Significance
Imaging of plant roots is severely limited by the opacity of soil media. Hydroponic (or gel) conditions provide transparency but nonphysiological root phenotypes. Here, we develop a “transparent soil” with high transparency, good mechanical stability, tunable pore sizes, low cost, and easy scalability. This porous media can support root growth in the presence of air, water, and nutrients, and allows for the imaging of unconstrained root systems in vivo by both photography and microscopy. Our study provides evidence that the roots of soybean developed in this medium are significantly more similar to those developed in real soil than those developed in hydroponic conditions and do not show signs of hypoxia.
Keywords: soil, transparent, hydrogels, plants, microbiome
Abstract
Root phenotypes are increasingly explored as predictors of crop performance but are still challenging to characterize. Media that mimic field conditions (e.g., soil, sand) are opaque to most forms of radiation, while transparent media do not provide field-relevant growing conditions and phenotypes. We describe here a “transparent soil” formed by the spherification of hydrogels of biopolymers. It is specifically designed to support root growth in the presence of air, water, and nutrients, and allows the time-resolved phenotyping of roots in vivo by both photography and microscopy. The roots developed by soybean plants in this medium are significantly more similar to those developed in real soil than those developed in hydroponic conditions and do not show signs of hypoxia. Lastly, we show that the granular nature and tunable properties of these hydrogel beads can be leveraged to investigate the response of roots to gradients in water availability and soil stiffness.
Growing plants for research is constrained by an apparently necessary compromise. On one hand, media that are representative of field soil (e.g., soil, sand) are opaque to most forms of radiation (1) and offer limited control over heterogeneities that affect the development of roots (e.g., gradients in water availability, nutrient concentrations, mechanical properties, porosity). On the other hand, transparent media (e.g., hydroponics, aeroponics, gels) do not provide field-relevant phenotypes and growing conditions (2).
Media that have air-filled, connected pores display several physiologically relevant characteristics of soil, such as aeration and physical interfaces (3). Unfortunately, these porous media are usually opaque to most electromagnetic radiation because each interface changes the direction of propagation of photons, due to refraction and reflection. The magnitude of these deflections increases with the difference between the refractive indices (a physical property of matter dependent on electronic density and susceptibility) of the medium and the material contained in the pores (4). Therefore, a porous medium can become transparent to light if it is fully saturated with a fluid whose refractive index matches that of the porous medium (5).
Index matching of granular materials, including hydrogels, was used successfully to study hydrology, soil physics, and fluid dynamics in porous media (6, 7). Nonetheless, the use of this approach to study root development is subject to numerous complex constraints that have made this task notoriously challenging. The medium must be (i) produced simply and inexpensively in large quantities (hectoliters), (ii) nontoxic to plants, (iii) transparent enough in common nutrient solutions to allow for the phenotyping of a whole root system in vivo, and (iv) strong enough to not collapse under its own weight. Furthermore, it should provide water and nutrition to the growing plant and have a fully connected porosity to prevent the formation of air pockets. A recent pioneering work by Downie et al. (5, 8) reported Nafion, a sulfonated tetrafluoroethylene-based fluoropolymer−copolymer, as a promising material for this purpose, given the similarity of its refractive index to that of water. Unfortunately, the material is currently very expensive (∼$1,000/kg), it must be chemically processed before use with plants, it does not absorb water or nutrients (some parts of the root system must be saturated with nutrient solution), and its index matching solution has significant concentrations of sorbitol (0 to 13%, wt/vol), which can cause osmotic stress in plants (9).
We here describe a porous medium that allows for the imaging of unconstrained root systems in vivo by both photography and microscopy and the time-resolved phenotyping of roots. The medium consists of interconnected pores that are surrounded by nutrient solution, held into spherical beads of hydrogel. These beads have controllable size and hardness and are produced simply, rapidly, and inexpensively by dropping a solution of gellan gum and alginate into a stirred solution of MgCl2. Temporary saturation with nutrient solution (a treatment comparable to rainfall or watering) makes this medium sufficiently transparent to allow imaging of a 20 × 20 × 20 cm volume. This medium outperformed hydroponics in producing field-relevant root phenotypes in Glycine max (six of seven key phenotypes were not significantly different from field soil’s, instead of two). Similarly, a key gene involved in response to root hypoxia [nonsymbiotic Hemoglobin (nsHB); Glyma.11G121800 (10)] was significantly overexpressed in hydroponics but was not overexpressed in “transparent soil” (TS) or soil. Lastly, this medium allows for the imaging of chemical changes (pH) in proximity to the roots, and of their development in response to designed heterogeneities (specifically gradients in hardness and water availability).
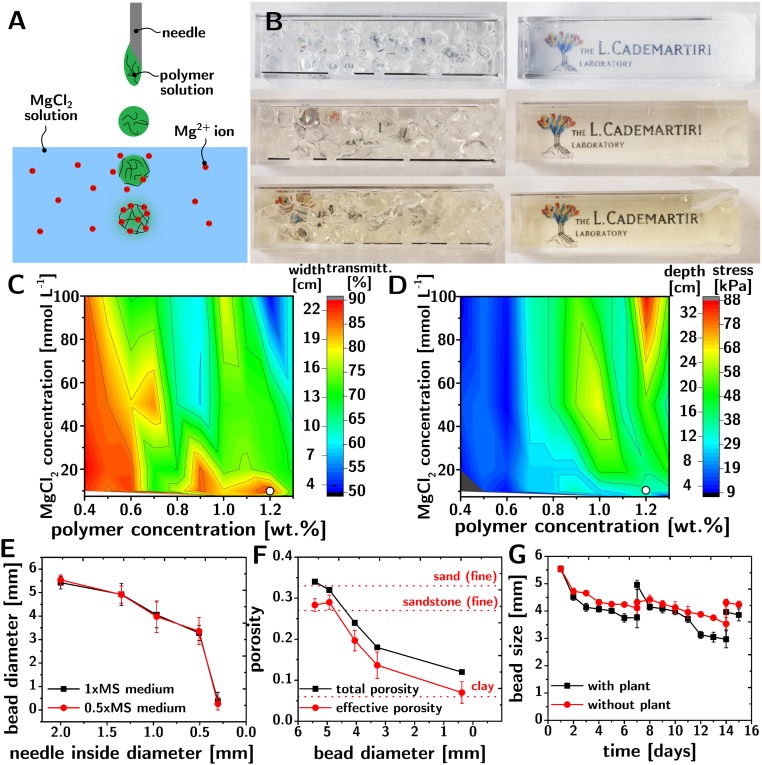
The hydrogel spherification process to create the TS is shown in Fig. 1A. Dropping a solution of alginate and gellan gum (1:4 ratio) into a stirred solution of MgCl2 rapidly gels the polymer into discrete spherical beads. The beads are then soaked into a nutrient solution [0.5× Murashige−Skoog (MS), Lysogeny broth, or soil extract] until equilibration, and then drained of excess liquid before introducing the organism of interest (cf. Fig. 1B). The medium is temporarily saturated with the nutrient solution to turn the medium transparent and enable phenotyping (cf. Fig. 1B). Since most of the opacity in this system is due to scattering, near-infrared (NIR) illumination provides significantly better transparency than visible light and prevents exposing the root to biologically active radiation (11) (cf. SI Appendix).
Fig. 1.
Fabrication and physical properties of hydrogel-based TS. (A) Sketch of the spherification process to make the hydrogel beads. (B) TS before (Left) and after (Right) saturation with nutrient growth media [0.5× Murashige and Skoog medium (Top), lysogeny broth (Middle), and soil extract (Bottom)]. The logo behind the cuvette is not visible before saturation but becomes clearly visible upon saturation of the TS. (C) Transmittance of TS (at 1,080 nm, in 0.5× MS) as a function of the concentration of the polymer and MgCl2 solutions used during spherification. The colormap also shows the length of the optical path that leads to 10% transmittance at 1,080 nm. (D) Collapse stress of TS (filled with 0.5× MS) as a function of the concentration of the polymer and MgCl2 solutions used during spherification. The colormap also shows the thickness of TS that would collapse at its bottom. (E) Bead size as a function of the inner diameter of the nozzle used during spherification. (F) Total and effective porosity of TS as a function of the size of the beads. (G) Shrinkage of the beads as a function of time (with and without plants) and their recovery upon saturation with media on days 7 and 14.
The design of this TS is based on the following rationale. Assemblies of spherical beads always have a connected porosity regardless of their arrangement. Alginate and gellan gum are already widely used for plant growth media. Alginate gels rapidly but is colored and makes relatively weak gels, while gellan gum is colorless and make strong gels but gels too slowly. The mixture of the two polymers synergically allows for the rapid spherification of strong, colorless hydrogel beads. Furthermore, alginate prevents the spontaneous gelling of gellan gum at the concentrations required in the polymer mixture. Compared with calcium, magnesium prevents the precipitation of sulfates from common nutrient solutions (e.g., MS), and yields a smaller increase in refractive index, but is similarly nontoxic to plants, even at high concentrations.
The transparency and mechanical properties of the TS determine the maximum volume of medium that can be phenotyped with it: Higher transparency enables wider containers, while higher strength enables deeper containers. Increasing the concentration of the polymer or MgCl2 solutions increases the strength of the TS at the expense of its transparency (Fig. 1 C and D), but there are optimized conditions (e.g., 1.2 wt% of polymer and 10 mM MgCl2 when using 0.5× MS as a nutrient solution) that provide a useful compromise (white dot in Fig. 1 C and D). In these conditions, this TS in a 22-cm-wide and 20-cm-deep container transmits 10% of 1,080-nm light (highest wavelength detectable with Si-based photodetectors found in commercial digital cameras), and does not collapse under its own weight.
The size of the pores and the effective porosity are important characteristics of soil that affect gas permeation and, therefore, the distribution of the roots and of the soil microbiome (12). The inner diameter of the nozzle used to drop the polymer solution determines the size of the beads (cf. Fig. 1E) from 0.5 mm to 5.5 mm. Due to the formation of menisci at the points of contact between the beads, the effective porosity of the TS depends on the beads’ size (cf. Fig. 1F) and can be controlled between 0.06 (similar to clay’s) and 0.28 (similar to sandstone’s). The volume fraction of inaccessible pores (i.e., the difference between the effective porosity and the total porosity) is only ∼4%. The size of the beads can change slightly over the course of days due to the settling of the gel and uptake of water by the roots. Nonetheless, saturating the TS with fresh medium reswells the beads close to their original size (cf. Fig. 1G).
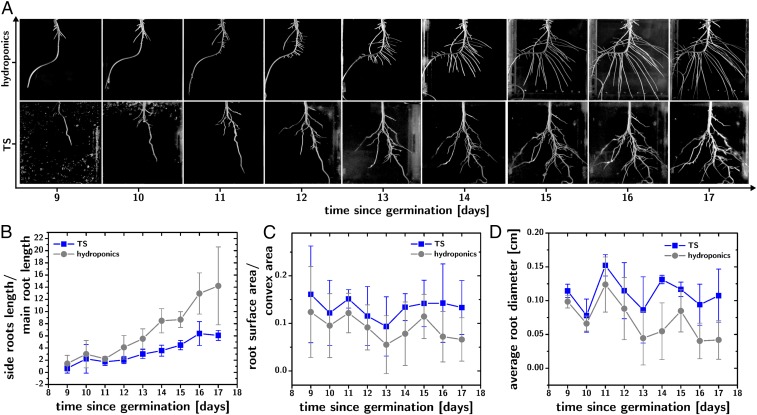
This TS provides water, nutrients, and aeration to the growing roots and becomes transparent upon saturation with fresh medium, allowing for time-resolved root phenotyping in vivo by photography. Fig. 2A compares the time-lapse phenotyping (24-h intervals) of the roots of soybean plants [G. max (L.) Merr., IA2102, n = 5] in hydroponics (Fig. 2A, Top) and TS (Fig. 2A, Bottom) grown in highly controlled, aerated habitats (13). Fig. 2 B–D shows the temporal evolution of root phenotypes in G. max that were selected for their ability to describe architectural differences (rather than size differences) between roots grown in hydroponics (gray) and TS (blue). The ratio between the length of the side roots and those of the primary root (Fig. 2B) shows that the root phenotype diverged at day 12 from germination, with hydroponically grown roots developing a significantly larger fraction of side roots (the length of the main root was similar in the two treatments; cf. SI Appendix). On the other hand, the ratio between the total root surface area and the convex area (Fig. 2C) shows a consistently higher value for the TS-grown roots, indicating the formation of a more efficiently space-filling root system. This finding can be easily explained by the physical support provided to the roots by the TS: Hydroponically grown roots are unsupported and lean down due to their weight and concentrate on the bottom of the container, while, in TS, the roots are supported and their development is guided by tropisms but not by their weight. The average root diameter (Fig. 2D) also shows higher values for the TS-grown plants, and an oscillatory behavior in time that is interestingly synchronized across the two treatments. The larger average root diameter for TS-grown plants could be, in part, due to their smaller fraction of side roots, while the oscillatory behavior of this phenotype over time is consistent with oscillations in the rate of root growth.
Fig. 2.
Root phenotyping in TS. (A) Time lapse (24-h interval) in vivo root phenotyping of G. max growing in hydroponic conditions (Top) and in TS (Bottom) between day 9 and day 17 from germination. Soil extract was used as a nutrient medium for both treatments. (B–D) Comparisons of the temporal evolutions of root phenotypes (n = 5) for the TS (blue) and hydroponic (gray) treatments: B plots the ratio between the length of the side roots and main root, and shows accelerated development of the side roots in hydroponic conditions; C plots the ratio between the surface area and the convex area of the root system, and shows increased space-filling of the TS-grown roots; and D plots the average root diameter, and shows larger diameter of for the TS-grown roots.
To assess to what extent this TS mimics field soil, we compared the root phenotypes of soybean plants (12 d old, n = 9) grown in hydroponics, TS, and sterilized field soil. We used soil extract as a nutrient solution in the hydroponic and TS treatments to ensure similar nutrient compositions across the different treatments. Nutrient concentrations in the soil extract are not limiting growth (shoot biomass produced by G. Max in soil extract was not significantly different from in MS media; SI Appendix, Fig. S5, P = 0.63, n = 4).
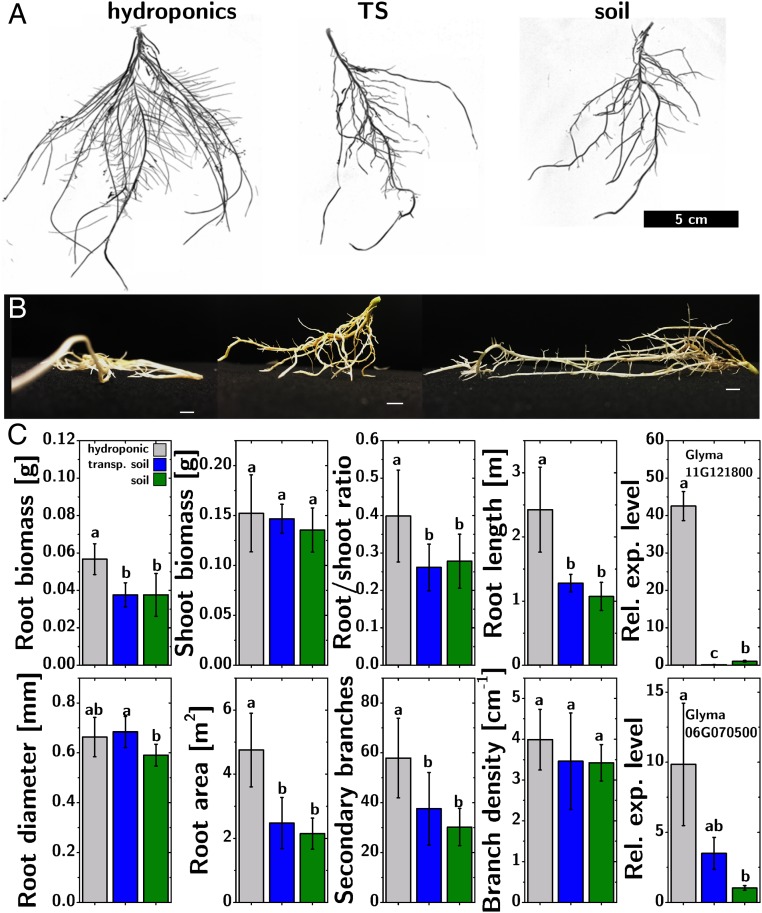
The root structure developed in TS was visibly more similar to that developed in field soil than that developed in hydroponics [cf. Fig. 3A, Left (hydroponics), Middle (TS), and Right (field soil)]. The ex vivo analysis of the roots from the three treatments revealed a very significant difference in the mechanical stiffness of the roots. Fig. 3B shows the side view of roots of G. max when lying on a flat surface. While the hydroponically grown roots collapsed under their own weight, both the TS-grown and the soil-grown roots were, instead, self-supporting. The larger stiffness of the TS-grown plants is consistent with the larger average diameter of the roots shown in Fig. 2D.
Fig. 3.
Root phenotyping in TS. (A and B) Comparison of (A) G. max roots and (B) their mechanical stiffness, after growth in hydroponics (Left), TS (Center), and sterilized field soil (Right). (Scale bar in B, 1 cm.) (C) Comparison of biomass, root morphology traits, and gene expression (Glyma.11G121800 and Glyma.06G070500) in G. max plants grown in hydroponics, TS, and sterilized field soil. Error bars for biomass and root morphology traits indicate SD (n = 9); error bars for gene expression represent SD (n = 4). Different letters above the histograms (a, b, c) indicate significant differences between treatments (P < 0.05, one-way ANOVA followed by Tukey’s test).
Quantification of fundamental traits (root and shoot biomass, RB and SB; root-to-shoot ratio, RSR; total root length, TRL; root diameter, RD; root surface area, RSA; secondary root number, SRN; secondary root density, SRD) validates the similarity between TS-grown and soil-grown roots, compared with hydroponically grown roots. Out of these eight phenotypes, only one (RD) shows a significant (P = 0.014) difference between TS and field soil. Five of the remaining phenotypes (RSR, TRL, RSA, SRN, RB) show a significant difference (P < 0.05) between hydroponics and both TS and field soil, while two (SB, SRD) show no significant difference across all three treatments (cf. Fig. 3C).
Real-time PCR analysis was used to investigate whether the TS medium provides adequate access to nutrients and oxygen, and how it compares to hydroponics and field soil. We characterized the expression of five genes that have been associated with abiotic stress [N deficiency, Glyma.12g221500 (14); P, Fe, and Zn deficiency, Glyma.08g053500 (15); drought, Glyma.04G203300 (16); hypoxia, Glyma.11G121800 and Glyma.04G240800 (10, 17)] and three genes that have been associated with the development of lateral roots and root tips (Glyma.06G070500, Glyma.08G133800, Glyma.02G043400) in plants (root tissues of G. max, IA2102, 12 d old, n = 4) grown in hydroponics, TS, and field soil, using soil extract as a nutrient medium, as described for the phenotyping study. Relative expression was calculated by the 2−ΔΔCt method by considering soil treatment as the calibrator (18). Genes associated with nutrient or water deficiency (Glyma.12g221500, Glyma.08g053500, Glyma.04G203300) were not differentially expressed across treatments, suggesting that the TS does not deprive plants of nutrients or water, compared with either field soil or hydroponics. Two of the genes associated with root development (Glyma.08G133800, Glyma.02G043400) were also not differentially expressed in the three treatments.
Two genes were found to be differentially expressed (cf. Fig. 3C). The first (Glyma.11G121800, nsHB) is strongly associated with response to hypoxia in soybean through the synthesis of leghemoglobin, and was strongly overexpressed in the hydroponic treatment (42-fold compared with field soil, P < 0.001) and underexpressed in the TS treatment (0.14-fold compared with field soil, P < 0.001), strongly indicating that TS, in contrast to hydroponics, does not cause hypoxic stress in roots. The lower hypoxic stress in TS and field soil would be consistent with the increased curvature observed in the root phenotypes (19). The other differentially expressed gene (Glyma.06G070500, GTP-binding nuclear protein Ran) is associated with signal transduction and stress response and was overexpressed in hydroponics (10-fold compared with field soil, P = 0.007). This gene is found to be highly expressed in lateral roots, and the expression qualitatively mimics the dependency of SRN on the treatments, suggesting that the observed differential expression is not due to a stress response but due to a different root structure.
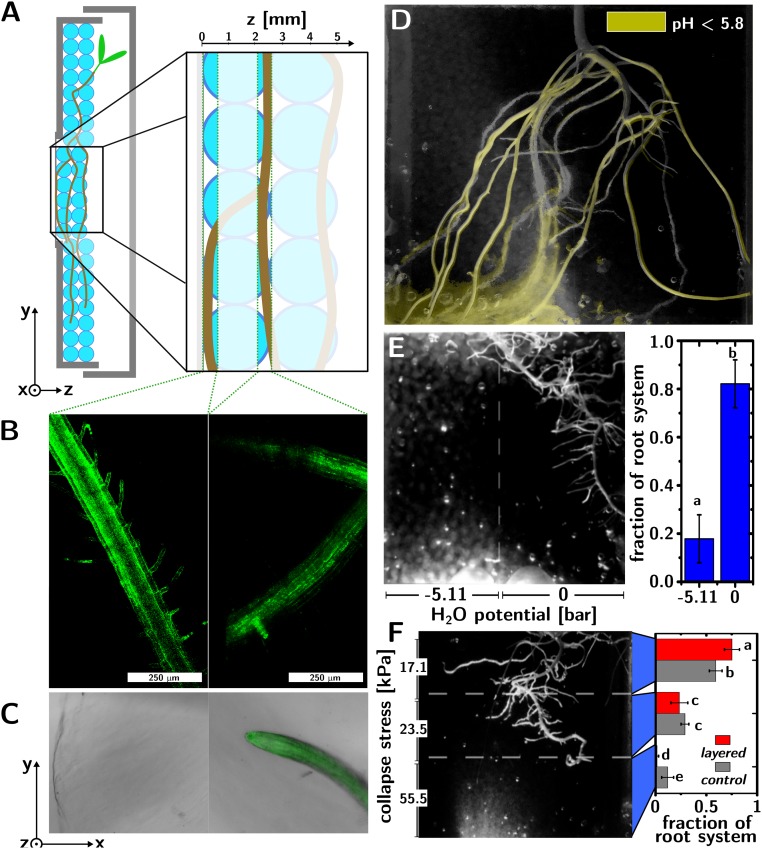
This TS is also compatible with in vivo microscopy of plant roots. Arabidopsis thaliana plants (with a GFP-tagged plasma membrane) were grown in Petri dishes filled with TS and fitted with a glass coverslip window (cf. Fig. 4A). Confocal microscopy (cf. Fig. 4B) shows root segments at different depths (0 and 2.4 mm, behind a hydrogel bead) into the TS. Similarly, fluorescence microscopy allows optical and fluorescence imaging of roots behind hydrogel beads, several millimeters (2.9 mm) inside the TS (cf. Fig. 4C).
Fig. 4.
Applications of TS. (A) Sketch of the in vivo phenotyping of A. thaliana roots in TS in a modified Petri dish. (B) In vivo confocal microscopy of A. thaliana roots at different depths into the TS [0 mm (Left), 2.4 mm (Right)]. (C) In vivo fluorescence microscopy at different depths into the TS [0 mm (Left), showing the surface of a TS bead; 2.9 mm (Right), showing visible and fluorescence overlay of the A. thaliana root behind the bead]. (D) In vivo pH mapping in TS obtained by overlaying a root image (obtained in the NIR from a G. max plant) with a yellow colormap obtained from a visible photograph of the same root system after isolating the color change caused in a pH indicator in the TS by acidification. (E) Growth of a G. max root system in a TS medium with a graded water potential. The left half of the TS medium contains 200 g⋅L−1 PEG (−5.11 bar water potential), while the right half contains TS without PEG (0 bar water potential). The root develops significantly (P = 10−11, n = 10; different letters above the histogram indicate significant differences) toward the region of higher water availability. (F) Growth of a G. max root system in a TS medium with graded stiffness (collapse stresses of 17.12, 23.52, and 55.52 kPa from the top down; different letters above the histogram indicate significant differences). The graded TS leads to the development of a shallower root system with a significantly higher fraction of projected root area (P < 0.001) in the top layer of soil, compared with a homogeneous control.
Lastly, this TS can be easily modified to visualize chemical changes caused by the roots or study the effect of soil heterogeneities on root development in vivo. Fig. 4D shows G. max roots growing in TS, where a yellow overlay indicates local acidification (pH < 5.8) of the medium caused by the roots (20). The map is obtained by overlaying two images. The background image is an NIR photograph of the root system in TS. The yellow colormap in the foreground was obtained from a visible photograph of the same root system after isolating the color change caused by acidification in a pH indicator (Bromocresol purple changing from purple to yellow between 6.8 and 5.2) that was introduced into the TS. Since the medium supports plant growth, the pH change can be monitored in time (Movie S3).
Differently from hydroponics, it is easy to create designed heterogeneities in TS by placing beads with different properties in different parts of the medium. Fig. 4E shows G. max roots growing in a medium where the left half is filled with TS containing polyethylene glycol (PEG, MW = 8,000, 200 g/L), which reduces water availability for the plant (21), while the right half is TS devoid of PEG. The root system develops asymmetrically toward the region of high water availability (82 ± 9% of the projected root area is located in the half of the medium with high water availability, P = 10−11, n = 10). Fig. 4F shows instead G. max roots growing in a medium consisting of three layers of TS of increasing hardness (collapse stresses of 17.12, 23.52, and 55.52 kPa from the top down). The roots developed more shallowly in the layered medium than in the homogeneous control, as demonstrated by the root fractions in each layer of soil (histogram in Fig. 3F; P = 0.001 in the top layer and P = 0.01 in the bottom layer, n = 3).
This TS medium is complementary to other nondestructive phenotyping approaches that rely on solid-penetrating radiation [e.g., X-ray Computed Tomography (CT) (1), Magnetic Resonance Imaging (MRI) (1), Positron Emission Tomography (22)], or on confining roots to a transparent surface [e.g., rhizotrons (23), GLO-Roots (24)]. The throughput achievable with photography in TS is much higher than what is currently possible in X-ray CT or MRI [where one instrument can characterize between one and two dozen plants a day (1, 25, 26), generating significant amounts of data (GB per scan), which must be processed extensively and segmented expertly to avoid artifacts (1)]. Our approach can easily characterize thousands of plants for a much lower cost in the same amount of time by implementing very simple automation (e.g., cameras on rails coupled with peristaltic pumps for watering/draining).
Root phenotypes are nonlinear functions of large numbers of correlated input variables. Therefore, their study is a statistics-dependent problem where data quantity and quality are key. Using a GxE screen as an example, our TS medium could enable the initial screening of relevant traits over very large genotype/environment sets, and allow a highly statistically informed decision on what genotypes to explore in an X-ray CT/MRI, data/time/expertise/cost-intensive manner.
X-ray CT is often regarded as the state-of-the-art technique for root phenotyping, but it is not devoid of limitations. Besides throughput and cost, X-ray tomography (i) generally requires moving the plant to the instrument [which can affect plants (27)], (ii) requires small pot sizes [∼8 cm in diameter, according to the review by Metzner et al. (1)], (iii) has difficulty identifying all roots [it reliably identifies 60 to 70% of the root system, according to Metzner et al. (1)], and (iv) is significantly affected by the moisture, type, and heterogeneity of soil (28).
These limitations affect its natural-substrate relevance: Limiting the pot size can expose plants to thigmotropic responses with the walls of the container (1, 28, 29), using soils suited for CT limits the ability to test the phenotypes in other soils, and missing 20 to 30% of the root system could add unknown systematic errors to phenotype characterization (e.g., in the characterization of average root length for genotypes with different average root diameter). In summary, when all methodologies have limitations, it is usually best to develop complementary techniques with different limitations. In this sense, this TS fills a much-needed niche in the spectrum of techniques available for root phenotyping.
Of course, as outlined above, this TS medium has limitations. (i) The transparency and the mechanical properties limit the size of the root volume to about 20 × 20 × 20 cm. (ii) The size of the beads is currently limited to between 0.5 mm and 5 mm. (iii) Methods to produce hectoliter amounts of TS are not detailed here (a scale-up approach to produce 50 L/d from a single setup is outlined in SI Appendix). (iv) The surface chemistry of the TS beads is significantly different from that of soil.
There are reasons to believe that many of the above limitations can be overcome: Hydrogels have been a material of choice for tissue engineering and drug delivery, due to the endless possibilities they offer for functionalization and control of mass transport (30–33). We expect that this application of hydrogels will provide, through synthetic polymers and their functionalization, broad possibilities for the quantitative study of GxE interactions in root development and the modeling of the rhizosphere, as well as in other fields of science such as animal science (34–36), robotics (37), soft matter physics (38), and biomechanics (39).
Supplementary Material
Acknowledgments
We thank Xinyi Xu and Asheesh Singh for help and assistance with gene expression analysis, Brian Scott for help and assistance with field soil sampling, and Souvik Banerjee for assistance in data collection. Preliminary work on this project was sponsored by the Plant Science Institute at Iowa State University through a Faculty Scholar Award (to L.C. and B.G.). Later work was sponsored by the United States Department of Agriculture through Award 2017-67007-25946.
Footnotes
Conflict of interest statement: L.M. and L.C. are inventors on a patent application (US 16/107,512) submitted by Iowa State University Research Foundation, Inc. that covers methods of making hydrogel-based transparent soil.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820334116/-/DCSupplemental.
References
- 1.Metzner R., et al. , Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: Potential and challenges for root trait quantification. Plant Methods 11, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuijken R. C., van Eeuwijk F. A., Marcelis L. F., Bouwmeester H. J., Root phenotyping: From component trait in the lab to breeding. J. Exp. Bot. 66, 5389–5401 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Unger P. W., Kaspar T. C., Soil compaction and root growth: A review. Agron. J. 86, 759–766 (1994). [Google Scholar]
- 4.Skaar J., Fresnel equations and the refractive index of active media. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 73, 026605 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Downie H., et al. , Transparent soil for imaging the rhizosphere. PLoS One 7, e44276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budwig R., Refractive index matching methods for liquid flow investigations. Exp. Fluids 17, 350–355 (1994). [Google Scholar]
- 7.Iskander M., Bathurst R., Omidvar M., Past, present, and future of transparent soils. Geotech. Test. J. 38, 557–573 (2015). [Google Scholar]
- 8.Downie H. F., Valentine T. A., Otten W., Spiers A. J., Dupuy L. X., Transparent soil microcosms allow 3D spatial quantification of soil microbiological processes in vivo. Plant Signal. Behav. 9, e970421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J.-K., Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama T. J., et al. , Insights into soybean transcriptome reconfiguration under hypoxic stress: Functional, regulatory, structural, and compositional characterization. PLoS One 12, e0187920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokawa K., Kagenishi T., Kawano T., Mancuso S., Baluška F., Illumination of Arabidopsis roots induces immediate burst of ROS production. Plant Signal. Behav. 6, 1460–1464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowell D. L., Soil Science: Methods & Applications (Routledge, 2014). [Google Scholar]
- 13.Siemianowski O., et al. , HOMEs for plants and microbes–A phenotyping approach with quantitative control of signaling between organisms and their individual environments. Lab Chip 18, 620–626 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Li D., Jiang J., Dong Z., Ma Y., Soybean NAC gene family: Sequence analysis and expression under low nitrogen supply. Biol. Plant. 61, 473–482 (2017). [Google Scholar]
- 15.Zeng H., Zhang Y., Zhang X., Pi E., Zhu Y., Analysis of EF-Hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 8, 877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neves-Borges A. C., et al. , Expression pattern of drought stress marker genes in soybean roots under two water deficit systems. Genet. Mol. Biol. 35 (Suppl 1), 212–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valliyodan B., et al. , Expression of root-related transcription factors associated with flooding tolerance of soybean (Glycine max). Int. J. Mol. Sci. 15, 17622–17643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Eysholdt-Derzsó E., Sauter M., Root bending is antagonistically affected by hypoxia and ERF-mediated transcription via auxin signaling. Plant Physiol. 175, 412–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaillard B., Plassard C., Hinsinger P., “Measurements of H+ fluxes and concentrations in the rhizosphere” in Handbook of Soil Acidity, Rengel Z. Ed. (CRC, 2003), pp. 231–266. [Google Scholar]
- 21.Lawlor D., Absorption of polyethylene glycols by plants and their effects on plant growth. New Phytol. 69, 501–513 (1970). [Google Scholar]
- 22.Hubeau M., Steppe K., Plant-PET scans: In vivo mapping of xylem and phloem functioning. Trends Plant Sci. 20, 676–685 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Taylor H., Huck M., Klepper B., Lund Z., Measurement of soil-grown roots in a rhizotron 1. Agron. J. 62, 807–809 (1970). [Google Scholar]
- 24.Rellán-Álvarez R., et al. , GLO-roots: An imaging platform enabling multidimensional characterization of soil-grown root systems. eLife 4, e07597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian S., Han L., Dutilleul P., Smith D. L., Computed tomography scanning can monitor the effects of soil medium on root system development: An example of salt stress in corn. Front. Plant Sci. 6, 256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Dusschoten D., et al. , Quantitative 3D analysis of plant roots growing in soil using magnetic resonance imaging. Plant Physiol.,170, 1176–1188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braam J., Davis R. W., Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 60, 357–364 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Zappala S., et al. , Quantifying the effect of soil moisture content on segmenting root system architecture in X-ray computed tomography images. Plant Soil 370, 35–45 (2013). [Google Scholar]
- 29.Atkinson J. A., Pound M. P., Bennett M. J., Wells D. M., Uncovering the hidden half of plants using new advances in root phenotyping. Curr. Opin. Biotechnol. 55, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slaughter B. V., Khurshid S. S., Fisher O. Z., Khademhosseini A., Peppas N. A., Hydrogels in regenerative medicine. Adv. Mater. 21, 3307–3329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K. Y., Mooney D. J., Alginate: Properties and biomedical applications. Prog. Polym. Sci. 37, 106–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoare T. R., Kohane D. S., Hydrogels in drug delivery: Progress and challenges. Polymer (Guildf.) 49, 1993–2007 (2008). [Google Scholar]
- 33.Omenetto F. G., Kaplan D. L., New opportunities for an ancient material. Science 329, 528–531 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maladen R. D., Ding Y., Li C., Goldman D. I., Undulatory swimming in sand: Subsurface locomotion of the sandfish lizard. Science 325, 314–318 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Sharpe S. S., Kuckuk R., Goldman D. I., Controlled preparation of wet granular media reveals limits to lizard burial ability. Phys. Biol. 12, 046009 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Dorgan K. M. The biomechanics of burrowing and boring. J. Exp. Biol. 218, 176–183 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Aguilar J., et al. , A review on locomotion robophysics: The study of movement at the intersection of robotics, soft matter and dynamical systems. Rep. Prog. Phys. 79, 110001 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Dijksman J. A., Brodu N., Behringer R. P., Refractive index matched scanning and detection of soft particles. Rev. Sci. Instrum. 88, 051807 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Wendell D., Luginbuhl K., Guerrero J., Hosoi A., Experimental investigation of plant root growth through granular substrates. Exp. Mech. 52, 945–949 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.