Significance
The ontogeny of B cells in the bone marrow is regulated by a complex network of proteins and signaling pathways. Regulation of gene expression through transcriptional activation and repression is necessary for establishing and maintaining appropriate transcriptomes in cellular development. Here we show that CHD4, a protein that combines epigenetic reader and nucleosome mobilizing functions, is indispensable for early B cell lineage specification and progression. CHD4 is required for IL-7 receptor signaling and efficient V(D)J recombination. Furthermore, CHD4 is essential for repression of non-B lineage gene transcription during B lymphopoiesis. Together, our findings define the importance of a key epigenetic regulator in B cell development and gene expression.
Keywords: B lymphopoiesis, B lymphocyte development, transcriptional programming, transcriptional repression, chromatin remodeling complexes
Abstract
Cell lineage specification is a tightly regulated process that is dependent on appropriate expression of lineage and developmental stage-specific transcriptional programs. Here, we show that Chromodomain Helicase DNA-binding protein 4 (CHD4), a major ATPase/helicase subunit of Nucleosome Remodeling and Deacetylase Complexes (NuRD) in lymphocytes, is essential for specification of the early B cell lineage transcriptional program. In the absence of CHD4 in B cell progenitors in vivo, development of these cells is arrested at an early pro-B-like stage that is unresponsive to IL-7 receptor signaling and unable to efficiently complete V(D)J rearrangements at Igh loci. Our studies confirm that chromatin accessibility and transcription of thousands of gene loci are controlled dynamically by CHD4 during early B cell development. Strikingly, CHD4-deficient pro-B cells express transcripts of many non-B cell lineage genes, including genes that are characteristic of other hematopoietic lineages, neuronal cells, and the CNS, lung, pancreas, and other cell types. We conclude that CHD4 inhibits inappropriate transcription in pro-B cells. Together, our data demonstrate the importance of CHD4 in establishing and maintaining an appropriate transcriptome in early B lymphopoiesis via chromatin accessibility.
A complex network of intrinsic and extrinsic factors regulates the differentiation of hematopoietic stem cells (HSCs) into diverse blood cell lineages, including B lymphocytes. B cell lineage specification is a function of cell- and stage-specific transcription, which is tightly controlled during B lymphopoiesis in the bone marrow. Master transcriptional regulators of B cell specification and commitment have been studied in depth. These factors, including Early B cell factor 1 (EBF1), TCF3 (E2A proteins E12 and E47), Ikaros (IKZF1), FoxO1, Pax5, and IRF4, are sequence-specific DNA binding proteins that bind regulatory modules in key genes to activate the B cell transcriptional program and repress non-B cell-specific genes (1–3). These factors recruit epigenetic regulators that control chromatin accessibility and transcriptional priming at specific gene loci to initiate B cell programming (4–7). In this regard, nucleosome mobilization by chromatin remodeling complexes (CRCs) is emerging as critical for the dynamic regulation and interconvertibility of active, poised, or silent gene loci in cell type specification and subsequent differentiation.
Nucleosome Remodeling and Deacetylase (NuRD; also known as Mi-2) was first characterized as a multiprotein complex that combines both ATP-dependent nucleosome mobilization and histone deacetylase activities (8–10). NuRD has been implicated in the regulation of transcriptional programs in both B and T lymphocytes (11). Of central importance for its function, NuRD components Chromodomain Helicase DNA-binding 4 (CHD4; also known as Mi-2β), CHD3 (Mi-2α; encoded by a separate gene), and CHD5 (restricted to the central nervous system), are members of the SNF2/RAD54 helicase family that unite ATP-dependent nucleosome remodeling functions with “reader” domains that bind histone tails. Each of these proteins comprises a core ATPase/helicase domain flanked by two Plant Homeodomain motifs (PHD fingers) that recognize modifications of histone tails, tandem chromodomains, and carboxyl-terminal domains necessary for transcriptional repression (12–16). Studies also identified components of NuRD complexes that assemble in a combinatorial fashion. CHD3, CHD4, and CHD5 each associate with five other core subunits that assemble as preformed higher order complexes (17–19). In this manner, CHD proteins combine their activities with HDAC1/HDAC2, MBD2/MBD3, WD40 repeat proteins RBBP4/RBBP7; metastasis-associated proteins MTA-1, MTA-2, or MTA-3; and the GATAD2A/B zinc finger proteins. Together, these complexes focus an extensive array of activities for chromatin remodeling and epigenetic regulation.
Importantly, CHD4-NuRD complexes do not recognize specific DNA sequences. Instead, they are recruited to sites by interactions with sequence-specific DNA binding proteins (e.g., BCL6 and Ikaros) (20–26). Subsequently, local chromatin structure is modified by mobilization of nucleosomes, histone deacetylation, and subsequent “decommissioning” of promoters (25, 27, 28). Thus, CHD4-NuRD has been revealed as an integral driver of appropriate transcription during cell type specification. Complementing its roles in CHD4-NuRD complexes, CHD4’s ability to be recruited independently of NuRD enables additional functions in DNA damage repair and cell cycle progression (29–33), signal transduction (34), and overall genome maintenance (35).
We previously demonstrated the importance of CHD4-NuRD complexes in regulating chromatin accessibility in B cell-specific transcription (14, 16, 36). To further understand how CHD4 contributes to specification of the B cell lineage, we assessed functions of CHD4 in vivo. In mice lacking CHD4 specifically in B cell progenitors, development is arrested at an early pro–B-like stage. CHD4-deficient pro-B cells do not proliferate in response to IL-7 and exhibit decreased phosphorylated Signal Transducer and Activator of Transcription 5A (STAT5A). The cells also feature greatly reduced frequencies of distal VH gene segment usage in V(D)J recombination, with undetectable expression of cytoplasmic μ-chains. Notably, CHD4-deficient pro-B cells expressed hundreds of non-B cell lineage genes out of context. Expression of these genes was due, in part, to greatly increased chromatin accessibility at intronic and intergenic elements. Together, our observations confirm that CHD4 is essential for B cell specification through its roles in cell homeostasis, V(D)J recombination, and repression of inappropriate transcription in early B lymphopoiesis.
Results
B Cell Development Is Arrested at an Early Pro-B Stage in the Absence of CHD4.
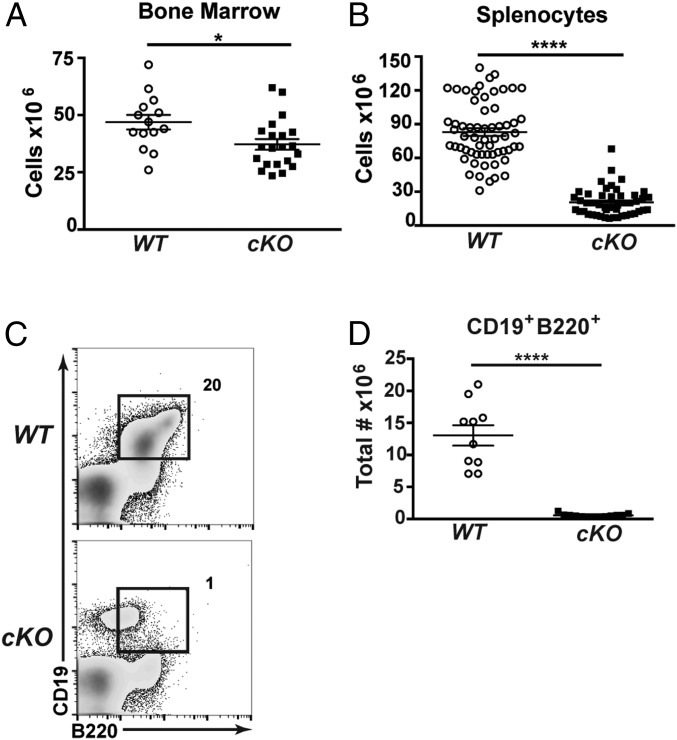
To assess roles of CHD4 in B cell development in vivo, we bred Chd4-floxed (Chd4fl/fl) mice (37) with transgenic mice that express Cre recombinase under the direction of the early B cell-specific CD79a gene (CD79a-CreTg/+) (38, 39). In resulting Chd4 conditional knock-out (Chd4cKO/cKO) mice, Cre expression and Chd4 deletion occur at the prepro-B to pro-B cell transition during B cell development in the bone marrow (SI Appendix, Fig. S1 A and B). Total cellularity of bone marrow and spleens was decreased in Chd4cKO/cKO mice compared with littermate controls (Fig. 1 A and B). Chd4cKO/cKO mice exhibited a 90% reduction in the frequency and number of B cells (B220+CD19+) in the bone marrow (Fig. 1 C and D). Notably, a B220loCD19+ population was only observed in Chd4cKO/cKO mice (Fig. 1C).
Fig. 1.
CHD4 depletion results in a total decrease of bone marrow and spleen cellularity. (A) Total cell counts of bone marrow cells harvested from 4- to 6-wk-old Cd79afl/flCd79a-CreTg/+ (cKO; n = 21) mice and WT (n = 14) mice. Each dot represents an individual mouse. (B) Total cell counts of splenocytes harvested from 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ (n = 47) and WT (n = 61) mice. Each dot represents an individual mouse. (C) Flow cytometry and (D) total numbers of B220+CD19+ cells isolated from bone marrow of Chd4fl/flCd79a-CreTg/+ (n = 13) and WT (n = 10) mice. Each dot represents an individual mouse. Asterisks indicate statistical significance compared with WT littermate controls as unpaired, two-tailed Student’s t test without Welch’s correction. *P < 0.05, ****P < 0.0001. Graphs represent arithmetic mean with ±SEM. All data are representative of at least three independent experiments.
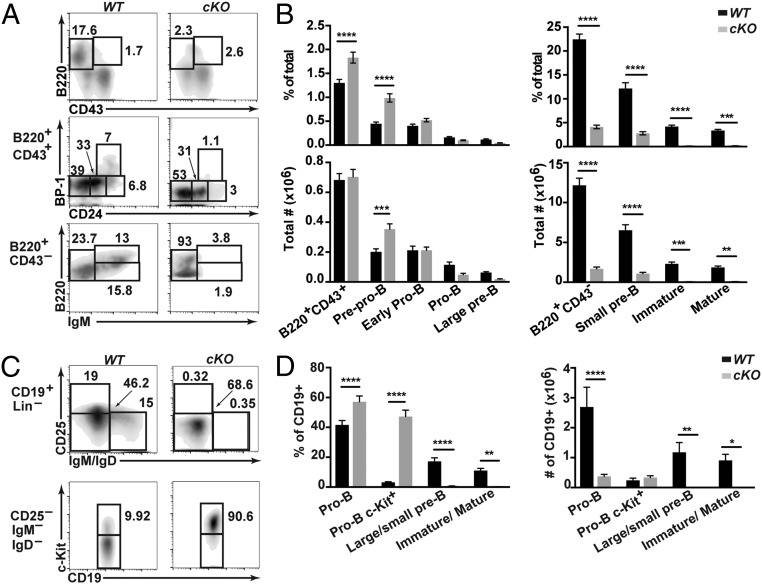
To characterize effects of CHD4 depletion on B cell differentiation in the bone marrow, we analyzed the Hardy phenotypic fractions of bone marrow B cells using flow cytometry (Fig. 2) (40, 41). Characterization of phenotypic fractions used were prepro-B cells (fraction A; B220+CD43+BP1−CD24low/−), a distinct population (not included in B cell fractions characterized by Hardy and colleagues) encompassing early pro-B cells (fraction A/B; B220+CD43+BP1−CD24int), pro-B cells (fraction B/C; B220+CD43+BP1−CD24+), large pre-B cells (fraction C′; B220+CD43+BP1+CD24+), small pre-B cells (fraction D; B220+CD43−IgM−), immature B cells (fraction E; B220+CD43−IgM+), and recirculating mature B cells (fraction F; B220hiCD43−IgM+). In the absence of CHD4, percentages of the earliest B cell progenitor populations (prepro-B and early pro-B) are increased, while later stages including pro-B through immature B cells and mature recirculating B cells are greatly decreased in mutant bone marrow (Fig. 2). Additionally, the lack of positive staining for cell surface markers BP-1+, CD25+, and IgM+ cells defines the developmental arrest as occurring before differentiation to pre-B cells (Fig. 2) (41). Interestingly, pro-B cells from Chd4cKO/cKO mice exhibited increased surface c-Kit (CD117) expression (Fig. 2 C and D). In addition, spleens isolated from Chd4cKO/cKO mice were approximately one half the size of wild-type (WT) control spleens (SI Appendix, Fig. S2A). Spleens of Chd4cKO/cKO mice exhibited a drastic reduction of IgM+- and IgD+-expressing B cells (SI Appendix, Fig. S2B). These data suggest that CHD4 deficiency impairs B cell development and differentiation in the bone marrow, which effectively prevents the establishment of mIg+ B cell populations in the periphery.
Fig. 2.
B cell development is arrested at an early pro–B-like stage of development in the absence of CHD4. Quantitation of B cell subsets in bone marrow. (A) Flow cytometry and (B) frequencies and numbers of various B cell populations (Hardy fractions) from bone marrow of 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ (cKO; n = 29) and WT (n = 24). (C) Flow cytometry and (D) frequencies (cKO, n = 9; WT, n = 10) and numbers (cKO, n = 4; WT, n = 4) of CD19+ and CD19+ c-Kit+ cells from bone marrow of 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ and WT mice. Asterisks indicate statistical significance compared with WT littermate controls using two-way ANOVA with Sidak’s multiple comparisons test. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001. Graphs represent arithmetic mean with ±SEM. All data are representative of at least three independent experiments.
CHD4-Deficient Pro-B Cells Have Impaired Responses to IL-7 Signaling.
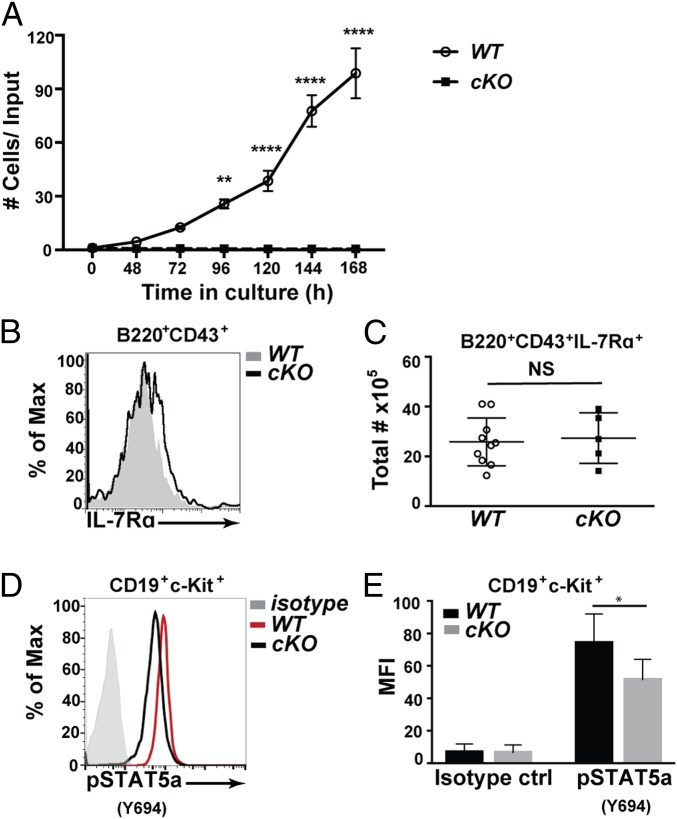
Our flow cytometry studies indicated that CHD4 deficiency results in the arrest of bone marrow B cells at an early pro–B-like stage of development. Interleukin-7 receptor (IL-7R) signaling is essential for early B cell differentiation, survival, and proliferation (42, 43). Therefore, we hypothesized that the developmental arrest due to the loss of CHD4 is due, in part, to the inability of B cell progenitors to proliferate in response to IL-7. To test this hypothesis, IL-7–dependent proliferation and cell survival were assayed. Purified pro-B cells from WT and Chd4cKO/cKO mice were stimulated with IL-7 and proliferation was measured by counting live cells over time (Fig. 3A). CHD4-deficient pro-B cells did not increase in number in the presence of IL-7, unlike WT pro-B cells, which actively proliferated.
Fig. 3.
CHD4 is required for proliferation of pro-B cells downstream of IL-7R signaling. (A) Pro-B cells (B220+CD43+CD24+BP1+IgM–) were sorted from Chd4fl/flCd79a-CreTg/+ mice (cKO; n = 3) and WT littermate controls (n = 3) for culture in IL-7 (10 ng/mL). Cell counts were taken daily for 7 d, represented as the number of cells over the initial cell count. Asterisks indicate statistical significance compared with WT littermate controls using two-way ANOVA with Sidak’s multiple comparisons test. **P < 0.005, ****P < 0.0001. Graphs represent arithmetic mean with ±SEM. All data are representative of at least three independent experiments. (B) Representative flow cytometry of CD127 (IL-7Rα) expression on B220+CD43+ cells isolated from Chd4fl/flCd79a-CreTg/+ and WT littermate controls. (C) Total numbers of B220+CD43+CD127+ cells from the bone marrow of Chd4fl/flCd79a-CreTg/+ (n = 5) and WT (n = 10) mice. Each dot represents an individual mouse. (D) Representative flow cytometry of intracellular phosphor-STAT5a in c-Kit+ pro-B cells (Lin–CD19+c-Kit+CD25–IgM–IgD–) isolated from Chd4fl/flCd79a-CreTg/+ and WT littermate controls. (E) Detection of pSTAT5A in CD19+ c-Kit+ pro-B cells isolated from Chd4fl/flCd79a-CreTg/+ (n = 3) and WT (n = 3) mice. The lineage (Lin) stain included: CD3ε, CD11b, CD11c, Gr-1, NK1.1, Ly6C, CD317, and Ter119. Asterisks indicate statistical significance compared with WT littermate controls as unpaired, two-tailed Student’s t test without Welch’s correction. *P < 0.05, **P < 0.005, ****P < 0.0001, NS, not significant. Graphs represent arithmetic mean ± SD. All data are representative of at least three independent experiments.
To determine whether the lack of proliferation was due to decreased IL-7R signaling, we measured levels of surface CD127 (IL-7Rα) on B220+CD43+ bone marrow cells from Chd4cKO/cKO mice and WT littermate controls (Fig. 3 B and C). The frequencies of CD127+ cells, as well as the level of CD127 expression (measured as mean fluorescence intensity, or MFI) were unaltered by CHD4 deficiency. However, we noted that downstream IL-7R signaling is reduced in CHD4-deficient pro-B cells. We examined intracellular Signal Transducer and Activator of Transcription 5A (STAT5A) phosphorylation after stimulation with IL-7 in vitro, because STAT5A is an essential downstream effector of IL-7R signaling in early B cell development and differentiation (44, 45). Intracellular flow cytometry detected decreased total phosphorylated STAT5A in CHD4-deficient pro-B cells (Fig. 3 D and E). These data suggest that IL-7R signaling is defective in CHD4-deficient pro-B cells.
CHD4 Is Required for Distal VH to DJH Usage and Igh Locus Contraction.
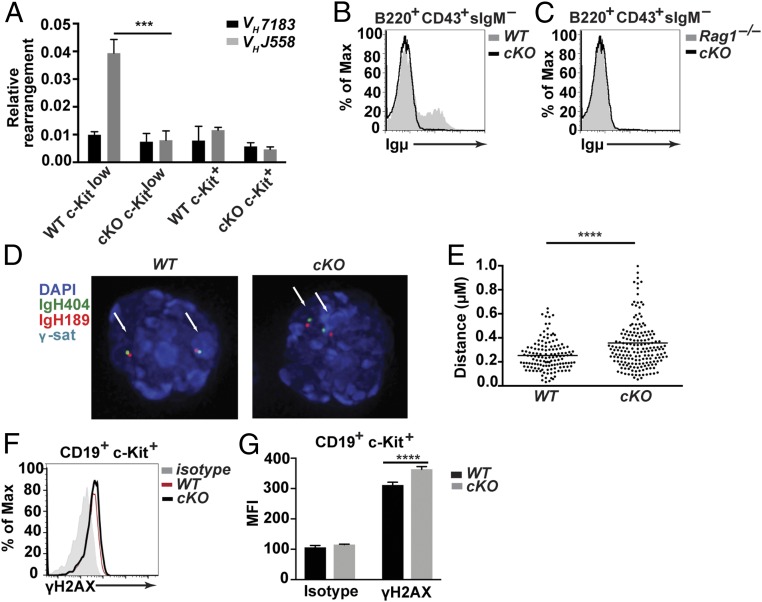
V(D)J recombination depends on the accessibility of recombination signal sequences (RSSs) to the recombinase machinery (46). It has been reported that IL-7R signaling plays a vital role in modulating the accessibility of chromatin during these events (47, 48). Therefore, we assessed the status of V(D)J recombination in cells lacking CHD4 and IL-7R signaling using genomic qPCR (Fig. 4A). We sorted c-Kit+ and c-Kitlow pro-B cells from Chd4cKO/cKO and WT control mice and quantified frequencies of DJH rearrangements with distal VHJ558 family versus proximal VH7183 family gene segments to determine accessibility of these sites. Genomic qPCR detected similar utilization of distal VHJ558 and proximal VH7183 segments in rearranged alleles from CHD4-deficient c-Kit+ pro-B cells, CHD4-deficient c-Kitlow pro-B cells, and WT c-Kit+ pro-B cells. In contrast, utilization of distal VHJ558 gene segments in c-Kitlow pro-B cells isolated from CHD4-deficient mice was decreased by fourfold. To further explore the mechanisms contributing to defective V(D)J recombination in CHD4-deficient mice, we purified pro-B cells from bone marrow of Chd4cKO/cKO and WT control mice and stained cells to detect cytoplasmic µ-chains (Fig. 4B). CHD4-deficient B cells lacked cytoplasmic µ-chains, similar to B cells from Rag1 knockout mice (Fig. 4C). Other genes associated with Igh recombination, including Pax5-activated intergenic repeat 4 (Pair4), Pair6, and µ0, which produce noncoding germline transcripts, and Cµ constant region mRNA (from DJH rearranged loci) (49–52), were detected at WT levels in CHD4-deficient pro-B cells (SI Appendix, Fig. S3A).
Fig. 4.
Loss of CHD4 results in reduced VH to DJH rearrangements and increased DNA damage in pro-B cells. (A) qPCR analysis of rearranged proximal (VH7183) and distal (VHJ558) gene segments from genomic DNA isolated from sorted c-Kit+ pro-B cells (Lin–CD19+c-Kit+CD25–IgM–IgD–) and c-Kitlow pro-B cells (Lin–CD19+c-KitlowCD25–IgM–IgD–) from bone marrow of 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ mice (n = 3) and WT littermate controls (n = 3). (B) Flow cytometry of cytoplasmic Ig µ-expression in sorted B220+CD43+sIgM– B cells isolated from bone marrow from 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ (n = 2) and WT (n = 2) mice. (C) Flow cytometry of cytoplasmic Ig µ-expression in sorted B220+CD43+sIgM– B cells isolated from bone marrow from 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ (n = 2) and a Rag1 knockout mouse (n = 1). Data are derived from one experiment. (D) The 3D-FISH confocal images of hybridized probes in CD19+c-Kit+ pro-B cell (as described above) nuclei isolated from bone marrow of Chd4fl/flCd79a-CreTg/+ and WT mice. The IgH404 proximal probe (RP23-404D8) was labeled with Alexa Fluor 488 (AF488); the IgH189 distal probe (RP23-189H12) was labeled with AF568; major satellite repeats (γ-sat) were conjugated to AF647; and nuclei were counterstained with DAPI. (E) Distributions of spatial distances (in micrometers) separating BAC probes, located at each end of the Igh locus (IgH404 vs. IgH189) in pro-B cells isolated from bone marrow of Chd4fl/flCd79a-CreTg/+ (n = 92 cells) and WT (n = 69 cells). Each dot represents an individual allele. (F) Representative flow cytometry and (G) MFI of c-Kit+ pro-B cells stained for intracellular γ-H2AX from bone marrow of Chd4fl/flCd79a-CreTg/+ (n = 4) and WT (n = 3) mice. The asterisks indicate statistical significance compared with the WT littermate controls as unpaired, two-tailed Student’s t test with equal SD, Mann–Whitney rank test or two-way ANOVA with Sidak’s multiple comparisons test. **P < 0.0005, ****P < 0.0001. All data are representative of at least three independent experiments.
The decreased usage of distal VH segments in CHD4-deficient pro-B cells could be due to an inability to generate long-range interactions between VH and d-JH loci. Therefore, we assessed the topology of Igh loci in early pro-B cells using 3D fluorescence in situ hybridization (3D-FISH) with labeled BAC probes (Fig. 4 D and E). The 3D-FISH detected greater distances between hybridized proximal (IgH404) and distal VH (IgH189) probes (median distance 0.30 µm) in CHD4-deficient versus WT pro-B cells (median distance 0.23 µM) with high significance (P < 0.0001). Taken together, our results suggest that CHD4 has a role in locus contraction of Igh loci, but further details of this mechanism remain to be determined.
CHD4-Deficient Pro-B Cells Have Higher Frequencies of DNA Damage.
Impairment of locus contraction and the inability to efficiently complete Igh rearrangements in CHD4-deficient pro-B cells could involve dysregulation of DNA damage response (DDR) pathways. V(D)J recombination is intimately coupled with DNA double-stranded break (DSB) repair. Cleavage of RSSs by Recombination Activating Gene (RAG-1 and RAG-2)-mediated DSBs activate Ataxia-Telangiectasia Mutated (ATM) kinase, which in turn initiates DDR (reviewed in ref. 53). CHD4 has been extensively studied as a key component of DDR pathways, where it is phosphorylated by ATM and recruited during the repair of DSBs (33). To assess DNA damage due to the loss of CHD4 directly, we measured amounts of intracellular γ-H2AX (phosphorylated on Ser139) in pro-B cells using flow cytometry. Pro-B cells lacking CHD4 exhibited a significant increase in the intracellular staining of γ-H2AX (Fig. 4 F and G). The results indicate that CHD4 deficiency increases the accumulation of DNA damage due to impaired DDR. This deficiency may be due to RAG DSBs failing to initiate the transcriptional program necessary to carry out these functions due to the loss of CHD4. Alternatively, we cannot rule out the initiation of damage by Cre-mediated DSBs.
CHD4 Is Essential for Repression of Inappropriate Transcription in Early Pro-B Cells.
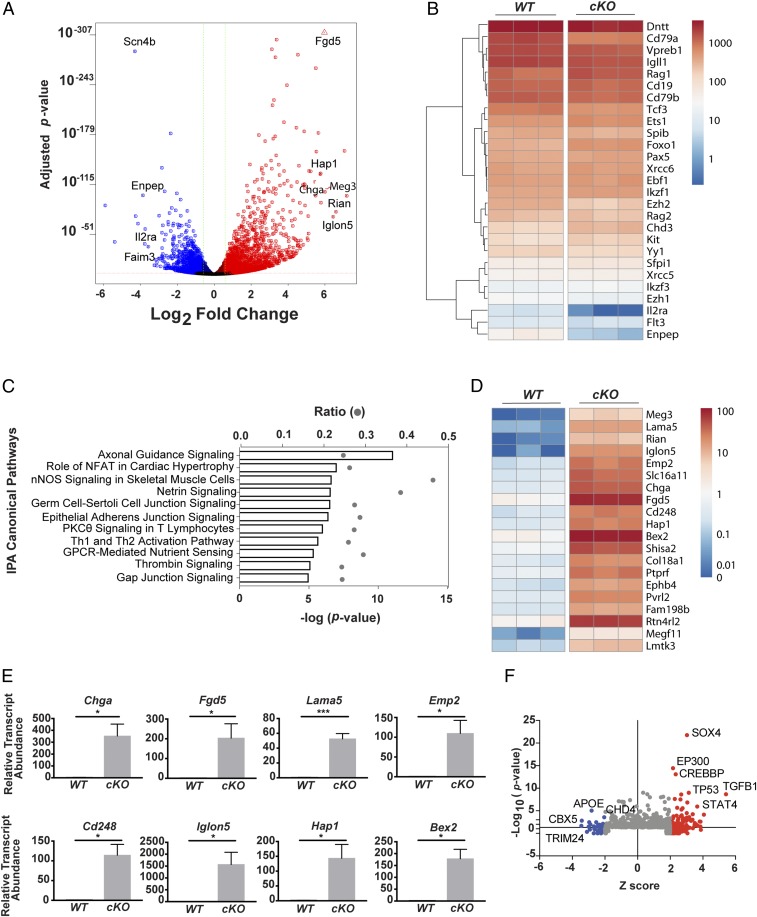
Correct transcriptome expression is essential for normal B cell developmental progression. Because CHD4-NuRD is a known corepressor of transcription, we sought to determine whether CHD4-deficient early pro-B cells exhibit altered transcriptomes relative to their WT counterparts. We employed RNA sequencing (RNA-seq) to interrogate genome-wide changes in transcription. Purity of sorted populations was assessed and principal component (PC) analysis of RNA-seq was performed to determine relationships between biological replicates (SI Appendix, Fig. S4 A and B). A total of 3,318 unique genes were differentially regulated between CHD4-deficient and WT pro-B cells (≥1.5 fold, Padj < 0.05) (Dataset S1). In the absence of CHD4, 1,241 genes were significantly down-regulated and 2,077 genes were significantly up-regulated (Fig. 5A).
Fig. 5.
CHD4 is required for transcriptional repression of non-B lineage genes. (A) Volcano plot of expression in transcripts per million (TPM) of genes in Chd4fl/flCd79a-CreTg/+ c-Kit+ pro-B cells. Genes significantly increased (red) and decreased (blue) greater than 1.5-fold (Padj ≤ 0.05) are highlighted. (B) Heatmap depicting TPM of select B-lineage genes. (C) IPA enrichment for the top 10 most significantly enriched canonical pathways. (D) Heatmap of TPM values for non-B lineage genes. (E) Validation (qRT-PCR) of select up-regulated non-B lineage genes in CHD4-deficient and WT pro-B cells. Statistical significance of RT-PCR data was validated using the Mann–Whitney rank test: *P < 0.05, ***P < 0.0005. (F) IPA upstream regulators predicted as activated (red) and inhibited (blue).
Phenotypic analysis demonstrated the loss of lineage progression in Chd4cKO/cKO B cells. Therefore, we assessed whether key genes necessary for early B cell development were differentially expressed in Chd4cKO/cKO pro-B cells (Fig. 5B). Transcripts associated with B cell progenitor specification and commitment, including Ebf1, Pax5, and Cd19, as well as the expression of other transcription factors, including Ikzf1, Ets1, Spi1, and Spib, were unchanged in CHD4-deficient pro-B cells. Loss of CHD4 resulted in increased Kit transcripts (1.7-fold), which are normally down-regulated before differentiation to canonical pro-B (fraction B) cells. Loss of CHD4 expression in pro-B cells did not affect the expression of Flt3 together with components of the pre-BCR, including Cd79b, Igll1, and Vpreb1. We detected reduced expression of Cd79a (reduced by 30%), which is not unexpected in the presence of the Cd79a-Cre allele. It is notable that Foxo1 and Rag1, which are necessary for V(D)J recombination, are up-regulated 1.5-fold in CHD4-deficient pro-B cells. Up-regulation of Rag1 is consistent with findings that IL-7R signaling and Rag1 expression are inversely correlated (48, 54).
Genes that constitute normal transcriptional signatures of subsequent progenitor stages, i.e., late pro-B cells and pre-B cells, including Il2ra and Enpep (encoding CD25 and BP-1), are activated first at low levels in WT pro-B cells. Transcripts of these late pro-B and pre-B cell genes were significantly reduced in CHD4-deficient pro-B cells, indicating that these cells fail to activate genes associated with the transition to subsequent stages of differentiation. In conjunction with our phenotypic analysis, these data further support requirements for CHD4 in the transition from early pro-B cells to late pro-B/pre-B cells.
We next identified affected pathways in CHD4-deficient pro-B cells using ingenuity pathway analysis (IPA) (Fig. 5C). CHD4 deficiency largely resulted in the enrichment of genes representing pathways in that feature prominently in cell populations other than B cells, including T cells, neuronal cells, heart and skeletal muscle cells, and other nonhematopoietic cells. Specifically, pathways activated in CHD4-deficient pro-B cells include NFAT signaling in cardiac hypertrophy, PKCθ signaling in T lymphocytes, Thrombin Signaling, and CREB signaling in neurons. In fact, many genes that are not normally expressed in B cells, but are expressed in other cell lineages, are significantly up-regulated in CHD4-deficient pro-B cells (Fig. 5 D and E). The most highly up-regulated genes in CHD4-deficient pro-B cells include Iglon5, encoding a neural cell adhesion protein (55), Chromogranin A (Chga), a glycoprotein expressed in the pancreas and neuronal cells, and FYVE, RhoGEF, and PH Domain containing 5 (Fgd5), which is a highly specific diagnostic marker of hematopoietic stem cells (56). Significant promiscuous expression of genes of other lineages, including endocrine genes and the lung and neuronal genes was noted. These genes include glucagon receptor/Gcgr, which is normally expressed in the liver, Huntingtin-associated protein 1/Hap1, expressed in the CNS, and multiple lung-expressed genes including laminin α5 (Lama5). Expression of these genes in CHD4-deficient pro-B cells indicates that CHD4 contributes to B cell lineage specification by repressing transcription of nonlineage gene programs.
To identify potential upstream regulators of the gene expression signature, we utilized IPA upstream regulator analysis (Fig. 5F). Our results indicate that many transcriptional regulators, including TGFB1, TP53, STAT4, SOX4, and EP300, are predicted to activate gene transcription in CHD4-deficient pro-B cells. Notably, regulators important for the regulation or transcriptional silencing, including TRIM24, CBX5, and CHD4 itself, are predicted to be inhibited.
CHD4 Modulates Chromatin Accessibility in Early Pro-B Cells.
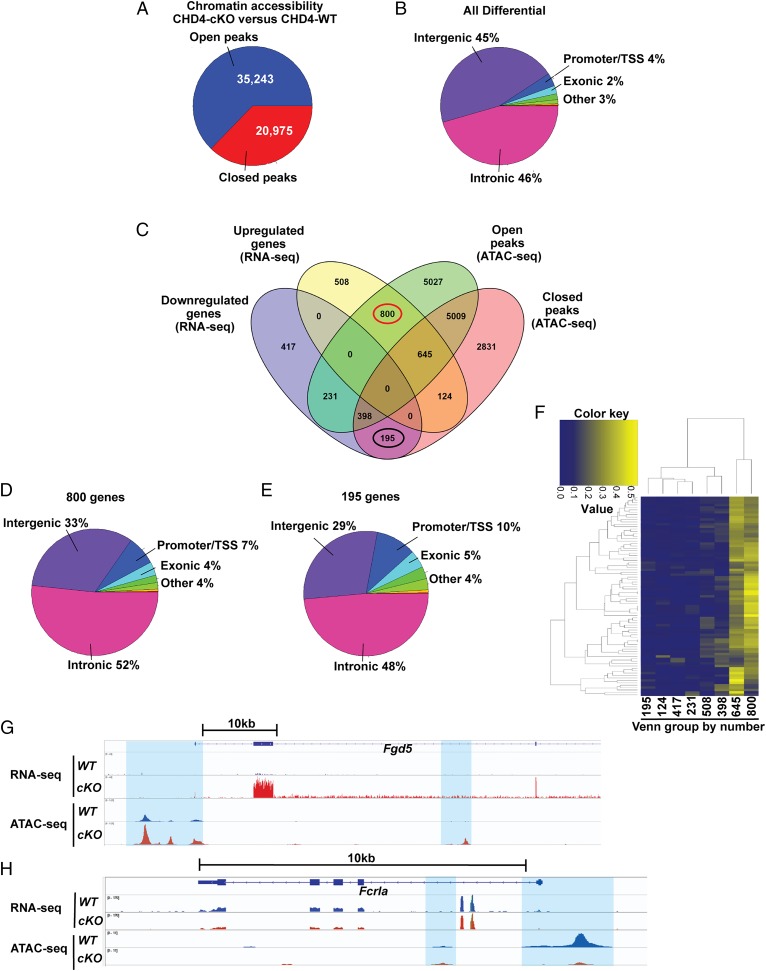
Given that CHD4 is a repressor of transcription and known to limit chromatin accessibility of individual genes (36), we employed the assay for transposase-accessible chromatin-coupled with next generation sequencing (ATAC-seq) to determine whether chromatin landscape alterations occur in parallel with the significantly changed transcriptional pattern observed in CHD4-deficient pro-B cells. We compared ATAC-seq peaks between CHD4-deficient and WT pro-B cells. Consistent with CHD4’s role as a chromatin remodeler, we observed ∼35,000 more accessible peaks in CHD4-deficient pro-B cells (Fig. 6A). CHD4-deficient pro-B cells also contained ∼20,000 fewer accessible peaks vs. WT cells. The majority of these differential ATAC-seq peaks localize to intergenic or intronic regions, while ∼4% reside within promoter/TSS chromatin regions based on HOMER annotation (Fig. 6B). To interrogate the potential functional links between the altered chromatin landscape and transcriptional programming governing B cell development, we integrated the differential RNA- and ATAC-seq datasets comparing CHD4-deficient vs. WT pro-B cells (Fig. 6C). The majority (2,393 genes or ∼72%) of differentially expressed genes determined via RNA-seq (1.5-fold cutoff), also exhibited altered chromatin accessibility in CHD4-deficient pro-B cells. Not surprisingly, complex patterns of increased and decreased accessibility mark genes with both increased and decreased gene expression in CHD4-deficient pro-B cells. Specifically, of the 2,077 genes with increased expression in CHD4-deficient pro-B cells, ∼70%, or 1,445 genes, exhibited increased chromatin accessibility (Fig. 6C). In contrast, of the 1,241 genes with decreased expression in CHD4-deficient pro-B cells, less than half, or 593 genes, exhibited decreased accessibility. In addition, evidence linking local repressor activity of CHD4 to increased accessibility and gene expression in CHD4-deficient pro-B cells is further supported by comparing our datasets with previously reported ChIP-seq analysis of CHD4 DNA binding (7). Of the 34,219 CHD4 binding signals detected in both replicates of RAG-deficient pro-B cells, 7,198 signals (21%) were detected within 10 kB of TSS in genes that change expression in our CHD4-deficient pro-B cells (estimated with 1.5-fold cutoff of significance). Together, these data largely support a role for CHD4 as a corepressor that controls chromatin accessibility in the early B cell developmental program.
Fig. 6.
CHD4 modulates chromatin accessibility in early pro-B cells. (A) Comparison of filtered, statistically called ATAC-seq peaks between CHD4-deficient and WT pro-B cells. (B) Analysis of HOMER annotated regions within differential peaks called between CHD4-deficient and WT pro-B cells. (C) Integration and comparison of differential RNA-seq and ATAC-seq datasets between CHD4-deficient and WT pro-B cells. The 800 gene set (up-regulated with only open peaks) and 195 gene set (down-regulated with only closed peaks) are circled red and black, respectively. (D) Assessment of chromatin distribution within 800 gene set (RNA-seq/ATAC-seq integration) between CHD4-deficient and WT pro-B cells. (E) Assessment of chromatin distribution within 195 gene set (RNA-seq/ATAC-seq integration) between CHD4-deficient and WT pro-B cells. (F) Heatmap of GO pathway analysis for the total set of RNA-seq differentially expressed genes compared with the number of shared genes between each Venn subgroup and the total genes within each GO pathway. (G) Visualization of Fgd5 gene landscape (integration of RNA-seq and ATAC-seq datasets) between CHD4-deficient and WT pro-B cells. (H) Visualization of Fcrla gene landscape (integration of RNA-seq and ATAC-seq datasets) between CHD4-deficient and WT pro-B cells.
Given the ability of CHD4 to regulate transcriptional machinery at promoter regions, we assessed the chromatin distribution for the least confounded integration of ATAC-seq data with RNA-seq data. Specifically, we analyzed the 800 peaks with only increased accessibility and increased gene expression and the 195 peaks with only decreased accessibility and decreased gene expression (Fig. 6 D and E). Compared with the total differential ATAC-seq peak distribution comparing CHD4-deficient vs. WT pro-B cells (Fig. 6B), the 800 ATAC-seq peaks linked to increased gene expression displayed an almost twofold increase (7% vs. 4%) in promoter/TSS distribution and a specific loss of distribution to intergenic regions (33% vs. 45%) (Fig. 6D). Similarly, the 195 ATAC-seq peaks linked to decreased gene expression displayed a greater than twofold increase (10% vs. 4%) in promoter/TSS distribution and a specific loss of intergenic distribution (29% vs. 45%) (Fig. 6E). The enhanced promoter/TSS localization and decrease in intergenic regions suggests that CHD4 regulates transcription in pro-B cells via direct regulation of chromatin accessibility. To further investigate this hypothesis, we examined the DNA binding motifs associated with promoter-localized ATAC-seq peaks using HOMER (Dataset S2). Analysis of the promoter annotated ATAC-seq peaks within the 800 gene set Venn subgroup revealed enrichments in E26 Transforming Sequence (ETS), zinc finger (Zf), and Myocyte Enhancer Factor 2 (MEF2) transcription factor binding sites. In contrast, analysis of the promoter annotated ATAC-seq peaks within the 195 subgroup revealed enrichment in Pit1-Oct1-Unc86 (POU) domain protein binding sites.
To further examine the biological significance of the CHD4 chromatin-regulated program in pro-B cells, we employed gene ontology (GO) analysis to identify the contributions of specific chromatin-regulated Venn gene sets (Fig. 6C) to transcriptionally driven biological pathways. GO pathway analysis was performed for the total set of 3,318 RNA-seq differentially expressed genes. The contribution of each Venn gene set to the expression-based biological pathways was determined by comparing the number of shared genes between each Venn subgroup and the total genes within each GO pathway. We then generated a heatmap based on the ratio of the gene number comparison for each Venn subgroup (Fig. 6F). This analysis reveals that the biological programs identified via the RNA-seq analysis of the CHD4-deficient pro-B cells is being driven by the 800 and 645 gene sets, which link increased accessibility with gene transcription. The shared GO terms for those two gene subsets distribute among immune and neural development, cell morphology, and cell signaling pathways (Dataset S3). These data support the role of CHD4 in enforcing transcription of a biological program, while repressing other programs in B cells.
Finally, to visualize the coordinated chromatin and transcriptional changes in CHD4-deficient vs. WT pro-B cells, we integrated ATAC- and RNA-seq data for specific genes using the integrative genomics viewer (IGV) (Fig. 6 G and H). We compared visualization of the inappropriately activated (increased 63.7-fold) Fgd5 locus (Fig. 6G), with the down-regulated (decreased by 67%) Fcrla locus (Fig. 6H). With loss of CHD4, the Fgd5 locus is characterized by increased chromatin accessibility and gene expression in mutant pro-B cells, while the Fcrla locus features decreased chromatin accessibility. These data are consistent with the hypothesis that CHD4 represses prior hematopoietic programs (including Fgd5) as B cell differentiation proceeds. Conversely, Fcrla is expressed in more mature B cells following further differentiation. Decreased Fcrla expression may be due to the lack of progression accompanying developmental arrest in CHD4-deficient pro-B cells.
Discussion
The network of transcription factors that orchestrate B cell development has been defined extensively. However, less understood are the functions of epigenetic regulators including chromatin remodeling complexes, which likely play pivotal roles in specifying patterns of gene expression in developing lymphocytes. A key group of these complexes is encompassed as NuRD, which while largely implicated as corepressors may regulate an array of transcriptional outcomes (27). In this study, we investigated functions of a catalytic ATPase/helicase subunit of NuRD complexes, CHD4. Mice lacking CHD4 in the early B cell compartment exhibit a developmental blockade at an early pro–B-like (c-Kit+CD19+) stage of development. CHD4-deficient pro-B cells do not proliferate in response to IL-7, are defective in V(D)J recombination, and accumulate DNA damage. Strikingly, pro-B cells lacking CHD4 have lost the ability to progress to later B cell stages, together with inappropriate expression of hundreds of non-B lineage genes. These defects result in the failure of CHD4-deficient early pro-B cells to differentiate into antibody-producing B cells. Developmental arrest occurs despite relatively normal expression of transcription factors that are essential for B cell lineage specification and commitment, including EBF1 and Pax5.
Loss of CHD4 in pro-B cells changes the relative expression of more than 3,000 genes. Thus, CHD4 and CHD4-NuRD complexes are widely employed to establish appropriate transcriptional patterns in B cells. Genes that are dependent on CHD4 for normal expression participate in key pathways necessary for B cell development, including IL-7 signaling and V(D)J recombination. The extensive list of affected genes related to these pathways makes it difficult to define a single mechanism that drives developmental arrest. This process involves both direct and indirect mechanisms that influence chromatin dynamics and transcription. Given this multifactorial regulation of lineage specification, further studies will be required to delineate the contributions of specific factors, such as CHD4, for the regulation of transcriptional and epigenetic programs defining B cell differentiation. However, a common underlying theme in each of these pathways is the impact of defective IL-7R signaling, which blocks proliferation. These effects may be exacerbated by the cells’ inability to undergo DNA damage repair and complete V(D)J recombination.
The most distinguishing characteristic of B cells is the productive rearrangement of Ig genes, which generates a diverse repertoire of antigen-specific receptors. Our data indicate that V(D)J recombination is impaired in CHD4-deficient pro-B cells. This conclusion is evidenced by defects in distal VH rearrangements which may be due, in part, to defective Igh locus contraction. Consistent with other studies, our RNA-seq data indicate that a number of components necessary for V(D)J recombination are expressed, and even up-regulated in CHD4-deficient pro-B cells, including Rag1, Foxo1, and Foxo3a transcripts, while IL-7R signaling is attenuated. In normal pro-B cells, transcription of these elements reflects increased chromatin accessibility at Igh loci (50). As evidenced by our 3D-FISH studies, the inefficient completion of distal VH gene rearrangements in CHD4-deficient pro-B cells may be due to an inability to establish long-range genomic interactions necessary for recombination with distant d-JH loci. Transcripts encoding multiple factors implicated in locus contraction, including Pax5, Ezh2, Brg1, Ikzf1, Tfe3, and Yy1 (57–62), were unchanged in pro-B cells isolated from CHD4-deficient vs. WT pro-B cells, suggesting that CHD4 may function downstream of proteins encoded by these genes. Rearrangements of distal VHJ558 segments may also be reduced in CHD4-deficient pro-B cells due to defects in IL-7R signaling, which recruits STAT5 to distal VH segments and increases accessibility of local chromatin (63). However, it is difficult to distinguish whether reduced Igh locus contraction is due to defects in intrinsic mechanisms of locus contraction, or whether developmental arrest occurs before the initiation of distal VH rearrangements in the mutant mice.
CHD4 has been linked with the fidelity of lineage-appropriate transcriptional programs (27, 64). Here, we determined that expression of CHD4 is important for establishing the pro-B cell program: CHD4-deficient pro-B cells promiscuously express transcripts of genes that are normally restricted to other lineages. Interestingly, NuRD complexes are important regulators of pluripotency and self-renewal in embryonic stem cells (reviewed in ref. 65). Two recent reports that examined CHD4-deficient embryos described changes in lineage commitment, with promiscuous expression of lineage marker genes that are related to a wide range of fate decisions (66, 67). Another study of requirements for cardiac striated muscle development reported that loss of CHD4 resulted in inappropriate expression of skeletal muscle gene expression; conversely, loss of CHD4 in skeletal muscle led to expression of key striated muscle genes (68). Thus, our data complement other studies demonstrating that CHD4 acts as a “gatekeeper” of lineage specification by controlling stem cell- and differentiation-associated genes in multiple tissues.
The question arises: Why are genes selectively up-regulated in the absence of CHD4? Certainly, the loss of CHD4 depletes pro-B cells of CHD4-NuRD, which can repress chromatin accessibility at genes that are normally silenced in these cells. We hypothesize that the loss of CHD4-dependent repression of genes that characterize other tissues, including the CNS, pancreas, and lung, makes these genes susceptible to activation by factors that are shared in common between pro-B and multiple cell types, but are normally insufficient to activate transcription in the presence of CHD4. Notably, HOMER analysis detected binding sites for ETS family proteins and other transcription factors frequently in new ATAC-seq peaks of 800 up-regulated genes (Fig. 6C and Dataset S2). These factors may activate transcription in synergy with other CRCs that colocalize at sites with CHD4-NuRD (e.g., Brg1; ref. 64). Alternatively, the absence of CHD4 may derepress expression of transcription factors necessary to “pioneer” non-B cell transcriptional programs. In non-B cell lineages, these drivers of other programs may overcome barriers imposed by CHD4-NuRD (or related CRCs) for transcriptional activation of lineage-specific genes. Conversely, the loss of ATAC-seq peaks and transcript expression of 195 genes correlated with binding sites for POU domain transcription factors within those peaks. We hypothesize that POU transcription factors are necessary for activation of these genes during developmental progression. Interestingly, half of the genes that fail to be activated in CHD4-deficient pro-B cells maintained their patterns of accessibility, as detected by ATAC-seq. This set of genes may be poised for transcriptional activation but are unable to be expressed due to the absence of a factor(s) necessary for developmental progression. Alternatively, small changes in the positioning of nucleosomes may block the binding of an essential activator(s) in the absence of CHD4 (27).
Loss of CHD4 function contributes to human diseases, but the full scope of its involvement is unknown. The partial or complete loss of CHD4 due to mutations is a driver of cancer (69, 70). Loss of CHD4 leads to unrepaired DNA DSBs (and microsatellite instability) that lead to deletions and chromosomal translocations (71). Interestingly, leukemogenesis is induced, in part, by redistribution of CHD4-NuRD complexes to new gene targets in lymphocytes in the absence of IKZF1 (Ikaros) (26). Thus, loss of function or inappropriate recruitment of CHD4-NuRD complexes each promote tumorigenesis. Additionally, the control of lineage specification by CHD4 in B cells suggests potential deleterious effects of its loss of function. If mature B cells with mutations in Chd4 genes (resulting in reduced CHD4 expression, but not cell death) persist at peripheral sites, inappropriate expression of non-B cell proteins in professional antigen-presenting cells may result in the display of novel antigenic determinants (i.e., neoantigens) that initiate autoimmune responses. Importantly, a series of de novo missense mutations in CHD4 genes have been detected in humans, resulting in intellectual disabilities and distinctive dysmorphisms (72).
Together, our studies demonstrated how CHD4, a key component of NuRD CRC, is required for the early development and lineage progression of B cells. In the future, additional studies will be necessary to integrate functions of CHD4 within a transcriptional network of regulators of B cell development and function.
Materials and Methods
Materials, including mouse models used and generated by this work, are available in SI Appendix. Mice were bred and housed in the Biological Resources Center at National Jewish Health. All experiments were performed under a protocol (AS2558-04-20) approved by the Institutional Animal Care and Use Committee at National Jewish Health. Reagents and methods used for cell preparation, flow cytometry, and cell culture; methods for analysis of STAT5 phosphorylation, qPCR analysis of IgH VH usage, 3D-FISH, confocal microscopy, and analysis of DNA damage are described in SI Appendix. Preparation of libraries, RNA-seq, and ATAC-seq were performed at the Center for Genes and Environment at National Jewish Health and are detailed in SI Appendix. Bioinformatic analysis of RNA-seq and ATAC-seq, including comparison with CHD4 ChIP-seq, and other statistical analysis, are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank J. Svaren and J. Cambier for the gifts of the Chd4 floxed and Cd79a-CreTg/+ mice; H. Huang and L. Wysocki for the constitutively active Stat5a and Igµ plasmids, respectively; R. Torres, C. Porter, A. Bhandoola, and E. Oltz for their insights; H. Lei and F. Han for excellent technical assistance; S. Welsh, K. Lukin, and J. Ramírez for helpful scientific discussions; Mark Dell’Aringa for critically reading the paper and helping with illustrations; and J. Loomis (National Jewish Flow Cytometry core) for excellent technical assistance. J.R.H. acknowledges NIH Awards R01AI081878, R01AI098417, R21AI115696, and a generous grant from the Wendy Siegel Fund for Leukemia and Cancer Research. T.A. was supported by the Victor W. Bolie and Earleen D. Bolie Graduate Scholarship Fund. C.D. was supported by a Cancer Research Institute Predoctoral Emphasis Pathway in Tumor Immunology Fellowship. A.B. was supported by the NIH, under Award F32 GM106631. C.J.F. was supported by the NIH Institutional National Service Award 2T32AI07449. A.J.F. was funded by NIH Grants R01AI21286, R21AI107343, and R03AI1154861. C.M. was funded by NIH Grant R01AI109599. B.P.O. acknowledges support by NIH Grant R01HL127461.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-seq and ATAC-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) under accession number GSE123504.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821301116/-/DCSupplemental.
References
- 1.Abdelrasoul H, Werner M, Setz CS, Okkenhaug K, Jumaa H (2018) PI3K induces B-cell development and regulates B cell identity. Sci Rep 8:1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyai T, et al. (2018) Three-step transcriptional priming that drives the commitment of multipotent progenitors toward B cells. Genes Dev 32:112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Revilla-I-Domingo R, et al. (2012) The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J 31:3130–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li R, et al. (2018) Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev 32:96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maier H, et al. (2004) Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol 5:1069–1077. [DOI] [PubMed] [Google Scholar]
- 6.McManus S, et al. (2011) The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J 30:2388–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwickert TA, et al. (2014) Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 15:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917–921. [DOI] [PubMed] [Google Scholar]
- 9.Xue Y, et al. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2:851–861. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279–289. [DOI] [PubMed] [Google Scholar]
- 11.Dege C, Hagman J (2014) Mi-2/NuRD chromatin remodeling complexes regulate B and T-lymphocyte development and function. Immunol Rev 261:126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatchalian J, et al. (2017) Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun 8:1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musselman CA, et al. (2009) Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J 423:179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musselman CA, et al. (2012) Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc Natl Acad Sci USA 109:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tencer AH, et al. (2017) Covalent modifications of histone H3K9 promote binding of CHD3. Cell Rep 21:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramírez J, Dege C, Kutateladze TG, Hagman J (2012) MBD2 and multiple domains of CHD4 are required for transcriptional repression by Mi-2/NuRD complexes. Mol Cell Biol 32:5078–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low JK, et al. (2016) CHD4 is a peripheral component of the nucleosome remodeling and deacetylase complex. J Biol Chem 291:15853–15866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nitarska J, et al. (2016) A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep 17:1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen HF, Wade PA, Kutateladze TG (2013) The NuRD architecture. Cell Mol Life Sci 70:3513–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita N, et al. (2004) MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119:75–86. [DOI] [PubMed] [Google Scholar]
- 21.Jaye DL, et al. (2007) The BCL6-associated transcriptional co-repressor, MTA3, is selectively expressed by germinal centre B cells and lymphomas of putative germinal centre derivation. J Pathol 213:106–115. [DOI] [PubMed] [Google Scholar]
- 22.Arenzana TL, Schjerven H, Smale ST (2015) Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev 29:1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottardi S, et al. (2014) The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis. PLoS Genet 10:e1004827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, et al. (1999) Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10:345–355. [DOI] [PubMed] [Google Scholar]
- 25.Liang Z, et al. (2017) A high-resolution map of transcriptional repression. eLife 6:e22767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, et al. (2011) Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bornelov S, et al. (2018) The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol Cell 71:56–72.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada T, et al. (2014) Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron 83:122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Shaughnessy A, Hendrich B (2013) CHD4 in the DNA-damage response and cell cycle progression: Not so NuRDy now. Biochem Soc Trans 41:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP (2010) Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J 29:3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith R, Sellou H, Chapuis C, Huet S, Timinszky G (2018) CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly(ADP-ribose)-dependent chromatin relaxation. Nucleic Acids Res 46:6087–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen DH, et al. (2010) The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 190:731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smeenk G, et al. (2010) The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtis CD, Griffin CT (2012) The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Mol Cell Biol 32:1312–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urquhart AJ, Gatei M, Richard DJ, Khanna KK (2011) ATM mediated phosphorylation of CHD4 contributes to genome maintenance. Genome Integr 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao H, et al. (2009) Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci USA 106:11258–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams CJ, et al. (2004) The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20:719–733. [DOI] [PubMed] [Google Scholar]
- 38.Pelanda R, et al. (2002) Cre recombinase-controlled expression of the mb-1 allele. Genesis 32:154–157. [DOI] [PubMed] [Google Scholar]
- 39.Hobeika E, et al. (2006) Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA 103:13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K (1991) Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 173:1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy RR, Kincade PW, Dorshkind K (2007) The protean nature of cells in the B lymphocyte lineage. Immunity 26:703–714. [DOI] [PubMed] [Google Scholar]
- 42.Peschon JJ, et al. (1994) Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med 180:1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Freeden-Jeffry U, et al. (1995) Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med 181:1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fistonich C, et al. (2018) Cell circuits between B cell progenitors and IL-7+ mesenchymal progenitor cells control B cell development. J Exp Med 215:2586–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goetz CA, Harmon IR, O’Neil JJ, Burchill MA, Farrar MA (2004) STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol 172:4770–4778. [DOI] [PubMed] [Google Scholar]
- 46.Jung D, Alt FW (2004) Unraveling V(D)J recombination; insights into gene regulation. Cell 116:299–311. [DOI] [PubMed] [Google Scholar]
- 47.Chowdhury D, Sen R (2003) Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity 18:229–241. [DOI] [PubMed] [Google Scholar]
- 48.Clark MR, Mandal M, Ochiai K, Singh H (2014) Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol 14:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebert A, et al. (2011) The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity 34:175–187. [DOI] [PubMed] [Google Scholar]
- 50.Verma-Gaur J, et al. (2012) Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci USA 109:17004–17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lennon GG, Perry RP (1985) C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature 318:475–478. [DOI] [PubMed] [Google Scholar]
- 52.Thompson A, Timmers E, Schuurman RK, Hendriks RW (1995) Immunoglobulin heavy chain germ-line JH-C mu transcription in human precursor B lymphocytes initiates in a unique region upstream of DQ52. Eur J Immunol 25:257–261. [DOI] [PubMed] [Google Scholar]
- 53.Bednarski JJ, Sleckman BP (2012) Lymphocyte development: Integration of DNA damage response signaling. Adv Immunol 116:175–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson K, et al. (2012) IL-7 functionally segregates the pro-B cell stage by regulating transcription of recombination mediators across cell cycle. J Immunol 188:6084–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honorat JA, et al. (2017) IgLON5 antibody: Neurological accompaniments and outcomes in 20 patients. Neurol Neuroimmunol Neuroinflamm 4:e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gazit R, et al. (2014) Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med 211:1315–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bossen C, et al. (2015) The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat Immunol 16:775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuxa M, et al. (2004) Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev 18:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H, et al. (2007) Yin Yang 1 is a critical regulator of B-cell development. Genes Dev 21:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reynaud D, et al. (2008) Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol 9:927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seet CS, Brumbaugh RL, Kee BL (2004) Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med 199:1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su IH, et al. (2003) Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 4:124–131. [DOI] [PubMed] [Google Scholar]
- 63.Bertolino E, et al. (2005) Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol 6:836–843. [DOI] [PubMed] [Google Scholar]
- 64.Giles KA, et al. (2019) Integrated epigenomic analysis stratifies chromatin remodellers into distinct functional groups. Epigenetics Chromatin 12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu G, Wade PA (2012) NuRD and pluripotency: A complex balancing act. Cell Stem Cell 10:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Shaughnessy-Kirwan A, Signolet J, Costello I, Gharbi S, Hendrich B (2015) Constraint of gene expression by the chromatin remodelling protein CHD4 facilitates lineage specification. Development 142:2586–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao H, et al. (2017) The chromatin remodeler Chd4 maintains embryonic stem cell identity by controlling pluripotency- and differentiation-associated genes. J Biol Chem 292:8507–8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gómez-Del Arco P, et al. (2016) The chromatin remodeling complex chd4/NuRD controls striated muscle identity and metabolic homeostasis. Cell Metab 23:881–892. [DOI] [PubMed] [Google Scholar]
- 69.Le Gallo M, et al. ; NIH Intramural Sequencing Center (NISC) Comparative Sequencing Program (2012) Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 44:1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar R, Li DQ, Müller S, Knapp S (2016) Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene 35:4423–4436. [DOI] [PubMed] [Google Scholar]
- 71.Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH (2011) Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 58:660–668. [DOI] [PubMed] [Google Scholar]
- 72.Weiss K, et al. ; DDD Study (2016) De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am J Hum Genet 99:934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.