Significance
Huntington’s disease (HD) is a devastating neurodegenerative disorder caused by expansion of a polyglutamine repeat within the huntingtin (HTT) protein. A normal function of HTT is that of a scaffold for selective autophagy, a mechanism of protein and organelle degradation by the lysosome required for neuronal health. Here, we show that the inflammatory IκB kinase (IKK) kinase subunit IKKβ may function in vivo to regulate autophagy through direct phosphorylation of HTT at serine 13 and through the activation of autophagy gene expression. IKKβ is required to slow HD behavioral progression and to suppress neurodegeneration and microglial activation in HD transgenic mice. Our work suggests that the early activation of IKK may be protective to activate autophagy, thereby slowing HD progression.
Keywords: huntingtin, autophagy, Huntington’s disease, neurodegeneration, IκB kinase
Abstract
Neuroinflammation is an important contributor to neuronal pathology and death in neurodegenerative diseases and neuronal injury. Therapeutic interventions blocking the activity of the inflammatory kinase IKKβ, a key regulator of neuroinflammatory pathways, is protective in several animal models of neurodegenerative disease and neuronal injury. In Huntington’s disease (HD), however, significant questions exist as to the impact of blocking or diminishing the activity of IKKβ on HD pathology given its potential role in Huntingtin (HTT) degradation. In cell culture, IKKβ phosphorylates HTT serine (S) 13 and activates HTT degradation, a process that becomes impaired with polyQ expansion. To investigate the in vivo relationship of IKKβ to HTT S13 phosphorylation and HD progression, we crossed conditional tamoxifen-inducible IKKβ knockout mice with R6/1 HD mice. Behavioral assays in these mice showed a significant worsening of HD pathological phenotypes. The increased behavioral pathology correlated with reduced levels of endogenous mouse full-length phospho-S13 HTT, supporting the importance of IKKβ in the phosphorylation of HTT S13 in vivo. Notably, many striatal autophagy genes were up-regulated in HD vs. control mice; however, IKKβ knockout partially reduced this up-regulation in HD, increased striatal neurodegeneration, and enhanced an activated microglial response. We propose that IKKβ is protective in striatal neurons early in HD progression via phosphorylation of HTT S13. As IKKβ is also required for up-regulation of some autophagy genes and HTT is a scaffold for selective autophagy, IKKβ may influence autophagy through multiple mechanisms to maintain healthy striatal function, thereby reducing neuronal degeneration to slow HD onset.
The development of effective treatments for neurodegenerative diseases critically depends on an understanding of the sources of pathology. In Huntington’s disease (HD), the genetically based primary cause of pathology is expansion of a CAG repeat encoding polyglutamine (polyQ) within the huntingtin (HTT) protein (1). While the physical properties of the polyQ repeat are a key element in HD pathology, polyQ expansion can also have an important impact on the functional properties of HTT itself.
We previously found that phosphorylation of HTT at serines (S) 13 and 16 can activate HTT clearance in cells (2), reduce aggregate accumulation and block HD progression in HD mice (3). This phosphorylation is impaired by polyQ expansion (2), suggesting that this posttranslational modification may be critical to HD pathogenesis. We further showed that the inflammatory IκB kinase complex (IKK) activates phosphorylation of HTT S13 and S16 and enhances HTT clearance (2). IKK is a cytokine-induced serine kinase that controls the activation of NF-κB, a ubiquitous transcription factor closely associated with inflammation (4). While inflammation has been implicated in neurodestructive outcomes in late stages of HD, Alzheimer’s disease (AD), and Parkinson’s disease (PD), immunoactivities and inflammatory processes may also be neuroprotective early in disease progression (5, 6). IKK is activated by acute cellular stress and has previously been shown to contribute to the induction of autophagy (7–9). To prevent continuous activation of IKK, autophagic degradation of IKK itself occurs through a negative feedback loop (10, 11). As autophagy declines with aging (12), an accumulation of IKK over time can cause its excessive activation and harmful responses such as tumorigenesis and inflammation (13).
IKK induces expression of several autophagy-related genes independent of its activation of NF-κB and has been proposed to play a more direct role in autophagy regulation through the phosphorylation of autophagy proteins (8, 9). The IKK kinase complex is composed of three subunits, α, β, and γ; α and β are homologous catalytic subunits and γ is a regulatory subunit. IKKα and IKKβ can directly phosphorylate HTT S13 and activate phosphorylation of S16 (2). In turn, phosphorylation at these residues increases HTT clearance in cells by the proteasome and lysosome and reduces mutant HTT exon 1 protein-mediated cellular toxicity (2). Extending these studies, we showed that mimicking S13 and S16 phosphorylation (S13,16D) in the context of full-length mutant HTT expression in vivo in BACHD mice completely blocked HD progression and aggregate formation (3), supporting a possible direct association between phosphorylation of HTT and mutant HTT clearance. IKK is activated in cell culture and mouse models of HD chronically expressing mutant HTT (14), potentially reflecting an initial compensatory effect that may help phosphorylate HTT. These data are consistent with the innate immune activation that is observed in presymptomatic HD patients at least a decade before overt neurological manifestation of disease (15). Over time, IKK activation and HTT phosphorylation would be predicted to be less efficient in inducing HTT degradation because of reduced function of the proteasome and lysosome with aging (16) and might contribute to the chronic inflammation associated with neuronal cell death paralleling other hallmarks of HD pathogenesis (2, 5, 6).
In this work we tested the relationship of IKKβ to HD pathogenesis. We determined that HD behavioral phenotypes are further impaired in R6/1 HD mice upon IKKβ knockout, paralleling striatal neurodegeneration and an activated microglial response. We found that the kinase IKKβ is responsible in vivo for phosphorylation of HTT S13, the regulation of striatal WT full-length HTT abundance in nontransgenic (NT) control and R6/1 (HD) mice, and the activation of transcription of several autophagy genes, potentially linking inflammatory signal transduction pathways to the induction of HTT’s autophagic scaffold function and autophagy in general. Thus, we find that IKKβ is required in vivo to slow the early stages of HD progression in R6/1 mice and suggest that the impact of IKKβ expression early in HD disease progression is due to IKKβ in vivo phosphorylation of HTT S13 and to the effect of IKKβ expression on autophagy.
Results
Generation of R6/1 HD Mice with Conditional Tamoxifen-Inducible IKKβ Knockout.
The R6/1 mouse line is a widely employed HD model that expresses a mutant human HTT exon 1 transgene (17) and demonstrates a progressive decline in motor abilities and cognitive and social behaviors starting around 12 to 14 wk of age (18–21) accompanied by striatal atrophy and ventricular enlargement (17, 18, 22). To investigate the in vivo relationship of IKKβ to HTT S13 phosphorylation and HD progression, we created conditional tamoxifen-inducible IKKβ knockout mice. Transgenic mice with loxP-flanked alleles of IKKβ were crossed with mice expressing the tamoxifen-inducible Cre recombinase under control of the Wfs1, the Wolfram syndrome 1 homolog (human) promoter/enhancer regions (23, 24). Wfs1 promotes expression in neurons and astrocytes in several brain regions (25, 26). Efficient conditional IKKβ knockout was achieved in adult tissue including striatum and liver, which were evaluated at the completion of the study at 16 wk of age. Cre+, loxP-flanked alleles of IKKβ mice were crossed with R6/1 HD mice and IKKβ knockdown was induced with tamoxifen vs. vehicle (oil) administered at week 10, at a time that mice do not have demonstrable phenotypes, in R6/1 (HD) vs. NT WT controls. In this study, 9/10 of the female NT mice did not survive 1 wk past tamoxifen injection, and therefore only male mice were used for this work (Tamoxifen Protocol).
IKKβ Knockout Exacerbates Behavioral Phenotypes in the R6/1 Mouse Model of HD.
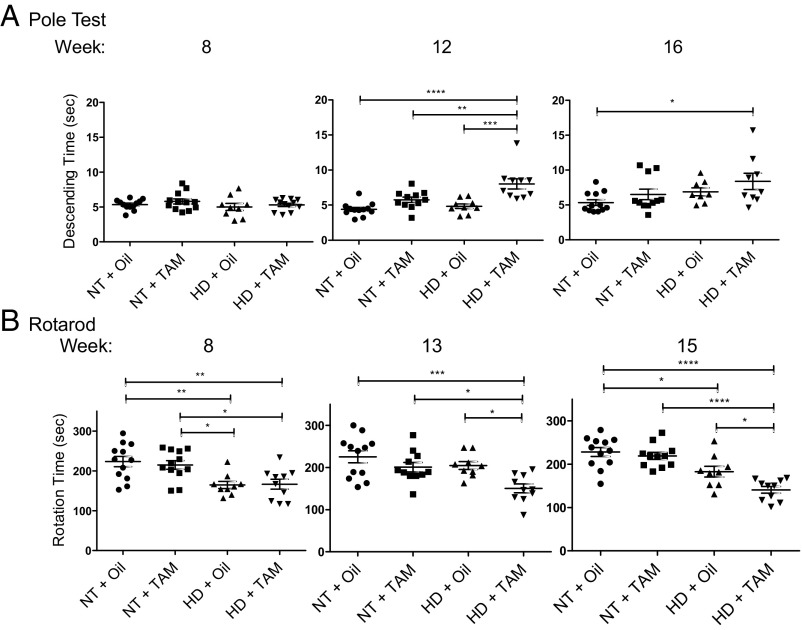
R6/1 mice begin losing weight and show a decline in behavioral tests beginning between 12 and 14 wk (20, 21), and our results are consistent with those findings. No treatment effect on weight or grip strength was observed for tamoxifen-induced IKKβ knockout in either R6/1 (HD) mice or NT WT controls (SI Appendix, Fig. S1 A and B), and tamoxifen-induced IKKβ knockout did not affect pole test or rotarod performance in the NT controls (Fig. 1). Behavioral assessments demonstrated significantly impaired pole test descending times (60% slower) following tamoxifen-induced IKKβ knockout in HD mice at 12 wk of age compared with oil-treated HD controls (Fig. 1A). However, the increase in impairment was not significant at the 16-wk time point. HD mice with tamoxifen-induced IKKβ knockout exhibited significantly impaired time (27% and 23% less time) on rotating rod before fall compared with oil-treated HD controls on weeks 13 and 15, respectively (Fig. 1B). The impairment of rotarod and pole test tasks was not due to a toxicity of tamoxifen treatment independent of IKKβ knockout, as a separate cohort of tamoxifen-treated standard R6/1 HD mice, without the Cre or floxed alleles of IKKβ, did not demonstrate further impairment in these pole test or rotarod tasks (SI Appendix, Fig. S2). Taken together, these results demonstrate that tamoxifen-induced IKKβ knockout exacerbates neurological phenotypes of R6/1 HD mice beyond the effect of mHTT transgene expression alone. There were no significant differences across the battery of behavior assessments with IKKβ knockout in NT control mice, supporting a selective role for IKKβ in the disease setting.
Fig. 1.
Effects of IKKβ knockout on behavior in male R6/1 mice. Pole test (8, 12, and 16 wk) and rotarod (8, 13, and 15 wk) are shown for tamoxifen (TAM)- vs. oil-treated R6/1 (HD) and NT controls (n = NT-Oil, 12; NT-TAM, 12; HD-Oil, 9; HD-TAM, 12 at week 8, and NT-Oil, 12; NT-TAM, 11; HD-Oil: 9, HD-TAM, 10 following injections). HD mice performed significantly worse on 12-wk pole test (A) and on weeks 13 and 15 rotarod (B) with TAM-induced IKKβ knockout than oil-treated HD control mice. No significant effect was observed in NT control mice with TAM-induced IKKβ knockout. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 values represent means ± SEM. Statistical significance was determined by one-way ANOVA with Bonferroni posttesting.
IKKβ Is Required for Phosphorylation of HTT S13 in Vivo.
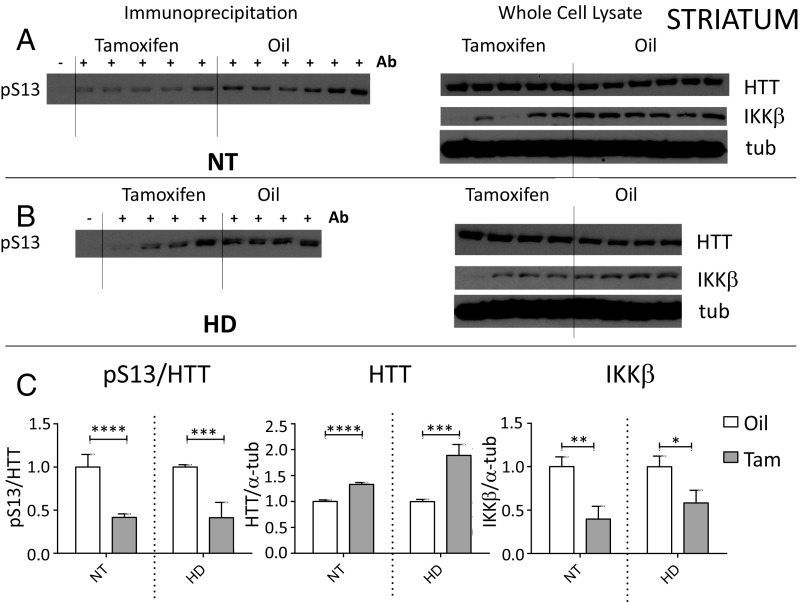
We hypothesized that the increased pathology observed in the IKKβ knockout HD mice may be the consequence of failure of HTT phosphorylation at amino acid S13 in this setting. We had previously found that IKKβ can directly phosphorylate HTT S13 using in vitro kinase assays, and that HTT S13 phosphorylation is increased with overexpression of IKKβ or induction of IKKβ with TNF-α or IL-1β treatment in cell culture, corresponding to an activation of HTT clearance (2). We now designed studies to evaluate whether IKKβ is a relevant HTT S13 kinase in vivo and whether it regulates HTT levels. After behavioral testing ending at week 16, IKKβ knockout and control mice were killed, and liver and striatal tissue were collected for Western analysis to measure levels of IKKβ, S13 phosphorylated full-length HTT, total full-length HTT, and transgenic mutant HTT exon 1 protein. S13 phosphorylation is quantitated using an immunoprecipitation (IP) method that does not show nonspecific binding of HTT to the IP beads (Materials and Methods), and total full-length HTT and transgenic mutant HTT exon 1 protein are each quantitated using whole-cell lysates. We evaluated protein levels in striatum of tamoxifen- vs. oil-treated NT control and HD mice, all expressing the tamoxifen-inducible Cre with the floxed alleles of IKKβ. Consistent with the hypothesis that reduced IKKβ levels impacted the phosphorylation at S13 and degradation of full-length HTT, tamoxifen treatment significantly reduced IKKβ and increased levels of full-length HTT in whole striatal cell extract and reduced relative levels of immunoprecipitated full-length S13 phosphorylated HTT in both NT and HD striatum (Fig. 2).
Fig. 2.
Levels of S13-phosphorylated HTT are reduced while total full-length (∼350 kDa) mouse HTT abundance is increased with IKKβ knockout in striatum, demonstrating that IKKβ is a relevant striatal HTT S13 kinase in vivo. Male R6/1 (HD) and NT WT controls, both containing the tamoxifen-inducible Cre and floxed alleles of IKKβ, were treated with tamoxifen or oil vehicle control during week 10 and killed at week 16. IKKβ normalized to loading control α-tubulin was significantly reduced in 16-wk striatum of HD and NT mice with tamoxifen treatment over oil control in whole-cell lysate (A–C). Anti-HTT phosphoserine 13 (pS13) antibody was used to immunoprecipitate phosphorylated full-length mouse HTT which was then detected by Western blot with anti-total HTT antibody. Levels of pS13-HTT were significantly reduced relative to total HTT with IKKβ knockout in HD and NT controls, while levels of total HTT normalized to α-tubulin were significantly increased (A–C). Western images (A and B) were quantitated using Scion software (C). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 values represent means ± SEM. Statistical significance was determined by paired t test.
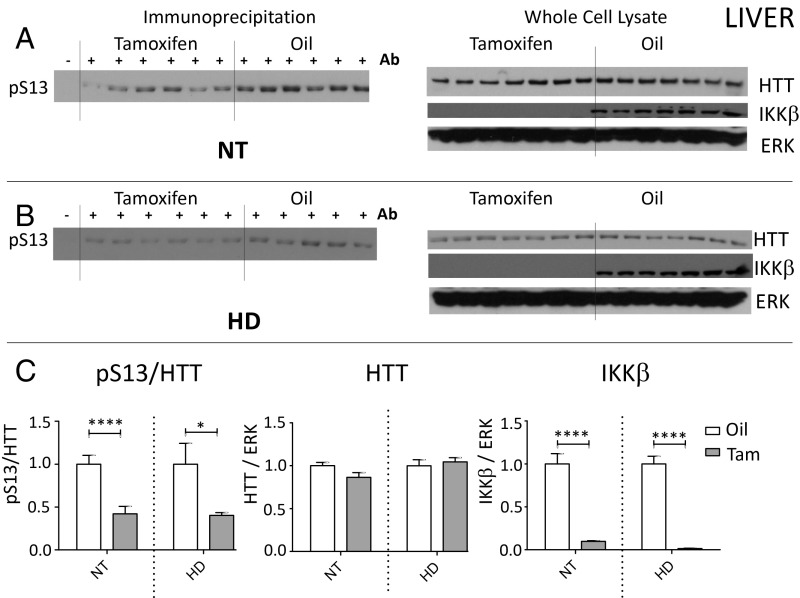
Liver dysfunction has been observed in manifest and premanifest HD patients and in HD mouse models (27, 28); we therefore chose to also examine IKKβ knockout liver tissue in our HD and NT control mice. Similar to striatum, IKKβ was significantly reduced in liver homogenates from these animals following tamoxifen treatment in both HD and NT control liver, which tracked with reduced relative levels of S13 phosphorylated HTT (Fig. 3), demonstrating the involvement of IKKβ in the phosphorylation of HTT S13 in liver in vivo. However, levels of full-length mouse HTT were not significantly altered in liver with IKKβ knockout, unlike the significant increase in total full-length HTT we observed in the striatum of the IKKβ knockout mice.
Fig. 3.
IKKβ is required for efficient S13 phosphorylation of ∼350-kDa full-length mouse HTT in liver. Male R6/1 (HD) and NT WT controls, both containing the tamoxifen-inducible Cre and floxed alleles of IKKβ, were treated with tamoxifen or oil vehicle control at week 10 and tissue was taken at week 16 at the completion of the study. IKKβ normalized to loading control ERK1/2 was significantly reduced in liver of HD and NT mice with tamoxifen treatment over oil control in whole-cell lysate (A–C). Anti-HTT phosphoserine 13 (pS13) antibody was used to immunoprecipitate phosphorylated full-length mouse HTT which was then detected by Western blot with anti-total HTT antibody. Levels of pS13-HTT were significantly reduced relative to total HTT with IKKβ knockout in HD and NT controls, while levels of total HTT normalized to ERK1/2 were not affected (A–C). Western images (A and B) were quantitated using Scion software (C). *P < 0.05, ****P < 0.0001 values represent means ± SEM. Statistical significance was determined by paired t test.
In both striatum and in liver, levels of transgenic mutant HTT exon 1 protein were evaluated using immunofluorescence analysis to determine numbers of HTT aggregates and by Western analysis to specifically detect human mutant HTT exon 1 protein high-molecular-weight species with anti-HTT MAB5492, which does not detect endogenous full-length mouse HTT. No significant changes in HTT aggregate numbers or transgenic human mutant HTT exon 1 protein levels were observed with IKKβ knockout in either striatum or liver (SI Appendix, Fig. S3). We did observe an accumulation of lipid droplets in HD liver with IKKβ knockout and higher liver IKKβ protein levels in oil-treated HD vs. NT controls (SI Appendix, Fig. S4) consistent with increased liver stress in R6/1 induced by expression of transgenic human mutant HTT exon 1 protein.
We find that in both striatum and liver, knockout of IKKβ reduces levels of S13-phosphorylated full-length HTT in NT and HD mice, demonstrating IKKβ is a relevant kinase for HTT S13 in vivo, but that levels of transgenic mutant HTT exon 1 protein and aggregate numbers were unaffected by IKKβ knockout in striatum and liver. This is consistent with our previous findings that IKKβ does not efficiently phosphorylate mutant HTT exon 1 protein with expanded polyglutamine repeats in cell culture (2). Liver IKKβ knockout (Fig. 3) was much more extensive than that in striatum (Fig. 2). The Wfs1 promoter we used to drive the tamoxifen-inducible Cre expression needed for IKKβ knockout expresses well in medium spiny neurons and astrocytes of the striatum, but not in microglia (SI Appendix, Fig. S5A) (29). The residual levels of IKKβ detected by Western analysis in striatum in tamoxifen-treated mice may reflect IKKβ expression in microglia and may explain why the IKKβ knockout was less efficient in striatum than in liver. Using qPCR analysis, we did not observe any great alteration in levels of IKKβ expression in cortex or cerebellum with tamoxifen treatment, so these tissues were not further examined (SI Appendix, Fig. S5B).
IKKβ Regulates Expression of Autophagy-Related Genes in Vivo.
A link has been found between the activation of the IKK complex and the induction of autophagy by an NF-κB–independent mechanism, which may at least partially be the result of increased autophagy gene expression (7, 8). To determine whether IKKβ knockout in male striatum and liver might modulate expression of autophagy genes in HD vs. NT control mice, we selected 20 genes encoding proteins with known autophagy functions, in addition to IKKβ itself, to evaluate by qRT-PCR analysis using RNA prepared from frozen striatal and liver tissue collected at the end of the study at week 16 (SI Appendix, Fig. S6).
In striatum, we observed a significant increase in expression of 13 out of 20 tested autophagy genes in HD vs. NT controls, showing that expression of transgenic human mutant HTT exon 1 protein independent of IKKβ knockout may globally increase striatal autophagy gene expression (SI Appendix, Fig. S7), consistent with previously published RNA-sequencing data showing up-regulated transcription of HTT, Atg14, Atg7, and p62 in R6/1 vs. NT control mice (30). Of the 13 up-regulated autophagy genes, 2 were significantly reduced in HD striatum with tamoxifen-induced IKKβ knockout, Atg14 and GABARAPL1, suggesting IKKβ expression may activate transcription of these autophagy genes in HD striatum.
In contrast with striatum, we did not observe the global up-regulation of autophagy genes in oil-treated HD vs. NT liver tissue independent of tamoxifen-induced IKKβ knockout. In fact, we saw a significant down-regulation of mAtg13, LC3A, LAMP-2A, and LAMP-2C in oil-treated HD liver vs. oil-treated NT control liver. Tamoxifen-induced IKKβ knockout significantly reduced expression of several liver autophagy genes in both HD and NT controls (SI Appendix, Fig. S7B): HTT, Atg14, GABARAPL1, Atg16L, Atg9A, and TFEB were significantly down-regulated in HD IKKβ knockout liver, and HTT, mAtg13, Atg14, LC3A, GABARAPL1, p62, and LAMP-2A were significantly down-regulated in NT IKKβ knockout liver. This transcriptional analysis may suggest that IKKβ has a greater impact on the basal expression of autophagy genes in liver (independent of mutant HTT exon 1 protein expression), whereas in striatum IKKβ knockout diminishes the stress-induced up-regulation of Atg14 and GABARAPL1 caused by mutant HTT exon 1 protein expression but has no effect on basal autophagy gene expression in NT controls.
IKKβ Knockout and Mutant HTT Exon 1 Protein Expression Impact Levels of Autophagy Proteins in Vivo.
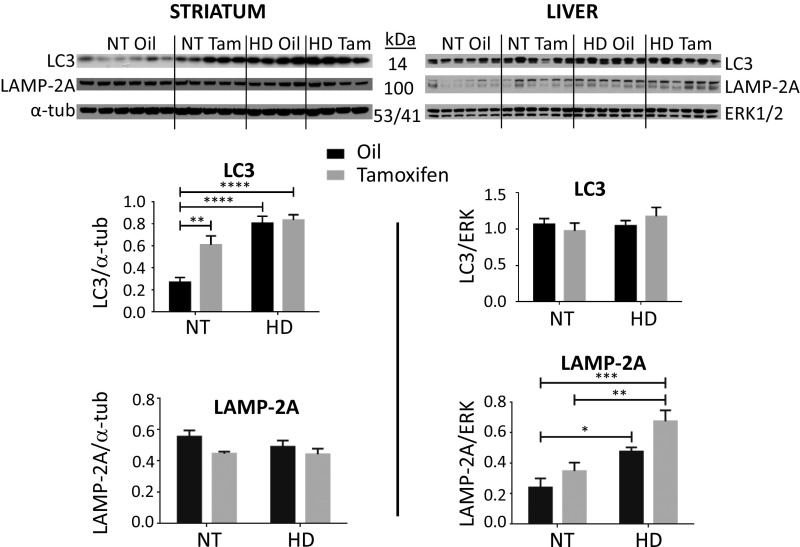
Mutant HTT exon 1 protein expression can cause a compensatory activation of autophagy in vivo but can also ultimately block autophagy, resulting in accumulation of several autophagy proteins and autophagic cargo (31–34). In striatum, expression of mutant HTT exon 1 protein results in increased levels of the LC3 protein, a mammalian Atg8 whose lipidation or accumulation is commonly assayed as an indicator of autophagic flux (31, 35). We examined levels of LC3 protein in 16-wk striatal and liver soluble fractions (Fig. 4) and only detected unlipidated LC3 I by Western analysis in striatum, while detecting both LC3 I and lipidated LC3 II in liver, similar to others who have examined LC3 in brain vs. liver soluble fractions (35). In R6/1 HD striatum, with or without IKKβ knockout, LC3 I protein accumulates in the soluble fraction, suggestive of a blockage of autophagic flux, while in liver no differences are observed in LC3 I or II, suggestive of functional basal autophagy. Interestingly, we saw a significant increase in striatal LC3 I levels in NT controls with IKKβ knockout, demonstrating that a loss of IKKβ activity in striatum may block autophagic flux independent of mutant HTT exon 1 protein expression. We conclude that mutant HTT exon 1 protein and IKKβ knockout may both independently contribute to LC3-dependent autophagy blockage in striatum. At 16 wk, levels of LC3 I were high in HD striatum, and IKKβ knockout did not further increase these levels; similarly, as mutant HTT exon 1 protein is cleared by autophagy, this may suggest why IKKβ knockout did not further alter levels of aggregates in HD striatum (SI Appendix, Fig. S3A). In liver tissue, LC3 I and II proteins were detected and were unchanged with IKKβ knockout or with mutant HTT exon 1 protein expression (Fig. 4), consistent with active autophagic clearance of mutant HTT exon 1 protein reflected by low levels of aggregates (SI Appendix, Fig. S3B), highlighting a difference in autophagy between striatum and liver. We previously found in cell culture that lysosomal membrane protein LAMP-2A overexpression could reduce levels of HTT exon 1 protein (2). We therefore examined levels of LAMP-2A protein, which were unchanged at 16 wk in striatum but were significantly increased in HD vs. NT controls in liver. This may suggest that increased LAMP-2A–mediated autophagy may help to reduce levels of mutant HTT exon 1 protein in liver cells capable of proliferation, but not in postmitotic neurons, keeping liver aggregate levels lower than those observed in striatum (SI Appendix, Fig. S3).
Fig. 4.
IKKβ knockout and mutant HTT exon 1 protein expression impact levels of autophagy proteins in vivo. R6/1 (HD) or NT WT control male mice containing the tamoxifen-inducible Cre and floxed alleles of IKKβ were treated with tamoxifen or oil vehicle control for 1 wk starting at 10 wk to knock out IKKβ in striatum and liver. At the termination of the study at 16 wk, Western analysis of NT and HD soluble fractions was used to examine levels of autophagy proteins LC3 and LAMP-2A relative to loading controls α-tubulin (striatum) or ERK1/2 (liver). LC3 I was detected in striatum, while LC3 I and II were observed in liver. LC3 I was quantitated for both tissues and was found in striatum, but not in liver, to be significantly increased with IKKβ knockout or with transgene expression. LAMP-2A levels were unchanged in striatum but were significantly increased in HD mouse liver. Western images, shown, were quantitated; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 values represent means ± SEM. Statistical significance was determined by one-way ANOVA with Bonferroni posttesting.
IKKβ Knockout Increases Neuronal Degeneration and Microglial Activation.
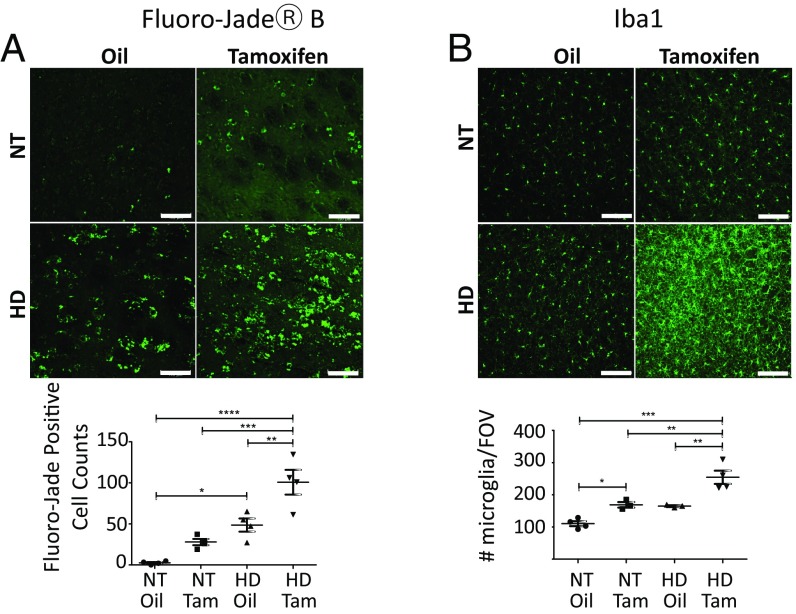
Since striatal neurodegeneration is a hallmark of HD (1), we asked whether overt striatal degeneration was impacted by IKKβ knockout in R6/1 HD mice using Fluoro-Jade B staining, a polyanionic fluorescein derivative which sensitively and specifically binds to degenerating neurons. We quantitated Fluoro-Jade B-positive cells in brain sections from oil vs. tamoxifen-treated HD and NT control mice (Fig. 5A). As expected, Fluoro-Jade B-positive striatal cells were robustly increased (∼50 fold) in oil-treated HD mice vs. NT controls, depicting the neurodegeneration in the striatum of R6/1 HD mice. With tamoxifen-induced IKKβ knockout, HD mice showed an overall enhancement of the Fluoro-Jade B signal (approximately twofold), consistent with an IKKβ knockout-mediated increase in striatal neurodegeneration in R6/1 HD mice.
Fig. 5.
IKKβ knockout increases neurodegeneration and microglial activation in striatum. Male R6/1 (HD) mice containing the tamoxifen-inducible Cre and floxed alleles of IKKβ were treated with tamoxifen or oil vehicle control for 1 wk starting at 10 wk. At the termination of the study at 16 wk, consecutive coronal brain sections containing striatum were stained against Fluoro-Jade B, a neuronal death marker (A) or Iba1, a microglia marker (B). Images (20×) show that R6/1 tamoxifen-treated mice had significant increases in both Fluoro-Jade B- (A) and Iba-1– (B) positive cells in the striatum compared with R6/1 (HD) oil-treated mice (representative images shown). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 values represent means ± SEM. Cells were counted using Bitplane’s Imaris microscopy image analysis software and number of microglia per field of vision (FOV) graphed. Statistical comparisons of results were performed by performing one-way ANOVA analysis followed by Bonferroni’s multiple comparison tests. n = 4 per treatment. (Scale bars, 100 μm.)
Microglia are the resident macrophages of the central nervous system that respond to brain insults including neurodegenerative disease (36). Microglia in striatum are increased and activated in response to HD-associated neurodegeneration and may play both protective and pathogenic roles in disease progression (1, 36). Upon activation of microglia, antiionized calcium binding adaptor molecule 1 (Iba1), a marker for microglia in mouse tissue, is up-regulated allowing the discrimination between surveying and activated microglia (37). In both HD and NT control mice, tamoxifen-induced IKKβ knockout resulted in significantly elevated relative steady-state levels of striatal Iba1, demonstrating that even in the context of healthy control striatum a reduction in neuronal IKKβ resulted in a significant activated microglial response (Fig. 5B).
Astrocytes are star-shaped glial cells in the central nervous system that express glial fibrillary acidic protein (GFAP), a type of astrocyte-enriched intermediate filament protein, and respond to all forms of injury, trauma, and infection in the brain. Astrocyte dysfunction has previously been found to accompany and contribute to neuronal deficits in HD model mice (38). We examined levels of GFAP by immunohistochemistry in striatum in NT and HD mice with and without IKKβ knockout and noted that staining of GFAP was greatly enhanced by IKKβ knockout in NT controls to similar levels as observed in HD mice. IKKβ knockout did not further enhance staining of GFAP beyond the HD effect (SI Appendix, Fig. S8). The increase in GFAP-positive astrocytes in control NT IKKβ knockout mice may reflect striatal neuronal stress due to a loss of IKKβ function.
Discussion
We previously showed that IKKβ directly phosphorylates HTT S13 in vitro and in cell culture and that this phosphorylation is impaired with expansion of the HTT polyQ repeat in cells (2). Further, it was shown that mimicking this phosphorylation in mutant HTT exon 1 protein was protective in an acute striatal slice culture model of HD as well as in the context of full-length mutant HTT in BACHD mice (2, 3). The current study was initiated to evaluate whether IKKβ is a relevant kinase for HTT S13 in vivo and to examine whether this IKKβ-mediated phosphorylation is consistent with a neuroprotective function in HD progression in vivo.
We now report that IKKβ knockout in both liver and striatum in male R6/1 mutant (HD) mice and in NT WT controls reduces phosphorylation of WT full-length mouse HTT S13, demonstrating that it is a relevant S13 kinase in vivo. In the current study, IKKβ knockout in R6/1 worsened the HD behavioral phenotype in these mice at specific time points but did not affect NT behavior, consistent with the predicted delay in R6/1 progression from IKKβ-mediated full-length HTT S13 phosphorylation as we observed in BACHD using phosphomimetic full-length mutant HTT (3). Interestingly, exacerbation of phenotypes appears to be modest and transient for the pole task, only showing significance at the 12-wk time point but not at the 16-wk time point. We speculate that this may be due to changes in phosphorylation of HTT S13 at various time points, to variations in IKKβ regulation of some autophagy genes over time, or by progressive inhibition of autophagy by transgenic mutant HTT exon 1 protein expression with aging, but more studies will be required to clarify these results. In the current study, we find histologically that IKKβ knockout increases overt neuronal degeneration and microglial activation in R6/1 striatum, suggesting that IKKβ may be essential for striatal health in mice expressing mutant HTT exon 1 protein in vivo.
In 16-wk-old male R6/1 HD mice, levels of transgenic mutant HTT exon 1 protein and aggregates were unchanged in striatum and liver with IKKβ knockout. Using cell culture, we previously found that WT HTT fragment clearance was increased by S13 phosphorylation and IKKβ overexpression. In contrast, mutant HTT fragment clearance was not greatly altered upon IKKβ overexpression and mutant HTT S13 phosphorylation was reduced compared with WT fragment (2). Therefore, we did not expect transgenic mutant HTT exon 1 protein levels to be reduced with IKKβ knockout in vivo, which proved to be the case. This is in contrast to the reduction in aggregation we observed when mimicking constitutive phosphorylation of HTT S13 and S16 in full-length mutant HTT in BACHD mice where numbers of aggregates were vastly reduced (3). HTT neuronal aggregate appearance and disease onset are observed much earlier in R6/1 than in BACHD (22), which may reflect a more rapid impairment of neuronal proteostasis and lysosomal function in the R6/1 model due to mutant HTT exon 1 protein expression vs. the mutant full-length HTT expression in BACHD. S13 phosphorylation in the context of polyQ expansion is inefficient (2), and in the current study in R6/1 we saw no evidence of transgenic mutant exon 1 protein phosphorylation in vivo, in contrast to the endogenous full-length WT HTT which required IKKβ for efficient phosphorylation. In R6/1 HD and NT control striatum, levels of full-length WT HTT increased and HTT S13 phosphorylation was reduced with IKKβ knockout suggestive of a reduced flux of WT full-length mouse HTT, but in liver WT full-length HTT levels were unchanged with IKKβ knockout despite a reduction in its S13 phosphorylation. Given that liver also has far fewer mutant HTT exon 1 protein aggregates than striatum, proteosomal and lysosomal protein clearance mechanisms for degradation of both WT full-length endogenous mouse HTT and transgenic mutant HTT exon 1 protein may be different and/or more efficient in liver than in striatum, thus resulting in reduced mutant HTT-mediated toxicity in liver compared with striatum. IKKβ-mediated phosphorylation of WT full-length mouse HTT S13 may therefore be more important for the health of the striatum than the liver and other tissues to maintain normal HTT function and proper proteostasis. We previously showed that HTT interacts with the mammalian Atg8 autophagy protein LC3 (39). We observed a significant LC3 protein accumulation in striatum, but not in liver, upon IKKβ knockout or with mutant HTT exon 1 protein expression, consistent with the hypothesis that IKKβ may regulate an LC3-mediated type of autophagy important for striatum but not for liver in R6/1 HD mice. In cell culture, we previously showed that overexpression of the lysosomal membrane protein LAMP-2A resulted in clearance of HTT exon 1 protein (2). In liver, but not in striatum, we saw increased levels of LAMP-2A protein in R6/1 HD mice, suggesting that activated LAMP-2A–mediated, LC3-independent autophagy may clear mutant HTT exon 1 protein and full-length WT HTT in liver but not in striatum, reducing aggregate numbers and contributing to liver health in HD.
Recently it was elucidated that incomplete transcription of the mutant HTT gene results in expression of toxic mutant HTT exon 1 protein in HD knock-in mouse models and in patients (40, 41). Further, mutant HTT exon 1 protein expression can activate the IKK complex (14), and IKKβ can induce expression of several autophagy genes and up-regulate autophagy by an NF-Kβ–independent mechanism (7, 8). Mutant HTT exon 1 protein is an autophagic substrate and its expression has been found by several groups to induce autophagy that activates its clearance (32–34). Our laboratory and another have independently demonstrated that at least one of HTT’s normal functions is that of a scaffold for selective autophagy (39, 42), a function that we propose may be induced by IKKβ-mediated phosphorylation of HTT S13. HTT may be degraded through its own selective autophagic scaffold function, a function that may be impaired by expansion of the polyQ repeat, resulting in the observed accumulation of mutant HTT exon 1 protein and autophagic substrates in inclusions of diseased neurons (43). Therefore, clearance of this toxic mutant exon 1 protein fragment may occur indirectly through IKK activation and be one mechanism of slowing HD progression in striatum. In the R6/1 HD mouse model, however, striatal transgenic mutant exon 1 protein is expressed at such high levels compared with those found in HD knock-in or patient striatum that IKKβ knockout ultimately does not impact its striatal accumulation, or that of LC3, suggesting that autophagic flux is blocked in striatum at 16 wk in R6/1, and at this late stage attempted activation of autophagy may be futile.
Transcriptional analysis shows that IKKβ may activate expression of several basal autophagy genes in NT WT liver. As striatum severely degenerates in HD but liver is less impacted, an increase in autophagy gene expression in striatum may represent a compensatory activation in response to the cellular stress caused by the chronic expression of mutant HTT, a response that may not be required in the liver. This is consistent with the hypothesis that mutant exon 1 protein clearance is more efficient in the liver and may potentially be mediated by the observed increase in liver LAMP-2A protein levels. Our data also suggest that early expression of IKKβ may prolong striatal health in HD partially through the activation of stress-induced autophagy that may be required for degradation of mutant HTT exon 1 protein in striatum in early stages of disease. Compensatory activation of IKKβ may therefore slow disease progression as long as lysosomal function is intact (2, 7).
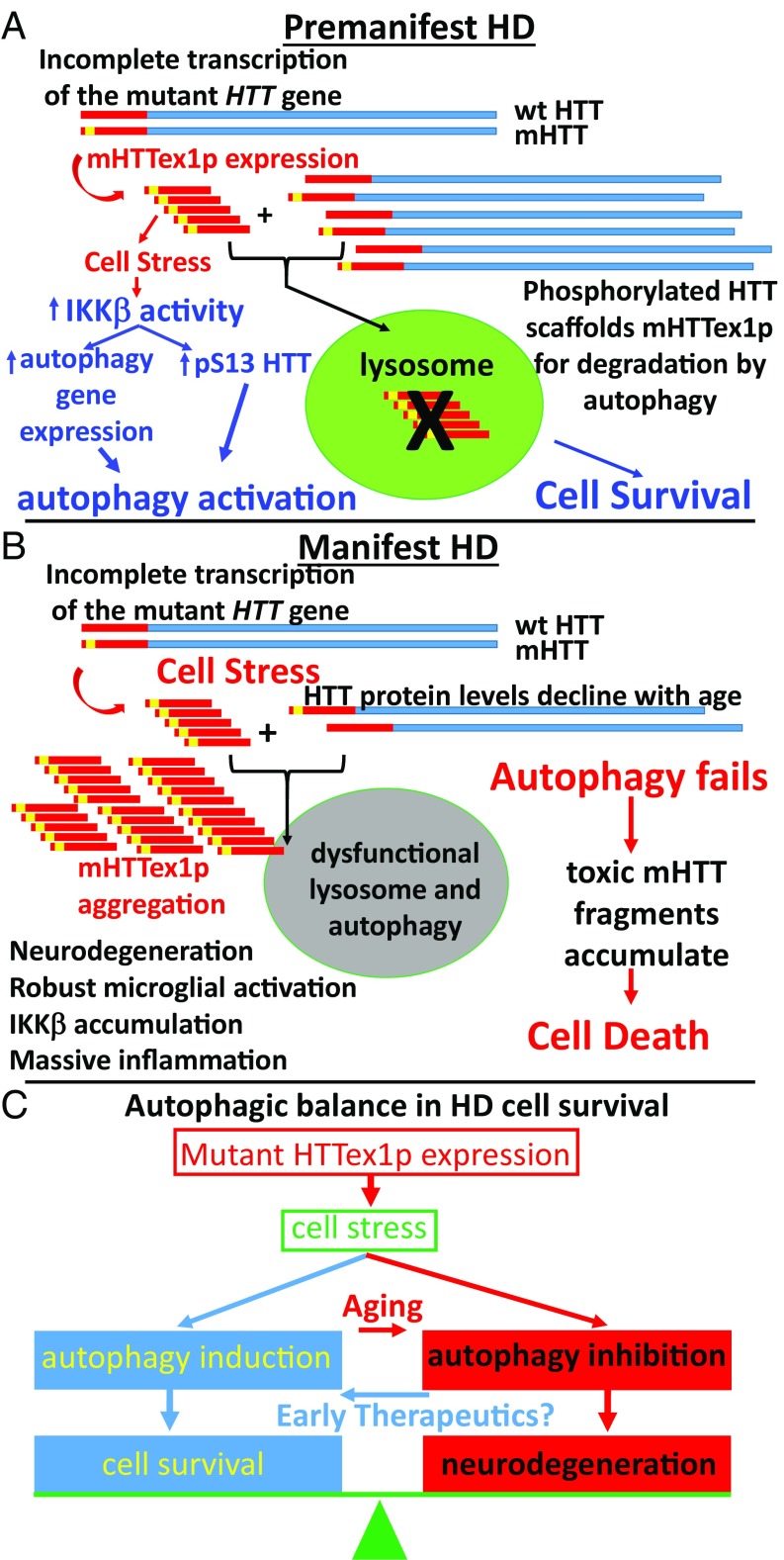
We suggest that autophagic balance regulates cell survival vs. neurodegeneration in HD and propose the following hypothetical model (Fig. 6). In premanifest HD, mutant HTT exon 1 protein is expressed due to incomplete transcription of the mutant HTT gene (40, 41). This creates a cellular stress that activates the IKK complex (14) to increase autophagy gene (7, 8) and HTT expression (44) and activates phosphorylation of HTT S13 (2), to induce autophagy important for clearance of mutant HTT exon 1 protein (32–34) and a mitigation of the cellular stress, allowing proteostasis to exist in a healthy balance (Fig. 6A). In manifest HD, lysosomal function and autophagy become impaired with aging in association with polyQ expansion of HTT and reduced HTT protein levels (31, 45), resulting in reduced full-length HTT phosphorylation, and mutant HTT exon 1 protein and other autophagy cargos accumulate and aggregate, or may be expelled from the cell for prion-like propagation to neighboring cells (46–48). The ensuing cellular stress causes a robust microglial activation (49) and inflammatory pathway induction and accumulation of IKKβ that can no longer activate autophagy (13), resulting in cellular dysfunction and neurodegeneration (Fig. 6B). Therapeutics that improve lysosomal and autophagic function in young individuals may keep mutant HTT exon 1 protein levels reduced and slow HD progression (Fig. 6C). However, if the lysosome is unable to degrade autophagic cargo, inhibition of autophagy may then be neuroprotective and reduce inflammation (50–52).
Fig. 6.
A model for the progression of HD pathogenesis dependent on the autophagic balance of the patient. In premanifest HD (A), mutant HTT exon 1 protein is expressed in striatum due to incomplete transcription of the mutant HTT gene, and accumulation of mutant HTT exon 1 protein (mHTTex1p) causes a cellular stress, which activates the IKK complex. IKKβ activates autophagy gene expression and increases phosphorylation of HTT S13 to induce HTT’s role as an autophagic scaffold protein. This results in autophagic clearance of mHTTex1p, ultimately reducing inflammation, and the cell survives. In manifest HD (B), levels of HTT and other autophagy proteins have declined with age and the ability of the lysosome to degrade autophagic cargos is reduced. With polyQ expansion, mutant HTT is less well phosphorylated reducing its function as an autophagic scaffold. mHTTex1p expressed from incomplete transcription of the mutant HTT gene accumulates and is not cleared by the lysosome, causing an up-regulation of IKK that is unable to further activate autophagy. This results in a robust microglial activation, massive inflammatory pathway activation, cellular dysfunction, and neurodegeneration. Therapies to treat HD need to be designed with the autophagic balance of the patient in mind (C). Early therapies to activate autophagy and IKKβ to degrade mHttex1p and reduce cellular stress may be protective as long as lysosomal function is still intact, but later therapies to inhibit autophagy and block IKKβ may be useful if the aging lysosome can no longer degrade autophagic cargo.
The development of therapeutic approaches to treat neurodegenerative diseases involving an imbalance in proteostasis, including AD, PD, ALS, HD, and other polyQ-repeat diseases, by correcting proteostatic imbalance is an attractive avenue for therapeutic development for these diseases. However, the results presented here provide a note of caution. We suggest that therapies for these diseases may need to be developed based on whether the patient’s lysosomal function is still intact, as early and late therapies may be very different between the two stages. A challenge which must be met, we believe, is an assessment of the status of the autophagic balance of each patient at the time therapy is initiated and the possibility that a “one-size-fits-all” therapeutic strategy may have to be replaced by a strategy which reflects the assessment of the autophagic balance of the patient at the time therapy is initiated and a modulation of therapeutic approach in relation to changes in the autophagic balance of the patient over time.
Materials and Methods
Experimental Animals.
All experiments were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals (53) and an approved animal research protocol by the Institutional Animal Care and Use Committee at the University of California, Irvine, an American Association for Accreditation of Laboratory Animal Care-accredited institution, as previously published in our laboratory (54). All efforts were made to minimize research animal suffering throughout the duration of the study. A transgenic mouse model with lox-P flanked alleles of IKKβ (23) was used for this study. Wfs1-Tg3-CreERT2 and R6/1 mice were obtained from Jackson Laboratories (Research Resource Identifiers IMSR_JAX: 009103 and 006471, respectively). Mice were cross-bred to generate Wfs-CreERT2/+;IKKβflox/flox mice+/− the R6/1 transgene. All mice were in C57BL/6 background. Mice were mix-housed under a 12-h light/dark cycle in groups of up to five animals per cage with food and water administered for ad libitum consumption. We performed CAG repeat sizing of R6/1 tail snips and a subset of striatal tissue punches (Laragen). The average transgenic CAG repeat number from R6/1 mice in this study was 132.
PCR on tail samples was used to genotype for IKKβ floxed alleles (sense oligo: 5′-GTTCAGAGGTCCAGTCCATTATC-3′, antisense oligo: 5′-TAGCCTGCAAGAGACAATACG-3′). To justify experimental group and trial sizes, we used G Power analysis based on published results (54–58) and our previous experience and for HD mouse model studies. Based on these analyses, we used n = 10 per group for behavioral studies and n = 4 for biochemistry Mice were killed by pentobarbital overdose and perfused with 0.01 M PBS. Liver was collected, striatum and cortex were dissected out of the left hemisphere, and tissues were flash-frozen for RNA or protein isolation as described below. One piece of liver and the other halves of brain were postfixed in 4% paraformaldehyde, cryoprotected in 30% sucrose, and cut at 40 μm on a sliding vibratome for immunohistochemistry.
Tamoxifen Protocol.
To acutely delete IKKβ we used a published protocol (59). Tamoxifen (T5648; Sigma) was suspended at a concentration of 20 mg/mL, in a mixture of 98% sunflower oil (700011-932; VWR) and 2% ethanol. Tamoxifen (200 μg/g) or sunflower oil control were injected intraperitoneally once per day for 5 d into 10-wk-old mice. Mice were dosed with tamoxifen at a volume based on weight (i.e., 180 μL of concentration 20 μg/μL for an 18-g mouse). Higher levels were tested and did not provide greater deletion efficiency. During the study, 9/10 of the NT-tamoxifen females, 3/9 of the HD-tamoxifen females, 1/12 of the NT-tamoxifen males, and 2/12 of the HD-tamoxifen males died following tamoxifen treatment. No death was observed with oil treatment in any of the groups. Because of the enhanced lethality of tamoxifen/IKKβ knockout particularly in NT females, our analysis was carried out exclusively with male mice.
Behavioral Timeline and Assessment.
All behavioral assessments were performed as previously described (54, 60). Specifically, body weights were measured weekly at 8 to 15 wk of age. The latency to fall from the rod was recorded over three trials then averaged and analyzed for significance. A pole test was used to examine the ability for mice to descend a vertical pole (1 cm in diameter, 60 cm high) at 8, 12, and 16 wk of age. Mice were habituated to the task 1 d before testing. Mice were tested in this assay by placing them facing down on the vertical pole and total time to descend until reaching the base was measured. This task was performed four times with 30-s rest periods provided between trials. Using the total time to descend from placement on the pole as the starting point, four trials were averaged and analyzed for statistical significance. For grip strength measurements, we used an IITC Life Science digital force transducer instrument to measure forelimb grip force, which gives readings in 1-g increments. Using this instrument, each mouse was lowered toward a wire mesh by the tail and the peak force applied on a digital display grip strength meter was measured. Mice were gently pulled back until they released their grip from the mesh. Five consecutive trials were measured, with the four strongest pulls averaged and analyzed for statistical significance.
Tissue Lysis, IP, Western Analysis, and Antibodies.
Whole male mouse striatum or liver tissue, taken at the end of the study at 16 wk, was dounced 20 times on ice in modified RIPA buffer (50 mM Tris⋅HCl, pH 7.4, 1% Nonidet P-40, 0.25% Na-doxycholate, 150 mM NaCl, and 1 mM EDTA) containing a complete protease/phosphatase-inhibitor pellet (Thermo), 1 μg/mL leupeptin and aprotenin, phosphatase inhibitors 2 and 3 (Sigma-Aldrich), and 1 mM PMSF. Homogenate lysates were centrifuged at 16.1 × g for 15 min at 4 °C. Protein concentration was determined on the supernatant by Bradford assay. IP was done as previously described (3), summarized as follows: 500 μg of supernatant was incubated ± 1 μL pS13 antibody for 1 h on ice in a total volume of 30 μL. Thirty microliters of Protein-G Dynabeads (Invitrogen) were preequilibrated and washed in 100 mM phosphate buffer (pH 5.0), and the buffer was removed using a magnetic rack. The beads were resuspended in the supernatant/antibody mixture and rocked for 45 min at 4 °C. They were then washed using a magnetic rack three times in 200 μL of 100 mM phosphate buffer (pH 5.0). The beads were resuspended in 25 μL of loading buffer (Invitrogen), incubated at 70° C for 10 min, and run on 3 to 8% Tris-acetate gels (Invitrogen). These gels were blotted to PVDF (Millipore), blocked with SuperBlock BlockingBuffer (ThermoScientific), and subjected to Western analysis using PICO reagent (Thermo) exposed to film for images. Protein quantification was performed by Scion Image analysis software. Band densities from whole-cell lysates were normalized to α-tubulin for striatum and ERK1/2 for liver. Antibodies used were anti-HTT phosphoserine 13 pS13 (2) 1:1,000, anti-HTT VB3130 (N17 epitope) to detect full-length ∼350 kDa mouse WT HTT levels (Viva Bioscience) 1:1,000, anti-α-tubulin (Sigma-Aldrich) 1:5,000, anti-ERK1/2 (Cell Signaling) 1:1,000, and anti-IKKβ (Abcam 10AG2) 1:500. Anti-LC3 antibody (MBL) was used at 1:1,000 and anti-LAMP-2A antibody (Zymed) at 1:500. Whole-cell supernatant was run at concentrations 30 μg per lane for striatum and 50 μg per lane for liver and subjected to Western analysis.
Analysis of Transgenic Mutant HTT Exon 1 Protein Levels by Western Analysis.
For Western analysis, striatal tissue was the same as that used for the above IP studies. Liver tissue was dounced 30 times in lysis buffer (10 mM Tris, pH 7.4, 1% Triton X-100, 150 mM NaCl, 10% glycerol, 20 mM N-ethylmaleimide, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 20 mM NaF), allowed to rest on ice for 1 h, then centrifuged at 15,000 × g at 4 °C for 20 min and the resulting supernatant was quantitated by Lowry assay. Thirty micrograms per lane of striatal and liver soluble whole-cell extracts were run on 4 to 12% Bis-Tris MidiGels (Invitrogen) using Mops running buffer and transferred onto 0.45-μm nitrocellulose membranes. We detected transgenic human mutant HTT exon 1 protein smears in the soluble fraction by Western analysis using anti-human HTT MAB5492 (Millipore) at 1:1,000, which does not recognize endogenous mouse HTT, quantitated using Scion Image analysis software, and performed statistical analyses with GraphPad Prism 6, version 6.01. This liver tissue lysis protocol and gel system was also used to compare levels of IKKβ in oil-treated R6/1 (HD) vs. NT animals using anti-IKKβ (10AG2; Abcam) 1:500. Loading controls for striatum and liver were measured using anti–α-tubulin (Sigma-Aldrich) at 1:5,000 and anti-ERK1/2 (Cell Signaling) at 1:1,000, respectively.
Quantitation of Mutant HTT Aggregates in Striatum.
Half-brain coronal sections, 40 μm each, four sections per 16-wk-old male mouse, were used for quantitation. Sections were picked from bregma 1.34, every 12 sections, to bregma −0.1, from male R6/1 mice containing the Cre and floxed alleles of IKKβ (HD) and treated with tamoxifin or oil vehicle control, four mice per treatment group. Fluorescent immunolabeling was done with anti-HTT clone mEM48 (MAB5374; Millipore) to label transgenic human mutant HTT exon 1 protein aggregates and TO-PRO-3 Iodide stain (T3605; Thermo Fisher Scientific) to label nuclei. Images were captured using Leica confocal microscope (DM2500) and Leica camera TCS SPE (Leica Microsystems Inc.). Images were taken at 40×, with z-volume set at 25 to 30 μm at 1 μm per step. Three striatal areas, upper, middle and lower, were imaged with consistent settings for laser intensity, exposure time, and gain through all images. Imaris x64 quantitation software (Bitplane Scientific Software) was used to evaluate the number of aggregates in each area normalized to the number of nuclei, calculated as the percent of cells expressing aggregates.
Quantitation of Mutant HTT Aggregates in Liver.
Paraffin-embedded liver sections, 40 μm per section, from male 16-wk-old R6/1 mice containing the Cre and floxed alleles of IKKβ (HD), treated with tamoxifin (n = 5) or oil vehicle control (n = 4), were used. One section per mouse was analyzed. Fluorescent aggregate immunolabeling was done with anti-HTT MW8 (AB528297; Developmental Studies Hybridoma Bank) (recommendation from Jeffrey Carroll for liver HTT, Western Washington University, Bellingham, WA) and TO-PRO-3 Iodide stain (T3605; Thermo Fisher Scientific) was used to label nuclei. Images were captured using Leica confocal microscope (DM2500) and Leica camera TCS SPE (Leica Microsystems Inc.). All images were taken at 20×, two areas per section, with consistent settings for laser intensity, exposure time, and gain through all images. In each area, the number of aggregates was counted by eye without normalization, as aggregate numbers were very low and visible per field.
Quantitation of Liver Lipid Droplets.
Bodipy 493/503 (D3922; Thermo Fisher) was used to stain neutral lipids in paraformaldehyde-fixed 40-μm male liver slices. Bodipy 493/503 was stored as stock of 1 mg/mL in DMSO. Three PBS washes of tissue were followed by staining with bodipy diluted to 10 μg/mL in PBS for 30 min at room temperature. This was followed by 1-min incubation in 1:5,000 Hoechst reagent to stain nuclei, followed by three PBS washes and mounting of tissue using fluoromount. Z-stack images were taken using a confocal microscope and surface area and volume of lipids determined using Imaris x64 quantitation software (Bitplane Scientific Software). The analysis was done on 16-wk-old male mouse liver using two images per liver section, three sections per animal, four animals per group. Quantification was performed using Imaris image software to calculate surface area of lipid droplets and cell number for normalization. Cell number was quantified using DAPI staining.
Analysis of Microglia in Striatum.
For immunohistochemical assessments, 40-μm sections of postfixed half-brains from four 16-wk-old male mice per treatment group were processed for immunohistochemistry and imaged via confocal microscopy. The following primary antibody was used: anti-Iba1 (27030; Wako Pure Chemicals Industries). Alexa fluorescent-conjugated secondary antibody was used (A21428; Thermo Fisher Scientific). Coronally sectioned stained tissue was processed and mounted on slides with Fluoromount-G (SouthernBiotech). Images were captured using Leica confocal microscope (DM2500) and Leica camera TCS SPE (Leica Microsystems Inc.). Five comparable representative images per treatment group were taken at 10×, 20×, and 63× z stack, with consistent settings for laser intensity, exposure time and gain through all images. Automatic analyses and cell quantitation from acquired images was processed using Imaris x64 quantitation software (Bitplane Scientific Software).
Fluoro-Jade B Analysis of Neurodegeneration in Striatum.
Analysis was done using four male postfixed half-brains for each treatment group, with five 40-μm representative sections prepared from each. Stack images (20×) were obtained using confocal microscopy at comparable sections for each animal. Images were captured using Leica confocal microscope (DM2500) and Leica camera TCS SPE (Leica Microsystems Inc.). Striatal sections were mounted on Shandon Polysine slides (Thermo Scientific), allowed to dry completely, and immersed in a series of washes: (i) 3 min in 80% ethanol, (ii) 2 min in 70% ethanol, (iii) 2 min in a 1:200 dilution of acetic acid to MilliQ water, (iv) 10 min gently rocking in 0.06% KMnO4 solution, (v) 2 min in a 1:200 dilution of acetic acid to MilliQ water, (vi) 15 min gently rocking in 0.0004% Fluoro-Jade B (AG310; Millipore Sigma) solution, and (vii) three sequential 1-min rinses in a 1:200 dilution of acetic acid to MilliQ water. Slides were allowed to dry, immersed in three sequential 1-min washes with 100% xylene, allowed to dry again, and coverslipped with DPX mounting medium (Electron Microscopy Sciences).
GFAP Free-Floating Immunohistochemistry Protocol.
Sections were incubated in 1% hydrogen peroxide in Tris-buffered saline (TBS; 0.05 M Tris, 145 mM NaCl, pH 7.6) for 30 min followed by three washes in TBS. After incubation in 15% (vol/vol) normal goat serum (S-1000; Vector Laboratories) in TBST (TBS with 0.3% Triton X-100) for 30 min, sections were then incubated in GFAP primary antibody (1:400 dilution, Z033429-2; Dako) in 10% (vol/vol) normal goat serum in TBST overnight. After three washes in TBS, sections were incubated with biotinylated secondary antibody (1:1,000 dilution, BA-1000; Vector Laboratories) in 10% (vol/vol) normal goat serum in TBST for 2 h. Sections were then washed three times in TBS and stained using Vectastain Elite ABC HRP Kit (PK-6101; Vector Laboratories) (1:1,000 dilution reagent A, 1:1,000 dilution reagent B) for 2 h. After two more washes in TBS, sections were then stained using DAB Peroxidase Substrate Kit (SK-4100; Vector Laboratories). After 1.5-min incubation in DAB substrate solution, sections were washes two times in ice-cold TBS, mounted on gelatin-coated slides, and coverslipped using Permount mounting medium (SP15-100; Fisher Scientific). Sections were viewed on an Olympus BX53 microscope, images with identical capture settings were acquired using a QImaging QIClick CCD camera and Q-Capture Pro-7 software, and brightness/contrast adjustments were made in parallel (via adjustment layer) in Photoshop.
RNA Extraction and qPCR.
Striatal and liver tissues were homogenized in TRIzol (Invitrogen) and total RNA was isolated using an RNEasy Mini kit (QIAGEN) with supplemental DNase treatment to remove residual DNA. Reverse transcription was performed using oligo dT primer sets and 1 μg total RNA using SuperScript III First-Strand Synthesis System (Invitrogen) as previously described (54). The qPCR was performed as previously described (61) and CT values for each gene of interest (GOI) were normalized against RPLPO to calculate dCT values. For statistical analysis ddCT values were used in a two-way ANOVA for each GOI. Post hoc analyses using Tukey’s honestly significant difference test with Bonferroni correction were used to test statistical significance between each group interaction contrast. Adjusted P values are reported. While the Wfs1 promoter can also affect cortex and cerebellum, we did not observe major qPCR changes in IKKβ expression in male cortex or cerebellum in either HD or NT mice with tamoxifen treatment. We therefore did not further evaluate these tissues.
Primer sequences (5′–3′) used were as follows:
HTT.
Forward sequence: GCAGGGAAAGAGCTTGAGACAC
Reverse sequence: CCTCATTCTCCTTGTGGCACTG
Beclin1.
Forward sequence: CAGCCTCTGAAACTGGACACGA
Reverse sequence: CTCTCCTGAGTTAGCCTCTTCC
Atg5.
Forward sequence: CTTGCATCAAGTTCAGCTCTTCC
Reverse sequence: AAGTGAGCCTCAACCGCATCCT
Atg7.
Forward sequence: CCTGTGAGCTTGGATCAAAGGC
Reverse sequence: GAGCAAGGAGACCAGAACAGTG
LC3A.
Forward sequence: CTGCCTGTCCTGGATAAGACCA
Reverse sequence: CTGGTTGACCAGCAGGAAGAAG
GABARAPL1.
Forward sequence: GTGGAGAAGGCTCCTAAAGCCA
Reverse sequence: AGGTCTCAGGTGGATCCTCTTC
p62/SQSTM1.
Forward sequence: GCTCTTCGGAAGTCAGCAAACC
Reverse sequence: GCAGTTTCCCGACTCCATCTGT
Atg16L.
Forward sequence: GGACACTCATCCTGCTTCTGGT
Reverse sequence: GCTTCCCAAAGTTTCACCCTGC
mAtg13.
Forward sequence: AGAGACTGGTGATGCACATGCC
Reverse sequence: CCGTCCTTCACTGCTGTTAGAC
IKKβ.
Forward sequence: GCAGACTGACATTGTGGACCTG
Reverse sequence: ATCTCCTGGCTGTCACCTTCTG
RPL18A.
Forward sequence: GAAGGTGGAAGAGATTGCAGCTG
Reverse sequence: TCTTGGTGGTGAAGCGTGGCTT
Atp5b.
Forward sequence: CTCTGACTGGTTTGACCGTTGC
Reverse sequence: TGGTAGCCTACAGCAGAAGGGA
Atg9A.
Forward sequence: GTTAGCTGTGGAACACGTCCTC
Reverse sequence: GCAAGAATCACTCGGAGCAGCT
Atg14.
Forward sequence: CAGCAAGCAGAAACTGACACGC
Reverse sequence: GACCAAGTGCATCAGGTTCCTG
BNIP3.
Forward sequence: GCTCCAAGAGTTCTCACTGTGAC
Reverse sequence: GTTTTTCTCGCCAAAGCTGTGGC
NIX (BNIP3L).
Forward sequence: GCATGAGGAAGAGTGGAGCCAT
Reverse sequence: AAGGTGTGCTCAGTCGTTTTCCA
FIP200 (RB1CC1).
Forward sequence: GGAATCTCTGGTCAGGAAGTGC
Reverse sequence: GTCCAAGGCATACAGCCGATCT
LAMP-2A.
Forward sequence: AGGTGCTTTCTGTGTCTAGAGCGT
Reverse sequence: AGAATAAGTACTCCTCCCAGAGCTGC
LAMP-2B.
Forward sequence: ATGTGCTGCTGACTCTGACCTCAA
Reverse sequence: TGGAAGCACGAGACTGGCTTGATT
LAMP-2C.
Forward sequence: ACCACCAATCTAAGAGCAGGACT
Reverse sequence: GGTGCTGGTCTTTCAGGCTTGATT
TFEB.
Forward sequence: CCACCCCAGCCATCAACAC
Reverse sequence: CAGACAGATACTCCCGAACCTT
TG2.
Forward sequence: GAAGGAACACGGCTGTCAGCAA
Reverse sequence: GATGAGCAGGTTGCTGTTCCTGG
Atg10.
Forward sequence: GAGACCTTGACACCACATGCCA
Reverse sequence: GCAGGTCTCGTCACTTCAGAATC
RPLPO.
Forward sequence: TGGTCATCCAGCAGGTGTTCGA
Reverse sequence: ACAGACACTGGCAACATTGCGG
Supplementary Material
Acknowledgments
We thank Manolis Pasparakis (University of Cologne) for generously providing mice with floxed alleles of IKKβ for this work, Dr. Jeffrey Carroll (Western Washington University) and Dr. Ali Khoshnan (California Institute of Technology) for helpful discussion and technical advice, and Dr. Eileen White (Rutgers) for her tamoxifen protocol before its publication. This work was supported by the Hereditary Disease Foundation and CHDI (J.S.S.); the 2017 HDSA Berman-Topper Family HD Career Development Fellowship (to S.H.); California Institute for Regenerative Medicine Grant DISC2-09569 (2017-19) (to L.M.T. and E.S.M.); and NIH Grants NS072453 (to J.S.S.), AG016573 (to J.S.S.), T32AG000096 (to G.F., M.K., and S.H.), NS091046-01 (to R.G.L.), NS052789 (to L.M.T.), and NS090390 (to L.M.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814246116/-/DCSupplemental.
References
- 1.Ross CA, Tabrizi SJ (2011) Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol 10:83–98. [DOI] [PubMed] [Google Scholar]
- 2.Thompson LM, et al. (2009) IKK phosphorylates huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol 187:1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu X, et al. (2009) Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron 64:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Häcker H, Karin M (2006) Regulation and function of IKK and IKK-related kinases. Sci STKE 2006:re13. [DOI] [PubMed] [Google Scholar]
- 5.Steffan JS. (2010) Does huntingtin play a role in selective macroautophagy? Cell Cycle 9:3401–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sochocka M, Diniz BS, Leszek J (2017) Inflammatory response in the CNS: Friend or foe? Mol Neurobiol 54:8071–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Criollo A, et al. (2010) The IKK complex contributes to the induction of autophagy. EMBO J 29:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comb WC, Cogswell P, Sitcheran R, Baldwin AS (2011) IKK-dependent, NF-κB-independent control of autophagic gene expression. Oncogene 30:1727–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen N, Debnath J (2013) IκB kinase complex (IKK) triggers detachment-induced autophagy in mammary epithelial cells independently of the PI3K-AKT-MTORC1 pathway. Autophagy 9:1214–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JE, et al. (2010) Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal 22:1645–1654. [DOI] [PubMed] [Google Scholar]
- 11.Niida M, Tanaka M, Kamitani T (2010) Downregulation of active IKK beta by Ro52-mediated autophagy. Mol Immunol 47:2378–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinsztein DC, Mariño G, Kroemer G (2011) Autophagy and aging. Cell 146:682–695. [DOI] [PubMed] [Google Scholar]
- 13.Salminen A, Hyttinen JM, Kauppinen A, Kaarniranta K (2012) Context-dependent regulation of autophagy by IKK-NF-κB signaling: Impact on the aging process. Int J Cell Biol 2012:849541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khoshnan A, et al. (2004) Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J Neurosci 24:7999–8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björkqvist M, et al. (2008) A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med 205:1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilchez D, Saez I, Dillin A (2014) The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun 5:5659. [DOI] [PubMed] [Google Scholar]
- 17.Mangiarini L, et al. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506. [DOI] [PubMed] [Google Scholar]
- 18.Rattray I, et al. (2013) Correlations of behavioral deficits with brain pathology assessed through longitudinal MRI and histopathology in the R6/1 mouse model of Huntington’s disease. PLoS One 8:e84726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naver B, et al. (2003) Molecular and behavioral analysis of the R6/1 Huntington’s disease transgenic mouse. Neuroscience 122:1049–1057. [DOI] [PubMed] [Google Scholar]
- 20.Anglada-Huguet M, et al. (2014) Prostaglandin E2 EP1 receptor antagonist improves motor deficits and rescues memory decline in R6/1 mouse model of Huntington’s disease. Mol Neurobiol 49:784–795. [DOI] [PubMed] [Google Scholar]
- 21.Lebreton F, Cayzac S, Pietropaolo S, Jeantet Y, Cho YH (2015) Sleep physiology alterations precede plethoric phenotypic changes in R6/1 Huntington’s disease mice. PLoS One 10:e0126972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crook ZR, Housman D (2011) Huntington’s disease: Can mice lead the way to treatment? Neuron 69:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasparakis M, et al. (2002) TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature 417:861–866. [DOI] [PubMed] [Google Scholar]
- 24.Madisen L, et al. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawano J, Tanizawa Y, Shinoda K (2008) Wolfram syndrome 1 (Wfs1) gene expression in the normal mouse visual system. J Comp Neurol 510:1–23. [DOI] [PubMed] [Google Scholar]
- 26.Luuk H, et al. (2008) Distribution of Wfs1 protein in the central nervous system of the mouse and its relation to clinical symptoms of the Wolfram syndrome. J Comp Neurol 509:642–660. [DOI] [PubMed] [Google Scholar]
- 27.Stüwe SH, et al. (2013) Hepatic mitochondrial dysfunction in manifest and premanifest Huntington disease. Neurology 80:743–746. [DOI] [PubMed] [Google Scholar]
- 28.Carroll JB, Bates GP, Steffan J, Saft C, Tabrizi SJ (2015) Treating the whole body in Huntington’s disease. Lancet Neurol 14:1135–1142. [DOI] [PubMed] [Google Scholar]
- 29.Mancarci BO, et al. (2017) Cross-laboratory analysis of brain cell type transcriptomes with applications to interpretation of bulk tissue data. eNeuro 4:ENEURO.0212-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Achour M, et al. (2015) Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington’s disease mice. Hum Mol Genet 24:3481–3496. [DOI] [PubMed] [Google Scholar]
- 31.Pircs K, et al. (2018) Huntingtin aggregation impairs autophagy, leading to Argonaute-2 accumulation and global microRNA dysregulation. Cell Rep 24:1397–1406. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto A, Cremona ML, Rothman JE (2006) Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol 172:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravikumar B, et al. (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36:585–595. [DOI] [PubMed] [Google Scholar]
- 34.Iwata A, Riley BE, Johnston JA, Kopito RR (2005) HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 280:40282–40292. [DOI] [PubMed] [Google Scholar]
- 35.Klionsky DJ, et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraft AD, Kaltenbach LS, Lo DC, Harry GJ (2012) Activated microglia proliferate at neurites of mutant huntingtin-expressing neurons. Neurobiol Aging 33:621.e17–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong HK, Ji K, Min K, Joe EH (2013) Brain inflammation and microglia: Facts and misconceptions. Exp Neurobiol 22:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khakh BS, et al. (2017) Unravelling and exploiting astrocyte dysfunction in Huntington’s disease. Trends Neurosci 40:422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochaba J, et al. (2014) Potential function for the huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci USA 111:16889–16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neueder A, et al. (2017) The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci Rep 7:1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sathasivam K, et al. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci USA 110:2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rui YN, et al. (2015) Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol 17:262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Vicente M, et al. (2010) Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci 13:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bečanović K, et al. ; REGISTRY Investigators of the European Huntington’s Disease Network (2015) A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nat Neurosci 18:807–816. [DOI] [PubMed] [Google Scholar]
- 45.Franich NR, et al. (2018) Striatal mutant huntingtin protein levels decline with age in homozygous Huntington’s disease knock-in mouse models. J Huntingtons Dis 7:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponpuak M, et al. (2015) Secretory autophagy. Curr Opin Cell Biol 35:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearce MM, Spartz EJ, Hong W, Luo L, Kopito RR (2015) Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat Commun 6:6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang BL. (2018) Unconventional secretion and intercellular transfer of mutant huntingtin. Cells 7:E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang HM, Yang S, Huang SS, Tang BS, Guo JF (2017) Microglial activation in the pathogenesis of Huntington’s disease. Front Aging Neurosci 9:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Button RW, Roberts SL, Willis TL, Hanemann CO, Luo S (2017) Accumulation of autophagosomes confers cytotoxicity. J Biol Chem 292:13599–13614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilhelm T, et al. (2017) Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. elegans. Genes Dev 31:1561–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudnick ND, et al. (2017) Distinct roles for motor neuron autophagy early and late in the SOD1G93A mouse model of ALS. Proc Natl Acad Sci USA 114:E8294–E8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Research Council (2011) Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC: ), 8th Ed. [Google Scholar]
- 54.Ochaba J, et al. (2016) PIAS1 regulates mutant huntingtin accumulation and Huntington’s disease-associated phenotypes in vivo. Neuron 90:507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carter RJ, et al. (1999) Characterization of progressive motor deficits in mice transgenic for the human Huntington’s disease mutation. J Neurosci 19:3248–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickey MA, Gallant K, Gross GG, Levine MS, Chesselet MF (2005) Early behavioral deficits in R6/2 mice suitable for use in preclinical drug testing. Neurobiol Dis 20:1–11. [DOI] [PubMed] [Google Scholar]
- 57.Hockly E, Woodman B, Mahal A, Lewis CM, Bates G (2003) Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res Bull 61:469–479. [DOI] [PubMed] [Google Scholar]
- 58.Stack EC, et al. (2005) Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington’s disease transgenic mice. J Comp Neurol 490:354–370. [DOI] [PubMed] [Google Scholar]
- 59.Karsli-Uzunbas G, et al. (2014) Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4:914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morozko EL, et al. (2018) Longitudinal biochemical assay analysis of mutant huntingtin exon 1 protein in R6/2 mice. J Huntingtons Dis 7:321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vashishtha M, et al. (2013) Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci USA 110:E3027–E3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.