Significance
Self-cleaving ribozymes are RNAs that catalyze position-specific cleavage of their phosphodiester backbone. The cleavage site of the newly discovered hatchet ribozyme is located at the very 5′ end of its consensus secondary structure motif. Here we report on the 2.1-Å crystal structure of the hatchet ribozyme in the product state, which defines its intricate tertiary fold and identifies key residues lining the catalytic pocket. This in turn has allowed us to propose a model of the precatalytic state structure and a role in catalysis for a conserved guanine. This study therefore provides a structure-based platform toward an improved understanding of the catalytic mechanism of hatchet ribozymes.
Keywords: hatchet, ribozyme, cleavage, catalysis, noncoding RNA
Abstract
Small self-cleaving ribozymes catalyze site-specific cleavage of their own phosphodiester backbone with implications for viral genome replication, pre-mRNA processing, and alternative splicing. We report on the 2.1-Å crystal structure of the hatchet ribozyme product, which adopts a compact pseudosymmetric dimeric scaffold, with each monomer stabilized by long-range interactions involving highly conserved nucleotides brought into close proximity of the scissile phosphate. Strikingly, the catalytic pocket contains a cavity capable of accommodating both the modeled scissile phosphate and its flanking 5′ nucleoside. The resulting modeled precatalytic conformation incorporates a splayed-apart alignment at the scissile phosphate, thereby providing structure-based insights into the in-line cleavage mechanism. We identify a guanine lining the catalytic pocket positioned to contribute to cleavage chemistry. The functional relevance of structure-based insights into hatchet ribozyme catalysis is strongly supported by cleavage assays monitoring the impact of selected nucleobase and atom-specific mutations on ribozyme activity.
Catalytic noncoding RNAs, termed ribozymes, are involved in many vital cellular reactions ranging from tRNA processing to intron splicing, protein synthesis, and regulation of gene expression (1). Nucleolytic ribozymes are small RNAs that adopt compact folds capable of site-specific cleavage/ligation reactions (2–4). Nine unique nucleolytic ribozymes have been identified to date, including recently discovered twister, pistol, twister-sister, and hatchet ribozymes that were identified based on application of comparative sequence and structural algorithms (5, 6). The structure/function characterization of such ribozymes would provide mechanistic insights into ribozyme activity and its modulation.
Nucleolytic ribozymes adopt an SN2-like mechanism that results in site-specific phosphodiester bond cleavage. In general, an activated 2′-OH of the ribose 5′ to the scissile phosphate adopts an in-line alignment to target the adjacent to-be-cleaved P-O5′ phosphodiester bond, resulting in formation of 2′,3′-cyclic phosphate and 5′-OH groups. To date, X-ray crystallographic structural studies on the hammerhead (7, 8), hairpin (9, 10), GlmS (11, 12), hepatitis delta virus (HDV) (13, 14), Varkud satellite (15), and pistol ribozymes (16, 17) have defined the overall RNA fold, the catalytic pocket arrangement, the in-line alignment, and the key residues that contribute to the cleavage reaction. By contrast, there is less clarity to date on the cleavage mechanism of twister (18–20) and twister-sister (21, 22) ribozymes, given distinct catalytic conformations reported from structural studies for these ribozymes, and a resolution must await additional structural studies of transition state vanadate mimics of these ribozymes.
We now report on structural studies of the hatchet ribozyme product supplemented by structure-based cleavage assays monitoring the impact of selected nucleobase and atom-specific mutations on ribozyme activity. These structure/function studies identify the tertiary fold of the ribozyme, the alignment of conserved residues lining the catalytic pocket, and the impact of site-specific mutations on ribozyme activity, with implications for modeling the precatalytic fold and insights into cleavage chemistry.
Results
The hatchet ribozyme is composed of four base-paired stems labeled P1–P4, in which P1 and P2 are linked by three highly conserved residues, while P2, P3, and P4 are bridged by two internal loops L2 and L3 (Fig. 1A). Most of the highly conserved residues (shown in red rectangles in Fig. 1A) are dispersed and positioned within loop L2. The cleavage site is located at the 5′ end of stem P1, a unique feature of the hatchet ribozyme, that contrasts it from the internal cleavage sites observed for the pistol, twister, and twister-sister ribozymes (5). The secondary structure of the hatchet ribozyme has been validated from covariation and mutation studies (23), but insights into the catalytic mechanism require information on both the tertiary fold and alignment of catalytic residues mediating in-line cleavage chemistry.
Fig. 1.
Schematic and tertiary structure of the hatchet ribozyme. (A) The predicted secondary structure of the env10 hatchet ribozyme. The sequence is color coded according to helical segments observed in the tertiary structure. The highly conserved residues are shown in red rectangles. (B) A schematic representation of hatchet ribozyme product secondary structure highlighting long-range interactions. (C) The tertiary fold of the HT-GAAA hatchet ribozyme product dimer. The red thick dashed line divided the dimer structure as two new monomer molecules termed A′ and B′. (D) A schematic representation of the tertiary structure of HT-GAAA hatchet ribozyme product dimer. Two hatchet ribozyme product molecules form a dimer through swapping of the 3′ ends of the pairing strand. Long-range interactions observed in the tertiary structure are labeled with solid lines. The cleavage site is indicated by a yellow star. To simplify the structural analysis, we swapped the 3′ end of the pairing strand of the two molecules in the dimer and termed them as the new monomer molecules A′ and B′ as shown in the dashed rectangles. (E) The tertiary fold of molecule A′ of the HT-GAAA ribozyme product structure. The color coding is similar to that in Fig. 1A. The cleavage site is labeled with a yellow star. (F) The residues G8, A9, and G10 from stem-loop L1 are stacked on each other on the top of stem P1, while U39 extruded from loop L3 forms extensive interaction with L1 and stabilizes the long-range interaction. (G) U7 and A11 formed a trans-Watson–Crick Hoogsteen pair in L1. The sugar pucker of A11 adopts a C2′-endo conformation, whereas U7 adopts a C3′-endo conformation. (H) Two consecutive canonical base pairs A36-U58 and U37-A57 form on zippering-up L3; A12 and U13 that are extruded from L1 interact with the minor groove edge of A36-U58 and U37-A57, thereby forming two stacked base triples involving long-range interactions. (I) The compounds 1-NH and 2-NH2 of G8 form hydrogen bonds with the phosphate oxygen of A11 in L1; 2′-OH of G8 hydrogen bonds with the Hoogsteen side of G10; the extruded residue U39 from L3 intercalates between G10 and A11, and its Watson–Crick edge pairs with the minor groove edge of G8; the Watson–Crick edge of G10 and the Watson–Crick edge of A11 are also involved in the stacking interaction of the long helix H12. (J) U59 formed a stable base triple with the Watson–Crick A3-U17 pair aligned along the major groove edge of stem P1, with the sugar pucker of U59 adopting a 2′-endo conformation. Note that the dashed lines indicate distances <3.5 Å and their number can exceed the possible number of hydrogen bonds formed by an atom.
Crystallization of the Hatchet Ribozyme Product.
For crystallization trials, we screened a large number of chemically synthesized (one- and two-stranded constructs) and in vitro transcribed constructs of the hatchet ribozyme. For the in vitro transcribed constructs, cleavage during transcription of the full-length hatchet ribozyme resulted in generation of product shown in Fig. 1B. Single-stranded in vitro transcripts of the env10 hatchet ribozyme with either a P4 stem closing GAAA or UUCG tetraloop (to facilitate crystal packing) yielded diffraction quality crystals of the cleaved hatchet ribozyme product. The sequence and secondary structure model are shown in Fig. 1A. In the following text, we named the cleaved product with GAAA tetraloop as HT-GAAA hatchet ribozyme and the product with UUCG tetraloop as HT-UUCG hatchet ribozyme. The phases for crystal structure determination were solved using the single-wavelength anomalous diffraction (SAD) method, based on crystals that were soaked with Ir(NH3)63+ (for the HT-UUCG structure). Molecular replacement (MR) was then applied to solve the HT-GAAA structure using the HT-UUCG structure as a model (X-ray statistics listed in SI Appendix, Table S1).
The structure of HT-GAAA hatchet ribozyme was refined at 2.1-Å resolution with Rwork/Rfree values of 0.19/0.23, while the structure of HT-UUCG hatchet ribozyme was refined at 2.6-Å resolution with Rwork/Rfree values of 0.20/0.23 (SI Appendix, Table S1). Both structures were stabilized by common long-range tertiary contacts as shown schematically for HT-GAAA in Fig. 1B. Unexpectedly, both ribozyme constructs formed dimers in the crystal lattice as shown for HT-GAAA in Fig. 1 C and D and for HT-UUCG in SI Appendix, Fig. S1 A and B (a direct comparison of HT-GAAA and HT-UUCG dimers is shown in SI Appendix, Fig. S2 A and B). When the solution state of the hatchet ribozyme product was tested by size-exclusion chromatography, we found that both hatchet ribozyme constructs (HT-GAAA and HT-UUCG) existed as an equilibrium of dimers and monomers in solution (SI Appendix, Fig. S3).
Tertiary Fold of the Hatchet Ribozyme Product.
We focus in the following sections on the higher resolution structure of the HT-GAAA hatchet ribozyme. Two molecules of the HT-GAAA hatchet ribozyme product formed a pseudosymmetric dimer in the asymmetric unit (space group: P212121), with both monomers exhibiting well-defined electron density. A schematic of the tertiary fold of the dimeric hatchet ribozyme is shown in Fig. 1D, while its 3D structure is shown in a ribbon representation in Fig. 1C.
The fold of each molecule of the HT-GAAA hatchet ribozyme is comprised of four stems P1, P2, P3, and P4, in which stem P1 coaxially stacks on P2 and forms the long H12 helix (SI Appendix, Fig. S4A). Further, H12 was aligned in parallel to another long helix formed by stem P3, parts of the internal loops (L2 and L3), and stem P4, termed long H34 helix (SI Appendix, Fig. S4B). Notably, the stem loop L1 of P1 formed long-distance interaction with L3 between stems P3 and P4. The conserved three-nucleotide linkage between stems P1 and P2 formed long-distance pairing interactions with L2 between stems P2 and P3 and the stem of P2 (Fig. 1 B–D). The cleavage site labeled with a yellow star in Fig. 1 C and D is positioned in the junctional region of stems P1 and P2, adjacent to stem P3 and loop L2.
The palindromic nucleotides from A67 to U70 (ACGU of loop L2) in molecule A formed a symmetric helix with the corresponding nucleotides from U70′ to A67′ in molecule B (Fig. 1 C and D and SI Appendix, Fig. S4C), which likely triggered dimer formation. To simplify the structural analysis, we swapped the 3′-end tertiary structure of molecules A and B at the pseudosymmetric site between C68 and G69 (Fig. 1 C and D) and we refer to the “monomers” Mol A′ and B′ as shown in Fig. 1 C and D.
To experimentally support this simplification, we designed cleavage assays for hatchet ribozyme variants that lacked the palindromic sequence portion by mutation of A67-C68-G69-U70 (SI Appendix, Fig. S5A) to U67-U68-U69-U70 (SI Appendix, Fig. S5B). For a second control experiment, we placed a UUCG tetraloop between C68 and G69 with the intention to support intramolecular base pairing of A67-U70 and C68-G69 via UUCG hairpin formation (SI Appendix, Fig. S5C). Size-exclusion chromatography indeed confirmed that the first mutant (A67U/C68U/G69U) exclusively exists as a monomer and that the second mutant (UUCG insertion) predominantly exists as a monomer (>85%) in solution (SI Appendix, Fig. S5 E and F). Importantly, we observed efficient cleavage of these mutated constructs (SI Appendix, Fig. S5 B and C) with activities comparable to the wild-type ribozyme (SI Appendix, Fig. S5A). Moreover, a hatchet ribozyme assembled from the “swapping” 3′-terminal RNA fragment (nucleosides 69–82) added in trans to the “complementary” fragment (nucleotides 1–68) was also highly active (SI Appendix, Fig. S5D). Taken together, these experiments suggest that the hatchet ribozyme does not necessarily need to act as dimer as implied by the crystal structure but can adopt the cleavage-active conformation as monomer in solution (corresponding to folds A′ and B′).
Long-Range Tertiary Interactions Involving L1 and L3.
For practical purposes, the structural analysis below focuses on monomeric Mol A′ of the HT-GAAA ribozyme as shown in Fig. 1E. Long-range pairing was observed between loops L1 and L3 on formation of the tertiary fold of the hatchet ribozyme (Fig. 1 B–D). The U7 to A11 segment of L1 together with inserted U39 formed a hairpin loop (Fig. 1F) closed by a trans-Watson–Crick Hoogsteen U7•A11(C2′-endo) pair (Fig. 1G) that stacked on the terminal Watson–Crick G6-C14 pair of stem P1. L3 zippered up by forming three consecutive Watson–Crick A36-U58, U37-A57, and A38-U56 pairs, with opposing bases U35 and U59 flipped out, thereby connecting stem P3 and P4 to form the long H34 helix (Fig. 1 B–D and SI Appendix, Fig. S4B).
A12 extruded from L1 and formed an A-minor base triple with the Watson–Crick A36-U58 pair in L3 (Fig. 1H), while extruded U13 formed a base triple with the minor groove edge of the Watson–Crick U37-A57 pair (Fig. 1H). Thus, interactions between L1 and L3 were stabilized through formation and mutual stacking of A12•(A36-U58) and U13•(U37-A57) minor groove base triples (Fig. 1H). Additional long-range interactions include a network of base-base, base-sugar, and base-phosphate hydrogen bonds between G8 and G10 on L1 with the A38-U39 step on L3, characterized by U39 pairing with G8 through formation of a Watson–Crick-minor groove U39•G8 pair and stacking with G10 (Fig. 1I), as well as between extruded U59(C2′-endo pucker) on L3 and the major groove edge of the Watson–Crick A3-U17 pair on stem P1 (Fig. 1J).
Pairing Alignment of Conserved Residues.
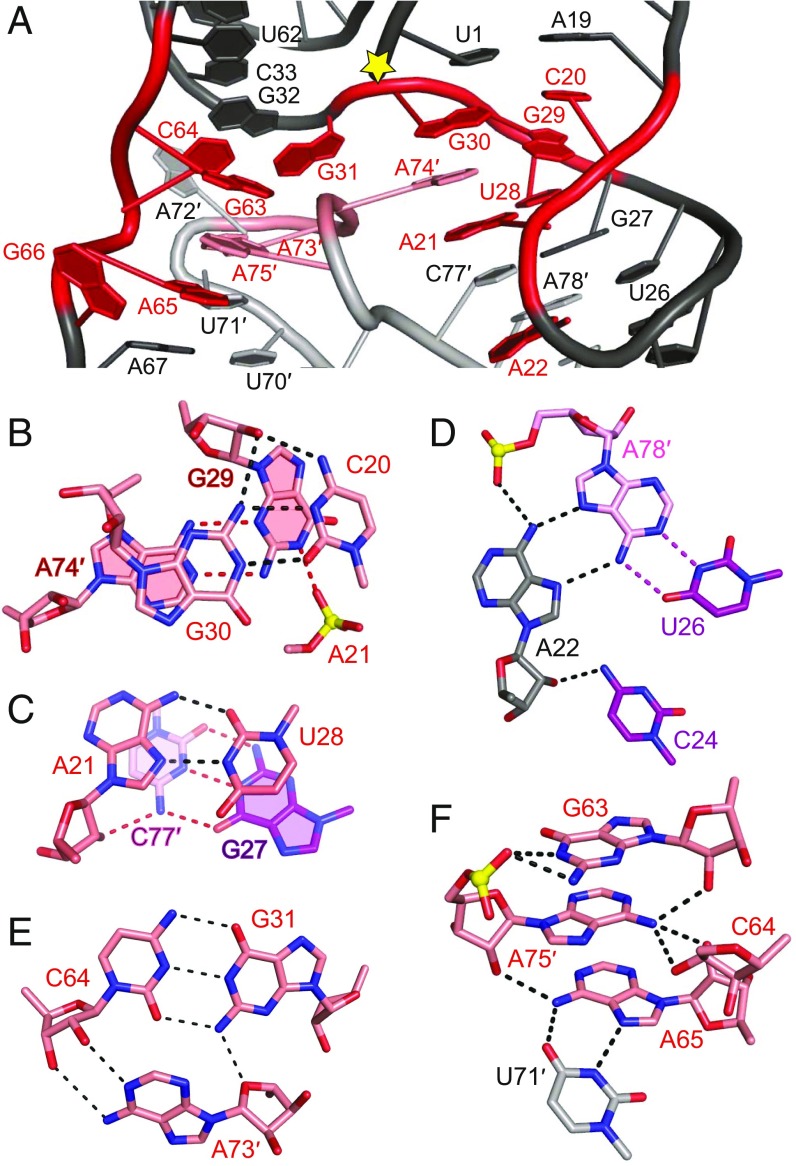
The conserved residues of the HT-GAAA hatchet ribozyme are highlighted in red in SI Appendix, Fig. S6A and also labeled in an expanded version in Fig. 2A. The highly conserved but sequence dispersed C20-A22, U28-G31, G63-G66, and A73-A75 segments of the hatchet ribozyme (shown in red rectangles in Fig. 1 A, B, and D) are clustered through pairing alignments (shown in red in Fig. 2A and SI Appendix, Fig. S6A) flanking the cleavage site (yellow star in Figs. 1E and 2A and SI Appendix, Fig. S6A).
Fig. 2.
Structural alignment of highly conserved residues in the hatchet ribozyme. (A) Highly conserved residues (in red) are brought into proximity near the cleavage site (labeled with a yellow star) through pairing and hydrogen bonding interactions in the tertiary structure. (B) C20 forms a trans-Watson–Crick base pair with G30 adjacent to the terminal part of stem P1, in which G30 adopts a C2′-endo sugar pucker. The sugar edge of G29 formed a sheared pairing interaction with the Watson–Crick edge of A74′, with the Watson–Crick edge of G29 forming additional hydrogen bonds with the nonbridging phosphate oxygen of A21, resulting in a stable interaction plane. The 2′-OH of G29 is pointed outwards from the plane and forms hydrogen bonds with the above G30-C20 base pair. Notably, both the sugar pucker of G29 and A74 adopted C2′-endo conformations. (C) The Hoogsteen edge of A21 forms a trans-pairing interaction with the Watson–Crick edge of U28; the 2′-OH of A21 forms one hydrogen bond with the adjacent stacked base pair G27-C77′ from stem P2. The sugar pucker of A21 adopts a C2′-endo conformation. (D) Highly conserved A22 forms a major groove-aligned base triple interaction with the Watson–Crick U26-A78′ base pair. The 2′-OH of A22 forms an additional hydrogen bond with 4-NH2 of C24. The sugar pucker of A22 adopts a C2′-endo conformation. (E) The minor groove-aligned base triple A73′•(G31-C64) involves highly conserved residues A73′, G31, and C64. (F) In molecule A′ of the HT-GAAA structure, the base G63 hydrogen bonds with the phosphate oxygen of A75′. The 6-NH2 of A75′ forms hydrogen bonds with the sugars of three residues G63, C64, and A65. The 2′-OH of A75′ hydrogen bonds with 6-NH2 of A65. A65 formed a cis-Hoogsteen Watson–Crick base pair with U71′. Note that the dashed lines indicate distances <3.5 Å and their number can exceed the possible number of hydrogen bonds formed by an atom.
The long-range interactions between C20-A21 and U28-G30 segments are shown in SI Appendix, Fig. S6B. C20 is involved in a trans-Watson–Crick C20•G30(C2′-endo) pair adjacent to the terminal part of stem P1, which in turn stacks over a sugar edge-Watson–Crick G29(C2′-endo)•A74′(C2′-endo) pair (Fig. 2B). A21 is involved in a trans-Hoogsteen Watson–Crick A21(C2′-endo)•U28 pair (Fig. 2C), which stacks over the Watson–Crick G27-C77′ pair (Fig. 2C). The long-range interaction of A22 with A78′ is shown in SI Appendix, Fig. S6C, where the Hoogsteen edge of A22(C2′-endo) pairs with the major groove edge of A78′ to form a A22•(A78′-U26) triple (Fig. 2D). Thus, consecutive stacking between three highly conserved C20•G30, G29•A74′, and A21•U28 noncanonical pairs bridge stems P1 and P2 to form the long stable H12 helix (SI Appendix, Fig. S6D).
The two conserved residues G30 and G31 adopt a splayed-apart conformation being positioned opposite the splayed-apart A73′-A75′ segment (SI Appendix, Fig. S6D). G30 is involved in the stacking interaction bridging stems P1 and P2, while G31 participates in long H34 helix formation composed of stems P3 and P4. G31 forms a canonical Watson–Crick G31-C64 pair with conserved residue C64 as part of a minor groove aligned A73′•(G31-C64) triple (Fig. 2E and SI Appendix, Fig. S6D), thereby extending the length of stem P3 (Fig. 1 B and E).
We noticed that the long-range interactions of Mol A′ of HT-GAAA ribozyme defined by a network of hydrogen bonding alignments involving conserved residues G63, A65-G66, and A74-A75 (SI Appendix, Fig. S7A) are slightly different in the pseudosymmetry-related Mol B′ (SI Appendix, Fig. S7B) of the HT-GAAA ribozyme, as well as molecule C′ of HT-UUCG ribozyme (SI Appendix, Fig. S7C). Further structural details are available in the caption to SI Appendix, Fig. S7. Notably, the sugar puckers of G63, A65, G66, A73′, A74′, and A75′ all adopt a C2′-endo conformation for Mol A′ of the HT-GAAA ribozyme.
Structural Alignment and the Modeling of the Cleavage Site.
The cleavage site located at the very 5′-end of the secondary structure (yellow star in Fig. 1A) is positioned in the center of the 3D fold of the hatchet ribozyme. It is surrounded by conserved residues originating from corresponding termini of stems P1, P2, and P3, and the zippered up segment of L2 (Fig. 1 B and C). Our structure represents the cleavage product of the hatchet ribozyme, which defines the overall fold and the conformation of O5′ of U1 (leaving group). U1 forms a canonical Watson–Crick pair with A19, representing the first base pair of stem P1, which in turn stacks on the conserved long-range trans-Watson–Crick G30•C20 pair (Fig. 3A and SI Appendix, Fig. S8A). The O5′ of U1 (leaving group) is extruded from the pairing segment and directed toward the major groove of the junctional stems constituted by the terminal Wobble G32•U62 pair of stem P3 and the first zippered-up highly conserved Watson–Crick G31-C64 pair of L2 (Fig. 3A and SI Appendix, Fig. S8A). In addition, three other conserved nucleotides G63, A75′, and A65 are stacked on each other and reside at the bottom of the long H34 helix (Figs. 1B and 3A and SI Appendix, Fig. S8A). G63, which is coordinated with the phosphate oxygen between A75 and A74, forms a platform below the cleavage site (Fig. 3A and SI Appendix, Fig. S8A). Notably, there is space formed by the alignments of U62, the highly conserved Watson–Crick G31-C64 base pair and G63 in molecule A′ of the HT-GAAA ribozyme generating sufficient space to form a pocket to accommodate C(−1) and the scissile phosphate linking C(−1) with U1 (Fig. 3A). The (2Fo-Fc) electron density map of the cleavage site is shown in SI Appendix, Fig. S8 C and D.
Fig. 3.
Structural alignment at the 5′-OH leaving group of U1 and modeling of the cleavage site of the hatchet ribozyme. (A) Base stacking with U1 at the cleavage site and alignment of junctional structure. U1 is paired with A19 in stem P1 and stacked above the conserved reversed Watson–Crick G30-C20 pair. G30 and G31 adopt a splayed-apart conformation and are involved in stacking with parallel helixes H12 and H34, respectively. A74′ and A75′ are also splayed apart adjacent to the cleavage site and stacked in H12 and H34, respectively. (B) A surface representation of the hatchet ribozyme product with U1 shown in a stick representation. A cavity is formed within the hatchet ribozyme product adjacent to the leaving group 5′-OH of U1. The dimensions of the cavity appear to be of sufficient size so as to accommodate the cleavage step of the hatchet ribozyme. (C) The base stacking interaction of the modeled cleavage site between C(−1) and U1, in which C(−1) was stacked partially between G32-U62 from stem P3 and two conserved residues G63 and A75′, whereas U1 was stacked between the conserved G30-C20 base pair and the termini of stem P1. (D) A surface representation of the model of the hatchet ribozyme with the cleavage step between C(−1) and U1 shown in stick representation. C(−1) was modeled based on the shape of the cavity on the hatchet ribozyme surface. C(−1) and U1 adopt a splayed-apart conformation. (E) The proposed model of the cleavage site of the hatchet ribozyme, in which C(−1) forms extensive hydrogen bonds with nearby residues. The modeled in-line alignment conformation indicates the potential nucleotides that may contribute to general base and general acid catalysis in the cleavage process.
We undertook modeling experiments to place C(−1) and the scissile phosphate linkage into our hatchet ribozyme product structure (Fig. 3 C and D and SI Appendix, Fig. S8B). The model was generated by superposing the cleavage site from the hammerhead ribozyme (PDB code: 2OEU) with U1 from the hatchet ribozyme in a similar way as was done for the HDV ribozyme (24). No clash with any part of the hatchet ribozyme product structure was observed. Then, C(−1) was manually rotated around the C5′-C4′ bond of U1 by 54° to optimally fit into the predicted cleavage site pocket, which was further optimized by energy minimization of the residue C(−1) in Schrodinger software (25) under OPLS force field to compute the final model (Fig. 3 C–E and SI Appendix, Fig. S8B). The C(−1) and U1 bases positioned in the resulting modeled cleavage pocket adopted a splayed-apart conformation, in which C(−1) appeared to be stabilized by forming a major-groove-aligned base triple with the Watson–Crick G31-C64 pair (Fig. 3E) and further partially stacked between the Wobble G32•U62 pair and G63 (Fig. 3C and SI Appendix, Fig. S8B). In this modeled alignment, the 4-NH2 of C(−1) forms two hydrogen bonds with the phosphate oxygen of U62 and G63 (Fig. 3E). The distance between the modeled 2′-O [C(−1)] and the scissile phosphate is ∼2.8 Å, while the angle from 2′-O [C(−1)] to P-O5′ [at C(−1)-U1 step] is ∼152°, which is consistent with an in-line attack conformation needed to obtain the pentavalent phosphorane transition state. Both C(−1) and U1 adopt anti-alignments at their glyosidic bonds with U1 adopting a C3′-endo sugar pucker conformation (Fig. 3E).
Conserved bases G31 and G30 are located close to the splayed-apart C(−1)-U1 step at the cleavage site (Fig. 3C and SI Appendix, Fig. S8B). The distance between the nucleophilic 2′-O of modeled C(−1) and the O6 and N7 of G31 is estimated to be ∼2.7 Å and ∼3.6 Å, respectively. The distance between the 5′-O of leaving group U1 and the 2′-OH and N7 of G30 is estimated to be ∼3.4 Å (Fig. 3E). This suggests that G31 can potentially serve as a general base for activation of the 2′-O of modeled C(−1) and G30 can potentially serve as general acid for protonation and hence compensation of the generated negative charge on 5′-O of U1 following cleavage of the C(−1)-U1 bond. To clarify if these spatial correlations are functionally relevant, we conducted a series of cleavage assays with ribozyme mutants (nucleobase and/or single atom substitutions) as described below.
Cleavage Assays on Hatchet Ribozyme Mutants.
Long-range tertiary interactions involving L1 and L3.
Although nucleobase identities of L1 and L3 are not highly conserved according to phylogenetic analysis, the tight interaction between these loops is crucial for stabilization of the overall fold. Thus, the L3 extruding U39 and U59 bases intercalate into L1, establishing an alignment that dominantly relies on base stacking, 2′-OH, and phosphate interaction networks. This is consistent with the observation that a ribozyme mutant that lacked the extruded residues in L3 and only formed the consecutive Watson–Crick base pairs between stem P3 and P4, completely lacked cleavage activity (SI Appendix, Fig. S9 A and B).
Pairing alignment of conserved residues.
The rigid “northern” scaffold of the hatchet ribozyme pocket is composed of the conserved trans-Watson–Crick C20•G30 and Watson–Crick G31-C64 pairs. Not unexpectedly, compared with wild type (Fig. 4A), inversion of the conserved C20•G30 into G20•C30 rendered the ribozyme inactive (Fig. 4B), while weakening base pairing strength by mutation to U20•A30 made cleavage slower with decreased yields (Fig. 4C). Likewise, mutation of conserved G31-C64 into A31-U64 resulted in barely detectable cleavage (Fig. 4D). Also, disruption of the conserved G31-C64 base pair in the single mutants C64U, C64G, C64A, or C64c3C (which we tested on the related env214 hatchet RNA) abolished cleavage (SI Appendix, Fig. S9 C–F).
Fig. 4.
Cleavage activity assays of the env10 hatchet ribozyme and mutants. Secondary structure cartoons of the two-stranded construct used in the cleavage assays with crucial base interactions highlighted in red and respective mutants in blue. HPLC traces following cleavage activity of wild-type ribozyme (A) and mutants C20G G30C (B), C20U G30A (C), G31A C64U (D), G31c7G (E), G30c7G (F), and G30dG (G). R1 and R2 denote the substrate and ribozyme strands; C1 (orange) and C2 (green) denote cleavage products. Reaction conditions: 55 μM RNA each strand; 10 mM MgCl2, 100 mM KCl, 30 mM Hepes, pH 7.5, 23 °C. HPLC conditions: Dionex DNAPac column (4 × 250 mm2), 80 °C, 1 mL min−1, 0–60% buffer B in 45 min. Buffer A: Tris–HCl (25 mM), urea (6 M), pH 8.0. Buffer B: Tris–HCl (25 mM), urea (6 M), NaClO4 (0.5 M), pH 8.0.
Concerning the conserved residues A21 and A22, as well as A65, A73, A74, and A75 that form hydrogen bonds via their Hoogsteen face and/or are involved in stacking interactions, we tested for their individual replacements by 1,3-dideaza-adenosine (c1c3A), an analog that lacks Watson–Crick base-pairing propensities. The mutants were as active as the wild type or only slightly decreased in activity (SI Appendix, Fig. S10 A–G). This observation confirmed their significance in shaping the stacked interface between P1 and P2 to form H12 without the utilization of the Watson–Crick pairing mode (SI Appendix, Fig. S6A).
Nucleoside-65 is highly conserved as purine and therefore it was not surprising that we observed wild-type cleavage for the A65G mutant (SI Appendix, Fig. S11A); also the related pyrimidine mutants A65C and A65U were active (SI Appendix, Fig. S11 B and C). More stringent for activity appeared the conservation of G63 and G66; the mutants G63A or G66A showed hardly any cleavage (SI Appendix, Fig, S11D, E). In contrast, G66I was active (SI Appendix, Fig. S11F) which is consistent with its Hoogsteen face retaining pairing with G63 as seen in our structure (SI Appendix, Fig. S7 B and C).
Structural alignment and the modeling of the cleavage site.
The nucleobase identities of G30 and G31 associated with base pairs C20•G30 and G31-C64 are stringent. Mutations are hardly tolerated as apparent from cleavage experiments of the mutants C20G-G30C, C20U-G30A, and G31A-C64U (discussed above and shown in Fig. 4 B–D). Moreover, these guanines (G30, G31) come closest to the modeled scissile phosphate. The N7 of G31 therefore is in an almost ideal distance to activate the attacking (modeled) 2′-OH of C(−1) (Fig. 3E). We therefore synthesized a 7-deazaguanosine (c7G31)-modified hatchet ribozyme. Cleavage of the G31c7G mutant was completely abolished (Fig. 4E) and this observation strongly supports the hypothesis that G31 serves as a general base in catalyzing phosphodiester hydrolysis (γ-catalysis) (26, 27). We also point out that this mode of (G31)N7···HO-2′[(C(−1)] activation would be independent of the nucleotide-(−1) identity. Consistently, we found that both C(−1)U and C(−1)A mutants were cleaved comparably to wild type C(−1) (SI Appendix, Fig. S11 H and I). Also, C(−1)G was cleaved, although slower and to a less extent (SI Appendix, Fig. S11J).
Furthermore, we note that an alternative mechanistic scenario is conceivable by potentially involving a N7(G31)-coordinated hydrated Mg2+ ion that activates the attacking 2′-OH in the precatalytic and transition-state structures. As discussed further below, the crystallization of the hatchet ribozyme product required high concentrations of ammonium sulfate, which can interfere with localizing Mg2+ binding sites. Another possibility to consider is that the specific Mg2+ binding site is no longer available in the product structure.
Concerning the second conserved guanosine (G30) at the cleavage site, its N7 is located in hydrogen bond distance to the nonbridging oxygen of the scissile phosphate, while its 2′-OH is located close to the 5′-O leaving group of U1 (Fig. 3E). Thus, either protonated N7(G30) and/or hydrated Mg2+-coordinated N7(G30) (as identified for pistol ribozymes) (16, 28) are therefore candidates for general acid catalysis (δ-catalysis). Likewise, the 2′-OH (G30) could potentially stabilize the transition state (β-catalysis). However, neither the G30c7G nor the G30dG mutant showed decreased cleavage activity (Fig. 4 F and G). Therefore, the precise role of G30—if solely structural or if involved in catalysis by any other path than discussed above—remains to be explored.
Discussion
The hatchet motif is unusual because its cleavage site is located at the very 5′ end of the ribozyme (Fig. 1A). The only other ribozymes that are wholly downstream of their cleavage sites are the HDV family of ribozymes (29). The importance of this fact lies in the utility of these ribozymes—they are used in expression of cleaved RNAs in vivo for a variety of synthetic biology applications, e.g., for CRIPSPR/Cas9 gRNA production (30).
Unexpectedly, both HT-GAAA and HT-UUCG constructs formed pseudosymmetric and symmetric dimers, respectively, in the crystal lattice (SI Appendix, Figs. S2A and S2B). The size-exclusion experiment confirmed that the hatchet ribozyme product existed in solution as an equilibrium between dimer and monomer (SI Appendix, Fig. S3). The existence of the palindromic nucleotides from A67 to U70 (SI Appendix, Fig. S4C) most likely contributes to the swapping of the 3′-end segments between two molecules of the hatchet ribozyme. Such RNA dimerization resulting from structural exchange has also been reported previously in the crystal structure of the Varkud satellite (VS) ribozyme (15) and the tetrahydrofolate (THF) riboswitch (31).
Overall Topology of the Hatchet Ribozyme Product.
The proposed monomer structure of the hatchet ribozyme (Fig. 1E) is composed of a pair parallel-aligned long helixes H12 and H34. H12 resulted from axial stacking of stems P1, P2 and the intervening pairing segment (SI Appendix, Fig. S4A), while H34 resulted from axial stacking of stems P3, P4, the pairing segment of L3, and the zippered-up stem-forming segment of L2 (SI Appendix, Fig. S4B). The overall structure of the hatchet ribozyme was stabilized in part by coaxial stacking interactions, which remains a common feature of higher-order RNA structure (32). The long-distance interaction between L1 and L3 identified in the tertiary fold (Fig. 1 B–D), together with several bridged nucleotides in L2, appear to anchor the relative alignments of H12 and H34 (Fig. 1E).
Catalytic Pocket Lined by Conserved Residues.
The dispersed conserved nucleotides in the proposed secondary structure of the hatchet ribozyme (Fig. 1A) are brought into proximity and aligned around the cleavage site (Fig. 2A and SI Appendix, Fig. S6A), suggestive of these residues playing a vital role in the cleavage reaction. Most of these conserved nucleotides are involved in pairing alignments, stacking, and hydrogen-bonding interactions (Fig. 2 B–F and SI Appendix, Fig. S6 B–D). Notably, the local structure around G63, A65-G66, and A74-A75 adopts slightly different structures between the two molecules of the HT-GAAA construct, as well as the HT-UUCG construct (SI Appendix, Fig. S7 A–C), potentially reflective of flexibility within these segments of the hatchet ribozyme.
Binding Pocket and Modeling the Alignment of the Scissile Phosphate.
Despite the cleavage site being located at the 5′ end of the sequence (Fig. 1A), it is positioned in the center of the tertiary structure of the hatchet ribozyme product (shown by yellow star in Fig. 1 C and D). The highly conserved residue G30 formed a trans-Watson–Crick base pair with conserved C20 and is intercalated between U1-A19 and A74′ in H12. Highly conserved G31 and C64 form a Watson–Crick base pair and stack with the terminal base pair G32•U62 of stem P3. The leaving group 5′-OH of U1 points toward the major groove of the stacked G32•U62 and G31-C64 pairs (Fig. 3A and SI Appendix, Fig. S8A). Such alignments result in the generation of a pocket within the major groove capable of accommodating a modeled C(−1). Hydrogen bonding of highly conserved G63 with the nonbridging phosphate oxygens between A74′ and A75′ resulted in the formation of a platform below the 5′-OH of U1 (Figs. 2F and 3A), thereby capping the resulting compact pocket. The dimensions of the pocket are capable of accommodating both the modeled C(−1) and the scissile phosphate linking the C(−1)-U1 step (Fig. 3 B and D). As shown in Fig. 3 C and E, the modeled C(−1) can potentially form extensive hydrogen bonding and stacking interactions within the hatchet ribozyme. The model also accommodates a splayed-apart in-line attack conformation at the cleavage site and provides insights into potential candidates for general base and general acid catalysts facilitating scissile phosphate cleavage chemistry.
Role of Hydrated Divalent Cations.
It has been reported that Mg2+ is required for the hatchet ribozyme to initiate the self-cleavage reaction (5, 23). Additionally, a recent SHAPE probing study underlines the requirement of high Mg2+ concentrations for structuring of the hatchet ribozyme fold (33). However, we did not detect Mg2+ cations in the vicinity of the cleavage site in the structures of either the HT-GAAA or HT-UUCG constructs of the product ribozyme. This may reflect the high salt conditions (2.0–3.0 M ammonium sulfate) required for crystallization of the HT-GAAA and HT-UUCG constructs, which may prevent the binding of Mg2+ cations within the cleaved product of the hatchet ribozyme. Notably, the number of observed waters in the structures of the hatchet ribozyme products also appear to be less than what has been reported for other RNA structures at the same resolution level. It is also conceivable that a possible hydrated Mg2+ binding site in the precatalytic state of the hatchet ribozyme (potentially needed for catalysis) is no longer available in the product.
Insights from Studies of Hatchet Ribozyme Mutants.
We analyzed phosphodiester cleavage for a large number of hatchet ribozyme mutants that were selected based on the observed (and seemingly most crucial) interactions in the crystal structure. First, the functional importance of dimer formation was scrutinized by mutation of the nonconserved 4-nt palindromic ACGU segment in L2 that forms an intermolecular double helix in the crystal. Replacement of the ACGU by UUUU or insertion of an extrastable UUCG loop between C68 and G69 (at the pseudosymmetric site) so as to favor intramolecular hairpin and hence monomer formation, resulted in ribozymes that exhibit equal activity as the wild type (SI Appendix, Fig. S5 A–C). In addition, the bimolecular assembly of the 82-nt comprising ribozyme, resulting from cutting into two fragments at the pseudosymmetric site, also shows wild-type activity (SI Appendix, Fig. S5D). Together, these results support the assumption that the monomeric fold (corresponding to A′ or B′ as shown in Fig. 1 C–E) is fully functional and likely represents the cellular fold.
Rigidity Versus Flexibility in the Hatchet Ribozyme Product.
Importantly, our crystal structures of the hatchet ribozyme provide valuable information of the structurally rigid versus flexible regions. Almost identical in both folds A′ and B′ (and thus considered rigid) is the long-range L1–L3 tertiary interaction, which in turn stabilizes the parallel orientation of H12 and H34 (Fig. 2 A and B). This alignment further dictates the opposite directionality of the highly conserved C20•G30 and G31-C64 base pairs, with neighboring G30 and G31 adopting splayed-apart conformations (Fig. 2A). This northern part of the active site pocket locks the active site U1 that is paired to A19 and also sandwiched into the extended helix P1 of the hatchet ribozyme. In addition, the P1–P2 connecting interface is uniformly stacked in both molecules A′ and B′.
By contrast, A74′ in molecule A′ takes over the role of A75′ in molecule B′, thereby defining the flexible part of L2 nucleotides that also include G63, A65, G66, A73′, A74′, and A75′ in the southern and western parts of the pocket. Such alternative alignments are observed not only in A′ and B′ of the HT-GAAA construct, but also in the HT-UUCG construct (SI Appendix, Fig. S7). The different alignments may reflect the empty space that would otherwise be occupied by C(−1) that is absent in our structure of the hatchet ribozyme product. It may also reflect the adaptability of this part of the pocket (consisting of L2 nucleotides) to accommodate and cleave U, A, and G at the same position −1 of the cleavage site (SI Appendix, Fig. S11 H–J).
Notably, modeling of C(−1) into the active site pocket suggests N7 of G31 as a potential general base for activation of the 2′-OH of C(−1) for attack of the scissile phosphate (Fig. 3E). This hypothesis was evaluated by atomic mutagenesis using a hatchet variant with 7-deazaguanosine in position 31. This mutant was totally inactive (Fig. 4E), thereby supporting our proposal for a significant role of N7 G31 in γ-catalysis of the phosphodiester cleavage. Using the same atomic mutagenesis approach, participation of other functional groups in close proximity to the 5′-O leaving group of the scissile phosphate, namely N7 and 2′-OH of G30, appear not to play a role in chemical catalysis (Fig. 4 F and G).
Comparison of Hatchet and HDV Ribozyme.
The only other currently known ribozyme class with the cleavage site at the very 5′ end is the HDV family of ribozymes (29) and like for hatchet, the first structural information on HDV RNA was obtained from crystal structure analysis of the cleaved product (13). For comparison, we juxtapose the secondary structures, the overall tertiary folds, and active sites of both HDV and hatchet ribozymes in Fig. 5.
Fig. 5.
Comparison of secondary structures, tertiary folds, and cleavage sites of HDV and hatchet ribozyme products. (A and B) Secondary structure schemes of HDV (A) and hatchet (B) ribozyme products (cleavage sites are highlighted by yellow stars and catalytic pockets by light pink shadows). Residues crucial for the cleavage reaction (C75 in HDV, and G31 in hatchet) are shown in red. Note that the color code highlights comparable stacks between HDV and hatchet RNA, and not consecutive stem numbering. (C and D) Cartoon representations of the tertiary structures of HDV (PDB code: 1DRZ) (C) and hatchet (D) ribozyme products. Labeling of cleavage sites, catalytic nucleosides in the pocket, and helix colors are as in A and B. (E) Catalytic pocket of HDV ribozyme product (PDB code: 1DRZ). The cleaved G1 terminus is shown in yellow, paired to U37, and stacked between G2-C36 and G38-C22 base pairs. The key residue C75 is unpaired but held in place by two base layers G28-C19 and G25-U20 from stem P3 and the junction base A65. (F) Catalytic pocket of hatchet ribozyme product. The cleaved U1 terminus is shown in yellow, paired to A19, and stacked between U2-A18 and G30-C20 base pairs. The key residue G31 is paired with C64 and stacked partially between G32-C62 and the ribose of A74′. Note that A74′ is also stacked with G30-C20 below the U1 terminus.
The HDV ribozyme comprises five helical regions (P1, P1.1, P2, P3, and P4) that are arranged in a nested double pseudoknot, forming two coaxial stacks (P1–P1.1–P4 and P2–P3) (Fig. 5A). The parallel alignment of these two long stacks is similar to the hatchet ribozyme, although its helical composition is distinct (P1–P2 and L2–P3–L3–P4) (Fig. 5B). Furthermore, we note that the cleavage sites of both ribozymes (labeled with a yellow star in Fig. 5 A–D) were located in the center of each tertiary fold and the active site formation involved junctional regions (highlighted by a light pink shadow in Fig. 5 A and B). Distinct from the HDV nested double pseudoknot, the hatchet tertiary fold was mainly stabilized by the long-distance interaction between L3 and P1 (Fig. 5 B and D).
An obvious similarity of hatchet and HDV active sites is that their 5′-terminal nucleosides (nucleoside 3′ from the scissile phosphate which is G1 in HDV and U1 in hatchet) are involved in Watson–Crick base pair formation (wobble G1•U37 in HDV versus U1-A19 in hatchet). These base pairs are perfectly stacked within the long helical segments of P1–P1.1 (HDV) and P1–P2 (hatchet) (Fig. 5 E and F) which likely helps to mold and stabilize the individual pockets.
Already based on the first HDV ribozyme structure (i.e., product), residue C75 was recognized to play a key role in HDV catalysis. Based on the hatchet product structure, we allocate G31 in its active site to be significant for catalysis although its precise mode of action (e.g., general acid–base catalysis via N7 or via a putative N7 coordinated hydrated Mg2+) has yet to be determined. Both C75 and G31, respectively, are located in junctional regions of the two ribozymes (Fig. 5 A–F). Notably, the distance between C75 and the leaving group G1 is shorter in the HDV product compared with the distance between G31 and U1 in the hatchet product (Fig. 5 E and F).
Although no Mg2+ ions were found in either ribozyme products’ active sites, we know from the HDV case that follow-up crystal structures of the precleavage state of HDV revealed a crucial Mg2+ binding pocket in its active site (24, 34). The protonated form of HDV C75 is generally thought to be stabilized through interactions with the scissile phosphate (35) and may interact electrostatically with the metal ion bound in the active site (24, 34).
Future Challenges.
We plan to extend the current studies on the structure of the hatchet ribozyme product to that of its precatalytic conformation, and in the longer term, to its transition state mimic vanadate conformation. Such efforts should provide a more complete overview of the catalytic cycle of the hatchet ribozyme.
Materials and Methods
Details of the methods, including RNA preparation, crystallization, structure determination and modeling, and cleavage assays are presented in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of the BL-17U1, BL-17B, BL18U1, and BL-19U1 beamlines at the National Center for Protein Sciences Shanghai at Shanghai Synchrotron Radiation Facility and the staff at Northeastern Collaborative Access Team (NE-CAT) beamlines at the Advanced Photon Source. We thank Hong Wu [Life Sciences Institute (LSI), Zhejiang University] for her help in some crystallization solution preparations and acknowledge the use of the LSI core facility. The research was supported by grants from the Natural Science Foundation of China (91640104, 31670826, and 31870810), the Fundamental Research Funds for the Central Universities (2017QN81010), new faculty start-up funds from Zhejiang University (to A.R.), the Austrian Science Fund FWF (P27947 and P31691), Austrian Research Promotion Agency FFG (West Austrian BioNMR 858017) (to R.M.), NIH 1U19CA179564, funds from the office of the President of SUSTech (to D.J.P.), and NIH P30CA008748 Cancer Center Core Grant to Memorial Sloan-Kettering Cancer Center.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6JQ5 for HT_GAAA structure and 6JQ6 for HT_UUCG structure).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902413116/-/DCSupplemental.
References
- 1.Talini G, Branciamore S, Gallori E (2011) Ribozymes: Flexible molecular devices at work. Biochimie 93:1998–2005. [DOI] [PubMed] [Google Scholar]
- 2.Lilley DMJ. (2017) How RNA acts as a nuclease: Some mechanistic comparisons in the nucleolytic ribozymes. Biochem Soc Trans 45:683–691. [DOI] [PubMed] [Google Scholar]
- 3.Ren A, Micura R, Patel DJ (2017) Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. Curr Opin Chem Biol 41:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez RM, Polanco JA, Lupták A (2015) Chemistry and biology of self-cleaving ribozymes. Trends Biochem Sci 40:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg Z, et al. (2015) New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 11:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth A, et al. (2014) A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol 10:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martick M, Scott WG (2006) Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mir A, Golden BL (2016) Two active site divalent ions in the crystal structure of the hammerhead ribozyme bound to a transition state analogue. Biochemistry 55:633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupert PB, Ferré-D’Amaré AR (2001) Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410:780–786. [DOI] [PubMed] [Google Scholar]
- 10.Rupert PB, Massey AP, Sigurdsson ST, Ferré-D’Amaré AR (2002) Transition state stabilization by a catalytic RNA. Science 298:1421–1424. [DOI] [PubMed] [Google Scholar]
- 11.Klein DJ, Ferré-D’Amaré AR (2006) Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313:1752–1756. [DOI] [PubMed] [Google Scholar]
- 12.Cochrane JC, Lipchock SV, Strobel SA (2007) Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol 14:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferré-D’Amaré AR, Zhou K, Doudna JA (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature 395:567–574. [DOI] [PubMed] [Google Scholar]
- 14.Ke A, Zhou K, Ding F, Cate JH, Doudna JA (2004) A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature 429:201–205. [DOI] [PubMed] [Google Scholar]
- 15.Suslov NB, et al. (2015) Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol 11:840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren A, et al. (2016) Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage. Nat Chem Biol 12:702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen LA, Wang J, Steitz TA (2017) Crystal structure of pistol, a class of self-cleaving ribozyme. Proc Natl Acad Sci USA 114:1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren A, et al. (2014) In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat Commun 5:5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Wilson TJ, McPhee SA, Lilley DM (2014) Crystal structure and mechanistic investigation of the twister ribozyme. Nat Chem Biol 10:739–744. [DOI] [PubMed] [Google Scholar]
- 20.Eiler D, Wang J, Steitz TA (2014) Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc Natl Acad Sci USA 111:13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng L, et al. (2017) Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme. Nat Commun 8:1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Wilson TJ, Lilley DMJ (2017) The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nat Chem Biol 13:508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Lünse CE, Harris KA, Breaker RR (2015) Biochemical analysis of hatchet self-cleaving ribozymes. RNA 21:1845–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JH, et al. (2010) A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry 49:6508–6518. [DOI] [PubMed] [Google Scholar]
- 25.Anonymous (2018) Schrödinger Release 2018-2: Maestro (Schrödinger, LLC, New York).
- 26.Emilsson GM, Nakamura S, Roth A, Breaker RR (2003) Ribozyme speed limits. RNA 9:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breaker RR, et al. (2003) A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. RNA 9:949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuner S, et al. (2017) Atom-specific mutagenesis reveals structural and catalytic roles for an active-site adenosine and hydrated Mg2+ in pistol ribozymes. Angew Chem Int Ed Engl 56:15954–15958. [DOI] [PubMed] [Google Scholar]
- 29.Riccitelli N, Lupták A (2013) HDV family of self-cleaving ribozymes. Prog Mol Biol Transl Sci 120:123–171. [DOI] [PubMed] [Google Scholar]
- 30.He Y, et al. (2017) Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J Genet Genomics 44:469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L, Ishibe-Murakami S, Patel DJ, Serganov A (2011) Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch. Proc Natl Acad Sci USA 108:14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batey RT, Rambo RP, Doudna JA (1999) Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl 38:2326–2343. [DOI] [PubMed] [Google Scholar]
- 33.Gasser C, Gebetsberger J, Gebetsberger M, Micura R (2018) SHAPE probing pictures Mg2+-dependent folding of small self-cleaving ribozymes. Nucleic Acids Res 46:6983–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golden BL. (2011) Two distinct catalytic strategies in the hepatitis δ virus ribozyme cleavage reaction. Biochemistry 50:9424–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong B, Chen JH, Bevilacqua PC, Golden BL, Carey PR (2009) Competition between Co(NH(3)(6)3+ and inner sphere Mg2+ ions in the HDV ribozyme. Biochemistry 48:11961–11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.