Significance
In this work, we developed an imaging-based pooled-library CRISPR screening approach that provides readouts of both phenotype and genotype of individual cells by high-resolution, high-content imaging. This approach promises to substantially expand the phenotype space accessible to pooled genetic screening by allowing the probing of complex cellular phenotypes, such as cell morphology and subcellular organization of different molecular species, as well as their dynamics. Applying this approach to screen for genetic factors involved in nuclear RNA localization, we identified both positive and negative regulators that control lncRNA localization to nuclear speckles.
Keywords: high-throughput screening, MERFISH, CRISPR, nuclear compartments, RNA localization
Abstract
Pooled-library CRISPR screening provides a powerful means to discover genetic factors involved in cellular processes in a high-throughput manner. However, the phenotypes accessible to pooled-library screening are limited. Complex phenotypes, such as cellular morphology and subcellular molecular organization, as well as their dynamics, require imaging-based readout and are currently beyond the reach of pooled-library CRISPR screening. Here we report an all imaging-based pooled-library CRISPR screening approach that combines high-content phenotype imaging with high-throughput single guide RNA (sgRNA) identification in individual cells. In this approach, sgRNAs are codelivered to cells with corresponding barcodes placed at the 3′ untranslated region of a reporter gene using a lentiviral delivery system with reduced recombination-induced sgRNA-barcode mispairing. Multiplexed error-robust fluorescence in situ hybridization (MERFISH) is used to read out the barcodes and hence identify the sgRNAs with high accuracy. We used this approach to screen 162 sgRNAs targeting 54 RNA-binding proteins for their effects on RNA localization to nuclear compartments and uncovered previously unknown regulatory factors for nuclear RNA localization. Notably, our screen revealed both positive and negative regulators for the nuclear speckle localization of a long noncoding RNA, MALAT1, suggesting a dynamic regulation of lncRNA localization in subcellular compartments.
The development of CRISPR-based gene editing systems has greatly advanced our ability to manipulate genes and probe molecular mechanisms underlying cellular functions through genetic perturbations (1, 2). Facilitated by the ability to generate high-diversity nucleic acid libraries, CRISPR-based pooled-library screening can substantially accelerate the discovery of genes involved in cellular processes (3–5). However, the phenotypes accessible in pooled-library screenings are limited primarily to cell viability and marker expression. Recently, single-cell RNA sequencing and mass cytometry have been combined with CRISPR screening to expand the phenotype space accessible to pooled-library screening, allowing for genetic screening based on the single-cell profiles of RNA and protein expression (6–10).
Many important cellular phenotypes, however, remain beyond the reach of high-throughput pooled-library screening. These include the morphology of cellular structures and intracellular molecular organization, as well as their dynamics, which can be measured only by high-resolution imaging. High-content imaging further allows the simultaneous measurement of these properties for many molecular species in a parallelized manner; for example, the recent development of single-cell transcriptome imaging methods has increased the number of molecular phenotypes that can be imaged in individual cells in a single experiment to the genomic scale (11–14).
Despite the power of imaging in assessing cellular phenotypes, imaging-based pooled-library screening remains challenging, primarily because of the difficulty associated with determining the genotypes of individual phenotype-imaged cells in a pooled-library screening. Approaches have been developed to allow genotype determination by sequencing after physically isolating cells with certain phenotypes (15, 16). However, determining the full genotype-phenotype correspondence requires an all-imaging–based pooled-library screen approach in which both genotypes and phenotypes are imaged for individual cells in situ.
In this work, we report an approach for all-imaging–based pooled-library CRISPR screening in mammalian cells. This approach allows both high-content phenotype imaging of multiple molecular targets in individual cells and high-accuracy identification of the genotype of each cell, the latter achieved by associating each sgRNA with unique barcodes and reading out the barcodes using multiplexed error-robust fluorescence in situ hybridization (MERFISH) (12). To illustrate the power of this approach, we performed a genetic screen for factors regulating RNA localization in nuclear compartments. Various nuclear RNAs, such as small nuclear RNAs (snRNAs), small nucleolar RNA (snoRNAs), and long noncoding RNAs (lncRNAs), are associated with nuclear compartments formed by liquid-liquid phase separation, such as nucleoli and nuclear speckles (17–21). Insight into the spatial regulation of these RNAs is critical to understand how they orchestrate diverse nuclear activities and functions, including transcription regulation, transcript processing, and genome stability (22–25). We thus screened the effect of 162 sgRNAs (targeting 54 genes) on the localizations of six RNA targets, including the lncRNA MALAT1, the U2 snRNA, and the noncoding RNA 7SK, all of which are known to localize to nuclear speckles (26, 27); the nascent preribosomal RNA and the noncoding RNA MRP, both of which are known to localize to nucleoli (28); and the poly-A–containing RNAs. Our results revealed a number of regulators for nuclear RNA localization. In particular, we identified both positive regulators that are essential for the nuclear speckle localization of MALAT1 and negative regulators that reduce the nuclear speckle localization of MALAT1, suggesting a dynamic regulation of lncRNA localization.
Results
High-Throughput, High-Accuracy Barcode Imaging in Mammalian Cells.
In situ imaging-based pooled-library screening, in which the genotypes of individual cells are identified through multiplexed FISH imaging of barcodes associated with the genetic variants, has recently been reported in bacteria by us and others (29, 30). Because of the small volume of bacterial cells, the diffuse signals from barcode RNAs in individual cells are sufficiently strong and can be readily measured. However, mammalian cell volumes are approximately 1,000 times larger than those of bacteria, making it difficult to achieve a sufficiently high concentration of barcode RNAs to allow for reliable measurement. Thus, a new barcode expression and detection scheme is needed to both increase the barcode signal and reduce the background for mammalian cells.
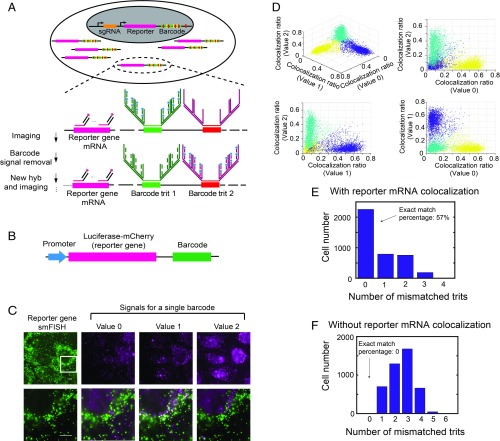
To achieve this goal, we expressed sgRNAs and a reporter gene using two independent promoters in the same vector and incorporated a 12-digit ternary barcode in the 3′ untranslated region (UTR) of the reporter gene (Fig. 1A). Each digit of the ternary barcode (referred to as a trit hereinafter) is composed of one of three different readout sequences [30 nucleotide (nt) long] specific to that digit, corresponding to the three possible trit values: 0, 1 and 2. Twelve trits have the capacity to encode a total of 312 = 531,441 barcodes. Because there are a total of 36 different trit sequences (three different sequences for each of the 12 trits), we read out the barcodes using sequential rounds of hybridization to form images with 36 pseudocolor channels (18 rounds of hybridization with two-color imaging per round, one pseudocolor channel per trit sequence), providing a highly multiplexed detection. To increase the signal from the barcodes, we used a branched DNA amplification scheme to amplify the signal for each trit sequence (Fig. 1A). To reduce interference from background, we costained the mRNA sequence of the reporter gene and detected the reporter gene mRNA with single-molecule fluorescence in situ hybridization (smFISH) (31, 32), so that only the barcode signals that colocalized with the reporter gene signals were considered (Fig. 1A). For each specific trit, the trit value (0, 1, or 2) was assigned based on the pseudocolor channel that exhibited the highest fraction of reporter mRNA smFISH signal colocalized with the trit signal. This detection scheme reduced background signals arising from nonspecific binding of barcode FISH probes, which is essential for decoding accuracy, as shown below.
Fig. 1.
Imaging-based barcode detection for genotype determination in mammalian cells. (A) Strategy for high-accuracy imaging-based barcode detection aiming for genotype determination. sgRNA and a reporter gene with an imaging-based barcode are codelivered into the genome of host cell. The reporter gene portion of mRNA is detected by smFISH, and the barcode is detected by MERFISH, with sequential rounds of hybridization to detect each digit (trit) of the ternary barcode. The barcode signal is amplified using a 4 × 4 branched DNA amplification scheme. (B) Construct design of the reporter gene-barcode library for probing barcode identification accuracy. (C, Upper) Example images showing reporter mRNA smFISH signal (green) and the signals for each of the three trit values (0, 1, and 2) for a single trit in the barcode (magenta). (C, Lower) Enlarged views of the white-boxed region in the upper images, with the reporter gene signal shown on the left and the overlay between the reporter gene signal and the barcode trit signals on the right. Trit value 1 has a high colocalization ratio for this cell, whereas Trit values 0 and 2 do not have high colocalization ratios. (Scale bars: 10 µm.) (D) Colocalization ratios of the three trit values measured for an example trit for all cells. Each spot in the plots corresponds to a single cell. The colocalization ratio is defined as the number of reporter gene smFISH spots that are colocalized with trit signal spots divided by total number of reporter gene smFISH spots within the cell. Cells are partitioned into three clusters (shown in different colors) based on their colocalization ratios using a k-means clustering algorithm. Each cluster corresponds to cells that have a specific trit value. (E) Histogram of the number of cells with different numbers of mismatched trits in the decoded barcodes compared with the valid barcodes in the library. The barcodes are decoded as described above using reporter gene signal and trit signal colocalization. (F) Same as E but with the barcodes decoded by using the number of measured trit signal spots only, without considering reporter gene signal and trit signal colocalization.
To test this barcode identification scheme, we cloned a library of vectors, each of which contains a common reporter gene, luciferase-mCherry, and a unique barcode under the control of the same promoter, in a pooled manner (Fig. 1B and SI Appendix, Fig. S1). Although the total number of possible barcodes exceeds 500,000, we restricted the library to only approximately 2,000 vectors for error-detection purposes (29) (as described below) and determined the barcodes in the library by sequencing. The library was delivered into the genome of U-2 OS cells using lentivirus at a low multiplicity of infection (MOI), so that most transfected cells received only one barcode. We then measured the barcode signals for individual cells using the multiplexed detection scheme as described above. After each round of hybridization, we observed clear barcode signals colocalizing with the smFISH signals of the reporter gene (luciferase-mCherry) mRNA (Fig. 1C).
For each trit detection, three trit values were separately probed (in different pseudocolor channels as described earlier), and three distinct populations of cells were observed, representing cells expressing barcodes with three different trit values (Fig. 1D and SI Appendix, Fig. S2). We used a k-means clustering algorithm to separate the three populations of cells, and a trit value assigned to each population based on which of the three pseudocolor channels assigned to this trit exhibited the highest fraction of reporter gene mRNA spots that were colocalized to the trit signal. The detection of 12 trits using 36 pseudocolor channels allowed us to assign a barcode to each cell. The decoded barcodes for the majority (∼57%) of cells matched the ∼2,000 barcodes in the library determined by sequencing (Fig. 1E), and cells with mismatching barcodes were discarded.
To assess the improvement in barcode detection accuracy using this reporter gene colocalization approach, we also assigned the barcode to each cell based on the number of FISH spots detected for the barcode signal alone (without considering colocalization with the reporter gene signal). We found that no decoded barcodes matched the actual barcodes in the library in this case (Fig. 1F), presumably due to background signals introduced by nonspecific FISH labeling, illustrating substantially improved decoding accuracy with the reporter gene colocalization approach.
The bottlenecking strategy that we used—limiting the total number of vectors in the library to ∼2,000, representing only <0.4% of the total possible number of 12-digit ternary barcodes—allowed for error detection (29), since a readout error of any digit would most likely generate an invalid barcode not present in the library. Quantitatively, since only 0.4% of all possible barcodes were present in the libraries, the probability that any erroneously detected barcode would match the barcodes in the libraries is only 0.4%. Thus, among the 57% exact-matched barcodes, only 0.3% could arise from barcode misidentification (SI Appendix, Materials and Methods).
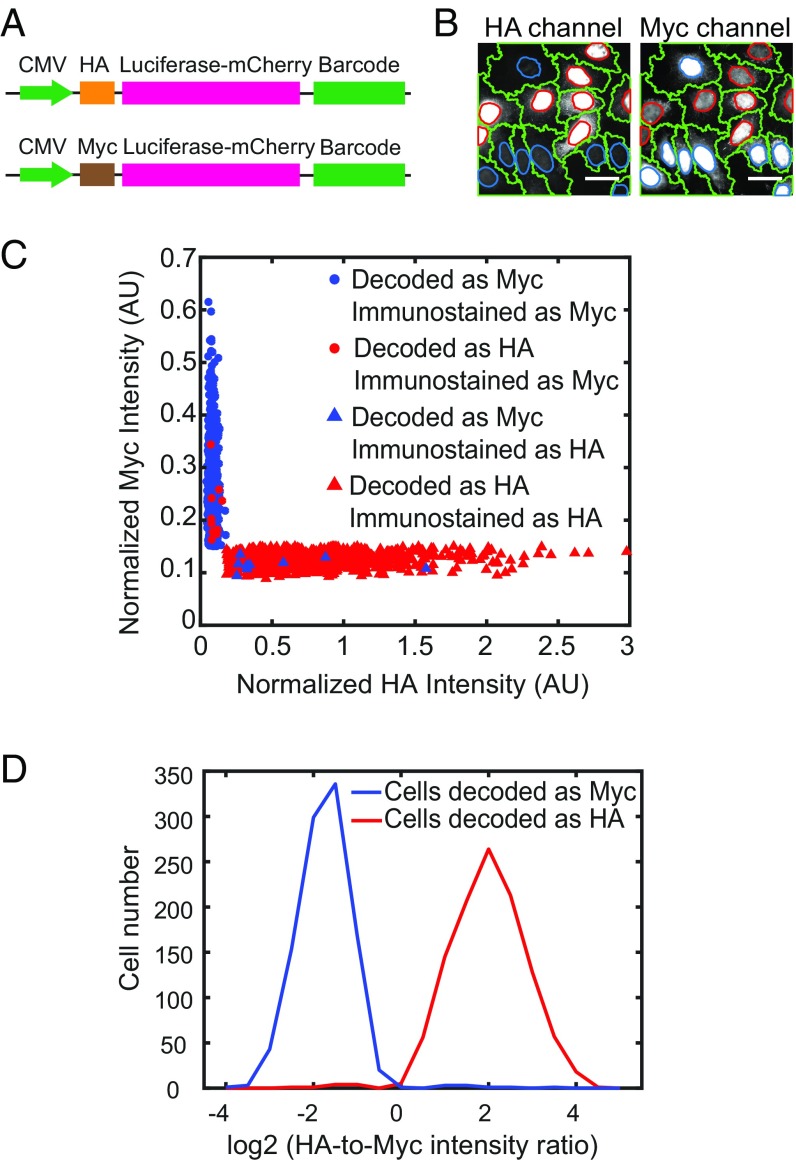
To further validate our low misidentification rate, we designed two reporter gene-barcode libraries, each expressing a reporter gene luciferase-mCherry with a distinct epitope tag (HA tag or Myc tag) fused to a library of barcodes as described above (Fig. 2A) and cloned the two libraries separately. We bottlenecked each library to contain <0.2% of total possible barcodes, so that the same barcodes were highly unlikely to appear in both libraries, and determined the barcode identities associated with each epitope-tagged reporter gene by sequencing. We introduced the two libraries separately in U-2 OS cells and then pooled the two libraries of cells together in roughly equal numbers. We then imaged the phenotype of each cell (i.e., expression of an HA or Myc tag), using immunofluorescence (Fig. 2B) and imaged the barcode associated with each cell using the multiplexed detection scheme as described above. Our rationale was that determining the phenotype of each cell would allow us to deduce the barcode identity of that cell from the sequencing results, and then a comparison with the barcode determined by imaging would allow us to determine the fraction of barcodes that were misidentified. Only approximately 1% of the cells had misidentified barcodes, as determined by barcode-phenotype mismatch (Fig. 2 C and D). Even this small error was largely due to errors in cell segmentation, which in turn caused phenotype determination errors, further supporting the very low barcode misidentification rate in our experiments.
Fig. 2.
Evaluation of the barcode misidentification rate using cells with known phenotype-barcode correspondence. (A) Constructs used to evaluate barcode detection accuracy. The reporter gene luciferase-mCherry is tagged with either a HA tag or a Myc tag to define two phenotypes, as well as a nuclear localization signal to concentrate HA and Myc signals in the nucleus to facilitate detection. The barcodes are placed at the 3′ UTR of the reporter gene, and the correspondence between the barcodes and the HA or Myc tag is determined by sequencing. The reporter gene is driven by a CMV promoter. (B) Images showing HA and Myc immunostaining signals in two different channels. The nuclei with strong HA signals have weak Myc signals and vice versa. The cell boundaries are labeled in green. The nucleus boundaries of HA expression cells and Myc-expressing cells are labeled in red and blue, respectively. (Scale bars: 50 µm.) (C) Scatterplot of the HA and Myc immunostaining intensities of individual cells. Cells assigned to the HA or Myc library based on imaging-based barcode determination are shown in red or blue, respectively. Cells classified as positive on HA or Myc immunostaining (SI Appendix, SI Materials and Methods) are denoted by triangles or circles, respectively. Only 10 out of 1,105 cells with HA-corresponding barcodes are observed as positive in Myc immunostaining, and only 9 out of 1,034 cells with Myc-corresponding barcodes are observed as positive in HA immunostaining, indicating a low barcode misidentification rate. A small fraction of cells (197 out of 2,336 cells) have both HA and Myc immunostaining signals below a threshold value or both HA and Myc immunostaining signals above a threshold value and thus cannot be unambiguously identified as HA-positive or Myc-positive (SI Appendix, SI Materials and Methods). These cells are excluded from analysis. (D) Histogram of the ratio of HA intensity over Myc intensity for individual cells that were decoded to contain HA tag reporter (shown in red) or Myc tag reporter (shown in blue) by barcode imaging.
Lentiviral Delivery System with Reduced Recombination Effect for Accurate sgRNA Identification.
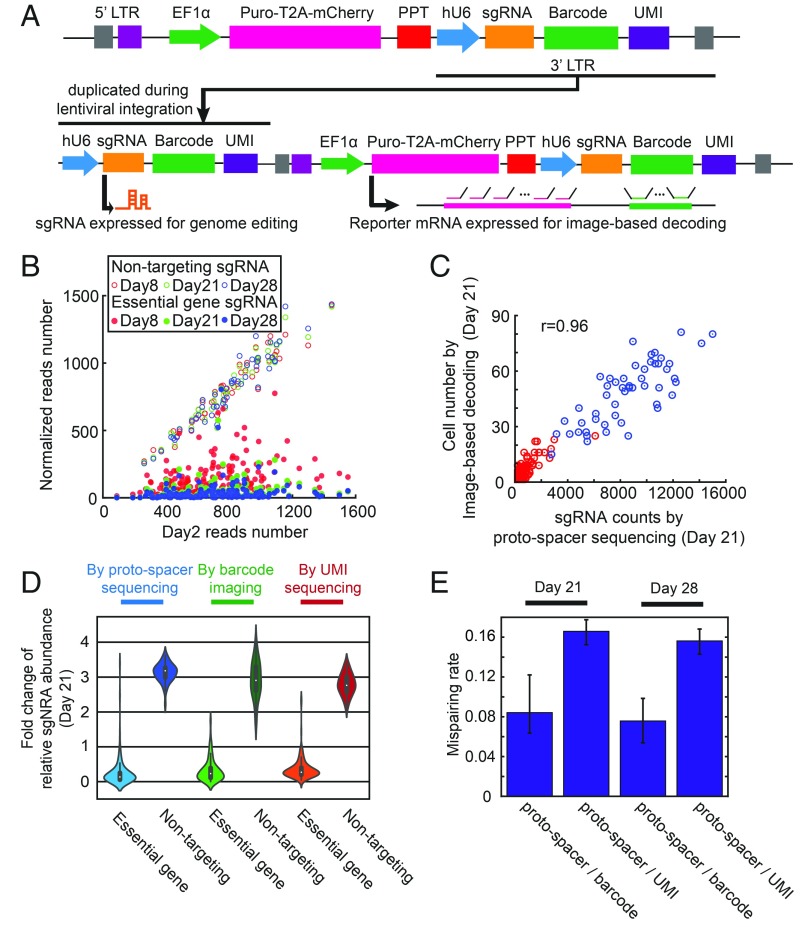
Another challenge in sgRNA identification by pooled-barcode imaging arises from the viral system for delivering the vector containing sgRNA, reporter gene, and barcode into the mammalian cells. Lentivirus is a preferred delivery system for mammalian cells because it allows for stable genome integration of the vector and the introduction of one sgRNA per cell by transduction at a low MOI. However, lentivirus has two single-stranded RNA genomes and is prone to recombination, which could lead to mispairing of sgRNAs and barcodes during viral transduction (33–35). The recombination rate of lentivirus is approximately one event per kilobase (36). Because of the need to separately express the sgRNA and the reporter gene-barcode combination under two independent promoters, the barcode and sgRNA sequences would be separated by a large genomic distance (>1 kb), and thus the probability of recombination-induced barcode-sgRNA mispairing would be substantial (33–35).
We devised a strategy, modified from the CROP-seq approach (9), to overcome this recombination problem. Specifically, we placed the report gene (puro-T2A-mCherry) under a strong Pol II promoter (EF1α) and placed the sgRNA under a separate promoter (hU6), together with the barcode, downstream of the polypurine tract in the lentiviral genome (Fig. 3A). This way, the proto-spacer of sgRNA, a ∼20-nt sequence for specific gene targeting, and the barcode sequence can be separated by a minimal genomic distance (∼100 nt). Although the expression of the sgRNA downstream of the reporter gene could be impaired due to interference from the strong EF1α promoter for reporter gene expression, the sgRNA expression cassette is duplicated to the 5′ LTR of the proviral genome during genome integration, resulting in an additional functional unit to express sgRNAs that is free of the interference from the EF1α promoter (Fig. 3A). The transcription of reporter gene only stops at 3′ end of the 3′ LTR, so the barcode should be expressed in the reporter mRNA 3′ UTR for imaging-based barcode identification (Fig. 3A).
Fig. 3.
Design of the lentiviral delivery approach with a low rate of recombination-induced sgRNA-barcode mispairing. (A) Lentiviral constructs used to deliver sgRNA and barcode for sgRNA identification. An sgRNA cassette (hU6 promoter with sgRNA) and barcode array is placed downstream of the polypurine tract (PPT). A strong Pol II promoter (EF1α) drives expression of the reporter gene, puro-T2A-mCherry. After genome integration, the sgRNA cassette is duplicated into the 5′ LTR for sgRNA expression, while the barcode is expressed with the reporter gene at 3′ UTR for barcode imaging. UMI, unique molecular identifier. (B) Proto-spacer counts of each sgRNA at days 8, 21, and 28 after lentivirus transduction are plotted against the proto-spacer counts measured at day 2 after transduction. The proto-spacer counts at days 8, 21, and 28 are normalized by factors, so that the mean counts for the nontargeting sgRNAs for these conditions are the same as the mean counts for the nontargeting sgRNAs at day 2. The proto-spacer counts are determined by sequencing. As expected, the cells expressing sgRNAs targeting essential ribosomal genes are strongly depleted over time, and thus the counts of sgRNAs targeting essential genes are significantly reduced compared with the nontargeting control sgRNAs. (C) Correlation between the number of cells expressing certain sgRNAs as measured by imaging-based barcode detection and the sgRNA counts measured by proto-spacer sequencing at day 21 after lentivirus transduction. sgRNAs targeting essential genes are shown in red, and nontargeting control sgRNAs are shown in blue. (D) Violin plots showing the median fold change of the relative sgRNA abundance between day 21 and day 2 after lentivirus transduction measured by proto-spacer sequencing, imaging-based barcode detection and UMI sequencing. The relative abundance of a certain sgRNA is defined as the fraction of total sgRNA reads that correspond to this specific sgRNA [i.e., the proto-spacer (or UMI) reads determined by sequencing for this particular sgRNA normalized by the total proto-spacer (or UMI) reads for all sgRNAs, or the number of cells expressing the barcode corresponding to this sgRNA determined by imaging normalized by the total cell number]. As expected, the relative abundance of sgRNAs targeting essential genes reduced over time and the relative abundances of nontargeting sgRNAs increased. Due to the recombination, the fold changes determined by barcode imaging is slightly smaller than those determined by proto-spacer sequencing, and the fold change determined by UMI sequencing is slightly smaller than that determined by barcode imaging. (E) The median mispairing rates between the proto-spacers and barcodes and between proto-spacers and UMI due to recombination, determined at 21 and 28 d after lentivirus transduction. The error bars represent 95% CIs.
To evaluate whether our construct design supports functional lentiviral infection and sgRNA expression, we constructed a library containing both sgRNAs targeting genes essential for cell survival and nontargeting control sgRNAs. An efficient sgRNA expression would cause depletion of cells that express sgRNAs targeting essential genes. We chose 159 sgRNAs targeting 53 essential ribosomal proteins (3 sgRNAs for each gene), as well as 51 nontargeting sgRNAs as controls (Dataset S1) (37) and generated a lentivirus library containing these 210 sgRNAs, together with the reporter gene (puro-T2A-mCherry) and barcodes, by pooled cloning (Fig. 3A and SI Appendix, Fig. S3). We then infected U-2 OS cells stably expressing Cas9-BFP with this lentivirus library. At day 2 after lentiviral infection, we sorted cells that were both infected by the library and expressed a high level of Cas9, based on mCherry and BFP fluorescence, respectively, and kept these cells for experiments at different time points postinfection. We then determined the abundance of cells expressing various sgRNAs by sequencing the genomic DNA. As expected, cells containing sgRNAs targeting essential genes were largely depleted compared with cells containing nontargeting sgRNAs, and the degree of depletion depended on the elapsed time after lentiviral infection (Fig. 3B), indicating that our viral system can support the expression of functional sgRNAs. In addition, we measured the abundance of cells containing different sgRNAs by imaging-based barcode identification, as described above. The abundance of cells containing individual sgRNAs measured by imaging-based barcode identification correlated closely with the cell abundance measured by direct sgRNA proto-spacer sequencing (Fig. 3C), further supporting accurate barcode detection.
We next used this experiment to evaluate the recombination rate of our constructs. If recombination occurs, the barcodes assigned to sgRNAs of essential genes can recombine with nontargeting sgRNAs, which should lead to a higher cell abundance measured by barcode imaging than by proto-spacer sequencing. Similarly, the barcodes assigned to nontargeting sgRNAs can recombine with sgRNAs targeting essential genes, leading to a lower cell abundance measured by barcode imaging. We thus measured the fold changes of relative cell abundance between day 2 and day 21 after lentiviral transduction for cells containing sgRNAs targeting essential genes and cells containing nontargeting sgRNAs. As expected, compared with day 2, at day 21 the relative abundance of cells containing sgRNAs targeting essential genes was greatly reduced, whereas the relative abundance of cells containing nontargeting sgRNAs was substantially increased (Fig. 3D). Compared with results obtained by sgRNA sequencing, the fold changes determined by barcode imaging were slightly smaller, due to recombination (Fig. 3D).
This difference allowed us to quantify the recombination-induced mispairing rate (SI Appendix, Materials and Methods), which we determined to be ∼8% between the sgRNA proto-spacers and barcodes (Fig. 3E). In addition, we measured the recombination-induced mispairing rate between the sgRNA proto-spacer and a unimolecular identifier (UMI), a 20-nt sequence placed ∼500 bases downstream from the proto-spacer (Fig. 3A). As expected, due to the larger genomic distance between the UMI and the proto-spacer (∼500 nt), compared with the genomic distance between the barcode and proto-spacer (∼100 nt), the recombination-induced mispairing rate for the region between the proto-spacer and UMI was larger, ∼16% (Fig. 3 D and E). We note that for a random pair of barcodes, the probability that these barcodes share the same sequence at any giving trit position is approximately 33%, because there are three possible sequences for any given trit and because the barcodes in the bottlenecked library compose a randomly selected subset of all possible barcodes. Thus, we estimate that the recombination rate in the barcode region should be roughly one-third the recombination rate for the fully homologous sequence of the same length. Based on the ∼8% recombination rate that we measured for the ∼100-nt genomic region between the barcode and proto-spacer (the common sequence of sgRNAs), we estimate the recombination rate in the ∼400-nt barcode region to be roughly (400/100) × 8%/3 = 10.7%, which would give a recombination rate of ∼8% + 10.7% = 18.7% for the genomic region between the proto-spacer and UMI, consistent with our measured value of ∼16%. Furthermore, since our barcode library was bottlenecked, the recombination that occurred within the barcode region is unlikely to generate a new barcode that matches with the valid barcodes in the library, and thus unlikely to lead to barcode misidentification.
Together, the low error rate in barcode imaging (<1%) and the low mismatching rate between sgRNA and barcode induced by recombination (∼8%) allowed for high accuracy in sgRNA identification by barcode imaging, which in turn enabled an all imaging-based pooled-library CRISPR screening. We note that although the remaining 8% mismatch rate between sgRNA and barcode can potentially generate false-positives and -negatives in the screening, the error rate would be minimal because we typically probed hundreds of cells carrying the same sgRNA to determine whether an sgRNA had a statistically significant effect; moreover, we probed three sgRNAs targeting each gene and only considered a gene a hit when two of the three sgRNAs exhibited a statistically significant effect. Any remaining false-positives can be readily identified by validation experiments.
Pooled CRISPR Screening for Factors Regulating Nuclear RNA Localization.
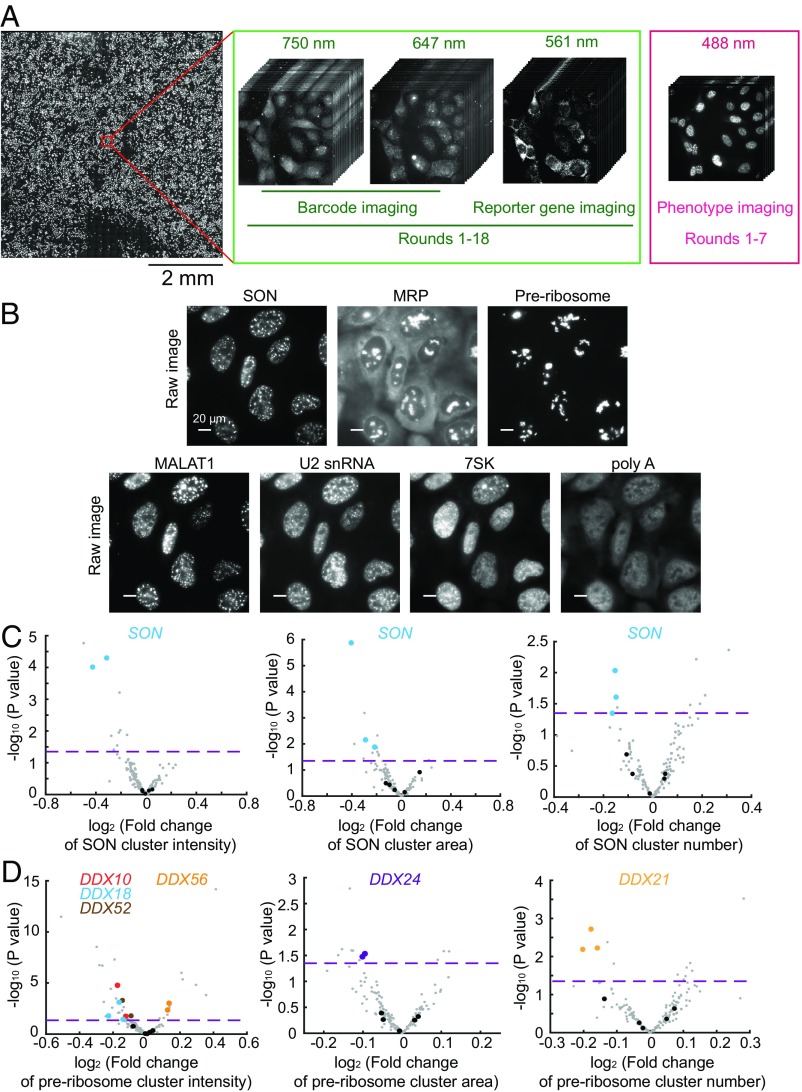
To illustrate the power of this screening method, we screened for potential regulators of RNA localization in the nucleus (Fig. 4A). We selected 54 candidate genes involved in nuclear RNA regulation, including hnRNP family proteins, DExD/H box RNA helicases, and genes involved in RNA modification (Dataset S2). We designed a library of 167 sgRNAs, containing 3 sgRNAs for each of the 54 genes and 5 nontargeting sgRNAs as controls, and generated a lentivirus library containing these sgRNAs, together with the reporter gene (puro-T2A-mCherry) and barcodes, by pooled cloning (SI Appendix, Fig. S3). To demonstrate the ability of this method to assess complex phenotypes, we imaged the spatial distributions of five specific RNA species—the lncRNA MALAT1, the U2 snRNA, 7SK, MRP, and the nascent preribosome—as well as the poly-A–containing RNAs, using FISH. In addition, we also included in the phenotype imaging a nuclear speckle protein, SON, using immunolabeling with an oligonucleotide-conjugated antibody. We imaged these RNA and protein targets, along with barcode imaging, using sequential rounds of hybridization with three to four different color channels per round (Fig. 4A). SI Appendix, Materials and Methods presents a detailed description of the imaging procedure.
Fig. 4.
Imaging-based pooled CRISPR screening for regulators of nuclear RNA localization. (A) The scheme of imaging-based screening. Cells infected with lentiviruses expressing sgRNAs, barcodes, and the reporter gene are fixed and imaged. The barcodes are imaged by MERFISH using 647-nm and 750-nm color channels in 18 rounds of hybridization (rounds 1–18). To increase the accuracy of barcode imaging, the reporter gene mRNA is imaged in every round (rounds 1–18) using the 561-nm color channel to allow the determination of colocalization between barcode and reporter gene mRNA signals. The seven protein and RNA targets for phenotype measurements are imaged in the 488-nm color channel in the first seven rounds (rounds 1–7). The mosaic on the left contains 900 fields of view from a single screen. (B) Phenotype images of SON, MRP, preribosome, MALAT1, U2 snRNA, 7SK, and poly-A–containing RNAs. SON marks nuclear speckles, and preribosome and MRP mark subnucleolar structures. For SON, preribosome, and MRP, the cluster numbers, cluster areas, and cluster intensities are quantified. For MALAT1, U2 snRNA, 7SK, and poly-A–containing RNAs, their enrichments in nuclear speckles are quantified. (Scale bars: 20 µm.) (C) Volcano plots for the effect of each sgRNA on SON cluster intensity, cluster area, and cluster number. (D) Volcano plots for the effect of each sgRNA on preribosome cluster intensity, cluster area, and cluster number. In C and D, the fold change induced by each sgRNA is calculated as the mean value from all cells containing this sgRNA divided by the mean value from all cells containing nontargeting sgRNAs. The horizontal dashed lines indicate the P value (0.05) used to define a hit of the screen. The data points of the indicated hits (i.e., two of the three sgRNAs targeting the gene show statistically significant fold changes; P < 0.05) are shown in colors matching the colors of the gene names shown in the legend, data points for other gene-targeting sgRNAs are shown in gray, and data points for nontargeting sgRNAs are shown in black. Additional hits are shown in Dataset S3.
As expected, SON exhibited a clustered distribution that marked the nuclear speckles, and the MRP and preribosome signals marked the subnucleolar compartments (Fig. 4B). Based on these images, we identified the boundaries of these structures and determined their numbers, the areas they cover, and their mean signal intensities (i.e., total signals localized within the identified cluster boundaries divided by total area covered by these clusters) in individual cells. We next quantified the enrichment of MALAT1, U2, 7SK, and poly-A–containing RNAs in the nuclear speckles identified by SON staining (SI Appendix, SI Materials and Methods).
For each of these feature quantifications, we compared the values determined for cells harboring a targeting sgRNA with the values measured from cells harboring nontargeting control sgRNAs to determine the fold change. We performed four biological replicates of experiments and decoded ∼30,000 cells, then determined hits based on the criterion that at least two of three sgRNAs targeting the gene exhibited a statistically significant fold change (Dataset S3).
As a positive control, we detected statistically significant decreases in cluster signal intensity, cluster area, and cluster number associated with the SON stain in cells expressing sgRNAs targeting SON (Fig. 4C). In addition, sgRNAs for several DExD/H box RNA helicases (DDX10, DDX18, DDX21, DDX24, DDX52, and DDX56) caused statistically significant changes in various features of the nascent preribosome stain (Fig. 4D), consistent with the known functions of these genes in ribosome biogenesis (38–40). We note that the magnitudes of change in these phenotype features were moderate (Fig. 4 C and D), possibly because not all cells expressing the sgRNAs underwent genome editing. Thus, our quantifications allowed the identification of genetic perturbations that had a statistically significant effect, but the magnitudes of the phenotype changes were less informative. We also noticed that the perturbation of several genes in the hnRNP family caused significant changes in the preribosome and MRP signals in the nucleoli (Dataset S3), potentially due to indirect effects.
Factors Involved in the Regulation of MALAT1 Nuclear Speckle Localization.
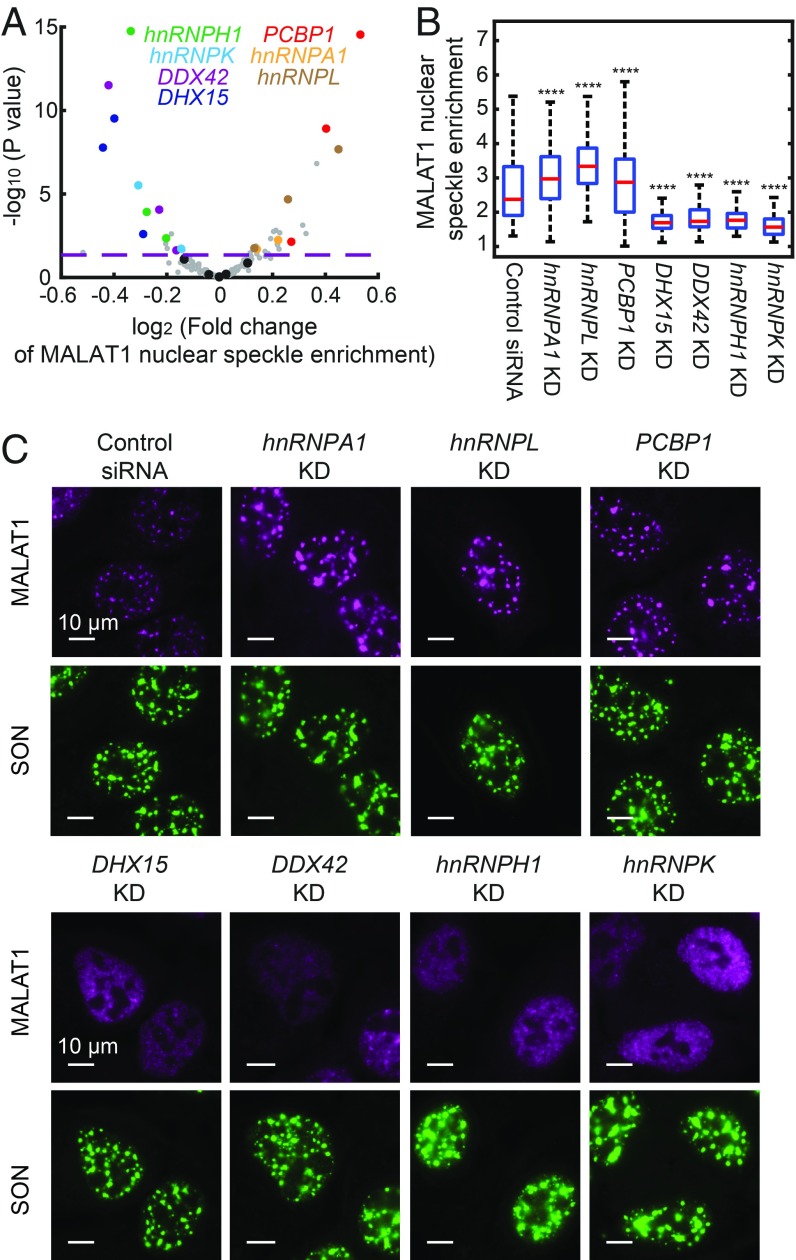
Our screening revealed genes involved in regulation of nuclear speckle localization of different RNA species (Dataset S3). Compared with 7SK, U2 snRNA, and poly-A–containing RNAs, we identified more genes that regulate MALAT1 localization, and we focus our discussion here on MALAT1. Of note, we identified two groups of genes that regulate the nuclear speckle localization of MALAT1 in opposite directions (Fig. 5A and Dataset S3), which were validated for all but one gene (hnRNPH3) by siRNA-mediated knockdown (Fig. 5 B and C). We were not able to confirm whether the siRNA for hnRNPH3 was effective, due to the lack of an effective antibody for this protein. Depletion of the first group of genes—DHX15, DDX42, hnRNPK, and hnRNPH1—caused a statistically significant reduction in the enrichment of MALAT1 in nuclear speckles (Fig. 5 A–C), suggesting that these genes up-regulate the nuclear speckle localization of MALAT1. DHX15 and DDX42 are involved in spliceosome recycling and assembly, respectively (41, 42), consistent with the involvement of mRNA splicing factors in recruiting MALAT1 into nuclear speckles (23, 43). Involvement of the hnRNP family proteins hnRNPH1 and hnRNPK in the up-regulation of nuclear speckle localization of MALAT1 has not been anticipated previously. These two genes were also found to affect the localization of other RNA species, including the U2 snRNA, poly-A–containing RNAs, preribosome RNA, and MRP (Dataset S3), which could imply a global effect of the perturbations of these two genes.
Fig. 5.
Genetic factors involved in the regulation of MALAT1 nuclear speckle localization. (A) Volcano plot for the effect of each sgRNA on MALAT1 nuclear speckle enrichment. The fold change is calculated as described in Fig. 4. The horizontal dashed line indicates the P value (0.05) used to define hit of the screen. The hits confirmed by siRNA knockdown are highlighted in colors matching the colors of the gene names shown in the legend, data points for other gene-targeting sgRNAs are shown in gray, and data points for nontargeting sgRNAs are shown in black. (B) Boxplots showing the effect of siRNA knockdown of the seven hit genes on MALAT1 localization alongside data for a control, nontargeting siRNA. The red lines show the median, the boxes show the 25th to 75th quartiles, and the whiskers show the maximum and minimum values. Between 100 and 300 cells are quantified for each condition. Student’s t tests are performed for each condition in comparison with control. ****P < 0.0001. (C) Images of MALAT1 localization on siRNA knockdown of the seven hit genes. Data from a control nontargeting siRNA are also shown. MALAT1 staining is shown in magenta, and SON staining is shown in green. (Scale bars: 10 µm.)
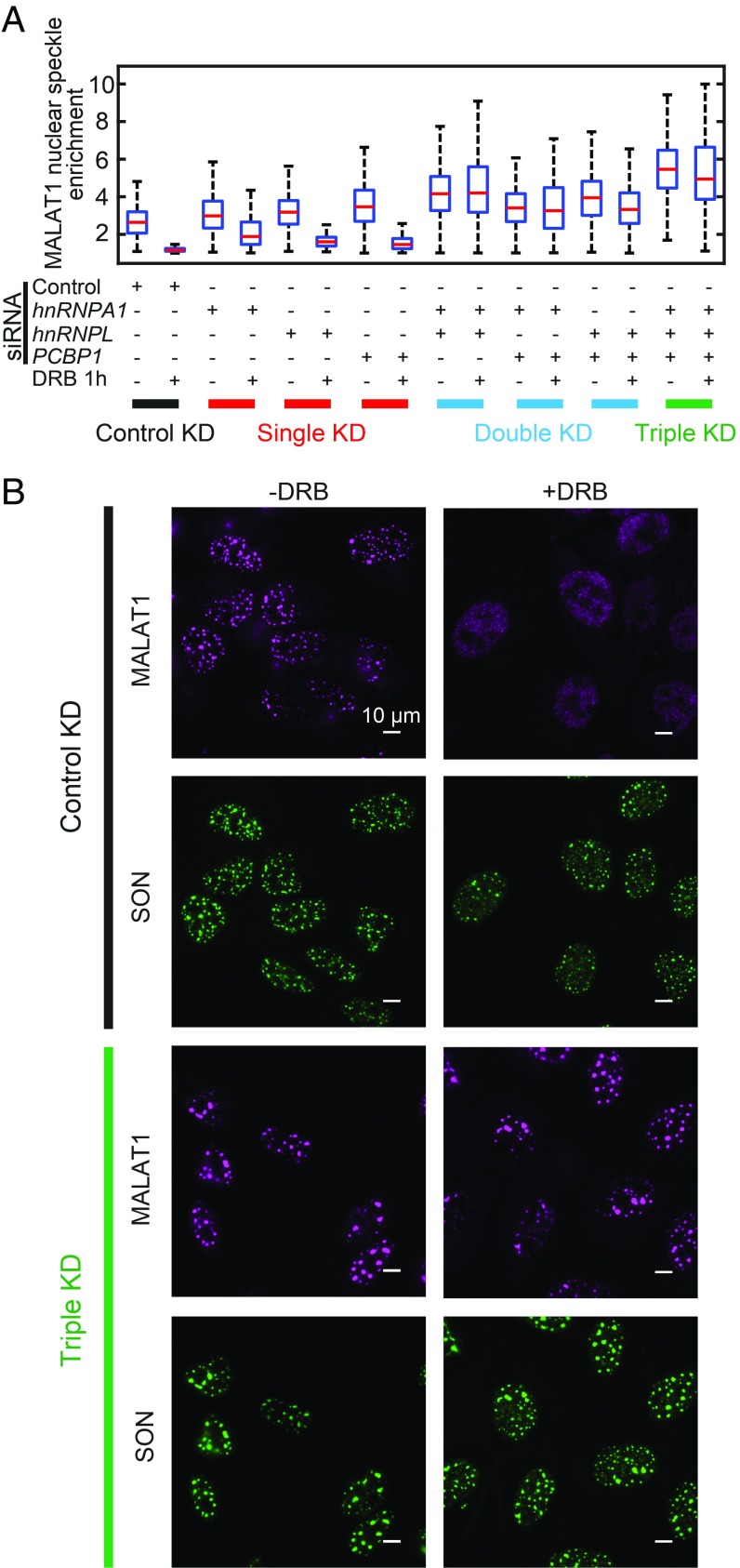
Unexpectedly, we also identified three factors—hnRNPA1, hnRNPL, and PCBP1—that negatively regulate the nuclear speckle localization of MALAT1. Their depletion by sgRNA or siRNA induced a statistically significant increase in MALAT1 enrichment in nuclear speckles (Fig. 5 A–C). The fold change of the MALAT1 enrichment induced by siRNA could be an underestimation due to incomplete knockdown. Combined knockdown of all three factors further increased MALAT1 enrichment in nuclear speckles (SI Appendix, Fig. S4A), which, interestingly, also resulted in enlargement of a fraction of nuclear speckles (SI Appendix, Fig. S4 B and C). The composition of each nuclear speckle, measured by the ratio of MALAT1 and SON levels in the nuclear speckle, also became more heterogeneous in the triple-knockdown sample; some speckles had a reduced MALAT1-to-SON ratio, whereas some had an increased ratio (SI Appendix, Fig. S4D). These results indicate that the enhanced nuclear speckle localization of MALAT1 due to the knockdown of the three negative regulators is associated with changes in nuclear speckle morphology and composition. This suggests a role of MALAT1 in regulating nuclear speckle structures, which is consistent with the observation that MALAT1 knockdown can lead to a reduction in nuclear speckle size (44).
It has been shown previously that nuclear speckle localization of MALAT1 can be impaired under transcription inhibition (45). However, the genetic factors involved in this process are largely unclear. Thus, we tested whether these three negative regulators play a role in this process. To this end, we added the drug 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) to inhibit transcription and observed a substantial reduction in MALAT1 enrichment in nuclear speckles. Single knockdown of hnRNPA1, hnRNPL, and PCBP1 did not substantially rescue the DRB-induced dissociation of MALAT1 from nuclear speckles (Fig. 6A and SI Appendix, Fig. S5). On the other hand, double knockdown of two of these three factors or triple knockdown of all three factors largely rescued this DRB-induced dissociation effect (Fig. 6 and SI Appendix, Fig. S5), suggesting that these hnRNP family proteins are important for transcription inhibition-induced disassociation of MALAT1 from nuclear speckles, and that these factors likely play redundant roles in this process.
Fig. 6.
hnRNPA1, hnRNPL, and PCBP1 are required for transcription inhibition-induced dissociation of MALAT1 from nuclear speckles. (A) Quantifications of MALAT1 nuclear speckle enrichment with or without transcription inhibitor DRB treatment (50 µM for 1 h) for cells transfected by different combination of siRNAs. Between 100 and 300 cells are quantified for each condition. The transcription inhibition-induced dissociation of MALAT1 from nuclear speckles is not rescued by single knockdowns of hnRNPA1, hnRNPL, and PCBP1 but is rescued by the double-knockdown and triple-knockdown of these factors. (B) Images showing that in cells transfected by control siRNAs, MALAT1 dissociates from nuclear speckles on transcription inhibition; whereas in cells cotransfected by siRNAs targeting hnRNPA1, hnRNPL, and PCBP1, transcription inhibition fails to dissociate MALAT1 from nuclear speckles. The MALAT1 staining is shown in magenta, and SON staining is shown in green. (Scale bars: 10 µm.)
Our results provide a potential mechanism for the dissociation of MALAT1 from nuclear speckles by transcription inhibition. During transcription inhibition, RNA-binding proteins such as hnRNPA1 and hnRNPL are freed from nascent mRNA transcripts to allow their binding to other RNA species (46, 47). It is thus possible that the freed hnRNPA1 and hnRNPL could bind to MALAT1, which may compete with factors that recruit MALAT1 to nuclear speckles, thereby preventing the nuclear speckle localization of MALAT1 under transcription inhibition.
Discussion
In this work, we developed an imaging-based pooled-library CRISPR screening method that allows the establishment of genotype-phenotype correspondence for individual cells and enables high-throughput screening of mammalian cells based on complex phenotypes that are previously inaccessible to pooled-library screening. This imaging-based screening is enabled by sgRNA identification through MERFISH-based barcode detection, and we demonstrated a barcode misidentification rate as low as ∼1%. We further devised a lentiviral delivery scheme with a reduced rate of recombination-induced mispairing of sgRNAs and barcodes (mispairing rate <10%). Together, these provide high-accuracy sgRNA identification through barcode imaging. Our approach substantially expands the phenotype space accessible for pooled-library screening. Compared with imaging-based screening using the arrayed format in which individual genetic perturbations are assayed separately in individual wells, a major advantage of performing pooled screening is that the reagents for genetic perturbations, such as the DNA plasmids and lentiviruses, can be prepared in a pooled manner with standard molecular biology procedures with reduced labor and at lower cost, which is particularly beneficial for large-scale custom-designed libraries. Reagent preparation for arrayed screening typically requires a costly multiwell robotic processing system and more complicated procedures (48). Another advantage of the pooled approach is that the variation in experimental conditions for different perturbations can be minimized since the measurements for all genetic perturbations are performed in the same experiment. This is particularly desirable when the cells should be treated with concentration or time sensitive conditions. Moreover, the pooled format can also simplify multiplexed phenotype measurements that require sequential rounds of staining and signal removal through buffer exchange. On the other hand, when generating individual genetic perturbation reagents is not especially demanding (e.g., for relatively small-scale screens) and when the phenotype measurement is not very sensitive to variations in sample treatment conditions, arrayed screening could be preferred, because the MERFISH barcode readout process increases the complexity of the imaging procedure.
Our current 12-digit ternary barcode library contains more than 500,000 barcodes. Even with a stringent 1% bottlenecking strategy to enable error-robust barcode detection, more than 5,000 distinct sgRNAs can be included in each library and this capacity can be readily increased by adding more digits to the barcodes. A current limitation on the number of sgRNAs that can be screened is the time required to image a large number of cells. Our current imaging system uses a high-magnification (60×) objective to read out the FISH signal on individual single mRNA molecules for barcode detection, limiting the number of cells that can be imaged in each field of view. However, the imaging speed could be substantially improved by (i) using greater amplification for the barcode signal, which would in turn allow each field of view to be captured with a faster frame rate and/or allow more cells to be imaged in each field of view by using lower-magnification objectives or (ii) using multiple cameras for detection, which would allow simultaneous detection of fluorescence signals in different color channels. With these improvements, we anticipate substantial increase in the number of cells and genotypes that can be screened per experiment.
To demonstrate the power of our approach for screening complex phenotypes of mammalian cells, we imaged subcellular localizations of seven different molecular species, including six RNAs and a protein. Our screening experiments revealed previously unknown regulators of nuclear RNA localization. Interestingly, we identified both positive and negative regulators of the nuclear speckle localization of the lncRNA MALAT1. The positive regulators include DExD/H box RNA helicases DHX15 and DDX42 and hnRNP family genes hnRNPH1 and hnRNPK, and the negative regulators include hnRNPA1, hnRNPL, and PCBP1. RNAs can be localized to cellular compartments formed by phase separation via two mechanisms (20): (i) RNAs can act as a scaffold, which could facilitate the nucleation of phase separation, such as mRNAs in P body and stress granules (49) and preribosome RNAs in nucleoli (50), and (ii) RNAs can be recruited to the phase-separated bodies as clients, which has been shown to be responsible for the localization of MALAT1 in nuclear speckles (23, 43, 51). It is possible that the negative regulators discovered in our screening could compete with the factors that recruit MALAT1 to nuclear speckles, thereby preventing the nuclear speckle localization of MALAT1. We further identified a role of these negative regulators in the dissociation of MALAT1 from nuclear speckles induced by transcription inhibition. These results suggest that lncRNA localization could be dynamically regulated by protein factors.
Our results demonstrate the ability of this imaging-based screening method to reveal molecular factors involved in cellular processes that can be assessed only by high-resolution imaging. This screening method should be broadly applicable to interrogating genetic factors controlling or regulating a broad spectrum of phenotypes, including morphological features, molecular organizations, and dynamics of cellular structures, as well as cell–cell interactions. We also anticipate that this screening approach can be combined with highly multiplexed DNA, RNA, and protein imaging approaches, including genomic-scale imaging approaches, to profile factors involved in gene regulation and other genomic functions in a high-throughput manner.
Materials and Methods
Details of the protocols for all methods used in this work are provided in SI Appendix, Materials and Methods. All reagents and software codes are available upon request.
The cloning of the reporter gene-barcode libraries and sgRNA-reporter-barcode libraries were performed in pooled manner using oligos ordered from IDT (Datasets S1, S2, and S4). These libraries were cloned into the lentiviral vector pFUGW as described in SI Appendix, Materials and Methods (SI Appendix). The identities of barcodes present in the libraries and the barcode-sgRNA correspondence were established using high-throughput sequencing. Lentivirus was produced in LentiX cells (632180; Takara) using Lenti-X Packaging Single Shots (VSV-G, 631276; Takara).
The lentiviral libraries were used to infect the U-2 OS cells at a low MOI, so that only 10–20% of the cells were infected. The infected cells were sorted based on mCherry expression and Cas9-BFP expression. The sorted cells were fixed, permeabilized, and stained for imaging as described in SI Appendix, Materials and Methods. All primary and secondary amplification probes for barcode staining, smFISH probes for imaging reporter mRNA, oligonucleotide probes for nuclear RNA staining, and the oligonucleotide for antibody labeling were obtained from IDT. All dye-labeled readout probes based on disulfide linkage were obtained from Bio-Synthesis. The sequences for all these oligonucleotides are provided in Dataset S5.
A custom microscope built around a Nikon Ti-U microscope body with a Nikon CFI Plan Apo Lambda 60× oil immersion objective with 1.4 NA was used for imaging. For sequential rounds of hybridization and imaging, a peristaltic pump (MINIPULS 3; Gilson) pulled liquids (TCEP buffer for dye cleavage, hybridization buffer with readout probes or hybridization buffer for sample wash) into a Bioptechs FCS2 flow chamber with sample coverslips, and three valves (Hamilton, MVP, and HVXM 8-5) were used to select the input fluid (details provided in SI Appendix, Materials and Methods).
The barcode decoding and phenotype quantification based on collected images are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health. C.W. received support from a Jane Coffin Childs Memorial Fund fellowship. X.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
Conflict of interest statement: The authors are applying for a patent based on the technology reported in this paper.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903808116/-/DCSupplemental.
References
- 1.Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, Doudna JA (2016) Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34:933–941. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert LA, et al. (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Wei JJ, Sabatini DM, Lander ES (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalem O, et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamson B, et al. (2016) A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167:1867–1882.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit A, et al. (2016) Perturb-Seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167:1853–1866.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaitin DA, et al. (2016) Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell 167:1883–1896.e15. [DOI] [PubMed] [Google Scholar]
- 9.Datlinger P, et al. (2017) Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 14:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wroblewska A, et al. (2018) Protein barcodes enable high-dimensional single-cell CRISPR screens. Cell 175:1141–1155.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JH, et al. (2014) Highly multiplexed subcellular RNA sequencing in situ. Science 343:1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X (2015) Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah S, et al. (2018) Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174:363–376.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. (2018) Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361:eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien MP, Werley CA, Farhi SL, Cohen AE (2015) Photostick: A method for selective isolation of target cells from culture. Chem Sci (Camb) 6:1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piatkevich KD, et al. (2018) A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat Chem Biol 14:352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao YS, Zhang B, Spector DL (2011) Biogenesis and function of nuclear bodies. Trends Genet 27:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batista PJ, Chang HY (2013) Long noncoding RNAs: Cellular address codes in development and disease. Cell 152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Brangwynne CP (2015) Nuclear bodies: The emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 34:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato M, McKnight SL (2018) A solid-state conceptualization of information transfer from gene to message to protein. Annu Rev Biochem 87:351–390. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa Y, et al. (2010) The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19:469–476. [DOI] [PubMed] [Google Scholar]
- 23.Tripathi V, et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasanth KV, et al. (2010) Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell 21:4184–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu HP, et al. (2017) TERRA RNA antagonizes ATRX and protects telomeres. Cell 170:86–101.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galganski L, Urbanek MO, Krzyzosiak WJ (2017) Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res 45:10350–10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spector DL, Lamond AI (2011) Nuclear speckles. Cold Spring Harb Perspect Biol 3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldfarb KC, Cech TR (2017) Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev 31:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emanuel G, Moffitt JR, Zhuang X (2017) High-throughput, image-based screening of pooled genetic-variant libraries. Nat Methods 14:1159–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson MJ, et al. (2017) In situ genotyping of a pooled strain library after characterizing complex phenotypes. Mol Syst Biol 13:947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Femino AM, Fay FS, Fogarty K, Singer RH (1998) Visualization of single RNA transcripts in situ. Science 280:585–590. [DOI] [PubMed] [Google Scholar]
- 32.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sack LM, Davoli T, Xu Q, Li MZ, Elledge SJ (2016) Sources of error in mammalian genetic screens. G3 (Bethesda) 6:2781–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill AJ, et al. (2018) On the design of CRISPR-based single-cell molecular screens. Nat Methods 15:271–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie S, Cooley A, Armendariz D, Zhou P, Hon GC (2018) Frequent sgRNA-barcode recombination in single-cell perturbation assays. PLoS One 13:e0198635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlub TE, Smyth RP, Grimm AJ, Mak J, Davenport MP (2010) Accurately measuring recombination between closely related HIV-1 genomes. PLOS Comput Biol 6:e1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doench JG, et al. (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin R, Straub AU, Doebele C, Bohnsack MT (2013) DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol 10:4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calo E, et al. (2015) RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells GR, et al. (2017) The ribosome biogenesis factor yUtp23/hUTP23 coordinates key interactions in the yeast and human pre-40S particle and hUTP23 contains an essential PIN domain. Nucleic Acids Res 45:4796–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Will CL, et al. (2002) Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J 21:4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimoto R, Kataoka N, Okawa K, Ohno M (2009) Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res 37:891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyagawa R, et al. (2012) Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18:738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fei J, et al. (2017) Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 130:4180–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernard D, et al. (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29:3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diribarne G, Bensaude O (2009) 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol 6:122–128. [DOI] [PubMed] [Google Scholar]
- 47.Giraud M, et al. (2014) An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc Natl Acad Sci USA 111:1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Groot R, Lüthi J, Lindsay H, Holtackers R, Pelkmans L (2018) Large-scale image-based profiling of single-cell phenotypes in arrayed CRISPR-Cas9 gene perturbation screens. Mol Syst Biol 14:e8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Treeck B, Parker R (2018) Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernandez-Verdun D. (2011) Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clemson CM, et al. (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.