Significance
The mechanisms of eukaryotic circadian clocks rely on transcriptional-translational feedback loops (TTFLs), but components of TTFLs from different phylogenetic lineages are thought to be evolutionarily diverse. Posttranslational modification is also required for clock function, but those within the plant clock are less studied, likely due to genetic redundancy. Here, we identified a small synthetic molecule that lengthened the Arabidopsis circadian period. Using an affinity probe, we found that the molecule inhibited multiple members of the casein kinase I (CK1) family, which is also essential in animal, fungal, and algal clocks. The CK1 family modulated plant-specific clock-associated transcriptional repressors. With other studies, our results established the prominent role of CK1 family to control circadian clocks among vastly divergent phylogenetic lineages.
Keywords: circadian clock, Arabidopsis, small molecule, posttranslational regulation
Abstract
The circadian clock provides organisms with the ability to adapt to daily and seasonal cycles. Eukaryotic clocks mostly rely on lineage-specific transcriptional-translational feedback loops (TTFLs). Posttranslational modifications are also crucial for clock functions in fungi and animals, but the posttranslational modifications that affect the plant clock are less understood. Here, using chemical biology strategies, we show that the Arabidopsis CASEIN KINASE 1 LIKE (CKL) family is involved in posttranslational modification in the plant clock. Chemical screening demonstrated that an animal CDC7/CDK9 inhibitor, PHA767491, lengthens the Arabidopsis circadian period. Affinity proteomics using a chemical probe revealed that PHA767491 binds to and inhibits multiple CKL proteins, rather than CDC7/CDK9 homologs. Simultaneous knockdown of Arabidopsis CKL-encoding genes lengthened the circadian period. CKL4 phosphorylated transcriptional repressors PSEUDO-RESPONSE REGULATOR 5 (PRR5) and TIMING OF CAB EXPRESSION 1 (TOC1) in the TTFL. PHA767491 treatment resulted in accumulation of PRR5 and TOC1, accompanied by decreasing expression of PRR5- and TOC1-target genes. A prr5 toc1 double mutant was hyposensitive to PHA767491-induced period lengthening. Together, our results reveal posttranslational modification of transcriptional repressors in plant clock TTFL by CK1 family proteins, which also modulate nonplant circadian clocks.
The circadian clock is a biological timekeeping system that generates genetic, metabolic, behavioral, and physiological rhythms in many organisms, enabling them to predict and adapt to the day-night cycle. Although the fundamental properties of circadian rhythms (self-sustaining oscillation, temperature compensation of period length, and entrainment by environmental time cues such as light or temperature) are common across many types of organisms, components of circadian clocks are assumed to be evolutionarily diverse among bacteria, fungi, animals, and plants (1). Cyanobacteria employ autonomous protein phosphorylation-dephosphorylation oscillations as a clock timekeeping system, whereas eukaryotes utilize transcriptional-translational feedback loops (TTFLs) for clock function (2–4). In addition to TTFLs, posttranslational modifications of components in TTFLs are crucial for clock in eukaryotes (3). CASEIN KINASE (CK) 1 is an evolutionarily conserved kinase that regulates circadian periodicity in fungi, animals, and algae, but the substrates of CK1 differ greatly across lineages. CK1 phosphorylates FREQUENCY (FRQ) in fungi (5) and PERIOD in mice (animals) (6), but the substrates of CK1 in algae are as yet unknown (7, 8).
In the TTFLs of the terrestrial plant Arabidopsis thaliana (Arabidopsis), dawn-expressed CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), morning-to-evening-expressed PSEUDO-RESPONSE REGULATOR [PRR9, PRR7, PRR5, TIMING OF CAB EXPRESSION 1 (TOC1)], and evening-to-night-expressed EARLY FLOWERING (ELF) 3, ELF4, and LUX ARRHYTHMO (LUX) repress genes expressed during earlier phases (4). REVEILLE 8 (RVE8) and NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED GENE 1 (LNK1) form a complex and activate PRR5 and TOC1 (4). In addition to TTFLs, some posttranslational regulation is also required for proper clock function. Phosphorylation of CCA1 and LHY by CK2 are related to DNA-binding activities of CCA1 and LHY, influencing clock pace (4). ZEITLUPE (ZTL) encodes an ubiquitin E3 ligase for PRR5 and TOC1, and its mutation resulted in accumulation of PRR5 and TOC1, and long period (4). Phosphorylated PRR5 and TOC1 are recognized by ZTL (9), but no kinase has yet been identified for PRR5 and TOC1 degradation-related phosphorylation.
Genetic and biochemical studies have been used to demonstrate the presence of TTFLs in Arabidopsis, but a whole-genome duplication event followed by local duplications during the evolution of Arabidopsis makes it difficult to identify clock-associated factors due to the presence of paralogous genes (10). In addition, genes involved in essential or fundamental biological processes possibly contribute to clock control. Chemical genetics approaches can often circumscribe the problems posed by genetic redundancy or lethality by inducing a phenotype, or phenotypes that would not be possible by introducing a single genetic mutation. Chemical compounds can also be applied in dose-dependent, time-dependent, or growth stage-conditional manners, allowing stringent controls to be employed for each of the biological processes of interest. Chemical-genetic strategies, including the use of natural compounds that affect actin-associated processes and clock-associated gene expression, have therefore become important for deciphering which genes encode clock-associated factors (11–13).
To reveal possible targets of biologically active molecules, several studies have identified mutants that are insensitive to the inhibitory molecules used in previous work (11). For example, discovery of a selective ABA agonist, pyrabactin, and identification of pyrabactin-insensitive mutants revealed highly redundant ABA receptors (14). However, genes that are responsible for insensitivity do not necessarily encode the direct target of molecule but may encode intermediate components in a regulatory cascade or pathway.
To identify the target or targets of a biologically active molecule, affinity purification using molecular probes is a more direct approach, and this technique has been used successfully in circadian biology in animal cells. For example, affinity-proteomics approaches with the mammal circadian clock modulators longdaysin and KL001, identified targets of the molecules in the mammalian clock (15, 16). However, affinity-proteomics approaches in plant research lag behind those of other organisms. The only successful case so far using this methodology was the identification of exocytosis-related EXO70 directly targeted by Endosidin2 (17).
As a complementary strategy to previous studies for plant clock, here we report a strategy that makes use of small synthetic molecules to alter the molecular mechanisms underlying Arabidopsis circadian clock functions. One such chemical, PHA767491, lengthens circadian period. Although PHA767491 was previously known as an inhibitor of mammalian cell division cycle (CDC) 7/cyclin-dependent kinase (CDK) 9 kinases, affinity-proteomics data suggest that PHA767491 binds to 13 members of the Arabidopsis CK1 family (CASEIN KINASE 1 LIKE, CKL). The inhibition of CK1 family protein activity by PHA767491 treatment in vivo results in accumulation of PRR5 and TOC1, suggesting that CK1 family kinases control their turnover, which had been considered as crucial for period control.
Results
PHA767491 Lengthens Arabidopsis Circadian Period.
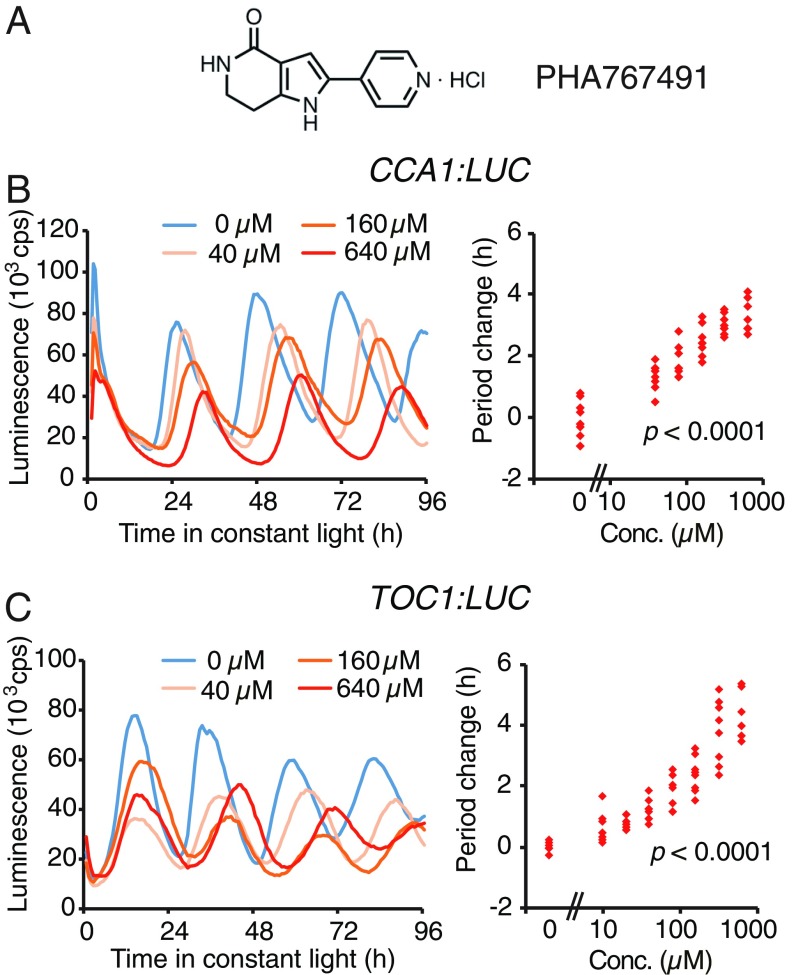
To identify small molecules that are capable of changing the circadian period in Arabidopsis, we treated seedlings with a Library of Pharmacologically Active Compounds that modulates a broad range of biological processes in mammals and microorganisms, and monitored circadian rhythms using a luciferase reporter driven by the CCA1 promoter (CCA1:LUC), whose expression peaks during the early morning. Of the 90 compounds in the library, we found that continuous treatment with PHA767491, an inhibitor of CDC7 and CDK9 in mammals (18), resulted in longer circadian periods in a dose-dependent manner (Fig. 1 A and B). PHA767491 also lengthened period of evening-peaked TOC1:LUC reporter, indicating that PHA767491 modulates clock pace (Fig. 1C).
Fig. 1.
The small molecule PHA767491 lengthens circadian period in Arabidopsis. (A) Chemical structure of PHA767491. Circadian luciferase reporter CCA1:LUC (B) and TOC1:LUC (C) activity in Arabidopsis with PHA767491 treatment. Representative traces (Left), and increases in period length relative to untreated (0 µM) control, indicate a dose–response (n = 8 for each concentration, with one-way ANOVA P values showing statistical significance of PHA767491-treated samples compared with untreated, Right).
We also found that treatment of PHA767491 at higher concentrations (>1 mM) resulted in strong growth retardation (SI Appendix, Fig. S1). Given that alternations of the circadian clock by genetic mutations do not cause these types of phenotypic alterations, PHA767491 may not only affect the circadian clock, but it may also alter other growth and development processes at higher concentrations.
PHA767491 Inhibits Activity of Arabidopsis CKL.
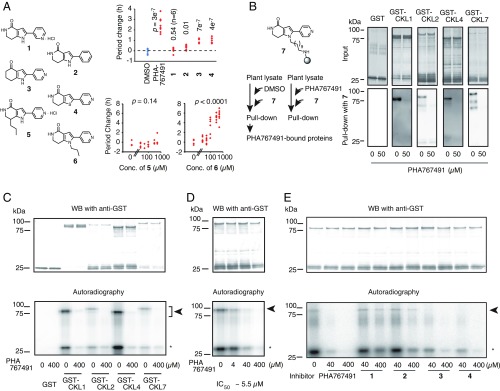
An affinity-based proteomic approach was used to identify the direct molecular targets of PHA767491 for period lengthening. We first performed a structure-activity relationship study using synthetic PHA767491 analogs to understand which moiety (or moieties) in PHA767491 could be linker-conjugated (Fig. 2A). Treatment of CCA1:LUC seedlings with analogs modified at the 4-pyridyl moiety failed to lengthen circadian period, suggesting that the 4-pyridyl group of PHA767491 is essential for period-lengthening activity (Fig. 2A, 1 and 2). Molecules altered at the γ-lactam and pyrrole moieties (Fig. 2A, 3 and 4) had weaker but significant period-lengthening activity, suggesting that the γ-lactam and pyrrole groups are not essential (Fig. 2A). Treatment with an analog substituted with an alkyl group at the γ-lactam ring did not lengthen the clock period (Fig. 2A, 5), but an alkyl group at the pyrrole position (Fig. 2A, 6) retained period-lengthening activity.
Fig. 2.
PHA767491 binds to and inhibits CKL family kinases. (A) Structure–activity relationship study of PHA767491 with period lengthening. Clock period change was determined compared with a DMSO treatment control [each concentration was 250 µM, n = 8, except for (1)], with Student’s t test P compared with DMSO control, Upper). Period changes by (5) or (6) (n = 8, with one-way ANOVA P, Lower). (B) Structure of PHA beads (7) and the procedure for screening proteins bound by PHA beads (Left), and binding between PHA beads and recombinant CKLs in vitro (Right). PHA767491 was added as competitor at 50 μM. GST-fusion proteins in PHA-bead fractions were analyzed by Western blotting (WB) with anti-GST antibody. (C) Inhibition of CKL kinase activity by PHA767491. (D) The IC50 of PHA767491 on CKL4 was determined from three separate experiments. (See also SI Appendix, Fig. S3.) (E) Inhibition of CKL4 by PHA767491 analogs. Arrowheads and asterisks indicate phosphorylated GST-fused CKL proteins and phosphorylated casein, respectively, from C to E. Signals around 25 kDa in lanes loaded with GST-fused CKL analyzed by WB were considerable truncated GST-fused CKL from B to E.
PHA767491-conjugated agarose beads were synthesized from molecule 6, with an alkyl linker at the nitrogen in the pyrrole ring (PHA beads, Fig. 2B, 7) for screening direct targets of PHA767491. PHA beads were incubated with Arabidopsis seedling lysates with 60 µM or without PHA767491. Sixty micromolars is more than the effective concentration for period lengthening in vivo (40 µM PHA767491 lengthens the period in Fig. 1). Proteins that bound to the affinity resin were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Two independent experiments showed that protein kinases, including the CK1 family (CKL), the shaggy-related protein kinase 3 (GSK3) family, AT2G32850, 5-METHYLTHIORIBOSE KINASE1 (MTK1), and other proteins were bound by PHA beads (SI Appendix, Table S1). CK1 is involved in clocks other than in land plants (e.g., fungi, flies, animals, and algae) (5–8, 19), but CKL clock-related functioning in Arabidopsis was unknown, likely due to the limits of traditional genetic analysis with functional redundancy among CKL family that contains 13 members in Arabidopsis (SI Appendix, Fig. S2). Arabidopsis homologs of the mammalian CDC7/CDK9 [CDKC1 (AT5G10270) and CDKC2 (AT5G64960) (47% identity, and Expected {E} value lower than 1e−100 to human CDK9) (20), AT4G16970 (27% identity, and E value 4e−34 to human CDC7)] were not enriched by PHA-bead binding, despite the fact that PHA767491 is an inhibitor of CDC7/CDK9 in mammals.
We next examined whether recombinant CKL proteins directly interact with PHA beads in vitro. There are 13 CKLs in the Arabidopsis genome (SI Appendix, Fig. S2), and all of the CKLs bound to PHA beads in at least one trial (SI Appendix, Table S1). We attempted to generate recombinant proteins for each CKL and obtained CKL1, CKL2, CKL4, and CKL7. PHA beads bound to GST-fused CKL proteins (GST-CKL1, GST-CKL2, GST-CKL4, and GST-CKL7) but did not to control GST. Binding was competitively inhibited by PHA767491 (50 µM) (Fig. 2B), showing that there are direct and specific interactions between PHA767491 and CKL proteins. In vitro assays using high concentrations of PHA767491 (400 µM) confirmed that PHA767491 inhibits CKL kinase activity, both CKL autophosphorylation and phosphorylation of the model substrate casein (Fig. 2C). Because CKL4, among all of the recombinant CKL proteins, had the strongest kinase activity on casein, we tested CKL4 in more detail. PHA767491 inhibited CKL4 kinase activity with an IC50 around 5 µM (Fig. 2D and SI Appendix, Fig. S3), far lower than the concentration required for period lengthening in vivo (Fig. 1).
We also tested for inhibitory activity of several PHA767491 analogs on CKL4 in vitro. Compounds 3 and 4 inhibited CKL4 activity, but there was less inhibition by analogs 1 and 2 (Fig. 2E). The correlation of PHA767491 analog CKL4 inhibition (Fig. 2E) with period lengthening (Fig. 2A) suggests that PHA767491 lengthens Arabidopsis circadian period by inhibiting CKL activity.
Arabidopsis CKL Is Required for Maintaining Proper Period Length.
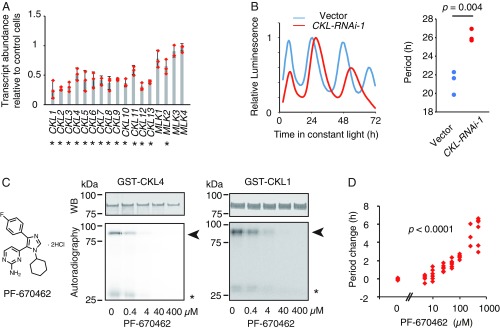
To investigate whether CKLs are crucial for maintaining proper period length, we examined circadian periodicity in plants in which CKLs were genetically modified. There are 13 CKLs and 4 kinase-encoding genes [MUT9-LIKE KINASE, MLK, also known as PHOTOREGULATORY PROTEIN KINASE (PPK)] that are the closest homologs to CKLs in the Arabidopsis genome (SI Appendix, Fig. S2). MLKs interact with the clock-associated proteins ELF3, ELF4, and LUX (21), thus controlling periodicity. In addition, MLKs interact with PHYTOCHROME-INTERACTING FACTOR (PIF) and CRYPTOCHROME (CRY), two proteins involved in the red- and blue-light signaling pathways (22, 23) that potentially function as input pathways to the clock. All of the CKLs were bound by PHA beads in at least one trial (SI Appendix, Fig. S2 and Table S1), whereas MLKs were not, suggesting that PHA767491 targets CKLs but not MLKs. CKL expression is ubiquitous in plant organs, and it is almost constant during diurnal and circadian cycles, except that CKL4 peaks in the morning and CKL5 peaks during nighttime (SI Appendix, Figs. S4 and S5). CKL expression is not greatly influenced by light or temperature (SI Appendix, Figs. S6 and S7). Similar expression patterns and biochemical activities among CKLs (Fig. 2 B and C) may suggest functional redundancy among CKLs, which makes genetic approaches to analyzing CKLs technically challenging. To investigate CKL clock involvement beyond genetic redundancy, we simultaneously knocked down genes belonging to CKL family in mesophyll cell protoplasts by transient transfection with a DNA vector harboring an RNA interference (RNAi) construct and a CCA1:LUC reporter, and measured the circadian period of transfected cells. To evaluate this approach, ZTL-RNAi and TOC1-RNAi constructs were cointroduced with CCA1:LUC into protoplasts for analysis of the circadian period (SI Appendix, Fig. S8A). Introduction of ZTL-RNAi resulted in longer periods of CCA1:LUC reporter expression compared with the empty vector control, whereas TOC1-RNAi shortened the period. Period-altered phenotypes of RNAi construct-transfected protoplasts were consistent with those of ztl and toc1 knockout mutants (24, 25), validating the method as reported (26).
Introduction of a CKL-RNAi construct (CKL-RNAi-1, SI Appendix, Fig. S8B) into mesophyll cell protoplasts resulted in reduced expression of all 13 CKL genes but did not reduce expression nearly as much for the four MLKs (Fig. 3A). The circadian periods of cells transfected with CKL-RNAi-1 were lengthened for the two reporters compared with the control (Fig. 3B and SI Appendix, Fig. S8C). The other RNAi construct (CKL-RNAi-2) lengthened the circadian period (SI Appendix, Fig. S8D), suggesting that CKL genes are required for maintaining period length. Furthermore, a potent and specific inhibitor of animal CK1, PF-670462 (27), inhibited Arabidopsis CKL1 and CKL4 in vitro, and lengthened the circadian period (Fig. 3 C and D), supporting the model that CKL kinase activity is involved in regulating clock function. We also tested the possibility that GSK3 is involved in the clock by using Bikinin, an inhibitor of GSK3 family proteins (28). Treatment with Bikinin did not cause period lengthening under our conditions (SI Appendix, Fig. S9), suggesting that GSK3 family kinases are not the likely target of PHA767491 for period lengthening.
Fig. 3.
CKL family is involved in the circadian clock. (A) Relative CKL and MLK expression in cells transfected with CKL-RNAi-1 compared with control cells (mean ± SEM, n = 3). Dots are mean values from three independent biological replicates. Asterisks indicate significant differences between control and CKL-RNAi-1 (Student’s t test P < 0.01, Left). (B) CCA1:LUC reporter activity codelivered with the CKL-RNAi-1 construct. Representative traces (Left), period lengths from three trials with Student’s t test P (Right). (C) Effect of a potent CK1 inhibitor PF-670462 on CKL4 and CKL1 kinase activity in vitro. Arrowheads indicate phosphorylated GST-fused CKL proteins, and asterisks show phosphorylated casein. (D) Effect of PF-670462 on period length of CCA1:LUC with one-way ANOVA P.
PHA767491 Down-Regulates Transcripts of Dawn- and Morning-Phase Genes.
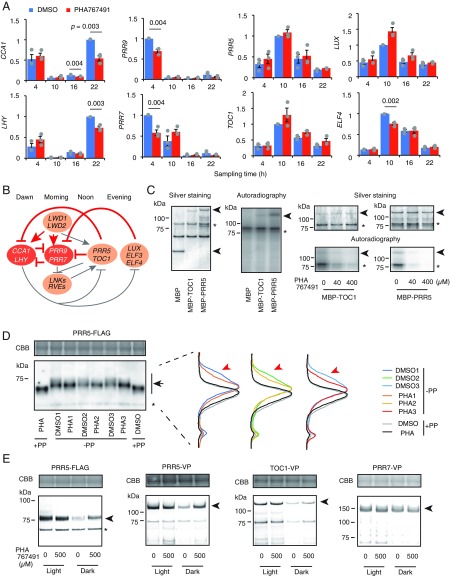
Although CK1 is involved in the control of circadian clocks in green algae, the mechanism of CK1 activity in clock control in plants is unknown (7, 8). To reveal possible action mechanisms of CK1 for clock control, we examined short-term (3 h) effects of PHA767491 on clock gene expression. This approach may allow us to see immediate effect on TTFL genes expression by the molecule and on the state of clock-transcription factors that regulate TTFL genes. Seedlings were germinated and grown under LD (12 h light/12 h dark) conditions, transferred to constant light conditions, and treated with PHA767491 at four time points (1, 7, 13, and 19 h after lights on). Plants were sampled 3 h after treatment (i.e., 4, 10, 16, and 22 h after lights on). Reverse transcription quantitative PCR (RT-qPCR) analyses indicated that except for ELF4, most evening gene expression was not strongly affected by PHA767491 (Student’s t test P > 0.01), but the dawn-expression genes CCA1 and LHY were decreased by treatment with PHA767491 at dawn (Fig. 4A, P < 0.01). The expression of morning genes PRR9 and PRR7 was also decreased by PHA767491 treatment during the morning hours, showing that there are PHA767491-sensitive and less-sensitive cyclic genes in TTFL (Fig. 4B). We observed time-dependent down-regulation of CCA1, LHY, PRR7, and PRR9 by PHA767491 (Fig. 4A). PHA767491 treatment was effective for down-regulating these genes during the time period before peak expression, but not after the peaks.
Fig. 4.
CKLs control PRR5 and TOC1 proteins. (A) Expression of clock-associated genes with 3-h treatment of PHA767491 (mean ± SEM, n = 3). Only P < 0.01 using Student’s t test are shown. Dots are mean values from three independent biological replicates. (B) PHA767491-sensitive genes in current model of clock TTFL. Genes colored red are PHA767491-sensitive genes, and red lines may be PHA767491-sensitive steps. (C) Phosphorylation of MBP-tagged PRR5 and TOC1 by GST-CKL4 (Left). Effect of PHA767491 on PRR5 and TOC1 phosphorylation by CKL4 (Right). Arrowheads and asterisks indicate substrates and GST-CKL4, respectively. (D) Band shifts of PRR5-FLAG by PHA767491 treatment in vivo with three biological replicates. Black arrows indicate PRR5-FLAG. The asterisks indicate nonspecific binding by anti-FLAG antibody. Intensity profiles of each PRR5-FLAG band are shown in the panel by normalizing the peak value to 1. Red arrowheads indicate electrophoretic mobility shift of PRR5-FLAG by PHA767491 treatment. PP is protein phosphatase treatment for extracted protein. (E) PRR5-FLAG, PRR5-VP, TOC1-VP, and PRR7-VP proteins in seedlings treated with PHA767491. Arrowheads indicate PRR-fusion protein bands. Two additional trials were performed with similar results (SI Appendix, Fig. S12). Upper are CBB-stained gels (D and E).
PHA767491 and CKL Regulates PRR5 and TOC1 Proteins.
Given that the PRR family directly represses expression of dawn and early morning clock genes CCA1, LHY, PRR7, and PRR9 (Fig. 4B) (4), we asked whether PHA767491 affects PRR through CKL activity for clock function. To detect possible interactions between CKL family and PRR family, we employed CKL4 as representative, because phosphorylation of CKL4 was strongest in our assay (Fig. 2). A yeast two-hybrid assay suggested that CKL4 interacts with PRR5 and TOC1, but less with PRR3, PRR7, or PRR9 (SI Appendix, Fig. S10). CKL4 phosphorylated MBP (Maltose-Binding Protein)-fused PRR5 and TOC1, but not MBP alone in vitro (Fig. 4C). PHA767491 inhibition of phosphorylation on PRR5 and TOC1 by CKL4 was dose dependent (Fig. 4C). Together, the yeast two-hybrid assays and in vitro phosphorylation indicate that there are direct, functional interactions among PHA767491, CKL4, PRR5, and TOC1.
We next examined PRR5 and TOC1 phosphorylation levels after PHA767491 treatment in vivo. To estimate the effect of PHA767491 on PRR5 protein expression without transcriptional controls on PRR5, transgenic plants overexpressing FLAG tag-fused PRR5 (35Spro:PRR5-FLAG) (29) were treated with PHA767491. Protein samples from 35Spro:PRR5-FLAG were precipitated with TCA (trichloroacetic acid) and analyzed with low concentration (6%) acrylamide gels to separate proteins as much as technically possible (Fig. 4D). PRR5-FLAG bands treated with phosphatase showed much faster mobilities than bands in samples without phosphatase treatment, confirming what has been described (9). We also found that PHA767491 treatment caused a shift of PRR5-FLAG to a position lower than the control treatment in vivo, suggesting that PHA767491 decreases phosphorylation of PRR5-FLAG (Fig. 4D). PHA767491-insensitive processes are also involved in PRR5 phosphorylation in vivo, since PHA767491 treatment resulted in relatively small shifts compared with those resulting by phosphatase treatment. We also tested whether PHA767491 causes a band shift of PRR5-VP in 35Spro:PRR5-VP, which has an opposite phenotype to 35Spro:PRR5-FLAG (29). PHA767491 caused lower band shift of PRR5-VP, suggesting that PHA767491 affects phosphorylation of PRR5 in vivo (SI Appendix, Fig. S11A). There was no shift of the TOC1-VP band as a result of PHA767491 treatment even in 6% acrylamide (SI Appendix, Fig. S11B). Although there was no apparent interaction between PRR7 and CKL4 in a yeast two-hybrid assay, we examined the effect of PHA767491 on PRR7 phosphorylation in vivo. An apparent band shift of PRR7-VP was likely due to partial dephosphorylation as a result of PHA767491 treatment (SI Appendix, Fig. S11C).
Phosphorylation of TOC1 is associated with multiple functions such as protein interactions, nuclear localization, stabilization, and degradation (9, 30), but the kinases involved in these processes have yet to be identified. Phosphorylation of PRR5 by an unknown kinase is related to degradation (9). To examine whether CKLs are involved in phosphorylation-dependent PRR5 degradation, CKLs were inhibited by PHA767491 treatment 35Spro:PRR5-FLAG, and PRR5-FLAG protein levels were measured using a hard gel (10–20% gradient acrylamide gel) to see any clear quantitative change (Fig. 4E and SI Appendix, Fig. S12). PRR5-FLAG protein was decreased in the dark due to ZTL-dependent degradation (31), but decreases in PRR5-FLAG expression under dark conditions were partly suppressed by PHA767491 treatment (Fig. 4E and SI Appendix, Fig. S12). PHA767491 treatment of 35Spro:PRR5-VP also suppressed PRR5-VP down-regulation in the dark (Fig. 4E and SI Appendix, Fig. S12), further supporting the PHA767491-dependent attenuation model of PRR5 down-regulation. Reduced levels of TOC1-VP protein by ZTL in the dark were found as expected (Fig. 4E and SI Appendix, Fig. S12) (32). This decrease in TOC1-VP was partly suppressed by PHA767491 treatment. PHA767491 treatment did not result in any changes in PRR7 protein amounts (Fig. 4E and SI Appendix, Fig. S12). These data show that PHA767491 alters the stability of PRR5 and TOC1. Increases in TOC1 expression result in period lengthening (32), and loss of function of TOC1 or PRR5 results in shorter periods (4), suggesting that PHA767491 lengthens clock periods through accumulation of these proteins.
PHA767491 Modulates the Circadian Clock Through PRR5 and TOC1 Proteins.
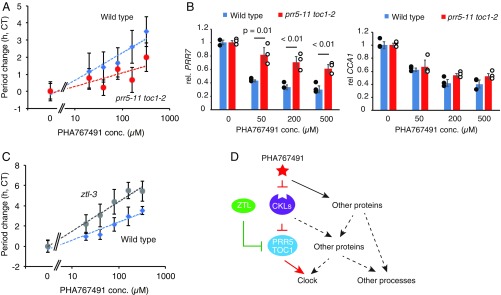
To evaluate the importance of PRR5 and TOC1 in PHA767491-dependent clock control, we analyzed the sensitivity of PHA767491-dependent period lengthening in prr5 toc1. In wild type, 40–100 µM PHA767491 was sufficient to lengthen the circadian period by 2 h [Circadian Time (CT)-corrected] (Fig. 5A). In the prr5 toc1 double mutant (prr5-11 toc1-2), 40–100 µM PHA767491 lengthened the circadian period by only 1 h CT and more than 200 µM PHA767491 was required for 2-h CT lengthening, indicating that there is hyposensitivity of the double mutant to PHA767941 for period lengthening (Fig. 5A). These results suggest that PRR5 and TOC1 are crucial mediators in CKL-dependent clock control, but that other pathways are also implicated.
Fig. 5.
PHA767491 and ZTL additively regulate PRR5 and TOC1 for clock control. (A) Period-lengthening effect of PHA767491 in prr5-11 toc1-2 double mutants. (B) PRR7 and CCA1 expression in prr5-11 toc1-2 treated with PHA767491. Dots indicate mean values from three independent biological replicates. Statistical differences between wild type and prr5-11 toc1-2 were determined by Student’s t test. (C) Period-lengthening effect of PHA767491 in ztl-3. (D) Action mechanisms of PHA767491 for clock control. ZTL as a part of ubiquitin E3 ligase targets TOC1 and PRR5 for degradation. CKLs phosphorylate PRR5 as part of the pathway for degradation of TOC1 and PRR5. PHA767491 targets proteins other than the CKLs (SI Appendix, Table S1), and CKLs phosphorylate other circadian clock-controlling proteins or proteins involve in other biological processes.
To further examine whether TOC1 and PRR5 mediate the effect of PHA767491 on gene expression, the expression of PHA767491 down-regulated genes CCA1 and PRR7 was analyzed in the prr5-11 toc1-2 double mutant. Seedlings were treated with PHA767491 at different concentrations at 19 h after lights-on and harvested after 3-h incubation. Suppression of PRR7 expression by 3 h of PHA767491 treatment was partly attenuated in the prr5 toc1 mutant (Fig. 5B). Suppression of CCA1 was not significantly attenuated in prr5 toc1 plants (Fig. 5B). These data support the hypothesis that PRR5 and TOC1 are involved in PHA767491-dependent regulation of TTFL genes, especially PRR7.
To further examine the involvement of PRR5 and TOC1 in CKL-dependent clock control, the sensitivity of PHA767491-dependent period lengthening in ztl, in which PRR5 and TOC1 degradation are attenuated, was examined. Although PRR5 and TOC1 are more accumulated in ztl compared with wild type due to a reduction in degradation, PRR5 and TOC1 are still degraded by ZTL homologs in ztl (33). In wild type, 40–100 µM PHA767491 was sufficient to lengthen the circadian period by 2 h CT (Fig. 5C). In the ztl mutants (ztl-3), the circadian period was lengthened 2 h CT by <20 µM PHA767491, and 40–100 µM PHA767491 lengthened the period by 4 h CT, showing hypersensitiveness of ztl-3 to PHA767491. This was interpreted as impairment of the PRR5 and TOC1 degradation pathway by the ztl-3 mutation results in further accumulation of PRR5 and TOC1 mediated by PHA767491 treatment and, thus, hypersensitivity of PHA767491 in the ztl-3 mutant compared with wild type.
To further explore the relationship between PHA767491 treatment and PRR5, TOC1, and ZTL on a genome-wide gene expression basis, we compared transcriptome data from wild-type plants treated with PHA767491, ztl-3 mutants, PRR5-VP–expressing plants (29), and toc1-2 mutants (34). Direct target genes of transcriptional repressors PRR5 and TOC1 tend to be expressed in the early morning (29, 35). Seedlings of wild-type and ztl-3 plants were harvested at Zeitgeber time 18 (ZT18, 18 h after lights are turned on) for RNAseq analysis. There were 127 genes significantly up-regulated and 452 genes down-regulated in ztl-3 compared with wild type [false discovery rate (FDR)-controlled q < 10−4, Dataset S1]. We noticed that both down-regulated genes and up-regulated genes in the ztl-3 mutants include cyclic genes under constant light conditions (LL12_LDHH, phaser, SI Appendix, Fig. S13A), supporting our supposition that RNAseq analysis detected alternation of clock output gene expression by ztl-3 mutation. Down-regulated genes in ztl-3 significantly overlapped with up-regulated genes in PRR5-VP and toc1-2 mutants, both of which contain target genes of PRR5 and TOC1, whereas up-regulated genes in ztl-3 did not (SI Appendix, Fig. S13). These data suggest that down-regulation of gene expression by the ztl-3 mutation is through accumulation of the transcriptional repressors TOC1 and PRR5.
RNAseq analysis revealed that 500 genes were up-regulated and 1,865 were down-regulated by PHA767491 treatment (FDR q < 10−4, Dataset S2). The group of genes that are up-regulated in PRR5-VP, including direct target genes of PRR5, significantly overlaps with the group of genes down-regulated by PHA767491 (Fisher’s exact P = 2e−16), but not up-regulated by PHA767491, suggesting that downstream genes of PRR5 are suppressed by PHA767491 (SI Appendix, Fig. S14). Down-regulated genes in PRR5-VP overlap with genes up-regulated by PHA767491 (P = 9e−15), suggesting that there is overlap between indirect gene regulation by PRR5-VP and PHA767491. Genes that are up-regulated by the toc1-2 mutation and the set of genes down-regulated by PHA767491 significantly overlapped (P = 1e−5), suggesting that the mechanism of action of PHA767491 on gene expression is related to PRR5 and TOC1 function.
Down-regulated genes in the ztl-3 mutant significantly overlapped with the set of genes whose expression was decreased by PHA767491 (P = 2e−16), but not with genes with increased expression by PHA767491 treatment (P = 0.9) (SI Appendix, Fig. S14). In addition, expression of 49% of the 452 down-regulated genes in the ztl-3 mutant was significantly decreased by PHA767491 (SI Appendix, Fig. S14 and Dataset S3). These results indicated that PHA767491 treatment and ZTL-dependent PRR5 and TOC1 degradation pathways have also overlapping effects on a genome-wide gene expression, especially genes in clock-output pathways.
Discussion
PHA767491 Treatment Indicates That There Is Involvement of the CK1 Family in Arabidopsis Circadian Clock Regulation.
We identified PHA767491, a CDC7/CDK9 inhibitor in mammals, as a CK1 inhibitor in Arabidopsis. We also found that PHA767491 inhibits mammalian CK1 in vitro, and PHA767491 treatment of cultured animal cells resulted in longer clock period (SI Appendix, Fig. S15). These results suggest that PHA767491 is in fact an inhibitor of mammalian CK1.
The structures of PHA767491 and ATP are somewhat similar. In addition, kinases harboring ATP-binding pocket were captured by PHA beads, suggesting that PHA767491 acts as a competitor at the ATP-binding pocket of target proteins. Interestingly, however, not all ATP-binding proteins (an estimated 2,070 proteins in the TAIR database) were captured by PHA beads. The MLK family in particular was not captured by PHA beads, despite their close resemblance to CKL family proteins, although MKL1 and MLK3 expression is comparable to CKL (SI Appendix, Figs S2 and S16).
We propose that PHA767491 modulates the circadian clock period by at least inhibiting CKL proteins, but PHA767491 beads also bound to other proteins, including GSK3. Our test suggests that the GSK3 family is not likely a factor in period lengthening due to PHA767491 treatment (SI Appendix, Fig. S9). However, there may be other proteins that bind to PHA767491 and affect clock period regulation, and full understanding for action mechanisms of PHA767491 needs further experiments.
We also found that PHA767491 not only modulates clock pace, but also influences other biological processes. PHA767491 treatment reduced the expression of gene related to primary metabolism (Dataset S2). PHA767491 binds to MTK, encoding a key enzyme involved in primary metabolism pathways, including the Yang cycle (36). Thus, it may be that PHA767491 affects Yang cycle through MTK and sequentially modulates expression of genes related to primary metabolism. We also found that genes involved in brassinosteroid biosynthesis are misexpressed in response to PHA767491 treatment (Dataset S2). This may explain why PHA767491 modulates GSK3 activity and eventually influences downstream genes, including the brassinosteroid biosynthesis genes (28), which may cause growth retardation observed when PHA767491 was treated in higher concentration (SI Appendix, Fig. S1).
Control of PRR5 and TOC1 Degradation by Phosphorylation.
Our observations of a phenotypic interaction between PHA767491 treatment and prr5-11 toc1-2 or ztl-3 mutations, and physical interactions among PHA767491, CKL4, PRR5, and TOC1, and hyperaccumulation of PRR5 and TOC1 proteins by PHA767491 in vivo, suggest that PHA767491 modulates the circadian clock via CKL activity on PRR5 phosphorylation at a minimum, that eventually results in PRR5 and TOC1 accumulation (Fig. 5D). Although PHA767491 treatment causes PRR5 accumulation (Fig. 4E), PHA767491 causes only few band shifts of the hyperphosphorylation form of PRR5 in vivo (Fig. 4D). Previous studies also reported few mobility differences of PRR5 that were critical for PRR5 degradation (9). Phosphorylation of TOC1 affects nuclear entry, degradation, and stabilization (30). In this study, we found that CKL4 phosphorylates TOC1 in vitro, and PHA767491 treatment resulted in an increase of TOC1 in vivo. However, there was no significant band shift of TOC1-VP after PHA767491 treatment (SI Appendix, Fig. S11). This result suggests either that reduced phosphorylation levels of TOC1 by PHA767491 is undetectable by mobility shift, or that PHA767491-dependent TOC1 accumulation is mediated by PRR5 dephosphorylation but not by TOC1 dephosphorylation. The later model may be compatible with previous studies suggesting PRR5-dependent control over TOC1 amounts (30, 33). Although the PRR7 band was shifted to a lower size by PHA767491 treatment (SI Appendix, Fig. S11), PRR7 amounts were unchanged (Fig. 4E), suggesting that control of PRR7 is unlikely a principal target for the PHA767491-inhibited mechanism. This supposition may be also supported by the evidence that PHA767491 hyposensitivity of the prr5-11 toc1-2 double mutant in which PRR7 is relatively high during the day (37). Phosphorylation of cryptochrome 2 by CKL3 and CKL4 also suggests that CKL controls the clock by modulating cryptochrome signaling as a light input pathway to the clock (38).
Identification of phosphorylation sites in PRR5 may be the first step toward a full understanding of multiple PRR5 phosphorylations, which is crucial for clock regulation. Control of negative factors by CK1 is crucial for regulating period length in mammals and fungi (6, 15, 39), which apparently have distinct molecular clock mechanisms from plants. Therefore, half-life control of negative factors, including transcriptional repressors, is critical for period determination from the point of view of a general design principle for cyclic biological phenomena.
Our proposed model, in which transcriptional repressors PRR5 and TOC1 are controlled by PHA767491 and CKLs (Fig. 5D), is consistent with the down-regulation of CCA1, LHY, and PRR7 in response to PHA767491 treatment before peak diurnal expression of these genes (Fig. 4B). This is likely because PRR5 and TOC1 proteins repress CCA1 and LHY expression during the time before these genes reach peak expression (35, 40). Sensitivity to PHA767491 in the prr5-11 toc1-2 double mutant for period lengthening and PRR7 suppression were attenuated, but not for CCA1 suppression (Fig. 5). Elevated PRR7 in prr5-11 toc1-2 may mask a direct effect of PHA767491 on CCA1, since PRR7 is a repressor of CCA1 transcription (40). We also found overlapping genes that are influenced as a group by PHA767491 treatment, in PRR5-VP, toc1-2, or ztl-3 mutants (SI Appendix, Fig. S14). Collectively, these gene expression analyses suggest that PHA767491 and CKL family proteins affect expression of dawn and morning clock-associated and clock-output genes by increasing PRR5 and TOC1.
Given that the circadian clock in plants regulates physiological processes such as photosynthesis, cell elongation, drought stress responses, and flowering time, all of which potentially affect biomass production or yield (41–43), further discovery of small molecules capable of altering the clock of flowering plants has profound implications for plant improvement.
Materials and Methods
Plant Material and Screening of Small Molecule Changing Circadian Period.
A. thaliana Col-0 harboring CCA1:LUC (44) was used for a reporter line for first screening small molecules changing circadian period. Experimental details are described in SI Appendix, SI Text.
Synthesis of PHA767491 Analogs and Target Identification of PHA767491.
Synthesis of PHA767491 and its analogs and target identification of PHA767491 are described in SI Appendix, SI Text.
In Vitro Kinase Assay.
In vitro kinase assays were done with isotope-labeled ATP. Protein purification, buffer, and conditions are described in SI Appendix, SI Text.
Gene Expression Analyses.
RT-qPCR and RNAseq were done as described (45). Plant materials and growth conditions were described in SI Appendix, SI Text.
Analyses of Proteins upon PHA767491 Treatment.
Western blotting analyses for proteins extracted seedlings were described (45). Detailed plant materials and growth conditions were described in SI Appendix, SI Text.
Supplementary Material
Acknowledgments
We thank Y. Machida for providing pYU501, Y. Tsuchiya and K. Miwa for technical advice, and I. Yamanaka and N. Ono for technical assistance. This study was supported partly by an Institute of Transformative Bio-Molecules research award (to T.N.U., S.T., J.Y., and N.N.); Grant-in-Aid for Scientific Research on Innovative Areas 16H01140 (to J.Y.), 15H05960 (to K.Y.-S.), and 15H05956 (to T.K., K.K., and N.N.); The Naito Foundation (J.Y.); a Research Program of Arid Land Research Center of Tottori University 28D2001; the Toyoake Foundation; Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research 17K19229 and 18H02136; and a JST Precursory Research for Embryonic Science and Technology Grant JPMJPR11B9 (to N.N.). Institute of Transformative Bio-Molecules is supported by the World Premier International Research Center Initiative, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing (RNAseq) data of plants treated with PHA767491, and RNAseq data of ztl mutant have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (accession nos. DRA006077 and DRA006078, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903357116/-/DCSupplemental.
References
- 1.Wijnen H., Young M. W., Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40, 409–448 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Nakajima M., et al. , Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 7, e62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nohales M. A., Kay S. A., Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 23, 1061–1069 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Q., et al. , CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20, 2552–2565 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowrey P. L., et al. , Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483–492 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt M., et al. , Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18, 1908–1930 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Ooijen G., et al. , Functional analysis of Casein Kinase 1 in a minimal circadian system. PLoS One 8, e70021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara S., et al. , Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283, 23073–23083 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Arabidopsis Genome Initiative , Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Dejonghe W., Russinova E., Plant chemical genetics: From phenotype-based screens to synthetic biology. Plant Physiol. 174, 5–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tóth R., et al. , Prieurianin/endosidin 1 is an actin-stabilizing small molecule identified from a chemical genetic screen for circadian clock effectors in Arabidopsis thaliana. Plant J. 71, 338–352 (2012). [DOI] [PubMed] [Google Scholar]
- 13.de Montaigu A., et al. , The root growth-regulating brevicompanine natural products modulate the plant circadian clock. ACS Chem. Biol. 12, 1466–1471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S. Y., et al. , Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirota T., et al. , High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol. 8, e1000559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirota T., et al. , Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C., et al. , Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl. Acad. Sci. U.S.A. 113, E41–E50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montagnoli A., et al. , A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat. Chem. Biol. 4, 357–365 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Kloss B., et al. , The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94, 97–107 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Cui X., Fan B., Scholz J., Chen Z., Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell 19, 1388–1402 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H., et al. , Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15, 201–217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q., et al. , Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 8, 15234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni W., et al. , PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8, 15236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somers D. E., Schultz T. F., Milnamow M., Kay S. A., ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Strayer C., et al. , Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Kim J., Somers D. E., Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 154, 611–621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badura L., et al. , An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther. 322, 730–738 (2007). [DOI] [PubMed] [Google Scholar]
- 28.De Rybel B., et al. , Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16, 594–604 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamichi N., et al. , Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U.S.A. 109, 17123–17128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., Fujiwara S., Somers D. E., PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29, 1903–1915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiba T., Henriques R., Sakakibara H., Chua N. H., Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19, 2516–2530 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Más P., Kim W. Y., Somers D. E., Kay S. A., Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Baudry A., et al. , F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22, 606–622 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legnaioli T., Cuevas J., Mas P., TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28, 3745–3757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W., et al. , Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Pommerrenig B., et al. , Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis. Plant Cell 23, 1904–1919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito S., et al. , Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 49, 201–213 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Tan S. T., Dai C., Liu H. T., Xue H. W., Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling. Plant Cell 25, 2618–2632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Görl M., et al. , A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 20, 7074–7084 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamichi N., et al. , PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodd A. N., et al. , Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Müller N. A., et al. , Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 48, 89–93 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Nakamichi N., Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol. 56, 594–604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T., PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46, 686–698 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Kamioka M., et al. , Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28, 696–711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.