Abstract
The small genome of sorghum (Sorghum bicolor L. Moench.) provides an important template for study of closely related large-genome crops such as maize (Zea mays) and sugarcane (Saccharum spp.), and is a logical complement to distantly related rice (Oryza sativa) as a “grass genome model.” Using a high-density RFLP map as a framework, a robust physical map of sorghum is being assembled by integrating hybridization and fingerprint data with comparative data from related taxa such as rice and using new methods to resolve genomic duplications into locus-specific groups. By taking advantage of allelic variation revealed by heterologous probes, the positions of corresponding loci on the wheat (Triticum aestivum), rice, maize, sugarcane, and Arabidopsis genomes are being interpolated on the sorghum physical map. Bacterial artificial chromosomes for the small genome of rice are shown to close several gaps in the sorghum contigs; the emerging rice physical map and assembled sequence will further accelerate progress. An important motivation for developing genomic tools is to relate molecular level variation to phenotypic diversity. “Diversity maps,” which depict the levels and patterns of variation in different gene pools, shed light on relationships of allelic diversity with chromosome organization, and suggest possible locations of genomic regions that are under selection due to major gene effects (some of which may be revealed by quantitative trait locus mapping). Both physical maps and diversity maps suggest interesting features that may be integrally related to the chromosomal context of DNA—progress in cytology promises to provide a means to elucidate such relationships. We seek to provide a detailed picture of the structure, function, and evolution of the genome of sorghum and its relatives, together with molecular tools such as locus-specific sequence-tagged site DNA markers and bacterial artificial chromosome contigs that will have enduring value for many aspects of genome analysis.
Sorghum (Sorghum bicolor L. Moench. [SB]) is a leading cereal in arid and semi-arid agriculture, ranking fifth in importance among the world's grain crops (Doggett, 1988), and is typically grown on 10 to 14 million acres per year in the USA. In addition to its importance as a crop, the Sorghum genus also includes Sorghum halepense, Sorghum almum, and hybrids of these species to SB, which are collectively referred to as “Johnson grass” and comprise one of the more noxious weeds affecting U.S. and world agriculture.
As a grass genome model, the small genome of SB (approximately 760 Mb; Arumunganathan and Earle, 1991) is the most logical complement to that of rice (Oryza sativa; approximately 440 Mb; Arumunganathan and Earle, 1991), a distant relative (tribe Oryzeae) that will be the first grass genome to be completely sequenced. Sorghum is much more closely related than rice to maize (Zea mays), sugarcane (Saccharum spp.), and other tropical grasses that are among the world's leading crops. Sorghum, maize, and sugarcane are all members of the same tribe, Andropogoneae, within the grass family Poaceae (Clayton, 1987). Sorghum and maize (approximately 2,500 Mb; Arumunganathan and Earle, 1991) may have diverged from a common ancestor about 24 million years ago (Thomasson, 1987) and retain similar chromosome organization (Hulbert et al., 1990; Whitkus et al., 1992; Paterson et al., 1995a). In contrast, rice and the maize/sorghum lineage may have diverged from a common ancestor about 66 million years ago (Linder, 1987), and show much greater levels of chromosome structural rearrangement (compare with Paterson et al., 1995a). Sorghum and sugarcane (Saccharum spp.; approximately 2,547–4,183 Mb; Arumunganathan and Earle, 1991) may have shared a common ancestor as recently as 5 million years ago (Sobral et al., 1994), retain very similar gene order (Ming et al., 1998), and even produce viable progeny in some intergeneric crosses (deWet et al., 1976; P.L. Morrell, T.D. Williams-Coplin, J.E. Bowers, J.M. Chandler, and A.H. Paterson, unpublished data).
Over the past 10 years, the value of physically small genomes as “templates” for the larger genomes of major crops has been considerably strengthened by comparative genomics (for recent reviews, see Gale and Devos, 1998; Messing and Llaca, 1998). The discovery of unexpected levels of similarity of gene order among grasses (Hulbert et al., 1990; Bennetzen and Freeling, 1993; Shields, 1993) and more distantly related taxa (Paterson et al., 1996) led to the notion that phenotypes precisely mapped in large-genome species may be dissected by isolation of the underlying genes in related small-genome species. The possibility that genes underlying a qualitative trait in one genotype or species may account for quantitative trait loci (QTLs) in other genotypes or species (Robertson, 1985) has further stimulated interest in this notion, and has been supported by several comparative QTL mapping studies (e.g. Pereira et al., 1994; Lin et al., 1995; Paterson et al., 1995a).
Physical analysis of the sorghum genome is likely to facilitate cloning of genes and QTLs associated with many aspects of plant domestication and crop productivity in its larger genome relatives such as maize and sugarcane. Abundant DNA polymorphism between cultivated SB and wild Sorghum propinquum (Kunth.) Hitch. (SP) has expedited assembly of a map of more than 2,500 RFLP loci (Bowers et al., 2000; A.H. Paterson et al., unpublished data), with DNA markers at an average physical distance of about 150 kb from most sorghum genes. Genes and QTLs affecting many important traits have been mapped in sorghum (Lin et al., 1995; Paterson et al., 1995a, 1995b; Pereira and Lee, 1995; Tuinstra et al., 1997; Rami et al., 1998; Tao et al., 1998; Tuinstra et al., 1998; Bowers et al., 2000; Katsar et al., submitted for publication) and many of these correspond in chromosomal location to QTLs mapped in maize, rice, wheat (Triticum aestivum; Lin et al., 1995; Paterson et al., 1995a, b; C.S. Katsar, A.H. Paterson, G.L. Teetes, and G. Peterson, submitted for publication), sugarcane (Ming et al., 1998), and presumably other taxa.
We are engaged in the construction of a robust physical map of the sorghum genome, based on stable, large-insert (Shizuya et al., 1992) bacterial artificial chromosome (BAC) clones that contain alleles conferring many dominant and additive phenotypes associated with domestication (Lin et al., 1999), and that are suitable substrates for genomic DNA sequencing. By merging probe-to-BAC hybridization data with DNA fingerprint data, and using the BACRF method (Lin et al., 2000) to resolve the chromosomal origin of BAC clones detected by multiple-locus DNA probes, the robustness of the physical maps is improved. The “checks and balances” resulting from integration of individual BAC hybridization data from genetically mapped sequence-tagged sites (STSs), together with the use of reliable and reproducible restriction enzyme-based fingerprinting, offers numerous advantages over alternative approaches based on arbitrary primer PCR-based fingerprinting of complex DNA populations resulting from pooling of low-coverage BAC libraries (Klein et al., 2000).
The sorghum genetic map, and therefore the genetically anchored physical map, has been aligned to varying degrees with the genetic maps of wheat, rice, sugarcane, maize, and Arabidopsis and with QTLs mapped in these taxa. Marked variations in the level of allelic polymorphism in wild and cultivated sorghums suggest the action of selection in particular gene pools and the locations of cytological features that are common to each gene pool. Advances in cytology promise to align such features to the genetic, physical, and diversity maps. The completed sorghum physical map will provide a valuable new tool for structural, functional, and comparative genomic investigations of many aspects of grass biology.
RESULTS AND DISCUSSION
Physical Mapping
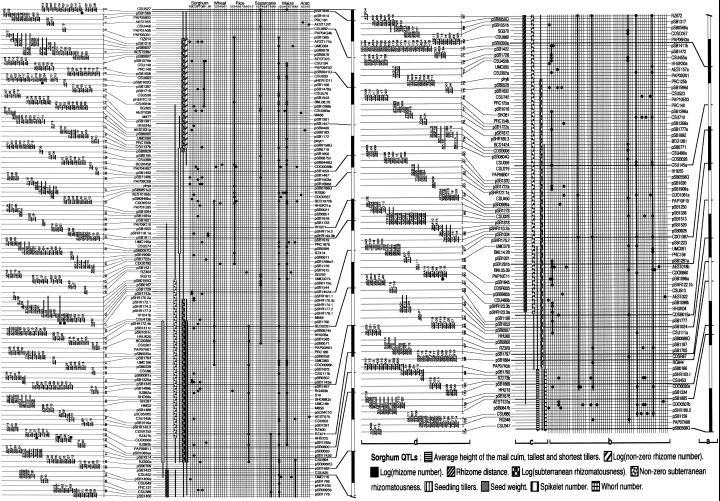
A robust physical map, comprised of large-insert DNA clones and anchored to the recombination-based genetic map by locus-specific STSs, is the centerpiece for a host of future directions in sorghum genome analysis. An example of progress toward an integrated genetic physical comparative QTL map for one sorghum chromosome is presented (Fig. 1). Determining the exact order of markers along a high-density map is most economically and efficiently accomplished by a prudent balance between genetic and physical approaches. The order of markers along this map was determined by first identifying a framework of 28 loci that could be genetically ordered with certainty in the small (56 individuals; Chittenden et al., 1994) mapping population, then placing the remaining markers into the correct intervals between framework markers (with some uncertainty near the framework markers themselves) in their “most likely” order (as determined by MapMaker, although in many cases the possibility of alternative orders cannot be precluded with statistical significance), and finally adjusting the local marker orders to be consistent with probe-to-BAC hybridization data. When the diagram was created, 156 probes had been used to anchor 550 BACs to LG C using the BACRF method. These clones were anchored at 164 loci and assembled into 103 contigs containing an average of 1.6 markers and 5.3 BACs. These results conform with computer simulations (see “Materials and Methods”) that predicted, with 164 loci, 101 to 123 contigs containing an average of 1.35 to 1.64 markers (intervals circumscribe 99% of the simulations). According to these simulations, the contig map presented in Figure 1 may cover 29.0 million bases (24.1–34.8 Mb) of genomic sequence. A slightly lower estimate of 23.5 Mb derives from probabilistic considerations that: (a) The set of BACs overlapping a single marker spans an of average 204 kb (based on the average insert size and effective redundancy of the library), and (b) the average distance between successive markers contained within the same contig should be about 52 kb for small contigs (based on the theoretical average for the absolute value of the difference between two uniform distributions describing the possible position of each marker along the contig).
Figure 1.
Aligned genetic, comparative, QTL, and BAC contig maps of sorghum linkage group (LG) C. The foundation for this figure is the full set of LG C loci that had been genetically mapped at the time it was made (in staggered rows). A total of 11 additional loci were detected by hybridization of these probes to the BACs but had not been genetically mapped due to lack of DNA polymorphism, and have been added to the figure based on cohybridization of mapped probes to the same BACs. a, Framework genetic map (28 loci, bar = 5cM). b, Comparative data; duplicated and/or heterologous loci in the sorghum, wheat, rice, sugarcane, maize, and Arabidopsis genomes are shown by black circles. LGs (sorghum; Chittenden et al., 1994), homoeologous groups (sugarcane; Ming et al., 1998), or chromosomes are indicated at top. Thick lines mark virtually uninterrupted sequences of such loci. In the case of sorghum, only duplicated loci are shown, not the original locus c, QTL map. Stars indicate the positions of sugarcane QTLs, whereas bars and lines show 1 and 2 log-of-odds ratio likelihood support intervals for sorghum QTLs. d, Contig map. The relative position of markers on BAC contigs are indicated by horizontal lines. Contigs are connected to the master list by their most informative marker. Black circles mark BACs that contain a copy of the pSB0880 repetitive sequence. Contig numbers (in order along the chromosome) are indicated to the right of the BACs.
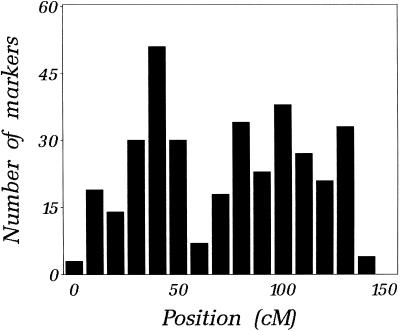
LG C accounts for 501 of 2,583 markers or 19.3% of the sorghum RFLPs, the largest of any LG (some of these markers that have been mapped recently are not shown in the figure). Although a direct estimate of the size of LG C is not available, electron microscopy observations (Bennett and Laurie, 1995) suggest that the three largest sorghum chromosomes contain 98, 90, and 85 Mb of DNA. These values fall between approximations of 147 and 80 Mb, which we obtained by weighting the size of the sorghum genome (760 Mb) with, respectively, the proportion of LG C markers in the sorghum map (501 of 2,583 markers or 19.3%), or with the proportion of genome length accounted for by LG C (121 of 1,140 cM or 10.6%, based on the present length of our genetic map; Chittenden et al., 1994; Bowers et al., 2000; A.H. Paterson et al., unpublished data). Using these estimates, the present physical map would cover 32.2% of the LG C and the average gap size would come to 570 kb. The application to the BACs of 345 additional probes known to map on LG C (as well as all probes mapping to other LGs) is well advanced, and is predicted (by the simulations described) to increase the coverage of the physical map to about 62.1 Mb (approximately 70% of the genomic DNA), with about 154 contigs. On average, each of these contigs would cover 403 kb of genomic sequence, comprise 3.25 markers, and leave gaps of 180 kb (simulated results).
Two approaches are being used to fill gaps in the contigs. The most direct means of gap filling is local alignment of BACs based on DNA fingerprints and integration of these alignments with BAC fingerprint data. An example is shown for the phyA region of sorghum LG C (Fig. 2). High-resolution fingerprint analysis of 31,820 clones from the SP BAC library (Lin et al., 1999) was carried out at the Clemson University Genomics Institute (G.G. Presting, R.A. Dean, A.H. Paterson, and R.A. Wing, unpublished data; www.genome.clemson.edu). A slightly modified protocol for high throughput fingerprint analysis of large-insert clones previously described by Marra et al. (1997) was employed, using HindIII-digested BAC DNA. Restriction fragments from the fingerprint of each clone were hand annotated using Image software (http://www.sanger.ac.uk/software/Image/). The fragments were transformed into a band file (for each clone) that was used as input for the Finger Print Contig program (version 4.5; Soderlund et al., 1997; http://www.sanger.ac.uk/software/fpc/), running on a Sun Ultra 10 workstation (Sun Microsystems, Palo Alto, CA). A comprehensive analysis of the fingerprint-based contigs will be published subsequently.
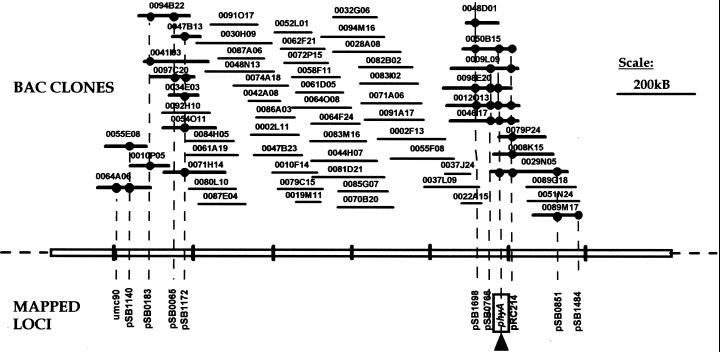
Figure 2.
Integrated physical and genetic map near phytochrome A (phy A, indicated by box and arrow) gene of sorghum. Contig 125 is approximately 1.2 MB and 5.5 cM, and consists of 63 BAC clones. A total of 11 loci were ordered into the contig based on direct and overgo hybridization, FPC fingerprint analysis (cutoff = 10−12 and tolerance = 7), and genetic mapping. Direct BAC hits are shown as black circles and connected to each locus by dashed line. Sizes (in kB) of each BAC clone are drawn to scale. Tick marks on the contig backbone represent 200 kB.
The phyA contig was built by integrating fingerprint contig (FPC)-based fingerprints (at cutoff of 10−12 and tolerance of 7) with hybridization data from direct (individual probes) or multiplex overgo hybridization (Cai et al., 1998). The contig includes 11 genetically mapped RFLP loci and 63 BAC clones, with an approximate physical size of 1.2 million bases (Mb, based on the lengths of nonoverlapping BAC clones) and genetic length of about 5.5 cM based on recombination distances from a high-density map of a cross between SB and SP (Chittenden et al., 1994; Paterson et al., 1995b; Ming et al., 1998; Bowers et al., 2000; Bowers et al., unpublished data). Both hybridization and fingerprint analysis showed that the depth of coverage of the contig is 6- to 12-fold, consistent with the estimated genomic coverage of the library (Lin et al., 1999). The linear order of the 11 genetically mapped markers based on FPC analysis of the contig agreed closely with their order in the high density genetic map of the sorghum (Bowers et al., 2000; Bowers et al., unpublished data). This permits us to determine the polarity (orientation) of the entire contig, including each individual BAC, relative to the chromosome. The ratio of genetic to physical distance in this region, approximately 0.2Mb/cM, is substantially lower than the predicted average ratio of 0.5 Mb/cM based on high density mapping data. Although the mapped loci nominally appear to cluster at the two ends of the contig, it is premature to assert that this particular clustering represents “islands” that are enriched in gene content because there are additional markers predicted to be in the area (based on the sorghum genetic map and comparative maps of other taxa) that are yet to be mapped to the BACs.
The robustness of the sorghum physical map is enhanced by the combination of high-density hybridization data with BAC fingerprint data. As is true of much high-throughput genomic data, each of these methods is subject to occasional anomalies. For example, BACs 41I03 (Fig. 2, to the left) and 9L09 (Fig. 2, to the right) each show two inconsistencies between the hybridization and fingerprint data, failing to hybridize with two mapped probes that are predicted to locate on the BACs. Among the 126 unambiguous LG C loci of the genetic map detected by homologous probes, failure to detect BAC clones occurred at only a single locus (0.79%), which is close to the 99.7% coverage estimated for the SP library. Taking account of 89, 28, 4, 3, and 1 BACs overlapping more than one locus (2–6, respectively), the average number of BACs per locus in the data shown here is 713/164 = 4.35, lower than the 6.06-genome-equivalent coverage estimated for the library (Lin et al., 1999). Hybridization data are subject to occasional false negatives due to variation in growth rates of individual BACs, variation in background signal levels, or other factors—these possible problems were accentuated by the fact that many of the grids used to screen these probes predated availability to us of a gridding robot; therefore, they were handmade. More recent results, applying overgo probes to grids prepared on our QBOT (Genetix, Christchurch, Dorset, UK), reduce the frequency of false negatives; however, they will never be completely eliminated. Fingerprint data (by the Marra et al. [1997] method used in this work, and also by other methods such as that of Klein et al. [2000]) is subject to formation of occasional false contigs due to DNA sequence duplications (but see below), low library coverage (probably not a problem in our case), short BACs that include too few bands to reliably determine overlaps, and artifacts due to arbitrarily primed PCR (in the case of Klein et al. [2000]). By careful analysis of merged hybridization and fingerprint data, robust first-generation physical maps can be constructed that can then be extended and refined by end sequencing of selected BACs (Venter et al., 1996) and hybridization analysis of BAC end-derived overgo markers in key regions such as the center of the phyA contig that lacks hybridization anchors, or the termini of present contigs to form new “joins.”
A particularly pervasive problem in physical mapping of many grasses, and surprisingly even occurring in plant genomes as “simple” as Arabidopsis (McGrath et al., 1993; Kowalski et al., 1994; Paterson et al., 2000) is the presence of large-scale duplications of DNA. Identification of BACs corresponding to a mapped DNA landmark by using “dot blots” or PCR amplification of short, well-conserved consensus sequences, usually fails to distinguish between BACs deriving from allelic or nonallelic loci, although quantitative variations in hybridization signal intensity are sometimes suggestive. High levels of duplication of genes or chromosomal segments increase the propensity for “false joins” among large DNA clones or contigs. Complex autopolyploids such as sugarcane are especially problematic because the many different homologous chromosomes that might be found in an individual often include six or more allelic variants at a locus (Ming et al., 1998). By using traditional fingerprinting methods (Marra et al., 1997; Klein et al., 2000) in autopolyploids, allelic differences between homologs at restriction sites are confounded with differences in the genomic DNA content of the underlying BAC clones. In such genomes, assembly of contigs that truly represent differences in the genomic DNA content of the underlying BAC (or other) clones is likely to require a prior knowledge that the clones derive from the same genetic locus.
The BACRF method (Lin et al., 2000) is a practical and reliable approach for using high-density RFLP maps to anchor sequence-ready BAC contigs in highly duplicated genomes. An example is depicted in Figure 3. BAC clones detected by the 33 multilocus probes screened that contain at least one locus on LG C were assigned to a total of 56 loci. So far, 27 of these have been assigned unambiguously to known RFLP loci. The position of five additional loci (suffix “u”), printed in italics on Figure 1, could be inferred by virtue of their membership in a contig anchored on the genetic map. The remaining 25 loci will require either BACRF analysis using a different restriction enzyme (this pilot experiment used only HindIII digests), or involve loci for which no RFLP variation has been found.
Figure 3.
BACRF analysis of pSB1742-hybridizing BACs. The sorghum genomic clone pSB1742 detects RFLPs that map to LGs B (allele pair indicated by a, one of which comigrates with BAC vector), C (single band designated b in SP), and G (single band designated c in SB), as well as several bands that could not be mapped (such as d). Lanes 1 through 13 are HindIII digests of pooled BAC DNA extracted from 384 clones each, according to Lin et al. (2000). The pSB1742b locus on LG C corresponds to a BAC in pool 13, proven by dot blots (not shown) to be BAC 13I24. A second BAC in pool 12 (12A22) corresponded to an unmapped locus. Other corresponding BACs are not shown.
BACRF analysis differs from fingerprint analysis in that it establishes a direct link between the genomic DNA composition of a particular BAC, and an allele that has been genetically mapped to a specific locus. The importance of this additional information is expected to vary widely among taxa. BACRF is likely to be of high value in recent polyploids that contain very similar subgenomes, or in heterozygous autopolyploids (such as many grasses) in which differences in BAC fingerprints can be due either to differences in genomic DNA coverage or to the presence of different alleles at restriction fragment loci. In ancient polyploids that show disomic inheritance and in which the respective sub-genomes are substantially diverged, traditional fingerprint analysis may be sufficient without BACRF to resolve locus-specific groups of BACs (although BACRF may still be useful to anchor such groupings to their corresponding locus).
Dispersed repetitive DNA probes also yield convenient landmarks to detect overlaps between adjacent contigs. This was illustrated by screening the sorghum BAC library with pSB0880, a clone from a dispersed repeat family found in both SB and SP. Among eight LG C BACs detected by pSB0880, three fell in adjacent contigs, supporting the notion that the terminal BACs of these contigs overlap. Two additional pSB0880-containing BACs were at one end of the same contig (both hybridizing to pSB1452a), and the remaining three were widely scattered. This strategy might be especially powerful for species whose genomes are comprised of a large amount of dispersed repetitive DNA. Cotton (Gossypium hirsutum), for example, is made of at least 24% dispersed repetitive DNA (Zhao et al., 1995), and about 80 individual families have been cloned and characterized (Zhao et al., 1998). By determining the BAC addresses for the majority of repetitive DNA elements, one might learn much about the “genomic environment” surrounding individual genes, identifying interesting features of genome organization and perhaps shedding light on factors influencing gene expression. The BACRF method also provides an elegant means to identify specific repeat units (or clusters) for a variety of analyses (Lin et al., 2000).
Physical mapping using integration of hybridization and fingerprint data is possible even in the absence of prior positional information, although a genetic map is clearly an asset. Such a process initially would only generate small, unordered contigs and screening of random probes would be an efficient approach. The expanding coverage of the genome would gradually enhance the chances of joining adjacent contigs. To resolve the inevitable conflicting data resulting from gene duplication or polyploidy, BACRF data (Lin et al., 2000) would likely to be needed. This kind of approach would be especially useful for species or cultivars like most trees or hybrid bananas whose genetic mapping is hampered by long generation intervals or by sterility concerns (Simmonds, 1962). If available, however, prior positional information adds enormous value to the physical map, aligning it with QTL locations and comparative data.
The “checks and balances” resulting from integration of individual BAC hybridization data from genetically mapped STSs, together with the use of reliable and reproducible restriction enzyme-based fingerprinting, offers numerous advantages over alternative approaches such as methods based on arbitrary primer PCR-based fingerprinting of complex DNA pools derived from low-coverage BAC libraries (Klein et al., 2000). The availability of nearly 2,600 low-copy STS loci mapped as RFLPs (Bowers et al., 2000; Bowers et al., unpublished data) obviates the need to identify BAC clones using arbitrary primer techniques that “… revealed an overall false positive rate of 15%” (Klein et al., 2000, pg 796). The finding that “… 25% of SAS-DNA markers were not useful as links between the genetic and physical maps … ” (Klein et al., 2000) raises further concerns about the quality and reliability of such data. Fingerprinting methods that represent virtually the entire genomic composition of a DNA clone, such as those used herein (Marra et al., 1997), have the further advantage of creating opportunities to develop fine-scale comparisons of closely related taxa for changes in genomic DNA composition—such investigations are planned for the SP BAC library (now expanded from 6.6 to >10×), and a >10× SB library that has been made (D. Begum, J. Tomkins, A.H. Paterson, and R. Wing, unpublished data) and fingerprinted (www.genome.clemson.edu). BACRF data (Lin et al., 2000) superimposed on fingerprint data may resolve discrepancies due to chromatin duplication, which appears to have had a substantial impact on sorghum evolution (Chittenden et al., 1994; Pereira et al., 1994; Lin et al., 1995; Paterson et al., 1995, 1996; Bowers et al., 2000).
Comparative Mapping
By use of tools that are suitable for comparative analysis of related taxa, we seek to exploit high-resolution genomic information from models such as rice to accelerate progress in sorghum, and to extend our results from the small sorghum genome to advancing progress in closely related large-genome taxa such as maize and sugarcane. The emerging physical map, integrating comparative data from heterologous probes (Whitkus et al., 1992; Paterson et al., 1995b; Ming et al., 1998), provides a means to accomplish these goals. In Figure 1, one arm of LG C parallels wheat homoeologous group-1 chromosomes, and the other parallels wheat group-4 chromosomes, each based on five informative loci. In rice, LG C paralleled much of chromosome 3 (based on 17 loci) and the putative centromeric region of chromosome 10 (based on four of seven informative loci). Sugarcane homologous groups 3 and 8, recently proposed to be different parts of a single homologous group (19), each corresponded to one arm of LG C. Conservation of alignments for maize involved chromosome 1 along all of LG C, plus chromosome 5 on one arm and chromosome 9 on the other arm. A few anchor loci provide a beginning for alignment of sorghum and Arabidopsis. Complete sequencing of all mapped sorghum probes is in progress—along with the Arabidopsis genomic sequence, this may provide the higher density of comparative data needed to identify small genome regions that may remain similar in these taxa (Paterson et al., 1996).
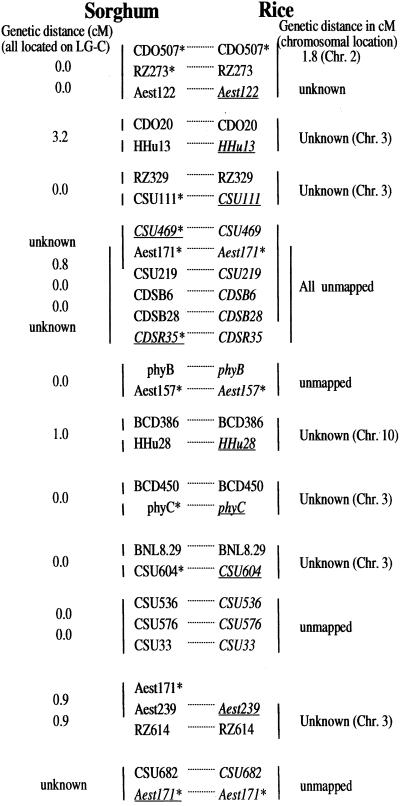
Comparative data from rice provide a means to help join sorghum contigs formed by hybridization and/or fingerprint data. Figure 4 summarizes genetic (if known) and physical (based on BAC hybridization) relationships in sorghum and rice, of 29 heterologous probes that were known a priori to be closely linked in sorghum. These probes have been hybridized to BACs from both sorghum (SP library) and rice (a rice cv Nipponbare BAC library provided by R. Wing). In many cases, loci that fall on different sorghum BACs fall on the same rice BAC(s; as summarized in Fig. 4). The rapidly coalescing physical map of rice (Mao et al., 2000; C. Soderlund, G. Presting, R. Dean, and R. Wing, personal communication) will be a powerful resource for closing gaps in contigs for sorghum and other grasses. As the rice sequence progresses, conserved sequences internal to rice BAC(s) may provide a targeted means to identify “linking probes” that will close gaps in the sorghum contigs. Further, a number of the probes studied had been genetically mapped only in one of the two taxa, but could be physically linked to mapped probes by cohybridization to one or more BACs. As BAC libraries become better characterized, they will increasingly become the reagent of choice (in preference to mapping populations) for determining the locus of DNA probes that have not been genetically mapped. Moreover, because hybridization analysis of BAC libraries is effectively a somatic cell genetic method and is not dependent on the identification of polymorphic alleles, all loci that can hybridize to a probe can be mapped.
Figure 4.
Density of DNA marker loci (RFLPs) along Sorghum LG C. The number of RFLP loci mapped in each consecutive 10-cM interval along the LG is plotted.
The coverage of the SP physical map may be further enhanced by using markers taken from corresponding regions of the chromosomes of other taxa. For example, among 26 maize and rice probes that have not been previously mapped in sorghum but are inferred (by comparative mapping) to lie on LG C, two (CSU490 and CSU413b) were included in existing contigs. BACs detected by the 24 other probes comprised 19 new contigs that have yet to link to anchored contigs, so may cover parts of the chromosome that were marker-poor in sorghum or may map to other chromosomes.
The benefits of comparative data in adding new probes to genetic maps, in “linking” contigs in large genomes by using information from orthologous loci in small genome, and in extrapolating detailed physical information from model taxa to the accelerated characterization of “orphan crops” are further advantages that distinguish our comparative approach from alternative physical mapping approaches based on arbitrary primer PCR (Klein et al., 2000).
QTL Mapping
Sixteen QTLs associated with domestication have been detected on LG C in a sorghum population of 370 F2 individuals from the same F1 plant as the reference mapping population (Lin et al., 1995; A.H. Paterson, unpublished data). As illustrated (Fig. 1), 90% likelihood support intervals for locations of a QTL may span an approximately 20-cM region corresponding to a 10- to 14-Mb interval. Fine mapping by using near-isogenic lines (Paterson et al., 1990; Dorweiler et al., 1993; Alpert and Tanksley, 1996) or selected backcrossed plants (Yamamoto et al., 1998) has demonstrated the possibility to map single QTLs down to sub-centimorgan intervals. The minimum tiling path for a 1-cM (about 500 kb) region of the sorghum genome might be only eight to 10 BACs and sequencing them with as little as 2-fold redundancy (Bouck et al., 1998) helps to find most exons in the region containing a QTL. Continuous improvements in DNA sequencing technology and in large-scale expression profiling of candidate exons (De Risi et al., 1997) may render this a viable approach to QTL discovery. In the last few years, the repertoire of QTLs detected on LG C-homologous maize chromosomes 1, 5, and 9 has increased by more than 100, involved in root number, drought tolerance (ABA content, water potential), ear and kernel number and size, anthesis and silking timing, plant size, yield under various stresses, flowering, maizin concentration, starch content, and disease resistance (Veldboom et al., 1994; Veldboom and Lee, 1994; Lebreton et al., 1995; Pereira and Lee, 1995; Agrama and Moussa, 1996; Austin and Lee, 1996a, 1996b; Byrne et al., 1996; Maroof et al., 1996; Ribaut et al., 1996; Veldboom and Lee, 1996a, 1996b; Bohn et al., 1997; Lubberstedt et al., 1997; Austin and Lee, 1998; Groh et al., 1998; Khairallah et al., 1998; Tuberosa et al., 1998). Economically important QTLs have also been detected in sugarcane, such as the number of stalks (Sills et al., 1995) and sugar content (R. Ming, S.-C. Liu, J.E. Irvine, and A.H. Paterson, submitted for publication). The possibility to initiate cloning of the transcripts underlying at least some of these QTLs in a facile genome encourages the completion of the physical map of LG C and the entire sorghum genome.
Diversity Mapping
There is growing awareness that levels and patterns of allelic diversity are related to the chromosomal context of a locus. “Diversity maps” showing the distribution(s) of allelic diversity across the chromosomes and genomes of a variety of organisms suggest association with chromosome structural features such as centromeres and telomeres and with selection in particular well-defined gene pools (Dvorak et al., 1998; Hamblin and Aquadro, 1999; Gaut et al., 2000).
Diversity analysis of individual genes promises to shed new light on crop productivity and evolutionary processes underlying plant domestication (Wang et al., 1999). Understanding of the genomic distribution of diversity (Dvorak et al., 1998; Hamblin et al., 1999; Gaut et al., 2000) will be important to build appropriate null hypotheses against which to test levels and patterns of diversity in individual genes. As a backdrop for study of levels of diversity in each member of large populations of genes, we have constructed diversity maps with genome-wide resolution based on neutral DNA markers for several gene pools in the Sorghum genus (P.L. Morrell, T.D. Williams-Coplin, J.E. Bowers, J.M. Chandler, and A.H. Paterson, unpublished data). Figure 6 illustrates one LG from a diversity map that has been made based on application of 160 previously mapped RFLP markers (Chittenden et al., 1994; Ming et al., 1998; Bowers et al., 2000; Bowers et al., unpublished data) to 89 Sorghum accessions, including 16 modern U.S. hybrids, 1 Sorghum nitidum (possible n = 5 progenitor), 51 exotic diploid SB accessions chosen (by Jeff Dahlberg, U.S. Department of Agriculture-Agricultural Research Service, Mayaguez, PR) to represent diversity within the species, three accessions of SP (all available), and 18 accessions of polyploid S. halepense (including “Johnson grass,” a highly heterogeneous form of S. halepense naturalized in the U.S.), and S. almum, all of which represent a worldwide sampling.
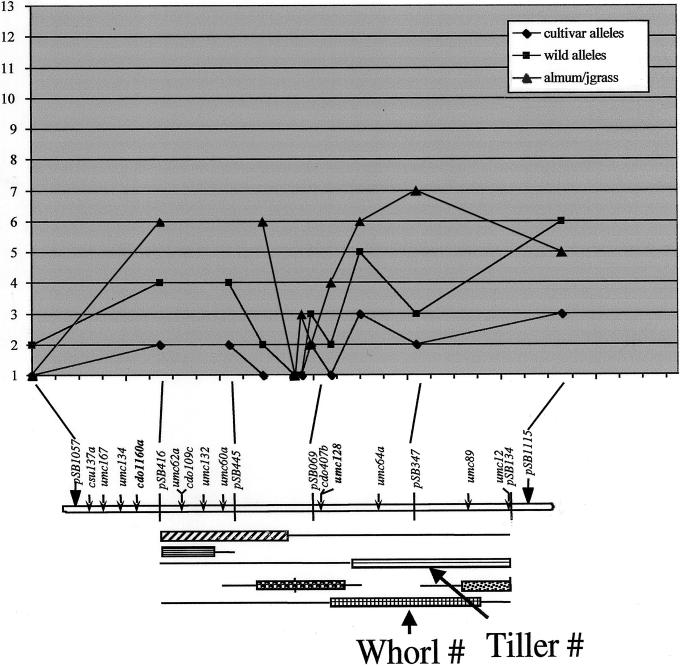
Figure 6.
Diversity map for LG G. The nature of each gene pool is described in the text. The total number of alleles in each gene pool, at each locus, has been plotted at the chromosomal location of each locus, and data for different gene pools has been coded (see inset). QTL data plotted along the map is from a population of 370 individuals from the same F1 plant that was used to make the primary linkage map, as previously described (Lin et al., 1995; Paterson et al., 1995a, 1995b).
The three gene pools show a number of common features and also some key differences. Each of the three gene pools show low levels of variation near the central region of the LG (ostensibly the centromere, but see below), and both termini of the LG. The cultivated sorghums show by far the lowest level of diversity of the three gene pools, despite the fact that virtually all are F1 hybrids, thereby doubling the number of alleles that they can potentially carry. The exotic diploid sorghums show intermediate diversity, and the polyploids show remarkably high levels of diversity, consistent with their likely interspecific origin from species that contain chromosomes that can pair and recombine (compare with Paterson et al., 1995a). In one region near the marker pSB347, the tetraploid gene pool shows an unusually high level of diversity, whereas the two diploid gene pools each show unusually low levels of diversity. It is curious that this marker lies near the center of a QTL likelihood interval that had been previously shown to affect tillering (Paterson et al., 1995a), a trait that tends to be adaptive in natural populations but is strongly selected against in cultivated types. U.S.-cultivated sorghums have traditionally been bred by selfing methods (hybrids being used only recently) from a very small founder population (among the 16 hybrids studied in the diversity map, the maximum number of alleles that we have found at any marker locus anywhere in the genome is four), and it is likely that relatively strong linkage disequilibrium may persist. The predominance of self-pollination (average outcrossing rates are usually less than 10% in natural populations) suggests that perhaps as little as a few hundred “effective meiotic cycles” have passed during the history of sorghum domestication—suggesting that chromosomal regions originally measuring 1 to 3 cM may remain in linkage disequilibrium (Wright, 1968). Although it is tantalizing to suggest that pSB347 is near the tillering gene and its level of variation in the cultivated gene pool may be constrained by linkage drag, finer resolution marker data and also more extensive phenotypic sampling will be needed to provide a robust test of this hypothesis.
With a map presently comprised of more than 2,500 DNA loci (Bowers et al., 2000; Bowers et al., unpublished data), it is possible in principle to develop diversity maps of average 0.4-cM resolution for sorghum gene pools, promising a host of new information about the consequences of natural selection, domestication, and polyploid formation. Efficient resequencing methods such as denaturing HPLC (Underhill et al., 1997) provide the means to identify virtually all DNA polymorphisms, including many that may escape detection by other techniques. Our colleagues (S. Kresovich, personal communication) are pursuing diversity maps of individual genes that are thought to be under especially strong selection, initially focusing on the phytochrome gene family.
Cytomolecular Mapping
Genetic, physical, and diversity maps provide considerable insight into the organization of DNA in genomes. However, in eukaryotic cells genomic DNA is always found within the context of chromosomes, and consequently any comprehensive understanding of how genomes function and evolve is dependent upon an understanding of the structure of chromosomes themselves. With regard to cytogenetics, research on sorghum has lagged far behind that of other grains (for review, see Doggett, 1988; Gómez et al., 1997). This appears to be due, in large part, to the relatively small size and morphological uniformity of sorghum mitotic metaphase chromosomes (Gómez et al., 1997). Attempts to generate a reliable karyotype of sorghum mitotic metaphase chromosomes based on relative chromosome length, arm ratio, and/or C-banding pattern have proven unsatisfactory or difficult to reproduce (Gu et al., 1984; Yu et al., 1991). As a consequence, to date none of the sorghum LGs have been conclusively assigned to a cytologically defined chromosome (Gómez et al., 1997).
In pachynema (a substage of prophase I of meiosis), homologous chromosomes are joined along their entire length by a proteinaceous structure known as the synaptonemal complex (SC; Moses, 1968). Each pair of synapsed homologs is called a bivalent (or a pachytene chromosome), and it is within the framework of bivalents that meiotic recombination is believed to occur (Sherman and Stack, 1995). Of particular note, bivalents are 5 to 15 times longer than corresponding metaphase chromosomes (Stack, 1984). As a consequence, small differences in relative chromosome length and/or arm ratio undetectable in metaphase preparations can be quite noticeable in pachytene chromosome preparations (Sherman and Stack, 1992; Peterson et al., 1999). Pachytene bivalents can be prepared for cytological analysis using several techniques (Peterson et al., 1999). In hypotonically spread bivalent sets, chromatin is dispersed laterally allowing visualization of SCs. Such preparations, known as SC spreads, have proven extremely useful in investigating chromosome structure and meiotic recombination (Loidl, 1991). In addition, SC spreads have been shown to be good substrates for repetitive and low-copy sequence fluorescence in situ hybridization (FISH; Moens and Pearlman, 1989, 1990; Heng et al., 1994; Solari and Dresser, 1995; Peterson et al., 1999).
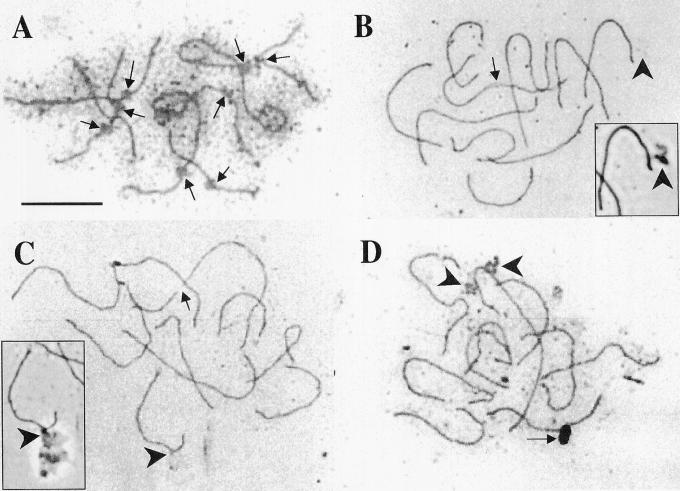
We recently developed a technique for generating SC spreads from sorghum (see “Materials and Methods”), and we are currently in the process of constructing a detailed SC karyotype for SB cv BT × 623. Initial results suggest that all 10 sorghum bivalents can be differentiated based on relative length and arm ratio. In addition, (a) Although kinetochores are not visible in most sorghum SC spreads (e.g. Fig. 7, B–D), some SC sets do possess distinctive kinetochores (Fig. 7A). In instances where kinetochores are not visible, centromeric locations presumably can be visualized on bivalents by performing FISH using a centromere-specific probe (see Jiang et al., 1996; Miller et al., 1998). (b) Sorghum SC sets are six to seven times longer than mitotic metaphase chromosome sets (Table I). (c) Although the nucleolus is generally dispersed during the SC spreading process, we have noticed that in some sorghum SC sets the longest SC is associated with what appears to be remnants of the nucleolus (Fig. 7D). In squashes of pachytene microsporocytes, Longley (1937) also observed that the longest sorghum bivalent is preferentially associated with the nucleolus. Sang and Liang (2000) recently used FISH to show that the nucleolus organizer region (i.e. the 18S-5.8S-26S rDNA sequence) is found on the longest SB mitotic metaphase chromosome (chromosome 1). These observations collectively suggest that the longest SC/bivalent corresponds to sorghum mitotic chromosome 1. (d) The sixth longest sorghum SC often appears to be closely associated with (or connected to) an amorphous structure of unknown identity (Figs. 7, B–C).
Figure 7.
SCs of SB. A through D, Silver-stained SC spreads. All photographs show bright-field images except where noted. A, Partial set of SCs in which kinetochores (arrows) are readily visible. B and C, Complete SC sets. In B, one SC contains a region of incomplete synapsis (arrow), whereas in C one SC appears to be broken into two pieces (arrow). In B and C, the sixth longest SC is associated with an amorphous structure of unknown origin (large arrowheads). These structures, most readily visualized by phase-contrast microscopy (see insets of B and C), may be part of the nuclear scaffold. In D, the longest chromosome appears to be associated with remnants of the nucleolus (large arrowheads). Although the dark oval object near the bottom of this bright-field image (arrow) looks like it could be a cellular structure, examination of this SC set by phase-contrast microscopy (photo not shown) suggests that the object is a small piece of glass. Scale bar = 10 μm.
Table I.
Comparison of length values (μm) for the mitotic metaphase chromosomes and SCs of sorghum
| Chromosmed | Metaphase, Gu et al. (1984)a
|
Metaphase, Yu et al. (1991)b
|
SC (Bivalent)c
|
|||
|---|---|---|---|---|---|---|
| Mean length | Relative length | Mean length | Relative length | Mean length | Relative length | |
| % | % | % | ||||
| 1 | 5.6 ± 0.06 | 17.17 | 4.80 ± 0.36 | 16.79 | 29.73 ± 1.19 | 15.34 |
| 2 | 4.16 ± 0.05 | 12.76 | 4.04 ± 0.33 | 14.13 | 24.77 ± 0.93 | 12.78 |
| 3 | 3.77 ± 0.10 | 11.56 | 3.38 ± 0.30 | 11.82 | 22.31 ± 0.73 | 11.51 |
| 4 | 3.34 ± 0.01 | 10.24 | 2.98 ± 0.21 | 10.42 | 20.65 ± 0.91 | 10.65 |
| 5 | 3.02 ± 0.05 | 9.26 | 2.75 ± 0.12 | 9.62 | 17.74 ± 0.71 | 9.15 |
| 6 | 2.77 ± 0.03 | 8.49 | 2.57 ± 0.13 | 8.99 | 16.94 ± 0.581 | 8.74 |
| 7 | 2.64 ± 0.10 | 8.10 | 2.30 ± 0.08 | 8.04 | 16.46 ± 0.62 | 8.49 |
| 8 | 2.55 ± 0.12 | 7.82 | 2.12 ± 0.18 | 7.42 | 15.95 ± 0.67 | 8.23 |
| 9 | 2.49 ± 0.08 | 7.64 | 1.93 ± 0.28 | 6.75 | 15.24 ± 0.63 | 7.86 |
| 10 | 2.27 ± 0.13 | 6.96 | 1.72 ± 0.13 | 6.02 | 14.07 ± 0.67 | 7.26 |
| Set lengthe | 32.61 | – | 28.59 | – | 193.87 | – |
Mean SC set length ÷ mean metaphase set length from Gu et al. (1984) = 5.95. Mean SC set length ÷ mean metaphase set length from Yu et al. (1991) = 6.78.
Length data are from Gu et al. (1984). The no. of sets from which measurements were taken is listed as four to six.
Length data are from Yu et al. (1991). Five chromosome sets were measured.
Values are based on measurement of 11 complete SC sets.
For each SC set, the 10 bivalents were assigned a no. based on relative length (with “1” being the longest). For “chromosome 1”, the mean length was the average of the longest chromosome in each of the 11 SC sets examined, the mean length of “chromosome 2” was the average of the second-longest SC from each of the 11 SC sets, etc. Whether a particular SC always maintains a consistent relative length to the other SCs in a set has yet to be determined. Chromosome nos. assigned to mitotic metaphase chromosomes also reflect relative length, although other factors including relative arm ratio and C banding were used to differentiate chromosomes. It is possible that SC length and mitotic metaphase chromosome length are not directly correlated (e.g. metaphase chromosome 5 may actually contain the same linkage group as SC 9, etc.).
Average length of a complete (1C) chromosome set.
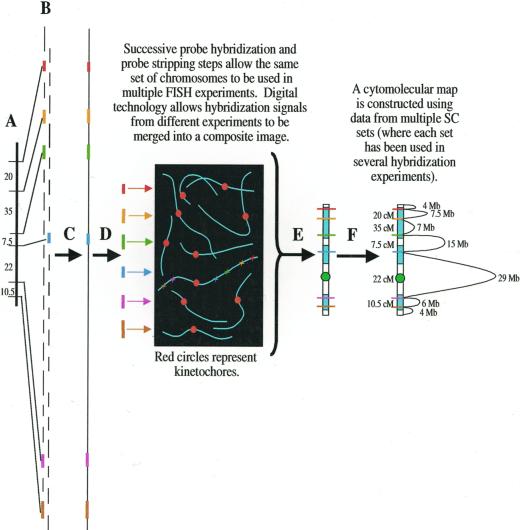
Once the SC karyotype is constructed, BACs associated with a specific LG will be pooled and used as a FISH-based “chromosome paint.” In this way, each sorghum LG will be coupled with its cytologically characterized bivalent. In addition, a series of single-copy molecular markers (or BACs containing molecular markers) from each sorghum LG will hybridized to SC/bivalent spreads allowing DNA markers to be positioned with respect to each other and chromosomal structures including kinetochores, telomeres, nucleolus organizer regions, heterochromatin, and euchromatin. This technique (known as “cytomolecular mapping”) has been successfully employed to relate molecular maps to chromosome structure in tomato (Lycopersicon esculentum; Peterson et al., 1999) and rice (Cheng et al., 2001). In species such as sorghum where physical maps are being constructed, cytomolecular mapping presumably can be used to position a complete chromosomal DNA molecule directly onto its corresponding SC. In such a case, each cytomolecular marker would serve as a point at which the DNA molecule would be “anchored” onto the framework of the chromosome (Fig. 8). Cytomolecular research promises to be instrumental in (a) identifying genomic regions and features related to marked deviations from the mean relationship between recombinational and physical distance, (b) comparing the locations of kinetochores, knobs, nucleolus organizers, and other cytological features of different grasses such as maize and sorghum at unparalleled levels of resolution, and (c) investigating the basis for unexpectedly high (“peaks”) or low (“valleys”) levels of variation in diversity maps.
Figure 8.
Using cytomolecular markers to anchor the physical map for a particular LG onto the actual structure of its pachytene chromosome. A, RFLP markers in the same LG are obtained via molecular mapping. B, Molecular markers are used to screen a BAC library. Positive BAC clones (colored lines) are isolated. C, A physical map of the LG is assembled. D, Insert DNA from BAC clones associated with particular linkage markers is used in FISH to SC spreads in which individual SCs and heterochromatin/euchromatin can be differentiated. The precise location(s) of each locus is determined. E, A cytomolecular map is constructed for the chromosome. In this example, heterochromatic regions are white, euchromatic regions are blue, and the kinetochore is represented by a red circle. F, The linkage map and physical map of the chromosome are superimposed directly onto the structure of the SC to produce a “cytophysical” map. Cytophysical mapping allows comparison of genetic linkage, chromatin configuration, and base pair distances.
Synthesis
Our long-term goal is to provide a detailed picture of the structure, function, and evolution of the sorghum genome, linked to a permanent and transferable set of modern molecular tools that will have enduring value for further study of many aspects of genome analysis in sorghum and other taxa. A robust physical map, comprised of large-insert DNA clones and anchored to the recombination-based genetic map by locus-specific STSs, is the centerpiece for these efforts. By using tools that are suitable for comparative analysis of related taxa, we seek to exploit high-resolution genomic information from models such as rice, extend our results from the small sorghum genome to closely related large-genome taxa such as maize and sugarcane, and evaluate the extent to which genomic features and specific genes/QTLs play common roles in diverse flora. The primary motivation for our work is to relate molecular-level variation to phenotypic diversity—early surveys of the levels and patterns of diversity at neutral DNA markers are an essential backdrop for future studies of diversity in large populations of candidate genes, using QTL information together with association approaches for narrowing the candidates to a small sampling that may be directly related to a specific phenotype. It has long been established that cytological features profoundly affect the relationship of recombination frequency to DNA content, and recently become clear that such features also affect the genomic distribution of allelic diversity—improved methods for visualizing chromosomal features and interleaving cytological and genetic/physical maps will better elucidate such relationships. Outcomes of these endeavors are expected to contribute to knowledge of plant genomic and phenotypic evolution, growth and development, and crop improvement.
MATERIALS AND METHODS
Materials
One-hundred fifty-six LG C-specific RFLP probes were used in the study. These comprised 127 homologous and 29 heterologous probes, detecting 128 and 31 LG C loci, respectively. Thirty-three were low-copy sequences that detected two or more loci in the sorghum (Sorghum bicolor [SB]) genome and the remainder are single copy probes. Probe inserts were isolated from low-melting-point gels after digestion with the appropriate enzyme and radiochemically labeled as described (Chittenden et al., 1994).
The Sorghum propinquum (SP) BAC library (Lin et al., 1999) comprises 38,016 clones with an average insert size of 126 kb and provides 6.06-genome-equivalent coverage. The theoretical probability of finding at least one BAC that contains a gene of interest is 99.7%.
Germplasm used for diversity mapping was provided by Jeff Dahlberg (exotic sorghums); Gil Lovell (U.S. Department of Agriculture-Agricultural Research Service, Griffin, GA; exotic sorghums); J. “Mike” Chandler (Johnson grass); and John Doebley (University of Wisconsin, Madison; S. nitidum), and the private breeding program reflected in cultivar designations.
BACRF Mapping and Library Screening
BAC pool DNA was prepared as described (Lin et al., 2000) for 99 pools of 384 BACs each. Ten micrograms of HindIII-digested SP genomic DNA and 150 ng or 330 ng of HindIII-digested BAC pool DNA were fractionated on 0.8% (w/v) agarose gels immersed in neutral electrophoresis buffer at 22 V cm−1 for 12 to14 h and transferred to nylon membranes. Membranes with 150- or 330-ng BAC pool DNA were used for hybridization with homologous and heterologous probes, respectively. The hybridization conditions were as described (Chittenden et al., 1994), except that 2.5% (w/v) dextran sulfate was used. BAC pools displaying one or more restriction fragments delineated subsets of the library containing putative BAC clones. These subsets were screened by colony hybridization, using 384-BAC dot blots handmade with a Nunc replicator. BACs detected by multilocus probes were assigned to their respective loci by comparing the restriction fragment(s) of their parent BAC pool with those of the SP genomic DNA. The BAC-data management system software (X. Draye and A.H. Paterson, unpublished data) was used for implementing the BACRF method, scoring films, archiving, and analyzing data. Overlapping BAC clones were assembled into contigs with FPC4.5 (Soderlund and Dunham, 1995).
Map Construction
The 156 probes used constitute a random subset of 346 RFLP probes, detecting 361 LG C loci, which had been mapped to sorghum LG C using 56 F2 individuals of an SB × SP cross (Chittenden et al., 1994; A.H. Paterson, personal communication). A framework genetic map of LG C comprised of 28 highly informative markers was established with MapMaker 3.0 (Lander et al., 1987) with an average interval between markers of 5.15 cM. The framework was used to place the most informative marker of each contig, so that BACs and markers included in BAC contigs could be anchored on the map. The remaining markers were grouped if they were closer than 0.5 cM, then placed on the map considering their most likely order between two points in the framework. Screening of the BAC library also detected clones at unmapped loci (suffix “u”) whose corresponding bands on the RFLP gels were monomorphic or too faint for an accurate scoring. The positions of five of these loci were able to be inferred because they linked physically to an anchored contig. Map order for the loci with suffix “Q” was verified in a separate population of 370 F2 individuals (Lin et al., 1995) from the same F1 plant.
DNA markers for wheat (Triticum aestivum; GrainGenes Database, http://wheat.pw.usda.gov/), rice (Oryza sativa; Causse et al., 1994), sugarcane (Saccharum spp. Ming et al., 1998), maize (Zea mays; Maize Genome Database, http://www.agron.missouri.edu/), and Arabidopsis (Paterson et al., 1996) revealed that 12, 28, 29, 65, and 9 heterologous probes, respectively, were aligned with sorghum LG C loci. QTLs associated with: (a) average height of the main culm, tallest tiller, and shortest tiller; (b) logarithm of rhizome number; (c) logarithm of non-zero rhizome number; (d) rhizome distance; (e) logarithm of subterranean rhizomatousness; (f) non-zero subterranean rhizomatousness; (g) seedling tillers; (h) seed weight; (i) spikelet number and whorl number in sorghum (Lin et al., 1995; Paterson et al., 1995b; A.H. Paterson, personal communication); and (j) sugar content in sugarcane (Ming et al., unpublished data) were interpolated on the genetic map.
Prefixes of DNA markers and their sources are as follows: Arabidopsis cDNA: AEST (R. Scholl, Arabidopsis Biological Resources Center, Ohio State University, Columbus), AHD, and HMG (T. Thomas, Texas A&M University); barley cDNA: BCD (M.E. Sorrells and S.D. Tanksley, Cornell University, Ithaca, NY); Johnson grass rhizome cDNA: pHER and pSHR (Y. Si and A.H. Paterson, unpublished results); maize PstI genomic clones: BNL and UMC (E.H. Coe and M. McMullen, University of Missouri, Columbia); maize cDNA: CSU (E.H. Coe, M.D. McMullen); millet (Pennisetum americanum) PstI genomic clones: M (M.D. Gale, John Innes Center, Norwich, UK); oat cDNA: CDO (Sorrells, Tanksley); sorghum cDNA: HHU (Wyrich et al., 1998), HHUK (Annen et al., 1998); sorghum phytochrome genes: PHY (L.H. Pratt and M.-M. C-Pratt, University of Georgia); sorghum PstI genomic DNA: pSB and SHO (A.H.P.); sugarcane cDNA: CDSB and CDSR (P. Moore, Hawaiian Agricultural Research Center, Aiea); sugarcane genomic clones: SG (Sorrells); and rice genomic clones RG and cDNA: RZ (S. McCouch and S. Tanksley, Cornell).
Physical Map Simulations
Two hundred simulations were run using the statistical analysis system/IML software to predict the progression of contig assembly as a function of the number of available markers. During each simulation, a LG C BAC library and a marker map were randomly created. The library comprised 4,000 clones with an insert size of 126 kb and provided over LG C a coverage equivalent to that of the library used for the experiment. The marker map comprised 1,000 markers. The contig assembly was simulated with increasing numbers of markers, starting at 5 markers and progressing by increments of 5 markers. At each step, the clones overlapping the current set of markers were identified and assembled into contigs. The number and length of the contigs were derived and recorded. The results of the 200 simulations were combined to estimate the median and percentiles of the following parameters: number of contigs, total contig map length, average contig length, and average number of markers per contig.
Fingerprinting and FPC Analysis
A slightly modified protocol for high-throughput fingerprint analysis of large-insert clones previously described by Marra et al., 1997 was employed, using HindIII-digested BAC DNA. Restriction fragments from the fingerprint of each clone was hand annotated using Image software (http://www.sanger.ac.uk/software/Image/), transformed into a band file (for each clone), and analyzed using the Finger Print Contig program (version 4.5, Soderlund et al., 1997, http://www.sanger.ac.uk/software/fpc/), running on a Sun Ultra 10 workstation. Automated contig assembly was initially performed at a cutoff value of 1 × 10−1 and tolerance of 7. Questionable (“Q”) contigs were split into smaller contigs by performing the analysis at more stringent cutoff values (ranging from 1 × 10−12 to 1 × 10−14) until all Qs were eliminated.
SC Spreads
SC spreads were prepared as described by Peterson et al. (1999) with several modifications. Based on squash preparations, it was determined that sorghum cv BT × 623 anthers between 0.9 and 1.1 mm in length contain microsporocytes in pachynema. As a consequence, anthers in this size range were placed in 200 μL of sugar-salt medium (0.56 mm KH2PO4, 0.1 mm PIPES [1,4-piperazinediethanesulfonic acid], 0.2% [w/v] potassium dextran sulfate, 1 mm CaCl2, 0.7 m mannitol, 1% [w/v] polyvinylpyrrolidone [pH 4.1]) containing 3 mg desalted pectinase (Sigma, St. Louis) and 3 mg desalted cytohelicase (Sepracor, Marlborough, MA). The anthers were bisected transversely to their long axis, and incubated for 40 min in the dark at 20°C in a closed humid chamber. During this interval, new glass microscope slides were wiped clean with 45% (w/v) acetic acid. The cleaned slides then were made hydrophilic by wiping them with a Kimwipe doused in Archer Anti-Static Spray (catalog no. 64-3310, Radio Shack, Fort Worth, TX). Using dissecting needles, protoplasts were squeezed from several anthers. A 1.0-μL aliquot of the protoplast suspension was drawn up into a siliconized micropipet and gently blown into a 10-μL droplet of hypotonic bursting medium [0.05% (w/v) (octylphenoxy)polyethoxyethanol (IGEPAL CA-630) and 0.1% (w/v) bovine serum albumin] suspended from the end of a 200-μL pipet tip. The resulting droplet was immediately placed on the center of one of the clean, hydrophilic glass slides. The slide immediately was given 30 puffs of 4% (w/v) aqueous formaldehyde (pH 8.5) from a hand-held nebulizer (Fullam, Latham, NY). Slides were air dried, fixed in 4% (w/v) aqueous formaldehyde for 10 min, rinsed twice without agitation in aqueous 0.01% (w/v) Photoflo 200 (20 s each rinse), rinsed four times (20 s each rinse) in distilled water, and allowed to dry. SC sets were stained with 33% (w/v) silver nitrate solution (see Sherman and Stack, 1992) and photographed using bright-field and/or phase-contrast microscopy. Chromosome measurements were made using the computer program MicroMeasure (version 3.01, available at www.colostate.edu/Depts/Biology/MicroMeasure).
Figure 5.
Comparative physical mapping of rice and sorghum. Vertical lines to left of sorghum loci and to right of rice loci indicate the sets of closely linked loci that colocate on one or more BAC clones in each species. Loci in italics are previously genetically unmapped; the subset in underlined italic font are physically linked to a mapped locus, so now it can be located on the integrated genetic physical map. For genetically mapped loci, genetic distances in centimorgans are indicated to left (sorghum) or right (rice). An asterisk shows multilocus probes that hybridize to additional loci in addition to the locus shown.
ACKNOWLEDGMENTS
We thank Dr. Keith F. Schertz for his dedication to the advancement of the study of sorghum, including long-term mentoring and collaboration that fostered this work. We also thank many members of the Paterson lab for technical help and moral support.
Footnotes
This research was funded by the Belgian American Educational Foundation (to X.D.); by the Rockefeller Foundation (to X.Q. and A.H.P.); by the U.S. National Science Foundation Plant Genome Research Program (to R.A.W. and A.H.P.); by the U.S. Department of Agriculture National Research Initiative Plant Genome Program (to D.G.P. and A.H.P.); by the International Consortium for Sugarcane Biotechnology (to R.A.W. and A.H.P.); by the National Grain Sorghum Producers (to R.A.W. and A.H.P.); by the U.S. Department of Agriculture Biotechnology Risk Assessment Program; and by the Texas, Georgia (to A.H.P.), and South Carolina (to R.A.W.) Agricultural Experiment Stations.
LITERATURE CITED
- Agrama HAS, Moussa ME. Mapping QTLs in breeding for drought tolerance in maize (Zea mays L.) Euphytica. 1996;91:89–97. [Google Scholar]
- Alpert KB, Tanksley SD. High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit weight quantitative trait locus in tomato. Proc Natl Acad Sci USA. 1996;93:15503–15507. doi: 10.1073/pnas.93.26.15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annen F, Chang JL, Paterson AH, Stockhaus J. Characterization of 14 different putative protein kinase cDNA clones of the C-4 plant Sorghum bicolor. Mol Gen Genet. 1998;259:115–122. doi: 10.1007/s004380050795. [DOI] [PubMed] [Google Scholar]
- Arumunganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–219. [Google Scholar]
- Austin DF, Lee M. Comparative mapping in F-2:3 and F-6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor Appl Genet. 1996a;92:817–826. doi: 10.1007/BF00221893. [DOI] [PubMed] [Google Scholar]
- Austin DF, Lee M. Genetic resolution and verification of quantitative trait loci for flowering and plant height with recombinant inbred lines of maize. Genome. 1996b;39:957–968. doi: 10.1139/g96-120. [DOI] [PubMed] [Google Scholar]
- Austin DF, Lee M. Detection of quantitative trait loci for grain yield and yield components in maize across generations in stress and nonstress environments. Crop Science. 1998;38:1296–1308. [Google Scholar]
- Bennett MD, Laurie DA. Chromosome size in maize and sorghum using EM serial section reconstructed nuclei. Maydica. 1995;40:199–204. [Google Scholar]
- Bennetzen JL, Freeling M. Grasses as a single genetic system: genome composition, collinearity and compatibility. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- Bohn M, Khairallah MM, Jiang C, GonzalezdeLeon D, Hoisington DA, Utz HF, Deutsch JA, Jewell DC, Mihm JA, Melchinger AE. QTL mapping in tropical maize .2. Comparison of genomic regions for resistance to Diatraea spp. Crop Sci. 1997;37:1892–1902. [Google Scholar]
- Bouck J, Miller W, Gorrell JH, Muzny D, Gibbs RA. Analysis of the quality and utility of random shotgun sequencing at low redundancies. Genome Res. 1998;8:1074–1084. doi: 10.1101/gr.8.10.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Schertz KF, Abbey C, Anderson S, Chang C, Chittenden LM, Draye X, Hoppe AH, Jessup R, Lennington J. A high-density 2399-locus genetic map of Sorghum. San Diego: Plant and Animal Genome VIII Conference; 2000. http://www.intl-pag.org/pag/8/abstracts/pag8712.html , http://www.intl-pag.org/pag/8/abstracts/pag8712.html (February 1, 2001) (February 1, 2001) [Google Scholar]
- Byrne PF, McMullen MD, Snook ME, Musket TA, Theuri JM, Widstrom NW, Wiseman BR, Coe EH. Quantitative trait loci and metabolic pathways: genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc Natl Acad Sci USA. 1996;93:8820–8825. doi: 10.1073/pnas.93.17.8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WW, Reneker J, Chow CW, Vaishnav M, Bradley A. An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics. 1998;54:387–397. doi: 10.1006/geno.1998.5620. [DOI] [PubMed] [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu KS, Xiao JH, Yu ZH, Ronald PC, Harrington SE. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Presting GG, Buell CR, Wing RA, Jiang J (2001) Integration of genetic and cytogenetic maps reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics (in press) [DOI] [PMC free article] [PubMed]
- Chittenden LM, Schertz KF, Lin YR, Wing RA, Paterson AH. A detailed RFLP map of Sorghum bicolor x S. propinquum, suitable for high-density mapping, suggests ancestral duplication of sorghum chromosomes or chromosomal segments. Theor Appl Genet. 1994;87:925–933. doi: 10.1007/BF00225786. [DOI] [PubMed] [Google Scholar]
- Clayton WD. Andropogoneae. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, editors. Grass Systematics and Evolution. Washington, DC: Smithsonian Institution Press; 1987. pp. 307–309. [Google Scholar]
- De Risi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- deWet JMJ, Gupta SC, Harlan JR, Grassl CO. Cytogenetics of introgression from Saccharum into Sorghum. Crop Sci. 1976;16:568–572. [Google Scholar]
- Doggett H. Sorghum. Ed 2. New York: John Wiley and Sons, Inc.; 1988. [Google Scholar]
- Dorweiler J, Stec A, Kermicle J, Doebley J. Teosinteglume architecture (TGA1), a locus differentiating maize and teosinte. Science. 1993;262:233–235. doi: 10.1126/science.262.5131.233. [DOI] [PubMed] [Google Scholar]
- Dvorak J, Luo MC, Yang ZL. Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics. 1998;148:423–434. doi: 10.1093/genetics/148.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Plant comparative genetics after 10 years. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Le Thierry I' Enneguin M, Peek AS, Saukins MC. Maize as a model for the evolution of plant nuclear genomes. Proc Natl Acad Sci USA. 2000;97:7008–7015. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez MI, Islam-Faridi MN, Woo S-S, Schertz KF, Czeschin D, Jr, Zwick MS, Wing RA, Stelly DM, Price HJ. FISH of a maize sh2-selected sorghum BAC to chromosomes of Sorghum bicolor. Genome. 1997;40:475–478. doi: 10.1139/g97-063. [DOI] [PubMed] [Google Scholar]
- Groh S, GonzalezdeLeon D, Khairallah MM, Jiang C, Bergvinson D, Bohn M, Hoisington DA, Melchinger AE. QTL mapping in tropical maize: III. Genomic regions for resistance to Diatraea spp. and associated traits in two RIL populations. Crop Sci. 1998;38:1062–1072. [Google Scholar]
- Gu MH, Ma HT, Liang GH. Karyotype analysis of seven species in the genus Sorghum. J Hered. 1984;75:196–202. [Google Scholar]
- Hamblin MT, Aquadro CF. DNA sequence variation and the recombinational landscape in Drosophila pseudoobscura: a study of the second chromosome. Genetics. 1999;153:859–869. doi: 10.1093/genetics/153.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HHQ, Tsui L-C, Moens PB. Organization of heterologous DNA inserts on the mouse meiotic chromosome core. Chromosoma. 1994;103:401–407. doi: 10.1007/BF00362284. [DOI] [PubMed] [Google Scholar]
- Hulbert SH, Richter TE, Axtell JD, Bennetzen JL. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc Natl Acad Sci USA. 1990;87:4251–5. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Nasuda S, Dong F, Scherrer CW, Woo S, Wing RA, Gill BS, Ward DC. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci USA. 1996;93:14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairallah MM, Bohn M, Jiang C, Deutsch JA, Jewell DC, Mihm JA, Melchinger AE, GonzalezdeLeon D, Hoisington DA. Molecular mapping of QTL for southwestern corn borer resistance, plant height and flowering in tropical maize. J Plant Breed. 1998;117:309–318. [Google Scholar]
- Klein PE, Klein RR, Cartinhour SW, Ulanch PE, Dong J, Obert JA, Morishige DT, Schlueter SD, Childs KL, Ale M. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genome Res. 2000;6:789–807. doi: 10.1101/gr.10.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski SP, Lan TH, Feldmann KA, Paterson AH. Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics. 1994;138:499–510. doi: 10.1093/genetics/138.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lebreton C, LazicJancic V, Steed A, Pekic S, Quarrie SA. Identification of QTL for drought responses in maize and their use in testing causal relationships between traits. J Exp Bot. 1995;46:853–865. [Google Scholar]
- Lin Y-R, Draye X, Qian X, Ren S, Zhu L-H, Tomkins J, Wing R, Li Z, Paterson AH. Locus-specific contig assembly in highly-duplicated genomes, using the BAC-RF method. Nucleic Acids Res. 2000;28:e23. doi: 10.1093/nar/28.7.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YR, Schertz KF, Paterson AH. Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics. 1995;141:391–411. doi: 10.1093/genetics/141.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YR, Zhu L, Ren S, Yang J, Schertz KF, Paterson AH. A Sorghum propinquum BAC library, suitable for cloning genes associated with loss-of-function mutations during crop domestication. Mol Breed. 1999;5:511–520. [Google Scholar]
- Linder HP. The evolutionary history of the Poales/Restionales: a hypothesis. Kew Bull. 1987;42:297–318. [Google Scholar]
- Loidl J. Coming to grips with a complex matter: a multidisciplinary approach to the synaptonemal complex. Chromosoma. 1991;100:289–292. doi: 10.1007/BF00360526. [DOI] [PubMed] [Google Scholar]
- Longley AE. Morphological characters of teosinte chromosomes. J Agric Res. 1937;54:835–862. [Google Scholar]
- Lubberstedt T, Melchinger AE, Schon CC, Utz HF, Klein D. QTL mapping in testcrosses of european flint lines of maize 1: comparison of different testers for forage yield traits. Crop Sci. 1997;37:921–931. [Google Scholar]
- Mao L, Wood TC, Yu YS, Budiman MA, Tomkins J, Woo SS, Sasinowski M, Presting G, Frisch D, Goff S. Rice transposable elements: a survey of 73,000 sequence-tagged connectors. Genome Res. 2000;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof MAS, Yue YG, Xiang ZX, Stromberg EL, Rufener GK. Identification of quantitative trait loci controlling resistance to gray leaf spot disease in maize. Theor Appl Genet. 1996;93:539–546. doi: 10.1007/BF00417945. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JM, Jansco MM, Pichersky E. Duplicate sequences with a similarity to expressed genes in the genome of Arabidopsis thaliana. Theor Appl Genet. 1993;86:880–888. doi: 10.1007/BF00212616. [DOI] [PubMed] [Google Scholar]
- Messing J, Llaca V. Importance of anchor genomes for any plant genome project. Proc Natl Acad Sci USA. 1998;95:2017–2020. doi: 10.1073/pnas.95.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JT, Jackson SA, Nasuda S, Gill BS, Wing RA, Jiang J. Cloning and characterization of a centromere-specific repetitive DNA element from Sorghum bicolor. Theor Appl Genet. 1998;96:832–839. [Google Scholar]
- Ming R, Liu SC, Lin YR, da Silva J, Wilson W, Braga D, van Deynze A, Wenslaff TF, Wu KK, Moore PH. Detailed alignment of Saccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics. 1998;150:1663–1682. doi: 10.1093/genetics/150.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Pearlman RE. Satellite DNA I in chromatin loops of rat pachytene chromosomes and in spermatids. Chromosoma. 1989;98:287–294. doi: 10.1007/BF00327315. [DOI] [PubMed] [Google Scholar]
- Moens PB, Pearlman RE. In situ DNA sequence mapping with surface-spread mouse pachytene chromosomes. Cytogenet Cell Genet. 1990;53:219–220. doi: 10.1159/000132935. [DOI] [PubMed] [Google Scholar]
- Moses M. Synaptinemal complex. Annu Rev Genet. 1968;2:363–412. [Google Scholar]
- Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX, Katsar CS, Lan TH, Lin YR, Ming RG. Comparative genomics of plant chromosomes. Plant Cell. 2000;12:1523–1539. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Deverna JW, Lanini B, Tanksley SD. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics. 1990;124:735–742. doi: 10.1093/genetics/124.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Lan TH, Reischmann KP, Chang C, Lin YR, Liu SC, Burow MD, Kowalski SP, Katsar CS, DelMonte TA. Toward a unified genetic map of higher plants, transcending the monocot-dicot divergence. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lin YR, Li ZK, Schertz KF, Doebley JF, Pinson SRM, Liu SC, Stansel JW, Irvine JE. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science. 1995a;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Schertz KF, Lin YR, Liu SC, Chang YL. The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) pers. Proc Natl Acad Sci USA. 1995b;92:6127–6131. doi: 10.1073/pnas.92.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MG, Lee M. Identification of genomic regions affecting plant height in sorghum and maize. Theor Appl Genet. 1995;90:380–388. doi: 10.1007/BF00221980. [DOI] [PubMed] [Google Scholar]
- Pereira MG, Lee M, Bramelcox P, Woodman W, Doebley J, Whitkus R. Construction of an RFLP map in sorghum and comparative mapping in maize. Genome. 1994;37:236–243. doi: 10.1139/g94-033. [DOI] [PubMed] [Google Scholar]
- Peterson DG, Lapitan NLV, Stack SM. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH) Genetics. 1999;152:427–439. doi: 10.1093/genetics/152.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rami JF, Dufour P, Trouche G, Fliedel G, Mestres C, Davrieux F, Blanchard P, Hamon P. Quantitative trait loci for pain quality, productivity, morphological and agronomical traits in sorghum (Sorghum bicolor l. Moench) Theor Appl Genet. 1998;97:605–616. [Google Scholar]
- Ribaut JM, Hoisington DA, Deutsch JA, Jiang C, GonzalezdeLeon D. Identification of quantitative trait loci under drought conditions in tropical maize: II. Flowering parameters and the anthesis-silking interval. Theor Appl Genet. 1996;92:905–914. doi: 10.1007/BF00221905. [DOI] [PubMed] [Google Scholar]
- Robertson DS. A possible technique for isolating genic DNA for quantitative traits in plants. J Theor Biol. 1985;117:1–10. [Google Scholar]
- Sang Y, Liang GH. Comparative physical mapping of the 18S-5.8S–26S rDNA in three sorghum species. Genome. 2000;43:918–922. [PubMed] [Google Scholar]
- Sherman JD, Stack SM. Two-dimensional spreads of synaptonemal complexes from solanaceous plants: V. Tomato (Lycopersicon esculentum) karyotype and idiogram. Genome. 1992;35:354–359. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman JD, Stack SM. Two-dimensional spreads of synaptonemal complexes from solanaceous plants: VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum) Genetics. 1995;141:683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. Pastoral synteny. Nature. 1993;365:297–298. [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sills GR, Bridges W, Aljanabi SM, Sobral BWS. Genetic analysis of agronomic traits in a cross between sugarcane (Saccharum officinarum L.) and its presumed progenitor (S-robustum Brandes and Jesw ex grassl) Mol Breed. 1995;1:355–363. [Google Scholar]
- Simmonds NW. The Evolution of the Bananas. London: Longman; 1962. [Google Scholar]
- Sobral BWS, Braga DPV, LaHood ES, Keim P. Phylogenetic analysis of chloroplast restriction enzyme site mutations in the Saccharinae Griseb. subtribe of the Andropogoneae Dumort. tribe. Theor Appl Genet. 1994;87:843–853. doi: 10.1007/BF00221137. [DOI] [PubMed] [Google Scholar]
- Soderlund C, Dunham I. SAM: a system for iteratively building marker maps. Comput Appl Biosci. 1995;11:645–655. doi: 10.1093/bioinformatics/11.6.645. [DOI] [PubMed] [Google Scholar]
- Soderlund C, Longden I, Mott R. FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci. 1997;13:523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- Solari AJ, Dresser ME. High-resolution cytological localization of the XhoI and EcoRI repeat sequence in the pachytene ZW bivalent of the chicken. Chromosome Res. 1995;3:87–93. doi: 10.1007/BF00710668. [DOI] [PubMed] [Google Scholar]
- Stack SM. Heterochromatin, the synaptonemal complex, and crossing over. J Cell Sci. 1984;71:159–176. doi: 10.1242/jcs.71.1.159. [DOI] [PubMed] [Google Scholar]
- Tao YZ, Jordan DR, Henzell RG, McIntyre CL. Construction of a genetic map in a sorghum recombinant inbred line using probes from different sources and its comparison with other sorghum maps. Aust J Agric Res. 1998;49:729–736. [Google Scholar]
- Thomasson JR. Fossil grasses, 1820–1986 and beyond. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, editors. Grass Systematics and Evolution. Washington, DC: Smithsonian Institution Press; 1987. pp. 159–167. [Google Scholar]
- Tuberosa R, Sanguineti MC, Landi P, Salvi S, Casarini E, Conti S. RFLP mapping of quantitative trait loci controlling abscisic acid concentration in leaves of drought-stressed maize (Zea mays L.) Theor Appl Genet. 1998;97:744–755. [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough P. Evaluation of near-isogenic sorghum lines contrasting for qtl markers associated with drought tolerance. Crop Sci. 1998;38:835–842. [Google Scholar]
- Tuinstra MR, Grote EM, Goldsbrough PB, Ejeta G. Genetic analysis of post-flowering drought tolerance and components of grain development in sorghum bicolor (l.) Moench. Mol Breed. 1997;3:439–448. [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldboom LR, Lee M. Molecular-marker-facilitated studies cbf morphological traits in maize 2: determination of QTLs for grain-yield and yield components. Theor Appl Genet. 1994;89:451–458. doi: 10.1007/BF00225380. [DOI] [PubMed] [Google Scholar]
- Veldboom LR, Lee M. Genetic mapping of quantitative trait loci in maize in stress and nonstress environments 1: grain yield and yield components. Crop Sci. 1996a;36:1310–1319. [Google Scholar]
- Veldboom LR, Lee M. Genetic mapping of quantitative trait loci in maize in stress and nonstress environments 2: plant height and flowering. Crop Sci. 1996b;36:1320–1327. [Google Scholar]
- Veldboom LR, Lee M, Woodman WL. Molecular marker-facilitated studies in an elite maize population 1: linkage analysis and determination of QTL for morphological traits. Theor Appl Genet. 1994;88:7–16. doi: 10.1007/BF00222387. [DOI] [PubMed] [Google Scholar]
- Venter JC, Smith HO, Hood L. A new strategy for genome sequencing. Nature. 1996;381:364–366. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- Wang R, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Whitkus R, Doebley J, Lee M. Comparative genome mapping of sorghum and maize. Genetics. 1992;132:1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and the Genetics of Populations. 2: The Theory of Gene Frequencies. The University of Chicago Press; 1968. p. 12. [Google Scholar]
- Wyrich R, Dressen U, Brockmann S, Streubel M, Chang C, Qiang D, Paterson AH, Westhoff P. The molecular basis of c-4 photosynthesis in sorghum: isolation, characterization and RFLP mapping of mesophyll- andbundle-sheath-specific cDNAs obtained by differential screening. Plant Mol Biol Rep. 1998;37:319–335. doi: 10.1023/a:1005900118292. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kuboki Y, Lin SY, Sasaki T, Yano M. Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single mendelian factors. Theor Appl Genet. 1998;97:37–44. [Google Scholar]
- Yu H, Liang GH, Kofoid KD. Analysis of C-banding patterns of sorghum. Crop Sci. 1991;31:1524–1527. [Google Scholar]
- Zhao XP, Ji YF, Ding XL, Stelly DM, Paterson AH. Macromolecular organization and genetic mapping of a rapidly evolving chromosome-specific tandem repeat family (b77) in cotton (Gossypium) Plant Mol Biol Rep. 1998;38:1031–1042. doi: 10.1023/a:1006073116627. [DOI] [PubMed] [Google Scholar]
- Zhao XP, Wing RA, Paterson AH. Cloning and characterization of the majority of repetitive DNA in cotton (Gossypium L.) Genome. 1995;38:1177–1188. doi: 10.1139/g95-156. [DOI] [PubMed] [Google Scholar]