Significance
Genome-wide association studies (GWAS) identify many sequence variants associated with a trait or disease, but only a minority of these variants are likely causal. These variants are mostly noncoding, implying that variation in cis-regulatory elements (CREs) controlling gene expression is their basis. However, owing to background associations (linkage disequilibrium) between variants, identifying the causal variants that disrupt CRE function and influence gene expression is challenging. In this study, we advance an approach to dissect the multiple causal variants and CREs that influence cardiac expression of SCN5A and explain the electrocardiographic QT interval (trait) GWAS signal on chromosome 3p22.2.
Keywords: cis-regulatory variants, QT interval, SCN5A, GWAS, enhancers
Abstract
The rationale for genome-wide association study (GWAS) results is sequence variation in cis-regulatory elements (CREs) modulating a target gene’s expression as the major cause of trait variation. To understand the complete molecular landscape of one of these GWAS loci, we performed in vitro reporter screens in cardiomyocyte cell lines for CREs overlapping nearly all common variants associated with any of five independent QT interval (QTi)-associated GWAS hits at the SCN5A-SCN10A locus. We identified 13 causal CRE variants using allelic reporter activity, cardiomyocyte nuclear extract-based binding assays, overlap with human cardiac tissue DNaseI hypersensitive regions, and predicted impact of sequence variants on DNaseI sensitivity. Our analyses identified at least one high-confidence causal CRE variant for each of the five sentinel hits that could collectively predict SCN5A cardiac gene expression and QTi association. Although all 13 variants could explain SCN5A gene expression, the highest statistical significance was obtained with seven variants (inclusive of the five above). Thus, multiple, causal, mutually associated CRE variants can underlie GWAS signals.
Variable gene expression plays a major role in the phenotypic evolution across species (1) and interindividual trait (or disease risk) variation within species (2, 3). Although several biological elements regulate gene expression, the major contribution is from cis-regulatory elements (CREs) of gene expression, such as promoters, enhancers, and insulators (4). However, for most coding genes, these noncoding CREs are largely unidentified. Studies in model organisms indicate that multiple unique and redundant CREs per gene are necessary for precise spatiotemporal control of gene expression (5) and as a buffer against widespread disruptive sequence variation within CREs (6). Genome-wide association studies (GWAS) of human complex traits and disease often implicate multiple independent noncoding associations within a locus, consistent with multiple CREs per gene. However, even multiple correlated trait-associated variants may contribute to gene regulation. We have previously demonstrated that in Hirschsprung disease, where multiple independent and associated common CRE variants modulate RET gene expression and disease risk (7).
Large-scale studies, such as the ENCODE (8) and NIH RoadMap Epigenomics (9) projects, have made significant strides toward annotation of the human genome with respect to the molecular components of cis-regulation. However, these datasets cannot be comprehensive or complete for all circumstances requiring cis-regulation of a specific gene. Consequently, we present here a systematic approach for identification of all CREs for a given gene, whose activity is altered by disease/trait-associated common variants. We exemplify this CRE screen for SCN5A using common variants associated with the electrocardiographic QT interval (QTi).
QTi, a clinically relevant quantitative trait with ∼30% heritability and with age, gender, and heart rate as covariates (10), measures the time taken by the cardiac ventricles to depolarize and repolarize in every heartbeat. Its prolongation or shortening is associated with an increased risk of cardiovascular morbidity and mortality, primarily in the form of ventricular tachycardia and ventricular fibrillation leading to sudden cardiac death (SCD) (11). Extremes of the QTi are hallmarks of Mendelian long-QT (LQTS) and short-QT syndromes, which are associated with elevated risk for cardiac arrhythmias and SCD, and arise from rare, high-penetrance coding mutations in nearly a dozen genes encoding ion channels and associated proteins (12, 13), including SCN5A. SCN5A encodes a voltage-gated sodium channel alpha subunit, with rare coding mutations in type 3 LQTS and Brugada syndrome (14). Further, at least one of the 35 loci identified by QTi GWAS in the general population encompasses SCN5A on chromosome 3p22.2 (15). Thus, both coding and noncoding variants in SCN5A affect QTi variation and SCD risk. Significantly, GWAS of other electrocardiographic traits, the PR interval (16) and the QRS duration (17), as well as conduction defects (18), atrial fibrillation (19), and Brugada syndrome (20), have also mapped common variants to this locus. Thus, although understanding SCN5A regulation is important per se, we limited our SCN5A CRE screen here to only the QTi-associated common variants at the 3p22.2 locus.
We performed an unbiased in vitro reporter screen for CREs overlapping all common variants in linkage disequilibrium (LD) with five QTi-associated GWAS hits at the SCN5A-SCN10A locus to assess their potential causality, provided that an assay could be developed (88%). We identified multiple causal CRE variants, using reporter assays in human AC16 (21) and mouse HL1 (22) cardiomyocyte cell lines, correlating with SCN5A cardiac gene expression and capable of explaining the QTi associations. Finally, we assessed the adequacy of AC16 and HL1 as in vitro cardiomyocyte cellular models, using RNA-sequencing (RNA-seq) (23) and assay for transposase-accessible chromatin using sequencing (ATAC-seq) (24) analyses. This study indicates that multiple causal variants, genetically independent or not, within CREs are contributory to trait association by varying their target gene expression.
Results
Identification of QTi-Associated Candidate CRE Variants at SCN5A.
rs6793245 is the primary sentinel variant at the SCN5A-SCN10A locus identified in a QTi GWAS meta-analysis (QT-International GWAS Consortium) in European ancestry subjects (SI Appendix, Fig. S1) (15). Four additional independent associations at the locus, rs11708996, rs11710077, rs6599234, and rs6801957, based on low LD (r2 < 0.05) to other genome-wide significant variants were also identified (15). Three of these variants map to introns of SCN5A (rs6793245, rs11708996, and rs11710077), one to an intron of SCN10A (rs6801957), and one (rs6599234) to their intergenic region (SI Appendix, Fig. S1). First, under the hypothesis that these five variants, or their LD surrogates, are the causal factors underlying the associations, we defined the target regions by their nearest upstream and downstream recombination hotspots (recombination rate > 10 cM/Mb): four contiguous target regions covered ∼450 kb (25) (SI Appendix, Fig. S1 and Table S1). Second, we used 1000 Genomes Project data (26) from European ancestry subjects (n = 379) to identify 121 common variants [minor allele frequency (MAF) > 5%] in the target and in moderate to high LD (r2 > 0.3) with the five index variants (Fig. 1 and Dataset S1). We assumed that all of these variants were causal candidates.
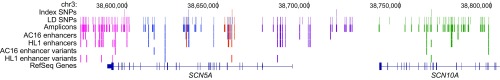
Fig. 1.
Genomic map of common variants at the QTi-associated SCN5A-SCN10A GWAS locus on chromosome 3p22.2. A 224-kb genomic segment is annotated with tracks, showing (from top) the five independent GWAS hits (Index SNPs); all common (MAF > 5%) SNPs in 1000 Genomes Project European ancestry samples in moderate to high LD (r2 > 0.3) with the five index SNPs (LD SNPs); amplicons encompassing the LD SNPs that were cloned and evaluated in reporter assays (Amplicons); amplicons that were cis-regulatory enhancers by in vitro reporter assays in the human cardiomyocyte cell line AC16 (AC16 enhancers) and the mouse cardiomyocyte cell line HL1 (HL1 enhancers); the cis-regulatory enhancers that displayed significant allelic difference in reporter activities in AC16 (AC16 enhancer variants) and HL1 (HL1 enhancer variants); and the protein-coding SCN5A and SCN10A (RefSeq) genes. The five independent GWAS hits are marked in color (pink, blue, brown, purple, and green) in the Index SNPs track, while features in other tracks are color-coded based on the highest LD with these five index variants. The genomic map was generated using custom tracks in the UCSC Genome Browser.
In Vitro Reporter Assays Identify 12 Candidate Causal CRE Variants.
We retrieved genomic sequences flanking these variants (±325 bp) from the UCSC Genome Browser and used Primer3 (27) to design variant-centered amplicons. A total of 104 amplicons passed in silico primer design (Dataset S2), with 97 covering one, 6 covering two, and 1 covering three variants each (256 bp–617 bp, median = 397 bp; 0.25–0.63 guanine–cytosine content, median = 0.49). We cloned these amplicons into linearized pGL4.23 vector, immediately upstream of a minimal promoter driving expression of firefly luciferase reporter gene; sequences (15–20 bases) homologous to the vector backbone cloning sites were added to the 5′ ends of the forward and reverse primers (Dataset S2). PCR amplification was performed on genomic DNA from selected 1000 Genomes Project samples (26) that were homozygous for either the reference or alternate alleles at each site. Genomic DNA samples for multivariant amplicons were also selected to capture all-reference or all-alternate alleles in separate amplicons. Of 112 variants that passed primer design across 104 amplicons, we successfully cloned both alleles at 106 variants within 98 amplicons (Fig. 1 and Dataset S2). All clones were sequence-verified to ensure that other sequence differences were absent.
At each site, reference and alternate alleles were evaluated for CRE activity using transient reporter assays in the human AC16 (21) and mouse HL1 (22) cardiomyocyte cell lines. We compared log10-transformed mean normalized reporter activity for each construct in each cell line, and found high correlation for all reference (r = 0.79) and all alternate (r = 0.81) alleles (SI Appendix, Fig. S2 and Dataset S3). To assess CRE activity, each test construct was compared with empty vector to calculate standardized z-scores for log2-transformed relative firefly activities. In HL1, at least one allele at 30 constructs behaved as an enhancer (average z-score > 2.326) and at least one allele at 51 constructs behaved as a suppressor (average z-score < −2.326), while 17 constructs had intermediate values for both alleles and were neutral (Fig. 2, SI Appendix, Fig. S3, and Dataset S4). The corresponding numbers in AC16 were 35, 47, and 16 constructs, respectively (Fig. 2, SI Appendix, Fig. S3, and Dataset S4). The overall concordance in CRE effect between the two cell lines was high (70%) and led to identification of 40 enhancers across the two cell lines (Dataset S4). We considered any test element with significant allelic enhancer activity difference in either cell line as a CRE variant. In HL1 and AC16, eight of 30 (27%) and five of 35 (14%) constructs, respectively, showed allelic difference (P < 0.05), corresponding to a total of 12 (30%) unique elements on 13 common QTi-associated variants (Figs. 1 and 2).
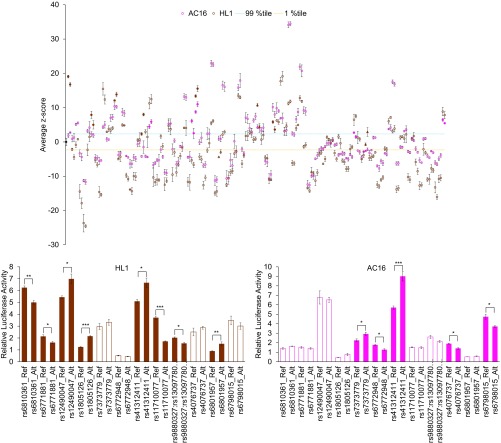
Fig. 2.
In vitro activity variation in QTi-associated CREs at the SCN5A-SCN10A locus in cardiomyocyte cell lines. (Top) Average standardized reporter activity (z-score) of all evaluated variant-centered amplicons (both alleles) is shown for assays performed in AC16 (pink) and HL1 (brown) cells; the empty vector activity (control) is shown as a black circle. All tested amplicons (both alleles: first reference and then alternate) are arranged in their genomic order (Dataset S2) along the x axis. The 99th and first-percentile z-scores from the empty vector controls are shown by the blue and orange dashed lines, respectively. Variant-centered amplicons are shown as colored circles, except for the five index variant-centered amplicons, shown as triangles. Variant-centered amplicons with significant allelic difference in enhancer activity are shown as filled circles or triangles; all other variant-centered amplicons are shown as empty circles or triangles. Error bars are the SD of z-scores. (Bottom) Bar plots of average relative luciferase activities of the 12 selected variant-centered amplicons (13 variants; both alleles) that displayed enhancer activity with significant allelic difference in either HL1 (Left) and/or AC16 (Right) cell lines. Alt, alternate; Ref, reference. Filled and empty bars indicate variant-centered amplicons with and without significant allelic difference in reporter activity, respectively. Asterisks indicate P values from the Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). Error bars are SEMs.
In Vitro Binding Assays and In Silico Binding Predictions for Observed CRE Variants.
For additional experimental support of these 13 variants, we first performed electrophoretic mobility shift assays (EMSAs) using both alleles at each variant, along with a 5-bp deletion centered on each variant (Dataset S5) and nuclear extracts from HL1 and AC16. For six variants (rs12490047, rs7373779, rs41312411, rs1171007, rs9880327, and rs13097780), we found evidence of differential binding (stronger binding at reference alleles for rs7373779, rs41312411, and rs13097780; stronger binding at alternate alleles for rs12490047, rs1171007, and rs9880327) by factors present in both HL1 and AC16 nuclear extracts (Fig. 3). Importantly, the observed DNA–protein complexes were lost in the deletion probes (Fig. 3). We did not observe any evidence of binding to probes representing the remaining seven variants in either cell line. Note that signals observed using AC16 nuclear extracts were consistently weaker than those from HL1, despite the former’s human origin, leading us to suspect that AC16 may be a less optimal cellular model than HL1 for these studies. Second, we also evaluated the 13 candidate variants by in silico transcription factor (TF) binding prediction. Twelve SNPs (except rs6810361) have at least one significant match with either the reference or alternative allele, or both (Dataset S6). Some SNPs are predicted to disrupt binding sites of known cardiac TFs and are consistent with EMSA results. For example, the alternate alleles of rs7373779 and rs41312411 are predicted to disrupt GATA and SP1 binding sites, respectively, while the reference allele of rs12490047 is predicted to disrupt PPAR binding.
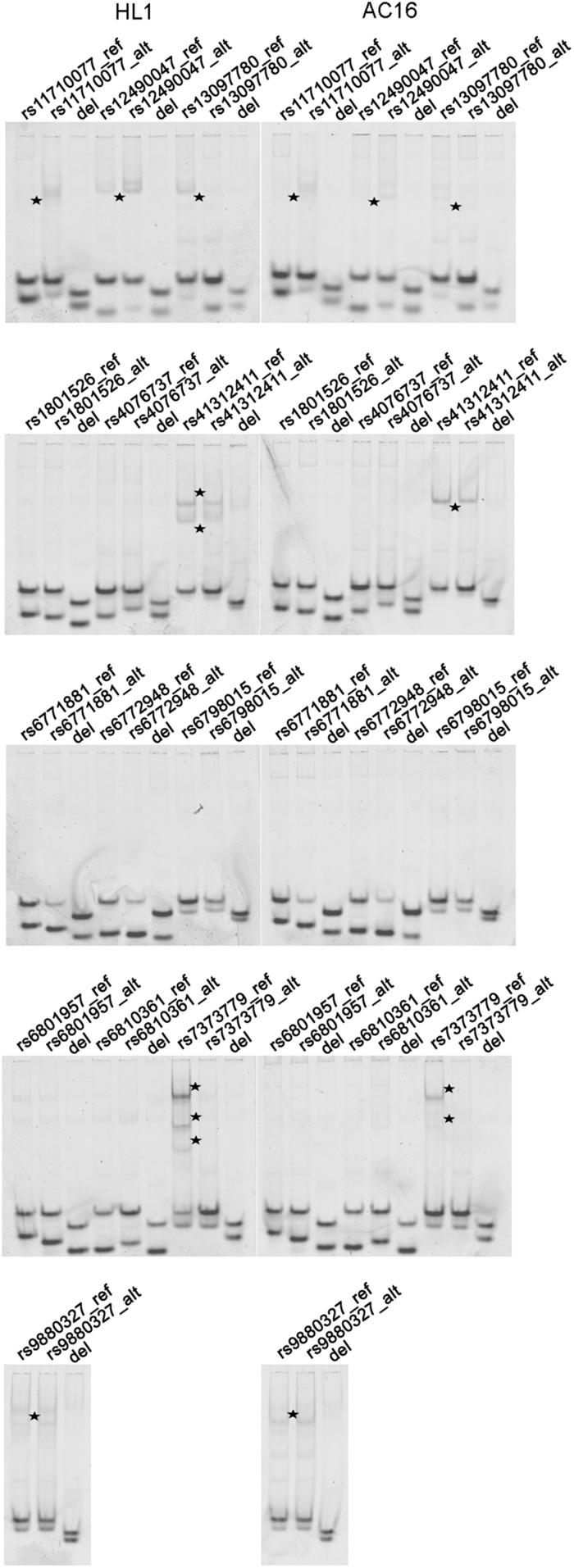
Fig. 3.
Differential binding of AC16 and HL1 nuclear proteins to cis-regulatory enhancer variants. EMSAs, using 25-bp Cy5-labeled probes containing the reference (ref) or alternate (alt) alleles at 13 enhancer variants show evidence for differential binding (black stars) of nuclear factors from HL1 (Left) and AC16 (Right) cells at six enhancer variants but are abrogated in 20-bp Cy5-labeled probes carrying a 5-base deletion centered on the variant of interest (del).
High Specificity and Sensitivity of Reporter Screens for Enhancers.
We sought to assess the specificity of our reporter screen by evaluating the 40 enhancer CREs we identified with data on human open-chromatin regions (8, 9) using DNase hypersensitive sites (DHSs), a hallmark of cis-regulation (28). We divided all human tissues and cells for which DHS data are publicly available (8, 9) into two groups: a “cardiac” group from adult heart tissues only and a remaining “noncardiac” group. Of 40 CREs, nine and 31 elements mapped to cardiac and noncardiac DHSs, respectively (SI Appendix, Fig. S4 and Dataset S7), while of the 12 CREs with significant allelic differences, six and nine elements, respectively, mapped to these groups (SI Appendix, Fig. S4 and Dataset S7). Compared with expectations based on DHS overlap with the 40 CREs, the subset of 12 with significant allelic differences represents a 2.2-fold enrichment for the cardiac group but none for the noncardiac group (cardiac: χ2 = 4.03, P = 0.04; noncardiac: χ2 = 0.01, P = 0.92). To assess sensitivity, we assessed the activity of 10 of the original set of 98 elements with >50% sequence overlap with adult cardiac DHSs (9). In HL1, eight and two of these elements acted as enhancers/suppressors and were neutral in reporter assays; in AC16, these numbers were nine and one, respectively (Dataset S7), demonstrating a sensitivity of 80–90%.
Multiple Sources of Evidence Identify High-Confidence Causal CRE Variants.
For each of five independent QTi-associated signals, we detected at least one candidate causal CRE variant (Fig. 1): five (rs6810361, rs6771881, rs12490047, rs1805126, and rs7373779) were in LD with the sentinel rs6793245; two others (rs6772948 and rs41312411) were in LD with the secondary hit rs11708996; the secondary hit rs1171007 was itself a CRE variant; two variants (rs9880327 and rs13097780) tested together were in LD with the secondary hit rs6599234; and the secondary hit rs6801957 was itself a CRE variant, as shown previously (29, 30), but also in LD with CRE variants rs4076737 and rs6798015. As biological evidence of a high-confidence causal CRE variant, we considered any that scored “positive” on two of three biological features: binding in EMSA, overlap with cardiac DHS peak, or high delta support vector machine (deltaSVM) score (>0.9) (31) (Table 1). Consequently, we found five high-confidence causal CRE variants, one for each of the five independent QTi-associated signals: rs7373779 for rs6793245 sentinel variant, rs41312411 for rs11708996 secondary signal, rs11710077 for itself, rs13097780 for rs6599234 secondary signal, and rs6801957 for itself (Table 1).
Table 1.
Features of the 13 putative causal enhancer variants identified in the CRE screen
| Putative CRE variant | EMSA binding | Cardiac DHS peak | deltaSVM score* | Index variant and LD |
| rs6810361 | N | Y | 0.12 | rs6793245; 0.32 |
| rs6771881 | N | Y | 0.77 | rs6793245; 0.60 |
| rs12490047 | Y | N | 0.03 | rs6793245; 0.57 |
| rs1805126 | N | N | 1.16 | rs6793245; 0.76 |
| rs7373779 | Y | Y | 1.55 | rs6793245; 0.94 |
| rs6772948 | N | N | 0.92 | rs11708996; 0.59 |
| rs41312411 | Y | Y | 1.13 | rs11708996; 0.94 |
| rs11710077 | Y | Y | 1.35 | rs11710077; 1.00 |
| rs9880327 | Y | N | 0.02 | rs6599234; 0.69 |
| rs13097780 | Y | N | 1.36 | rs6599234; 0.41 |
| rs4076737 | N | N | 0.19 | rs6801957; 0.88 |
| rs6801957 | N | Y | 1.42 | rs6801957; 1.00 |
| rs6798015 | N | N | 1.09 | rs6801957; 0.76 |
N, no; Y, yes.
deltaSVM scores >0.9 are considered statistically significant and are set in boldface.
Multiple Enhancer Variants Predict SCN5A Cardiac Gene Expression and QTi Variation.
Noncoding causal variants are expected to influence gene expression of a nearby gene(s) within the topologically associating domain (TAD) encompassing these variants (32). The TAD of interest containing SCN5A and SCN10A also contains three additional genes (ACVR2B, EXOG, and SCN11A). Gene expression in human cardiac left ventricular tissue from the Genotype Tissue Expression (GTEx) project (n = 268) (33) failed to detect any expression quantitative trait locus (eQTL) for any of these genes. Because this result could arise from the low statistical power of eQTL detection, given the limited sample size, we next evaluated whether multiple variants could be better predictors (7). We first used standard multiple linear regression models to demonstrate that SCN5A expression in left ventricles was significantly associated with the five GWAS index SNPs taken together (adjusted R2 = 0.04, P = 6.4 × 10−3) (Fig. 4A and Dataset S8). It is important to point out that the direction of effect for rs6801957 on SCN5A cardiac expression in the multiple linear regression model (Dataset S8) was consistent with that reported previously (30). A similar association was also observed for SCN10A expression (adjusted R2 = 0.04, P = 5.4 × 10−3), although SCN10A shows far lower expression than SCN5A [transcripts per million (TPM) in the GTEx project, version 7: 0.044 vs. 36.02]. Of the five genes, only three have median TPM greater than 0.1 in left ventricles, and of these, only SCN5A exhibits significant association (ACVR2B: adjusted R2 = 0.003, P = 0.33; EXOG: adjusted R2 = 0.005, P = 0.29) (Dataset S8).
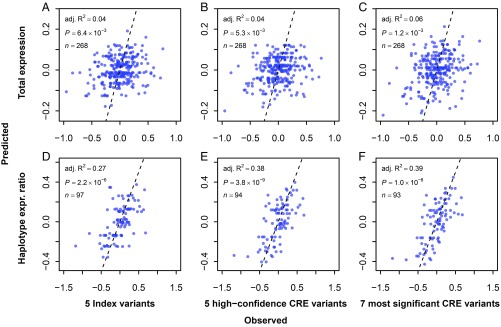
Fig. 4.
Cardiac gene expression of SCN5A is significantly associated with QTi GWAS and CRE variants. Normalized expression of SCN5A in human heart left ventricle tissue (n = 268; data from GTEx project, version 7) was compared with predicted expression from multivariable linear regression models using five independent index GWAS variants (A), five high-confidence causal CRE variants (B), and the most significant causal CRE variants that maximized the association (C). The circles represent normalized expression in each sample, with the regression fit as shown. (D–F) Repeated comparisons of the SCN5A haplotype expression (expr.) ratio to the predicted expression from multivariable linear regression models using the same sets of variants as A, B, and C, respectively.
Given multiple candidate causal variants and LD between SNPs, one can expect similar significant association for other combinations of CRE variants. Indeed, the five high-confidence causal CRE variants also achieved significant association with SCN5A comparable to the five sentinel SNPs (adjusted R2 = 0.04, P = 5.3 × 10−3) (Fig. 4B and Dataset S9). Thus, we asked whether any or all of the remaining eight candidate causal CRE variants can further improve the genotype–gene expression association. We included the five high-confidence causal CRE variants and then considered all 255 (i.e., 28−1) possible combinations of the remaining eight candidate causal CRE variants to build multivariable linear regression models and assess association. Of these, a set containing seven CRE variants (rs7373779, rs6772948, rs41312411, rs11710077, rs13097780, rs4076737, and rs6801957; henceforth referred to as the most significant causal CRE variants) achieved the highest statistical significance (adjusted R2 = 0.06, P = 1.2 × 10−3) (Fig. 4C and Dataset S9), suggesting that the two additional CRE variants, rs6772948 and rs4076737, also likely affect gene expression and the QTi phenotype.
We next assessed the CRE genotype and gene expression association by studying allelic imbalances of SCN5A cardiac expression. Thus, for each SCN5A heterozygous sample from the GTEx project, we estimated SCN5A expression by haplotype and calculated its ratio (“haplotype expression ratio”). We repeated the multivariable linear regression analysis using the five index variants, the five high-confidence causal CRE variants, and the seven most significant causal CRE variants. Interestingly, the five index SNPs are much more significantly associated with the haplotype expression ratio than with total expression (adjusted R2 = 0.27, P = 2.2 × 10−6; Fig. 4D). The five high-confidence causal CRE variants achieved an even better association (adjusted R2 = 0.38, P = 3.8 × 10−9; Fig. 4E) than the five index SNPs, but the set of seven causal CRE variants improved the association marginally (adjusted R2 = 0.39, P = 1.0 × 10−8; Fig. 4F). Importantly, the estimated effects of these variants are largely concordant between the two analyses (SI Appendix, Fig. S5). When comparing the total expression-based and haplotype expression-based models for the effects of the seven most significant causal CRE variants, five of them showed the same direction of effect, while the remainder have near-zero beta values in the total expression-based model. Thus, the signals captured in the two models are largely shared even though their statistics are completely independent.
Finally, we asked whether SCN5A cardiac expression also correlates with QTi. Since no existing study has cardiac gene expression and QTi values in the same individuals, we instead predicted (or imputed) SCN5A expression from genotypes in the Atherosclerosis Risk in Communities (ARIC) study, where QTi phenotypes are also available (15, 34). Although this approach is similar to PrediXcan (35), we differ in that only potential regulatory variants are used as predictors. Both models for the seven most significant causal CRE variants achieved significant correlation between predicted SCN5A expression and QTi, but the haplotype expression-based model predicted QTi better than the total expression-based model (P < 1.6 × 10−7 versus P < 1.5 × 10−10). Note that the QTi variation explained by SCN5A expression variation (R2 = 0.005) is not trivial, given that many genetic, as well as environmental, factors influence the QTi (SI Appendix, Fig. S6).
Adequacy of AC16 and HL1 as Cardiac Cellular Models.
The genetic studies we conducted critically depend on cellular models that can serve as adequate surrogates for human tissue transcriptional systems. To assess AC16 (human) and HL1 (mouse) as cardiomyocyte models, we performed RNA-seq (23) and ATAC-seq (24) to generate gene expression and open-chromatin maps, respectively, and systematically compared them with available data from various tissues and primary cells of human and mouse origin. Gene expression profiles of both cell lines were moderately to highly correlated to many matched human and mouse tissues, including the heart (36, 37) (SI Appendix, Fig. S7 A, B, E, and F and Table S2). We suspect that these widespread correlations arise from the many genes commonly expressed in multiple tissues. Thus, we repeated this analysis after excluding all commonly expressed genes (defined as expression >1 TPM in at least 50% of samples), significantly reducing the correlation for all tissues in both cell lines (one-tailed P < 2.2 × 10−16 for all tissues in both cell lines based on Fisher r-to-z transformation). Surprisingly, the HL1 gene expression profile showed the highest correlation to mouse heart tissue, while AC16 did not, suggesting that HL1 may be physiologically more relevant to cardiac analysis than AC16 (SI Appendix, Fig. S7 C, D, G, and H). To further probe this aspect, we compared AC16 gene expression with publicly available human primary cell gene expression data (8) (SI Appendix, Table S3). Here again, reasonably high Pearson correlation coefficients with all primary cells were observed (SI Appendix, Fig. S8A), but removal of commonly expressed genes revealed that AC16 was most significantly correlated with fibroblasts, followed by skeletal muscle myoblasts (no data were available from primary cardiomyocytes) (SI Appendix, Fig. S8B). These results support the fact that AC16 cells, generated by fusing SV40-transformed human skin fibroblasts lacking mitochondria with nonproliferating ventricular primary cardiomyocytes (21), transcriptionally appear fibroblast-like even though they express many well-established cardiac markers.
Finally, we compared open-chromatin maps of AC16 and HL1 with available data on human and mouse tissues and primary cells (9, 37) (SI Appendix, Tables S4 and S5). In the top-scoring 50,000 open-chromatin regions in all tissues and primary cells evaluated, consistent with the above data, we also discovered that HL1 most significantly overlaps features in the mouse heart (SI Appendix, Fig. S9A), while AC16 does so with human skin fibroblasts (SI Appendix, Fig. S9B). Thus, for in vitro studies of enhancer function in cardiomyocytes, the mouse HL1 appears to be a better transcriptional cell line model than the human AC16.
Discussion
Significant variants from GWAS are commonly annotated using epigenomic data from genome-wide analyses, such as in the ENCODE (8) and NIH RoadMap Epigenomic (9) projects. This is because the major assumption underlying GWAS is the presence of one or more variants disrupting CREs controlling a target gene’s expression and, thereby, a downstream phenotype. However, such annotations are not comprehensive because (i) many of these CRE effects are cell type-specific and (ii) enhancer effects can be unique or redundant effects (shadow enhancers), as well as continuous or stage-specific, situations not well represented in a single genome-wide survey (38). Also, trait associations can arise from multiple variants that are functionally, but not necessarily genetically, independent (7, 39). Thus, deep studies of individual genes/loci are an important adjunct to understand the diversity of enhancers, their variants, and their phenotypic effects, exemplified here for SCN5A using QTi-associated common variants. The method we outline in this study is one of several approaches one can take. Further, additional in-depth work is necessary to understand the role of multiple causal CRE variants.
In this study, in contrast to other investigations, we comprehensively examine the DNA containing every QTi-associated common variant for enhancer function, using three biological criteria: allele-specific CRE activity, EMSA binding, and predicted effects on chromatin openness (DNaseI hypersensitivity). We identify five high-confidence causal CRE variants collectively associated with SCN5A cardiac gene expression to the same level as the five independent index GWAS variants. However, the best statistical model for explaining SCN5A gene expression includes two additional CRE variants, beyond the above five, based on functional, rather than statistical, data. Thus, at this association locus, we have identified a set of seven CRE variants that can explain the QTi GWAS signal on chromosome 3p22.2. The alleles that prolong QTi increase SCN5A expression for all five GWAS hits except one (rs11708996) (Dataset S10), likely a false-negative finding owing to the small sample size in eQTL analysis. However, the QTi-prolonging alleles of functional CRE variants are not necessarily concordant with the direction of enhancer activity or SCN5A gene expression. This feature is not unexpected, because variant effects on the trait (SCN5A or QTi) are from the ensemble of causal variants, rather than an individual SNP.
As opposed to most GWAS loci, where the target gene is unknown, we selected to identify CREs and their variants for SCN5A using QTi-associated GWAS variants because of SCN5A’s role in QTi regulation (12, 13). However, we also evaluated the four other genes within the TAD encompassing the QTi GWAS signals at this locus. Immediately downstream of SCN5A is SCN10A, a gene that encodes a nociceptor-associated, voltage-gated sodium channel alpha subunit. Based on our eQTL analyses, we cannot completely rule out SCN10A as an additional target gene regulating QTi, but its role in PR and QRS intervals is debatable and remains mechanistically unclear (40). The three other genes are unrelated to QTi.
As shown here, GWAS associations can arise from multiple CRE variants that are functionally, but may not be genetically, independent. At both the population and individual levels then, the genetic effects on the phenotype need to be understood as arising from an ensemble of TFs that bind to multiple CREs (7). As a consequence, a specific gene expression level can arise from many different CREs, different CRE genotypes, and therefore different TF occupancy profiles (5). A corollary is that some combinations create a bigger gene expression effect than others, leading to an ascertainment bias of detecting CREs with strong allelic activity differences when assessed through a trait association. Comprehensive CRE detection, such as performed here, can therefore lead to uncovering a more unbiased set of variants and CREs (SI Appendix, Fig. S10). Gene expression regulation is largely (∼60% of variation) through variable CRE activity; however, genetic variations at transregulatory sites, microRNA (miR) binding sites, and sequences regulating posttranscriptional and translational processing are additional causal factors. Consider that rs1805126, with enhancer activity in HL1 cells (Table 1), is also a site for common SCN5A synonymous variation associated with cardiac electrophysiological parameters and modulating miR-24–driven regulation of SCN5A gene expression (41). This suggests that our CRE-based screen may have failed to detect other causal variants that do not also alter CRE activity.
Our in vitro reporter assays for CRE detection used two cardiomyocyte-like cell lines, because cardiomyocytes are the primary cells underlying cardiac impulse generation and propagation (42), rather than published epigenomic information only. As we show by their genomic characterization, the human cell line AC16 is less cardiomyocyte-like than the mouse cell line HL1, justifying the use of both. Also within the SCN5A QTi GWAS locus we evaluated, we identified 22 ATAC-seq peaks in AC16 cells, of which 13 (59%) overlapped human cardiac DHS peaks and all but one (95%) overlapped human noncardiac DHS peaks (Dataset S11). Nevertheless, the functional data show high correlation for allelic activity and expression effect (enhancer/suppressor/neutral) between AC16 and HL1. Since no single cell line is expected to be a perfect proxy for cardiomyocytes in vivo, using multiple cell lines can throw a wider net to identify relevant CREs. However, genomic characterization of the cell lines used is necessary.
This study focused on one locus and tested ∼100 sequence elements using the classical luciferase enzymatic readout for CRE detection; it also allowed us to test longer elements (median = 397 bp) that can capture the typical size of enhancers. However, expanding this approach to identify CREs across many genes/loci is only practical using higher throughput methods, such as the massively parallel reporter assay (MPRA) (43). However, due to current technical limitations in high-density oligonucleotide synthesis, current MPRA designs can only evaluate thousands of elements of ∼150 bp, limiting their use in CRE/enhancer screens. Therefore, for CRE identification, each of these approaches has its complementary advantages. However, both of these approaches suffer from technical artifacts. First, false-positive findings in reporter assays are not uncommon, which is why we require additional functional criteria (open-chromatin overlap, binding of nuclear factors, and predicted impact of variants on DNaseI sensitivity) for CRE identification. Second, reporter assays can also identify apparent suppressors of reporter activity. In this study, AC16 and HL1 identified 59 unique suppressor elements. Given that the reporter activity of each test element evaluated was in the context of a minimal TATA-box promoter in pGL4.23, observing this large number of suppressive elements was surprising. We hypothesize that this silencing effect is due to positional effects of heterochromatinized test elements. We believe this to be an artifact because 31 (53%) of the 59 suppressors overlap known DHS peaks in one or more human tissues/cells, close to its expectation of ∼42% (χ2 = 1.6, P = 0.21) (2). In contrast, of the 40 enhancers identified across AC16 and HL1, 32 (80%) overlap known DHS peaks in one or more human tissues/cells, a twofold enrichment over expectation (χ2 = 13.8, P = 2.0 × 10−4). Therefore, broader studies of the kind reported here are necessary to delineate how trait associations arise from noncoding genetic variation.
Materials and Methods
Variant Selection.
Our analysis focused on the sentinel variant, rs6793245, and four other independent association signals, rs11708996, rs11710077, rs6599234, and rs6801957, at the SCN5A-SCN10A locus; these were associated with the QTi in a GWAS meta-analysis of individuals of European ancestry (15). We defined target regions around these variants using recombination hotspots (recombination rate >10 cM/Mb) identified in the HapMap phase II genetic map (25) (ftp://ftp.ncbi.nlm.nih.gov/hapmap/recombination/2011-01_phaseII_B37/). Within this region, we selected all common (MAF > 5%) variants observed in the 1000 Genomes Project (26) European ancestry samples (n = 379; ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/) that were in moderate to high LD (r2 > 0.3) with any of the five variants. LD was calculated by VCFtools (vcftools.sourceforge.net/) using the 1000 Genome Project phased genotypes.
Amplicon Design.
Genomic sequences flanking selected variants (±325 bp) were obtained from the UCSC Genome browser (https://genome.ucsc.edu/cgi-bin/hgGateway; hg19) and used as input for primer design. Primers were designed using Primer3 (27) (bioinfo.ut.ee/primer3-0.4.0/) in batch mode (all default settings except the following: mispriming library human; primer size minimum 20, optimum 25, maximum 27; primer Tm minimum 55, optimum 65, maximum 70; maximum Tm difference 5; primer GC minimum 30, optimum 50, maximum 60; primer maximum polynucleotide 3; primer GC clamp 0/1). Primer pairs were evaluated for specificity against the human reference genome template using the Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to retain only those that mapped to a single target. For In-Fusion cloning of amplicons into the pGL4.23 vector (Promega) between the KpnI and XhoI sites, TGGCCTAACTGGCCGGTACC vector homologous sequence was added to the 5′ end of all forward primers and TCTTGATATCCTCGAG vector homologous sequence was added to the 5′ end of all reverse primers.
Amplification and Cloning.
All steps of amplification and cloning were performed in 96-well and 24-well formats. Variant-centered amplicons were PCR-amplified using genomic DNA from 1000 Genomes Project samples (26) that were homozygous for the reference or alternate allele at each variant. PCR was performed in a 50-μL volume containing 50 ng of genomic DNA, 0.2 mM (each) dNTP, 500 nM forward primer, 500 nM reverse primer, 1 unit of Phusion High-Fidelity DNA Polymerase (New England Biolabs), and 1× Phusion HF buffer (New England Biolabs). Thermal cycling was performed as follows: initial denaturation at 98 °C for 1 min, 35 cycles of denaturation at 98 °C for 10 s, annealing at 65 °C for 15 s, extension at 72 °C for 20 s, and final extension at 72 °C for 5 min. An aliquot (5 μL) of PCR product was visualized by 2% agarose TAE (Tris base, acetic acid, and EDTA) gel electrophoresis. PCR products were purified using AMPure XP magnetic beads (Beckman Coulter) following the manufacturer’s recommendations, and the final elution was performed using 30 μL of 10 mM Tris pH 8.0. The pGL4.23 vector (Promega) was linearized by double digestion with KpnI-HF (New England Biolabs) and XhoI (New England Biolabs) and gel-purified using a QIAquick Gel Extraction Kit (Qiagen). Purified PCR products were cloned into linearized pGL42.3 vector using an In-Fusion HD EcoDry Cloning Kit (Clontech) following the manufacturer’s recommendations. Briefly, 10–50 ng of purified PCR product (2 μL postelution) was mixed with 50 ng of linearized pGL4.23 vector in a total volume of 10 μL, added to the In-Fusion HD EcoDry pellet, and mixed by pipetting. The In-Fusion reaction mixture was incubated at 37 °C for 15 min, followed by 50 °C for 15 min, and was then cooled on ice. Following a 10-fold dilution with 10 mM Tris pH 8.0, 1 μL of the diluted In-Fusion reaction mixture was used to transform 20 μL of Stellar competent cells (Clontech). Of the 100-μL transformation mixture [after addition of 80 μL of SOC medium (Thermo Fisher Scientific)], 2 μL was diluted with 23 μL of SOC medium, spread on selective medium [LB agar + ampicillin (50 µg/mL)], and incubated overnight at 37 °C. Bacterial colonies were inoculated in LB broth + ampicillin, incubated at 37 °C and 225 rpm overnight, and then harvested for glycerol stock and plasmid preparations. Positive clones for each allele of each variant were identified by restriction digestion (KpnI and XhoI) and Sanger sequencing of plasmid DNA using pGL4.23 vector backbone primers.
Cell Culture.
Mouse cardiomyocyte HL1 cells (a gift from William C. Claycomb, Louisiana State University, New Orleans) were maintained in Claycomb medium (Sigma) (22). Human cardiomyocyte AC16 cells (Mercy P. Davidson, Columbia University, New York) were maintained in DMEM/F-12 (Thermo Fisher Scientific) as described (21).
Reporter Assays.
To assess cis-regulatory activity, each test element (variant-centered amplicon) was cloned upstream of a minimal promoter driving firefly luciferase gene in the pGL4.23 vector (Promega). Reporter constructs were transfected into HL1 and AC16 cells, grown in 24-well plates at ∼90% confluency, using FuGENE HD Transfection Reagent (Promega) following the manufacturer’s recommendations. pRLSV40 (Promega), expressing Renilla luciferase, was cotransfected to normalize for transfection efficiency. Twenty-four hours after transfection, cells were harvested and lysed, and firefly and Renilla luciferase activities were measured on the Infinite 200Pro multiplate reader (Tecan) using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s protocols. Luciferase activity from each test construct was measured in three replicates, except for the empty vector construct, which was measured in six replicates. For each measurement, the observed firefly luciferase reading was divided by the observed Renilla luciferase reading to get relative firefly activity. Relative firefly activity was divided by the average relative firefly activity from empty vector, and then averaged across replicates, to obtain the mean normalized reporter activity for each test construct. To compare reporter assays between two cell lines, the log10-transformed mean normalized reporter activity for the reference and alternate allele in one cell line was compared with the log10-transformed mean normalized reporter activity for the reference and alternate allele in the other cell line. To assess enhancer, suppressor, or neutral activity, the relative firefly activity from each measurement was log2-transformed and a standardized z-score was calculated based on the mean and SD of log2-transformed relative firefly activity from empty vector. The z-scores across replicates of a test construct were averaged: constructs with an average z-score value greater than 2.326 (99th percentile of a standard normal distribution) were called enhancers, and constructs with an average z-score value less than −2.326 (analogous first percentile) were called suppressors; other elements were neutral. The significance of the allelic difference for enhancer constructs was evaluated by comparing the log2-transformed relative firefly activities of the two alleles using a Student’s t test.
EMSAs.
Nuclear extracts from HL1 and AC16 cells were prepared with NE-PER Nuclear and Cytoplasmic Extraction Kits (Thermo Fisher Scientific) following the manufacturer’s instructions. Twenty-five–base–long sense (3′ Cy5-labeled) and antisense oligos centered on the variants of interest (for both alleles), along with 20-base-long sense (3′ Cy5-labeled) and antisense oligos carrying a 5-base deletion centered on the variant of interest, based on the reference human genome sequence (UCSC Genome Browser, https://genome.ucsc.edu/cgi-bin/hgGateway), were synthesized (Integrated DNA Technologies). Equimolar amounts of complementary oligonucleotides were mixed and annealed in 10 mM Tris pH 7.4 using a thermal cycler [95 °C for 5 min, −1 °C per cycle with a 1-min incubation at each cycle (70 cycles), and 25 °C for 5 min] to generate double-stranded fluorescent-labeled probes. For binding assays, 2 μL of labeled probe (10 nM) was incubated with 4 μL of the nuclear extract and 1 μg of poly(deoxyinosinic–deoxycytidylic) in a buffer containing 10 mM Tris pH 8.0, 0.1 mg/mL BSA, 50 μM ZnCl2, 100 mM KCl, 10% glycerol, and 0.1% IGEPAL CA-630 in a 20-μL reaction, and incubated in the dark for 1 h at 4 °C. Protein–DNA complexes were resolved by running them on a nondenaturing 8% polyacrylamide 1× Tris-glycine gel (prerun for 1 h at 100 V) for ∼30 min at 200 V, with fluorescence detected using a Typhoon 9400 Imager (GE Amersham).
Overlap with DHS Datasets and DeltaSVM Scores.
To evaluate overlap of each cis-element with published DHS data, we used all available DNase-sequencing (DNase-seq) data (n = 799) from the ENCODE (8) (https://www.encodeproject.org/) and NIH Roadmap Epigenomics (9) (www.roadmapepigenomics.org/) projects. We also included DNase-seq data from two adult human heart samples we generated recently (44). For each experiment, DHS peaks were called using MACS2 (version 2.1.1) (45) and an overlap was declared when >50% of a test element was part of a DHS peak. Overlap analysis was performed against DHS peaks called in (i) samples from adult heart tissues only (cardiac set; n = 5) and (ii) samples from all human tissues and cells except adult heart tissues (noncardiac set; n = 796) as described (44). We also used deltaSVM scores for selected variants from the generic cardiac gkm-SVM model in the same study (44).
In Silico Prediction of TF Binding Sites.
For each of the 13 candidate causal CRE variants, we generated two 21-base-long sequences centered at the SNP, corresponding to the reference and alternate alleles. We scanned these 26 sequences using FIMO (46) against the JASPAR CORE vertebrates (nonredundant) database (2018; meme-suite.org/db/motifs), with default settings. The database contains 579 manually curated and nonredundant motifs.
GTEx eQTL Analyses.
Raw read counts per transcript for 272 heart left ventricle samples (GTEx_Analysis_2016-01-15_v7_RSEMv1.2.22_transcript_expected_count.txt.gz) were obtained from the GTEx portal (33) (https://www.gtexportal.org/home/; version 7), as were the first three principal components of genotypes, gender, and genotyping platform. Theses raw counts were aggregated to obtain raw read counts per gene, which were subjected to variance stabilizing transformation (VST) as implemented in DESeq2 (47). Genes with at least three VST-normalized read counts in at least 20% of the samples (at least 55 samples) were retained. We then used the probabilistic estimation of expression residuals (PEER) method (48) to account for hidden covariates (i.e., batch effects) in the expression data. We obtained the first 40 PEER factors using the VST-normalized read counts as expression input and the three principal components from genotypes, gender, and genotyping platform as covariates. After removing four outliers (>3 SD from the mean of the VST-normalized read counts), we regressed out the 45 covariates (40 PEER factors, three principal components, gender, and platform) and performed eQTL analysis using residuals as normalized gene expression values. We built standard multivariable linear regression models using genotypes from the GTEx dbGaP database via authorized access (accession no. phs000424.v7.p2). All genotypes were identified by whole-genome sequencing and coded for alternate alleles. Therefore, the sign of beta values (effect sizes) is always relative to their alternative alleles.
To calculate haplotype expression ratios, allele-specific expression (ASE) data for SCN5A in the 220 heart left ventricle samples that have at least one heterozygous ASE SNP were extracted from the GTEx dbGaP database via authorized access. Since phased genotypes from whole-genome sequencing data are not yet available, we instead obtained phased (and imputed) genotypes from the array data for 152 of the 220 samples. We further filtered out 12 additional samples in which no phased genotypes were available for their ASE SNPs, resulting in 140 samples. Then, for each of these samples, we aggregated the read counts of the SCN5A ASE SNPs by haplotype to estimate haplotype expression levels and calculated the ratio between the two haplotypes. We excluded samples that significantly deviate from the mean of the expression ratio (>3 SD). As for the independent variables, we calculated the difference in allele identities for each test SNP between the two haplotypes using phased genotypes. We note that any samples with unphased genotypes for the independent variables could not be used in our analysis and were removed. Similar to the previous total expression-based analysis, we built multivariable linear regression models to predict haplotype expression ratio from the same sets of variants. To evaluate the effect of noise in the ASE read counts, we repeated the regression analysis using different thresholds for the minimum haplotype read counts (0, 30, 50, and 100) (SI Appendix, Fig. S11) and determined that 100 read counts were an optimal threshold.
Comparison of QTi and SCN5A Expression Using the ARIC Study Data.
We studied 8,046 European ancestry subjects in whom we had access to both QTi phenotype and imputed genotype data. For sample quality control, we adopted an established pipeline from our previous study (15, 49). We corrected the raw QTi ECG measure for three covariates: heart rate (calculated from RR interval on ECG), age and gender, as previously described (15, 49); regression residuals were used as the “normalized QTi.” We then calculated the predicted gene expression using the multivariable regression model of the seven most significant causal CRE variants.
Cell Line RNA-Seq.
Total RNA was isolated from HL1 and AC16 cells using a RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions, including the on-column DNase treatment using the RNase-Free DNase set (Qiagen). RNA quality was assessed using a Bioanalyzer RNA Kit (Agilent Technologies) before preparing sequencing libraries. An Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina) was used to generate indexed sequencing libraries following the manufacturer’s protocols. A sample from each library was used to assess library fragment size distribution by electrophoresis, using a BioAnalyzer High Sensitivity DNA Assay (Agilent Technologies), and to assess library concentration by qPCR, using a KAPA Library Quantification Kit (KAPA Biosystems). Equimolar amounts of libraries were pooled and sequenced on an Illumina HiSeq 2500 instrument using standard protocols for paired-end 100-bp reads with a desired sequencing depth of ∼75 million paired-end reads per library. RNA-seq for both cell lines was performed in technical duplicates.
Cell Line RNA-Seq Analyses.
We estimated the abundance of mRNA transcripts for each of the replicates using Kallisto (50). For estimating gene and transcript expression levels, we used the GENCODE human transcript (release 25 mapped to GRCh37) and mouse transcript (release 12 mapped to GRCm38) sequences as references (gencode.v25lift37.transcripts.fa.gz and gencode.vM12.transcripts.fa.gz; https://www.gencodegenes.org/). TPM values were estimated using Kallisto (50) directly from the raw reads, with default settings, and aggregated to obtain gene-level TPMs for downstream analysis. To compare gene expression profiles of AC16 and HL1 cell lines with human and mouse tissues and primary cells, we calculated the Pearson correlation coefficients between gene level TPMs using public RNA-seq data for a broad set of human tissues/primary cells and mouse tissues downloaded from the mouse ENCODE project (37) (www.mouseencode.org/). Specifically, we obtained the raw read FASTQ RNA-seq files for matched human and mouse tissues (36) and quantified gene level TPMs using the same pipeline as above. For primary human cells, we obtained gene level TPMs from the ENCODE project (8) (https://www.encodeproject.org/). We applied the log2 transformation to TPMs with one pseudocount, and only considered protein coding genes for correlation analysis. Average TPM values of the AC16 and HL1 technical replicates were used for analysis. We also repeated the correlation analysis after removing commonly expressed genes, defined as genes with TPM > 1 in at least 50% of samples (six for tissues and 13 for primary cells). All HL1 and AC16 RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and are accessible at accession no. GSE109716.
ATAC-Seq and Analysis.
ATAC-seq was performed on ∼50,000 fresh unfixed HL1 and AC16 cells each, using published protocols (24). A sample from each indexed library was used to assess library fragment size distribution by 2% agarose TAE gel electrophoresis and to assess library concentration by qPCR using the KAPA Library Quantification Kit (KAPA Biosystems). Equimolar amounts of libraries were pooled and sequenced on an Illumina HiSeq 2500 instrument using standard protocols for 50-bp paired-end sequencing, with a desired sequencing depth of ∼150 million paired-end reads per library.
For primary data processing and peak calling, we adopted existing workflows (24, 51) with some modifications. Briefly, we aligned the paired-end reads to the reference genome (hg19 for human and mm9 for mouse) using Bowtie2 (version 2.2.3) (52) with options “-X 2000 --dovetail --no-mixed --no-discordant -t” after trimming the adapter sequences (CTGTCTCTTATACACATCT) from the raw reads using Cutadapt (version 1.12) (53) with parameters “-q 3,3 -m 35.” Duplicated reads were removed using SAMtools (version 1.3.1) (54) with the “rmdup” command, and only properly paired and mapped reads with a mapping quality score >30 were retained for peak calling. All reads mapping to the + or − strand were offset by +5 bp and −4 bp, respectively, to account for the 9-bp duplication of the target site by Tn5 transposase (55). Next, we identified peaks using MACS2 (version 2.1.1) (45) with the option “--nomodel --shift -50 --extsize 100 --keep-dup all” and defined open-chromatin regions as 600-bp regions centered at the summits of MACS2 peaks; overlapping regions were merged. To compare these regions from AC16 and HL1 cell lines with human and mouse tissues and primary cells, we used public DNase-seq data from the NIH Roadmap Epigenomics (9) (www.roadmapepigenomics.org/) and mouse ENCODE (37) (www.mouseencode.org/) projects. Specifically, we obtained mapped reads of DNase-seq experiments from diverse human and mouse tissues and primary cells, and called DHS peaks and defined open-chromatin regions as above. For a fair comparison, we further selected the top 50,000 regions from each of the datasets based on MACS2 P values. We evaluated the similarities between the open-chromatin regions of AC16 and HL1 cell lines and human/mouse tissues and primary cells using the Jaccard index (number of bases in their intersection over their union). All HL1 and AC16 ATAC-seq data have been deposited in the NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) and are accessible at accession no. GSE109716.
Supplementary Material
Acknowledgments
We thank William C. Claycomb (deceased; Louisiana State University) for providing HL1 cells. We thank the staff and participants of the ARIC study for their important contributions. AC16 cells were received under an institutional material transfer agreement with Mercy P. Davidson (Columbia University). Illumina sequencing was performed at the Genetic Resources Core Facility (Johns Hopkins Institute of Genetic Medicine). This work was supported by US NIH Grant R01 GM104469 (to A.C.). The ARIC study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, NIH, Department of Health and Human Services, under contract (Contracts HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, and HHSN268201700004I), Grant TR01HL087641, and Grant R01HL086694; National Human Genome Research Institute Contract U01HG004402; and NIH Contract HHSN268200625226C. Infrastructure was partly supported by Grant UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research.
Footnotes
Conflict of interest statement: E.Z.S., M.L.G., E.B., D.E.A., and A.C. have coauthored multiple papers with C.R.B. and K.L.M. in the past 48 months. E.Z.S., M.L.G., E.B., D.E.A., and A.C. and the reviewers coauthored these papers as part of a consortium; they did not collaborate directly on the papers.
Data deposition: All HL1 and AC16 RNA-sequencing and ATAC-sequencing (assay for transposase-accessible chromatin using sequencing) data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database https://www.ncbi.nlm.nih.gov/geo (accession no. GSE109716).
See Profile on page 10608.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808734116/-/DCSupplemental.
References
- 1.Wray GA. (2007) The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8:206–216. [DOI] [PubMed] [Google Scholar]
- 2.Maurano MT, et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert FW, Kruglyak L (2015) The role of regulatory variation in complex traits and disease. Nat Rev Genet 16:197–212. [DOI] [PubMed] [Google Scholar]
- 4.Maston GA, Evans SK, Green MR (2006) Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet 7:29–59. [DOI] [PubMed] [Google Scholar]
- 5.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U (2008) Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451:535–540. [DOI] [PubMed] [Google Scholar]
- 6.Asthana S, et al. (2007) Widely distributed noncoding purifying selection in the human genome. Proc Natl Acad Sci USA 104:12410–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee S, et al. (2016) Enhancer variants synergistically drive dysfunction of a gene regulatory network in Hirschsprung disease. Cell 167:355–368.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kundaje A, et al. ; Roadmap Epigenomics Consortium (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton-Cheh C, et al. (2005) QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: The Framingham Heart Study. Heart Rhythm 2:277–284. [DOI] [PubMed] [Google Scholar]
- 11.Dekker JM, Crow RS, Hannan PJ, Schouten EG, Folsom AR; ARIC Study (2004) Heart rate-corrected QT interval prolongation predicts risk of coronary heart disease in black and white middle-aged men and women: The ARIC study. J Am Coll Cardiol 43:565–571. [DOI] [PubMed] [Google Scholar]
- 12.Priori SG, Napolitano C (2004) Genetics of cardiac arrhythmias and sudden cardiac death. Ann N Y Acad Sci 1015:96–110. [DOI] [PubMed] [Google Scholar]
- 13.Arking DE, Chugh SS, Chakravarti A, Spooner PM (2004) Genomics in sudden cardiac death. Circ Res 94:712–723. [DOI] [PubMed] [Google Scholar]
- 14.Veerman CC, Wilde AA, Lodder EM (2015) The cardiac sodium channel gene SCN5A and its gene product NaV1.5: Role in physiology and pathophysiology. Gene 573:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arking DE, et al. ; CARe Consortium; COGENT Consortium; DCCT/EDIC; eMERGE Consortium; HRGEN Consortium (2014) Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet 46:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeufer A, et al. (2010) Genome-wide association study of PR interval. Nat Genet 42:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotoodehnia N, et al. (2010) Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet 42:1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers JC, et al. (2010) Genetic variation in SCN10A influences cardiac conduction. Nat Genet 42:149–152. [DOI] [PubMed] [Google Scholar]
- 19.Jabbari J, et al. (2015) Common and rare variants in SCN10A modulate the risk of atrial fibrillation. Circ Cardiovasc Genet 8:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bezzina CR, et al. (2013) Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet 45:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson MM, et al. (2005) Novel cell lines derived from adult human ventricular cardiomyocytes. J Mol Cell Cardiol 39:133–147. [DOI] [PubMed] [Google Scholar]
- 22.Claycomb WC, et al. (1998) HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA 95:2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. [DOI] [PubMed] [Google Scholar]
- 24.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frazer KA, et al. ; International HapMap Consortium (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.1000 Genomes Project Consortium , et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Untergasser A, et al. (2012) Primer3–New capabilities and interfaces. Nucleic Acids Res 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross DS, Garrard WT (1988) Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57:159–197. [DOI] [PubMed] [Google Scholar]
- 29.van den Boogaard M, et al. (2012) Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest 122:2519–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Boogaard M, et al. (2014) A common genetic variant within SCN10A modulates cardiac SCN5A expression. J Clin Invest 124:1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee D, et al. (2015) A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 47:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao SS, et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.GTEx Consortium (2015) Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The ARIC Investigators (1989) The Atherosclerosis Risk in Communities (ARIC) Study: Design and objectives. The ARIC investigators. Am J Epidemiol 129:687–702. [PubMed] [Google Scholar]
- 35.Gamazon ER, et al. ; GTEx Consortium (2015) A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S, et al. (2014) Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci USA 111:17224–17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue F, et al. ; Mouse ENCODE Consortium (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature 515:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakravarti A, Turner TN (2016) Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. BioEssays 38:578–586. [DOI] [PubMed] [Google Scholar]
- 39.Boyle EA, Li YI, Pritchard JK (2017) An expanded view of complex traits: From polygenic to omnigenic. Cell 169:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park DS, Fishman GI (2014) Nav-igating through a complex landscape: SCN10A and cardiac conduction. J Clin Invest 124:1460–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, et al. (2018) A common variant alters SCN5A-miR-24 interaction and associates with heart failure mortality. J Clin Invest 128:1154–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nerbonne JM, Kass RS (2005) Molecular physiology of cardiac repolarization. Physiol Rev 85:1205–1253. [DOI] [PubMed] [Google Scholar]
- 43.Patwardhan RP, et al. (2012) Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol 30:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee D, et al. (2018) Human cardiac cis-regulatory elements, their cognate transcription factors, and regulatory DNA sequence variants. Genome Res 28:1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. (2008) Model-based analysis of ChIP-seq (MACS). Genome Biol 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grant CE, Bailey TL, Noble WS (2011) FIMO: Scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stegle O, Parts L, Piipari M, Winn J, Durbin R (2012) Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 7:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapoor A, et al. (2016) Rare coding TTN variants are associated with electrocardiographic QT interval in the general population. Sci Rep 6:28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. [DOI] [PubMed] [Google Scholar]
- 51.Mo A, et al. (2016) Epigenomic landscapes of retinal rods and cones. eLife 5:e11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. [Google Scholar]
- 54.Li H, et al. ; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reznikoff WS. (2008) Transposon Tn5. Annu Rev Genet 42:269–286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.