Significance
This study expands understanding of opsin 3 (OPN3) function and of skin pigmentation. The findings presented here reveal that the nonvisual OPN3 modulates the pigmentation of human epidermal melanocytes—the melanin-producing cells of the skin—by controlling the activity of the main pigmentation receptor, melanocortin 1 receptor (MC1R). The study identifies an OPN3 function in regulating human skin pigmentation via a unique molecular mechanism; it also reveals a regulatory mechanism for MC1R in melanocytes. These results advance our understanding of nonvisual opsins and their extraocular role; they also represent a paradigm for OPN3 function via modulation of MC1R’s activity. These findings set the stage for future investigations of OPN3 function in other tissues.
Keywords: opsin3, encephalopsin, melanocytes, melanocortin 1 receptor, pigmentation
Abstract
Opsins form a family of light-activated, retinal-dependent, G protein-coupled receptors (GPCRs) that serve a multitude of visual and nonvisual functions. Opsin 3 (OPN3 or encephalopsin), initially identified in the brain, remains one of the few members of the mammalian opsin family with unknown function and ambiguous light absorption properties. We recently discovered that OPN3 is highly expressed in human epidermal melanocytes (HEMs)—the skin cells that produce melanin. The melanin pigment is a critical defense against ultraviolet radiation (UVR), and its production is mediated by the Gαs-coupled melanocortin 1 receptor (MC1R). The physiological function and light sensitivity of OPN3 in melanocytes are yet to be determined. Here, we show that in HEMs, OPN3 acts as a negative regulator of melanin production by modulating the signaling of MC1R. OPN3 negatively regulates the cyclic adenosine monophosphate (cAMP) response evoked by MC1R via activation of the Gαi subunit of G proteins, thus decreasing cellular melanin levels. In addition to their functional relationship, OPN3 and MC1R colocalize at both the plasma membrane and in intracellular structures, and can form a physical complex. Remarkably, OPN3 can bind retinal, but does not mediate light-induced signaling in melanocytes. Our results identify a function for OPN3 in the regulation of the melanogenic pathway in epidermal melanocytes; we have revealed a light-independent function for the poorly characterized OPN3 and a pathway that greatly expands our understanding of melanocyte and skin physiology.
Unlike most mammals, which have melanin-producing melanocytes predominantly in the hair follicle bulb (1), humans are uniquely equipped with melanocytes in the outermost layer of the skin, the epidermis (2, 3). These neural crest-derived melanocytes are the only source of the photoprotective pigment melanin in human skin, and thus are critical for the defense against solar ultraviolet radiation (UVR)-induced genotoxic damage (4–6).
Solar UVR at the surface of the earth is composed of ∼5% short-wavelength UVB rays and ∼95% long-wavelength UVA rays. Much of our current knowledge about melanogenesis in epidermal melanocytes stems from the well-characterized UVB-induced melanin pathway (7). UVB elicits DNA damage in epidermal keratinocytes, triggering facultative skin darkening through increased melanin production in neighboring melanocytes (8). UVB-irradiated keratinocytes and melanocytes locally secrete α-melanocyte stimulating hormone (α-MSH), an agonist of the Gαs-coupled melanocortin 1 receptor (MC1R) that is primarily expressed on melanocytes (8, 9). MC1R has a pivotal role in determining pigmentation, as several naturally occurring loss-of-function MC1R variants are associated with the redhead phenotype (10, 11) characterized by a pale complexion and increased sensitivity to UVR (12). Downstream, α-MSH–induced MC1R activation leads to stimulation of adenylyl cyclase (AC) and production of cyclic adenosine monophosphate (cAMP). Accumulation of cAMP, through several molecular steps, induces up-regulation of microphthalmia-associated transcription factor (MITF)—the master transcription factor in melanocytes—leading to increased expression of melanogenic enzymes like tyrosinase (TYR) (13).
In addition to the UVB pathway, we recently characterized a UVA-induced melanogenic pathway in human epidermal melanocytes (HEMs). This retinal-dependent phototransduction pathway is mediated by Gαq/11 activation, resulting in a rapid increase in intracellular Ca2+ and elevated cellular melanin levels (14–16). The identity of the putative G protein-coupled receptor (GPCR) that mediates the UVA phototransduction cascade remains unknown, but members of the retinal-dependent, light-sensitive opsin family are ideal candidates. Coincidentally, in HEMs, we and others have found expression of messenger RNA (mRNA) corresponding to several opsins (16–20): Among these, opsin 3 (OPN3), which has an unknown physiological function, has significantly higher mRNA expression than any other detected opsin (18).
The scarcity of functional data on OPN3, discovered nearly 20 y ago (21, 22), may derive from its unique structure and widespread expression ranging from deep brain regions (21, 22) to peripheral tissues (23). OPN3 has a unique, long carboxyl (C) terminus with no sequence homology to any known GPCR. Recent studies have determined the photoreceptive properties for several nonmammalian OPN3 homologs (24–26). Zebrafish, pufferfish, and chicken OPN3 absorb blue light (wavelength of maximum absorbance, λmax = 465 nm) (25). Mosquito OPN3 forms a bistable photopigment with 13-cis, 11-cis, and 9-cis retinal; absorbs blue-green light (λmax = 490 nm); and activates G proteins Gαi/o in a light-dependent manner (24, 26). However, the light sensitivity, G protein coupling, and function of human OPN3 remain unknown.
Here, we show that OPN3 is a negative regulator of melanogenesis in human melanocytes. OPN3 does not mediate the UVA-evoked Ca2+ response of HEMs, and it does not modulate the sensitivity of these cells to visible light, despite being able to bind 11-cis and all-trans retinal. OPN3 couples to Gαi to negatively regulate the α-MSH–induced cAMP response of MC1R. In addition, OPN3 and MC1R colocalize to the same subcellular microdomains and can form a physical complex. Our data identify a melanogenic regulatory mechanism and a key function of human OPN3 in melanocytes, both of which expand our knowledge of melanocyte physiology.
Results
OPN3 Does Not Mediate Ca2+-Dependent UVR Phototransduction in HEMs.
Physiological doses of UVR induce a retinal- and phospholipase C-beta (PLC-β)–dependent transient increase in cytosolic Ca2+ mediated by activation of Gαq/11 via an unknown putative GPCR (14–16). Because mosquito OPN3 activates Gαi/o subunits of G proteins in a light-dependent manner (24) and the Gβγ subunits that dissociate from Gαi could activate PLC-β and cause a Ca+2 response, we reasoned that OPN3 may be the GPCR that mediates UVR phototransduction in HEMs. Like all opsins, OPN3 possesses a lysine in the seventh transmembrane domain (K299) and a negatively charged counter-ion in the third transmembrane domain (D117) (Fig. 1A), both involved in the binding of the retinal chromophore to the opsin apoprotein (27) that confers light sensitivity.
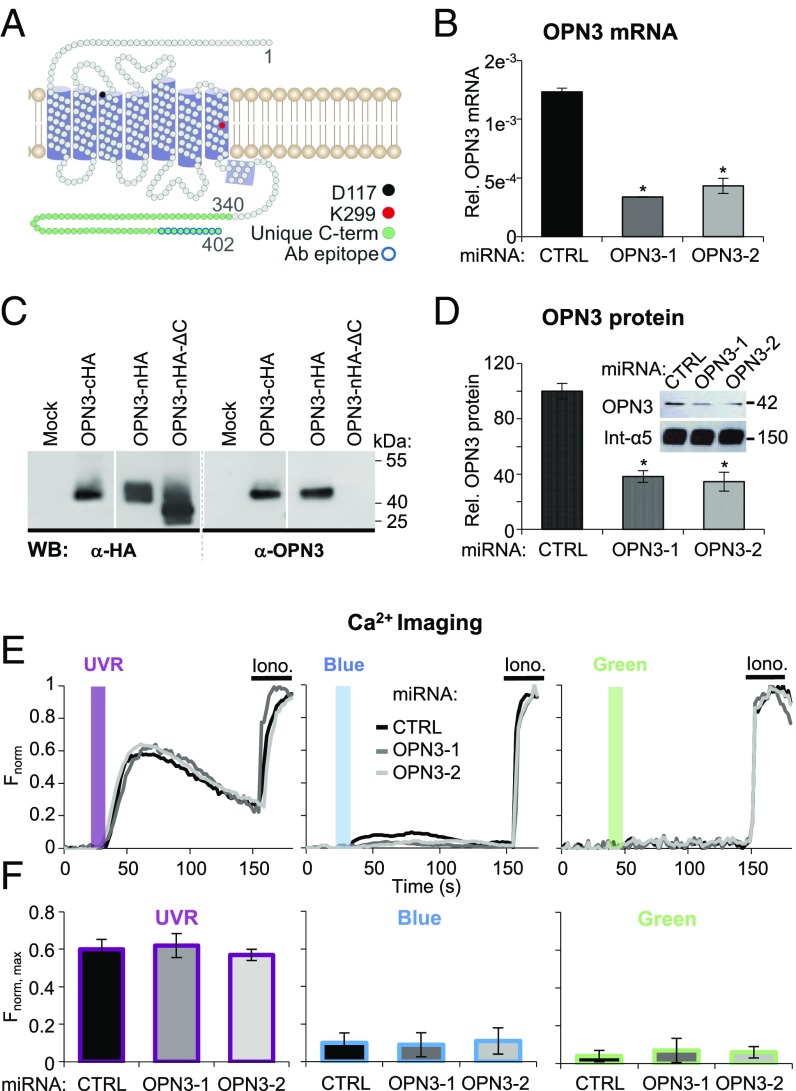
Fig. 1.
Human OPN3 does not mediate light-induced Ca2+ responses in HEMs. (A) Two-dimensional model of human OPN3 (hOPN3). hOPN3, similar to OPN2, has seven predicted transmembrane domains followed by a short intracellular helix (purple cylinders) and a unique C terminus (amino acids 340–402, green) containing the epitope for the anti-OPN3 antibody (Ab) (blue outline) (58). Each circle represents one amino acid. The conserved lysine K299 (red) is involved in retinal binding, which forms a Schiff base with counter-ion D117 (black). (B) OPN3 mRNA levels in HEMs expressing CTRL or OPN3-targeting miRNA (OPN3-1 or OPN3-2). A qPCR analysis of OPN3 relative (Rel.) to actin mRNA levels was performed. OPN3-1 and OPN3-2 miRNA-expressing HEMs have decreased OPN3 mRNA levels by ∼70% and ∼60%, respectively, compared with CTRL miRNA-transduced HEMs (n = 3 independent experiments, mean ± SEM; *P < 0.05). (C) Specificity of the anti-OPN3 antibody. OPN3 tagged with HA at the C terminus (OPN3-cHA) or N terminus (OPN3-nHA), N-terminal HA-tagged OPN3 mutant missing the last 10 amino acids (OPN3-nHA-ΔC), or empty vector (Mock) were expressed in HEK293 cells. Immunoblots using anti-HA or anti-OPN3 antibodies show the same size band (∼42 kDa) corresponding to OPN3 (calculated molecular mass of 45 kDa) for both full-length constructs but not for the ΔC mutant, which resulted in an ∼40-kDa band using anti-HA antibody and no band using anti-OPN3 antibody (representative of n = 4 independent experiments). WB, Western blot. (D) OPN3 protein levels in HEMs expressing CTRL or OPN3-targeting miRNA (OPN3-1 or OPN3-2). (Inset) Densitometric analysis of HEMs expressing CTRL, OPN3-1, or OPN3-2 miRNA and immunoblotted with anti-OPN3 or anti-integrin α5 antibody. Bars represent OPN3 protein level normalized to integrin α5 (n = 3 independent experiments, mean ± SEM; *P < 0.05). (E and F) Light-induced Ca2+ signaling in HEMs is not dependent on OPN3 expression. (E) Fluorescent Ca2+ imaging of HEMs expressing CTRL, OPN3-1, or OPN3-2 miRNA and stimulated with 200 mJ/cm2 UVR (λmax = 360 nm), blue (λmax = 450 nm), or green (λmax = 550 nm) light, normalized to the maximal Fluo4-AM Ca2+ response obtained with ionomycin (Iono). Each trace represents the average of 10–20 cells from one coverslip. (F) Average amplitude of Ca2+ responses of HEMs under the conditions shown in E (n = 5 independent experiments for each bar, mean ± SEM). max, maximum.
To determine if OPN3 mediates UVR phototransduction, we reduced OPN3 mRNA levels in HEMs using two OPN3-targeted microRNAs (miRNAs; OPN3-1 and OPN3-2). Each miRNA reduced the level of OPN3 mRNA by more than 60% compared with control scrambled (CTRL) miRNA (Fig. 1B). To confirm that OPN3 protein levels were also reduced, we first validated an antibody against the unique C terminus of OPN3 (Fig. 1C). Using this antibody, we showed that OPN3 protein levels are also reduced by more than 60% in HEMs expressing OPN3-1 or OPN3-2 miRNA (Fig. 1D). We monitored intracellular Ca2+ levels using the fluorometric Ca2+ indicator Fluo-4 AM in HEMs preincubated with all-trans retinal and expressing OPN3-1, OPN3-2, or CTRL miRNA. Exposure to UVR (200 mJ/cm2) led to a synchronized and transient Ca2+ response of similar amplitude in both HEMs expressing CTRL miRNA or OPN3-1 or OPN3-2 miRNA (Fig. 1 E and F, Left), indicating that OPN3 is not required for the UVR-induced Ca2+ response.
We next determined if OPN3 mediates blue or green light-induced Ca2+ changes in HEMs. Although we have previously shown in HEMs that the UVR-Ca2+ photocascade is UVR-specific and no Ca2+ responses could be evoked by similar doses of visible light (16), we questioned if human OPN3 could act to suppress the Ca2+ response to blue or green light in HEMs. In this case, reducing OPN3 expression may uncover a measurable Ca2+ response to blue or green light. This idea is plausible, given that OPN3 homologs are sensitive to blue-green light in other organisms (24). We monitored the Ca2+ responses of HEMs expressing CTRL or OPN3-targeted miRNAs, preincubated with all-trans retinal and exposed to 200 mJ/cm2 of blue (λmax = 450 nm) or green (λmax = 550 nm) light. HEMs expressing CTRL miRNA did not have a significant Ca2+ response to blue or green light, and neither did HEMs expressing OPN3-targeted miRNAs (Fig. 1 E and F, Center and Right).
Because the miRNAs partially reduced OPN3 expression and the residual OPN3 could be sufficient for the UVR response, we used CRISPR/Cas9 to eliminate OPN3. Primary HEMs with CRISPR/Cas9-induced mutations did not survive the selection of clonal lines; we used immortalized HEMs (Hermes 2b), which express similar levels of MC1R and OPN3 mRNA as HEMs (SI Appendix, Fig. S1A), to generate clonal CRISPR/Cas9 lines. Hermes 2b cells containing a mutation introduced in exon 1 of OPN3 lack OPN3 expression (SI Appendix, Fig. S1B). We performed Ca2+ imaging experiments in Hermes 2b cells (wild type or lacking OPN3 expression) and obtained similar results to those shown in Fig. 1 E and F: The UVR-induced Ca2+ response was not affected by the absence of OPN3 expression, and no responses to blue or green light were measured with or without OPN3 (SI Appendix, Fig. S1 D and E). These results further bolster our conclusion that OPN3 does not mediate Ca2+-dependent phototransduction of UVR, blue light, or green light in melanocytes.
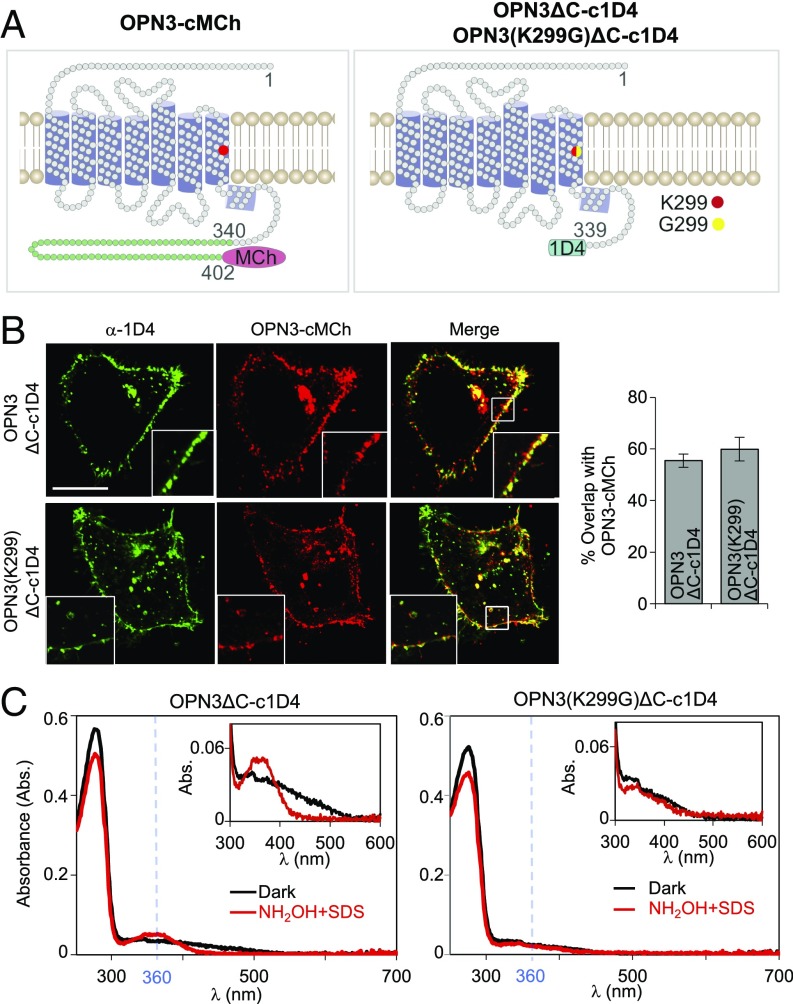
These findings raised an interesting question: Does human OPN3 form a photopigment? Spectroscopic analyses of recombinant OPN3 homologs indicated that they can bind 11-cis retinal and have an absorption maximum at ∼470 nm (24, 25). To determine if human OPN3 and retinal form a photopigment, we expressed C-terminal–truncated, 1D4-tagged human OPN3 (OPN3ΔC-c1D4) (28) in HEK293-GnTI− cells. We also expressed a variant in which the retinal-binding residue K299 was mutated to glycine [OPN3(K299G)ΔC-c1D4] (Fig. 2A, Right), a mutation shown to abolish retinal binding and light sensitivity of rhodopsin (OPN2) and other opsins (29, 30). Neither the OPN3 C-terminal truncation nor the K299G mutation affected the cellular localization of OPN3 in a heterologous system compared with C-terminal–tagged, full-length OPN3-MCherry (OPN3-cMCh) (Fig. 2A, Left and Fig. 2B). UV-visible spectroscopy using purified OPN3ΔC-c1D4 protein or the K299G mutant protein revealed a significant protein peak for both proteins, but no detectable absorption peak at λmax > 300 nm for either one (Fig. 2C). To test if retinal was bound to human OPN3, we treated the purified proteins with a mixture of hydroxylamine (NH2OH) and sodium dodecyl sulfate (SDS) (Fig. 2C, Insets). When retinal is bound to the apoprotein, SDS denatures the protein to expose the Schiff base that reacts with NH2OH to release retinal oxime, which absorbs at λmax = 360 nm (31, 32). Upon treatment with NH2OH and SDS, a retinal oxime absorption peak was observed for OPN3ΔC-c1D4 treated with 11-cis retinal (Fig. 2C, Left) or all-trans retinal (SI Appendix, Fig. S2A), but not for OPN3(K299G)ΔC-c1D4 (Fig. 2C, Right). These data suggest that OPN3, unlike the K299G mutant, can bind 11-cis retinal and all-trans retinal. Nevertheless, the reduced amplitude of the retinal oxime peak, compared with the protein peak (λmax = 280 nm) and purity of protein samples (SI Appendix, Fig. S2C), suggests that OPN3 binds retinal in a weak manner.
Fig. 2.
OPN3 requires the K299 residue for retinal binding. (A) Schematic representation of OPN3 variants. We purified 1D4-tagged OPN3 with a partially truncated C terminus variant (OPN3ΔC-c1D4) or a K299G mutation of this variant known to inhibit retinal binding [OPN3(K299G)ΔC-c1D4]. (B) OPN3ΔC-c1D4 and OPN3(K299G)ΔC-c1D4 maintain the cellular localization of full-length OPN3. Confocal images of HeLa cells coexpressing OPN3-cMCh and OPN3ΔC-c1D4 or OPN3(K299G)ΔC-c1D4 and immunostained with anti-1D4 antibody show that OPN3ΔC variants have similar cellular localization as OPN3-cMCh. Quantitative analysis of OPN3-cMCh colocalization with OPN3ΔC-c1D4 or OPN3(K299)ΔC-c1D4, measured as percent overlap between the two fluorescent signals, shows significant colocalization (bar graph) (n = 30 cells from three independent experiments). (Calibration bar: 10 μm.) (C) UV-visible absorption spectra of OPN3ΔC and OPN3(K299G)ΔC. Absorption spectra of purified OPN3ΔC-c1D4 and OPN3(K299G)ΔC-c1D4 incubated with 11-cis retinal were measured in the dark (black) and after hydroxylamine (NH2OH) + SDS treatment (red). Absorption spectra measured in the dark have similar protein peaks at λmax = 280 nm for the two OPN3 variants. NH2OH + SDS treatment of OPN3ΔC-c1D4, but not OPN3(K299G)ΔC-c1D4, led to a peak at λmax = 360 nm corresponding to retinal oxime. (Insets) Retinal oxime peak of OPN3ΔC-c1D4 was ∼10-fold smaller than the protein peak. Traces are representative of n = 3 independent experiments.
OPN3 Is a Negative Regulator of Melanin Levels in Human Melanocytes.
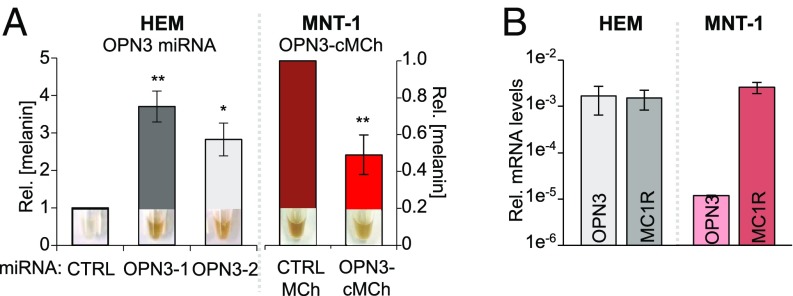
We observed that HEMs with reduced OPN3 expression (OPN3-1 and OPN3-2 miRNAs) appeared to be more pigmented than CTRL miRNA-expressing HEMs (Fig. 3A, Left Insets). Quantification of cellular melanin revealed that HEMs with reduced OPN3 had significantly higher melanin levels than CTRL (Fig. 3A, Left graph). Similar to miRNA-treated HEMs, Hermes 2b cells lacking OPN3 have significantly higher melanin levels than CTRL cells (SI Appendix, Fig. S1C). We then tested the reciprocal: Do elevated OPN3 levels lead to reduced melanin? Because we were not able to efficiently express exogenous OPN3 in HEMs, we used the pigmented melanoma cell line MNT-1. MNT-1 cells are highly pigmented and have been used extensively as a model system for melanocyte function because they preserve the majority of primary melanocyte signaling and trafficking pathways. MNT-1 cells have MC1R levels comparable to HEMs, but lower OPN3 mRNA levels (Fig. 3B). We transfected MNT-1 cells with OPN3-cMCh or with only MCherry (MCh) and quantified cellular melanin levels. MNT-1 cells expressing OPN3-cMCh have an approximately 60% reduction in melanin levels compared with MNT-1 expressing MCh (Fig. 3A, Right). These results indicate that OPN3 negatively modulates the melanin levels of human melanocytes.
Fig. 3.
OPN3 expression inversely correlates with cellular melanin concentration. (A) Effect of OPN3 levels on melanin. HEMs expressing OPN3-1 or OPN3-2 miRNA have significantly higher melanin levels compared with CTRL miRNA-expressing cells (Left), while MNT-1 cells expressing OPN3-MCh have reduced melanin compared with CTRL MCh-expressing cells (Right). (Insets) Representative pellets from each condition reflecting melanin levels (n = 3 independent experiments, mean ± SEM). Rel., relative. (B) Endogenous OPN3 and MC1R mRNA levels in HEM and MNT-1. mRNA levels of OPN3 and MC1R in HEMs and MNT-1 cells were measured by qPCR relative to actin. MNT-1 cells have similar MC1R levels as HEMs, but significantly lower OPN3 expression (n = 3 independent experiments, mean ± SEM; *P < 0.05, **P < 0.01).
OPN3 Is a Negative Regulator of MC1R-Mediated Signaling in Human Melanocytes.
Basal melanin levels are regulated by MC1R; α-MSH stimulates Gαs-coupled MC1R, which increases cAMP levels. Ultimately, this cascade up-regulates the transcription factor MITF, which increases levels of the main melanogenic enzyme TYR and results in increased cellular melanin. Because mosquito OPN3 couples to Gαi/o subunits of G proteins and Gαi/o signals by decreasing cellular cAMP, we tested if the negative effect of OPN3 on pigmentation is due to its inhibition of MC1R-mediated cAMP signaling.
To measure changes in cellular cAMP levels, we used the validated fluorescence resonance energy transfer (FRET)-based genetic cAMP sensor Epac H187 (33) (SI Appendix, Fig. S3). MNT-1 cells transfected with Epac H187 and either OPN3-cMCh or MCh were stimulated with α-MSH. MNT-1 cells expressing MCh had a 50% increase in cellular cAMP, measured as the change in FRET ratio [cyan fluorescent protein (CFP)/yellow fluorescent protein (YFP)] normalized to the maximum cAMP signal elicited by a mixture of the AC activator forskolin (FSK) and phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX) (Fig. 4 A, i, red trace and Fig. 4 A, iv, red bar graphs). In contrast, MNT-1 cells expressing exogenous OPN3-MCh and stimulated with α-MSH had a significantly smaller increase in cellular cAMP levels (Fig. 4 A, i, dark blue trace and Fig. 4 A, iv, dark blue bar graphs), suggesting that OPN3 attenuates MC1R-mediated cAMP signaling in MNT-1 cells. We performed the same cAMP experiments in Hermes 2b cells expressing C-terminal YFP-tagged OPN3 (OPN3-cYFP) or YFP alone, along with the red fluorescence-based cAMP indicator R-FlincA (34). Because Hermes 2b cells have lower levels of MC1R (SI Appendix, Fig. S1A), we used the more sensitive cAMP indicator R-FlincA instead of Epac H187. Hermes 2b cells expressing YFP (CTRL YFP) exhibit a significant α-MSH–induced cAMP response, while expression of OPN3-cYFP leads to a negligible cAMP response under similar conditions (Fig. 4 C, i, yellow vs. light green traces and Fig. 4 C, ii). These results confirm that OPN3 negatively modulates α-MSH–induced MC1R cAMP signaling.
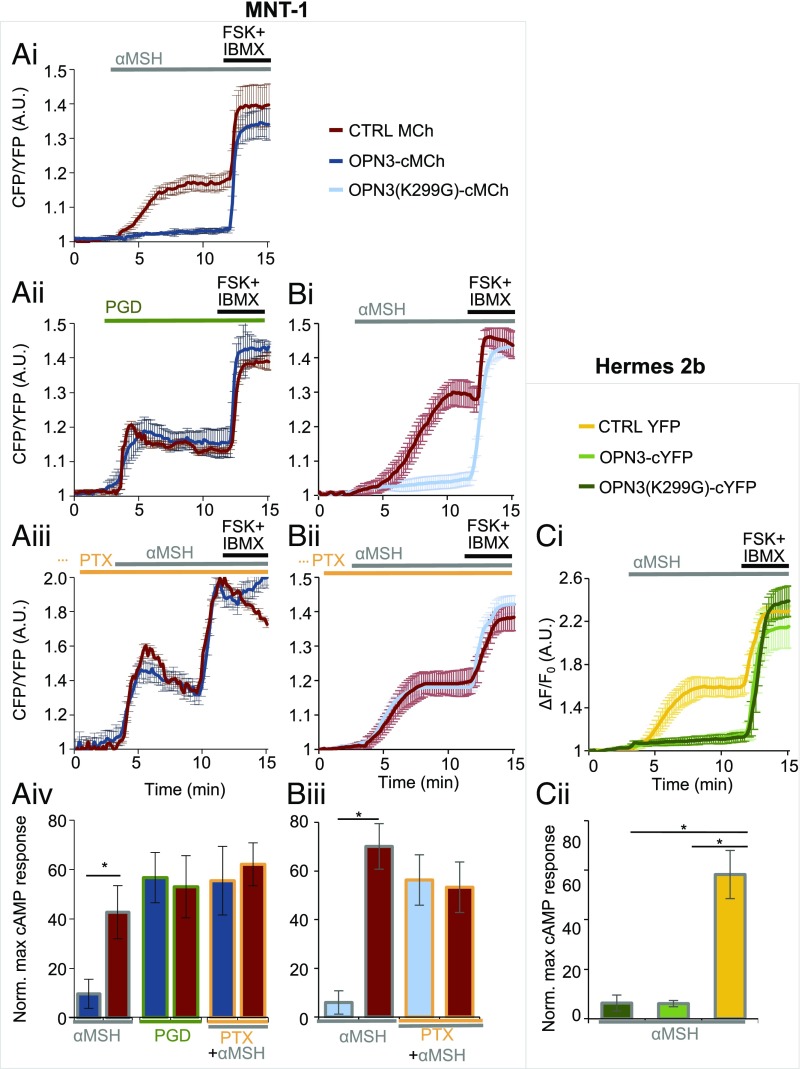
Fig. 4.
OPN3 modulates MC1R signaling. (A) OPN3 inhibits the MC1R-evoked cAMP response via a PTX-sensitive mechanism in MNT-1 cells. (A, i) OPN3 inhibits the α-MSH–induced cAMP responses of MC1R. MNT-1 cells expressing the FRET-based cAMP indicator Epac H187 and OPN3-cMCh or MCh alone (CTRL) were stimulated with α-MSH (1 μM). The cAMP response of individual cells was monitored as the ratio of CFP and YFP fluorescence intensities and represented as a function of time. FSK and IBMX were added to elicit a maximal cAMP response, used for normalization. α-MSH elicits a significant cAMP response in cells expressing MCh (CTRL, red trace), but not in cells expressing OPN3-cMCh (dark blue trace) (n = 5–10 cells, mean ± SEM). A.U., arbitrary units. (A, ii) OPN3 specifically attenuates the MC1R-mediated cAMP response. Prostaglandin (PGD; 5 μM)-mediated activation of the endogenous prostaglandin E2 receptor leads to an increase in cellular cAMP (red trace) that is not attenuated in the presence of OPN3-cMCh (dark blue trace) (n = 5–10 cells, mean ± SEM). (A, iii) OPN3-mediated attenuation of MC1R signaling is PTX-sensitive. MNT-1 cells treated with PTX (200 ng/mL, 4 h), which specifically inhibits the Gαi subunit of G proteins, and stimulated with α-MSH exhibited a similar cAMP response both in cells expressing MCh (red trace) and OPN3-cMCh (dark blue trace) (n = 5–10 cells, mean ± SEM). (A, iv) OPN3 inhibits MC1R signaling specifically and in a Gαi-dependent manner. Average normalized amplitudes of α-MSH– or PGD-induced cAMP responses in the presence or absence of PTX show that OPN3 expression significantly reduces the amplitude of cAMP responses to α-MSH, but not to PGD, and this effect is prevented by blocking Gαi activation with PTX (n = 3 independent experiments per condition, mean ± SEM; *P < 0.05). Norm. max, normalized maximum. (B) OPN3 inhibits MC1R-evoked cAMP responses independent of its retinal-binding ability. (B, i) OPN3-mediated inhibition of MC1R-induced cAMP responses does not require retinal binding. MNT-1 cells expressing OPN3(K299G)-cMCh (light blue trace) and stimulated with α-MSH (1 μM) exhibited significantly lower cAMP responses compared with MCh-expressing cells (red trace) (n = 5–10 cells, mean ± SEM). (B, ii) OPN3(K299G)-mediated attenuation of MC1R signaling is PTX-sensitive. MNT-1 cells treated with the Gαi inhibitor PTX (200 ng/mL, 4 h) and stimulated with α-MSH exhibited a similar cAMP response both in cells expressing MCh (red trace) and OPN3(K299G)-cMCh (light blue trace) (n = 5–10 cells, mean ± SEM). (B, iii) OPN3(K299G) inhibits MC1R signaling in a Gαi-dependent manner. Average normalized amplitudes of α-MSH–induced cAMP responses of MNT-1 cells expressing either OPN3(K299G)-cMCh or MCh. OPN3(K299G)-cMCh expression significantly reduces the amplitude of cAMP responses (n = 3 independent experiments per condition, mean ± SEM; *P < 0.05). (C) OPN3 inhibits MC1R-evoked cAMP responses in immortalized Hermes 2b HEMs independent of retinal binding. (C, i) OPN3 inhibits the α-MSH–induced cAMP responses of MC1R in Hermes 2b melanocytes in a retinal-independent manner. Hermes 2b cells expressing the red fluorescent cAMP indicator R-FlincA and OPN3-cYFP, OPN3(K299G)-cYFP, or YFP alone (CTRL) were stimulated with α-MSH (1 μM). α-MSH elicits a significant cAMP response in cells expressing YFP (CTRL, yellow trace), but not in cells expressing OPN3(K299G)-cYFP (light green trace) or OPN3-cYFP (dark green trace) (n = 5–10 cells per condition, mean ± SEM). ΔF/F, change in fluorescence over baseline fluorescence. (C, ii) OPN3 and OPN3(K299G) inhibit MC1R signaling in Hermes 2b cells. Average normalized amplitudes of α-MSH–induced cAMP responses of Hermes 2b cells expressing OPN3-cYFP or OPN3(K299G)-cYFP are significantly lower than for YFP alone (CTRL) (n = 2 independent experiments per condition, mean ± SEM; *P < 0.01).
To determine whether OPN3 modulation is specific to MC1R-mediated cAMP signaling, we stimulated the endogenous Gαs-coupled prostaglandin E2 receptor (EP2) in MNT-1 cells expressing MCh or OPN3-cMCh. Stimulation of EP2 with prostaglandin led to ∼60% increase in cAMP both in MNT-1 cells expressing OPN3-cMCh and in those expressing MCh (Fig. 4 A, ii and iv). This suggests that OPN3 does not modulate all Gαs-coupled receptors, but specifically regulates MC1R-mediated cAMP signaling.
We next sought to determine how OPN3 negatively regulates MC1R signaling. Gαs signaling can be inhibited by the signaling of Gαi-coupled GPCRs (35). To test if OPN3 couples to Gαi to reduce MC1R-mediated cAMP accumulation, we treated MNT-1 cells expressing MCh or OPN3-cMCh with pertussis toxin (PTX), an inhibitor of Gαi signaling (8). PTX-treated MNT-1 cells expressing MCh had a robust cAMP response to α-MSH, as expected. PTX-treated MNT-1 cells expressing OPN3-cMCh had a similarly robust response to α-MSH, unlike the negative regulatory effect of OPN3 displayed in the absence of PTX (Fig. 4 A, iii compared with i and iv). Since PTX-mediated inhibition of Gαi signaling abolishes the negative regulation OPN3 has on MC1R cAMP signaling, this suggests that OPN3 is likely coupled to Gαi. Interestingly, the lysine mutation of OPN3(K299G) did not affect the OPN3-mediated negative modulation of MC1R signaling, and PTX abolished the effect of OPN3(K299G) in MNT-1 and Hermes 2b (Fig. 4 B, i–iii and C, i and ii). This indicates that OPN3 has a regulatory influence over MC1R independent of its ability to bind retinal.
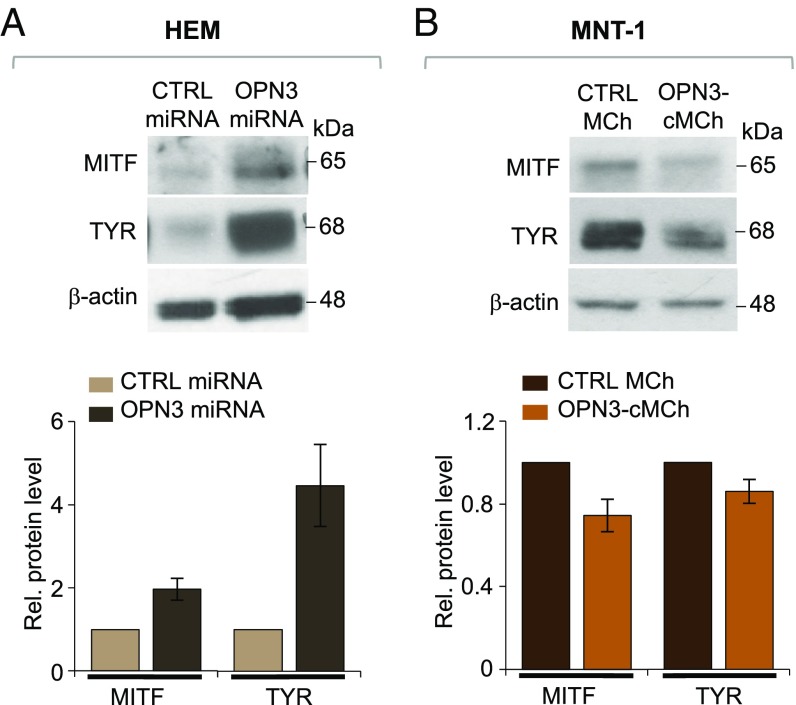
After MC1R-mediated cAMP levels increase, several molecular steps, including increased expression of MITF and up-regulation of TYR, lead to an increase in melanin levels (36). If OPN3 negatively regulates this pathway, as our cAMP data suggest, reducing OPN3 expression should increase MC1R-mediated signaling, increasing MITF and TYR expression. Indeed, HEMs expressing OPN3-2 miRNA (the more efficient of the two OPN3-targeted miRNAs) have higher levels of MITF and TYR compared with HEMs expressing CTRL miRNA (Fig. 5A). Conversely, MNT-1 cells expressing OPN3-cMCh had reduced levels of both MITF and TYR, compared with MNT-1 cells expressing MCh (Fig. 5B). These results indicate that OPN3 is a negative regulator of MC1R-meditated melanogenic signaling in human melanocytes.
Fig. 5.
OPN3 modulates MITF and TYR expression. (A) Reduced expression of OPN3 leads to increased MITF and TYR protein levels in HEMs. Representative Western blots of HEMs expressing CTRL or OPN3-targeted miRNA show increased MITF and TYR expression in cells with reduced levels of OPN3 compared with CTRL miRNA. MITF or TYR protein levels measured relative (Rel.) to β-actin were approximately twofold higher for MITF and approximately fivefold higher for TYR in HEMs with reduced OPN3 expression, compared with CTRL miRNA-expressing cells (n = 3 independent experiments for each condition, mean ± SEM). (B) Increased expression of OPN3 leads to reduced MITF and TYR levels in MNT-1 cells. Representative Western blots of MNT-1 cells stably expressing MCh (CTRL) or OPN3-cMCh show decreased MITF and TYR expression in OPN3-cMCh–expressing cells, compared with CTRL. MITF and TYR levels measured relative to β-actin were reduced by ∼30% for MITF and by ∼20% for TYR in cells expressing OPN3-cMCh, as compared CTRL (n = 3 independent experiments for each condition, mean ± SEM).
MC1R and OPN3 Are Able to Form a Complex.
Because OPN3 specifically modulates the signaling of MC1R, we proposed that OPN3 and MC1R could form a molecular complex. This would explain why OPN3 modulates MC1R cAMP signaling and not EP2 signaling despite MC1R and EP2 both being Gαs-coupled. OPN3, MC1R, and EP2 are all expressed on the plasma membrane, but OPN3 and MC1R, but not EP2, could be localized within the same microdomains.
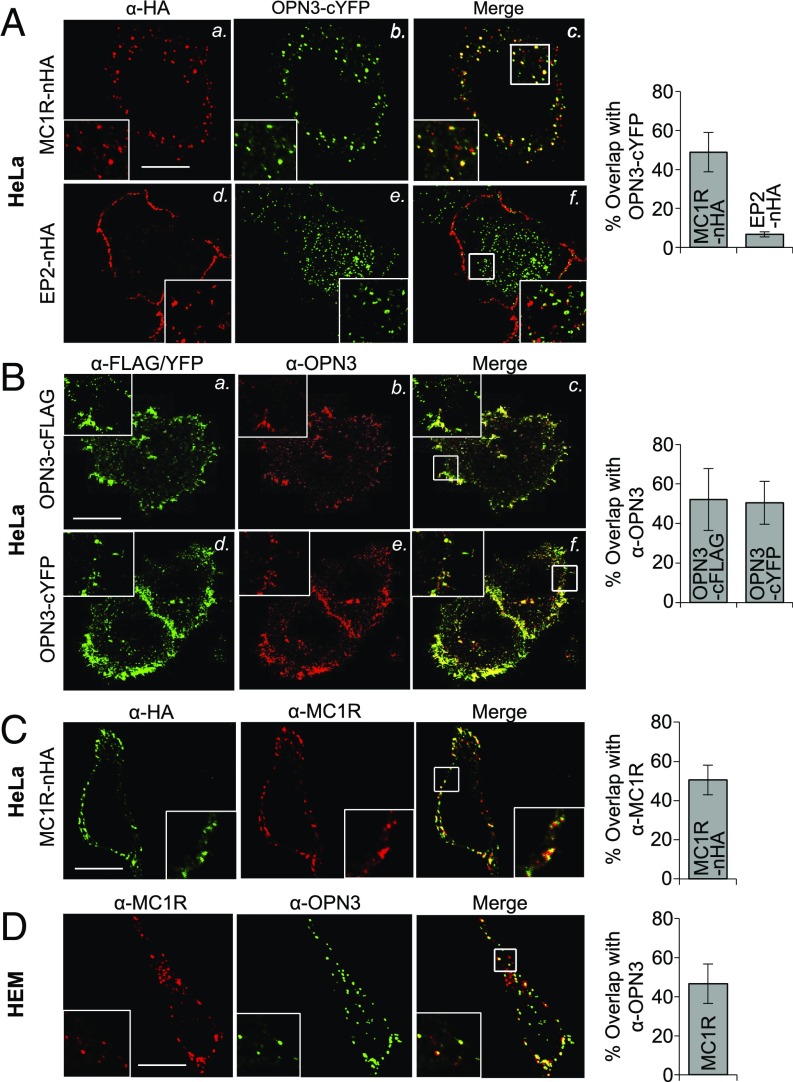
Several recent reports indicate that GPCRs can form functional and physical interactions (37, 38). Members of the opsin family, in particular, have recently been shown to form hetero- and homomeric complexes (39, 40). To determine whether OPN3 and MC1R colocalize to the same cellular compartments, we expressed epitope-tagged OPN3 and MC1R in HeLa cells. OPN3-cYFP localized to the plasma membrane, as well as to intracellular structures that have partial overlap with the Rab11 marker for recycling endosomes (SI Appendix, Fig. S4). To determine if OPN3 colocalization with MC1R is specific to MC1R, we measured OPN3 colocalization with EP2, which is also Gαs-coupled. In HeLa cells, we coexpressed OPN3-cYFP and either N-terminal hemagglutinin (HA)–tagged MC1R (MC1R-nHA) or nHA-tagged EP2 (EP2-nHA), immunostained with an anti-HA antibody, and quantified colocalization of YFP and HA. Interestingly, MC1R-nHA and OPN3-cYFP were significantly colocalized (>50%) in intracellular structures, while EP2-nHA, localized primarily to the plasma membrane and had <10% overlap with OPN3-cYFP (Fig. 6A).
Fig. 6.
OPN3 and MC1R are localized to the same cellular compartments. (A) OPN3-cYFP is colocalized with MC1R, but not with EP2. Representative fluorescence confocal images of HeLa cells coexpressing OPN3-cYFP and either MC1R-nHA or EP2-nHA and immunostained with anti-HA antibody are shown. OPN3-cYFP shows significant colocalization with MC1R-nHA, but not with EP2-nHA. As the bar graph illustrates, the percent overlap between the fluorescent signals of OPN3-cYFP and MC1R-nHA is ∼50%, compared with less than 10% for EP2-nHA (n = 20 cells from three independent experiments, mean ± SEM). (B) Anti-OPN3 antibody specifically recognizes OPN3-cFLAG and OPN3-cYFP. Representative fluorescence confocal images of HeLa cells expressing either OPN3-cFLAG or OPN3-cYFP and immunostained with anti-FLAG and anti-OPN3 antibodies are shown. As the bar graph illustrates, the percent overlap between anti-OPN3 fluorescence and anti-FLAG or YFP fluorescent signal is ∼50%, suggesting significant colocalization of anti-OPN3 with both anti-FLAG and YFP signals (n = 20 cells from three independent experiments, mean ± SEM). (C) Anti-MC1R antibody specifically recognizes MC1R-nHA. Representative fluorescence confocal images of HeLa cells expressing MC1R-nHA and coimmunostained with anti-HA and anti-MC1R antibodies are shown. As the bar graph illustrates, the percent overlap between anti-MC1R and anti-HA fluorescent signals is >50%, indicating significant colocalization (n = 25 cells from three independent experiments, mean ± SEM). (D) Endogenously expressed OPN3 and MC1R colocalize in HEMs. Representative fluorescence confocal images of HEMs coimmunostained with anti-OPN3 and anti-MC1R antibodies are shown. As the bar graph illustrates, the percent overlap between anti-OPN3 and anti-MC1R fluorescent signals is ∼50%, indicating significant colocalization (n = 20 cells from three independent experiments, mean ± SEM). (Scale bar: 10 μm.)
To confirm that endogenous OPN3 and MC1R colocalize in HEMs, we first validated anti-OPN3 and anti-MC1R antibodies. Cells expressing OPN3-cYFP or OPN3-cFLAG had significant (>50%) overlap between the fluorescent signals obtained with anti-OPN3 antibody or with the respective tags (YFP or FLAG) (Fig. 6B). Similarly, cells expressing MC1R-nHA had significant (>50%) overlap between the fluorescent signals of anti-MC1R and anti-HA antibodies (Fig. 6C), suggesting that both anti-OPN3 and anti-MC1R antibodies are specific. Immunostaining HEMs with these antibodies revealed that endogenous OPN3 and MC1R are present both at the plasma membrane and in intracellular compartments and exhibit significant colocalization (Fig. 6D).
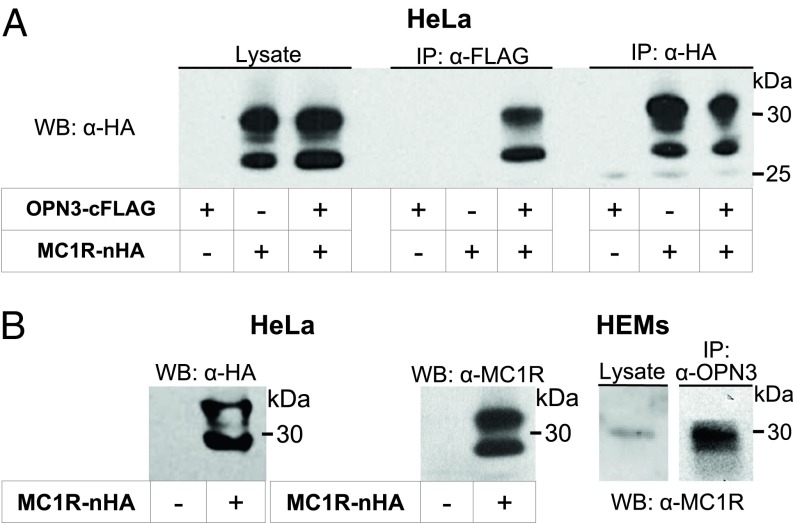
We next determined if OPN3 and MC1R could form a physical complex. We first tested if expressed OPN3-cFLAG and MC1R-nHA interact in HeLa cells. Immunoprecipitation with anti-FLAG antibody and immunoblotting with anti-HA antibody revealed a band corresponding to the molecular weight of MC1R-nHA, but only when both receptors were expressed (Fig. 7A). Similarly, immunoprecipitation with anti-HA antibody and immunoblotting with anti-FLAG antibody revealed a band corresponding to OPN3-cFLAG, but only when both receptors were expressed (SI Appendix, Fig. S5). These results suggest that in HeLa cells, OPN3-cFLAG and MC1R-nHA can form a complex. To test that endogenous OPN3 and MC1R can also interact in HEMs, we confirmed that the anti-MC1R antibody detects the same bands as the anti-HA antibody in HeLa cells expressing MC1R-nHA (Fig. 7B, Left). When HEM lysate is immunoprecipitated with anti-OPN3 antibody and immunoblotted with the anti-MC1R antibody, a band corresponding to the molecular weight of MC1R is detected (Fig. 7B, Right), indicating that OPN3 and MC1R can form a complex in melanocytes.
Fig. 7.
OPN3 and MC1R can form a molecular complex. (A) OPN3-cFLAG and MC1R-nHA coimmunoprecipitate. HeLa cells expressing OPN3-cFLAG, MC1R-nHA, or both were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-HA antibody. A band corresponding to MC1R in the cell lysates (Left) was detected in the anti-FLAG immunoprecipitation (IP) only when both OPN3 and MC1R were expressed (Center). The same band was detected by IP with anti-HA antibody (Right). Findings are representative of n = 3 independent experiments. WB, Western blot. (B) Endogenously expressed OPN3 and MC1R coimmunoprecipitate. WB analysis of HeLa cells expressing MC1R-nHA and immunoblotted with anti-MC1R antibody reveal the same size and pattern of bands as detected with the anti-HA antibody in A (Left). HEM lysates coimmunoprecipitated with anti-OPN3 antibody and immunoblotted with anti-MC1R antibody reveal the same band corresponding to MC1R, suggesting that endogenously expressed OPN3 and MC1R can form a complex in HEMs. Findings are representative of n = 3 independent experiments for each condition.
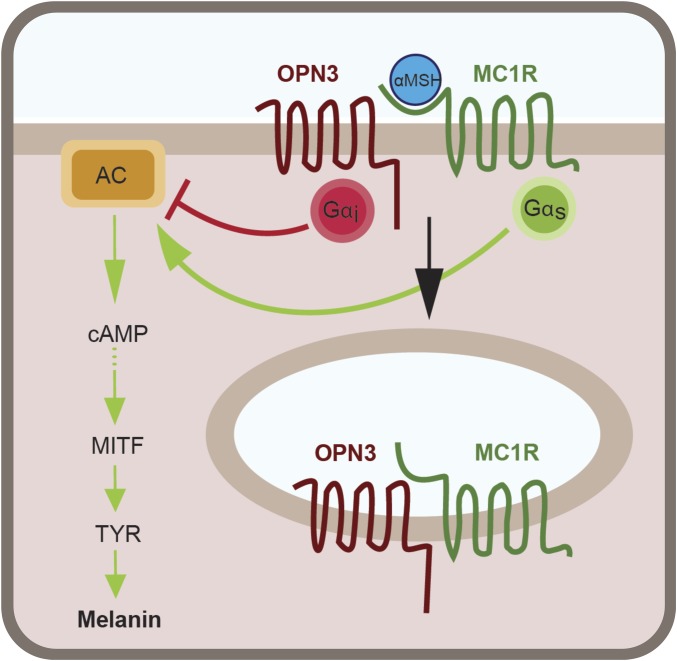
Taken together, our results reveal a molecular mechanism by which Gαi-coupled OPN3 negatively regulates the cAMP signal resulting specifically from the α-MSH–induced activation of MC1R. In addition to the functional interaction, OPN3 and MC1R can form a physical complex and colocalize at the plasma membrane and in intracellular structures. The reduced MC1R-mediated cAMP production in the presence of OPN3 leads to reduced activation of MITF and reduced levels of TYR, and ultimately results in decreased melanin production in melanocytes (Fig. 8).
Fig. 8.
OPN3 and MC1R interact functionally and physically to regulate melanin levels in melanocytes. In melanocytes, α-MSH binds to and activates the Gαs-coupled MC1R, leading to stimulation of AC and a transient increase in cAMP levels, which, through a series of molecular steps, leads to activation and up-regulation of the MITF. MITF controls the expression of TYR, the main melanogenic enzyme; higher MITF levels will result in more TYR that will generate a higher amount of melanin. Our data suggest that OPN3, via Gαi activation, reduces the amount of cAMP generated by MC1R activation, causing decreased transcription of MITF and, in consequence, of TYR. OPN3 and MC1R can form a complex, and OPN3 may additionally alter the function of MC1R by enhancing its internalization and lowering the number of receptors available to bind α-MSH at the plasma membrane.
Discussion
In this study, we have determined the cellular function of OPN3 in human melanocytes. We initially hypothesized that OPN3 functioned as the UVR photoreceptor in the UVR-Ca2+ signaling pathway previously characterized by our laboratory (14–16). Surprisingly, our results disproved this hypothesis: We showed that reducing the mRNA and protein levels of OPN3 in melanocytes (Fig. 1 B–D and SI Appendix, Fig. S1B) did not alter their Ca2+-mediated responses to physiological levels of UVR, blue, or green light (Fig. 1 E and F and SI Appendix, Fig. S1 D and E). Moreover, when we tested whether purified human OPN3 can respond to visible light, similar to its homologs (24, 25), we found no evidence of visible light absorption in the OPN3 UV-visible spectrum (Fig. 2 and SI Appendix, Fig. S2B) despite weakly binding both 11-cis and all-trans retinal (Fig. 2C and SI Appendix, Fig. S2A).
We were puzzled by the finding that human OPN3, despite being able to bind retinal, did not have an absorption peak in the visible range (Fig. 2C and SI Appendix, Fig. S2A). This was particularly intriguing considering that OPN3 homologs absorb light in the visible spectrum (24–26). In addition, unlike two recent reports suggesting that human OPN3 functions in a blue light-dependent manner in human skin (41) and in rat and human pulmonary vasorelaxation (42), we did not detect OPN3 sensitivity to blue light by either spectrophotometric analysis or monitoring Ca2+ levels in melanocytes. It is worth noting that Ca2+ flux in these other studies was measured in response to a total exposure to 50–62.5 J/cm2 of blue-violet (415 nm) or blue (465 nm) light (41), a dose more than 250-fold higher than the physiological dose used in our experiments (200 mJ/cm2). The high dose of visible light was administered at a similar rate as in our experiments (∼20 mW/cm2), but over a much longer time period; exposure time was 33–66 min (1,980–3,960 s), compared with 10 s in our studies. The prolonged light exposure could cause a gradual increase in temperature and could generate extracellular or intracellular radicals affecting membrane permeability to Ca2+ from the extracellular medium or from intracellular stores like the endoplasmic reticulum or mitochondria.
One possible scenario explaining our results is that OPN3 absorbs light outside the range covered by conventional UV-visible spectroscopy (200–700 nm) or that its absorption overlaps with protein absorption (λmax = 280 nm) and is masked by the large protein peak (43). Alternatively, OPN3 may be light-insensitive and function in a similar manner as retinal G protein-coupled receptor (RGR), an opsin receptor that binds retinal but lacks light-dependent activation. It was shown that RGR, independent of light, accelerates the conversion of retinyl esters to 11-cis–retinal by modulating isomerohydrolase activity (44). Thus, it is conceivable that OPN3 is not light-sensitive and retinal binding serves another purpose in OPN3 folding, trafficking, or signaling. Future studies are needed to characterize human OPN3 photosensitivity and to understand the role of retinal in OPN3 function.
Reducing or eliminating OPN3 expression in melanocytes led to a surprising observation: These melanocytes had a significantly higher melanin content (Fig. 3A and SI Appendix, Fig. S1C). Because melanin production in melanocytes is controlled by the Gαs-coupled MC1R, which signals via cAMP, we used genetically encoded cAMP indicators to measure the effect of OPN3 on MC1R-mediated signaling. We found that OPN3 inhibits the MC1R-mediated cAMP response in a Gαi-dependent manner (Fig. 4), indicating that MC1R and OPN3 functionally interact. The functional interaction between OPN3 and MC1R is similar to the mechanism described for the cross-regulation of the G protein-coupled estrogen receptor (GPER) and steroid hormone adipoQ receptor 7 (PAQR7), in which PAQR7 regulates skin pigmentation through Gαi-mediated down-regulation of GPER activity (45). The ability of OPN3 to negatively regulate MC1R signaling does not appear to require retinal binding to OPN3, as the OPN3(K299G) mutant is able to modulate MC1R-mediated signaling despite not binding retinal (Fig. 4), further supporting the idea that OPN3 functions in a light-independent manner in melanocytes.
Human OPN3 is likely to be coupled to Gαi, similar to its homologs (24–26). Indeed, using PTX to prevent Gαi activation eliminated the inhibitory effect of OPN3 on MC1R-mediated cAMP signaling (Fig. 4 A, iii and iv). These results indicate that OPN3 is coupled to Gαi constitutively or, alternatively, light might turn off OPN3 that is coupled to Gαi. In this case, exposure to light would uncouple OPN3 from Gαi, resulting in an increase in the baseline cAMP. Initially, we were not able to measure light-induced OPN3-dependent changes in cAMP in melanocytes because the FRET indicator Epac H187, used to monitor cAMP, requires illumination of the cells with visible light in the blue-green range (∼400–530 nm). Thus, if the light used for Epac H187 imaging was sufficient to inactivate OPN3, the increase in cAMP could occur before starting image acquisition. We circumvented this problem by using a recently developed genetically encoded red-fluorescent cAMP indicator, R-FlincA (excitation wavelength, λex = 587 nm) (34). To determine if blue, green, or UVR elicit an OPN3-dependent change in baseline cAMP levels, we used MNT-1 cells that have low endogenous levels of OPN3 (Fig. 3B) to express R-FlincA together with either OPN3-cYFP or YFP. We monitored R-FlincA fluorescence emission before, during, and after stimulation with 200 mW/cm2 of blue, green, or UVR (SI Appendix, Fig. S6). We did not measure any light-induced or OPN3-dependent changes in cAMP levels, suggesting that the amount of UVR, blue, or green light applied does not modulate OPN3 activity. This would suggest that OPN3 is more likely to have full or partial constitutive activity. The constitutive activity of OPN3 might require conformational changes induced through direct or indirect interaction with MC1R. In fact, MC1R has unusually high partial constitutive activity, which is responsible for the baseline pigmentation in our skin and hair. A potential constitutive activity of OPN3, coupled to Gαi, could be responsible for regulating both baseline cellular cAMP levels in the absence of MC1R agonists and cAMP levels upon MC1R activation.
Our colocalization (Fig. 6) and coimmunoprecipitation (Fig. 7) data indicate that OPN3 and MC1R reside in the same microdomains where they could form a molecular complex. The presence of both OPN3 and MC1R in the same microdomains could explain the specific OPN3-mediated regulation of MC1R, but not EP2, signaling (Fig. 6). The microdomain localization might allow OPN3 to become activated by binding to MC1R directly or via other proteins in the complex. Alternatively, the microdomain might contain the specific AC that is activated by MC1R and inhibited by OPN3, or a particular phosphodiesterase that could be modulated by OPN3. In all these scenarios, OPN3 would not be able to modulate signaling via cAMP evoked by Gαs-coupled receptors, like EP2, that are not localized to the same microdomains.
Heteromeric receptors have been described for other GPCRs and have been shown to have altered ligand binding, signaling, and internalization properties compared with the individual GPCRs (37, 38). For example, in the brain, the melanocortin 3 receptor (MC3R), of the same melanocortin family as MC1R, and the ghrelin receptor form heteromers with altered melanocortin- and ghrelin-induced intracellular responses to regulate energy metabolism (46, 47). Interestingly, OPN3 was initially identified in deep regions of the brain (hence the initial name “encephalopsin”) (21), including the hypothalamus, where MC3R is also expressed (48–50). This raises the intriguing question of whether OPN3 functions in the brain similarly to how it does in melanocytes. OPN3 is not the only opsin that functions as part of a dimeric complex; a functional homomeric complex has also been reported for OPN2 and cone opsin (OPN1) (40). These complexes are mediated by residues within the fifth transmembrane domain of human red and green OPN1 (39), and by transmembrane domain 1 and helix 8 for OPN2 dimerization (40). Whether OPN3 and MC1R directly interact through domain coupling or are part of a larger complex will be determined by future studies.
Our immunostaining results of both expressed and endogenous OPN3 and MC1R show that the two receptors colocalize at the plasma membrane as well as in intracellular compartments. Because recent studies showed that activated GPCRs are internalized and could continue to signal in endosomal compartments (51), it is intriguing to hypothesize that MC1R and OPN3 continue to signal even when they are no longer at the plasma membrane. We wondered if the colocalization of OPN3 and MC1R in melanocytes depends on the activation state of MC1R. We compared the localization of OPN3 and MC1R in growth media not containing α-MSH or 1, 3, or 6 h after α-MSH stimulation (SI Appendix, Fig. S7). We found that α-MSH–mediated activation of MC1R did not cause a significant shift in the fraction of MC1R at the plasma membrane vs. intracellular organelles and, implicitly, did not alter the fraction of MC1R colocalized with OPN3, suggesting that both active and inactive forms of MC1R are associated with OPN3.
The findings presented here expand our understanding of OPN3 function and its role as an extraocular opsin in human skin. We have uncovered a light-independent function of OPN3 in regulating melanin levels in human melanocytes. Taking into consideration that retinoic acid, a derivative of vitamin A, is widely used in skin treatments for conditions such as hyperpigmentation (52–54) and that OPN3 can bind retinal, OPN3 could yield a novel therapeutic target for skin pigmentation disorders such as melasma. The complex that can form between OPN3 and MC1R may reveal novel molecular mechanisms for opsin function and for regulating melanin production in melanocytes.
Materials and Methods
Cell Culture and Transfection.
All cell culture reagents were purchased from Thermo Fisher Scientific, unless otherwise stated.
HeLa cells, used for immunostaining and biochemical analyses of expressed tagged OPN3 and MC1R, were maintained at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum (FBS) and 100 units/mL penicillin/streptomycin and transiently transfected using Lipofectamine 2000 according to the manufacturer’s recommendations.
HEK293-GnTl− cells, used for expression and purification of OPN3 variants and OPN2 (as a control) for UV-visible spectroscopy, were maintained under standard conditions in DMEM supplemented with 10% FBS and 100 units/mL penicillin/streptomycin and transfected using calcium phosphate precipitation, as previously described (55).
We used three types of melanocytes:
-
i)
Normal primary neonatal HEMs. HEM lines derived from at least three individuals were purchased and maintained under standard conditions in Medium 254 supplemented with human melanocyte growth supplement (HMGS-2) and 100 units/mL penicillin/streptomycin. For miRNA experiments, HEMs were transduced with either OPN3-targeted or CTRL miRNA using the BLOCK-IT Lentiviral RNAi expression system according to the manufacturer’s protocol. The lentiviral transduction rates were ∼60% as detected by coexpression of MCh. HEMs expressing miRNAs were selected with blasticidin (4 μg/mL) for at least 14 d.
-
ii)
Immortalized normal HEMs (Hermes 2b), obtained from Wellcome Trust Functional Genomics Cell Bank (https://www.sgul.ac.uk/depts/anatomy/pages/Dot/Cell%20bank%20holdings.htm#Hermes), are HEMs immortalized with human telomerase reverse transcriptase and maintained at 37 °C and 10% CO2 in RPMI 1640 media supplemented with 20% FBS, 200 nM 12-O-tetradecanoylphorbol 13-acetate, 200 pM cholera toxin, 10 ng/mL human stem cell factor, 10 nM endothelin, and 100 units/mL penicillin/streptomycin. Hermes 2b cells were transiently transfected using Nucleofector Kits for Human Melanocytes (Lonza) according to the manufacturer’s instructions. For CRISPR/Cas9 knockout experiments, Hermes 2b cells were transduced with OPN3-targeted lentiCRISPR v2 (single-guide RNA: ggccacggctactgggacgg). Transduced Hermes 2b cells were selected with puromycin (10 μg/mL), and clonal lines were isolated and maintained.
-
iii)
Human pigmented melanoma cells (MNT-1) were maintained under standard conditions in DMEM supplemented with 18% FBS, 10% AIM-V, and 100 units/mL penicillin/streptomycin and transiently transfected using Nucleofector Kits for Human Melanocytes or magnetofection with PolyMag Neo magnetic beads (OZ Biosciences), according to the manufacturers’ instructions.
DNA Constructs.
cDNA encoding full-length human OPN3 was obtained by RT-PCR using RNA extracted from HEMs. Different OPN3 variants were cloned as summarized in Table 1. Mutations were introduced by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The human MC1R-n(3xHA) and EP2-n(3xHA) in pcDNA3.1 expression vectors were purchased from the cDNA Resource Center (https://www.cdna.org/). EEA1-nRFP, Rab7-nRFP, Rab11-nDsRed, Rab9-nDsRed, and MEM-nDsRed were obtained from Addgene (www.addgene.org). All constructs were confirmed by sequencing.
Table 1.
| Construct | Vector | Restriction sites |
| OPN3-cMCh | pcDNA4/TO-cMCh | HindIII-XhoI |
| OPN3-cHA | pcDNA4/TO-cHA | EcoRI-XhoI |
| OPN3-nHA | pcDNA4/TO-nGFP | EcoRI-XhoI |
| OPN3-nHA-ΔC | pcDNA4/TO-cGFP | EcoRI-XhoI |
| MCh CTRL | pcDNA4/TO-cMCh | — |
| OPN3-cFLAG | pcDNA3.1-cFLAG | HindIII-XhoI |
| OPN3-cYFP | pcDNA3.1-cYFP | BamHI-XhoI |
| OPN3(K299G)-cYFP | pcDNA3.1-cYFP | BamHI-XhoI |
| YFP CTRL | pcDNA3.1 c-YFP | — |
| miRNA OPN3-1 and OPN3-2 | pLenti V5-DEST | Attb1-Attb2 |
| OPN3-c1D4 | pMT3 | EcoRI-NotI |
| OPN3ΔC-c1D4 | pMT3 | EcoRI-NotI |
| OPN3(K299G)ΔC-c1D4 | pMT3 | EcoRI-NotI |
Immunoprecipitation and Western Blot.
Cells were rinsed with cold phosphate-buffered saline (PBS) before addition of 500 μL lysis buffer [300 mM NaCl, 50 mM Tris⋅HCl (pH 7.4), 1% Triton X-100, protease inhibitor mix (Roche)]. Cells were scraped and homogenized using a 22-gauge syringe needle. Lysates were rotated end-over-end for 1 h at 4 °C and then centrifuged at 14,000 rpm for 30 min at 4 °C to remove cell debris. Fifteen microliters of 50% (wt/vol) protein A/G or protein A beads (Santa Cruz Biotechnology) were added to the supernatant and rotated for 30 min to preclear the samples. Samples were centrifuged at 14,000 rpm for 5 min, and the agarose pellet was discarded. Samples were split into two aliquots and mixed with 25 μL of primary antibody conjugated to protein A or protein A/G beads and rotated overnight at 4 °C. Immunoprecipitates were collected by centrifugation at 7,000 rpm for 5 s, washed three times with wash buffer [300 mM NaCl, 50 mM Tris⋅HCl (pH 7.4), 0.1% Triton X-100], and solubilized with 10 μL of elution buffer (100 mM Tris⋅HCl, 1% SDS, 10 mM DTT) and 5 μL of 4× NuPAGE LDS sample buffer (Thermo Fisher Scientific). For Western blots and immunoprecipitation experiments, the following primary antibodies were used: anti-HA (11867423001; Roche), anti-FLAG (F1804; Sigma–Aldrich), anti-OPN3 (sc-98799; Santa Cruz Biotechnology), anti-OPN3 (A15803; ABclonal), anti-MC1R (sc-6875; Santa Cruz Biotechnology), anti-MITF (MA514146; Thermo Fisher Scientific), anti-TYR (sc-7833; Santa Cruz Biotechnology), anti–β-actin (MA515739; Thermo Fisher Scientific), and anti-integrin α5 (H-104; Santa Cruz Biotechnology). The primary antibodies were detected by incubation with horseradish peroxidase-conjugated goat anti-rat, goat anti-mouse, or donkey anti-goat secondary antibodies.
Immunofluorescence.
Cells seeded on glass coverslips were fixed with 4% paraformaldehyde at room temperature for 10 min and incubated with blocking solution (0.2% saponin, 0.1% BSA, 0.02% sodium azide) for 1 h, followed by overnight incubation with primary antibodies (1:200). For visualization, fluorescently labeled secondary antibodies (1:1,000; Life Technologies) were used. Confocal images were acquired using an Olympus FV3000 confocal microscope and analyzed using ImageJ software. The Pearson correlation coefficient (PCC) was calculated for each cell, as previously described (56). Two proteins were considered significantly colocalized if the overlap was higher than 50% and not colocalized if the overlap was lower than 10% (57). As a positive control for the colocalization analysis, we calculated the PCC for two well-characterized lysosomal proteins, TPC2 and LAMP1, expressed in HeLa cells, which showed ∼50% overlap (SI Appendix, Fig. S8).
Ca2+ Imaging and Light Stimulation.
Cells cultured on glass coverslips were incubated for 20 min with 12 μM all-trans retinal (Sigma–Aldrich) in Ringer’s solution [150 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1.2 mM MgCl2, 10 mM Hepes, 10 mM d-glucose (pH 7.4)], followed by a wash and 20-min incubation with 7.5 μM Fluo4-AM (Life Technologies) and 250 μM sulfinpyrazone (uridine 5′-diphospho-glucuronosyltransferase; Sigma–Aldrich) in Ringer’s solution. Ca2+ imaging was performed using an inverted microscope (Olympus IX71). Sequential images were acquired with a 20× objective every 2 s before, during, and after light stimulation with 200 mJ/cm2 UVR (λmax = 360 nm), 200 mJ/cm2 blue (λmax = 450 nm), or green (λmax = 550 nm) light. UVR was applied using 400-nm short-pass and 280-nm long-pass filters (Newport) attached to a 200-W Hg-Xe arc lamp (Newport) as previously described (16). For blue and green light, 450-nm and 550-nm light-emitting diode light sources (Prizmatix) were used. For all experiments, cells were exposed to 20 mW/cm2 of radiation for 10 s, resulting in a total dose of 200 mJ/cm2. Ionomycin (1 μM) was added at the end of each experiment to elicit a maximal Ca2+ response, used for normalization. Changes in fluorescence intensity of individual cells as a function of time were obtained using MetaMorph software, and then analyzed with MATLAB and Microsoft Excel.
cAMP Imaging.
The genetically encoded cAMP indicator mTurq2Del-EPAC(dDEPCD)Q270E-tdcp173Venus(d) EPAC-SH187 (Epac H187) [dissociation constant (Kd) = 4.0 µM] and red fluorescent cAMP indicator R-FlincA (Kd = 0.3 µM) were generous gifts from the Jalink Laboratory (Netherlands Cancer Institute) and Horikawa Laboratory (Tokushima University), respectively. MNT-1 cells were transfected with Epac H187, and ∼24 h after transfection cells were serum-starved in OPTI-MEM (Thermo Fisher Scientific) for another ∼24 h. Hermes 2b cells were transfected with R-FlincA, and ∼6 h after transfection cells were serum-starved for another ∼6 h. For PTX treatment, serum-starved cells were incubated with 200 ng/mL PTX for 4 h before the experiment. Coverslips were transferred to an imaging chamber with Ringer’s solution. For Epac H187 imaging, sequential fluorescence images were acquired with MetaMorph software on an inverted microscope every 10 s using CFP and FRET filter cubes (λex = 430 nm), and CFP and YFP emissions were detected simultaneously using 470 ± 20-nm and 530 ± 25-nm band-pass filters. For R-FlincA imaging, images were acquired every 10 s using an MCh filter cube (λex = 587 nm). For both Epac H187 and R-FlincA, after acquiring 18 baseline images (3 min), 1 μM [Nle4, D-Phe7] (NDP)–α-MSH (Sigma–Aldrich) or 5 μM prostaglandin (Sigma–Aldrich) was added. After 54 images (9 min), a mix of 25 μM FSK (Sigma–Aldrich) and 100 μM IBMX (Sigma–Aldrich) was added to elicit a maximal cAMP response, used for normalization. For MNT-1 cells expressing R-FlincA, cells were exposed to 200 mJ/cm2 UVR (λmax = 360 nm), blue (λmax = 450 nm), or green (λmax = 550 nm) light after 100 s baseline; after 5 min, FSK + IBMX was added. Fluorescence emission intensities for Epac H187 and R-FlincA were calculated as F = FCFP/FYFP and F = FMCh, respectively. Normalized fluorescence intensities were quantified using Fnorm(t) = (Fcell(t) − Fmin)/(FFSK+IBMX − Fmin), where t is the time point, Fcell is the fluorescence of an intracellular region of interest, FFSK+IBMX is the maximal fluorescence with FSK and IBMX, and Fmin is the baseline fluorescence before stimulation. Light-induced changes in fluorescence intensity were quantified using MetaMorph and Excel software (Microsoft). NDP–α-MSH, prostaglandin, FSK, and IBMX were solubilized in dimethyl sulfoxide (DMSO; Sigma–Aldrich) at >100-fold the final concentration, so that the final DMSO concentration in the imaging chamber remained <1% (vol/vol) for all experiments.
Melanin Assay.
Confluent melanocytes cultured in 35-mm dishes were lysed in 1% Triton X-100 and centrifuged at 14,000 rpm for 30 min at 4 °C to separate melanin from solubilized protein. Melanin pellets were solubilized in 1 M NaOH at 85 °C for 1+ h. The exact volumes of the solubilized protein and solubilized melanin were noted. Spectrophotometric analysis of melanin content was determined by measuring absorbance at 405 nm and using a calibration curve obtained with synthetic melanin, as previously described (16). Total melanin was determined as the product of the melanin concentration measured spectrophotometrically and the total volume of solubilized melanin. The protein content for each sample was measured using a bicinchoninic acid (BCA) protein assay kit (Pierce, Thermo Fisher Scientific). Total protein was determined as the product of the protein concentration measured with BCA and the volume of solubilized protein. Cellular melanin concentration was determined as total melanin/total protein for each condition; relative melanin content was calculated as the ratio of cellular melanin concentration for each experimental condition to control.
UV-Visible Spectroscopy.
HEK293-GnTl− cells plated on 100-mm culture dishes were transfected using calcium phosphate precipitation with OPN3ΔCc1D4 or OPN3(K299G)ΔC-c1D4 (the ΔC truncation of C-terminal amino acids 315–402 increased protein yield). After 3 d, cells were harvested, centrifuged at 3,500 rpm for 20 min, and resuspended in PBS. All subsequent steps were performed in the dark. Cells were treated with 4.8 mM all-trans or 11-cis retinal at 4 °C for 30 min, lysed with 1% n-Dodecyl-β-D-maltoside (DDM; Sigma–Aldrich) at 4 °C for 1 h, and then centrifuged at 3,500 rpm for 20 min. The supernatant containing the solubilized protein was incubated with preconjugated 1D4 antibody (P21940; Life Technologies)-Sepharose beads at 4 °C for 2 h and then run through a disposable plastic column (Thermo Fisher Scientific) and washed with 0.1% DDM. Proteins were eluted with 0.4 mM 1D4 peptide solution at 4 °C. Absorbance spectra were measured on a Cary 50-UV visible spectrometer between 200 and 800 nm as previously described (55). To test the presence of a Schiff-base bond between chromophore and the K299 residue of OPN3, 0.8% SDS and 80 μM NH2OH were added to create retinal oxime, which absorbs maximally at 360 nm (31).
Statistical Analysis.
For each tested condition, several replicate experiments were performed and results were averaged. All data are given as mean ± SEM. Statistical differences among the experimental groups were analyzed by a two-sided Student t test when comparing two experimental groups. Significance was defined as *P < 0.05 and **P < 0.01.
Supplementary Material
Acknowledgments
We thank members of the E.O. laboratory for technical assistance and insightful discussions. We thank Dr. K. Jalink (Netherlands Cancer Institute) and Dr. K. Horikawa (Tokushima University) for generously providing the cAMP indicators Epac H187 and R-FlincA, respectively. This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Disease Grant R01 AR066318 (to E.O.), National Institute of General Medical Sciences Training Grant T32 GM077995 (to L.E.O.), and the Brown University Institute for Brain Science/The Suna and Inan Kirac Foundation Fellowship (to R.N.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902825116/-/DCSupplemental.
References
- 1.Fitzpatrick T. B., Biology and Disease of Dermal Pigmentation (University of Tokyo Press, 1981). [Google Scholar]
- 2.Odland G. F., The fine structure of the interrelationship of cells in the human epidermis. J. Biophys. Biochem. Cytol. 4, 529–538 (1958). [PMC free article] [PubMed] [Google Scholar]
- 3.Clark W. H. Jr, Hibbs R. G., Electron microscope studies of the human epidermis: The clear cell of Masson (dendritic cell or melanocyte). J. Biophys. Biochem. Cytol. 4, 679–684 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikehata H., Ono T., The mechanisms of UV mutagenesis. J. Radiat. Res. 52, 115–125 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Heck D. E., Gerecke D. R., Vetrano A. M., Laskin J. D., Solar ultraviolet radiation as a trigger of cell signal transduction. Toxicol. Appl. Pharmacol. 195, 288–297 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Abdel-Malek Z. A., Kadekaro A. L., Swope V. B., Stepping up melanocytes to the challenge of UV exposure. Pigment Cell Melanoma Res. 23, 171–186 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Lin J. Y., Fisher D. E., Melanocyte biology and skin pigmentation. Nature 445, 843–850 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Cui R., et al. , Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell 128, 853–864 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Donatien P. D., et al. , The expression of functional MSH receptors on cultured human melanocytes. Arch. Dermatol. Res. 284, 424–426 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Valverde P., Healy E., Jackson I., Rees J. L., Thody A. J., Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat. Genet. 11, 328–330 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Beaumont K. A., Shekar S. N., Cook A. L., Duffy D. L., Sturm R. A., Red hair is the null phenotype of MC1R. Hum. Mutat. 29, E88–E94 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Palmer J. S., et al. , Melanocortin-1 receptor polymorphisms and risk of melanoma: Is the association explained solely by pigmentation phenotype? Am. J. Hum. Genet. 66, 176–186 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy C., Khaled M., Fisher D. E., MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12, 406–414 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Bellono N. W., Kammel L. G., Zimmerman A. L., Oancea E., UV light phototransduction activates transient receptor potential A1 ion channels in human melanocytes. Proc. Natl. Acad. Sci. U.S.A. 110, 2383–2388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellono N. W., Najera J. A., Oancea E., UV light activates a Gαq/11-coupled phototransduction pathway in human melanocytes. J. Gen. Physiol. 143, 203–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wicks N. L., Chan J. W., Najera J. A., Ciriello J. M., Oancea E., UVA phototransduction drives early melanin synthesis in human melanocytes. Curr. Biol. 21, 1906–1911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haltaufderhyde K. D., Oancea E., Genome-wide transcriptome analysis of human epidermal melanocytes. Genomics 104(6 Pt B), 482–489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haltaufderhyde K., Ozdeslik R. N., Wicks N. L., Najera J. A., Oancea E., Opsin expression in human epidermal skin. Photochem. Photobiol. 91, 117–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reemann P., et al. , Melanocytes in the skin–Comparative whole transcriptome analysis of main skin cell types. PLoS One 9, e115717–e115718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsutsumi M., et al. , Expressions of rod and cone photoreceptor-like proteins in human epidermis. Exp. Dermatol. 18, 567–570 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Blackshaw S., Snyder S. H., Encephalopsin: A novel mammalian extraretinal opsin discretely localized in the brain. J. Neurosci. 19, 3681–3690 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nissilä J., et al. , Encephalopsin (OPN3) protein abundance in the adult mouse brain. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 198, 833–839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halford S., et al. , Characterization of a novel human opsin gene with wide tissue expression and identification of embedded and flanking genes on chromosome 1q43. Genomics 72, 203–208 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Koyanagi M., Takada E., Nagata T., Tsukamoto H., Terakita A., Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc. Natl. Acad. Sci. U.S.A. 110, 4998–5003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugihara T., Nagata T., Mason B., Koyanagi M., Terakita A., Absorption characteristics of vertebrate non-visual opsin, Opn3. PLoS One 11, e0161215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terakita A., Nagata T., Functional properties of opsins and their contribution to light-sensing physiology. Zool. Sci. 31, 653–659 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Terakita A. The opsins. Genome Biol. 6, 213 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves P. J., Callewaert N., Contreras R., Khorana H. G., Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owens S. L., et al. , Ocular manifestations in autosomal dominant retinitis pigmentosa with a Lys-296-Glu rhodopsin mutation at the retinal binding site. Br. J. Ophthalmol. 78, 353–358 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhukovsky E. A., Robinson P. R., Oprian D. D., Transducin activation by rhodopsin without a covalent bond to the 11-cis-retinal chromophore. Science 251, 558–560 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Sieving P. A., et al. , Constitutive “light” adaptation in rods from G90D rhodopsin: A mechanism for human congenital nightblindness without rod cell loss. J. Neurosci. 21, 5449–5460 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jastrzebska B., Golczak M., Fotiadis D., Engel A., Palczewski K., Isolation and functional characterization of a stable complex between photoactivated rhodopsin and the G protein, transducin. FASEB J. 23, 371–381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klarenbeek J., Goedhart J., van Batenburg A., Groenewald D., Jalink K., Fourth-generation epac-based FRET sensors for cAMP feature exceptional brightness, photostability and dynamic range: Characterization of dedicated sensors for FLIM, for ratiometry and with high affinity. PLoS One 10, e0122513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta Y., Furuta T., Nagai T., Horikawa K., Red fluorescent cAMP indicator with increased affinity and expanded dynamic range. Sci. Rep. 8, 1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taussig R., Tang W.-J., Hepler J. R., Gilman A. G., Distinct patterns of bidirectional regulation of mammalian adenylyl cyclases. J. Biol. Chem. 269, 6093–6100 (1994). [PubMed] [Google Scholar]
- 36.D’Orazio J., Jarrett S., Amaro-Ortiz A., Scott T., UV radiation and the skin. Int. J. Mol. Sci. 14, 12222–12248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satake H., Matsubara S., Aoyama M., Kawada T., Sakai T., GPCR heterodimerization in the reproductive system: Functional regulation and implication for biodiversity. Front. Endocrinol. (Lausanne) 4, 100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes I., et al. , G protein-coupled receptor heteromers. Annu. Rev. Pharmacol. Toxicol. 56, 403–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jastrzebska B., et al. , A G protein-coupled receptor dimerization interface in human cone opsins. Biochemistry 56, 61–72 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T., et al. , Dimerization of visual pigments in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 9093–9098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Regazzetti C., et al. , Melanocytes sense blue light and regulate pigmentation through opsin-3. J. Invest. Dermatol. 138, 171–178 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Barreto Ortiz S., et al. , Opsin 3 and 4 mediate light-induced pulmonary vasorelaxation that is potentiated by G protein-coupled receptor kinase 2 inhibition. Am. J. Physiol. Lung Cell Mol. Physiol. 314, L93–L106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teale F. W., Weber G., Ultraviolet fluorescence of the aromatic amino acids. Biochem. J. 65, 476–482 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wenzel A., et al. , The retinal G protein-coupled receptor (RGR) enhances isomerohydrolase activity independent of light. J. Biol. Chem. 280, 29874–29884 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Natale C. A., et al. , Sex steroids regulate skin pigmentation through nonclassical membrane-bound receptors. eLife 5, e15104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rediger A., et al. , Heterodimerization of hypothalamic G-protein-coupled receptors involved in weight regulation. Obes. Facts 2, 80–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rediger A., et al. , MC4R dimerization in the paraventricular nucleus and GHSR/MC3R heterodimerization in the arcuate nucleus: Is there relevance for body weight regulation? Neuroendocrinology 95, 277–288 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Caruso V., Lagerström M. C., Olszewski P. K., Fredriksson R., Schiöth H. B., Synaptic changes induced by melanocortin signalling. Nat. Rev. Neurosci. 15, 98–110 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Erlanson-Albertsson C., How palatable food disrupts appetite regulation. Basic Clin. Pharmacol. Toxicol. 97, 61–73 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Breit A., et al. , Alternative G protein coupling and biased agonism: New insights into melanocortin-4 receptor signalling. Mol. Cell. Endocrinol. 331, 232–240 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Jalink K., Moolenaar W. H., G protein-coupled receptors: The inside story. Bioessays 32, 13–16 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Kligman A. M., Willis I., A new formula for depigmenting human skin. Arch. Dermatol. 111, 40–48 (1975). [PubMed] [Google Scholar]
- 53.Pathak M. A., Fitzpatrick T. B., Kraus E. W., Usefulness of retinoic acid in the treatment of melasma. J. Am. Acad. Dermatol. 15, 894–899 (1986). [DOI] [PubMed] [Google Scholar]
- 54.Rossi A. M., Perez M. I., Treatment of hyperpigmentation. Facial Plast. Surg. Clin. North Am. 19, 313–324 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Xie G., Gross A. K., Oprian D. D., An opsin mutant with increased thermal stability. Biochemistry 42, 1995–2001 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Dunn K. W., Kamocka M. M., McDonald J. H., A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q., et al. , A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: Analysis by quantitative immunocolocalization. J. Neurosci. 24, 4070–4081 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rost B., Yachdav G., Liu J., The PredictProtein server. Nucleic Acids Res. 32, W321–W326 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.