix, Fig. S2C). The mean of the total Irelative average load per group was considered for statistical comparison between groups. One-way ANOVA with Tukey’s multiple-comparisons test was used to confirm the significance of observed differences in the YFP intensity in GRASP55 mRNA-injected, and SNX27 mRNA-injected or uninjected groups.
Restoring Hair Bundle Localization of Mutant CLRN1N48K-YFP Protein Using SERCA Inhibitor Drugs.
TG (1 mg; T9033; Sigma-Aldrich) stock prepared in 100% DMSO was diluted in the E3 medium (zebrafish water) to make the required final concentration of the drug. Five concentrations that were earlier reported to induce ER stress and that are nontoxic or below lethal dose (LD50) in the zebrafish (0.1, 0.5, 1.0, 2.5, and 5 µM) were used in this study (47–49). Twenty-five larvae were used per dosage and per treatment duration of the genotypes: cwr1005 (WT; CLRN1-YFP) and cwr1006 (WT; CLRN1N48K-YFP). Larvae were no older than 72 hpf at the start of the experiment and were placed to a maximum of 10 larvae per well of a 12-well uncoated sterile culture plate (BD Falcon). For ER stress induction, larvae were incubated in different concentrations of TG, for 1 h or longer (as indicated in SI Appendix, Table S2) at 28.5 °C, followed by three washes in E3 medium at 28.5 °C (SI Appendix, Table S2). Hair cells from the cwr1005 and cwr1006 larvae were examined for the respective protein distribution pattern in the hair cell bundle and the soma. Live confocal images of otic (O) and middle (MI) neuromasts per larvae were captured using a Leica SP2 confocal microscope with 40× oil-immersion objective. Unadjusted 1-µm middle section images of four hair bundles and their somas per MI neuromast of larva from TG-treated and untreated groups were analyzed using ImageJ software, version 1.48. Quantification of YFP fluorescence pixel intensity was carried out, as described in the previous experiment for the unconventional trafficking of CLRN1N48K-YFP by GRASP55-dependent unconventional protein secretory pathway. One-way ANOVA with Tukey’s multiple-comparisons test was used to confirm the significance of the observed changes in YFP intensity in the hair bundles of TG-treated compared with untreated cwr1006 larvae.
ART (100 mg; 361593; Sigma-Aldrich) stock prepared in 100% DMSO was diluted in E3 medium to make the final concentrations. Five concentrations within the range reported to induce ER stress in zebrafish (0.1, 0.25, 0.4, 0.5, and 1 µM concentrations) were used in this study. Twenty-five larvae per treatment, no older than 72 hpf at the start of the experiment, were placed to a maximum of 10 larvae per well of a 12-well uncoated sterile culture plate (BD Falcon). For ER stress induction, larvae were exposed to different concentrations of ART for 1–10 h at 28.5 °C (SI Appendix, Table S3) and were then washed three times in E3 medium. Hair cells from the cwr1005 and cwr1006 larvae were examined for the respective protein distribution pattern in the hair bundle and soma. Live confocal Z-stack images of otic “O” and middle “MI” neuromasts were collected from larvae of the cwr1005 and cwr1006 genotypes from the following: the untreated group in the E3 medium; the 0.01% DMSO treatment group; and the ART-treated group, using a Leica SP2 confocal microscope with 40× oil-immersion objective. Hair cells from two O and MI neuromasts per larvae were examined for a change in YFP intensity in the hair bundle and soma. From the captured confocal images of these neuromast hair cells, unadjusted 1-µm middle section images of four hair bundles and their somas from MI neuromast per larva were used to quantify YFP intensity (pixels) in an unadjusted region of interest covering the middle section of each hair bundle and soma, using the ImageJ software. Quantification of YFP fluorescence intensity was carried out, as described in the previous experiment for the unconventional trafficking of CLRN1N48K-YFP by GRASP55 cargos. One-way ANOVA with Tukey’s multiple-comparisons test was used to confirm the significance of the observed changes in YFP intensity in the hair bundles of ART-treated larvae, compared with untreated and TG-treated cwr1006 larvae.
General Approaches Used for Quantification of YFP Intensity in Hair Cells.
On average, four hair bundles per MI neuromast per larva from each treatment group were used to quantify YFP intensity. The number of hair bundles considered for YFP intensity quantification were as follows: group 1, cwr1006 larvae treated with 1 µM TG plus 0.01% DMSO in E3 medium, (n = 46); group 2, cwr1006 larvae treated with 0.4 µM ART plus 0.01% DMSO in E3 medium (n = 44); group 3, cwr1006 larvae treated with 0.01% DMSO in E3 medium (n = 80); group 4, untreated cwr1006 larvae in E3 medium (n = 84); group 5, cwr1005 larvae in E3 medium (n = 30); group 6, cwr1005 larvae treated with 0.01% DMSO in E3 medium (n = 30); group 7, cwr1005 larvae treated with 1 µM TG plus 0.01% DMSO in E3 medium (n = 30); and group 8, cwr1005 larvae treated with 0.4 µM ART plus 0.01% DMSO in E3 medium (n = 30) (Fig. 4J). Similarly, the relative ratio of YFP pixel intensity in the hair bundle to YFP pixel intensity in the soma in these groups was analyzed as described in an earlier section (Fig. 4K).
Recording Microphonic Potentials.
Zebrafish larvae at 6–7 dpf were anesthetized using MS-222 (Sigma-Aldrich) dissolved in a standard bath solution containing 120 mM NaCl, 2 mM KCl, 10 mM Hepes, 2 mM CaCl2, and 0.7 mM NaH2PO4 adjusted to pH ∼7.2. The larvae were then secured in a recording chamber using dental floss tie downs (50). Their blood flow and heart rate were visually observed to assess viability using an upright Olympus BX51WI microscope. Recordings were made using a PC-505B amplifier (Warner Instruments), a SIM983 amplifier (set at 20×; Stanford Research), and a PCI-6221 digitizer (National Instruments). Images were taken with a Grasshopper3 CMOS camera (FLIR System) controlled by software provided by the manufacturer. To record the activity of hair cells from the lateral line neuromasts, we deflected the cupulae of neuromasts with a fluid jet placed ∼50 µm from the neuromast (51); this fluid jet was driven by an HSPC-1 (ALA Scientific Instruments) controlled by a jClamp (SciSoft), to deliver sinusoidal stimuli of 50 Hz. Microphonic potentials were recorded at room temperature (22 °C). For this purpose, borosilicate glass electrodes with a resistance of 5–6 MΩ were filled with a standard bath solution and were placed near the apical edges of the lateral line neuromasts. Microphonic potentials were recorded using a jClamp in current-clamp mode and low-pass filtered at 200 Hz. The data represent an average of at least 500 trials. GraphPad Prism 7.03 software was used for statistical analyses. ANOVA with Tukey’s multiple-comparisons test was used. Statistical significance was set at P < 0.05. A biological replicate was defined as an individual zebrafish. Values for each zebrafish represent an average measurement of maximal peak-to-peak amplitudes of microphonic potential responses produced from anterior and posterior deflections of cupula of the lateral line neuromast. Microphonic potentials were analyzed from at least three different clutches across three different experiments.
Supplementary Material
Acknowledgments
We acknowledge the use of the zebrafish core facility at Case Western Reserve University (CWRU). We also acknowledge the use of the microscopy module of the Leica SP8 confocal in the Genetics Department Imaging Facility at CWRU, made available through the Office of Research Infrastructure Programs (NIH-ORIP) Shared Instrumentation Grant S10 OD016164. This work was supported in part by funds from the Anthony J. Maniglia Endowed Chair, University Hospitals Cleveland Medical Center (to K.N.A.). S.R.G., B.M.M., and R.S. were supported by NIH/National Institute on Deafness and Other Communication Disorders Grants DC015621, DC009437, and DC015016, respectively.
Footnotes
Conflict of interest statement: S.R.G. and K.N.A. have filed a patent application (PCT/US17/55914) titled, “Methods of treating disorders associated with glycosylation defective proteins.”
This article is a PNAS Direct Submission. M.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817500116/-/DCSupplemental.
References
- 1.Petit C, Richardson GP (2009) Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci 12:703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dror AA, Avraham KB (2010) Hearing impairment: A panoply of genes and functions. Neuron 68:293–308. [DOI] [PubMed] [Google Scholar]
- 3.Van Camp G, Smith RJH (2019) Hereditary Hearing Loss Homepage. Available at https://hereditaryhearingloss.org. Accessed May 1, 2019.
- 4.Steel KP. (1995) Inherited hearing defects in mice. Annu Rev Genet 29:675–701. [DOI] [PubMed] [Google Scholar]
- 5.Ahituv N, Avraham KB (2000) Auditory and vestibular mouse mutants: Models for human deafness. J Basic Clin Physiol Pharmacol 11:181–191. [DOI] [PubMed] [Google Scholar]
- 6.Brown SDM, Hardisty-Hughes RE, Mburu P (2008) Quiet as a mouse: Dissecting the molecular and genetic basis of hearing. Nat Rev Genet 9:277–290. [DOI] [PubMed] [Google Scholar]
- 7.Jones SM, Jones TA (2014) Genetics of peripheral vestibular dysfunction: Lessons from mutant mouse strains. J Am Acad Audiol 25:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolson T. (2005) The genetics of hearing and balance in zebrafish. Annu Rev Genet 39:9–22. [DOI] [PubMed] [Google Scholar]
- 9.Blanco-Sánchez B, Clément A, Phillips JB, Westerfield M (2017) Zebrafish models of human eye and inner ear diseases. Methods Cell Biol 138:415–467. [DOI] [PubMed] [Google Scholar]
- 10.Pickett SB, Raible DW (2019) Water waves to sound waves: Using zebrafish to explore hair cell biology. J Assoc Res Otolaryngol 20:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ness SL, et al. (2003) Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet 40:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plantinga RF, et al. (2005) Serial audiometry and speech recognition findings in Finnish Usher syndrome type III patients. Audiol Neurotol 10:79–89. [DOI] [PubMed] [Google Scholar]
- 13.Tian G, et al. (2009) Clarin-1, encoded by the Usher syndrome III causative gene, forms a membranous microdomain: Possible role of clarin-1 in organizing the actin cytoskeleton. J Biol Chem 284:18980–18993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng R, et al. (2012) The mechanosensory structure of the hair cell requires clarin-1, a protein encoded by Usher syndrome III causative gene. J Neurosci 32:9485–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alagramam KN, et al. (2016) A small molecule mitigates hearing loss in a mouse model of Usher syndrome III. Nat Chem Biol 12:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopal SR, et al. (2015) Zebrafish models for the mechanosensory hair cell dysfunction in Usher syndrome 3 reveal that clarin-1 is an essential hair bundle protein. J Neurosci 35:10188–10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng R, et al. (2009) Usher syndrome IIIA gene clarin-1 is essential for hair cell function and associated neural activation. Hum Mol Genet 18:2748–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDermott BM Jr, et al. (2010) Transgenic labeling of hair cells in the zebrafish acousticolateralis system. Gene Expr Patterns 10:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimmel CB, Patterson J, Kimmel RO (1974) The development and behavioral characteristics of the startle response in the zebra fish. Dev Psychobiol 7:47–60. [DOI] [PubMed] [Google Scholar]
- 20.Gee HY, Noh SH, Tang BL, Kim KH, Lee MG (2011) Rescue of ΔF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell 146:746–760. [DOI] [PubMed] [Google Scholar]
- 21.Steinberg F, et al. (2013) A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol 15:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauffer BE, et al. (2010) SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J Cell Biol 190:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pahl HL. (1999) Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol Rev 79:683–701. [DOI] [PubMed] [Google Scholar]
- 24.Azvolinsky A. (2017) Repurposing existing drugs for new indications. Available at https://www.the-scientist.com/features/repurposing-existing-drugs-for-new-indications-32285. Accessed May 1, 2019.
- 25.Adato A, et al. (2002) USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur J Hum Genet 10:339–350. [DOI] [PubMed] [Google Scholar]
- 26.Schröder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74:739–789. [DOI] [PubMed] [Google Scholar]
- 27.Glozman R, et al. (2009) N-glycans are direct determinants of CFTR folding and stability in secretory and endocytic membrane traffic. J Cell Biol 184:847–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bas T, et al. (2011) Post-translational N-glycosylation of type I transmembrane KCNE1 peptides: Implications for membrane protein biogenesis and disease. J Biol Chem 286:28150–28159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sano R, Reed JC (2013) ER stress-induced cell mechanism. Biochim Biophys Acta Mol Cell Res 1833:3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinseth MA, et al. (2007) The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell 130:524–534. [DOI] [PubMed] [Google Scholar]
- 31.Schotman H, Karhinen L, Rabouille C (2008) dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev Cell 14:171–182. [DOI] [PubMed] [Google Scholar]
- 32.Rabouille C. (2017) Pathways of unconventional protein secretion. Trends Cell Biol 27:230–240. [DOI] [PubMed] [Google Scholar]
- 33.Heim R, Prasher DC, Tsien RY (1994) Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA 91:12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai T, et al. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Campbell RE, Ting AY, Tsien RY (2002) Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3:906–918. [DOI] [PubMed] [Google Scholar]
- 36.Taylor WR, White NJ (2004) Antimalarial drug toxicity: A review. Drug Saf 27:25–61. [DOI] [PubMed] [Google Scholar]
- 37.Mahalingam D, et al. (2016) Mipsagargin, a novel thapsigargin-based PSMA-activated prodrug: Results of a first-in-man phase I clinical trial in patients with refractory, advanced or metastatic solid tumours. Br J Cancer 114:986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haynes RK, Vonwiller SC (1994) Extraction of artemisinin and artemisinic acid: Preparation of artemether and new analogues. Trans R Soc Trop Med Hyg 88:S23–S26. [DOI] [PubMed] [Google Scholar]
- 39.Maude RJ, et al. (2009) Does artesunate prolong the electrocardiograph QT interval in patients with severe malaria? Am J Trop Med Hyg 80:126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isosomppi J, et al. (2009) Disease-causing mutations in the CLRN1 gene alter normal CLRN1 protein trafficking to the plasma membrane. Mol Vis 15:1806–1818. [PMC free article] [PubMed] [Google Scholar]
- 41.Nosengo N. (2016) Can you teach old drugs new tricks? Nature 534:314–316. [DOI] [PubMed] [Google Scholar]
- 42.Eckstein-Ludwig U, et al. (2003) Artemisinins target the SERCA of Plasmodium falciparum. Nature 424:957–961. [DOI] [PubMed] [Google Scholar]
- 43.Michelangeli F, East JM (2011) A diversity of SERCA Ca2+ pump inhibitors. Biochem Soc Trans 39:789–797. [DOI] [PubMed] [Google Scholar]
- 44.Nüsslein-Volhard C, Dahm R (2002) Zebrafish: A Practical Approach (Oxford Univ Press, New York: ), p 303. [Google Scholar]
- 45.Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pang Z, Laplante NE, Filkins RJ (2012) Dark pixel intensity determination and its applications in normalizing different exposure time and autofluorescence removal. J Microsc 246:1–10. [DOI] [PubMed] [Google Scholar]
- 47.Pyati UJ, et al. (2011) p63 mediates an apoptotic response to pharmacological and disease-related ER stress in the developing epidermis. Dev Cell 21:492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HC, et al. (2011) Transgenic zebrafish model to study translational control mediated by upstream open reading frame of human chop gene. Nucleic Acids Res 39:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas-Rivera D, et al. (2012) TMBIM3/GRINA is a novel unfolded protein response (UPR) target gene that controls apoptosis through the modulation of ER calcium homeostasis. Cell Death Differ 19:1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ricci AJ, Fettiplace R (1997) The effects of calcium buffering and cyclic AMP on mechano-electrical transduction in turtle auditory hair cells. J Physiol 501:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapani JG, Nicolson T (2010) Physiological recordings from zebrafish lateral-line hair cells and afferent neurons. Methods Cell Biol 100:219–231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.