Significance
The most important task of the mammalian zygote is to globally remodel its epigenome. Chromosomes that carried out sperm or oocyte functions are remodeled toward directing the unique totipotent state that characterizes the healthy preimplantation-stage embryo. The guiding differences in cell functions are marked on the chromosomes by interdependent epigenetic modifications including histone and DNA methylation, which both undergo global changes in the zygote. By performing genetic experiments, we found that two maternally supplied histone methyltransferase enzymes play important roles in the zygote in specifying the process of global DNA remodeling by 5-methylcytosine oxidation.
Keywords: maternal effect, 5-hydroxymethylcytosine, 5-methylcytosine, EHMT2, STEDB1
Abstract
Genome-wide DNA “demethylation” in the zygote involves global TET3-mediated oxidation of 5‐methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in the paternal pronucleus. Asymmetrically enriched histone H3K9 methylation in the maternal pronucleus was suggested to protect the underlying DNA from 5mC conversion. We hypothesized that an H3K9 methyltransferase enzyme, either EHMT2 or SETDB1, must be expressed in the oocyte to specify the asymmetry of 5mC oxidation. To test these possibilities, we genetically deleted the catalytic domain of either EHMT2 or SETDB1 in growing oocytes and achieved significant reduction of global H3K9me2 or H3K9me3 levels, respectively, in the maternal pronucleus. We found that the asymmetry of global 5mC oxidation was significantly reduced in the zygotes that carried maternal mutation of either the Ehmt2 or Setdb1 genes. Whereas the levels of 5hmC, 5fC, and 5caC increased, 5mC levels decreased in the mutant maternal pronuclei. H3K9me3-rich rings around the nucleolar-like bodies retained 5mC in the maternal mutant zygotes, suggesting that the pericentromeric heterochromatin regions are protected from DNA demethylation independently of EHMT2 and SETDB1. We observed that the maternal pronuclei expanded in size in the mutant zygotes and contained a significantly increased number of nucleolar-like bodies compared with normal zygotes. These findings suggest that oocyte-derived EHMT2 and SETDB1 enzymes have roles in regulating 5mC oxidation and in the structural aspects of zygote development.
The fertilized egg is the bottleneck between generations and also the gateway between the germ line and the soma. Germ-line–specific programs of cell function culminate in the male and female gametes and give way to the totipotency program after fertilization. Cell functions are governed by gene regulatory mechanisms that include DNA CpG methylation and histone covalent modifications. Both mechanisms undergo global remodeling in the mammalian zygote, together with structural changes (1). The maternally and paternally inherited genomes exhibit asymmetries from the start in the mammalian zygote as a consequence of the differential epigenetic reprogramming during oogenesis and spermatogenesis (2–8). The asymmetry continues after fertilization when the two half genomes are organized separately in the paternal and maternal pronuclei and treated differently by the epigenetic machinery deposited in the oocyte (9). The paternally inherited chromosomes that arrive from the sperm are packaged in protamines, replaced by transitory histones after fertilization, and then finally replaced by canonical histones after replication of the DNA. The maternally inherited chromosomes arrive from the oocyte packaged in nucleosomes and covalently modified histones. At the zygote stage, the two parental pronuclei can be distinguished by immunostaining against histone marks (10); for example, global H3K9 methylation levels show asymmetrical, highly parental-specific patterns. H3K9me2 and H3K9me3 immunostaining is detected exclusively in the maternal pronucleus (11–16). Another asymmetry is apparent at the level of DNA methylation: the chromosomes arrive from the sperm in a highly methylated form and from the oocyte with somewhat less 5mC (8). Global loss of DNA methylation takes place after fertilization in both the paternal and maternal genomes, but with different dynamics and a different level of contribution of active and passive processes (17–19). Removal of 5‐methylcytosine (5mC) in the maternal genome occurs slowly and predominantly by passive dilution during DNA replication. The paternal genome, however, undergoes a rapid loss of 5mC. We, and others, have reported that this genome-wide active DNA “demethylation” involves global TET3-mediated oxidation of 5mC to 5hmC, 5fC, and 5caC in the paternal pronucleus (20–25).
The asymmetry of 5mC oxidation between the two parental pronuclei depends on the asymmetry of H3K9 methylation according to microinjection experiments. When Nakamura and colleagues (26, 27) injected ectopic Jhdm2a (Kdm3a) mRNA into the zygote to achieve enzymatic demethylation of H3K9me2, they found that 5mC staining was reduced and that 5hmC increased in the maternal pronucleus. This finding was consistent with the possibility that global 5mC in the maternal pronucleus is protected from TET3-mediated oxidation by H3K9me2 enrichment. The role of a maternally deposited H3K9 histone methyltransferase (HMT) was suggested in this process, but the identity of this enzyme is unknown.
There are six known mammalian H3K9 HMT enzymes (28). EHMT2 (also known as G9A and KMT1C) and its heterodimer-forming partner EHMT1 (also known as GLP and KMT1D) catalyze H3K9 mono- and di-methylation in euchromatin (29, 30). The EHMT2/EHMT1 complex is the main functional H3K9 mono- and dimethyltransferase because the absence of either EHMT2 or EHMT1 strongly affects global H3K9me1/2 in postimplantation embryos (31, 32). Zygotic EHMT2 is required for the maintenance of DNA methylation in embryonic stem (ES) cells and in the embryo at some loci (32–36). SETDB1 (also known as ESET and KMT1E) is a euchromatic H3K9 di- and trimethyltransferase that interacts directly with DNMT3A (37) and is essential for embryo development (38). SETDB1 silences certain gene promoters (37) and is critical for maintaining DNA methylation at long terminal repeat retrotransposons in ES cells (39, 40). SETDB2 (also known as CLLD8 or KMT1F), a close homolog of SETDB1, contributes to the trimethylation of both interspersed repetitive elements and centromere-associated repeats in human cells (41). SUV39H1 (also known as KMT1A) and SUV39H2 (also known as KMT1B) are responsible for depositing H3K9me3 at constitutive heterochromatin (42). They are essential for targeting de novo DNA methylation to major satellites and pericentric heterochromatin regions in ES cells (43) and in the zygote (44).
Very recently, maternal effects were reported for mutations in the Setdb1 (45, 46) and Ehmt2 (47) genes. EHMT2 and SETDB1 are present at high levels in oocytes and zygotes, which persist through preimplantation development. SETDB1 controls global H3K9me2 levels in growing oocytes (46). EHMT2, but not EHMT1, was found by siRNA injection to be involved in the regulation of asymmetric H3K9me2 in mouse zygotes (15).
The role of H3K9 HMTs in affecting 5mC turnover in the zygote has not been addressed. Based on the activities of EHMT2 and SETDB1 to catalyze H3K9 methylation, and because they exhibit maternal effects, we hypothesized that EHMT2 and/or SETDB1 play a role in protecting the maternal pronucleus from TET3-mediated oxidation of 5mC. The goal of this study was to genetically test the role of these two enzymes in protecting DNA methylation in the zygote.
Results
Ehmt2 and Setdb1 Mutant Mouse Lines.
We generated an Ehmt2 knockout mouse line by gene targeting in 129S1/SvImJ ES cells (SI Appendix, Fig. S1). The conditional mutant Ehmt2f/f mice, used in the present study, carried a floxed SET domain and were viable and fertile. The Ehmt2−/− zygotic homozygous mutant embryos expressed a modified Ehmt2 transcript that lacked two exons encoding the SET domain (SI Appendix, Fig. S1E). The Ehmt2−/− SET domain deletion mutation resulted in an embryonic lethal phenotype by 11.5 d post coitum, corroborating another study (29) that eliminated EHMT2 protein expression in the mouse.
We obtained a Setdb1 mutant mouse line from the European Mouse Mutant Archive depository (SI Appendix, Fig. S2). We confirmed that the Setdb1−/− genotype was embryonic lethal and that the Setdb1f/f genotype, containing the floxed SET domain, was viable and fertile. This mouse model showed a delay in the germinal vesicle breakdown in Setdb1-depleted oocytes, reduced frequency of zygote formation, and failure to reach the blastocyst stage in Setdb1 maternal mutants, which agrees with previous reports that use the same mutant mouse line (45, 46).
We immunostained Ehmt2f/f and Setdb1f/f control zygotes using antibodies against 5mC and its oxidized forms (SI Appendix, Fig. S3). We found that control pronuclear stage 2 (PN2) zygotes already contained visible amounts of 5hmC in the paternal pronucleus inside the 5mC-rich perinuclear ring (SI Appendix, Fig. S3A). This matched our finding in wild-type zygotes (22). Zygotes at the PN4–5 stage exhibited a strong asymmetry of 5mC oxidation (SI Appendix, Fig. S3B), as expected, and the asymmetry persisted in the mitotic chromosomes (SI Appendix, Fig. S3C). In the present study, we selected stages PN4–5 for making comparisons between control and mutant zygotes.
Effect of EHMT2 Loss on H3K9 Methylation.
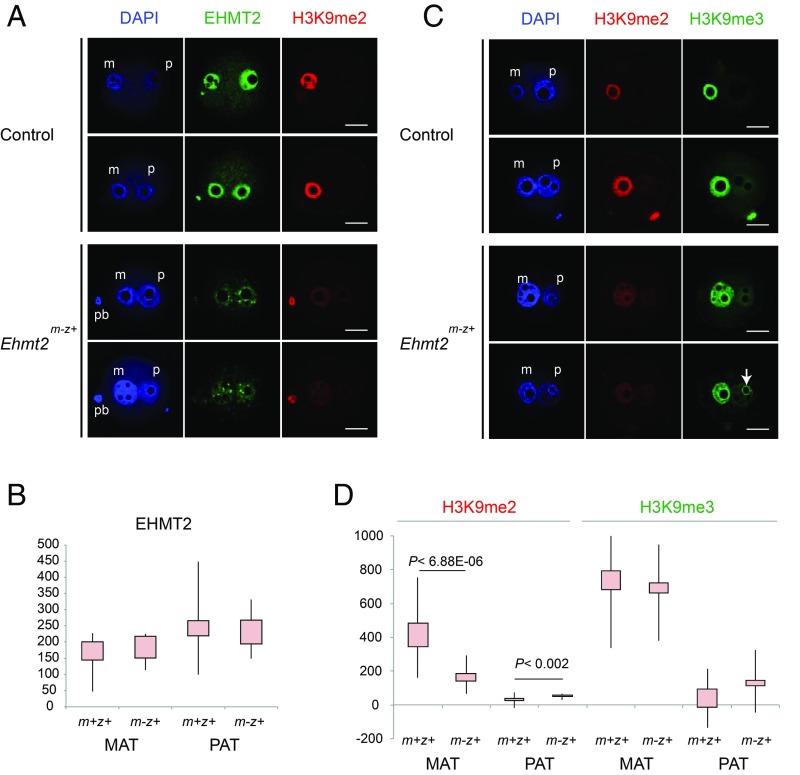
We crossed Ehmt2f/f; Zp3-creTg+ females with wild-type males to obtain maternal mutant Ehmt2m-z+ zygotes (SI Appendix, Fig. S4). These zygotes were derived from homozygous oocytes that lack the SET domain in the EHMT2 protein. The SET-domain–coding exons were excised in growing oocytes by cre recombinase, driven by the zona pellucida 3 (Zp3) promoter. Ehmt2m-z+ zygotes inherit a normal zygotic allele (z+) from the wild-type sperm. We generated control Ehmt2m+z+ zygotes by crossing Ehmt2f/f; Zp3-creTg- females with wild-type males. To see if EHMT2 protein can be detected after removing the SET domain, we immunostained the Ehmt2m-z+ and control zygotes with an antibody against EHMT2 and captured the resulting images using confocal microscopy. The epitope for raising this antibody was the N terminus of EHMT2. The staining was specific to the pronuclei in control zygotes (Fig. 1A). Ehmt2m-z+ zygotes appeared to have weaker signals than control zygotes (Fig. 1A). We quantified the staining intensities in the two pronuclei (SI Appendix, Fig. S5). We found that the difference of quantified EHMT2 intensities did not reach statistical significance (Fig. 1B). The staining pattern was speckled in the mutant zygotes. These findings suggest that deleting the catalytic SET domain did not eliminate the EHMT2 protein from the zygote, but did change its localization.
Fig. 1.
Decreased global H3K9me2 levels in Ehmt2m-z+ zygotes. (A) Confocal microscopy images of representative control and Ehmt2m-z+ zygotes immunostained with anti-EHMT2 and anti-H3K9me2 antibodies are shown in duplicates. DNA was counterstained with DAPI (blue). Paternal pronucleus (p), maternal pronucleus (m), and the polar body (pb) are marked when present in the focal plane view. (B) Immunostaining intensities are plotted for the MAT and PAT pronuclei in control (m+z+) and Ehmt2m-z+ maternal mutant (m-z+) zygotes against the EHMT2 antibody. (C) Confocal microscopy images of representative control and Ehmt2m-z+ zygotes immunostained with anti-H3K9me2 and anti-H3K9me3 antibodies are shown in duplicates. An arrow points to strongly staining circles that appear in the mutant paternal pronucleus. (D) Immunostaining intensities are plotted for the MAT and PAT pronuclei in control (m+z+) and Ehmt2m-z+ maternal mutant (m-z+) zygotes against the H3K9me2 and H3K9me3 antibodies. Statistically significant differences were calculated between the control and mutant samples using Wilcoxon rank-sum tests. Significance values are indicated in the plots. Total number of PN4- to PN5-stage zygotes used for quantification (control and mutant zygotes, respectively) were as follows: EHMT2 (n = 7, n = 8); H3K9me2 (n = 14, n = 16); and H3K9me3 (n = 7, n = 8). (Scale bar: 20 μm.)
We detected asymmetric H3K9me2 staining in the control zygotes (Fig. 1 A and C). H3K9me2 staining was strong and uniform in the maternal pronucleus, but it was very weak in the paternal pronucleus. The intensity of H3K9me2 staining was significantly reduced in the maternal pronucleus of the Ehmt2m-z+ zygotes (Fig. 1D). We concluded that EHMT2 is important for the global H3K9me2 markings in the zygote.
In control zygotes, we detected asymmetric H3K9me3 staining (Fig. 1C) as expected (16). H3K9me3 staining was strong in the maternal pronucleus but was only weakly visible in the paternal pronucleus. The overall staining intensity of H3K9me3 did not change significantly in the Ehmt2m-z+ zygotes (Fig. 1D). We observed that the pattern of H3K9me3 staining was less uniform in the mutant zygotes (Fig. 1C). Some areas became weaker, while a considerable amount of H3K9me3 was retained at circular structures, at the periphery of nucleolar-like bodies (NLBs) in the maternal and the paternal pronuclei (SI Appendix, Fig. S6). The periphery of NLBs is enriched in constitutive heterochromatin at satellite repeat regions (16). These findings suggest that EHMT2 contributes to global H3K9me3 patterns in the zygote.
Effect of EHMT2 on 5mC Oxidation.
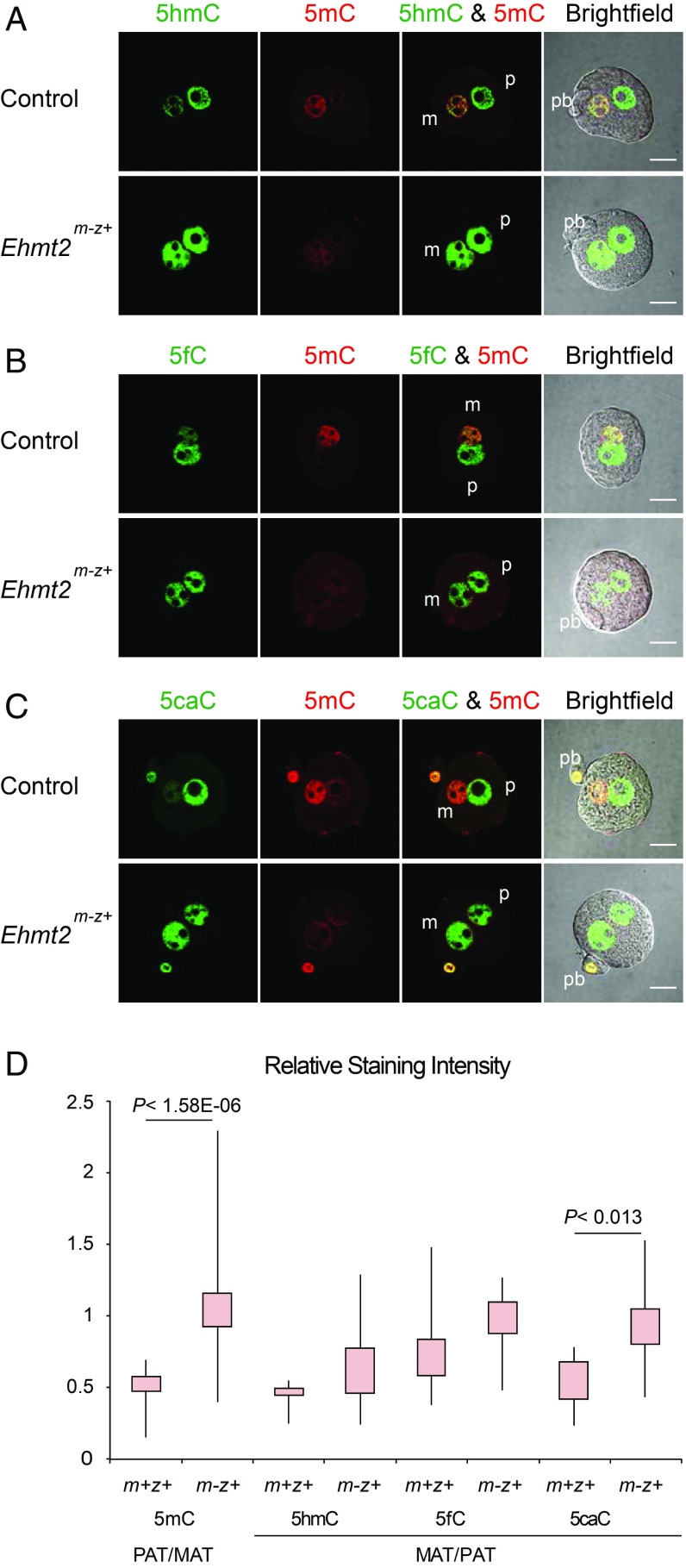
To test the effect of the maternal Ehmt2 mutation on zygotic 5mC oxidation, we immunostained Ehmt2m-z+ and control zygotes using antibodies against 5mC and its oxidized forms (Fig. 2). At the PN4–5 stage in control zygotes, the maternal pronucleus exhibited strong 5mC signals while the paternal pronucleus showed strong 5hmC signals, which matches the results found in Iqbal et al. (22). The paternal/maternal asymmetry of 5mC staining was lost in the Ehmt2m-z+ zygotes (Fig. 2 A–C), and this change was significant (Fig. 2D). The global 5mC of the maternal pronucleus was reduced in the Ehmt2m-z+ zygotes to the level of the paternal pronucleus, which undergoes rapid DNA demethylation. This can be best appreciated by comparing the 5mC staining of the maternal and paternal pronucleus with each other and with the polar body in control and mutant zygotes (Fig. 2C). There was an increased trend of the maternal/paternal (MAT/PAT) ratio for all of the oxidized nucleotides, and among them the 5caC change reached significance (Fig. 2D). These changes were due to increased 5hmC, 5fC, and 5caC in the maternal pronucleus (Fig. 2 A, B, and C, respectively), to a level similar to the paternal pronucleus. In summary, increased 5mC oxidation in Ehmt2m-z+ zygotes supports the role of maternally deposited EHMT2 in protecting global CpG methylation in the maternal pronucleus.
Fig. 2.
Increased 5mC oxidation in the maternal pronucleus of Ehmt2m-z+ zygotes. Distribution of 5mC and its oxidized derivatives is shown by immunostaining in control and Ehmt2m-z+ zygotes. Zygotes were stained at the PN4–PN5 stage with anti‐5mC (red) and one of the following: anti‐5hmC (A), anti‐5fC (B), or anti‐5caC (C) antibodies (green). (D) Quantification results of the relative staining intensities are shown. Total numbers of samples in each staining (5mC, 5hmC, 5fC, and 5caC) were 15, 4, 5, and 6 in the control and 36, 12, 12, and 12 in Ehmt2m-z+ zygotes, respectively. Other details are as described in Fig. 1. (Scale bar: 20 μm.)
Effect of SETDB1 on H3K9 Methylation.
To generate zygotes that lacked maternally inherited SETDB1 HMT function, we crossed Setdb1f/f; Zp3-creTg+ females with wild-type males. We also generated control zygotes by crossing Setdb1f/f; Zp3-creTg- females with wild-type males. We immunostained the resulting Setdb1m-z+ and control Setdb1m+z+ zygotes with an antibody against SETDB1. We found that the SETDB1 staining intensity did not change substantially (SI Appendix, Fig. S2B).
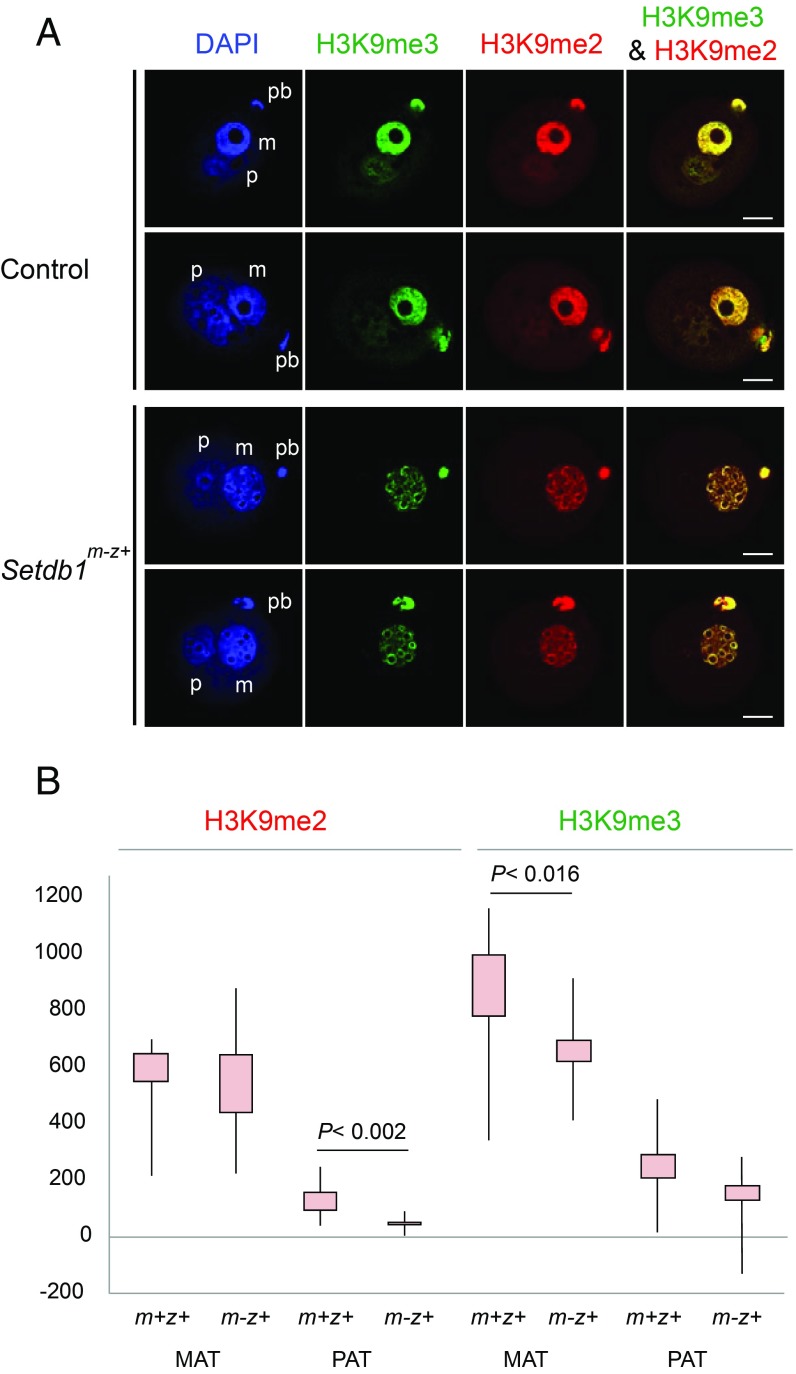
Next we tested the effect of the maternal Setdb1 mutation on global H3K9 methylation in the zygote (Fig. 3). When we assessed global H3K9me2 and H3K9me3 enrichment in control zygotes, we detected asymmetric distribution for both marks (Fig. 3A). The anti-H3K9me2 antibody strongly and uniformly stained the maternal pronucleus except for the NLBs. H3K9me2 was very weak in the paternal pronucleus. H3K9me2 staining intensity did not change in the maternal pronucleus of Setdb1m-z+ zygotes (Fig. 3B), but its distribution was different. The weak H3K9me2 staining was lost from the paternal pronucleus upon maternal Setdb1 mutation. H3K9me3 staining was strong in the maternal pronucleus with clear circles at the periphery of NLBs, but it was only weakly visible in the paternal pronucleus (Fig. 3A). The total level of H3K9me3 was significantly reduced in the maternal pronucleus of Setdb1m-z+ zygotes (Fig. 3B) with the exception of H3K9me3-rich circles at the periphery of NLBs. These findings suggest that SETDB1 is important for global H3K9me3 markings in the zygote.
Fig. 3.
Decreased global H3K9me3 levels in Setdb1m-z+ zygotes. (A) Global enrichment of H3K9me3 and H3K9me2 is shown in control and Setdb1m-z+ zygotes. Immunostaining was performed with anti-H3K9me2 (red) and anti-H3K9me3 (green) antibodies. (B) Quantification of the immunostaining intensities is plotted from n = 10 control and n = 17 mutant zygotes. Other details are as described in Fig. 1. (Scale bar: 20 μm.)
Effect of SETDB1 on 5mC Oxidation.
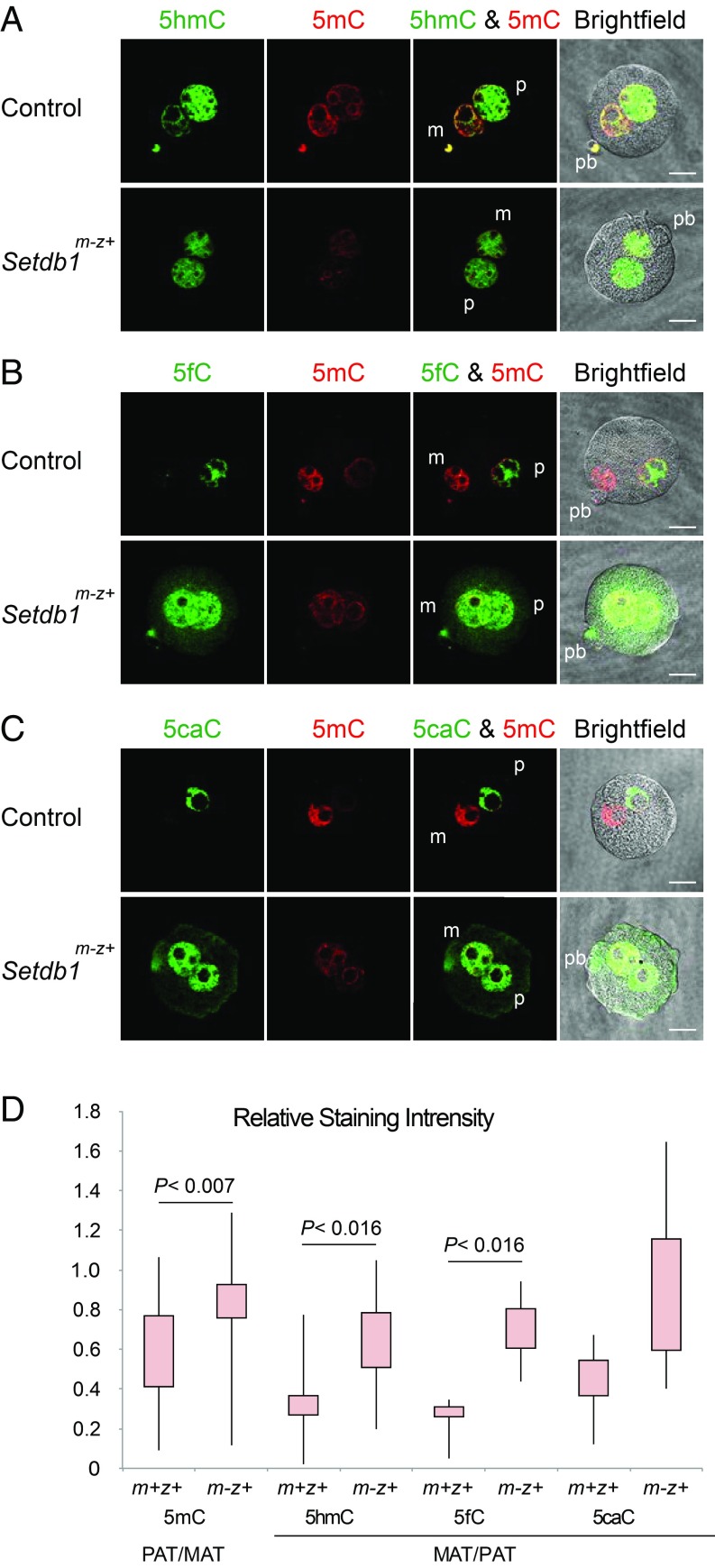
Next we tested the effect of the maternal Setdb1 mutation on global 5mC oxidation. We immunostained the Setdb1m-z+ and control zygotes using antibodies against 5mC, 5hmC, 5fC, and 5caC. We found a decreased 5mC signal and increased 5hmC, 5fC, and 5caC staining in the maternal pronucleus in the Setdb1m-z+ zygotes compared with Setdb1m+z+ controls (Fig. 4 A–C), resulting in the statistically significant loss of asymmetry between the two pronuclei (Fig. 4D). The 5mC-rich circle around the NLBs still remained clearly detectable, indicating the lack of 5mC oxidation at those DNA sequences. These results showed that, similar to EHMT2, maternally deposited SETDB1 plays a role in protecting global CpG methylation in the zygote.
Fig. 4.
Increased 5mC oxidation in the maternal pronucleus of Setdb1m-z+ zygotes. Distribution of 5mC (red) and its oxidized derivatives (green), 5hmC (A), 5fC (B), or 5caC (C), is shown by immunostaining in control and Setdb1m-z+ zygotes. (D) Quantification results of the relative staining intensities. Total numbers of quantified samples in each staining (5mC, 5hmC, 5fC, and 5caC) were 19, 8, 4, and 7 in the control and 22, 11, 5, and 6 in Setdb1m-z+ zygotes, respectively. Other details are as described in Fig. 1. (Scale bar: 20 μm.)
Effect of EHMT2 and SETDB1 on Pronuclear Structure.
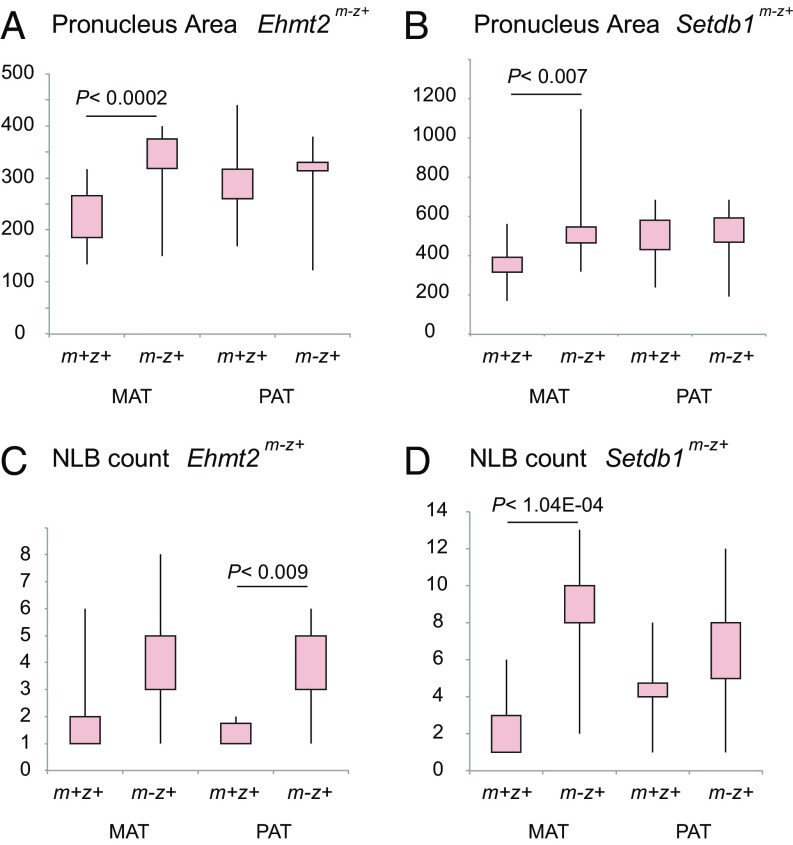
In wild-type zygotes, the paternal pronucleus is usually the larger one of the two pronuclei. We noted that this asymmetry was frequently lost in Ehmt2m-z+ and Setdb1m-z+ zygotes. This was apparent in each experiment, even when using different immunostaining methods (Figs. 1–4). When we measured the area of each pronucleus in its best-fitting Z-section, we found that the size of the maternal pronucleus was significantly larger in the Ehmt2m-z+ and Setdb1m-z+ zygotes than in their respective control zygotes, and they become, on average, equal in size with the paternal pronucleus of the same zygote (Fig. 5 A and B). We also noted that the number of NLBs increased in both pronuclei of the mutant zygotes (Fig. 5 C and D). These results revealed that EHMT2 and SETDB1 are important for structural aspects of zygote development. Because the catalytic SET domain was deleted, and the H3K9me2-H3K9me3 patterns and intensities were affected in the mutant zygotes, it is most likely that the histone methyltransferase function of these enzymes is required for properly structured pronucleus formation.
Fig. 5.
Structural changes in the Ehmt2m-z+ and Setdb1m-z+ zygotes. Quantification of the area of the maternal and paternal pronucleus is depicted in Ehmt2m-z+ (A) and Setdb1m-z+ (B) zygotes compared with control zygotes. NLB counts in the maternal and paternal pronucleus of Ehmt2m-z+ (C) and Setdb1m-z+ (D) zygotes are also shown. The Ehmt2m-z+ and Setdb1m-z+ results are derived from the immunostaining experiments depicted in Fig. 1 and Fig. 3, respectively.
The Distribution of TET3 in Maternal Mutant and Control Zygotes.
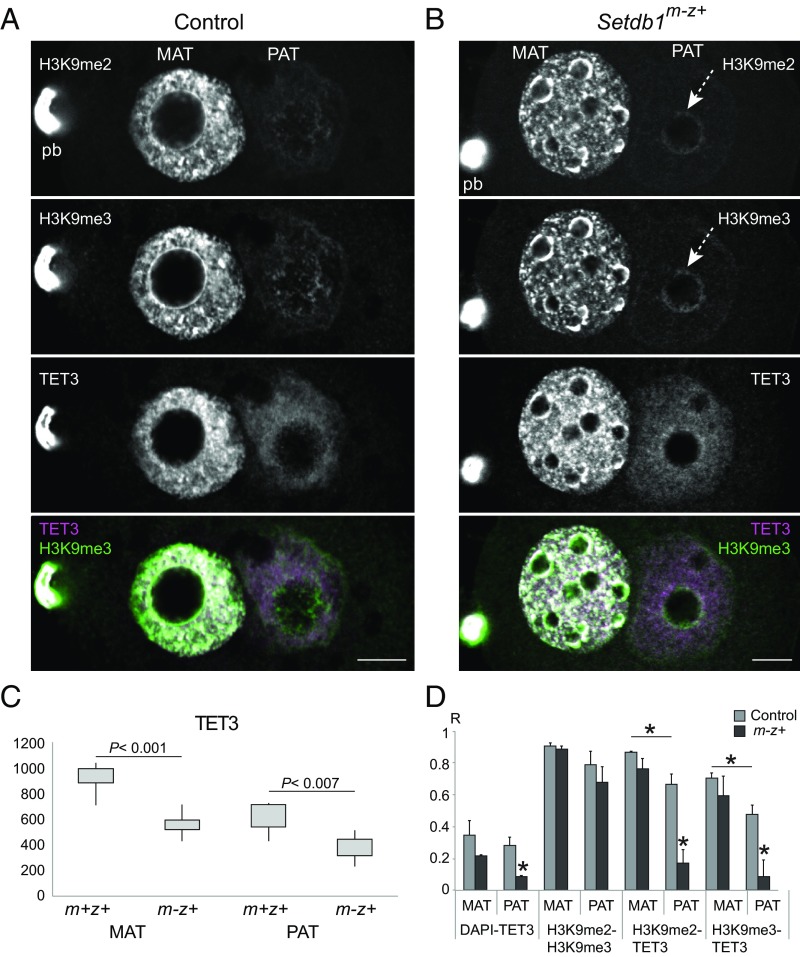
TET3 is the main enzyme responsible for oxidizing 5mC in the zygote (19, 20, 22, 48). It is important to find out whether TET3 localization (i) is asymmetric, (ii) depends on the H3K9 methylation due to EHMT2 or SETDB1, and (iii) contributes to the asymmetry of 5mC oxidation. TET3 localization to paternal and/or maternal pronuclei appears to depend on the antibody used and on the pronuclear stages with most reports showing localization in the paternal and maternal pronuclei at PN4–5 (19, 20, 25, 27, 48, 49). We found TET3 staining in both pronuclei in the control PN4–5 zygotes (SI Appendix, Figs. S6A and S7A). The maternal pronucleus showed TET3 spots that corresponded to many of the H3K9me2/H3K9me3 spots. The peripheries of the NLBs in the control maternal pronucleus (Fig. 6A and SI Appendix, Fig. S6A) and the mutant paternal pronucleus (SI Appendix, Fig. S7B) were rich in H3K9me2 and H3K9me3 but not in TET3. There was no change in TET3 levels in the Ehmt2m-z+ zygotes (SI Appendix, Fig. S6B), but TET3 levels were significantly reduced in the Setdb1m-z+ zygotes (Fig. 6C). We measured the colocalization of different signals and plotted the Pearson correlation coefficients (Fig. 6D). Colocalization of H3K9me2 and H3K9me3 was very high in both pronuclei. We found that H3K9me2 and H3K9me3 also colocalized with TET3 in the maternal and paternal pronucleus of control zygotes. TET3 colocalization with H3K9me2 and H3K9me3 was significantly stronger in the maternal than in the paternal pronucleus. TET3 colocalization with H3K9me2 and H3K9me3 was significantly reduced in the Setdb1m-z+ paternal pronucleus (Fig. 6D).
Fig. 6.
The relationship of H3K9me3 and TET3 patterns in the zygote. Confocal images are shown of a representative control (A) and Setdb1m-z+ (B) zygote, stained with anti-H3K9me2, anti-H3K9me3, and TET3 antibodies. A merged image shows the overlap of H3K9me3 and TET3 substructures (in white). Image brightness in this figure was optimized to aid in observing the substructures. For relative intensities between control and mutant zygotes, see Fig. 3, where the same Z-section of each zygote is displayed. The maternal pronucleus is in focus in these images. See SI Appendix, Fig. S7, to observe the paternal pronucleus in focus in the next Z-section of each zygote. (C) Quantification of the TET3 immunostaining intensities is shown in the maternal and paternal pronuclei of control, n = 5, and Setdb1m-z+ zygotes, n = 10. Statistically significant differences were calculated using Wilcoxon rank-sum tests. (D) Quantification of the colocalization (Pearson correlation coefficient) between the different signals is depicted in control and mutant zygotes. *P < 0.05 indicates statistically significant differences calculated by t tests. (Scale bar: 10 μm.)
These staining results collectively showed that maternally deposited EHMT2 and SETDB1 affect TET3 enrichment and localization in the zygote. However, TET3 levels and localization do not correlate with TET3-mediated oxidation in the control and mutant zygotes.
Discussion
We found that decreased levels of H3K9me2 or H3K9me3 in the maternal pronucleus of Ehmt2m-z+ or Setdb1m-z+ mouse zygotes resulted in greatly reduced asymmetry of 5mC oxidation between the parental pronuclei. The 5mC level was reduced and 5hmC, 5fC, and 5caC levels were increased in the mutant maternal pronuclei. We conclude that EHMT2- and SETDB1-dependent H3K9 methylation is important for protecting global CpG methylation in the maternal pronucleus from TET3-mediated 5mC oxidation. In addition, we show that the size and the structure of the maternal pronucleus are abnormal in Ehmt2m-z+ and Setdb1m-z+ mouse zygotes, revealing an additional role of both EHMT2 and SETDB1 in zygote development.
We achieved reduced H3K9me2 and H3K9me3 levels by genetically inactivating two histone methyltransferase enzymes responsible for placing these marks on the chromosomes at the oocyte and zygote stages. H3K9me2 and H3K9me3 marks were reduced globally in Ehmt2m-z+ and Setdb1m-z+ mutant zygotes, respectively, resulting in a global increase of oxidized 5mC products in both maternal mutants. Our findings suggest that both the H3K9me2 and H3K9me3 marks contribute to preventing TET3-mediated oxidation of 5mC in the maternal pronucleus. Reducing either one of these marks in the maternal pronucleus is sufficient to visibly change its resistance to global 5mC oxidation. The ratio of paternal/maternal 5mC and maternal/paternal 5hmC, 5fC, and 5caC approximated 1 in the mutant zygotes although it did not quite reach 1. This is likely due to the additive effect of the two enzymes that may protect some nonoverlapping parts of the genome. Future genome-wide mapping studies will shed further light on the nature of the protected regions.
Even though EHMT2 and SETDB1 can be detected equally in the two pronuclei, they methylate H3K9 effectively only in the maternal pronucleus. It will be interesting to discover the molecular mechanism by which the activity of maternally derived EHMT2 or SETDB1 is inhibited in the paternal pronucleus or why H3K9 residues in the paternal pronucleus are largely resistant to both of these enzymes.
NLBs are the sites for pericentromeric heterochromatin. We observed residual 5mC at the periphery of NLBs in the maternal pronucleus of Setdb1m-z+ mutant zygotes, where we also found H3K9me2 and H3K9me3 enrichment. This suggests that H3K9me3 protects satellite DNA from 5mC oxidation at the pericentric heterochromatin, but marking these regions by H3K9me2/me3 or protection of 5mC at these sequences does not require SETDB1. Another H3K9 HMT, SUV39H2, is likely responsible for this circular deposition of H3K9me3 in the maternal pronucleus, but such a feature has not been found in the paternal pronucleus. Interestingly, a weak but clear H3K9me3 staining also appeared at the periphery of NLBs in the paternal pronucleus of Ehmt2m-z+ (Fig. 1C) and Setdb1m-z+ zygotes (Fig. 6B). Such a response was observed in the paternal pronucleus in the zygotes that carried a maternal deletion of Lsd1/Kdm1A, which is known as an H3K4 demethylase that also demethylates H3K9 (16). However, increased H3K9 methylation at the circles was unexpected from the mutation of an H3K9me3 HMT. We speculate that, in the absence of EHMT2 or SETDB1, a SUV39H2-mediated H3K9me3 deposition is triggered at pericentric heterochromatin regions in the paternal pronucleus, regions which are normally repressed by a Polycomb-dependent mechanism (44).
We show that genetic disruption of H3K9 methylation in the egg leads to aberrant 5mC oxidation in the maternal pronucleus. However, the molecular mechanism of how asymmetric H3K9 methylation governs asymmetric 5mC oxidation dynamics between the two parental pronuclei is still unknown. TET3 is the main enzyme responsible for 5mC oxidation in the zygote, and it functions predominantly in the paternal pronucleus. We found that TET3 protein is abundant in both pronuclei and shows a brightly punctated pattern in the maternal pronucleus (Fig. 6 and SI Appendix, Fig. S6). Because of the overlap of the punctated patterns of H3K9me2/me3 and TET3 in the normal maternal pronucleus, we suggest that H3K9 methylation does not inhibit TET3 binding to the chromosomes. It rather appears that H3K9me2/me3–TET3 interaction takes place where TET3 is not catalytically active. TET3 is active in the control paternal pronucleus where H3K9me/me3 is weak and also in the Ehmt2m-z+ or Setdb1m-z+ mutant maternal pronucleus where H3K9 methylation is greatly reduced. In addition, colocalization of H3K9me2/me3 with TET3 is stronger in the control maternal than in the control paternal pronucleus (Fig. 6D). Based on the above staining patterns we find it likely that TET3-mediated oxidation takes place in the genomic regions that are devoid of H3K9 methylation and, conversely, that TET3-mediated oxidation is inhibited where TET3 is localized at H3K9 methylated regions. In support of this idea, we found a very strong anticorrelation between the TET reaction product 5hmC and H3K9me3 when we compared existing genome-wide mapping of 5hmC (50) and H3K9me3 (51) from mouse embryo neurons, suggesting that 5hmC is not generated where H3K9me3 is present (SI Appendix, Fig. S8). In addition, recent genome-wide results give support to this possibility. Peat et al. (24) used reduced representation bisulfite sequencing in Tet3 maternal mutant and control zygotes and determined the location of a small number of genomic regions where DNA demethylation in the maternal pronucleus depends on TET3. Analyzing the results of Peat et al. (24), we found that none of these 204 sites overlapped with any of the 844,422 H3K9me3 peaks, as had been mapped in zygotes using ChIP-seq. (52), suggesting that TET3 activity is not compatible with H3K9me3 localization. A direct TET3-H3K9me2/me3 interaction may inhibit TET3 catalytic activity. Alternatively, another protein factor may trap TET3 at H3K9me3-rich regions in the maternal pronucleus. Such a mechanism is consistent with the finding that PGC7 is attracted to H3K9me2 in the maternal pronucleus (27) and also that 5mC oxidation is enhanced in the maternal pronucleus in zygotes where PGC7 protein is absent (53). In future experiments, it will be important to characterize the TET3–H3K9me2/me3 interaction at the molecular level in vitro to find out whether H3K9 methylation directly inhibits the catalytic activity of TET3. Although challenging, future genome-wide mapping of TET3 binding may also help us to understand these events together with high-resolution mapping of 5mC, 5hmC, 5fC, 5caC, and unmethylated DNA in Ehmt2m-z+ or Setdb1m-z+ mutant zygotes.
It is not known whether the asymmetry of active DNA demethylation is important for development. TET3-mediated 5mC oxidation is not essential for the early stages of mouse development (20, 54–56), but whether maternal protection from global 5mC oxidation is essential for development has not been tested. It is conceivable that specific sequences need to remain methylated in the maternal pronucleus to ensure properly timed embryonic genome activation and/or early embryo development. Aberrant global 5mC oxidation in the zygote’s maternal pronucleus (Figs. 2 and 4) in the absence of maternal HMT function may contribute to the abnormal development of Setdb1m-z+ and Ehmt2m-z+ embryos. If the oxidation chain is completed and 5caC is replaced by unmethylated CpGs, this may lead to a premature active DNA demethylation of certain sequences and altered gene activities from the maternal chromosomes. At other sequences where the demethylation reaction is not completed, as can be seen from strong 5hmC, 5fmC, and 5caC enrichment in the chromosomes, the mutant maternal genome may still be interpreted very differently from a fully methylated maternal genome. It will be important to find out the normal physiological function of delaying 5mC oxidation in the maternal pronucleus and the specific consequences of failed 5mC protection in the Ehmt2m-z+ and Setdb1m-z+ zygotes.
Methods
Mice.
All animal experiments were performed according to the National Research Council’s Guide for the Care and Use of Laboratory Animals (57), with Institutional Care and Use Committee-approved protocols at Van Andel Research Institute (VARI).
An Ehmt2 conditional knockout mouse line (Ehmt2f/f) was generated in our laboratory by gene targeting in 129S1/SvImJ ES cells. A Setdb1 conditional knockout mouse line (Setdb1f/f) was generated from the line Setdb1tm1a(EUCOMM)Wtsi (European Mouse Mutant Archive). We removed the Pgk-neo positive selection cassette by crossing with an Flpe recombinase-expressing transgenic mouse B6.Cg-Tg(Pgk1-flpo)10Sykr/J purchased from the Jackson Laboratory (JAX#011065). Zp3-cre transgenic mice C57BL/6-Tg(Zp3-cre)93Knw/J (58), purchased from the Jackson Laboratory (JAX#003651), were used to excise the SET-domain–encoding exons from Ehmt2 or Setdb1 in growing oocytes. Ehmt2f/f; Zp3-creTg+ or Setdb1f/f; Zp3-creTg+ female mice were used as the experimental group. Ehmt2f/f or Setdb1f/f mice without the cre transgene were used as the control group. Ehmt2f/f, Setdb1f/f, and Zp3-cre mice were maintained on a 129S1/SvImJ, C57BL/6N, and C57BL/6J genetic background, respectively. Mice were genotyped by PCR using primers listed in SI Appendix, Table S1.
Isolation of Preimplantation Mouse Embryos.
Zygotes were collected from the oviducts of ∼4-wk-old female C57BL/6 mice or 6- to 8-wk-old female 129S1 mice. Female mice were superovulated by injecting 5 IU of human chorionic gonadotropin (hCG) 46 h after injecting 5 IU of pregnant mare serum gonadotropin and then mated with wild-type male mice. Zygotes were collected from the oviducts at 27–30 h post hCG.
Immunofluorescence Staining.
Cumulus cells were removed from zygotes with 1% hyaluronidase treatment. The zona pellucida was removed using acidic tyrode solution. After washing in M2 medium +0.3% BSA, zygotes were fixed in 4% paraformaldehyde in PBS at room temperature for 20 min. Zygotes were permeabilized in 0.2% Triton X-100 in PBS at room temperature for 10 min, blocked for 1 h at room temperature in blocking solution (1% BSA, 0.2% Triton X-100 in PBS), and incubated overnight at 4 °C with the primary antibody. The following day, the embryos were washed three times with PBST (0.05% Tween 20 in PBS), and staining was detected by incubating the embryos with the secondary antibody at room temperature for 1 h. The zygotes were washed three times with PBST before mounting on slides with ProLong Gold antifade reagent with DAPI (Molecular Probes). We used antibodies that show similar patterns in control zygotes to those previously published by others (16, 46). All of the primary and secondary antibodies are listed in SI Appendix, Table S2.
For staining of 5mC and its oxidative derivatives, permeabilized zygotes were incubated in 4 N HCl at room temperature for 10 min to denature DNA followed by neutralization in Tris⋅HCl, pH 8.0, for 10 min. The samples were blocked overnight at 4 °C in blocking solution. The following day, the zygotes were incubated with anti-5mC and anti-5hmC (or anti-5fC, anti-5caC) antibodies in blocking solution for 1 h at room temperature. After washing with PBST, the incubation with secondary antibodies and mounting on slides were performed as described above. We identified the maternal pronucleus by its proximity to the polar body. This was done by inspecting more than one Z section, because in many cases the polar body is located in a Z section that is not in the same focal plane as the pronuclei and is not visible in the actual image displayed in the figure. The experiments were performed multiple times and were presented from at least two independent mothers.
Microscope and Image Analysis.
Immunofluorescence was visualized using a Nikon A1plus-RSi laser scanning confocal microscope with a 63× oil objective. Images were acquired using the NIS-Elements Software (Nikon). Z sections were taken every 0.4 μm. Mean pronuclear intensities and profiles of immunofluorescence signals and pronuclear size measurements were quantified using the Fiji (ImageJ) software. Staining profile was measured by “profile” function and colocalization was measured by the “coloc2” function of Fiji. When making quantitative comparisons of image intensities between mutant and control zygotes, we used zygotes that were collected and stained at the same time, and the images were acquired using the same confocal settings. The displayed microscopic images of control and mutant samples were adjusted equally using the same brightness and contrast values per each antibody.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey R. Mann for providing the pCTV2 plasmid vector; Walter Tsark (City of Hope Transgenic Core Facility) for his help in developing the Ehmt2 mutant mouse line; the City of Hope Animal House and the VARI Vivarium for mouse maintenance; Corinne Esquibel (VARI Imaging Core Facility) for providing expert advice on confocal microscopy; and Ji Liao for her help with mouse breeding and genotyping. This work was supported by Grant R01GM064378 from the NIH and by VARI Funding (to P.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819946116/-/DCSupplemental.
References
- 1.Borsos M., Torres-Padilla M. E., Building up the nucleus: Nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 30, 611–621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh P., et al. , De novo DNA methylation in the male germ line occurs by default but is excluded at sites of H3K4 methylation. Cell Rep. 4, 205–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borgel J., et al. , Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi H., et al. , High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 23, 616–627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smallwood S. A., et al. , Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart K. R., et al. , Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 29, 2449–2462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veselovska L., et al. , Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 16, 209 (2015). Correction in: Genome Biol. 16, 271 (2015). [DOI] [PMC free article] [PubMed]
- 8.Seisenberger S., et al. , The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada Y., Yamaguchi K., Epigenetic modifications and reprogramming in paternal pronucleus: Sperm, preimplantation embryo, and beyond. Cell. Mol. Life Sci. 74, 1957–1967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Probst A. V., Almouzni G., Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 27, 177–185 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Arney K. L., Bao S., Bannister A. J., Kouzarides T., Surani M. A., Histone methylation defines epigenetic asymmetry in the mouse zygote. Int. J. Dev. Biol. 46, 317–320 (2002). [PubMed] [Google Scholar]
- 12.Burton A., Torres-Padilla M. E., Epigenetic reprogramming and development: A unique heterochromatin organization in the preimplantation mouse embryo. Brief. Funct. Genomics 9, 444–454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepikhov K., Walter J., Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev. Biol. 4, 12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H., Kim J. M., Aoki F., Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development 131, 2269–2280 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Ma X. S., et al. , The dynamics and regulatory mechanism of pronuclear H3k9me2 asymmetry in mouse zygotes. Sci. Rep. 5, 17924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ancelin K., et al. , Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. eLife 5, e08851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckersley-Maslin M. A., Alda-Catalinas C., Reik W., Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Wu X., Zhang Y., TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Guo F., et al. , Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Gu T. P., et al. , The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Inoue A., Zhang Y., Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iqbal K., Jin S. G., Pfeifer G. P., Szabó P. E., Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 108, 3642–3647 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wossidlo M., et al. , 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Peat J. R., et al. , Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 9, 1990–2000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amouroux R., et al. , De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura T., et al. , PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Nakamura T., et al. , PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486, 415–419 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Mohan M., Herz H.M., Shilatifard A., SnapShot: Histone lysine methylase complexes. Cell 149, 498–498.e1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tachibana M., et al. , G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tachibana M., et al. , Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19, 815–826 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zylicz J. J., et al. , Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. eLife 4, e09571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auclair G., et al. , EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res. 26, 192–202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monk D., et al. , Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res. 18, 1270–1281 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagano T., et al. , The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Wagschal A., et al. , G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell. Biol. 28, 1104–1113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xin Z., et al. , Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 278, 14996–15000 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Li H., et al. , The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Biol. Chem. 281, 19489–19500 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Dodge J. E., Kang Y. K., Beppu H., Lei H., Li E., Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478–2486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung D., et al. , Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc. Natl. Acad. Sci. U.S.A. 111, 6690–6695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karimi M. M., et al. , DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falandry C., et al. , CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J. Biol. Chem. 285, 20234–20241 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters A. H., et al. , Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Lehnertz B., et al. , Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Puschendorf M., et al. , PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40, 411–420 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Eymery A., Liu Z., Ozonov E. A., Stadler M. B., Peters A. H., The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development 143, 2767–2779 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Kim J., et al. , Maternal Setdb1 is required for meiotic progression and preimplantation development in mouse. PLoS Genet. 12, e1005970 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zylicz J. J., et al. , G9a regulates temporal preimplantation developmental program and lineage segregation in blastocyst. eLife 7, e33361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L., et al. , Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15, 459–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakatani T., et al. , Stella preserves maternal chromosome integrity by inhibiting 5hmC-induced γH2AX accumulation. EMBO Rep. 16, 582–589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hahn M. A., et al. , Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 3, 291–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato M., Takemoto K., Shinkai Y., A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat. Commun. 9, 1683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C., et al. , Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Bian C., Yu X., PGC7 suppresses TET3 for protecting DNA methylation. Nucleic Acids Res. 42, 2893–2905 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue A., Shen L., Matoba S., Zhang Y., Haploinsufficiency, but not defective paternal 5mC oxidation, accounts for the developmental defects of maternal Tet3 knockouts. Cell Rep. 10, 463–470 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsukada Y., Akiyama T., Nakayama K. I., Maternal TET3 is dispensable for embryonic development but is required for neonatal growth. Sci. Rep. 5, 15876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai H. Q., et al. , TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature 538, 528–532 (2016). [DOI] [PubMed] [Google Scholar]
- 57.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 58.de Vries W. N., et al. , Expression of cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis 26, 110–112 (2000). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.