Significance
Staphylococcus aureus is the most common cause of skin infections and is becoming increasingly resistant to antibiotics. If immune-based therapies are to provide an alternative to antibiotics, a better understanding of immunity to S. aureus skin infections is crucial. During an S. aureus skin infection in mice, a clonal Vγ6+Vδ4+ T cell population expressing a single complementarity-determining 3 (CDR3) region encoded by canonical TRGV6 and TRDV4 sequences expanded in the skin-draining lymph nodes, trafficked to infected skin, and promoted IL-17–mediated immune clearance by inducing neutrophil recruitment, inflammatory cytokines, and host defense peptides. Together, identification of a clonal T cell population in immunity to S. aureus skin infections provides a specific response to target for future vaccines and immunotherapies.
Keywords: Staphylococcus aureus, IL-17, T cells, neutrophils, skin
Abstract
T cell cytokines contribute to immunity against Staphylococcus aureus, but the predominant T cell subsets involved are unclear. In an S. aureus skin infection mouse model, we found that the IL-17 response was mediated by γδ T cells, which trafficked from lymph nodes to the infected skin to induce neutrophil recruitment, proinflammatory cytokines IL-1α, IL-1β, and TNF, and host defense peptides. RNA-seq for TRG and TRD sequences in lymph nodes and skin revealed a single clonotypic expansion of the encoded complementarity-determining region 3 amino acid sequence, which could be generated by canonical nucleotide sequences of TRGV5 or TRGV6 and TRDV4. However, only TRGV6 and TRDV4 but not TRGV5 sequences expanded. Finally, Vγ6+ T cells were a predominant γδ T cell subset that produced IL-17A as well as IL-22, TNF, and IFNγ, indicating a broad and substantial role for clonal Vγ6+Vδ4+ T cells in immunity against S. aureus skin infections.
The gram-positive extracellular bacterium Staphylococcus aureus causes the vast majority of skin infections in humans (1). In addition, S. aureus has become increasingly resistant to antibiotics, and multidrug-resistant community-acquired methicillin-resistant S. aureus (CA-MRSA) strains cause severe skin and invasive infections (e.g., cellulitis, pneumonia, bacteremia, endocarditis, osteomyelitis, and sepsis) in otherwise healthy individuals outside of hospitals, creating a serious public health concern (2, 3).
If immune-based therapies are to provide an alternative to antibiotics, an increased understanding of protective immunity against S. aureus skin infections is essential. This is imperative, because all prior S. aureus vaccines targeting antibody-mediated phagocytosis failed in human clinical trials (4). Notably, an S. aureus vaccine targeting the surface component iron surface determinant B against deep sternal wound infections and bacteremia following cardiothoracic surgery had a worse outcome, as individuals who suffered an S. aureus infection were five times more likely to die if they had received the vaccine rather than placebo (5).
As an alternative to antibody responses, there has been a recent focus on T cells in contributing to protective immunity against S. aureus infections. In humans, a variety of T cell subsets and cytokines has been implicated in host defense against S. aureus. For example, rare genetic diseases characterized by reduced IL-17–producing CD4+ T cells (i.e., Th17 cells) or IL-17–mediated immune responses (e.g., autosomal dominant hyper-IgE syndrome, IL-17F deficiency, and IL-17RA receptor deficiency) have an increased susceptibility to S. aureus skin infections (6–9). Similarly, in mouse models, IL-17 produced by γδ T cells and/or Th17 cells was found to be important in neutrophil recruitment and host defense against S. aureus skin and bacteremia infections (10–16). However, in vaccination attempts in mouse models of S. aureus skin and bacteremia infection, the IL-17–mediated protection was thought to be mediated by Th17 cells rather than γδ T cells (17–20). Additionally, IFNγ-producing CD4+ T cells (Th1 cells) were found to contribute to protection against S. aureus skin infections in patients with HIV disease as well as in S. aureus wound and bacteremia infections in mouse models (21–23). Another study found that the IFNγ produced by human CD8+ T cells contributed to antigen-induced immunity against S. aureus (24). We previously reported that IFNγ and TNF protected against a recurrent S. aureus skin infection in mice deficient in IL-1β (25). Finally, several studies have reported that IL-22 contributes to host defense peptide production and bacterial clearance of an S. aureus skin infection or mucosal colonization (10, 26–28).
Taken together, these findings in humans and mice suggest that different T cell subsets and their cytokine responses are involved in immunity against S. aureus infections. However, whether a predominant T cell subset and effector cytokine responses contribute to host defense against S. aureus skin infections is unclear. In particular, the studies in humans and mice suggest an important role for IL-17 responses in immunity against S. aureus, but the precise T cell sources and ensuing immune responses are not entirely understood. Therefore, we chose to determine the specific T cell subsets and mechanisms of IL-17–mediated immunity in cutaneous host defense in an in vivo mouse model of an S. aureus skin infection.
Results
Recruited Lymphocytes from Lymph Nodes Are Required for IL-17–Mediated Host Defense.
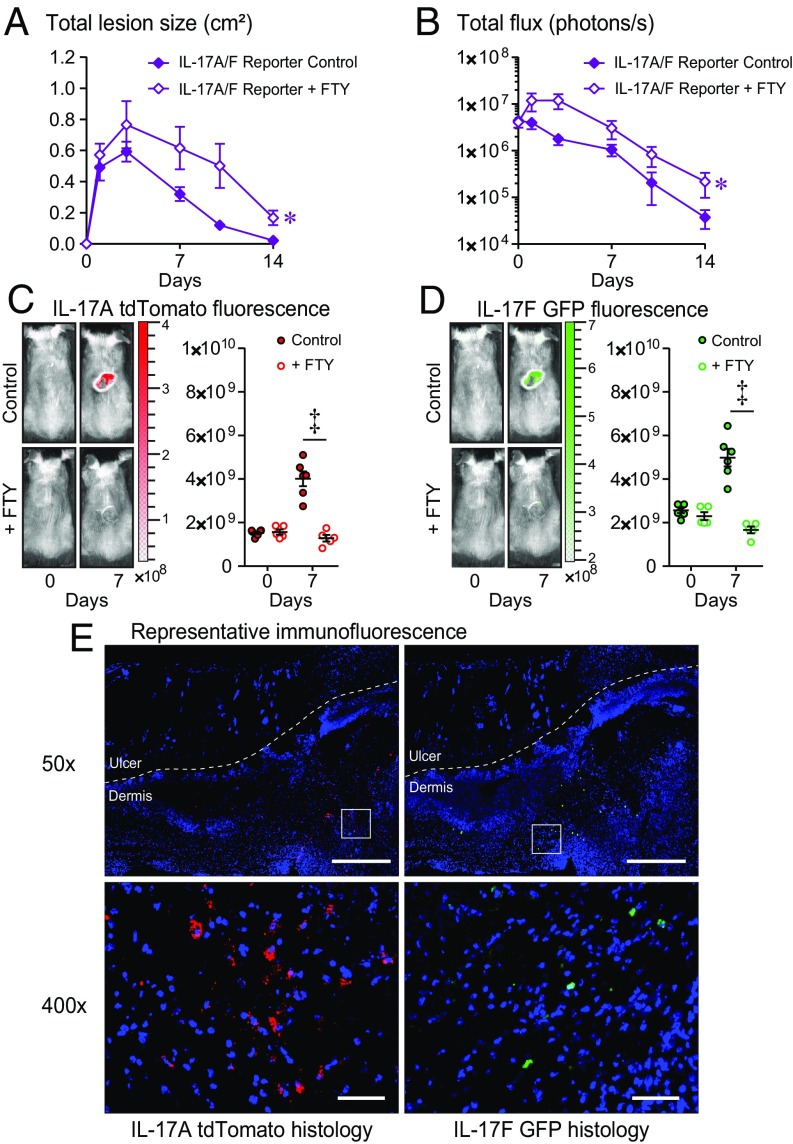
First, to determine whether the protective T cell immune response against an S. aureus skin infection was mediated by T cells residing in the skin or T cells recruited from lymph nodes, an intradermal (i.d.) S. aureus infection model was used (11, 25, 29–31) in which the bioluminescent CA-MRSA USA300 LAC::lux strain was injected intradermally into the back skin of mice ± FTY720 (administered on days −1, 0, and 1, and every other day thereafter until day 14 postinfection), which inhibits lymphocyte egress (including all T cells) from lymph nodes (25, 32). We chose to investigate the role of IL-17A and IL-17F because they are produced by many different T cell subsets and have been implicated in a variety of mouse models of S. aureus infection as being critical to host defense (10–16). For these experiments, we used an IL-17A-tdTomato/IL-17F-GFP dual-color reporter mouse strain, which is on a C57BL/6 background and produces normal levels of IL-17A and IL-17F. Before performing in vivo experiments, this reporter mouse strain was validated in vitro by culturing CD3+ T cells from skin-draining lymph nodes of IL-17A-tdTomato/IL-17F-GFP dual-color reporter mice in the presence of Th17/IL-17 polarizing conditions. We found that the expression of tdTomato and GFP by Th17 cells and γδ T cells closely corresponded to the expression of endogenous IL-17A and IL-17F protein levels using specific mAbs and intracellular flow cytometry (SI Appendix, Methods and Fig. S1).
Using this reporter mouse strain, we found that FTY720 treatment resulted in significantly increased skin lesion sizes and in vivo bioluminescence imaging (BLI) signals [which highly correlates with ex vivo CFUs harvested at different time points after the S. aureus i.d. skin infection (11, 25, 33)], compared with no treatment (P < 0.05) (Fig. 1 A and B). In vivo whole-animal fluorescence imaging (FLI) was sequentially employed (following in vivo BLI) to evaluate IL-17A expression (tdTomato fluorescence) and IL-17F expression (GFP fluorescence) in the S. aureus-infected skin in the anesthetized mice noninvasively over time. The in vivo FLI signals for tdTomato (IL-17A) and GFP (IL-17F) that peaked on day 7 were completely inhibited by treatment with FTY720 (P < 0.001) (Fig. 1 C and D). In addition, skin biopsy specimens of the S. aureus-infected skin obtained on day 7 were also evaluated by immunofluorescence microscopy for tdTomato-labeled IL-17A–producing cells and GFP-labeled IL-17F–producing cells, and these fluorescently labeled cells were found interspersed within the inflammatory infiltrate in the dermis underlying the necrotic epidermis and upper dermis (Fig. 1E). Taken together, IL-17A/F–producing cells that mediated host defense against the S. aureus skin infection in vivo were recruited from lymph nodes to the S. aureus-infected skin, and inhibition of this trafficking resulted in worsening of the infection.
Fig. 1.
Recruitment of IL-17A/F–producing T cells from lymph nodes to the skin is required for host defense against S. aureus skin infection. S. aureus skin infection was performed on IL-17A-tdTomato/IL-17F-GFP dual-color reporter mice (IL-17A/F reporter) ± FTY720 treatment (n = 5–10 mice per group). (A) Mean total lesion size (cm2) ± SEM. (B) Mean total flux (photons/s) ± SEM. (C and D) Representative in vivo fluorescence imaging signals and mean tdTomato (IL-17A) (C) or GFP (IL-17F) (D) total radiant efficiency ([p/s]/[μW/cm2]) ± SEM. (E) S. aureus-infected skin was harvested on day 7, and immunofluorescence microscopy labeling with anti-tdTomato and anti-GFP mAbs demonstrating localization of IL-17A/F production within cells in the dermis. [Scale bars, 500 μm (50×) and 50 μm (400×).] *P < 0.05, ‡P < 0.001, as calculated by a two-way ANOVA (A and B) or two-tailed Student’s t test (C and D). Data are representative of two independent experiments.
γδ T Cells Are the Major Cellular Source of IL-17.
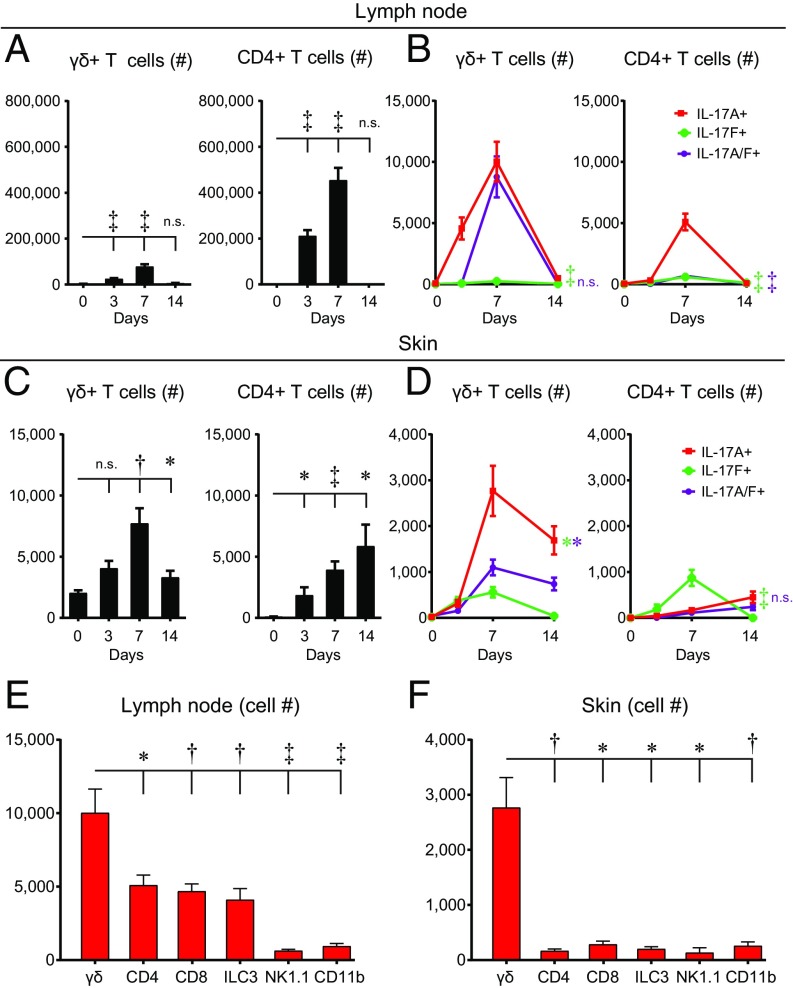
Next, to determine the T cell subset that produced the IL-17A and IL-17F, lymph nodes and skin samples from wild-type (wt) C57BL/6 mice were obtained on days 0 (naïve), 3, 7, and 14 during the S. aureus skin infection, and intracellular labeling for IL-17A+–, IL-17F+–, and IL-17A/F+–producing cells was performed and quantified by flow cytometry (Fig. 2 A–D). In the skin-draining lymph nodes, there were statistically greater total numbers of γδ T cells and CD4+ T cells on days 3 and 7, compared with naïve mice (day 0) (P < 0.001) (Fig. 2A). In addition, in the lymph nodes there were markedly increased numbers of IL-17A+ and IL-17A/F+ γδ T cells (which peaked on day 7) and barely detectable numbers of IL-17F+ γδ T cells (P < 0.001). There were also increased numbers of IL-17A+ CD4+ T cells (which also peaked on day 7) and to a much lesser extent IL-17F+ and IL-17A/F+ CD4+ T cells (P < 0.001) (Fig. 2B). In addition to differences in cell numbers, the mean fluorescence intensity (MFI) of IL-17A–expressing γδ T cells was fivefold higher than IL-17A–expressing CD4+ T cells, whereas the MFI of IL-17F–expressing CD4+ T cells was slightly higher than IL-17F–expressing γδ T cells (SI Appendix, Fig. S2), indicating that γδ T cells produced substantially higher amounts of IL-17A but similar amounts of IL-17F as CD4+ T cells.
Fig. 2.
γδ T cells are a predominant cellular source of IL-17A in the lymph nodes and S. aureus-infected skin. Skin-draining lymph nodes and skin specimens were harvested from wt mice on days 0 (naïve), 3, 7, and 14 after S. aureus skin infection, and flow cytometry was performed (n = 10 per group). (A and C) Mean number of γδ and CD4+ T cells ± SEM isolated from lymph nodes (A) or skin (C). (B and D) Mean number of IL-17A+, IL-17F+, and IL-17A/F+ γδ and CD4+ T cells ± SEM isolated from lymph nodes (B) or skin (D). (E and F) Mean cell number ± SEM of IL-17A+ cells, including γδ T cells, CD4+ T cells, CD8+ T cells, ILC3s (CD45+CD127+RORγt+, Lin−), natural killer cells (NK1.1+), and myeloid cells (CD11b+) isolated from skin-draining lymph nodes (E) and S. aureus-infected skin (F) on day 7. *P < 0.05, †P < 0.01, ‡P < 0.001, as calculated by a two-tailed Student’s t test. n.s., not significant. Data are a compilation of two independent experiments.
In the S. aureus-infected skin, there was a statistically greater total number of γδ T cells on days 7 and 14 (which peaked on day 7) (P < 0.01) and CD4+ T cells on days 3, 7, and 14 (P < 0.01), compared with naïve mice (Fig. 2C). In the S. aureus-infected skin, there was a substantial increase in the numbers of IL-17A+ γδ T cells (which peaked on day 7) and to a lesser extent IL-17F+ and IL-17A/F+ γδ T cells (P < 0.05). In the skin, there was also an increase in the numbers of IL-17F+ CD4+ T cells (which peaked on day 7), which were greater than the slightly increased numbers of IL-17A+ CD4+ T cells (P < 0.001), but the numbers of both IL-17A+ and IL-17A/F+ CD4+ T cells remained close to the low background numbers of naïve mice (Fig. 2D).
Next, total lymph node cells from day 7 of the S. aureus skin infection were obtained to determine the number of ex vivo IL-17A+ cells after PMA/ionomycin stimulation. γδ T cells represented the most abundant cellular source of IL-17A+, compared with CD4+ T cells, CD8+ T cells, innate lymphoid cells 3 (ILC3s), natural killer (NK) cells, and CD11b+ myeloid cells (Fig. 2E). In addition, immune cells from S. aureus-infected skin samples were obtained to determine the number of ex vivo IL-17A+ cells after PMA/ionomycin stimulation, especially since myeloid cells can be a source of IL-17A in other models of infection and inflammation (34). We found that γδ T cells were the most abundant cellular source of IL-17A, and there were fewer numbers of IL-17A+ CD11b+ cells and other cell types (Fig. 2F). Collectively, these data indicate that γδ T cells were the most abundant cellular source of IL-17 in the skin-draining lymph nodes and S. aureus-infected skin.
IL-17A and IL-17F Have Compensatory and Redundant Roles in Host Defense.
In other bacterial, fungal, and viral infection models, IL-17A and IL-17F have been shown to have either redundant or differential roles in immune responses (12, 35–37). In particular, Ishigame et al. (12) reported redundant activity of IL-17A and IL-17F in spontaneous S. aureus mucocutaneous infections that developed in their mouse colony in mice with constitutive genetic deletion of both IL-17A and IL-17F but not deletion of IL-17A or IL-17F alone. However, 16S rDNA sequencing revealed that S. aureus was not present in the skin microbiome of our mouse colony (38), and consequently our IL-17A/F–deficient mice did not spontaneously develop S. aureus mucocutaneous infections. This provided the opportunity for us to investigate the relative contribution and temporal dynamics of IL-17A and IL-17F responses that occurred during the course of an acute S. aureus skin infection, which would be more representative of how acute S. aureus skin infections commonly occur in humans. Therefore, the S. aureus i.d. infection model was performed in mice deficient in both IL-17A and IL-17F (IL-17A/F−/−) and wt C57BL/6 mice. IL-17A/F−/− mice developed significantly increased lesion sizes during the course of infection that peaked on day 7 compared with wt mice (P < 0.05) (SI Appendix, Fig. S3 A and B). In addition, IL-17A/F−/− mice had significantly increased bacterial burden during the course of infection as measured by in vivo BLI, especially from days 3–14 compared with wt mice (P < 0.01) (SI Appendix, Fig. S3 C and D). These data were similar to the increased lesion size data and in vivo BLI data observed in FTY720-treated mice (Fig. 1 A and B), providing further evidence that the IL-17A/F response was due to recruited γδ T cells. To confirm the in vivo BLI data, on day 7 the S. aureus-infected skin was homogenized and ex vivo CFUs were enumerated (SI Appendix, Fig. S3E). IL-17A/F−/− mice had statistically greater (i.e., approximately fourfold) ex vivo CFUs compared with wt mice (P < 0.01). Taken together, these results suggest that γδ T cell-derived IL-17A and/or IL-17F contributed to host defense during the S. aureus skin infection, especially at time points beyond day 3 after the bacterial skin inoculation.
To further evaluate the contribution of IL-17A versus IL-17F, wt mice were treated systemically (i.p. injection) with an anti–IL-17A neutralizing mAb, an anti–IL-17F neutralizing mAb, anti–IL-17A and anti–IL-17F neutralizing mAbs combined, or an isotype control mAb (SI Appendix, Fig. S3 F and G). Anti–IL-17A and anti–IL-17F neutralizing mAbs combined resulted in statistically increased lesion sizes (P < 0.001) and in vivo BLI signals (P < 0.001) during the course of infection compared with isotype mAb-treated wt mice. In contrast, there were no significant differences in lesion sizes or in vivo BLI signals with treatment of either anti–IL-17A mAb alone or anti–IL-17F mAb alone compared with isotype mAb-treated wt mice. To further evaluate the contribution of IL-17A versus IL-17F, IL-17A/F−/− mice were administered recombinant IL-17A (rIL-17A) or rIL-17F along with the i.d. bacterial inoculum in the skin (SI Appendix, Fig. S3 H and I). As in SI Appendix, Fig. S3 B and D, IL-17A/F−/− mice had significantly increased lesion sizes and bacterial burden as measured by in vivo BLI during the course of infection compared with wt mice (P < 0.001). However, administration of either rIL-17A or rIL-17F resulted in significantly reduced lesion sizes (P < 0.001) and lower in vivo BLI signals (P < 0.001 and P < 0.05, respectively) (similar to those observed in wt mice) compared with IL-17A/F−/− mice. Taken together, since neutralization of both IL-17A and IL-17F was required to observe an immune defect in wt mice and treatment with either rIL-17A or rIL-17F was sufficient to rescue the immune impairment in IL-17A/F−/− mice, these findings indicate that γδ T cell-derived IL-17A and IL-17F have redundant and compensatory roles in host defense against an S. aureus skin infection.
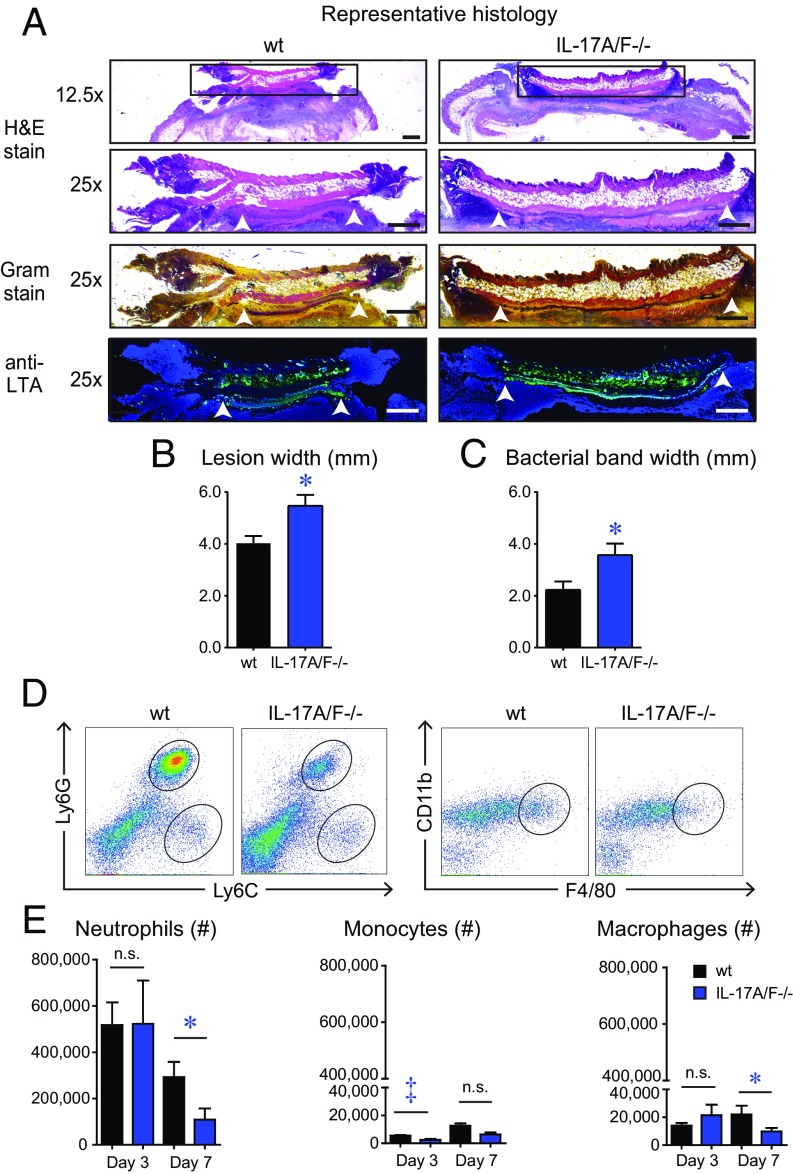
IL-17A/F–Deficient Mice Develop Increased Skin Necrosis and Bacterial Clusters and Impaired Neutrophil Recruitment.
Based upon histologic analysis on day 7 after infection, IL-17A/F−/− mice had significantly increased width of epidermal and dermal skin necrosis compared with wt mice (P < 0.05) (Fig. 3 A and B), which is consistent with the larger skin lesion sizes observed by gross morphology using digital photography (Fig. 3 A and B). In addition, in Gram stain and anti-S. aureus lipoteichoic acid (LTA) antibody-labeled sections, IL-17A/F−/− mice had a statistically significant increase in width of the horizontal band of bacterial clusters in the dermis (underlying the area of skin necrosis), compared with wt mice (Fig. 3A, white arrowheads and Fig. 3C), corroborating the increased bacterial burden as measured by in vivo BLI and ex vivo CFUs in IL-17A/F−/− mice compared with wt mice (SI Appendix, Fig. S3 C–E).
Fig. 3.
IL-17A/F–deficient mice develop increased skin necrosis and bacterial burden along with impaired neutrophil recruitment. S. aureus skin infection was performed on IL-17A/F−/− and wt mice and the infected skin tissue was harvested on day 7 for histology (n = 8 mice per group) and on days 3 and 7 for flow cytometry analysis (n = 10 mice per group). (A) Representative histology of skin biopsy specimens obtained on day 7 and stained with hematoxylin and eosin (H&E) stain or Gram stain and immunofluorescence labeling with an anti-LTA mAb. The three Lower panels (25×) are from the boxed area in the Upper panel (12.5×). (Scale bars, 500 μm.) White arrowheads indicate peripheral ends of the horizontal band of bacterial clusters in the dermis. (B) Mean skin necrosis width (mm) ± SEM. (C) Mean bacterial band width (mm) ± SEM. (D) Representative flow plots of neutrophils (Ly6GhiLy6Cint), monocytes (Ly6GloLy6Chi), and macrophages (CD11b+F4/80+) from S. aureus-infected skin on day 7. (E) Total number of cells ± SEM on days 3 and 7. *P < 0.05, ‡P < 0.001, as calculated by a two-tailed Student’s t test. n.s., not significant. Data in B, C, and E are a compilation of two independent experiments.
To evaluate whether the immune impairment in IL-17A/F−/− mice was due to defective neutrophil, monocyte, and/or macrophage recruitment, day 3 and 7 skin biopsies were harvested and the numbers of these myeloid cells were determined by flow cytometry, according to Ly6GhiLy6Cint (neutrophils), Ly6GloLy6Chi (monocytes), and CD11b+F4/80+ (macrophages) (Fig. 3 D and E). On day 3, there was no significant difference in neutrophil number between IL-17A/F−/− mice and wt mice. However, on day 7, IL-17A/F−/− mice had nearly a threefold statistically significant reduction in neutrophils compared with wt mice. In addition, IL-17A/F−/− mice also had significantly decreased monocyte numbers on day 3 and macrophage numbers on day 7 compared with wt mice. However, these slight differences in monocyte and macrophage numbers might not be as biologically relevant as the differences in neutrophil numbers, because there were more than ∼15- to 30-fold fewer monocytes and macrophages than neutrophils during the S. aureus skin infection.
IL-17A/F–Deficient Mice Express Decreased Proinflammatory Cytokines and Host Defense Peptides.
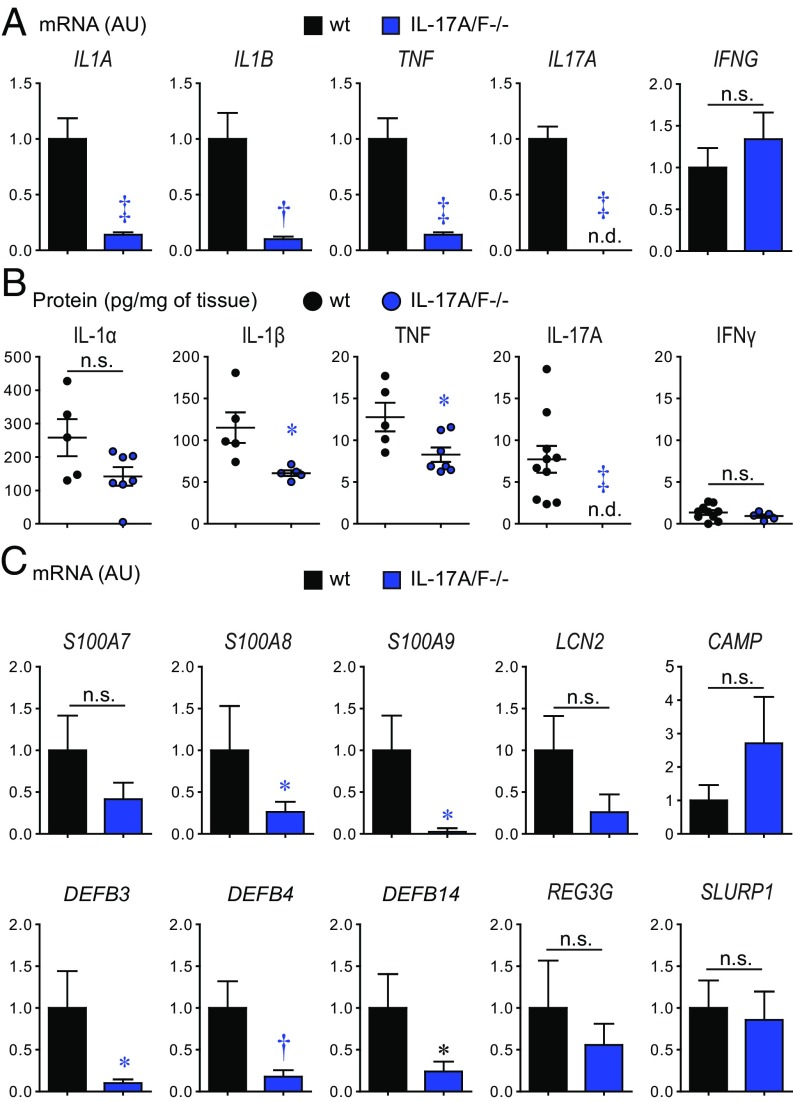
Next, to determine whether IL-17A and IL-17F contributed to expression of other proinflammatory cytokines, day 7 skin biopsies were evaluated for mRNA and protein levels of IL-1α, IL-1β, and TNF, which have each been implicated in immunity against S. aureus skin infections (25, 30, 31). IL-17A/F−/− mice had markedly decreased mRNA transcript expression of IL1A (P < 0.001), IL1B (P < 0.01), and TNF (P < 0.001), significantly decreased protein levels of IL-1β (P < 0.05) and TNF (P < 0.05), and a trend toward decreased protein levels of IL-1α (P = 0.06) (Fig. 4 A and B). As expected, IL-17A was not detected in IL-17A/F−/− mice. In contrast to these findings, there was no difference between mRNA or protein levels of IFNγ in IL-17A/F−/− mice and wt mice.
Fig. 4.
IL-17A/F regulates cytokine and host defense peptide expression in S. aureus-infected skin. S. aureus skin infection was performed on IL-17A/F−/− and wt mice and infected skin tissue was harvested on day 7 (n = 5–10 per group). (A) Mean mRNA levels of cytokines (arbitrary units; AU) ± SEM. (B) Mean protein levels (pg/mg of tissue) ± SEM. (C) Mean levels of host defense peptides (AU) ± SEM. *P < 0.05, †P < 0.01, ‡P < 0.001, as calculated by a two-tailed Student’s t test. n.d., not detected; n.s., not significant. Data are representative of two independent experiments.
In addition, several host defense peptides have either bacteriostatic or bactericidal activity against S. aureus in various different mouse models of S. aureus infection, including psoriasin (S100a7) (10), calprotectin (S100a8/S100a9) (39), β-defensins (mBD3, mBD4, and mBD14) (10, 40, 41), lipocalin 2 (42), cramp (10, 43, 44), Reg3γ (45), and Slurp1 (27), but whether the expression of these relevant host defense peptides is regulated by IL-17A or IL-17F during an in vivo S. aureus skin infection is incompletely understood. Therefore, we performed our S. aureus skin infection model in IL-17A/F−/− and wt mice, and skin biopsy specimens on day 7 were evaluated for mRNA levels of these host defense peptides. IL-17A/F−/− mice had a significant decrease in mRNA transcript expression of calprotectin (S100A8/S100A9, P < 0.05) and β-defensins (DEFB3, P < 0.05; DEFB4, P < 0.01; and DEFB14, P < 0.05) but not LCN2, CAMP, REG3G, or SLURP1, compared with wt mice (Fig. 4C). These results indicate that during an S. aureus skin infection, IL-17A/F predominantly regulated the expression of calprotectin and mouse β-defensins 3, 4, and 14.
Vγ6+Vγ4+ γδ T Cells Clonally Expanded in LNs in Response to S. aureus Skin Infection.
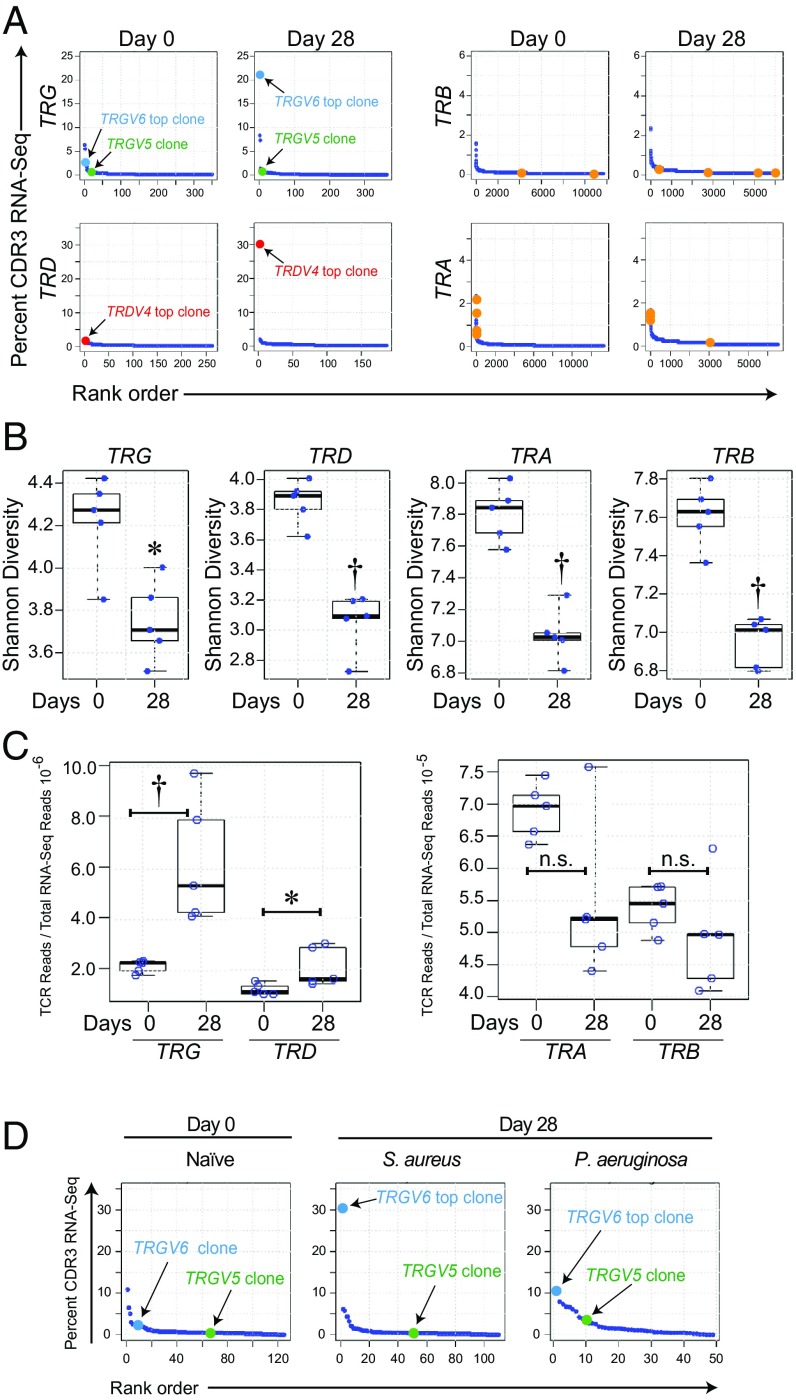
Since γδ T cells were the major cellular source of IL-17A in the lymph nodes and S. aureus-infected skin (Fig. 2), we set out to determine if there was clonotypic expansion of γδ T cells (in comparison with αβ T cells) in response to the S. aureus skin infection (Fig. 5). TCR nucleotide (nt) sequences encoding complementarity-determining region 3 (CDR3) amino acid sequences were mined from RNA-sequencing (RNA-seq) datasets of day 0 (naïve) and day 28 harvested wt lymph nodes following S. aureus skin infection. Results are reported here following International ImMunoGeneTics (IMGT) (http://www.imgt.org/) nomenclature (Fig. 5A). The lymph nodes of naïve wt mice had diverse αβ and γδ T cell repertoires with no expanded “public” (present in multiple mice) clones, although unexpanded TRG, TRD, TRA, or TRB clones were present in multiple animals (Fig. 5A). However, in response to the S. aureus skin infection (i.e., day 28), in all wt mice there was a single “top” (dominant) TRGV6 CDR3-encoding nt sequence that comprised 21% of all TRG CDR3-encoding nt sequences, with concomitant expansion of a single top TRDV4 CDR3-encoding nt sequence that comprised 30% of all TRD CDR3-encoding nt sequences (Fig. 5A, Table 1, and SI Appendix, Tables S1 and S2). This expanded TRGV6 nt sequence encodes the amino acid sequence CACWDSSGFHKVF, which in this case likely pairs with the expanded TRD4-encoded receptor CGSDIGGSSWDTRQMFF. These TCR chains are identical to those expressed by the previously described invariant Vγ6+Vδ4+ T cells (previously referred to as Vγ6+Vδ1+ T cells according to prior nomenclature) (46), which are resident in the female reproductive tract, lung, and peritoneum and have been shown to preferentially expand in a wide variety of inflammatory settings (47, 48). Interestingly, and as has been previously noted, the expanded TRGV6 CDR3 sequence CACWDSSGFHKVF has the exact same CDR3 amino acid sequence as the TRGV5-encoding CDR3 sequence expressed by the dendritic epidermal T cells (DETCs), which are Vγ5+Vδ4+ γδ T cells that normally reside in mouse epidermis (previously referred to as Vγ5+Vδ1+ T cells according to prior nomenclature) (49–51). Therefore, the lymph node RNA-seq dataset was also mined for the TRGV5 nt sequence of DETCs. Remarkably, even though the canonical TRGV5 nt sequence encoded the exact same CDR3 amino acid sequence (CACWDSSGFHKVF) as the S. aureus-expanded TRGV6, there was no expansion of this canonical TRGV5 nt sequence in response to the S. aureus skin infection, as the percentage of TRGV5 CACWDSSGFHKVF-encoding sequences was similar in wt and day 28 S. aureus-infected mice (0.7 and 0.9%, respectively) (Fig. 5A and Table 1). Thus, although CACWDSSGFHKVF can be encoded by either TRGV5 or TRGV6, only the TRGV6-encoded sequences expand in response to S. aureus. There were also public CDR3-encoding TRA and TRB nt sequences that were present at low frequencies in the lymph nodes of naïve mice (i.e., below 2.5% of the total respective CDR3-encoding nt sequences) (Fig. 5A). However, these sequences did not expand following the S. aureus skin infection on day 28. Moreover, on day 28, there were no public TRA and TRB CDR3-encoding nt sequence expansions that were greater than 2% of the total respective TRA and TRB CDR3-encoding nt sequences in response to the S. aureus skin infection.
Fig. 5.
Clonotypic T cell expansion in lymph nodes in response to S. aureus skin infection. TCR complementarity-determining region 3 sequences were mined from an RNA-seq dataset of skin-draining lymph nodes harvested from wt mice on days 0 (naïve) and 28 after S. aureus skin infection (n = 5 mice per group) (25). (A) Pooled results from all lymph node samples presented as the CDR3 nucleotide sequence rank (x axis) versus the percentage of the total TRG, TRD, TRA, and TRB CDR3-encoding nt reads occupied for each particular CDR3 nt sequence (y axis). Dark blue dots indicate each of the different CDR3-encoding nt reads. Light blue dots and red dots indicate top (dominantly expanded) TRGV6- and top TRDV4-encoding CDR3 nt sequence on day 28 lymph nodes, respectively. Green dots indicate TRGV5-encoding CDR3 nt sequence that encodes the exact same amino acid sequence as the top TRGV6-encoding CDR3 nt sequence and is expressed by DETCs. Orange dots indicate public (found in all samples) TRA- and TRB-encoded CDR3 nt sequences. (B) Shannon diversity index ± SEM (boxplot: 95% confidence interval). (C) Proportion of TRG, TRD, TRA, or TRB CDR3-encoding nt reads of total RNA-seq reads ± SEM (boxplot: 95% confidence interval). (D) RNA-seq was performed for TCR CDR3 sequences of skin-draining lymph nodes harvested from wt mice on days 0 (naïve) and 28 after S. aureus or P. aeruginosa skin infection (n = 6 mice per group) (86); data are reported as in A. *P < 0.05, †P < 0.01, as calculated by a Wilcoxon rank-sum test. n.s., not significant.
Table 1.
TRG nt sequence alignments
| Gene | V | J | CDR3 nt sequence | CDR3 amino acid sequence | Percentage of all CDR3 nt sequences | |||
| LN day 0 | LN day 28 | Skin day 0* | Skin day 7* | |||||
| TRG | V6 | J1 | tgtgcatgctgggatagctcaggttttcacaaggtattt | CACWDSSGFHKVF | 2.8 | 21.1 | 0 | 4.6 |
| TRG | V5 | J1 | tgtgcctgctgggatagctcaggttttcacaaggtattt | CACWDSSGFHKVF | 0.7 | 0.9 | 15.7 | 6.9 |
| TRD | V4 | J2 | tgtgggtcagatatcggagggagctcctgggacacccgacagatgtttttt | CGSDIGGSSWDTRQMFF | 1.6 | 30.0 | 3.8 | 10.9 |
TRGV6, TRGV5, and TRDV4 CDR3 nucleotide sequences, gene sequence alignments (underlining indicates nucleotide differences), and respective encoded CDR3 amino acid sequences of the different clonotypes from the TRG6, TRG5, and TRD4 CDR3 reads in Fig. 5A and Brady et al. (52). The percentage of each of the specific TRGV6, TRGV5, or TRDV4 CDR3 nt sequences of the total number of reads of all respective TRGV6, TRGV5, or TRDV4 CDR3 nt sequences is shown for lymph node (LN) specimens from days 0 (naïve) and 28 as well as for skin specimens of S. aureus-infected skin from days 0 and 7 from Brady et al. (52).
Brady et al. (52).
To determine whether there were similar expansions of the specific TRGV6 and TRDV4 nt sequences in S. aureus-infected wt mouse skin, we mined the RNA-seq dataset of Brady et al. (52), which included S. aureus-infected skin of wt mice on days 0 and 7 (Table 1). This revealed that the same TRGV6 nt sequence encoding the Vγ6 CDR3 amino acid sequence CACWDSSGFHKVF that expanded in the lymph nodes also expanded in the S. aureus-infected skin from 0% (day 0) to 4.6% (day 7). In addition, the same TRD4 nt sequence encoding the Vδ4 CDR3 amino acid sequence CGSDIGGSSWDTRQMFF that expanded in the lymph nodes also expanded in the S. aureus-infected skin from 3.8% (day 0) to 10.9% (day 7). Interestingly, the percentage of the TRGV5 nt sequence encoding the same CDR3 amino acid sequence as the TCR Vγ6 CDR3 amino acid sequence CACWDSSGFHKVF decreased in the S. aureus-infected skin from 15.7% (day 0) to 6.9% (day 7) (Table 1), which was likely in part due to the increased total TRG reads caused by the expansion in the clonotypic TRGV6 nt sequence. Importantly, the S. aureus-responsive TRGV6 clones that expanded in the lymph nodes were undetectable in naïve mouse skin, at the depth of sequencing performed. This supports the data in Figs. 1 and 2, which demonstrate that the immune protection is mediated by T cells that trafficked from the lymph nodes to the S. aureus-infected skin.
Next, the diversity of TRG, TRD, TRA, and TRB CDR3-encoding nt sequences was then evaluated by calculating their Shannon diversity indices, which demonstrated a statistically significant decrease in the diversity of all of the total respective CDR3-encoding nt sequences in response to the S. aureus skin infection on day 28, compared with naïve mice (Fig. 5B). These data indicate that the skin-draining αβ T cell repertoire was also altered by S. aureus skin infection, although there were no public αβ T cell clonal expansions. Importantly, there are relatively fewer TRG- and TRD-mapping reads in the skin-draining lymph nodes compared with TRA- and TRB-mapping reads; however, the proportional increase of TRG and TRD sequences (respective to total RNA-seq reads) was greater following S. aureus skin infection (Fig. 5C). This is strong evidence to support the importance of the γδ T cell clonal expansion in the S. aureus-induced immune response.
The Clonotypic T Cell Expansion Was More Specific to S. aureus Skin Infection.
To determine whether the clonotypic TRGV6 expansion was specific to S. aureus, we evaluated whether a similar expansion occurred in response to a gram-negative skin infection with Pseudomonas aeruginosa. P. aeruginosa induced skin lesions that were smaller than those of S. aureus (P < 0.001) (SI Appendix, Fig. S4 A and B). However, the in vivo BLI signals of P. aeruginosa and S. aureus peaked to a similar level on day 1 (SI Appendix, Fig. S4 C and D). P. aeruginosa signals then decreased more rapidly than those of S. aureus (P < 0.001). On day 28, the lymph nodes from the S. aureus- and P. aeruginosa-infected mice as well as naïve wt mice were harvested, RNA-seq was performed, and TCR nt sequences encoding CDR3 amino acid sequences were mined as in Fig. 5A. From these sequences, the percentage of TRGV6 and TRGV5 sequences encoding the same clonotypic CDR3 amino acid sequence (CACWDSSGFHKVF) was determined (Fig. 5D). In day 0 (naïve) mice, the clonotypic TRGV6 and TRGV5 CDR3-encoding nt sequences represented 2.3 and 0.43% of all TRG CDR3-encoding nt sequences, respectively. Similar to Fig. 5A, in response to S. aureus, the clonotypic TRGV6 sequence markedly expanded 13.3-fold to 30.3% of all TRG CDR3-encoding nt sequences, whereas TRGV5 had no expansion (i.e., 0.37%). In contrast, in response to P. aeruginosa, both clonotypic TRGV6 and TRGV5 modestly increased to 10.6 and 3.4%, respectively, of all TRG CDR3-encoding nt sequences. Thus, following S. aureus skin infection, the ratio of the clonotypic TRGV6 nt sequence to the clonotypic TRGV5 nt sequence in the skin-draining lymph nodes was 81.9 compared with 3.1 following P. aeruginosa skin infection. The latter was similar to the ratio observed in naïve animals (5.3). Taken together, the P. aeruginosa skin infection had less specificity, as there was a modest increase in both clonotypic TRGV6 and TRGV5 CDR3-encoding nt sequences, whereas the S. aureus skin infection induced a specific dominant expansion of only the clonotypic TRGV6 CDR3-encoding nt sequences.
Vγ6+ T Cells Are the Most Abundant γδ T Cell Subset in Lymph Nodes.
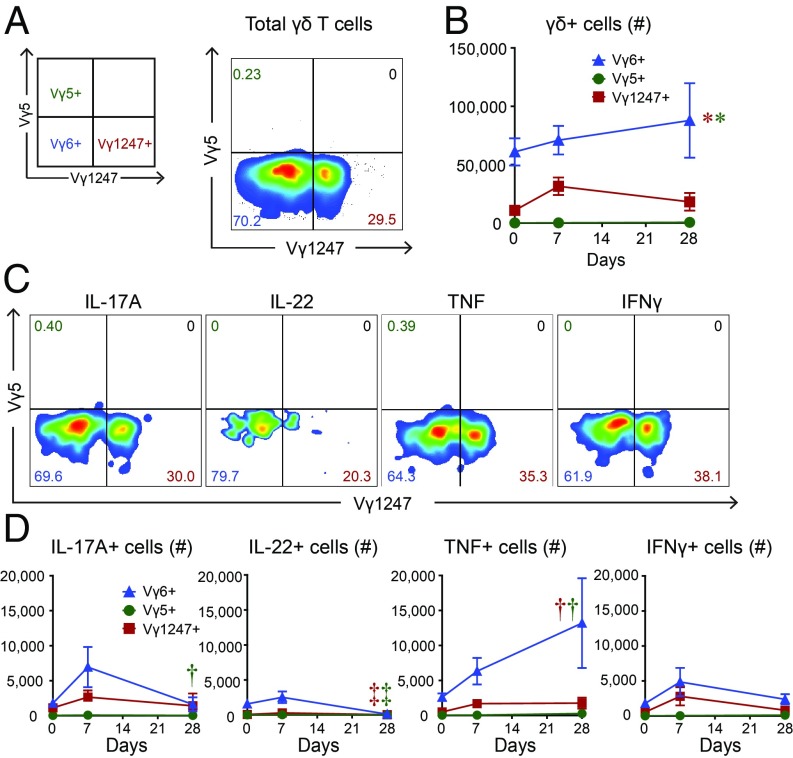
Flow cytometry was used to confirm the RNA-seq data, which indicated a marked expansion of a clonotypic population of Vγ6+Vδ4+ γδ T cells. This confirmation is important, because the canonical TRGV6 and TRGV5 nt sequences are known to pair with the canonical TRD4 sequence to encode the same CDR3 amino acid sequence of Vγ6+Vδ4+ γδ T cells and DETCs, respectively; however, only TRGV6 and TRDV4 but not TRGV5 CDR3-encoding nt sequences expanded. Skin-draining lymph nodes were obtained from wt mice on 0 (naïve), 7, and 28 d following the S. aureus skin infection, and the total numbers of Vγ6+ cells, Vγ5+ cells, and a combined group (denoted Vγ1247+) that utilize alternative mouse Vγ chains (i.e., Vγ1, Vγ2, Vγ4, and Vγ7) were determined by flow cytometry [Fig. 6A; Vγ3 was not assessed because it is a pseudogene in many mouse strains and, although it might theoretically be functional in C57BL/6 mice, it is so rare it can be disregarded (53, 54)]. The flow cytometry gating strategy first involved gating on live cells, followed by gating on CD3+ TCRγδ+ T cells. Since there is no specific mAb for Vγ6+ cells, γδ T cells were labeled with a mAb against Vγ5 (y axis, Fig. 6A; clone F536) versus γδ T cells labeled with a combination of mAbs specific for virtually all of the other Vγ chains except Vγ6 (Vγ1 and Vγ2, clone 4B2.9; Vγ4, clone UC3; and Vγ7, clone GL1.7) (x axis, Fig. 6A). Vγ6+ cells had significantly increased numbers (which steadily increased from day 0 to day 28), compared with either the Vγ1247+ cells (which peaked on day 7 but the numbers were ∼55–80% lower than the Vγ6+ cells) or Vγ5+ cells (which were barely detectable) (Fig. 6B). After ex vivo PMA/ionomycin stimulation, there were also significantly increased numbers of Vγ6+IL-17A+, Vγ6+IL-22+, Vγ6+TNF+, and Vγ6+IL-22+ cells (which all peaked on day 7, except for Vγ6+TNF+ cells that steadily increased and peaked on day 28), compared with either the Vγ1247+ cells or Vγ5+ cells (which were barely detectable) (Fig. 6 C and D). These data confirm that of the γδ T cell subsets present in the skin-draining lymph nodes, Vγ6+ cells represented the greatest number and the most abundant source of IL-17A as well as IL-22, TNF, and IFNγ after ex vivo stimulation. Further, there were barely detectable numbers of Vγ5+ cells in the lymph nodes at all time points evaluated, corroborating the RNA-seq data indicating that Vγ6+ cells rather than Vγ5+ cells were the γδ T cells that expanded in response to the S. aureus skin infection.
Fig. 6.
γδ T cell subsets that expand in response to S. aureus skin infection. Skin-draining lymph nodes were harvested from wt mice on days 0 (naïve), 7, and 28 after S. aureus skin infection and flow cytometry was performed (n = 5 mice). (A) Representative flow plot of γδ T cells (first gated on live cells, then CD3+ and TCRγδ+ cells) from day 7 labeled with mAbs against a combined group of Vγ1, Vγ2, Vγ4, and Vγ7 (i.e., Vγ1247+) (x axis) versus a mAb against Vγ5 (i.e., Vγ5+) (y axis). Vγ6+ γδ T cells are unlabeled (i.e., Vγ1247− Vγ5−) (Left Lower). (B) Total number of cells ± SEM. (C) Representative flow plots of IL-17A–, IL-22–, TNF-, and IFNγ-producing Vγ1247+, Vγ5+, and Vγ6+ γδ T cells. (D) Total number of cells ± SEM. *P < 0.05, †P < 0.01, ‡P < 0.001, as measured by a two-way ANOVA. Data are representative of two independent experiments.
Discussion
T cells and their cytokine responses have been implicated in host defense against S. aureus infections, but whether a predominant T cell subset can mediate protection is not entirely clear. In the present study, we employed a mouse model of S. aureus skin infection and found that recruited γδ T cells from lymph nodes to the S. aureus-infected skin were critical in mediating IL-17 immune responses, including induction of neutrophil recruitment, proinflammatory cytokines, and host defense peptides. Moreover, the primary T cell source of IL-17 was from a population of clonotypic Vγ6+Vδ4+ γδ T cells expressing a single TCR CDR3 amino acid sequence (generated from canonical TRGV6 and TRDV4 nt sequences). Taken together, the findings provide several new and important insights into the role and mechanisms of γδ T cells and ensuing IL-17 responses in host defense and resolution of S. aureus skin infections.
First, using an IL-17A/F dual-color reporter mouse strain, we determined that recruited rather than skin-resident T cells were required to mediate host defense against the S. aureus skin infection. These results are consistent with the increasing role of IL-17A/F–producing γδ T cells that are rapidly recruited by trafficking through the bloodstream to sites of infection and inflammation in the skin (32, 55–59), including to an S. aureus i.p. infection (60). Consistent with these data, flow cytometry revealed that γδ T cells primarily produced IL-17A in the skin-draining lymph nodes and in the S. aureus-infected skin to a much greater extent than CD4+ T cells (i.e., Th17 cells), CD8+ T cells, ILC3s, NK cells, or myeloid cells (Fig. 2 E and F). The trafficking of the γδ T cells to the skin was crucial for host defense, because naïve mouse skin (day 0) had virtually undetectable numbers of IL-17–producing γδ T cells (Fig. 2D), and provides an explanation for the impaired host defense in mice treated with FTY720 (Fig. 1 A and B). Of note, the impairments in lesion size and bacteria burden in FTY720-treated mice occurred earlier (beginning on day 1) compared with those of IL-17A/F−/− mice (beginning on day 3). This difference was likely due to effects of FTY720 other than lymphocyte egress from lymph nodes [reviewed elsewhere (61)], including monocyte egress from the bone marrow or neutrophil recruitment to the skin that might have contributed to host defense against S. aureus (62, 63). It should also be mentioned that we found varying expression of IL-17A+, IL-17F+, and IL-17A/F+ γδ and CD4+ T cells from lymph nodes and S. aureus-infected skin by flow cytometry (Fig. 2) and γδ and CD4+ T cells from lymph nodes of IL-17A/F dual-color reporter mouse strain in vitro after Th17/IL-17 polarizing conditions (SI Appendix, Fig. S1). These findings are consistent with single-cell sequencing of IL-17–producing γδ T cells and Th17 cells (64, 65) and prior results using IL-17 reporter mice created by other groups [e.g., Il17aCreR26ReYFP (66), IL-17F-CreEYFP (67), IL-17F reporter mice (Il17fThy1.1/Thy1.1) (68), and Smart-17A mice (surface marker for transcription-17A mice) (69)], which indicate that IL-17A and IL-17F are often coexpressed and can be differentially induced. In particular, differential expression of IL-17A versus IL-17F can be induced by the activity of certain transcription factors such as RORα versus RORγ (70), interleukin-2–inducible T cell kinase (Itk) (71), and STAT3 versus STAT5 (72).
Second, we found that IL-17A and IL-17F had compensatory and redundant roles in host defense during an S. aureus skin infection, which is consistent with prior reports in which IL-17A and IL-17F had redundant roles against a mucocutaneous S. aureus infection and an adenovirus liver infection (12, 35), and differ from other studies that found differential roles for IL-17A and IL-17F during S. aureus pneumonia, contact hypersensitivity, autoimmune encephalomyelitis, arthritis, and chemically induced colitis (12, 36, 37). We also found that γδ T cells primarily produced IL-17A in the S. aureus-infected skin (Fig. 2D), indicating that γδ T cell-derived IL-17A was critical in orchestrating numerous important cutaneous host defense mechanisms against the S. aureus skin infection, including inducing neutrophil recruitment, proinflammatory cytokine production (IL-1α, IL-1β, and TNF but not IFNγ), and host defense peptide production (calprotectin and β-defensins mBD3, mBD4, and mBD14 but not psoriasin, cramp, Reg3γ, or Slurp1).
Most importantly, mining of RNA-seq datasets of skin-draining lymph nodes and infected skin in response to an S. aureus skin infection (25, 52) provided new findings that add to the accepted views of mouse γδ T cells. During development, the initial mouse γδ T cells generated in the thymus have canonical TCRs with invariant γ- and δ-chains (46, 51). These invariant γδ T cells disseminate to specific anatomical sites based on the TCR γ-chain they express and are found resident in the female reproductive tract, lung, and peritoneum. They have also been shown to preferentially expand in a wide variety of inflammatory settings (47, 48). Indeed, invariant Vγ6+Vδ4+ γδ T cells migrate during development and normally reside in the liver, placenta, kidney, uterus, tongue, and other mucosal sites (47, 48). Additionally, Vγ6+Vδ4+ cells were found to normally reside in the dermis of mice (73). However, we found that in response to an S. aureus skin infection, invariant Vγ6+Vδ4+ γδ T cells expressing the same identical CDR3 amino acid sequence expanded in skin-draining lymph nodes and infected skin of wt mice. In particular, the canonical TRGV6 and TRDV4 nt sequences that encoded the invariant CDR3 amino acid sequence of the Vγ6+Vδ4+ γδ T cells were found at low frequency in the skin-draining lymph nodes before infection but expanded nearly 10- and 20-fold, respectively, in response to the S. aureus skin infection (Table 1). Moreover, the expanded Vγ6+Vδ4+ γδ T cells likely trafficked from the lymph nodes to the infected skin to mediate host defense against the S. aureus skin infection (0% on day 0 to 4.6% on day 7 of the total TRG CDR3 reads in S. aureus-infected skin) (Table 1). The canonical TRDV4 nt sequence is utilized by invariant Vγ6+Vδ4+ γδ T cells (47, 48) as well as Vγ5+Vδ4+ DETCs that reside in mouse epidermis (49–51). However, we found that both canonical TRGV6 and TRGV5 nt sequences were found in the TCR repertoire of lymph nodes in naïve wt mice, but only the TRGV6 nt sequence expanded in the day 28 lymph nodes and day 7 skin in response to the S. aureus skin infection. The mechanism by which Vγ6+Vδ4+ γδ T cells and Vγ5+Vδ4+ γδ T cells possessing an identical CDR3 amino acid sequence differentially expand in response to the S. aureus skin infection is unclear. This differential result could be due to recognition of different antigens. However, the antigen(s) that the CDR3 amino acid sequence of Vγ6+Vδ4+ γδ T cells recognizes is unknown (74), and DETCs are thought to recognize a stress-induced self-antigen derived from keratinocytes (51, 75). Nonetheless, since the CDR3 sequences are identical, the difference in expansion could also be due to differential signaling between the TCRs composed of Vγ6 versus Vγ5 chains or expression of costimulatory molecules, transcription factors, and other intrinsic factors between the Vγ6+Vδ4+ and Vγ5+Vδ4+ γδ T cells. Consistent with the potential differences, an alternative skin infection with P. aeruginosa resulted in modest increases of both clonotypic TRGV6 and TRGV5 nt sequences, suggesting that the single expansion of Vγ6+Vδ4+ T cells might be more specific to S. aureus skin infection.
In our recent published report studying IL-1β−/− mice (25), we found that the same CACWDSSGFHKVF CDR3 amino acid sequence expanded in skin-draining lymph nodes; however, it was unknown whether the expansion was encoded for by TRGV5 or TRGV6. At the time, we had presumed that both were expanded because the CACWDSSGFHKVF CDR3 sequence was encoded by both of these TCR gene segments. That manuscript also demonstrated that γδ T cells trafficked from skin-draining lymph nodes to the infected skin during a subsequent S. aureus skin infection to produce TNF and IFNγ (but not IL-17A) to mediate host defense (25), which differs from the major role of Vγ6+Vδ4+ γδ T cell-derived IL-17A in cutaneous host defense during an initial S. aureus skin infection in wt mice described in the present study.
To verify the RNA-seq data by an alternative method, we performed flow cytometry and provide conclusive evidence that Vγ6+ cells were the most abundant γδ T cell population in the skin-draining lymph nodes that produced IL-17A as well as IL-22, TNF, and IFNγ, which might have also contributed to host defense. The precise mechanisms by which the Vγ6+ γδ T cells were recruited to the S. aureus-infected skin are not entirely clear. However, Vγ6+IL-17A+ T cells were likely more responsive to chemokine-mediated recruitment, as a significantly higher percentage of these cells expressed CCR2, CCR5, and CCR6 compared with Vγ6+IL-17A− T cells (SI Appendix, Fig. S5). Our findings are likely broadly applicable to other types of bacterial and fungal infections, as Vγ6+ γδ T cells have been shown to strongly produce IL-17 and promote host defense at different sites of infection, including i.p. exposure to S. aureus (60, 76–78). Although it is unclear whether human γδ T cells (or other human T cell subsets) are the primary source of IL-17 that induces similar protective immunity against S. aureus skin infections in humans, the two major populations of human circulating γδ T cells, Vδ1+ and Vδ2+, are increasingly recognized to promote antigen-specific adaptive immunity against different microbial infections (79, 80). The data presented in this study suggest that future investigation into the role of human γδ T cells in host defense against S. aureus skin infections might be warranted.
Finally, the current findings add important mechanistic understanding to previous observations of protective immunity in experimental models of S. aureus skin infection in mice in which IL-17 responses were shown to play an important role (10, 11, 13–16, 20). Our current results strongly support these prior findings, and now provide specific knowledge that the protective IL-17A–producing T cells identified in these other studies were likely the same specific clonotypic Vγ6+Vδ4+ T cells that we identified. This has important implications in host defense against S. aureus infections in the skin, and it is unknown whether the same Vγ6+Vδ4+ T cells are also involved in protective IL-17 responses against S. aureus bloodstream infections or in other organs (17–19), which will be the subject of our future work. Since responses to a vaccine often differ from natural infection, it could also be that antigen-specific αβ T cells might traffic to the skin and provide similar IL-17–mediated protection. Indeed, αβ Th17 cells have been previously associated with vaccine-induced protection against skin and other S. aureus infections in mice (18, 20, 81–83). However, as an immune evasion mechanism, S. aureus inhibits Th17 and Th1 generation and responses (84), which is consistent with the observed lack of expansion of TRA or TRB CDR3 sequence reads (Fig. 5A). Therefore, it could be that the γδ T cell response, which was not inhibited by the S. aureus infection, could represent a more effective response to target in future vaccines and immunotherapies.
In summary, clonotypic Vγ6+Vδ4+ T cells trafficked from the lymph nodes to the S. aureus-infected skin and were critical in inducing IL-17–mediated host defense mechanisms, including neutrophil recruitment and production of proinflammatory cytokines and host defense peptides. These findings increase our mechanistic understanding of T cell responses in immunity to S. aureus skin infections and provide a specific clonotypic T cell subset that could be targeted in the development of future vaccines and immunotherapies against S. aureus skin infections.
Methods
Bacteria.
The bioluminescent S. aureus CA-MRSA strain USA300 LAC::lux was used in all S. aureus experiments and previously generated from the well-described USA300 LAC parent isolate obtained from a CA-MRSA skin infection outbreak in the Los Angeles County Jail (85). USA300 LAC::lux possesses a modified luxABCDE operon from Photorhabdus luminescens, transduced into the bacterial chromosome from the bioluminescent S. aureus strain Xen29 (PerkinElmer), and emits bioluminescent signals from live, actively metabolizing bacteria in all states of the S. aureus life cycle. The bioluminescent P. aeruginosa strain Xen41 (PerkinElmer) was previously generated from the well-characterized PAO1 reference strain.
Preparation of S. aureus and P. aeruginosa for Skin Inoculation.
See SI Appendix, Methods for details.
Mice.
Six- to 8-wk-old female mice on a C57BL/6 genetic background were used in all experiments. C57BL/6 wt mice were obtained from the Jackson Laboratory. IL-17A/F−/− mice were provided by Yoichiro Iwakura, University of Tokyo, Tokyo, and generated as previously described (12). IL-17A-tdTomato/IL-17F-GFP dual-color reporter mice on a C57BL/6 background were generated as described below.
Generation of IL-17A/F Dual-Color Reporter Mice.
A bacterial artificial chromosome was modified to introduce two fluorescent reporter genes into the Il17 locus, which includes IL-17A and IL-17F. By homologous recombination, the synthesis of the signal peptide of Il17a/f in the BAC was disrupted and the GFP gene with polyA was inserted immediately after the ATG start site of Il17f, replacing exon 1, while the tdTomato gene with poly(A) was inserted immediately after the ATG start site of Il17a. BAC end-sequencing and DNA-fingerprinting results showed that there is no rearrangement and deletion of the BAC construct with the reporter gene. Mice on a C57BL/6 background were generated that harbor the BAC construct. Description of the methods for validation of the reporter mouse strain can be found in SI Appendix, Methods.
Mouse Model of S. aureus Skin Infection and Lesion Size Quantification.
All experiments were approved by the Johns Hopkins Animal Care and Use Committee. This mouse model of intradermal S. aureus infection was performed as previously described (11, 25, 29–31). Briefly, mice were anesthetized (2% isoflurane), and the dorsal backs were shaved and injected intradermally with 3 × 107 CFUs/100 μL PBS of CA-MRSA strain USA300 LAC::lux using a 29-gauge insulin syringe. In some experiments, IL-17A/F dual-color reporter mice were injected intraperitoneally with FTY720 (Sigma-Aldrich), 1 mg/kg in 100 μL sterile water on days −1, 0, and 1, and every other day thereafter until the experiment was arbitrarily ended on day 14, according to previously described methods (25, 32). In some experiments, wt mice were treated intraperitoneally with anti–IL-17A mAb (clone 17F3; BioXCell), anti–IL-17F mAb (clone MM17F8F5.1A9; BioXCell), or combined anti–IL-17A and anti–IL-17F mAb each on days −1 and 0 (200 µg/100 µL) and every other day thereafter (100 µg/100 µL) until the experiment was arbitrarily ended on day 14, modified from previously described methods (10). In other experiments, IL-17A/F−/− mice were treated with recombinant IL-17A or rIL-17F (1,000 ng) that was included with the i.d. bacterial inoculum, modified from previously described methods (11). Total lesion size (cm2) was measured by analyzing digital photographs using ImageJ (https://imagej.nih.gov/ij/) and a millimeter ruler as a reference.
Quantification of S. aureus by in Vivo BLI and ex Vivo CFUs.
Mice were anesthetized via inhalation of isoflurane (2%), and in vivo BLI was performed using a Lumina III IVIS (PerkinElmer); total flux (photons/s) was measured within a 1 × 103-pixel circular region of interest using Living Image software (PerkinElmer) (limit of detection, 2 × 104 photons/s). The in vivo bioluminescent signals of USA300 LAC::lux closely approximate the ex vivo CFUs from homogenized skin obtained at different time points after infection (correlation coefficient, R2 = 0.9996) (33). Ex vivo CFUs were enumerated from overnight cultures of serially diluted 10-mm lesional skin punch biopsy specimens homogenized at 4 °C (Pro200 Series homogenizer; Pro Scientific).
Mouse Model of P. aeruginosa Skin Infection.
See SI Appendix, Methods for details.
Histology and Immunofluorescence Microscopy.
See SI Appendix, Methods for details.
Flow Cytometry.
For skin specimens, 10-mm skin punch biopsies were minced and placed in 3 mL RPMI containing 100 μg/mL DNaseI (Sigma-Aldrich) and 1.67 Wunsch units/mL Liberase TL (Roche). Skin was digested for 1 h at 37 °C and shaken at 140 rpm. For skin and lymph node specimens, single cells were isolated by manually pushing grinded skin or lymph nodes with a 3-mL syringe plunger through a 40-μm cell strainer and the cells were then washed in RPMI. For additional flow cytometry methods, see SI Appendix, Methods.
RNA Extraction and mRNA Quantification for Gene Expression Arrays.
See SI Appendix, Methods for details.
Cytokine Protein Levels.
Protein levels (pg/mg tissue weight) of IL-1α, IL-1β, TNF, IL-17A, and IFNγ were measured from homogenized 10-mm skin punch biopsies collected on days 0 and 7 following S. aureus skin infection by ELISA according to the manufacturer’s recommendations (R&D Systems).
In Vivo Fluorescence Imaging of IL-17A/F–Producing Cells.
Mice were anesthetized with inhalation isoflurane, and in vivo FLI was performed sequentially after in vivo BLI using a Lumina III IVIS (PerkinElmer). tdTomato fluorescence was measured using excitation 554 nm, emission 581 nm, and exposure time 0.5 s. GFP fluorescence was measured using excitation 488 nm, emission 507 nm, and exposure time 0.5 s. Data are presented on a color scale overlaid on a grayscale photograph of mice and quantified as total radiant efficiency ([photons/s]/[mW/cm2]) within a circular region of interest using Living Image software (PerkinElmer).
Analysis of Public RNA-Seq Datasets.
See SI Appendix, Methods and our previously described methods (25).
RNA Isolation, RNA-Seq and Analysis, Amplification of TCR CDR3, TCR Library Preparation, and TCR Library Sequencing and Analysis.
See SI Appendix, Methods, ref. 86, and our previously described methods (25).
Statistical Analysis.
For all data except the RNA-seq analyses, data for single comparisons were compared using a two-tailed Student’s t test and data for multiple comparisons were compared using a two-way ANOVA using Prism software (GraphPad). For the RNA-seq datasets, Shannon diversity indices were calculated using the “vegan” R package (87), and the Wilcoxon rank-sum test was used for all between-group comparisons and all pairwise comparisons. Values of P < 0.05 for all statistical comparisons were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Tammy Kielian (University of Nebraska) for providing the USA300 LAC::lux strain, Yoichiro Iwakura (University of Tokyo) for providing the IL-17A/F−/− mice, and AstraZeneca for providing the anti-LTA antibody. This work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR069502 and R01AR073665), grants from the National Institute of Allergy and Infectious Diseases (R21AI126896 to L.S.M.; U01AI124319 and R33AI111661 to M.R.Y.; and R01AI129302 to S.I.S.), a grant from the National Institute of Neurological Disorders and Stroke (R01NS054791 to X.D.), and federal funds from the National Cancer Institute under Contract HHSN261200800001E (to S.K.D.) and a grant from the Office of the NIH Director (1DP2OD008752 to E.M.) from the National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Conflict of interest statement: M.R.Y. is a cofounder of NovaDigm Therapeutics, which is developing novel vaccines and immunotherapeutics for infectious diseases, including S. aureus. L.S.M. has received grant support for work unrelated to the work reported in this manuscript from AstraZeneca, Pfizer, Regeneron Pharmaceuticals, Moderna Therapeutics, and Boehringer Ingelheim, is on the scientific advisory board for Integrated Biotherapeutics, and is a shareholder of Noveome Biotherapeutics, which are each developing vaccines and therapeutics against S. aureus and other pathogens.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing data reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA) (accession no. SRP194263).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818256116/-/DCSupplemental.
References
- 1.Miller LS, Cho JS (2011) Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol 11:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF (2010) Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr (2015) Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Missiakas D, Schneewind O (2016) Staphylococcus aureus vaccines: Deviating from the carol. J Exp Med 213:1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler VG, et al. (2013) Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: A randomized trial. JAMA 309:1368–1378. [DOI] [PubMed] [Google Scholar]
- 6.Ma CS, et al. (2008) Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med 205:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milner JD, et al. (2008) Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452:773–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puel A, et al. (2011) Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332:65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renner ED, et al. (2008) Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol 122:181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan LC, et al. (2015) Nonredundant roles of interleukin-17A (IL-17A) and IL-22 in murine host defense against cutaneous and hematogenous infection due to methicillin-resistant Staphylococcus aureus. Infect Immun 83:4427–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho JS, et al. (2010) IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest 120:1762–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishigame H, et al. (2009) Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30:108–119. [DOI] [PubMed] [Google Scholar]
- 13.Maher BM, et al. (2013) Nlrp-3-driven interleukin 17 production by γδT cells controls infection outcomes during Staphylococcus aureus surgical site infection. Infect Immun 81:4478–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montgomery CP, et al. (2014) Protective immunity against recurrent Staphylococcus aureus skin infection requires antibody and interleukin-17A. Infect Immun 82:2125–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myles IA, et al. (2013) Signaling via the IL-20 receptor inhibits cutaneous production of IL-1β and IL-17A to promote infection with methicillin-resistant Staphylococcus aureus. Nat Immunol 14:804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tkaczyk C, et al. (2013) Staphylococcus aureus alpha toxin suppresses effective innate and adaptive immune responses in a murine dermonecrosis model. PLoS One 8:e75103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi A, et al. (2012) Immunization with Staphylococcus aureus iron regulated surface determinant B (IsdB) confers protection via Th17/IL17 pathway in a murine sepsis model. Hum Vaccin Immunother 8:336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin L, et al. (2009) Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog 5:e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narita K, et al. (2010) Role of interleukin-17A in cell-mediated protection against Staphylococcus aureus infection in mice immunized with the fibrinogen-binding domain of clumping factor A. Infect Immun 78:4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeaman MR, et al. (2014) Mechanisms of NDV-3 vaccine efficacy in MRSA skin versus invasive infection. Proc Natl Acad Sci USA 111:E5555–E5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown AF, et al. (2015) Memory Th1 cells are protective in invasive Staphylococcus aureus infection. PLoS Pathog 11:e1005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLoughlin RM, Lee JC, Kasper DL, Tzianabos AO (2008) IFN-gamma regulated chemokine production determines the outcome of Staphylococcus aureus infection. J Immunol 181:1323–1332. [DOI] [PubMed] [Google Scholar]
- 23.Utay NS, et al. (2016) MRSA infections in HIV-infected people are associated with decreased MRSA-specific Th1 immunity. PLoS Pathog 12:e1005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uebele J, et al. (2017) Antigen delivery to dendritic cells shapes human CD4+ and CD8+ T cell memory responses to Staphylococcus aureus. PLoS Pathog 13:e1006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillen CA, et al. (2018) Clonally expanded γδ T cells protect against Staphylococcus aureus skin reinfection. J Clin Invest 128:1026–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhotra N, et al. (2016) IL-22 derived from gammadelta T cells restricts Staphylococcus aureus infection of mechanically injured skin. J Allergy Clin Immunol 138:1098–1107.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriwaki Y, et al. (2015) IL-22/STAT3-induced increases in SLURP1 expression within psoriatic lesions exerts antimicrobial effects against Staphylococcus aureus. PLoS One 10:e0140750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulcahy ME, Leech JM, Renauld JC, Mills KH, McLoughlin RM (2016) Interleukin-22 regulates antimicrobial peptide expression and keratinocyte differentiation to control Staphylococcus aureus colonization of the nasal mucosa. Mucosal Immunol 9:1429–1441. [DOI] [PubMed] [Google Scholar]
- 29.Miller LS, et al. (2006) MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity 24:79–91. [DOI] [PubMed] [Google Scholar]
- 30.Cho JS, et al. (2011) Noninvasive in vivo imaging to evaluate immune responses and antimicrobial therapy against Staphylococcus aureus and USA300 MRSA skin infections. J Invest Dermatol 131:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho JS, et al. (2012) Neutrophil-derived IL-1β is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog 8:e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramírez-Valle F, Gray EE, Cyster JG (2015) Inflammation induces dermal Vγ4+ γδT17 memory-like cells that travel to distant skin and accelerate secondary IL-17–driven responses. Proc Natl Acad Sci USA 112:8046–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, et al. (2013) In vivo bioluminescence imaging to evaluate systemic and topical antibiotics against community-acquired methicillin-resistant Staphylococcus aureus-infected skin wounds in mice. Antimicrob Agents Chemother 57:855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor PR, et al. (2014) Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORγt and dectin-2. Nat Immunol 15:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jie Z, et al. (2014) Intrahepatic innate lymphoid cells secrete IL-17A and IL-17F that are crucial for T cell priming in viral infection. J Immunol 192:3289–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibue Y, et al. (2019) Role of interleukin-17 in a murine community-associated methicillin-resistant Staphylococcus aureus pneumonia model. Microbes Infect 21:33–39. [DOI] [PubMed] [Google Scholar]
- 37.Chen F, et al. (2016) mTOR mediates IL-23 induction of neutrophil IL-17 and IL-22 production. J Immunol 196:4390–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Archer NK, et al. (2019) Injury, dysbiosis, and filaggrin deficiency drive skin inflammation through keratinocyte IL-1α release. J Allergy Clin Immunol 143:1426–1443.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbin BD, et al. (2008) Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965. [DOI] [PubMed] [Google Scholar]
- 40.Hinrichsen K, et al. (2008) Mouse beta-defensin-14, an antimicrobial ortholog of human beta-defensin-3. Antimicrob Agents Chemother 52:1876–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Röhrl J, Yang D, Oppenheim JJ, Hehlgans T (2008) Identification and biological characterization of mouse beta-defensin 14, the orthologue of human beta-defensin 3. J Biol Chem 283:5414–5419. [DOI] [PubMed] [Google Scholar]
- 42.Breyne K, Steenbrugge J, Demeyere K, Vanden Berghe T, Meyer E (2017) Preconditioning with lipopolysaccharide or lipoteichoic acid protects against Staphylococcus aureus mammary infection in mice. Front Immunol 8:833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braff MH, Zaiou M, Fierer J, Nizet V, Gallo RL (2005) Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect Immun 73:6771–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang LJ, et al. (2015) Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 347:67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi SM, et al. (2013) Innate Stat3-mediated induction of the antimicrobial protein Reg3γ is required for host defense against MRSA pneumonia. J Exp Med 210:551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itohara S, et al. (1990) Homing of a γδ thymocyte subset with homogeneous T-cell receptors to mucosal epithelia. Nature 343:754–757. [DOI] [PubMed] [Google Scholar]
- 47.Bonneville M, O’Brien RL, Born WK (2010) Gammadelta T cell effector functions: A blend of innate programming and acquired plasticity. Nat Rev Immunol 10:467–478. [DOI] [PubMed] [Google Scholar]
- 48.Muñoz-Ruiz M, Sumaria N, Pennington DJ, Silva-Santos B (2017) Thymic determinants of γδ T cell differentiation. Trends Immunol 38:336–344. [DOI] [PubMed] [Google Scholar]
- 49.Asarnow DM, Goodman T, LeFrancois L, Allison JP (1989) Distinct antigen receptor repertoires of two classes of murine epithelium-associated T cells. Nature 341:60–62. [DOI] [PubMed] [Google Scholar]
- 50.Asarnow DM, et al. (1988) Limited diversity of γδ antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell 55:837–847. [DOI] [PubMed] [Google Scholar]
- 51.Havran WL, Chien YH, Allison JP (1991) Recognition of self antigens by skin-derived T cells with invariant gamma delta antigen receptors. Science 252:1430–1432. [DOI] [PubMed] [Google Scholar]
- 52.Brady RA, Bruno VM, Burns DL (2015) RNA-seq analysis of the host response to Staphylococcus aureus skin and soft tissue infection in a mouse model. PLoS One 10:e0124877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayday AC, et al. (1985) Structure, organization, and somatic rearrangement of T cell gamma genes. Cell 40:259–269. [DOI] [PubMed] [Google Scholar]
- 54.Pereira P, et al. (1996) Rearrangement and expression of Vγ1, Vγ2 and Vγ3 TCR γ genes in C57BL/6 mice. Int Immunol 8:83–90. [DOI] [PubMed] [Google Scholar]
- 55.Cai Y, et al. (2011) Pivotal role of dermal IL-17-producing γδ T cells in skin inflammation. Immunity 35:596–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gray EE, Suzuki K, Cyster JG (2011) Cutting edge: Identification of a motile IL-17-producing gammadelta T cell population in the dermis. J Immunol 186:6091–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mabuchi T, Takekoshi T, Hwang ST (2011) Epidermal CCR6+ γδ T cells are major producers of IL-22 and IL-17 in a murine model of psoriasiform dermatitis. J Immunol 187:5026–5031. [DOI] [PubMed] [Google Scholar]
- 58.Pantelyushin S, et al. (2012) Rorγt+ innate lymphocytes and γδ T cells initiate psoriasiform plaque formation in mice. J Clin Invest 122:2252–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sumaria N, et al. (2011) Cutaneous immunosurveillance by self-renewing dermal gammadelta T cells. J Exp Med 208:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy AG, et al. (2014) Staphylococcus aureus infection of mice expands a population of memory γδ T cells that are protective against subsequent infection. J Immunol 192:3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cyster JG, Schwab SR (2012) Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu Rev Immunol 30:69–94. [DOI] [PubMed] [Google Scholar]
- 62.Shi C, et al. (2011) Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating Toll-like receptor ligands. Immunity 34:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun WY, et al. (2016) Topical application of fingolimod perturbs cutaneous inflammation. J Immunol 196:3854–3864. [DOI] [PubMed] [Google Scholar]
- 64.Gaublomme JT, et al. (2015) Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuberbuehler MK, et al. (2019) The transcription factor c-Maf is essential for the commitment of IL-17-producing γδ T cells. Nat Immunol 20:73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirota K, et al. (2011) Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol 12:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Croxford AL, Kurschus FC, Waisman A (2009) Cutting edge: An IL-17F-CreEYFP reporter mouse allows fate mapping of Th17 cells. J Immunol 182:1237–1241. [DOI] [PubMed] [Google Scholar]
- 68.Lee YK, et al. (2009) Late developmental plasticity in the T helper 17 lineage. Immunity 30:92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price AE, Reinhardt RL, Liang HE, Locksley RM (2012) Marking and quantifying IL-17A-producing cells in vivo. PLoS One 7:e39750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang XO, et al. (2008) T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity 28:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gomez-Rodriguez J, et al. (2009) Differential expression of interleukin-17A and -17F is coupled to T cell receptor signaling via inducible T cell kinase. Immunity 31:587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang XP, et al. (2011) Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol 12:247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cai Y, et al. (2014) Differential developmental requirement and peripheral regulation for dermal Vγ4 and Vγ6T17 cells in health and inflammation. Nat Commun 5:3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Willcox BE, Willcox CR (2019) γδ TCR ligands: The quest to solve a 500-million-year-old mystery. Nat Immunol 20:121–128. [DOI] [PubMed] [Google Scholar]
- 75.Komori HK, et al. (2012) Cutting edge: Dendritic epidermal γδ T cell ligands are rapidly and locally expressed by keratinocytes following cutaneous wounding. J Immunol 188:2972–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conti HR, et al. (2014) Oral-resident natural Th17 cells and γδ T cells control opportunistic Candida albicans infections. J Exp Med 211:2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Misiak A, Wilk MM, Raverdeau M, Mills KH (2017) IL-17-producing innate and pathogen-specific tissue resident memory γδ T cells expand in the lungs of Bordetella pertussis-infected mice. J Immunol 198:363–374. [DOI] [PubMed] [Google Scholar]
- 78.Okamoto Yoshida Y, et al. (2010) Essential role of IL-17A in the formation of a mycobacterial infection-induced granuloma in the lung. J Immunol 184:4414–4422. [DOI] [PubMed] [Google Scholar]
- 79.Davey MS, et al. (2017) Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat Commun 8:14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dimova T, et al. (2015) Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc Natl Acad Sci USA 112:E556–E565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bagnoli F, et al. (2015) Vaccine composition formulated with a novel TLR7-dependent adjuvant induces high and broad protection against Staphylococcus aureus. Proc Natl Acad Sci USA 112:3680–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lacey KA, et al. (2017) The Staphylococcus aureus cell wall-anchored protein clumping factor A is an important T cell antigen. Infect Immun 85:e00549-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang R, et al. (2018) Mechanisms of fibronectin-binding protein A (FnBPA110-263) vaccine efficacy in Staphylococcus aureus sepsis versus skin infection. Clin Immunol 194:1–8. [DOI] [PubMed] [Google Scholar]
- 84.Sanchez M, et al. (2017) O-acetylation of peptidoglycan limits helper T cell priming and permits Staphylococcus aureus reinfection. Cell Host Microbe 22:543–551.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thurlow LR, et al. (2011) Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol 186:6585–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meerlev AA. (2019) Clonal Vγ6+Vδ4+ T cells promote IL-17-mediated immunity against Staphylococcus aureus skin infection. NCBI Sequence Read Archive. Available at https://www.ncbi.nlm.nih.gov/sra?term=SRP194263. Deposited April 29, 2019.
- 87.Oksanen J, et al. (2019) vegan: Community Ecology Package. R Package Version 2.5-4. Available at https://cran.r-project.org/web/packages/vegan/index.html. Accessed April 29, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.