Significance
We sequenced the genomes of 15 skeletons from a 5,000-y-old mass grave in Poland associated with the Globular Amphora culture. All individuals had been brutally killed by blows to the head, but buried with great care. Genome-wide analyses demonstrate that this was a large extended family and that the people who buried them knew them well: mothers are buried with their children, and siblings next to each other. From a population genetic viewpoint, the individuals are clearly distinct from neighboring Corded Ware groups because of their lack of steppe-related ancestry. Although the reason for the massacre is unknown, it is possible that it was connected with the expansion of Corded Ware groups, which may have resulted in violent conflict.
Keywords: ancient DNA, archaeology, kinship, migration, violence
Abstract
The third millennium BCE was a period of major cultural and demographic changes in Europe that signaled the beginning of the Bronze Age. People from the Pontic steppe expanded westward, leading to the formation of the Corded Ware complex and transforming the genetic landscape of Europe. At the time, the Globular Amphora culture (3300–2700 BCE) existed over large parts of Central and Eastern Europe, but little is known about their interaction with neighboring Corded Ware groups and steppe societies. Here we present a detailed study of a Late Neolithic mass grave from southern Poland belonging to the Globular Amphora culture and containing the remains of 15 men, women, and children, all killed by blows to the head. We sequenced their genomes to between 1.1- and 3.9-fold coverage and performed kinship analyses that demonstrate that the individuals belonged to a large extended family. The bodies had been carefully laid out according to kin relationships by someone who evidently knew the deceased. From a population genetic viewpoint, the people from Koszyce are clearly distinct from neighboring Corded Ware groups because of their lack of steppe-related ancestry. Although the reason for the massacre is unknown, it is possible that it was connected with the expansion of Corded Ware groups, which may have resulted in competition for resources and violent conflict. Together with the archaeological evidence, these analyses provide an unprecedented level of insight into the kinship structure and social behavior of a Late Neolithic community.
In 2011, archaeological excavations near the village of Koszyce in southern Poland uncovered a ca. 5,000-y-old mass grave (Fig. 1) associated with the Globular Amphora culture and containing the remains of 15 men, women, and children who had been killed, but carefully buried with rich grave goods (1). Closer study of the skeletons (2) revealed that the individuals had all been killed by blows to the head, possibly during a raid on their settlement. To shed light on this Late Neolithic community and the events that unfolded at Koszyce 5,000 y ago, we sequenced their genomes to between 1.1- and 3.9-fold coverage (Table 1) and performed genome-wide analyses to explore their genetic ancestry and kinship relations. In addition, we obtained 16 radiocarbon dates (SI Appendix, section 4 and Dataset S1) to narrow down the date of the massacre to 2880–2776 BCE (SI Appendix, Fig. S5). We also provide a detailed description of the injuries (SI Appendix, section 3), and strontium isotope measurements of dental enamel provide information on mobility and residence patterns (SI Appendix, section 5). Together, the analyses enable us to draw up a remarkably detailed picture of this Late Neolithic community, including their genetic ancestry, physical appearance, kinship structure, and social organization.
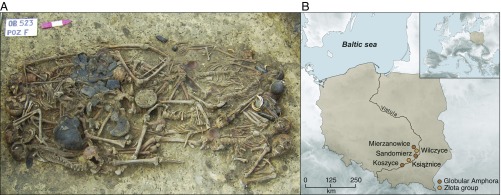
Fig. 1.
The mass grave at Koszyce, southern Poland. (A) Photograph of the 15 skeletons and grave goods buried at Koszyce site 3 (reproduced with permission from ref. 2). (B) Map of Poland showing the location of Koszyce and four other Globular Amphora/Złota group sites included in this study.
Table 1.
Sequencing results for the Koszyce individuals
| Individual no. | Age at death, y | Average depth of coverage | Genome coverage (%) | Chromosomal sex | 5′ C-T (%) | Contamination rate (%) | mtDNA haplogroup | ChrY haplogroup |
| 1 | 25–30 | 2.8× | 82 | XX | 19 | 0.3 | T2b | — |
| 2 | 1.5–2 | 1.9× | 75 | XY | 24 | 0.7 | T2b | I2a-L801 |
| 3 | 30–35 | 1.3× | 67 | XX | 24 | 0.1 | H27+16093 | — |
| 4 | 16–17 | 3.0× | 83 | XY | 21 | 0.1 | K1a1b1e | I2a-L801 |
| 5 | 20–25 | 3.9× | 85 | XY | 19 | 0.0 | HV0a | I2a-L801 |
| 6 | 13–14 | 1.1× | 60 | XX | 20 | 0.4 | K1a1b1e | — |
| 7 | 2–2.5 | 2.1× | 77 | XY | 21 | 0.0 | HV16 | I2a-L801 |
| 8 | 30–35 | 3.0× | 83 | XX | 22 | 0.2 | J1c3f | — |
| 9 | 15–16 | 2.5× | 81 | XX | 21 | 0.2 | J1c3f | — |
| 10 | 18–20 | 2.4× | 80 | XY | 20 | 0.1 | HV0a | I2a-L801 |
| 11 | 40–50 | 1.1× | 60 | XY | 26 | 0.0 | HV0a | I2a-L801 |
| 12 | 30–40 | 3.7× | 85 | XX | 22 | 0.4 | K1a1b1e | — |
| 13 | 5–6 | 2.6× | 81 | XY | 19 | 0.0 | J1c3f | I2a-L801 |
| 14 | 50–60 | 3.0× | 83 | XX | 24 | 0.3 | HV0a | — |
| 15 | 40–50 | 2.9× | 82 | XY | 21 | 0.0 | HV0a | I2a-L801 |
Age at death was estimated based on standard osteological methods (47, 48). Genetic sex estimates were obtained by assessing the reads mapping to the Y versus X chromosome (49). Deamination rates (5′ C-T) were estimated using MapDamage (34). Contamination rates are based on Schmutzi (35). For more information, see Materials and Methods.
Results and Discussion
DNA was isolated from teeth and petrous bones, using established protocols, and the libraries were sequenced on Illumina HiSeq 2500 platforms. The sequence data showed all the hallmarks of damaged ancient DNA, and modern contamination was estimated to be very low (Table 1). The human endogenous DNA contents ranged between 13% and 75% (Dataset S3). According to the sequencing data, eight of the individuals in the grave were males and seven were females (Table 1). This is consistent with previously published results based on skeletal traits, with the exception of one male (individual 4) who had previously been identified as a probable female and five juveniles who could not be sexed previously (2). Data on phenotypic traits based on imputed genotypes (Dataset S5) revealed that the individuals had mostly brown eyes, dark or dark-blonde hair, and intermediate to dark skin.
Genetic Affinities.
To investigate their genetic ancestry, we merged the 15 Koszyce genomes with the Human Origins data set (3), as well as 168 previously published ancient genomes (Dataset S6). In addition, we included genome-wide data for nine individuals from four contemporary, neighboring sites in southern Poland belonging to the Globular Amphora culture and its Złota group variant that we sequenced to between 0.2- and 1-fold coverage (Dataset S3). We then performed a principal component analysis and found that all 24 Globular Amphora/Złota group individuals clustered with other previously sequenced Globular Amphora individuals (4, 5) and other Neolithic groups (Fig. 2A). This confirms earlier suggestions that the Globular Amphora people belonged to the Neolithic gene pool of Europe, as typified by early Anatolian farmers (4, 5).
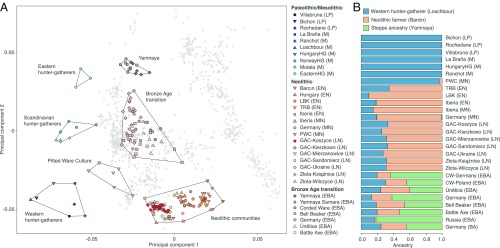
Fig. 2.
Genetic affinities of the Koszyce individuals and other GAC groups (here including Złota) analyzed in this study. (A) Principal component analysis of previously published and newly sequenced ancient individuals. Ancient genomes were projected onto modern reference populations, shown in gray. (B) Ancestry proportions based on supervised ADMIXTURE analysis (K = 3), specifying Western hunter-gatherers, Anatolian Neolithic farmers, and early Bronze Age steppe populations as ancestral source populations. LP, Late Paleolithic; M, Mesolithic; EN, Early Neolithic; MN, Middle Neolithic; LN, Late Neolithic; EBA, Early Bronze Age; PWC, Pitted Ware culture; TRB, Trichterbecherkultur/Funnelbeaker culture; LBK, Linearbandkeramik/Linear Pottery culture; GAC, Globular Amphora culture; Złota, Złota culture.
To further investigate the ancestry of the Globular Amphora individuals, we performed a supervised ADMIXTURE (6) analysis, specifying typical western European hunter-gatherers (Loschbour), early Neolithic Anatolian farmers (Barcın), and early Bronze Age steppe populations (Yamnaya) as ancestral source populations (Fig. 2B). The results indicate that the Globular Amphora/Złota group individuals harbor ca. 30% western hunter-gatherer and 70% Neolithic farmer ancestry, but lack steppe ancestry. To formally test different admixture models and estimate mixture proportions, we then used qpAdm (7) and find that the Polish Globular Amphora/Złota group individuals can be modeled as a mix of western European hunter-gatherer (17%) and Anatolian Neolithic farmer (83%) ancestry (SI Appendix, Table S2), mirroring the results of previous studies (4, 5).
Kinship and Consanguinity.
Analyses of ancient genomes can provide detailed information on the kinship structures and social organization of past communities (8–10). At Koszyce, mitochondrial DNA (mtDNA) analysis revealed the presence of six different maternal lineages, whereas analysis of the nonrecombining region of the Y chromosome showed that all males carried the same Y chromosome haplotype: I2a-L801 (Table 1). We then estimated genomic runs of homozygosity (ROH) and found that the Koszyce individuals were not particularly inbred. Although a slightly larger section of the Koszyce genomes is contained within ROH compared with typical modern European populations (SI Appendix, Fig. S7), this signal is mainly driven by an increased fraction of short ROH (<2 Mb), which is indicative of ancestral restrictions in population size rather than recent inbreeding. On the basis of genome-wide patterns of allelic identity-by-state (IBS), we computed kinship coefficients between all pairs of individuals and applied established cutoff values for possible kinship categories (Materials and Methods). We find that the Koszyce burial represents a large extended family connected via several first- and second-degree relationships (Fig. 3 and SI Appendix, Fig. S9).
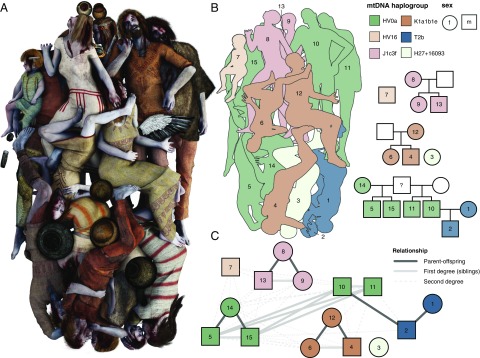
Fig. 3.
Kinship. (A) Artistic reconstruction of the Koszyce mass burial based partly on phenotypic traits inferred from the ancient genomes (reconstruction by Michał Podsiadło); (B) Schematic representation of the burial and pedigree plots showing kinship relations between the Koszyce individuals inferred from genetic data. (C) kinship network based on kinship coefficients inferred from IBS scores for pairs of Koszyce individuals showing first- and second-degree relationships. Kinship coefficients and R scores are reported in Dataset S7 and plotted in SI Appendix, Fig. S9.
Overall, we identified four nuclear families in the grave, which are for the most part represented by mothers and their children (Fig. 3). Closely related kin were buried next to each other: a mother was buried cradling her child, and siblings were placed side by side. Evidently, these individuals were buried by people who knew them well and who carefully placed them in the grave according to familial relationships. For example, individual 14, the oldest individual in the grave, was buried close to her two sons (individuals 5 and 15), whereas individual 8, a 30–35-y-old woman, was buried with her teenage daughter (individual 9) and 5-y-old son (individual 13). Using genome-wide patterns of IBS, we were also able to reconstruct more complex relationships: individuals 5, 10, 11, and 15 all appear to be brothers, and yet they do not have the same mother (individual 14 is the mother of individuals 5 and 15, but not 10 and 11), suggesting that they might be half-brothers. However, all four of them share the same mitochondrial DNA haplotype, suggesting that their mothers might also have been related.
Interestingly, the older males/fathers are mostly missing from the grave, suggesting that it might have been them who buried their kin. The only father present in the grave is individual 10, whose partner and son are placed together opposite him in the grave. In addition, there is a young boy (individual 7), aged 2–2.5 y, whose parents are not in the grave, but he is placed next to other individuals to whom he is closely related through various second-degree relationships. Finally, there is individual 3, an adult female, who does not seem to be genetically related to anyone in the group. However, her position in the grave close to individual 4, a young man, suggests that she may have been as close to him in life as she was in death. These biological data and burial arrangements show that the social relationships held to be most significant in these societies were identical with genetic and reproductive relationships. However, they also demonstrate that nuclear families were nested in larger, extended family groups, either permanently or for parts of the year.
Social Organization, Residence Patterns, and Subsistence Strategies.
The presence of unrelated females and related males in the grave is interesting because it suggests that the community at Koszyce was organized along patrilineal lines of descent, adding to the mounting evidence that this was the dominant form of social organization among Late Neolithic communities in Central Europe (11, 12). Usually, patrilineal forms of social organization go hand in hand with female exogamy (i.e., the practice of women marrying outside their social group). Indeed, several studies (11, 12) have shown that patrilocal residence patterns and female exogamy prevailed in several parts of Central Europe during the Late Neolithic. At Koszyce, there is no clear difference in enamel 87Sr/86Sr ratios between males and females (SI Appendix, Fig. S6) that would suggest that the females are nonlocal. However, the high diversity of mtDNA lineages, combined with the presence of only a single Y chromosome lineage, is certainly consistent with a patrilocal residence system.
Social organization is most often aligned with settlement and subsistence patterns, and several studies (13–15) suggest that Globular Amphora communities and other related groups specialized in animal husbandry, often with a main focus on cattle, and that they moved around the landscape to seek new pastures for their animals at different times of the year (see SI Appendix, section 1 for a more detailed discussion). This form of mobility is likely to have included fission-fusion dynamics in which a larger social unit, similar to the extended family, would split up into smaller groups, perhaps nuclear families, for certain purposes and parts of the year (16). This dynamic could explain the relatively high variation we observe in the 87Sr/86Sr isotope signatures at Koszyce. Similar to strongly patrilineal modes of social organization, such pastoral economic strategies have often been linked to Corded Ware groups that introduced steppe genetic ancestry into Europe (7, 17), and the two (social organization and economic strategy) are probably linked: Pastoral ways of life involve a high level of mobility within vaguely defined territories and with the groups’ main economic capital, their animal herds, exposed across the landscape, and thus harbor a significant potential for conflict with neighboring groups. One ethnographically known cultural response to this situation is to adopt an aggressive strategy toward competing groups in which male dominance, including patrilineal kin alliance, and warrior-like values prevail (18). Although we cannot be certain that the people at Koszyce shared these values, we show that they were organized around patrilineal descent groups, demonstrating that this form of social organization was already present in communities before the expansion of the Corded Ware complex in Central and Eastern Europe (13, 14).
Intergroup Conflict and Violence.
All individuals buried in the mass grave at Koszyce exhibit extensive evidence of perimortem injuries (SI Appendix, section 3). The most common injuries are cranial fractures (SI Appendix, Fig. S2), which indicate that the individuals were killed by blows to the head. Overall, the nature of the injuries and the near absence of so-called parry fractures (i.e., injuries sustained to the upper limbs) suggest that the individuals were captured and executed, rather than killed in hand-to-hand combat. The evidence for violence at Koszyce fits within a wider pattern of extensive, frequent violence during specific stages in European prehistory (19). Evidence from the Neolithic indicates that lethal violence and massacres prevailed during periods of population pressure, competition over resources, and/or the expansion of new groups into already-occupied territories (20, 21), a pattern also observed in well-known cases from the New World (22).
Neolithic cases of intergroup violence appear to fall into one of two categories, either targeting whole communities (11, 20) or aimed specifically at males from competing groups (21). The former strategy presumably aims at eradicating a competing group from a given territory, whereas in the latter case, it may be assumed that the women and/or children are taken as captives, a practice that is well documented ethnographically and historically for prestate societies (23). Although alternative scenarios (e.g., ritualistic violence or familicide) cannot be ruled out, it seems most plausible that the massacre at Koszyce falls in the former category. The fact that most adult males of the group are missing from the grave most likely reflects that they were away (or fled) when the raid occurred, leaving the remaining group vulnerable.
Although it is impossible to identify the culprits of the massacre that took place at Koszyce around 2880–2776 BCE, it is interesting to note that it occurred right around the time when the Corded Ware complex started to spread rapidly across large parts of Central Europe, and it seems plausible that the group from Koszyce fell victim to some violent intergroup conflict related to the territorial expansion of Corded Ware groups or another competing group in the area. If the general interaction between Globular Amphora people and neighboring, steppe-related cultures (including early Corded Ware) was primarily hostile, it would explain why Globular Amphora individuals carry no steppe ancestry and, in part, why Europe experienced such a dramatic reduction in Neolithic genomic ancestry at this time (7, 17).
Concluding Remarks
Brutal events such as the family massacre documented in the Koszyce burial may have been all too common in the unstable, tumultuous centuries at the beginning of the third millennium BCE. However, along with all the violence and aggression illustrated by the Koszyce find, our study also demonstrates the strong sense of family affiliation and cohesion that prevailed among this group of people. From the careful positioning of the bodies in the grave, it is clear that both nuclear and extended family relations were key to how people organized their lives, and that these relations represented major, normative values in Globular Amphora communities of this period. Although it has often been suggested that nuclear and/or extended family structures were important in many prehistoric societies, the archaeological and genomic data we have presented here provide actual proof that this was indeed the case. Furthermore, as we start to better understand the mating patterns and social structures of past populations, we can start to move beyond our often all too simplistic assumptions used to model population history and culture change (24). Ultimately, it will be through the integration of archaeological, anthropological, and biological data and theory that we will be able to produce more accurate representations of past social realities and population histories.
Materials and Methods
Samples.
The samples analyzed in this study (21 petrous bones and three teeth) stem from five different archaeological sites in southern Poland, all associated with the Globular Amphora culture or its Złota group variant (SI Appendix, section 1). For detailed site descriptions, see SI Appendix, section 2.
DNA Extraction, Library Preparation, and Sequencing.
DNA was extracted from petrous bones and tooth roots, using established protocols specifically designed for ancient DNA (SI Appendix, section 6). Double-stranded DNA libraries were built, using Illumina-specific adapters, and amplified and indexed using a dual-indexing protocol (25). The optimal number of PCR cycles was determined using qPCR. The indexed and amplified libraries were purified and quantified on an Agilent 2200 TapeStation before being pooled in equimolar amounts. The pooled libraries were sequenced (80 bp, single read) on Illumina HiSeq 2500 platforms at the Danish National High-throughput DNA Sequencing Centre.
Raw Read Processing and Alignment.
Base-calling was performed using CASAVA 1.8.2. Adapter sequences and leading/trailing stretches of Ns were trimmed using AdapterRemoval 2.1.3 (26). Reads of at least 30 bp were mapped to the human reference genome GRCh37 using bwa 0.7.10 (27) with the seed disabled. Duplicates and reads with MAPQ <30 were removed using samtools 1.3.1 (28). All reads were then merged to sample level and realigned using GATK-3.3.0 (29). The realigned reads had the md-tag updated and extended base alignment qualities calculated using samtools calmd. Read depth and coverage were determined using BEDtools 2.27.0 genomecov (30). For full details, see SI Appendix, section 6.
Genotype Likelihoods and Imputation.
Genotype likelihoods were determined using GATK 3.7.0 (31). We called biallelic sites present in the 1000 Genomes (–genotyping_mode GENOTYPE_GIVEN_ALLELES) filtering for transitions by setting the genotype likelihoods to 0 for all three genotypes (e.g., hom ref, het, and hom alt). Subsequently, the individual VCF (variant call format) files were merged using BCFtools 1.3.1. The combined VCF was then split into separate files containing 200,000 markers each and imputed separately using Beagle 4.0 (r1399) (32), using the 1000 Genomes (33) phase 3 map included with Beagle (*.phase3.v5a.snps.vcf.gz and plink.chr*.GRCh37.map) with input through the genotype likelihood option.
MapDamage Analysis.
We used mapDamage 2.0 (34) to assess the extent of DNA damage in our samples and found that all of them showed the features characteristic for ancient DNA, including short average fragment lengths, an increased occurrence of purines before strand breaks, and an increased frequency of apparent cytosine (C) to thymine (T) substitutions at 5′-ends (Dataset S3), which likely results from the deamination of cytosine residues that occur primarily in the single-stranded overhangs of DNA fragments.
Mitochondrial DNA.
We used Schmutzi (35) to determine the endogenous consensus mtDNA sequences. Reads were mapped to the Cambridge reference sequence and filtered for MAPQ ≥ 30. Haploid variants were called using the endoCaller program implemented in Schmutzi (35), and only variants with a posterior probability exceeding 50 on the PHRED scale (probability of error: 1/100,000) were retained. We then used Haplogrep 2.2 (36) to determine the mtDNA haplogroups, specifying PhyloTree (build 17) as the reference phylogeny (37).
Y Chromosome DNA.
To determine the Y chromosome haplogroups, we first called haploid genotypes for reads mapping (MAPQ ≥ 30) to the Y chromosome, using samtools/bcftools (27, 38). We then extracted genotypes for all individuals at SNPs included in the Y-DNA haplogroup tree version 13.57 from the International Society of Genetic Genealogy (https://isogg.org/). The full list of observed alleles defining subhaplogroups of I2a is listed in Dataset S4 and plotted in SI Appendix, Fig. S8.
Principal Component Analysis.
Principal component analysis was performed using smartPCA (39), using around 1,000 modern Eurasian individuals from the HO dataset (3) and 168 previously published ancient genomes (Dataset S6). The ancient genomes were projected onto the modern variation, using the option lsq project. Individuals with too much missing data (mind 0.1) were excluded from the analysis.
Model-Based Clustering.
We ran ADMIXTURE (6) in supervised mode on the same merged dataset, specifying Western hunter-gatherers (Loschbour), Neolithic farmers (Barcın), and Yamnaya as ancestral source populations. The resulting admixture proportions are plotted in Fig. 2B. The plot shows population averages, and to help visualize the results, only a selected number of ancient populations are shown.
qpAdm Analysis.
Admixture proportions were modeled using qpAdm (7), specifying Mesolithic Western European hunter-gatherers, Anatolian Neolithic farmers (Barcın), and either Yamnaya or Eastern hunter-gatherers as possible source populations. In most cases, a two-way model without steppe ancestry provides the best fit (SI Appendix, Table S2). The only exception is Książnice, for which a small amount of additional gene-flow from some steppe-related population provides a better fit than a simple two-way model. To determine the source of this additional gene-flow, we modeled the Książnice genomes as a mix of Western European hunter-gatherers, Neolithic farmers, and either Yamnaya or Eastern hunter-gatherer ancestry, and found that both provide an equally good fit, suggesting that this additional gene-flow is not necessarily Yamnaya related.
Phenotype.
To reconstruct the physical appearance of the Koszyce individuals, we first imputed missing genotypes for our low-coverage genomes and then selected 39 SNPs (40) to determine phenotypic traits, including skin, hair, and eye color. The allele counts were uploaded to the HIrisPlex-S website to obtain the phenotype predictions (Dataset S5).
Runs of Homozygosity.
We identified runs of homozygosity using IBDseq (41) on a merge of the imputed genotypes of the Koszyce individuals and 214 individuals from two populations (IBS, TSI) of the 1000 Genomes Project (33). Genotypes for the Koszyce individuals with imputed genotype probability <0.99 were set to missing for this analysis. We ran IBDseq on the merged and filtered dataset with default parameters, except allowing for a higher genotype error rate (errormax = 0.005). Inferred tracts were filtered for LOD score ≥2, and then further processed by splitting tracts that spanned centromeres or those that spanned large assembly gaps with less than 1 Mb on both sides of the gaps. For each individual, we inferred the total length contained in short (<2 Mb) and long (>8 Mb) ROH by summing up individual tract lengths within those length categories, as done in ref. 42 (SI Appendix, Fig. S7).
Kinship Analysis.
Kinship analysis was performed using a recently described method based on pairwise sharing of alleles IBS (10, 43). Because of the low sequencing coverage, we first computed a matrix of expected pairwise IBS sharing for all pairs of individuals as the 2D site frequency spectrum, using the program realSFS from the ANGSD package (44). For each pair, the 2D-site frequency spectrum with the best likelihood across 10 replicate runs was retained and was used to calculate a number of relatedness estimators: the kinship estimator introduced by Manichaikul et al. (45), implemented in the KING package [equation (9) in ref. 45], and the “R0” and “R1” ratios described in Waples et al. (43)
We used the kinship coefficient ranges described in Manichaikul et al. (45) to classify pairs of individuals into first-, second-, and third-degree relatives. First-degree relatives were further distinguished using the R0 ratio, into parent–offspring (R0 ≤ 10–2) or sibling relationships (R0 > 10–2; SI Appendix, Fig. S9 and Dataset S7). The resulting relatedness networks of first- and second-degree relatives were visualized using Cytoscape version 3.4.0 (46), including annotations of individual sex and mitochondrial haplogroups.
Supplementary Material
Acknowledgments
We thank Jesper Stenderup and the staff at the Danish National High-throughput Sequencing Centre for technical assistance. We also thank Agata Ulanowska, Eva Andersson Strand, Małgorzata Siennicka, and Ulla Mannering for helpful discussion. The project was funded by a Villum Foundation Young Investigator grant (Grant 10120 to M.E.A.). The Centre for GeoGenetics is funded by the National Research Foundation of Denmark and the Lundbeck Foundation. H.S. was supported by HERA (Humanities in the European Research Area) and the European Union’s Horizon 2020 Research and Innovation Programme (Grant 649307). S.R. was supported by the Novo Nordisk Foundation (Grant NNF14CC0001). T.Z.T.J. is supported by the European Union’s EU Framework Programme for Research and Innovation Horizon 2020 under Grant Agreement 676154 (ArchSci2020). Work on the site’s radiocarbon chronology was funded by a grant from the Aarhus University Research Foundation (N.N.J.). The strontium isotope measurements were funded by the Carlsberg Foundation “Tales of Bronze Age Women” project (Grant CF15-0878 to K.M.F.). Publication fees were covered by the Materials, Culture and Heritage research program at Aarhus University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The aligned reads reported in this paper have been deposited in the European Nucleotide Archive and are available for download under accession no. PRJEB28451.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820210116/-/DCSupplemental.
References
- 1.Przybyła MM, Szczepanek A, Włodarczak P, editors. Koszyce stanowisko 3. Przemoc i rytuał u schyłku neolitu. Profil-Archeo; Kraków-Pękowice, Poland: 2013. [Google Scholar]
- 2.Konopka T, Szczepanek A, Przybyła MM, Włodarczak P. Evidence of interpersonal violence or a special funeral rite in the Neolithic multiple burial from Koszyce in southern Poland–a forensic analysis. Anthropol Rev. 2016;79:69–85. [Google Scholar]
- 3.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tassi F, et al. Genome diversity in the Neolithic Globular Amphorae culture and the spread of Indo-European languages. Proc Biol Sci. 2017;284:20171540. doi: 10.1098/rspb.2017.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathieson I, et al. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennett DJ, et al. Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat Commun. 2017;8:14115. doi: 10.1038/ncomms14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Sullivan N, et al. Ancient genome-wide analyses infer kinship structure in an Early Medieval Alemannic graveyard. Sci Adv. 2018;4:eaao1262. doi: 10.1126/sciadv.aao1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikora M, et al. Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science. 2017;358:659–662. doi: 10.1126/science.aao1807. [DOI] [PubMed] [Google Scholar]
- 11.Haak W, et al. Ancient DNA, strontium isotopes, and osteological analyses shed light on social and kinship organization of the later stone age. Proc Natl Acad Sci USA. 2008;105:18226–18231. doi: 10.1073/pnas.0807592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knipper C, et al. Female exogamy and gene pool diversification at the transition from the final Neolithic to the early Bronze Age in central Europe. Proc Natl Acad Sci USA. 2017;114:10083–10088. doi: 10.1073/pnas.1706355114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szmyt M. Between West and East: People of the Globular Amphora Culture in Eastern Europe; 2950-2350 BC. Institute of Prehistory, Adam Mickiewicz University; Poznan, Poland: 1999. [Google Scholar]
- 14.Woidich M. The Western Globular Amphora culture. A new model for its emergence and expansion. eTopoi. 2014;3:67–85. [Google Scholar]
- 15.Johannsen N, Laursen S. Routes and wheeled transport in late 4th–early 3rd millennium funerary customs of the Jutland Peninsula: Regional evidence and European context. Praehist Z. 2010;85:15–58. [Google Scholar]
- 16.Czebreszuk J, Szmyt M. 2011. Identities, differentiation and interactions on the Central European Plain in the 3rd millennium BC. Sozialarchäologische Perspektiven: Gesellschaftlicher Wandel 5000-1500 v. Chr. Zwischen Atlantik Und Kaukasus, Archäologie in Eurasien, eds Hansen S, Müller J (Philipp von Zabern, Mainz, Germany), pp 247–269.
- 17.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 18.Kelly RC. The Nuer Conquest: The Structure and Development of an Expansionist System. University of Michigan Press; Ann Arbor, MI: 1985. [Google Scholar]
- 19.Schulting RJ, Fibiger L. Sticks, Stones, and Broken Bones: Neolithic Violence in a European Perspective. Oxford Univ Press; Oxford: 2012. [Google Scholar]
- 20.Chenal F, Perrin B, Barrand-Emam H, Boulestin B. A farewell to arms: A deposit of human limbs and bodies at Bergheim, France, c. 4000 BC. Antiquity. 2015;89:1313–1330. [Google Scholar]
- 21.Meyer C, et al. Early Neolithic executions indicated by clustered cranial trauma in the mass grave of Halberstadt. Nat Commun. 2018;9:2472. doi: 10.1038/s41467-018-04773-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milner GR, Anderson E, Smith VG. Warfare in late prehistoric west-central Illinois. Am Antiq. 1991;56:581–603. [Google Scholar]
- 23.Cameron CM. Captives: How Stolen People Changed the World. University of Nebraska Press; Lincoln, NE: 2016. [Google Scholar]
- 24.Johannsen NN, Larson G, Meltzer DJ, Vander Linden M. A composite window into human history. Science. 2017;356:1118–1120. doi: 10.1126/science.aan0737. [DOI] [PubMed] [Google Scholar]
- 25.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browning BL, Browning SR. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renaud G, Slon V, Duggan AT, Kelso J. Schmutzi: Estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 2015;16:224. doi: 10.1186/s13059-015-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissensteiner H, et al. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58-63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Oven M. PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci Int Genet Suppl Ser. 2015;5:e392–e394. [Google Scholar]
- 38.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaitanya L, et al. The HIrisPlex-S system for eye, hair and skin colour prediction from DNA: Introduction and forensic developmental validation. Forensic Sci Int Genet. 2018;35:123–135. doi: 10.1016/j.fsigen.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Browning BL, Browning SR. Detecting identity by descent and estimating genotype error rates in sequence data. Am J Hum Genet. 2013;93:840–851. doi: 10.1016/j.ajhg.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder H, et al. Origins and genetic legacies of the Caribbean Taino. Proc Natl Acad Sci USA. 2018;115:2341–2346. doi: 10.1073/pnas.1716839115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waples RK, Albrechtsen A, Moltke I. Allele frequency-free inference of close familial relationships from genotypes or low-depth sequencing data. Mol Ecol. 2019;28:35–48. doi: 10.1111/mec.14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunningham C, Scheuer L, Black S. Developmental Juvenile Osteology. Elsevier Academic Press; Burlington, MA: 2016. [Google Scholar]
- 48.Ubelaker DH. Human Skeletal Remains: Excavation, Analysis, Interpretation. Taraxacum; Washington, DC: 1989. [Google Scholar]
- 49.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Arch Sci. 2013;40:4477–4482. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.