Significance
Condensin and cohesin are heteropentameric complexes containing two structural maintenance of chromosomes (SMC) subunits and three non-SMC subunits. SMC dimers form head and hinge domains connected by long coiled coils. Suppressor screening for head-associated non-SMC, and SMC hinge mutants of fission yeast, reveals that condensin is regulated by SUMOylation, ubiquitination, and phosphorylation, while cohesin is regulated by RNA elimination and chromatin remodeling and releasing factors. So, they are regulated by distinct pathways. However, hinge interface mutations are commonly suppressed by mutations in the kleisin N terminus. The results support a “hold and release” model, in which the head and hinge interact to form arched coiled coils that hold and release chromosomal DNAs. The head-kleisin and hinge may cooperate to regulate arched coiled coils’ orientation, which affects their interaction with DNAs.
Keywords: SMC head, SMC hinge, kleisin, SUMO, RNA elimination
Abstract
Cohesin and condensin play fundamental roles in sister chromatid cohesion and chromosome segregation, respectively. Both consist of heterodimeric structural maintenance of chromosomes (SMC) subunits, which possess a head (containing ATPase) and a hinge, intervened by long coiled coils. Non-SMC subunits (Cnd1, Cnd2, and Cnd3 for condensin; Rad21, Psc3, and Mis4 for cohesin) bind to the SMC heads. Here, we report a large number of spontaneous extragenic suppressors for fission yeast condensin and cohesin mutants, and their sites were determined by whole-genome sequencing. Mutants of condensin’s non-SMC subunits were rescued by impairing the SUMOylation pathway. Indeed, SUMOylation of Cnd2, Cnd3, and Cut3 occurs in midmitosis, and Cnd3 K870 SUMOylation functionally opposes Cnd subunits. In contrast, cohesin mutants rad21 and psc3 were rescued by loss of the RNA elimination pathway (Erh1, Mmi1, and Red1), and loader mutant mis4 was rescued by loss of Hrp1-mediated chromatin remodeling. In addition, distinct regulations were discovered for condensin and cohesin hinge mutants. Mutations in the N-terminal helix bundle [containing a helix–turn–helix (HTH) motif] of kleisin subunits (Cnd2 and Rad21) rescue virtually identical hinge interface mutations in cohesin and condensin, respectively. These mutations may regulate kleisin’s interaction with the coiled coil at the SMC head, thereby revealing a common, but previously unknown, suppression mechanism between the hinge and the kleisin N domain, which is required for successful chromosome segregation. We propose that in both condensin and cohesin, the head (or kleisin) and hinge may interact and collaboratively regulate the resulting coiled coils to hold and release chromosomal DNAs.
Isolation of extragenic suppressors is a convenient tool to search for genes with protein products that function in the same process as a gene of interest, or that physically interact with that gene’s protein product (1–4). Alternatively, extragenic suppressors often oppose the gene function that is impaired. For example, the loss of adenylate cyclase (resulting in reduced cAMP concentration) is compensated for by mutations in phosphodiesterase, which cause an increase in [cAMP] (5, 6). We previously developed an efficient and cost-effective suppressor mutation identification method using next-generation sequencing of a genomic DNA mixture to identify suppressor mutations produced spontaneously under restrictive conditions (7). The initial mutation is temperature-sensitive (ts), causing, for example, protein instability, and the extragenic suppressor mutation (the second mutation) can alleviate or cover the ts phenotype by stabilizing the protein or protein–protein interactions. For example, ts histone H2B mutant htb1-G52D fails to form colonies at 36 °C, and multiple htb1-G52D suppressors were identified in Spt-Ada-Gcn5-acetyl transferase (SAGA) complex genes (e.g., ubp8, gcn5) using the method (7). The SAGA complex contains deubiquitinating activity of histone H2B, which deubiquitinates and destabilizes H2B. Hence, ∆ubp8 and ∆gcn5 stabilized H2B and were able to rescue the ts phenotype. Two other examples of genetic suppression involving Cdc48-mediated proteasome-dependent destruction and the Eso1-Wpl1–mediated cohesion establishment/dissolution cycle have been demonstrated (7). This kind of approach, if employed systematically using numerous mutations, can be developed on a much more comprehensive scale, and will give us a systematic view of how complex molecular assemblies are organized (8).
Condensin and cohesin are two fundamental protein complexes required to generate functional chromosome structure. Both contain structural maintenance of chromosomes (SMC) subunits, which are composed of three domains, namely, the head, coiled coil, and hinge. Each SMC subunit comprises two head segments at the N and C termini, a hinge segment in the middle, and two 50-nm coiled coils linking the head and hinge segments (9, 10) (Fig. 1A). Condensin and cohesin contain additional essential subunits. In the fission yeast Schizosaccharomyces pombe, in addition to the heterodimeric Cut14/SMC2 and Cut3/SMC4 (11–13), three subunits are bound to the head region of condensin (Cnd1/NCAPD2, Cnd2/NCAPH, and Cnd3/NCAPG; NCAPD2, NCAPH, and NCAPG are their human homologs) (14, 15). For cohesin, heterodimeric Psm1/SMC1 and Psm3/SMC3 head domains associate with Rad21/RAD21, Psc3/STAG1–3, and Mis4/NIPBL subunits (RAD21, STAG1–3, NIPBL are their human homologs) (16, 17). SMC heads have ATPase activity (9, 18, 19). Together, the hinge segments form a doughnut-shaped structure with two (north and south) interfaces (20). Rad21 associates with the Psm1 head and the Psm3 coiled coil adjacent to the head (21–26). Separase Cut1 is activated when securin Cut2 is ubiquitinated by the anaphase-promoting complex/cyclosome complex and degraded by the 26S proteasome (27, 28), and Cut1 then cleaves residues 179R and 231R of Rad21 (which bridges the head domains of Psm1 and Psm3) during the transition from mitotic metaphase to anaphase (29–31).
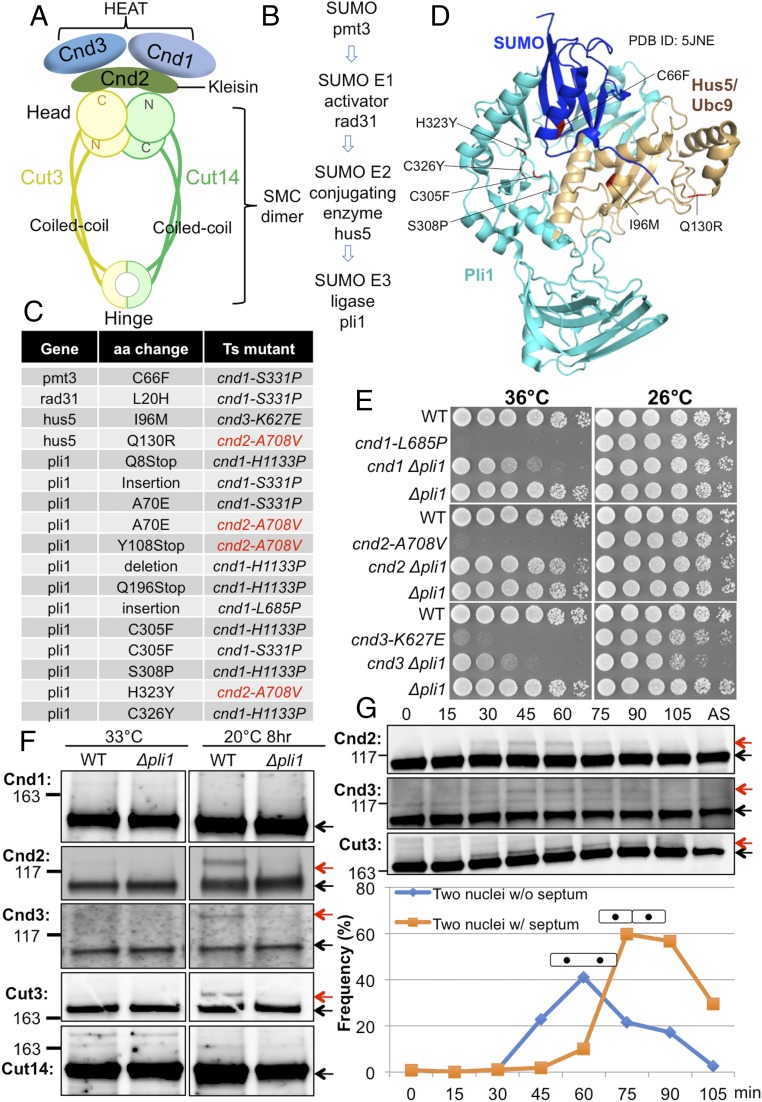
Fig. 1.
Loss of SUMOylation suppresses ts cnd1, cnd2, and cnd3 mutations. (A) Arrangement of the subunits in the fission yeast condensin complex. Cut3/SMC4 and Cut14/SMC2 are SMC proteins, while Cnd1, Cnd2/kleisin, and Cnd3 are the three non-SMC proteins that bind to the SMC head domain. (B) Genes identified as suppressors for cnd mutants form the SUMOylation pathway. (C) SUMOylation mutations that suppressed cnd1, cnd2, and cnd3 ts mutants. The majority of them are in the SUMO E3 ligase gene pli1. (D) Location of suppressor mutations in the 3D structure of SUMO, Hus5/Ubc9, and Pli1 (PDB ID code 5JNE) (Materials and Methods). (E) Spot tests showed extragenic suppression of cnd1, cnd2, and cnd3 mutants by deletion of the pli1 gene, which encodes SUMO E3 ligase. WT, wild type. (F) Immunoblotting of WT or ∆pli1 in the background of the β-tubulin mutant nda3-KM311 cultured at 33 °C (asynchronous culture) and 20 °C (restrictive temperature, 8 h; cells were arrested at prometaphase) was performed. The upper SUMO bands (red arrows) were detected for Cnd2, Cnd3, and Cut3 in the WT and were abolished in the deletion mutant ∆pli1. (G) Block and release experiment was done using the ts cdc25-22 mutant. These cells were blocked in late G2 phase and released synchronously into mitosis by a temperature shift from the restrictive temperature, 36 °C, to the permissive temperature, 26 °C. Aliquots were taken every 15 min for immunoblotting and measurement of the septation index. Cnd2, Cnd3, and Cut3 clearly produced upper bands (red arrows) only during mitosis. Cells of two nuclei without (w/o) septum are mitotic cells, while cells with (w/) septum are postmitotic, but before cytokinesis. The protein bands, that are not SUMOylated, were indicated by black arrows in F and G.
In the present study, we intended to examine the interrelationship between condensin and cohesin by isolating many ts cohesin and condensin suppressors in a systematic fashion. If condensin and cohesin share common suppressor genes, the same gene functions might be employed in their complex organization. As the SMC subunits in condensin and cohesin are similar, they may be under similar molecular control. We previously isolated spontaneous suppressors for the ts mutants of the separase/securin protease Cut1–Cut2 complex and found that separase protease is largely dispensable if the interfaces of cohesin subunits become unstable (8). Since the separase protease is specific for cohesin subunit Rad21, we have not yet found any common components that control organization of the cohesin and condensin protein complexes. In this study, we show various distinct suppressors of condensin and cohesin mutants, demonstrating their dissimilar regulation in modifications, recruitment, and protein level. On the other hand, we demonstrate that the N termini of kleisin-like Cnd2 of condensin and Rad21 of cohesin both interact with the hinge directly or indirectly, and this common interaction appears to play a critical role. These interactions may support the “hold and release” model, in which the head and hinge are proximal (8).
Results
Ts/Cold-Sensitive Mutants Selected and Suppressor Screening.
Multiple ts/cold-sensitive (cs) mutants in SMC hinge domains or non-SMC subunits (associated with SMC heads) are available for condensin and cohesin, and were selected for suppressor screening in this study. For condensin, ts mutations of three non-SMC subunits (Cnd1, Cnd2, and Cnd3) were isolated using error-prone mutagenesis (32). cnd2-1 [containing A114T in the N-terminal helix–turn–helix (HTH) motif] (33) was identified by screening for ts mutants exhibiting chromosome segregation defects. Twelve condensin ts mutants with a single amino acid substitution targeted to the hinge region were also isolated using site-directed mutagenesis (34). For cohesin, rad21-K1 (containing an effective I67F substitution mutation in the N-terminal HTH motif) (8, 35), psc3-407 (containing a T234I substitution) (36), and mis4-242 (containing a G1326E substitution) (37) were identified by screening for ts mutants exhibiting chromosome segregation defects. In addition, six ts and six cs cohesin hinge mutants with a single amino acid substitution were recently isolated (8). A suppressor screening method had been developed and it worked well (7, 8). Therefore, we were able to employ a number of cohesin and condensin mutants to isolate their suppressors.
SUMOylation Impairment Rescues ts cnd1, cnd2, and cnd3 Mutants.
We obtained suppressors for cnd1, cnd2, and cnd3 mutations in condensin non-SMC subunits (Cnd1/NCAPD2, Cnd2/NCAPH, and Cnd3/NCAPG) using previously isolated ts strains: cnd1-S331P, cnd1-H1133P, cnd2-A708V, and cnd3-K627E (32). The Cnd subunits associate with the head ATPase domain of condensin SMC subunits Cut3/SMC4 and Cut14/SMC2, as illustrated in Fig. 1A. Spontaneous revertants for cnd1, cnd2, and cnd3 mutants were isolated, and sites of the extragenic suppressor mutations were determined using whole-genome sequencing. We obtained four classes of suppressor genes, pmt3, rad31, hus5, and pli1, which turned out to form the SUMOylation pathway (Fig. 1 B–D), consisting of SUMO (Pmt3), SUMO E1-activator (Rad31), SUMO E2-conjugating enzyme (Hus5/Ubc9), and SUMO E3-ligase (Pli1). Suppressor mutation sites obtained, and original ts mutants (used for suppressor screening) are provided (Fig. 1C). The great majority of suppressors were obtained from the ligase gene. All four of the genes are essential for SUMOylation (38–41). All substituted amino acids involved are conserved among species (SI Appendix, Fig. S1). Thirteen independent suppressors were identified in pli1 and five of them are single amino acid substitutions. Except for A70E, all of the other four single amino acid substitution events are mapped in Pli1’s SP-RING domain (SI Appendix, Fig. S1 D–F). All nonsense mutations are located N-terminal to the SP-RING domain (SI Appendix, Fig. S1D). Thus, suppression of the condensin non-SMC mutants’ ts phenotype is mostly mediated by inactivation of the SUMOylation pathway.
The ligase deletion mutant ∆pli1 strongly suppressed the ts phenotype of cnd1-L685P, cnd2-A708V, and cnd3-K627E (Fig. 1E). A potentially SUMOylated band could be detected for FLAG-tagged Cnd2 (anti-FLAG antibody), Cnd3 (anti-Cnd3 antibody), and HA-tagged Cut3/SMC4 (anti-HA antibody) proteins only in mitotically arrested cells using nda3-KM311 (Materials and Methods and Fig. 1F). No upper band (potential SUMOylation band) could be detected for Cnd1 or Cut14/SMC2, however. Notably, the upper bands (for Cnd2, Cnd3, and Cut3) (Fig. 1F) disappeared in ∆pli1 deletion mutant cells. To confirm that condensin SUMOylation occurs in mitosis, cells containing the cdc25-22 ts mutation were blocked in late G2 phase and then released synchronously into mitosis by a temperature shift from the restrictive (36 °C) to the permissive (26 °C) temperature. Consistently, the appearance/disappearance of the upper SUMOylated bands coincided with the timing of progression from mitotic metaphase to anaphase (42) (Fig. 1G). These results suggested that while condensin SUMO function is apparently concealed in the wild type, ts cnd mutant defects were partly restored by deletion of SUMOylation. None of the cohesin non-SMC ts mutants (psc3-407, mis4-G1326E, and rad21-K1) could be rescued by Δpli1 (SI Appendix, Fig. S2A). No cohesin non-SMC protein band showed any change in Δpli1 mutant (SI Appendix, Fig. S2B).
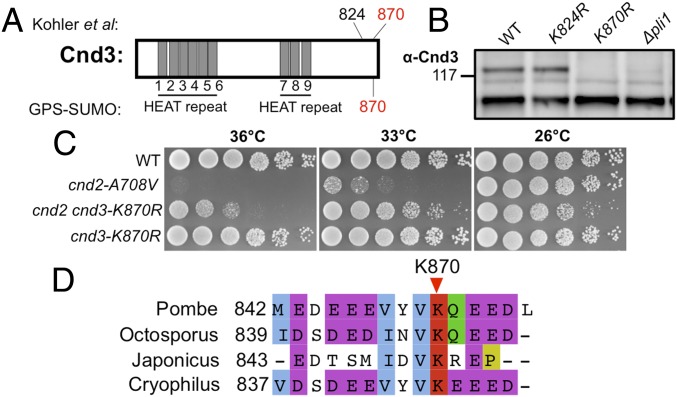
Cnd3 K870 Is the SUMOylation Site, and cnd3-K870R Mutant Rescues cnd2-A708V.
Køhler et al. (43) reported that Cnd3-K824 and Cnd3-K870 may be the sites of SUMOylation (Fig. 2A). We constructed two chromosomally integrated substitution mutants containing Cnd3-K870R or Cnd3-K824R, in which Cnd3 could not associate with SUMO. The upper SUMOylation band of Cnd3 was abolished in ∆pli1 and also in the cnd3-K870R chromosomally substituted mutant, but not in cnd3-K824R, indicating that K870 may be the actual SUMOylation site (Fig. 2B). Suppression of cnd2-A708V by cnd3-K870R indicated that the failure of C-terminal SUMOylation of Cnd3 in cnd3-K870R alleviates the cnd2-A708V ts phenotype (Fig. 2C). Therefore, loss of Cnd3 K870 SUMOylation partially resembles the effects of loss of SUMOylation on condensin. Cnd3 K870 is conserved among the four fission yeast species (Fig. 2D).
Fig. 2.
Cnd3 K870 is the responsible SUMOylation target. (A) Potential SUMOylation target sites in Cnd3 identified in a proteome-wide study (43) or predicted by GPS-SUMO software (86). Cnd3 K870 was identified by both methods. (B) K870 may be the sole SUMOylation target in Cnd3, as the Cnd3 SUMOylation band (upper band) disappeared in the cnd3-K870R mutant. The cs mutant, nda3-KM311, was used to arrest cells in mitosis (20 °C, 8 h). (C) cnd3-K870R rescues the cnd2-A708V ts mutant, which resembles those of ∆pli1 in Fig. 1E. (D) Conservation of Cnd3 K870 among four fission yeast species: S. pombe, Schizosaccharomyces octosporus, Schizosaccharomyces japonicus, and Schizosaccharomyces cryophilus.
Cnd2 May Have a Hooked Structure, and Its N Terminus May Interact with the Cut14 Head-Coiled Coil Junction.
Condensin kleisin-like subunit Cnd2 contains an HTH motif at its N terminus and a winged helix domain (WHD) at its C terminus (44) (SI Appendix, Fig. S3A). Two cnd2 ts mutants have been previously isolated. One of them contains an A114T substitution in the N-terminal HTH motif (33), and the other contains an A708V substitution in the C-terminal WHD domain (32) (SI Appendix, Fig. S3A). We were able to isolate three suppressors in cut14 and two in cnd2 for cnd2-A114T mutant (SI Appendix, Fig. S3B). In contrast, many more suppressors for cnd2-A708V were obtained, as shown in SI Appendix, Fig. S3C. One of them is extragenic cut3-S1292I, which is situated close to the original Cnd2-A708V mutation in the 3D structure (SI Appendix, Fig. S3E).
All three cut14 suppressor mutations for cnd2-A114T were mapped onto the 3D structure at the head-coiled coil junction, and they are situated close to the original mutation A114T site in the structure (21) (SI Appendix, Fig. S3D). The results are reminiscent of cohesin’s kleisin-like subunit mutant rad21-K1 (the responsible mutation, I67F, is located in the HTH motif of Rad21), suppressors of which were mapped in the Psm3 head-coiled coil junction (figure 3A of ref. 8). Again, gratifyingly, the cnd2-A708V suppressor mutation, Cut3-S1292I, was mapped to its head, close to Cnd2-A708, according to the structure determined by Bürmann et al. (21) (SI Appendix, Fig. S3E). These suppressors might restore the protein–protein interaction impaired by the original ts mutations; therefore, suppressor localizations of cnd2 ts mutants indicated that the N terminus of Cnd2 might interact with the Cut14 coiled coil at the head and that the C terminus of Cnd2 might interact with the Cut3 head domain (SI Appendix, Fig. S3F). In cohesin, the Rad21 N terminus interacts with the Psm3 coiled coil at the head and the Rad21 C terminus interacts with the Psm1 head, so judging from the modes how kleisins bind to SMC heads, Cut14 and Cut3 may be the counterparts of Psm3 and Psm1, respectively.
Except for extragenic suppressor mutations in the SUMOylation pathway, cnd2-A708V suppressors were mapped in the Cnd1 C terminus and Cnd2 itself (SI Appendix, Fig. S3 C and G). Four intragenic suppressors of cnd2-A708V were mapped to a narrow central region (aa 312–318) far from the original ts mutation site, located at the C terminus (SI Appendix, Fig. S3G). Therefore, Cnd2 may have a hooked structure (45). Since six suppressors of cnd2-A708V were mapped to the Cnd1 C terminus, the Cnd2 C terminus may interact with the Cnd1 C terminus (SI Appendix, Fig. S3G). A cartoon (SI Appendix, Fig. S3H) illustrates the possible structural organization of Cnd2 and its interaction with the Cnd1 C terminus.
Proteasome Deficiencies Rescue ts cnd3-L269P.
Then, using suppressor screening, we found that cnd3-L269P was rescued by any of 10 proteasome mutants (SI Appendix, Fig. S4A). Spot tests for cnd3-L269P are shown in SI Appendix, Fig. S4B, and cnd3-L269P was rescued by proteasome deletion mutants ∆pre9, ∆rpt4, and ∆rpn10. Therefore, the rather strong suppression of the ts cnd3 phenotype resulted from blocking proteasome-mediated proteolysis. Blocking ubiquitin-mediated protein destruction may alleviate the defect of cnd3-L269P. In the single cnd3 mutant, the mutant cnd3 protein band in SDS/PAGE was less intense than that of wild type (SI Appendix, Fig. S4C). Band intensity was restored in the double mutant (cnd3 ∆pre9) (SI Appendix, Fig. S4C) at both the permissive and restrictive temperatures, suggesting that suppression was due to an increase of cnd3 mutant protein, which failed to be destroyed in proteasome mutants. We provided evidence that Cnd3 mutant protein is unstable and that this instability was restored in proteasome mutants in the presence of the protein synthesis inhibitor cycloheximide (46) (SI Appendix, Fig. S4D).
Rescue of rad21 and psc3 Mutants by the Loss of RNA Elimination Factors.
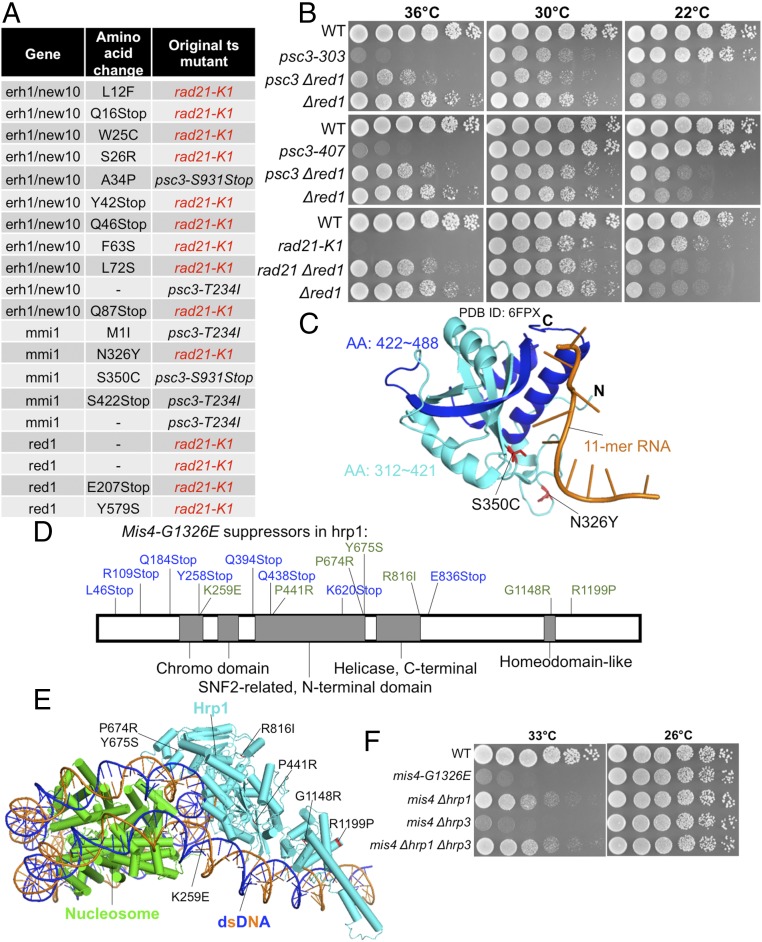
Suppressor screening and subsequent analysis of identified suppressors were conducted for the cohesin ts mutants rad21-K1, psc3-T234I (T234I is the responsible mutation of psc3-407), and psc3-S931Stop (S931Stop is the responsible mutation of psc3-303) (35, 36). We obtained 20 suppressors belonging to a group of mRNA catabolic (elimination) factors: erh1/new10, mmi1, and red1 (47–49) (Fig. 3A). For example, spot test results show suppression of the psc3 and rad21 mutants by Δred1 (Fig. 3B). Eight suppressors of rad21-K1 reside in the erh1/new10 gene (Fig. 3A). ERH is a small, highly conserved, but enigmatic protein implicated in heterochromatin domain assembly (50, 51). It seems to play an important role in the cell cycle through its transcript-splicing activity and is critically required for genomic stability and cancer cell survival (52). Two mmi1 suppressors in the RNA-binding YTH domain (47, 53) are shown in Fig. 3C, and the mutations may directly disrupt its ability to bind RNA. Thus, the loss of RNA elimination restores the mitotic sister chromatid cohesion in rad21 and psc3 mutants. However, how cohesion-defective mutations are rescued by the loss of RNA elimination is not well understood.
Fig. 3.
Suppressors of rad21-K1, psc3-T234I, and psc3-S931Stop reside in erh1/new10, mmi1, and red1 loci, all of which are involved in mRNA elimination. (A) Suppressors in erh1/new10, mmi1, and red1 that were obtained as spontaneous suppressors for ts rad21 and psc3. (B) Suppression of the ts phenotype of psc3 and rad21 by ∆red1 is shown. The ∆red1 is cs. WT, wild type. (C) Mmi1 mutations in an Mmi1 structure in complex with an 11-mer RNA (PDB ID code 6FPX) (Materials and Methods). Mmi1 contains a YTH domain at its C terminus that binds specific RNA sequences. Mmi1-S326 and Mmi1-S350 were located in Mmi1’s YTH domain. Mmi1-S326Y and Mmi1-S350C mutations may disrupt Mmi1’s ability to bind RNA directly. Mmi1-S422Stop causes loss of the Mmi1 C terminus (blue); therefore, it cannot bind RNA. AA, amino acid. (D) mis4-G1326E extragenic suppressors were mapped onto a chromosome remodeling factor gene, hrp1. (E) Hrp1 mutation in a nucleosome-Hrp1 structure (PDB ID code 5O9G) (Materials and Methods). (F) ∆hrp1 (but not another chromosome remodeling factor mutant, ∆hrp3) rescued mis4-G1326E at 33 °C too.
Cohesin Mutant mis4-G1326E was Rescued by the Loss of Chromatin Remodeling Factor Hrp1.
Suppressor screening was extended to a cohesin loading factor ts mutant, mis4-G1326E (37), and a number of suppressors were obtained and found to be derived from the hrp1 locus (Fig. 3D). The genetic interaction between the Mis4/NIPBL defective in cohesin loader and chromatin remodeling factor Hrp1 is highly selective, and of considerable interest. Mutations indicated in blue in Fig. 3D are nonsense mutations that introduced premature stop codons into the hrp1 gene, while those in green are substitutions, which were broadly distributed in the chromodomain, SNF2-like domain, helicase domain, and homeodomain. Suppression seems to be evoked by any one of many mutations. Hrp1 single amino acid substitutions are shown in a nucleosome–Hrp1 complex structure (54) (Fig. 3E). The deletion mutant Δhrp1 (but not Δhrp3) rescued mis4-G1326E too (Fig. 3F). Mis4 and Hrp1 were copurified with a heterochromatic protein, Swi6/HP1 (55, 56); therefore, Hrp1 may mediate the connection of cohesin with chromosomal nucleosome and heterochromatic proteins such as Swi6/HP1. In addition, Mis4 human homolog NIPBL is the causal gene of Cornelia de Lange syndrome (57, 58), and the suppression of mis4 by hrp1 mutations may offer clues to treat the disease.
Suppression of Condensin Hinge Mutants by Kleisin-Like cnd2 and SMC Head Mutations.
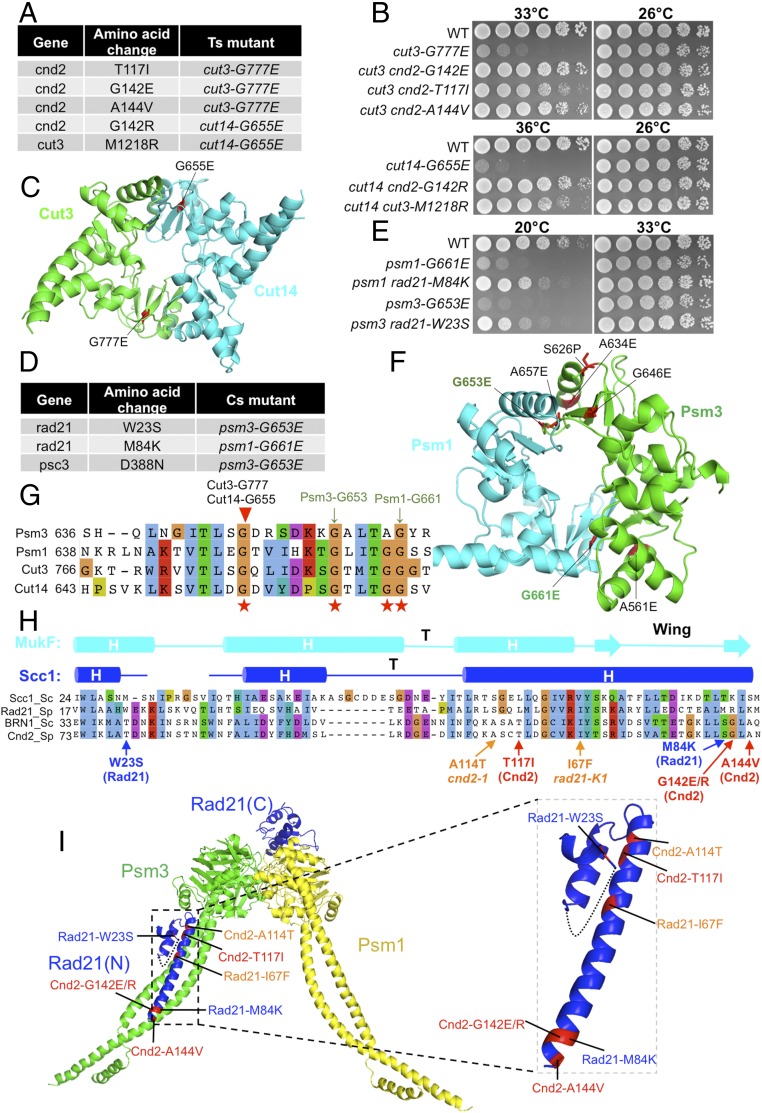
For screening suppressors of condensin hinge mutants, we employed nine of the 11 condensin hinge ts mutants (34). Only two strains, cut3-G777E and cut14-G655E (locations of the substitutions in a hinge structure are shown in Fig. 4C), yielded four suppressors in the non-SMC Cnd2 gene that rescued the ts phenotype of the hinge mutant cut3-G777E or cut14-G655E (Fig. 4 A and B). In addition, one SMC cut3 mutation, M1218R, rescued the hinge cut14-G655E mutation (Fig. 4A). Note that Cut14 G655 and Cut3 G777 are located at the same positions in the amino acid alignment, but in the 3D structure, they reside at different interfaces of the hinge. In the 3D hinge structure, locations of these two residues are symmetrical under 180° rotation, as the heterodimeric hinge has approximately twofold rotational symmetry (Fig. 4 C and G). Curiously, the hinge suppressor in Cut3/SMC4 (M1218R) resided in the head domain, while the other four were located in the N terminus of kleisin-like Cnd2 (T117I, G142R, G142E, and A144V) (Fig. 4A).
Fig. 4.
Suppressors of condensin hinge ts mutants and cohesin hinge cs mutants. (A) Suppressors in the cnd2 and cut3 head domain obtained from condensin hinge ts mutants (cut3-G777E and cut14-G655E). (B) Suppression of the condensin hinge ts mutants by the suppressors in A. WT, wild type. (C) Localization of Cut14-G655E and Cut3-G777E in the condensin hinge structure. Both mutations are located in hinge dimer interfaces. (D) Suppressors in rad21 and psc3 obtained from cohesin hinge cs mutants. (E) Suppression of cohesin hinge cs mutants by the suppressors in D. (F) Localization of Psm3-G653E and Psm1-G661E in the cohesin hinge structure. (G) Localization of the corresponding condensin hinge ts mutations in A and cohesin hinge cs mutations in D in a protein alignment of the hinges. (H) Localization of condensin hinge and cohesin hinge suppressors in a protein alignment of kleisin N termini. The secondary structure is predicted based on the structure of the S. cerevisiae Scc1 N terminus. Condensin hinge suppressors are shown in red, and cohesin hinge suppressors are shown in blue. In addition, responsible mutations of ts mutants cnd2-1 (A114T) and rad21-K1 (I67F) that are located in their N termini are shown (orange). (I) Localization of the mutations from H in the structure. All of them may directly affect kleisin’s interaction with the SMC head-coiled coil junction. Condensin hinge suppressors (Cnd2-T117I, Cnd2-G142E/R, and Cnd2-A144V) and cohesin hinge suppressors (Rad21-W23S and Rad21-M84K) may enhance kleisin’s interaction with the SMC head-coiled coil junction, while the cnd2-1 mutation A114T and rad21-K1 mutation I67F may disrupt this interaction.
We looked at Cnd2 substitutions, and found that Cnd2-T117, Cnd2-G142, and Cnd2-A144 are all located in the same helix of the conserved HTH motif at its N terminus (Fig. 4H and SI Appendix, Fig. S5A). Note that the cut3 and cnd2 suppressors contained bulkier side-chain residues (SI Appendix, Fig. S5B). Three of the cnd2 suppressors were only two residues apart (G142E/R and A144V). Two distinct mutations containing larger side-chain residues (E and R) at G142 suppressed the cut3-G777E and cut14-G655E mutations (discussed below). A simple hypothesis to explain this suppression is that destabilization of the hinge by Cut3-G777E or Cut14-G655E appeared to be compensated for by the second destabilizing mutation (e.g., G142E) in the amino terminus of kleisin-like subunit Cnd2. The hinge and Cnd2 N terminus may directly associate or indirectly interact through, for example, the mediation of DNA (Discussion).
Cohesin Hinge Mutants Are Rescued by Kleisin-Like rad21 and STAG-Like psc3 Mutations.
Similar suppressor screening was conducted for cohesin hinge mutants isolated previously (8). Only two cs mutants (psm3-G653E and psm1-G661E) yielded three distinct spontaneous suppressors in non-SMC cohesin subunits, Rad21 and Stag-like Psc3 (Fig. 4 D and E). Psm3-G653E and Psm1-G661E are located at different hinge interfaces (Fig. 4F) and are conserved in condensin Cut3/SMC4 and Cut14/SMC2 subunits too (Fig. 4G). All condensin and cohesin hinge mutations that were rescued by mutations in the SMC head or non-SMC subunits are substitutions of G residues (to E residues) in the conserved GX6GX3GG sequence motif, which is normally found in hinge dimerization interfaces. Both the Rad21-W23S and Rad21-M84K suppressors reside in the N-terminal domain (Fig. 4H and SI Appendix, Fig. S5C). The cs phenotypes of psm3-G653E and psm1-G661E were rescued by these suppressing mutations (Fig. 4E). Psc3 contains multiple HEAT repeats, and the mutation Psc3-D388N resides in the repeat (59). This mutation may affect the affinity of Psc3 for DNA binding (SI Appendix, Fig. S5D). It is clear that the two hinge residues Psm1-G661 and Psm3-G653, located at the hinge interfaces (Fig. 4F), and bulkier side-chain amino acids (from G to E), causing destabilization of the interfaces, were introduced. Suppressor residues showed the change from W→S and from M→K, significantly altered in their side-chain properties (from aromatic to hydrophilic and from hydrophobic to basic) (SI Appendix, Fig. S5E). It remains to be determined whether these changes restored the physically destabilized hinge.
Condensin and Cohesin Hinge Suppressors Reside in the N-Terminal Domain of Rad21 and Cnd2.
Cnd2 and Rad21 proteins are kleisin-like homologous subunits of condensin and cohesin, respectively. As suppressors of the hinge located in the N termini of Cnd2 and Rad21, we prepared the alignment of suppressor mutation sites. Rad21-W23S, Rad21-M84K, cnd2-T117I, cnd2-G142E/R, and cnd2-A144V are all arranged in the same N-terminal domain (Fig. 4H). Strikingly, Rad21-M84K and Cnd2-G142E/R differed at only one residue, strongly suggesting that the rescue of hinge defects by the suppressors in kleisin-like subunits might occur through highly similar mechanisms in condensin and cohesin, consistent with the hypothesis that hinge structure and function are coupled with the HTH structure (or helix bundle; Fig. 4I) formed by the N terminus of kleisin-like Cnd2 and Rad21. Implications of these findings are discussed below (Discussion).
Rescue of Cohesin Hinge Defects by wpl1 or pds5 Mutations.
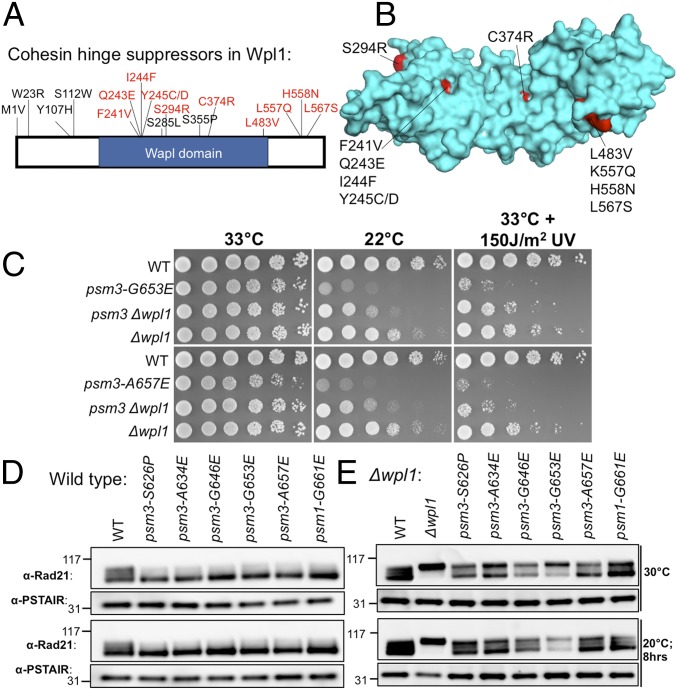
We found that cohesin cs hinge mutants were also rescued by mutations in Wpl1 and Pds5, which associate with the cohesin head and act as cohesin-releasing factors (60–62). This suppression occurs in both psm1 and psm3 hinge cs mutants. Single amino acid substitutions in the wpl1 gene that suppressed psm1 or psm3 cs mutants are shown in Fig. 5A. Two pds5 mutants could also suppress psm1 and psm3 hinge mutations, while ∼60 wpl1 suppressors were obtained for psm1 and psm3 hinge mutants. Since Wpl1 forms the complex with Pds5, this result suggested that the wpl1 mutant is the main extragenic suppressor gene for the psm1 and psm3 hinge. We mapped Wpl1 mutations onto the structure (Fig. 5B) and found that they are all located on the surface (62, 63). These mutations may disrupt the physical interaction between Wpl1 and Rad21 and further Wpl1’s association with Rad21. Fig. 5C and SI Appendix, Fig. S6A show spot tests of cohesin hinge cs mutants’ suppression by Δwpl1, while cohesin hinge ts mutants cannot be rescued (SI Appendix, Fig. S6B).
Fig. 5.
Suppression of cohesin hinge cs mutants by wpl1. (A) Localization of single amino acid substitutions in Wpl1 protein that rescued cohesin hinge cs mutants. Fifty-nine suppressors in wpl1 that suppressed cohesin hinge cs mutants were obtained, and some of them are nonsense mutations or indels. (B) Localization of the mutation sites on the Wpl1 structure (PDB ID code 3ZIK) (Materials and Methods). (C) Suppression of cohesin hinge cs mutants by ∆wpl1 (more spot results are shown in SI Appendix, Fig. S6A). These cohesin hinge cs mutants are hypersensitive to UV light. The UV sensitivity of these cs mutants was rescued by ∆wpl1 too (more spot results are shown in SI Appendix, Fig. S6A). WT, wild type. (D) Rad21 phosphorylation level in WT and cohesin hinge cs mutants detected using an anti-Rad21 polyclonal antibody (17, 64). Rad21 phosphorylation serves as an indicator of functional cohesin (8). (E) Rad21 phosphorylation level in WT, ∆wpl1, and hinge ∆wpl1 double mutants. Wpl1 and Pds5 bind the cohesin head and function as cohesin-releasing factors.
Rad21/Scc1 is hyperphosphorylated (17, 64), and it may serve as an indicator of functional cohesin (8). Immunoblotting using an anti-Rad21 polyclonal antibody indicates that Rad21 phosphorylation decreased greatly in cohesin hinge cs mutants (Fig. 5D); not only Rad21 phosphorylation but the Rad21 protein level also decreased greatly in cohesin hinge ts mutants (SI Appendix, Fig. S7A). Therefore, cohesin hinge cs mutants may disrupt sister chromatid cohesion. Cohesin hinge ts mutants may not only disrupt sister chromatid cohesion but also cause a decrease of cohesin protein levels. Actually, Rad21 is fully phosphorylated in Δwpl1, and the Rad21 phosphorylation level in cohesin hinge cs mutants is rescued by Δwpl1, while the loss of the Rad21 protein level in cohesin hinge ts mutants cannot be rescued by Δwpl1 (Fig. 5E and SI Appendix, Fig. S7B). These results explain Cut1/separase ts mutants’ suppression by these cohesin hinge mutants, as observed in the study by Xu et al. (8): Either loss of cohesion (in cohesin hinge cs mutants) or reduction of cohesin abundance (in cohesin hinge ts mutants) rescued defective cleavage of cohesin (and its release from chromatin) in Cut1/separase ts mutants.
Suppression of Condensin Hinge Mutants by Loss of Kinases and a Phosphatase.
Two condensin hinge ts mutants (cut14-L608P and cut14-G655E) were rescued by multiple mutations in kinase genes (sck1, sck2, and ksg1) and Ppe1/PP6 phosphatase complex genes (ppe1 and ekc1) (65) (SI Appendix, Fig. S8A). Ppe1 is similar to human PP6. Spot test results are shown in SI Appendix, Fig. S8B. Chromosome segregation defects of cut14-L608P were partially rescued by a phosphatase deletion mutant Δppe1: Sister chromatids were segregated but unequal, as large and small daughter nuclei were observed frequently (SI Appendix, Fig. S8C), suggesting that centromeric function was impaired in the double mutants, which is consistent with the Ppe1–Ekc1 phosphatase complex’s role in the centromere/kinetochore (66). Mutation localization mapping onto the protein sequence indicated that mutations were enriched in kinase domains of Sck1 and Ksg1 (SI Appendix, Fig. S8D); therefore, loss of their kinase activities rescued condensin hinge ts mutants. Sck1 mutations were located in the cleft that binds a nonhydrolyzable ATP analog 5′-adenylyl-imidodiphosphate (AMP-PNP) directly (67) (SI Appendix, Fig. S8E), while Ksg1 mutations might not affect ATP binding directly (68) (SI Appendix, Fig. S8F). Ksg1 is an essential gene and similar to human PDPK. Whether these kinases and phosphatase directly affect condensin hinge phosphorylation remains to be clarified.
Discussion
In this study, we employed many ts or cs cohesin and condensin mutants of SMC hinge domains and also head-associated non-SMC subunits, and obtained numerous spontaneous suppressors, genomic loci of which were determined by whole-genome sequencing. Presumed gene functions of suppressors suggested that distinct pathways are implicated in the rescue of ts or cs phenotypes of condensin and cohesin mutants. In condensin, SUMOylation pathway mutants and 26S proteasome protein destruction mutants suppressed mutants of head-interacting non-SMC subunits. Protein phosphorylation/dephosphorylation was involved in rescuing condensin hinge mutants, because mutations in protein kinases (Ksg1, Sck1, and Sck2) and a phosphatase complex (Ppe1 and its regulatory subunit Ekc1) were identified (Fig. 6A and SI Appendix, Fig. S8). ATPase-dependent autophosphorylation of the condensin hinge was previously shown to diminish the DNA-binding ability of the hinge (69), suggesting that hinge phosphorylation resulted in the decline of the DNA-binding ability of condensin. If Ppe1 phosphatase acts on the hinge and hinge phosphorylation were up-regulated by kinases, the loss of Ppe1 might enhance hinge phosphorylation.
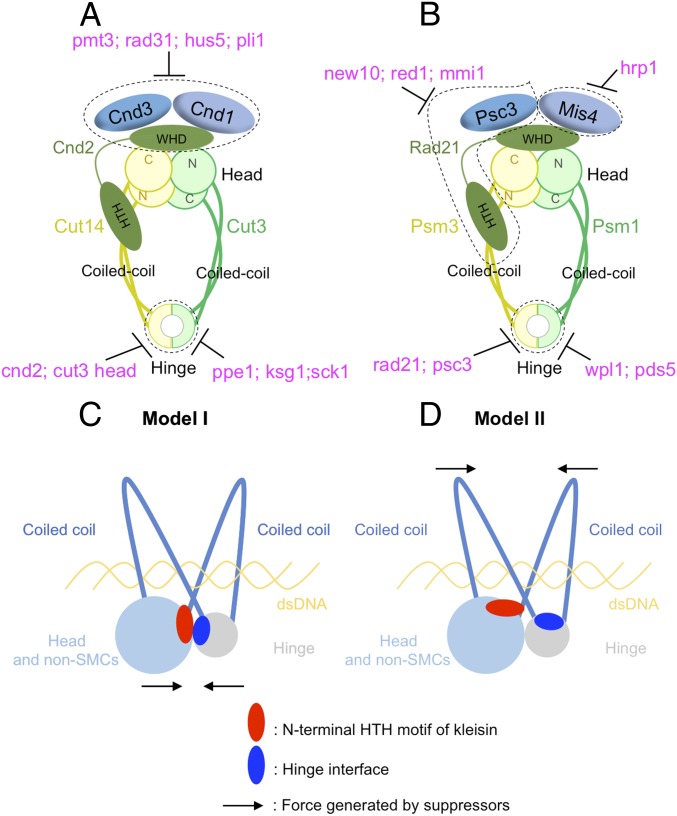
Fig. 6.
Condensin and cohesin are regulated differently, but they may adopt a similar organization in which the hinge and head interact. (A) Summary of condensin’s suppression by SUMOylation pathway mutants (pmt3, rad31, hus5, and pli1), kinase or phosphatase mutants (ppe1, ksg1, and sck1), and condensin mutants (cnd2 and cut3 head mutations). (B) Summary of cohesin’s suppression by RNA elimination pathway mutants (new10, red1, and mmi1), chromatin-remodeling factor mutants (hrp1), cohesin-releasing factor mutants (wpl1 and pds5), and cohesin non-SMC mutants (rad21 and psc3) (text). Two models were proposed in C and D to explain SMC hinge interface mutants’ suppression by mutations in the N-terminal HTH motif of kleisins. (C) SMC hinge interfaces and the N-terminal HTH motif of kleisins may interact directly to form arched coiled coils, which hold and release chromosomal DNA. SMC hinge interface mutations may impair head–hinge interaction, and suppressors in the N-terminal HTH motif of kleisins rescue the interaction. (D) SMC hinge interfaces and the N-terminal HTH motif of kleisins may not directly interact, but they both regulate coiled-coil orientation. SMC hinge interface mutations may widen the coiled-coil angle, thereby impairing the capacity of the coiled coils to hold chromosomal DNA. Suppressors in the N-terminal HTH motif of kleisins rescue the DNA-binding ability of the coiled coils.
In cohesin, on the other hand, mutants of RNA elimination pathways (new10, red1, and mmi1) or hrp1 mutant defective in chromatin remodeling suppressed the mutants of three head-interacting cohesin non-SMC subunits (rad21, psc3, and mis4). In addition, cohesin hinge cs mutants were rescued by mutations in cohesin-releasing factors (wpl1 and pds5) (Fig. 6B). Hence, although condensin and cohesin are similar in that both complexes contain SMC and non-SMC kleisin subunits, they are regulated by distinct pathways. We have not yet found suppressors of cohesin implicated in SUMOylation, ubiquitination, or phosphorylation/dephosphorylation. Judging from the putative roles of cohesin suppressors, cohesin may be regulated by protein [and possibly RNA (70)] loading/releasing rather than modification-reverse modification, as seen in condensin.
We showed that three condensin subunits (Cnd2, Cnd3, and Cut3) are SUMOylated during mitosis, and our results suggest that SUMOylation antagonizes the function of non-SMC subunits. K870 is the candidate SUMOylation site in Cnd3, as K870R mutation abolishes the band of putative SUMO-bound Cnd3. The rescue of cnd2-A708V by cnd3-K870R indicates that the K870R mutant resembles the SUMOylation loss phenotype. Therefore, SUMOylation at Cnd3 K870 may weaken non-SMC subunits’ function; thus, temperature sensitivity was rescued in cnd ∆SUMO double-mutant cells. To examine the effects of SUMOylation loss on condensin, we observed phenotypes of the cnd3-K627E single mutant and cnd3-K627E Δpli1 double mutant at permissive (26 °C) and restrictive (36 °C) temperatures (SI Appendix, Fig. S9A). The cnd3-K627E exhibited typical segregation defects observed in condensin mutants. This defect was rescued in the cnd3-K627E Δpli1 double mutant, but large and small daughter nuclei, which are typical phenotypes of centromere mutants, were newly observed (SI Appendix, Fig. S9A). Therefore, loss of SUMOylation partially rescued condensin ts mutants’ segregation defects but, at the same time, caused a unique centromeric segregation defect, suggesting that SUMOylation might protect centromeric function in mitosis. Consistently, mitotic condensin is bound to active genes as well as to central centromeric chromatin (71–73), and condensin non-SMC subunits may have a kinetochore/centromere function (32). Actually, although the temperature sensitivity of cnd3-K627E was rescued by Δpli1, double mutants’ sensitivity to thiabendazole (TBZ, a microtubule destabilizing drug) was additive (SI Appendix, Fig. S9B). These results suggested that SUMOylation may act distinctively at chromosome arms and centromeres during mitosis. In addition to cohesin and condensin, there is one more SMC complex, the Smc5–Smc6 complex, and it affects kinetochore protein SUMOylation (74). Therefore further experiments are needed to test if the Smc5–Smc6 complex is affected by Δpli1 and to clarify whether the centromeric defects observed in cnd3-K627E Δpli1 were due to loss of condensin SUMOylation.
For condensin and cohesin hinge mutants, suppressors were obtained in kleisin-like Cnd2 and Rad21, respectively (Fig. 4 A and D). This would indicate that fission yeast kleisin homologs play a common hinge-interacting role in the organization of cohesin and condensin complexes (75). Cnd2 and Rad21 are bound to the SMC heads. Hence, the interaction resulting in suppression may occur between the head and hinge. This presumed head–hinge interaction is consistent with a model of arched coiled coils for DNA binding/dissociation proposed for cohesin (8). The present suppressor screening further revealed the importance of the HTH motif in the N terminus of kleisin, which appears to mediate kleisin homolog’s interaction with the SMC (Cut14 in condensin and Psm3 in cohesin) head-coiled coil junction (Fig. 4I). This HTH domain may be required for interaction with the hinge and/or DNA.
Surprisingly, condensin ts mutations (cut3-G777E and cut14-G655E) actually resided at the same positions in the alignment of heterodimeric (Cut3/SMC4 and Cut14/SMC2) hinge sequences (Fig. 4G). These mutations, located in β-structures at the central hinge interfaces, presumably destabilize the interfaces, as substitutions were from G to much larger E residues. Four cnd2 (and one cut3 head mutation) suppressors were obtained for these two hinge mutants, all located in the amino-terminal HTH-containing helix bundle of Cnd2 (Fig. 4I). Three mutations (Cnd2-G142E, Cnd2-G142R, and Cnd2-A144V) resided very closely and were located in the same α-helix (Fig. 4I, Right). These results strongly suggested the presence of a tightly coupled mechanism to restore hinge mutations by second mutations in the helix bundle of N-terminal Cnd2. To understand the restoration mechanism, further investigation is required. Direct interaction between the hinge and Cnd2 may exist, or, alternatively, indirect interaction mediated by DNA, which may be sandwiched between the arched coiled coils, may exist.
For cohesin, the hinge mutants cs psm3-G653E and psm1-G661E produced two suppressors, rad21-W23S and rad21-M84K, located at the HTH in the N terminus, as in the case of cnd2 suppressors. Strikingly, all four hinge mutants (cut3-G777E, cut14-G655E, psm3-G653E, and psm1-G661E) have the same substitutions from G to E located in the same GX6GX3GG sequence motif at the conserved G residues (Fig. 4G). This G-rich motif, conserved even in homodimeric prokaryote SMC, is required for hinge dimerization (76–78). We speculate that the helix bundle of the kleisin, which interacts with the SMC head-linked coiled coil, may be important in regulating the coiled-coil orientation. These mutations may restore the capacity of DNA binding, which is defective in hinge mutants, so that the orientation of arched coiled coils might alter the properties of its association/dissociation cycle with chromosomal DNA.
Evidence that interactions between the HTH motif in the helix bundle and the hinge are critical for the role of cohesin and condensin is provided below. Mutations of rad21-I67F (8) and cnd2-A114T (33) reside in the same HTH of kleisin homologs (Fig. 4 H and I). These two mutations not only exhibited severe defects in mitotic chromosome segregation but were also highly sensitive to DNA-damaging agents at the permissive temperature. The great majority of SMC hinge suppressors resided in kleisin’s HTH motif. Furthermore, hinge interface mutants residing at similar positions generated such HTH suppressor mutations. The actual mechanism of suppression remains to be clarified, while understanding the modes of association and dissociation of DNA with cohesin and condensin is imperatively needed.
In the hold and release model, the hinge interfaces are probably important in regulating coiled-coil orientation, and these G-to-E mutations may weaken the coiled coils’ ability to associate with DNA, possibly by widening the angle of the arched coils. Notably, second mutations that suppressed hinge mutations were found in the amino terminal region of kleisin-like Cnd2 and Rad21. Surprisingly, among six suppressors, four of them (cnd2-A144V, cnd2-G142E, Cnd2-G142R, and rad21-M84K) reside closely at the end of the second helix of the HTH motif, close to the ATPase head domain in the 3D structure (Fig. 4 H and I). The remaining two suppressors, Cnd2-T117I and Rad21-W23S, also reside very closely. One possible explanation for this finding is that the kleisin HTH motif may be critical to the capacity for DNA binding.
In addition to the hold and release model, Skibbens (79) proposed a C-clamp conformation of cohesin, in which SMC coiled coils can fold over into a “C” shape to promote head–hinge association and DNA is entrapped into the C-clamp. Rad50 binds to dsDNA, and its structure (80–82) resembles the SMC Psm1-Psm3 head-coiled coil region (8). From the model of DNA interaction with Rad50 (80–82), in which two coiled coils and a head hold DNA inside, one may speculate that DNA binds to the basic residues on the inner sides of the cohesin coiled coils and head as proposed in the hold and release model (8). We consider two models to explain the mechanism by which hinge mutants were rescued by mutations in the N-terminal HTH motif of kleisins (Fig. 6 C and D). In the first model, to form arched coiled coils that bind DNA, as in the case of Rad50, the head and hinge need to interact. However, how the head of the holocomplex interacts with the hinge is unclear yet. Head–hinge interaction may require the kleisin N-terminal HTH motif and hinge interfaces, judging from the locations of original mutations and their suppressors in the 3D structure (8, 20–26). Hinge interface mutations may destabilize head–hinge interaction, and suppressor mutations in the kleisin N-terminal HTH motif may restore the interaction. This restoration occurs by direct interaction between the hinge interface and the N-terminal HTH motif of kleisin that binds to the SMC head region (model I, Fig. 6C). In the second model, the N-terminal HTH motif of kleisin subunits and hinge interfaces may not be involved directly in head–hinge interaction, but may instead regulate the orientation of coiled coils that enables them to hold and release DNA. Both coiled coils emerging from head and hinge are required to hold DNA, and they work collaboratively in regulating DNA binding. Hinge interface mutations may weaken the DNA-binding activity of coiled coils at the hinge, but suppressor mutations in the kleisin N-terminal HTH motif may enhance the DNA-binding activity of coiled coils at the head by changing the angle made by arched coiled coils (Fig. 6D), therefore balancing DNA binding by the coiled coils.
Materials and Methods
Strains, Plasmids, and Media.
Parental S. pombe ts strains of cnd1, cnd2, cnd3, rad21, mis4, and the 12 cohesin hinge mutants (six cs and six ts) used for suppressor screens have been described previously (8, 32). Briefly, the responsible ts mutations were reintegrated into the S. pombe haploid wild-type strain 972 h− by site-directed PCR-based mutagenesis to obtain ts mutants with a wild-type background (7). The cnd3-K824R and cnd3-K870R were constructed using the same method as described above. The Δpli1, Δred1, Δerh1, Δhrp1, and Δhrp3 were constructed in a similar way: ∼500-bp sequences before and after the corresponding ORFs were cloned and ligated into pBluescript plasmids with the hygromycin antibiotic resistance gene (hygR) in between; plasmids were linearized and were chromosomally integrated into corresponding endogenous loci of the wild-type strain 972 h−. Hygromycin-resistant colonies were then picked, and deletion of the responsible genes was verified by PCR. Proteasome complex mutants (Δpre9, Δrpn10, and Δrpt4) and Δwpl1, which contain the KanMX4 selection marker and are resistant to G418, were obtained from an S. pombe haploid deletion mutant library (Bioneer Corporation). Parental S. pombe strains used for immunoblotting of Cnd1-3FLAG, Cnd2-3FLAG, Cut3-3HA6His, Cut14-3FLAG, Rad21-3FLAG, Psc3-3FLAG, and Mis4-3FLAG have also been described previously (15, 17, 33, 64, 71). YPD (1% yeast extract, 2% polypeptone, 2% d-glucose) and Edinburgh minimal medium 2 were used to culture S. pombe strains, and malt extract agar medium was used for sporulation (83).
Suppressor Screening, Next-Generation Sequencing, and Suppressor Identification.
An efficient suppressor screening method, which applied genomic DNA mixtures for next-generation sequencing to identify suppressor mutations, was developed (7). Suppressor screening, next-generation sequencing of suppressor genomic DNA mixtures, and suppressor mutation identification followed the same procedure described in that paper by Xu et al. (7).
Synchronous Culture and Temperature Shift Experiments.
To arrest cells in mitosis, nda3-KM311 (a cs β-tubulin mutant)–containing strains were used. The nda3-KM311 cells fail in mitotic spindle assembly and arrest in prometaphase due to spindle checkpoint activation (84). Cells were first cultured at a permissive temperature of 30 °C (to 4–5 × 106 cells per milliliter), and were then shifted to a restrictive temperature (20 °C) for 8 h. For the block and release experiment with the cdc25-22 mutant (85), cells were grown in YPD at 26 °C (to 3 × 106 cells per milliliter, 100 mL) and then shifted to 36 °C for 4 h to block cells in late G2 phase. Cells were then released to 26 °C. The time point of release was treated as the start point (0 min). Then, aliquots (10 mL) were taken every 15 min for immunoblotting and measurement of the septation index.
Fluorescence Microscopy.
Cells were cultured to 4–5 × 106 cells per milliliter, fixed with 2% glutaraldehyde, stained with DAPI (a fluorescent probe for DNA), and observed under an all-in-one microscope BZ9000 (Keyence).
Immunochemistry.
For trichloroacetic acid (TCA) precipitation, 10 mL of S. pombe cell culture (containing ∼1 × 108 cells) was mixed with a 1:4 volume (2.5 mL) of ice-cold 100% TCA. The resulting mixture was centrifuged, and pellets were washed with 10% TCA, followed by cell disruption with glass beads in 10% TCA. After centrifugation at 8,000 rpm (Tomy, MX-301) for 10 min at 4 °C, washed precipitates were resuspended in SDS sample buffer containing 1 mM PMSF and boiled at 70 °C for 10 min. After centrifugation at 14,000 rpm (Tomy, MX-301) for 10 min, supernatants were loaded onto custom-made 3–8% gradient Tris-acetate gels (NuPAGE; Invitrogen). Antibodies against FLAG (Sigma), Rad21 (17, 29, 64), Cnd3 (33), tubulin (TAT1; a gift from Keith Gull, University of Oxford, Oxford), and Cdc2 (PSTAIR; a gift from Yoshitaka Nagahama, National Institute for Basic Biology, Okazaki, Japan) were employed as primary antibodies. Anti-mouse–HRP and anti-rabbit–HRP were used as secondary antibodies.
Mutational Analysis of Suppressors in Protein Structures.
Atomic models of S. pombe cohesin and condensin (Fig. 4 C, F, and I) were generated from existing crystal structures of cohesin and condensin from other organisms using homology modeling (8, 69). Mmi1 mutations in Fig. 3C were mapped onto a crystal structure of S. pombe Mmi1 in complex with 11-mer RNA [Protein Data Bank (PDB) ID code 6FPX]. The following structures used in this study are from other organisms; therefore, structural analysis was based on protein sequence alignment results. SUMO-Hus5-Pli1 mutations in Fig. 1D were mapped onto a crystal structure of Saccharomyces cerevisiae E2-SUMO-Siz1/E3-SUMO-PCNA complex, based on protein sequence alignment results (PDB ID code 5JNE). Hrp1 mutations in Fig. 3E were mapped onto a structure of nucleosome–Chd1 complex (PDB ID code 5O9G). Wpl1 mutations in Fig. 6B were mapped onto the structure of the Wpl1 protein (PDB ID code 3ZIK). Cut14 and Cnd2 N-terminal mutations in SI Appendix, Fig. S3D were mapped onto the structure of the kleisin-N SMC interface in prokaryotic condensin (PDB ID code 3ZGX). Cut3 and Cnd2 C-terminal mutations in SI Appendix, Fig. S3E were mapped onto the structure of the kleisin-C SMC interface (PDB ID code 4I99). The Psc3 mutation in SI Appendix, Fig. S5D was mapped onto the structure of Psc3 bound to a fragment of the Rad21 kleisin subunit and DNA (PDB ID code 6H8Q). Sck1 mutations in SI Appendix, Fig. S8E were mapped onto the structure of SGK1 in complex with AMP-PNP (PDB ID code 2R5T). Ksg1 mutations in SI Appendix, Fig. S8F were mapped onto the structure of human PDK1 catalytic domain (PDB ID code 1H1W).
Supplementary Material
Acknowledgments
We thank Dr. Man-Wah Tsang and Dr. Haifeng Zhang for their help in condensin SUMOylation detection by immunoblotting, Dr. Norihiko Nakazawa and Dr. Ryuta Kanai for technical support and valuable discussions, and Dr. Steven D. Aird for technical editing. Generous support from the Okinawa Institute of Science and Technology Graduate University is acknowledged.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database (accession nos. PRJNA450289 and PRJNA525996).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902699116/-/DCSupplemental.
References
- 1.Hayles J, Beach D, Durkacz B, Nurse P (1986) The fission yeast cell cycle control gene cdc2: Isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol Gen Genet 202:291–293. [DOI] [PubMed] [Google Scholar]
- 2.Oliver SG. (1996) From DNA sequence to biological function. Nature 379:597–600. [DOI] [PubMed] [Google Scholar]
- 3.van Leeuwen J, Pons C, Boone C, Andrews BJ (2017) Mechanisms of suppression: The wiring of genetic resilience. BioEssays 39:1700042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Leeuwen J, et al. (2016) Exploring genetic suppression interactions on a global scale. Science 354:aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsuzawa H. (1993) Responsiveness to exogenous cAMP of a Saccharomyces cerevisiae strain conferred by naturally occurring alleles of PDE1 and PDE2. Genetics 135:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Griffiths K Jr, Zhang YH, Ivey FD, Hoffman CS (2005) Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics 171:1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Wang L, Yanagida M (2018) Whole-genome sequencing of suppressor DNA mixtures identifies pathways that compensate for chromosome segregation defects in Schizosaccharomyces pombe. G3 (Bethesda) 8:1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, et al. (2018) Suppressor mutation analysis combined with 3D modeling explains cohesin’s capacity to hold and release DNA. Proc Natl Acad Sci USA 115:E4833–E4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano T, Mitchison TJ (1994) A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79:449–458. [DOI] [PubMed] [Google Scholar]
- 10.Strunnikov AV, Larionov VL, Koshland D (1993) SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol 123:1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koshland D, Strunnikov A (1996) Mitotic chromosome condensation. Annu Rev Cell Dev Biol 12:305–333. [DOI] [PubMed] [Google Scholar]
- 12.Saka Y, et al. (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J 13:4938–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strunnikov AV, Hogan E, Koshland D (1995) SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev 9:587–599. [DOI] [PubMed] [Google Scholar]
- 14.Hirano T, Kobayashi R, Hirano M (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511–521. [DOI] [PubMed] [Google Scholar]
- 15.Sutani T, et al. (1999) Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev 13:2271–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao WC, et al. (2017) Structure of the cohesin loader Scc2. Nat Commun 8:13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomonaga T, et al. (2000) Characterization of fission yeast cohesin: Essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev 14:2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano T, Mitchison TJ, Swedlow JR (1995) The SMC family: From chromosome condensation to dosage compensation. Curr Opin Cell Biol 7:329–336. [DOI] [PubMed] [Google Scholar]
- 19.Kimura K, Hirano T (1997) ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell 90:625–634. [DOI] [PubMed] [Google Scholar]
- 20.Haering CH, Löwe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9:773–788. [DOI] [PubMed] [Google Scholar]
- 21.Bürmann F, et al. (2013) An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol 20:371–379. [DOI] [PubMed] [Google Scholar]
- 22.Diebold-Durand ML, et al. (2017) Structure of full-length SMC and rearrangements required for chromosome organization. Molecular cell 67:334–347.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gligoris TG, et al. (2014) Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science 346:963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haering CH, et al. (2004) Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell 15:951–964. [DOI] [PubMed] [Google Scholar]
- 25.Huis in ’t Veld PJ, et al. (2014) Characterization of a DNA exit gate in the human cohesin ring. Science 346:968–972. [DOI] [PubMed] [Google Scholar]
- 26.Kamada K, Su’etsugu M, Takada H, Miyata M, Hirano T (2017) Overall shapes of the SMC-ScpAB complex are determined by balance between constraint and relaxation of its structural parts. Structure 25:603–616.e4. [DOI] [PubMed] [Google Scholar]
- 27.Funabiki H, et al. (1996) Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature 381:438–441. [DOI] [PubMed] [Google Scholar]
- 28.Funabiki H, et al. (1997) Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J 16:5977–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagao K, Adachi Y, Yanagida M (2004) Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature 430:1044–1048. [DOI] [PubMed] [Google Scholar]
- 30.Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37–42. [DOI] [PubMed] [Google Scholar]
- 31.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103:375–386. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Nakazawa N, Yanagida M (2015) Condensin HEAT subunits required for DNA repair, kinetochore/centromere function and ploidy maintenance in fission yeast. PLoS One 10:e0119347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M (2002) Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197–202. [DOI] [PubMed] [Google Scholar]
- 34.Xu X, Yanagida M (March 26, 2019) Isolation of fission yeast condensin temperature-sensitive mutants with single amino acid substitutions targeted to hinge domain. G3 (Bethesda), 10.1534/g3.119.400156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatebayashi K, Kato J, Ikeda H (1998) Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: Possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics 148:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuasa T, et al. (2004) An interactive gene network for securin-separase, condensin, cohesin, Dis1/Mtc1 and histones constructed by mass transformation. Genes Cells 9:1069–1082. [DOI] [PubMed] [Google Scholar]
- 37.Furuya K, Takahashi K, Yanagida M (1998) Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev 12:3408–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.al-Khodairy F, Enoch T, Hagan IM, Carr AM (1995) The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J Cell Sci 108:475–486. [DOI] [PubMed] [Google Scholar]
- 39.Shayeghi M, Doe CL, Tavassoli M, Watts FZ (1997) Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res 25:1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka K, et al. (1999) Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol 19:8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B (2004) Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J 23:3844–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dasso M. (2008) Emerging roles of the SUMO pathway in mitosis. Cell Div 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Køhler JB, et al. (2015) Targeting of SUMO substrates to a Cdc48-Ufd1-Npl4 segregase and STUbL pathway in fission yeast. Nat Commun 6:8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schleiffer A, et al. (2003) Kleisins: A superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell 11:571–575. [DOI] [PubMed] [Google Scholar]
- 45.Kschonsak M, et al. (2017) Structural basis for a safety-belt mechanism that anchors condensin to chromosomes. Cell 171:588–600.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doughty TW, Arsenault HE, Benanti JA (2016) Levels of Ycg1 limit condensin function during the cell cycle. PLoS Genet 12:e1006216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harigaya Y, et al. (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442:45–50. [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama T, Sugioka-Sugiyama R (2011) Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J 30:1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita A, Takayama T, Iwata R, Yamamoto M (2013) A novel factor Iss10 regulates Mmi1-mediated selective elimination of meiotic transcripts. Nucleic Acids Res 41:9680–9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krzyzanowski MK, Kozlowska E, Kozlowski P (2012) Identification and functional analysis of the erh1(+) gene encoding enhancer of rudimentary homolog from the fission yeast Schizosaccharomyces pombe. PLoS One 7:e49059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugiyama T, et al. (2016) Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol Cell 61:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng MT, Luo J (2013) The enigmatic ERH protein: Its role in cell cycle, RNA splicing and cancer. Protein Cell 4:807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stowell JAW, et al. (2018) A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding. J Biol Chem 293:9210–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farnung L, Vos SM, Wigge C, Cramer P (2017) Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer T, et al. (2009) Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA 106:8998–9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Motamedi MR, et al. (2008) HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krantz ID, et al. (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36:631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, et al. (2018) Structural basis for Scc3-dependent cohesin recruitment to chromatin. eLife 7:e38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murayama Y, Uhlmann F (2015) DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell 163:1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouyang Z, Yu H (2017) Releasing the cohesin ring: A rigid scaffold model for opening the DNA exit gate by Pds5 and Wapl. BioEssays 39:1600207. [DOI] [PubMed] [Google Scholar]
- 62.Ouyang Z, et al. (2013) Structure of the human cohesin inhibitor Wapl. Proc Natl Acad Sci USA 110:11355–11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatterjee A, Zakian S, Hu XW, Singleton MR (2013) Structural insights into the regulation of cohesion establishment by Wpl1. EMBO J 32:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adachi Y, Kokubu A, Ebe M, Nagao K, Yanagida M (2008) Cut1/separase-dependent roles of multiple phosphorylation of fission yeast cohesion subunit Rad21 in post-replicative damage repair and mitosis. Cell Cycle 7:765–776. [DOI] [PubMed] [Google Scholar]
- 65.Shimanuki M, et al. (1993) Isolation and characterization of the fission yeast protein phosphatase gene ppe1+ involved in cell shape control and mitosis. Mol Biol Cell 4:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goshima G, Iwasaki O, Obuse C, Yanagida M (2003) The role of Ppe1/PP6 phosphatase for equal chromosome segregation in fission yeast kinetochore. EMBO J 22:2752–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao B, et al. (2007) Crystal structure of the kinase domain of serum and glucocorticoid-regulated kinase 1 in complex with AMP PNP. Protein Sci 16:2761–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biondi RM, et al. (2002) High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J 21:4219–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Akai Y, et al. (2014) ATPase-dependent auto-phosphorylation of the open condensin hinge diminishes DNA binding. Open Biol 4:140193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanagida M. (2009) Clearing the way for mitosis: Is cohesin a target? Nat Rev Mol Cell Biol 10:489–496. [DOI] [PubMed] [Google Scholar]
- 71.Nakazawa N, et al. (2008) Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J Cell Biol 180:1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakazawa N, et al. (2015) RNA pol II transcript abundance controls condensin accumulation at mitotically up-regulated and heat-shock-inducible genes in fission yeast. Genes Cells 20:481–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sutani T, et al. (2015) Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun 6:7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yong-Gonzales V, Hang LE, Castellucci F, Branzei D, Zhao X (2012) The Smc5-Smc6 complex regulates recombination at centromeric regions and affects kinetochore protein sumoylation during normal growth. PLoS One 7:e51540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skibbens RV. (2019) Condensins and cohesins–One of these things is not like the other! J Cell Sci 132:jcs220491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirano M, Anderson DE, Erickson HP, Hirano T (2001) Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J 20:3238–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hirano M, Hirano T (2002) Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J 21:5733–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirano M, Hirano T (2006) Opening closed arms: Long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell 21:175–186. [DOI] [PubMed] [Google Scholar]
- 79.Skibbens RV. (2016) Of rings and rods: Regulating cohesin entrapment of DNA to generate intra- and intermolecular tethers. PLoS Genet 12:e1006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y, et al. (2016) ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J 35:743–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rojowska A, et al. (2014) Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. EMBO J 33:2847–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner KP (2016) Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J 35:759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hiraoka Y, Toda T, Yanagida M (1984) The NDA3 gene of fission yeast encodes beta-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39:349–358. [DOI] [PubMed] [Google Scholar]
- 85.Moreno S, Hayles J, Nurse P (1989) Regulation of p34cdc2 protein kinase during mitosis. Cell 58:361–372. [DOI] [PubMed] [Google Scholar]
- 86.Zhao Q, et al. (2014) GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42:W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.