Significance
IBD is one of the most common early manifestations of LRBA deficiency and has been attributed to impaired regulatory T cell function. However, whether other immune cell types also contribute has not been comprehensively tested. We found that, in LRBA-deficient mice, DCs contribute significantly to colitis in the DSS model. We also showed that blocking innate immune signaling from the endosomal TLRs, TLR3, TLR7, and TLR9, in Lrba−/− mice dramatically reduced their susceptibility to DSS-induced colitis. Our data indicate a role for LRBA in limiting endosomal TLR signaling and suggest that elevated IRF3 and IRF7 activation leading to increased expression of inflammatory chemokines promotes excessive intestinal inflammation in LRBA-deficient mice.
Keywords: Toll-like receptor, dendritic cells, IRF3, IRF7, inflammatory bowel disease
Abstract
LPS-responsive beige-like anchor (LRBA) protein deficiency in humans causes immune dysregulation resulting in autoimmunity, inflammatory bowel disease (IBD), hypogammaglobulinemia, regulatory T (Treg) cell defects, and B cell functional defects, but the cellular and molecular mechanisms responsible are incompletely understood. In an ongoing forward genetic screen for N-ethyl-N-nitrosourea (ENU)-induced mutations that increase susceptibility to dextran sodium sulfate (DSS)-induced colitis in mice, we identified two nonsense mutations in Lrba. Although Treg cells have been a main focus in LRBA research to date, we found that dendritic cells (DCs) contribute significantly to DSS-induced intestinal inflammation in LRBA-deficient mice. Lrba−/− DCs exhibited excessive IRF3/7- and PI3K/mTORC1-dependent signaling and type I IFN production in response to the stimulation of the Toll-like receptors (TLRs) 3, TLR7, and TLR9. Substantial reductions in cytokine expression and sensitivity to DSS in LRBA-deficient mice were caused by knockout of Unc93b1, a chaperone necessary for trafficking of TLR3, TLR7, and TLR9 to endosomes. Our data support a function for LRBA in limiting endosomal TLR signaling and consequent intestinal inflammation.
Mutations of LRBA have recently been linked to a recessive immune deficiency disorder (OMIM #614700) characterized by hypogammaglobulinemia, hepato/splenomegaly, respiratory infections, autoimmune disease, and inflammatory bowel disease (IBD) (1–4). Analysis of immune cells isolated from the blood of LPS-responsive beige-like anchor (LRBA)-deficient patients demonstrated generally normal counts of T cells and natural killer (NK) cells (1–4). B cells were reduced in 42–75% of patients; however, hypogammaglobulinemia and a reduction or absence of class-switched memory B cells were typically observed, which may contribute to the high frequency and recurrence of respiratory infections (1–4). Decreased regulatory T (Treg) cell numbers, inhibitory function, and expression of Treg markers [including forkhead box P3 (FOXP3), IL-2 receptor subunit α (IL-2RA), cytotoxic T lymphocyte-associated protein 4 (CTLA4), and Helios] were also reported; these defects have been proposed to promote the development of autoimmunity and IBD (1, 2, 4, 5). Current treatment for immune dysregulation in LRBA-deficient patients is symptomatic and varied (4), reflecting an incomplete understanding of the molecular etiology of the disease.
Several findings support the idea that LRBA may regulate vesicle trafficking in the endolysosomal pathway. LRBA contains conserved Pleckstrin homology (PH) and Beige and Chediak-Higashi (BEACH) domains commonly found in proteins that associate with and regulate membrane and vesicle trafficking (6). LRBA has been localized in the endoplasmic reticulum, endocytic vesicles, lysosomes, and trans-Golgi network (6, 7). Moreover, electron microscopic analysis of B cells from LRBA-deficient patients showed increased areas of Golgi apparatus and accumulation of autophagosomes; decreased autophagy was measured in B cells (3). Finally, an interaction between LRBA and the T cell inhibitory receptor CTLA4 in recycling endosomes and the trans-Golgi network has been demonstrated in Treg cells (1). This interaction was proposed to sequester CTLA4 from binding to the adaptor protein complex 1 (AP-1), the clathrin-associated adaptor protein complex implicated in the shuttling of CTLA4 to lysosomes (8). Thus, decreased expression of CTLA4 in LRBA-deficient Treg cells (1, 7) may result from excessive lysosomal degradation of CTLA4 (1). Abatacept, a CTLA4-Ig fusion protein that inhibits T cell responses by competing for costimulatory ligands, has been used effectively in some LRBA-deficient patients to reverse infiltrative inflammatory and autoimmune phenotypes (1). However, it is unclear whether all patients would respond (4), and in at least one patient, abatacept monotherapy failed to resolve chronic diarrhea (1).
We encountered LRBA in a forward genetic screen for mutations that enhance susceptibility to DSS-induced colitis in mice. We found that conventional dendritic cells (cDCs) and plasmacytoid DCs (pDCs) contribute to the intestinal inflammatory phenotype in LRBA-deficient mice. Strikingly, genetic deletion of Unc93b1 moderated to near wild-type levels the susceptibility of LRBA-deficient mice to DSS-induced colitis, implicating excessive TLR3, TLR7, and TLR9 signaling in intestinal inflammation caused by LRBA deficiency.
Results
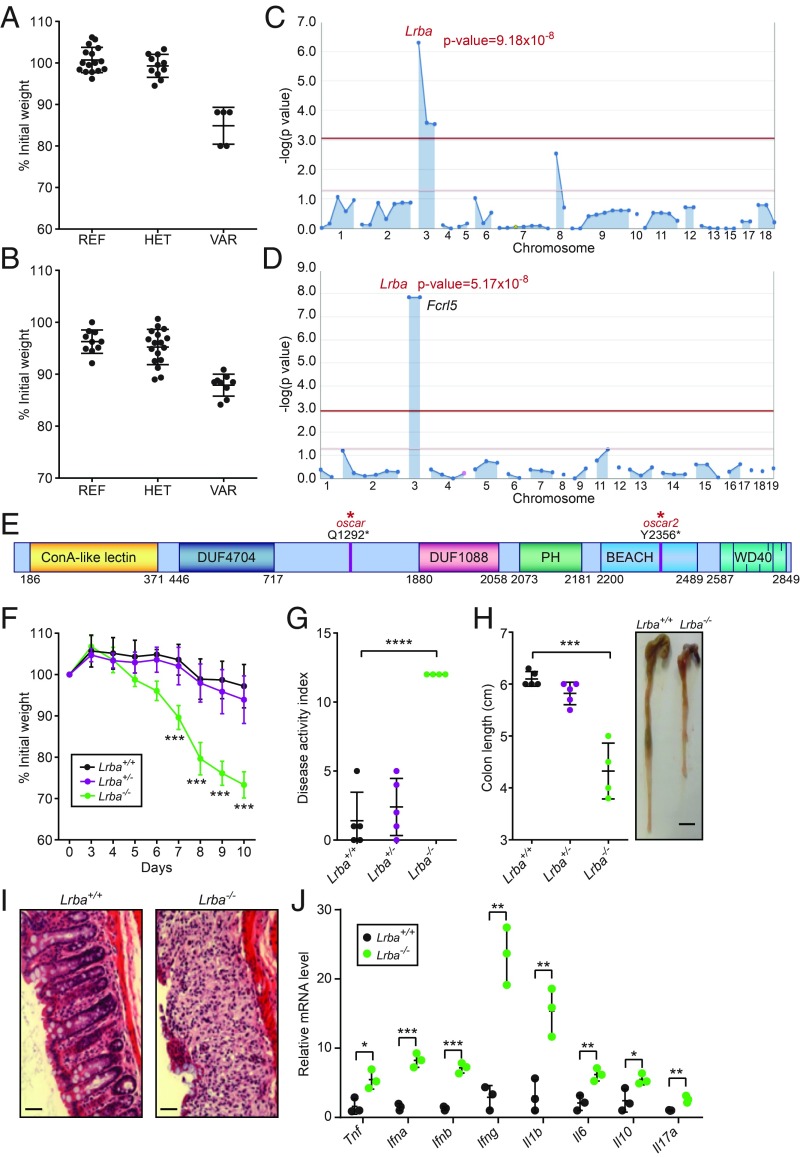
Susceptibility to DSS-Induced Colitis Caused by Nonsense Mutations of Lrba.
To identify genes necessary for intestinal homeostasis, a forward genetic screen was performed in which C57BL/6J mice carrying ENU-induced mutations were tested for susceptibility to DSS-induced colitis (9). Mice were treated with 1.3% DSS in the drinking water for 7 d, and body weight was measured on day 0, 7, and 10 after initiation of DSS treatment. Several mice from two unrelated pedigrees exhibited severe body weight loss by day 7, and the phenotypes were named oscar and oscar2 (Fig. 1 A and B). Linkage analysis using a recessive model of inheritance (10) showed that both phenotypes were associated with distinct nonsense mutations in Lrba (Fig. 1 C and D): Q1292* (oscar) and Y2356* (oscar2) (Fig. 1E). The oscar2 mutation was also linked to a cosegregating mutation in Fcrl5 (Fig. 1D). Mice homozygous for a clustered regularly interspersed short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9)-targeted null allele of Lrba (Lrba−/−) recapitulated the oscar and oscar2 weight loss phenotypes (Fig. 1F), confirming that mutations in Lrba cause increased susceptibility to DSS-induced colitis. Lrba−/− mice treated with DSS also displayed an elevated disease activity index (DAI), including weight loss, severe diarrhea, and colonic bleeding, and greater shortening of the colon compared with wild-type (Lrba+/+) mice (Fig. 1 G and H). Histological analysis showed increased infiltration of inflammatory cells and destruction of the epithelial cell architecture in the Lrba−/− colon relative to the Lrba+/+ colon (Fig. 1I). Moreover, increased transcript expression levels of proinflammatory cytokines, including IL-1β, IFN-α, IFN-β, IFN-γ, TNF, and IL-6, were detected in the Lrba−/− colon after DSS treatment (Fig. 1J). However, frequencies of major immune cell populations were normal in the blood of Lrba−/− mice (SI Appendix, Fig. S1). These data indicate that LRBA is necessary for normal resistance to DSS-induced colitis in mice.
Fig. 1.
Mapping of the oscar and oscar2 mutations in Lrba. (A) Body weight (% relative to weight on day 0) of the oscar pedigree on day 10 after initiation of DSS treatment. REF, Lrba+/+ (n = 16); HET, Lrbaoscar/+ (n = 11); VAR, Lrbaoscar/oscar (n = 5). (B) Body weight (% relative to weight on day 0) of the oscar2 pedigree on day 7 after initiation of DSS treatment. REF, Lrba+/+ (n = 10); HET, Lrbaoscar2/+ (n = 17); VAR, Lrbaoscar2/oscar2 (n = 9). (C and D) Manhattan plots showing P values of association between the DSS-induced weight loss phenotype and the mutations identified in the oscar (C) and oscar2 (D) pedigrees calculated using a recessive model of inheritance. The −log10 P values are plotted versus the chromosomal positions of mutations. Horizontal red and purple lines represent thresholds of P = 0.05 with or without Bonferroni correction, respectively. The P values for linkage of Lrba mutations are indicated. (E) Protein domain diagram of LRBA with the oscar and oscar2 mutations indicated. DUF, domain of unknown function; WD40, WD40 repeat domain. (F) Body weight (% relative to weight on day 0) of Lrba+/+ (n = 5), Lrba+/− (n = 5), and Lrba−/− (n = 4) mice on the indicated day after initiation of DSS treatment. (G) DAI and (H) colon length of Lrba+/+ (n = 5), Lrba+/− (n = 5), and Lrba−/− (n = 4) mice on day 10 after initiation of DSS treatment. The side panel in H shows a representative image of colons. (Scale bar, 5 mm.) (I) Colon sections stained with hematoxylin and eosin on day 10 after initiation of DSS treatment. (Scale bars, 80 μm.) (J) RT-qPCR analysis of the indicated mRNAs in colons from Lrba+/+ (n = 3) and Lrba−/− (n = 3) mice on day 6 after initiation of DSS treatment. Each symbol (A, B, G, H, and J) represents an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 for comparison of the indicated genotype with Lrba+/+ (two-tailed Student’s t test). Data are representative of three independent experiments (mean ± SD in A, B, F, G, H, and J).
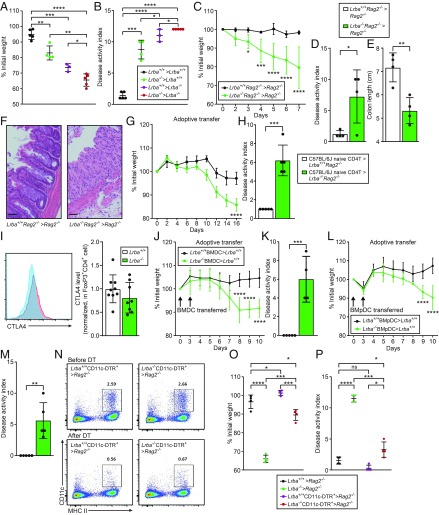
DCs Promote DSS-Induced Colitis in Lrba−/− Mice.
To investigate whether hematopoietic or nonhematopoietic cells were required for the development of DSS-induced colitis in Lrba−/− mice, reciprocal bone marrow transplantation (BMT) was performed. Chimeric mice, irrespective of whether Lrba deficiency originated in the donor or the recipient, showed more body weight loss after the DSS challenge compared with Lrba+/+ recipients of Lrba+/+ BM (Fig. 2 A and B), indicating that both hematopoietic and nonhematopoietic Lrba−/− cells contributed to the development of DSS-induced colitis. In another BMT experiment, we tested the requirement of B cells and T cells for the colitis phenotype of Lrba−/− mice. Rag2−/− mice that received Lrba−/−Rag2−/− BM exhibited increased body weight loss (Fig. 2C), DAI score (Fig. 2D), colonic shortening (Fig. 2E), and histologic evidence of colitis (Fig. 2F) induced by DSS treatment compared with those that received Rag2−/− BM. This finding suggests that Lrba−/− T cells and B cells are not necessary for the development of DSS-induced colitis and that Lrba−/− nonadaptive immune cell population(s) are sufficient to cause disease.
Fig. 2.
DCs promote DSS-induced colitis in Lrba−/− mice. (A–F) BM transplantation was performed using mice of the indicated genotypes (donor > recipient). (A and C) Body weight (% relative to weight on day 0) on day 10 (A) or on the indicated day (C) after initiation of DSS treatment. Lrba+/+ > Lrba+/+ (n = 5), Lrba−/− > Lrba+/+ (n = 4), Lrba+/+ > Lrba−/− (n = 3), Lrba−/− > Lrba−/− (n = 5) for A. Lrba+/+Rag2−/− > Rag2−/− (n = 4), Lrba−/−Rag2−/− > Rag2−/− (n = 4) for C. (B and D) DAI and (E) colon length on day 10 after initiation of DSS treatment. (F) Colon sections stained with hematoxylin and eosin on day 7 after initiation of DSS treatment. (Scale bars, 80 μm.) (G and H) CD4+ T cell transfer model of colitis. Naive CD4+ T cells from donor mice were transferred to recipient mice (donor > recipient). (G) Body weight (% relative to weight on day 0) on the indicated day after initiation of DSS treatment. C57BL/6J > Lrba+/+Rag2−/− (n = 5), C57BL/6J > Lrba−/−Rag2−/− (n = 5). (H) DAI on day 16 after naive CD4+ T cell transfer. (I, Left) Flow cytometry of colon lamina propria cells from Lrba+/+ and Lrba−/− mice, assessing expression of CTLA4 in CD3+CD4+FoxP3+ cell population. (I, Right) Quantification of CTLA4 protein expression in colon lamina propria CD3+CD4+FoxP3+ cells (mean fluorescence intensity normalized to wild type). (J–M) Adoptive transfer of in vitro differentiated BMDCs (J and K) or BMpDCs (L and M) from donor mice to recipient mice (donor > recipient). (J and L) Body weight (% relative to weight on day 0) on the indicated day after initiation of DSS treatment. Lrba+/+ BMDCs > Lrba+/+ (n = 5), Lrba−/− BMDCs > Lrba+/+ (n = 4) for J and K. Lrba+/+ BMpDCs > Lrba+/+ (n = 5), Lrba−/− BMpDCs > Lrba+/+ (n = 5) for L and M. (K and M) DAI on day 10 after initiation of DSS treatment. (N) Flow cytometry of peripheral blood cells from BM chimeric mice (donor > recipient) before and after diphtheria toxin (DT) treatment (three doses, day −4, −2, and 2 relative to DSS initiation), assessing expression of CD11c and MHC II. Numbers above boxed regions represent percent cells in each. (O) Body weight (% relative to weight on day 0) and (P) DAI on day 10 after initiation of DSS treatment of BM chimeric mice (donor > recipient) treated with DT to ablate DCs. All mice were treated with DT. Each symbol (A, B, D, E, H, I, K, M, O, and P) represents an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (two-tailed Student’s t test for C–E and G–M; one-way ANOVA with post hoc Tukey multiple comparisons test for A, B, O, and P). Data are representative of three independent experiments (mean ± SD in A–E, G–M, O, and P).
We also used the CD4+ T cell transfer model of colitis to test the impact of Lrba-deficient nonadaptive immune cells (e.g., innate immune cells: macrophages and DCs) on colitis susceptibility. Adoptive transfer of wild-type C57BL/6J naive CD4+ T cells resulted in more weight loss (Fig. 2G) and elevated DAI scores (Fig. 2H) in Lrba−/−Rag2−/− recipient mice compared with Rag2−/− recipients, suggesting that Lrba−/− innate immune cells may potentiate the development of colitis induced by CD4+ T cell transfer; defective epithelial cells may also contribute to colitis in these experiments.
However, we found that expression of CTLA4 was normal in FoxP3+ cells isolated from the lamina propria of Lrba−/− mice (Fig. 2I), consistent with a role for innate immune cells rather than Treg cells in intestinal inflammation. CTLA4 expression in Lrba−/− FoxP3+ cells from the spleen was reduced (SI Appendix, Fig. S2) as reported previously (1, 4, 5). In addition, Lrba−/− mice showed normal antibody responses to immunization as previously reported (7) (SI Appendix, Fig. S3). Taken together, the above data strongly suggest that LRBA function in innate immune cells is necessary for the recovery and restoration of intestinal homeostasis after DSS injury.
Both cDCs and pDCs can exacerbate acute DSS-induced colitis (11–13), and we compared the effects of adoptive transfer of wild-type DCs versus Lrba−/− DCs on DSS-induced colitis in wild-type recipient mice. BM cells were differentiated in vitro to yield BMDCs and BMpDCs, which were transferred to wild-type mice on day 0 and day 3 of the DSS challenge. Mice that received Lrba−/− BMDCs or BMpDCs showed increased weight loss and DAI scores compared with mice that received the corresponding type of Lrba+/+ DCs (Fig. 2 J–M). To confirm the effect of Lrba−/− DCs, we generated Lrba−/− mice carrying the CD11c-diphtheria toxin receptor (DTR) transgene, which encodes a DT receptor controlled by the CD11c promoter; injection of DT results in depletion of DC populations (14). BM chimeras were created by transplantation of BM from transgenic or nontransgenic mice into irradiated Rag2−/− recipients, which were subsequently injected i.p. with DT 4 and 2 d before and 2 d after initiation of DSS treatment (Fig. 2N). We found that, on average, similar body weights were recorded on day 10 after DSS initiation for mice that received wild-type BM and in which DCs were either ablated or left intact (Fig. 2 O and P). In contrast, for mice that received Lrba−/− BM, those in which DCs were left intact lost a greater amount of body weight by day 10 compared with those in which DCs were ablated (Fig. 2 O and P). These data indicate that Lrba−/− cDCs and pDCs promote intestinal inflammation in response to DSS treatment. By contrast, tolerogenic CD103+ DCs were reduced in Lrba−/− lamina propria (SI Appendix, Fig. S4); this defect may favor intestinal inflammation induced by DSS treatment.
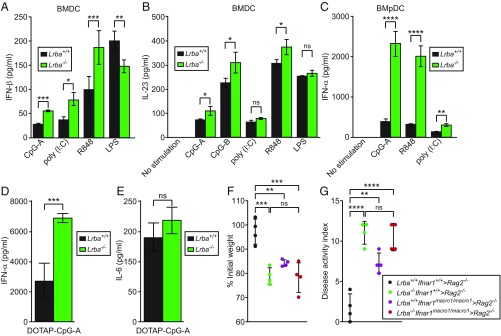
Increased Type I IFN Responses to Endosomal TLR Stimulation in Lrba−/− DCs.
To investigate how LRBA-deficient DCs might contribute to intestinal inflammation, we tested the cytokine responses of Lrba−/− BMDCs or BMpDCs to the TLR ligands. Stimulation with poly(I:C) (TLR3 ligand), R848 (TLR7 ligand), or CpG-oligodeoxynucleotides type A [(CpG-A); TLR9 ligand] in the culture medium resulted in increased production of IFN-β by Lrba−/− BMDCs relative to Lrba+/+ BMDCs, whereas LPS (TLR4 ligand) induced slightly less IFN-β production by Lrba−/− compared with Lrba+/+ BMDCs (Fig. 3A). IFN-α production by Lrba−/− BMDCs was also increased relative to Lrba+/+ BMDCs in response to CpG-A (SI Appendix, Fig. S5A). Lrba−/− BMDCs produced slightly greater amounts of IL-23 than Lrba+/+ BMDCs in response to R848, CpG-A, and CpG-type B (CpG-B) (Fig. 3B). Lrba−/− BMpDCs produced elevated IFN-α in response to TLR3, TLR7, or TLR9 stimulation (Fig. 3C). The proinflammatory cytokines TNF and IL-6 were produced at similar levels by Lrba−/− and Lrba+/+ BMDCs or BMpDCs (SI Appendix, Fig. S5 B–E) in response to all TLR ligands tested. In vivo, serum levels of IFN-α but not IL-6 were elevated in Lrba−/− mice compared with wild-type littermates in response to i.v. injection of CpG-A encapsulated in the liposomal transfection reagent N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate (DOTAP) (Fig. 3 D and E). TNF and IFN-β production by Lrba−/− BM-derived macrophages (BMDMs) and peritoneal macrophages was normal in response to all TLR ligands tested (SI Appendix, Fig. S6). These data indicate that Lrba deficiency resulted in exaggerated type I IFN responses by DC to endosomal TLR stimulation in vitro. However, stimulation of Lrba−/− BMDCs or BMpDCs with type I IFN or type II IFN led to phosphorylated signal transducer and activator of transcription 1 (STAT1) levels comparable to those observed in the corresponding Lrba+/+ cells, indicating that STAT1 signaling was unaffected in Lrba−/− DCs (SI Appendix, Fig. S7).
Fig. 3.
Increased type I IFN responses to endosomal TLR stimulation in Lrba−/− DCs. (A–E) In vitro differentiated BMDCs or BMpDCs were stimulated with TLR ligands for 16 h. Concentration of (A) IFN-β or (B) IL-23 in the culture supernatant of BMDCs after stimulation with different TLR ligands (n = 4 independent cultures of each genotype from separate mice). (C) Concentration of IFN-α in the culture supernatant of BMpDCs after stimulation with the indicated ligands (n = 3 independent cultures of each genotype from separate mice). (D and E) Serum concentration of (D) IFN-α and (E) IL-6 4 h after injection of DOTAP-encapsulated CpG-A into mice (n = 4 mice per genotype). (F and G) BM transplantation was performed using mice of the indicated genotypes (donor > recipient). (F) Body weight (% relative to weight on day 0) and (G) DAI on day 10 after initiation of DSS treatment. Lrba+/+Ifnar1+/+ > Rag2−/− (n = 4), Lrba−/−Ifnar1+/+ > Rag2−/− (n = 4), Lrba+/+Ifnar1macro−1/macro−1 > Rag2−/− (n = 4), and Lrba−/−Ifnar1macro−1/macro−1 > Rag2−/− (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (two-tailed Student’s t test for A–E; one-way ANOVA with post hoc Tukey multiple comparisons test for F and G). Data are representative of three (B, F, and G) or four (A and C–E) independent experiments (mean ± SD).
To test whether the elevated DSS-induced colitis susceptibility of Lrba−/− mice was attributable to increased type I IFN production, we crossed Lrba−/− mice to mice with the macro-1 mutation of Ifnar1 (15). Lrba−/−Ifnar1macro−1/macro−1 mice displayed a similar amount of weight loss as Lrba−/− mice after the DSS challenge (Fig. 3 F and G), demonstrating that excessive type I IFN was not responsible for DSS susceptibility of LRBA-deficient mice.
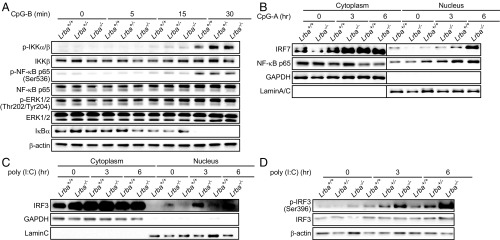
Hyperactivation of IRF3, IRF7, and PI3K/AKT/mTOR Signaling in Lrba−/− DC.
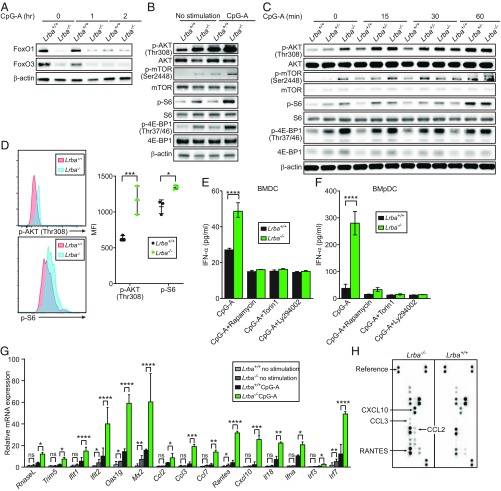
Consistent with their normal TNF and IL-6 responses, we found that the levels and kinetics of IKKα/β, NF-κB p65, and ERK phosphorylation were normal in Lrba−/− BMDCs in response to CpG-B (Fig. 4A). In contrast, the enhanced type I IFN production of Lrba−/− BMDCs in response to endosomal TLR stimulation suggested hyperactivation of the IRF3- and IRF7-dependent pathways (16–18). We found that levels of IRF7 but not NF-κB p65 (Fig. 4B) and IRF3 (Fig. 4C) were elevated in the nuclear fractions of Lrba−/− BMpDCs stimulated with CpG-A or poly (I:C) compared with those induced in wild-type BMpDCs for at least 6 h poststimulation. Moreover, the active phosphorylated form of IRF3 was also increased in Lrba−/− BMpDCs relative to wild-type BMpDCs stimulated with poly (I:C) (Fig. 4D).
Fig. 4.
Enhanced activation of IRF3 and IRF7 in Lrba−/− cells stimulated with TLR3 or TLR9 ligands. (A) Immunoblot analysis of phosphorylated (p) or total IKKα/β, NF-κB p65, ERK1/2, and IκBα in Lrba+/+, Lrba+/−, and Lrba−/− BMDCs after stimulation with CpG-B. (B and C) Immunoblot analysis of (B) IRF7, NF-κB p65, and (C) IRF3 in nuclear and cytoplasmic fractions of lysates of Lrba+/+ and Lrba−/− BMpDCs after stimulation with (B) CpG-A and (C) poly(I:C). (D) Immunoblot analysis of phosphorylated (p) and total IRF3 in Lrba+/+, Lrba+/−, and Lrba−/− BMpDCs after stimulation with poly(I:C). Data are representative of three independent experiments.
IRF3 and IRF7 activation depend on PI3K and mTORC1 in pDCs, and a blockade of these pathways results in decreased type I IFN responses to TLR stimulation (19–22). Conversely, FOXO1 and FOXO3, respectively. limit IRF3 and IRF7 function in response to viral infection or TLR3 stimulation (23–25). We found that FOXO1 and FOXO3 levels were diminished in unstimulated Lrba−/− BMpDCs or after stimulation with CpG-A (Fig. 5A). Moreover, Lrba−/− BMDCs and BMpDCs had increased basal and CpG-A-induced levels of the activated phosphorylated forms of mTOR, AKT, S6, and 4E-BP1 relative to wild-type BMDCs (Fig. 5 B and C). The increased expression of phospho-AKT and phospho-S6 were confirmed in vivo by flow cytometric analysis of primary unstimulated DCs from Lrba−/− mice (Fig. 5D). Treatment of Lrba−/− BMDCs or BMpDCs with the PI3K inhibitor Ly294002 or either of two mTORC1 inhibitors, rapamycin or Torin1, resulted in normalization of CpG-A-induced IFN-α production (Fig. 5 E and F).
Fig. 5.
Enhanced PI3K/AKT/mTOR signaling in Lrba−/− BMDCs and BMpDCs. (A) Immunoblot analysis of FOXO1 and FOXO3 in Lrba+/+ and Lrba−/− BMpDCs after stimulation with CpG-A. (B and C) Immunoblot analysis of phosphorylated and total mTOR, AKT, S6, and 4E-BP1 in Lrba+/+, Lrba+/−, and Lrba−/− (B) BMpDCs and (C) BMDCs stimulated with CpG-A for 16 h (B) or for the indicated times (C). (D, Left) Flow cytometry of Lrba+/+ and Lrba−/− splenocytes, assessing expression of p-AKT and p-S6 by CD11c+ DCs. (D, Right) Quantification of p-AKT and p-S6 mean fluorescence intensity (MFI) on CD11c+ DCs. (E and F) Concentration of IFN-α in the culture supernatant of (E) BMDCs or (F) BMpDCs pretreated with PI3K and mTOR inhibitors for 3 h, then stimulated with CpG-A for 16 h (n = 3 independent cultures of each genotype from separate mice). (G) RT-qPCR analysis of the indicated mRNAs in Lrba+/+ and Lrba−/− BMpDCs after stimulation with CpG-A for 16 h. (H) Protein array analysis of the culture supernatant from BMpDCs stimulated with CpG-A. Each symbol (D) represents an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (two-tailed Student’s t test). Data are representative of two or three independent experiments (mean ± SD in D–G).
We examined the effects of augmented IRF3 and IRF7 activation on the transcriptional responses of several IRF-dependent chemokines and IFN-stimulated genes (ISGs). Quantitative RT-PCR analysis of Lrba−/− BMpDCs showed elevated CpG-A-induced expression of the chemokines Ccl2, Ccl3, Rantes, and Cxcl10 as well as the ISGs Oas1g, Mx2, and Ifit2 relative to Lrba+/+ BMpDCs (Fig. 5G). Lrba−/− BMpDCs also secreted greater amounts of CCL2, CCL3, RANTES, and CXCL10 after CpG-A stimulation compared with Lrba+/+ BMpDCs (Fig. 5H). To summarize, these data suggest that enhanced PI3K/AKT/mTOR signaling mediates hyperactivation of IRF3 and IRF7 in response to endosomal TLR stimulation in Lrba−/− DCs, leading to enhanced production of type I IFN, ISG, and IRF-dependent chemokines.
Knockout of UNC93B1 Partially Rescues DSS-Induced Colitis in Lrba−/− Mice.
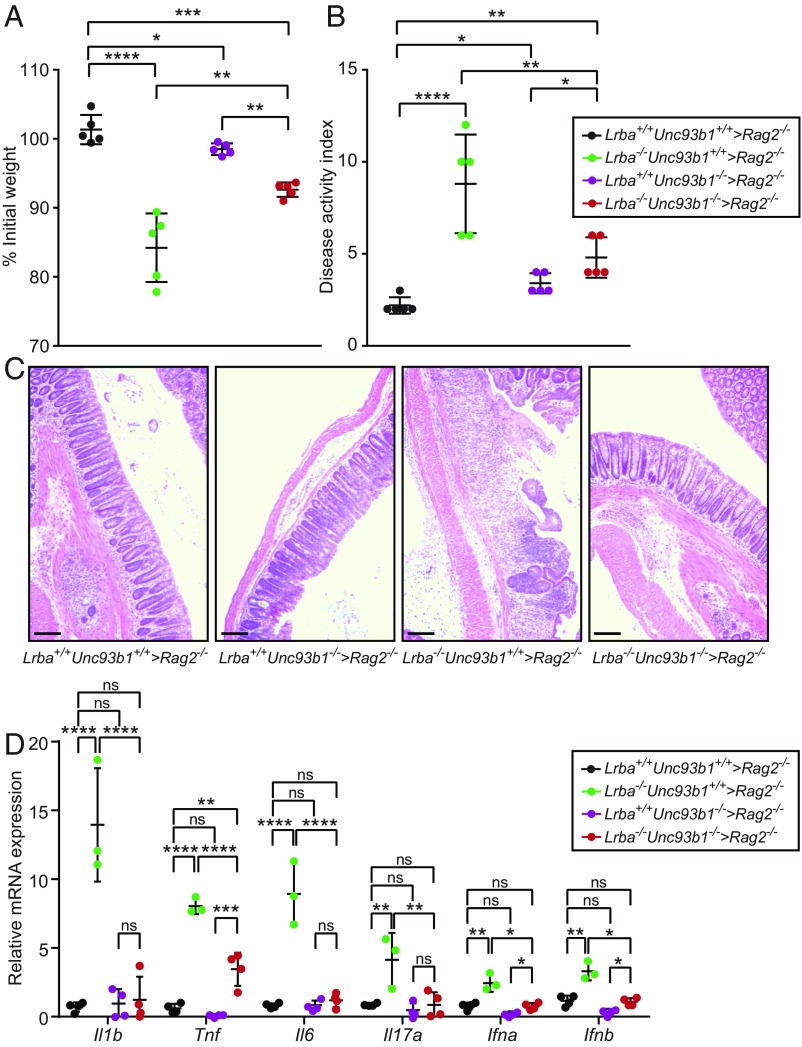
The chaperone protein UNC93B1 mediates translocation of TLR3, TLR7, and TLR9 from the endoplasmic reticulum to endolysosomes (26, 27) in which these TLRs encounter ligands and propagate signaling. Deficiency of UNC93B1 results in abrogation of TLR3, TLR7, and TLR9 signaling in mice and humans (28, 29). To determine whether endosomal TLR signaling contributed to the DSS-induced colitis susceptibility of Lrba−/− mice, we bred Lrba−/− mice onto the Unc93b1−/− background, generated hematopoietic chimeras, and tested for sensitivity to DSS. Rag2−/− mice transplanted with Lrba−/−Unc93b1−/− BM lost significantly less body weight after DSS treatment than mice that received Lrba−/−Unc93b1+/+ BM, although full rescue to wild-type levels (i.e., similar to mice receiving either wild-type or Lrba+/+Unc93b1−/− BM) was not achieved (Fig. 6A). For DAI scores, a similar degree of rescue was achieved by knockout of Unc93b1 (Fig. 6B). Moreover, histologic examination of the colons showed decreased infiltration of inflammatory cells and decreased structural damage in those mice receiving BM from Lrba−/−/Unc93b1−/− mice compared with those receiving Lrba−/−Unc93b1+/+ BM (Fig. 6C). Transcript expression levels of Il1b, Tnf, Il6, Il17a, Ifna, and Ifnb in the colons of Lrba−/− mice after DSS treatment were reduced to relatively normal levels by Unc93b1 knockout (Fig. 6D). In addition, the frequency of splenic IFN-γ-producing T cells that may promote intestinal inflammation was reduced to normal in mice that received Lrba−/−Unc93b1−/− BM (SI Appendix, Fig. S8). These findings suggest that elevated signaling from one or more endosomal TLRs in Lrba−/− mice results in increased production of inflammatory cytokines and enhanced susceptibility to DSS-induced colitis.
Fig. 6.
Knockout of UNC93B1 partially rescues DSS-induced colitis in Lrba−/− mice. BM transplantation was performed using mice of the indicated genotypes (donor > recipient). (A) Body weight (% relative to weight on day 0), (B) DAI, and (C) hematoxylin and eosin staining of colon sections on day 10 after initiation of DSS treatment. (Scale bars, 25 μm.) (D) RT-qPCR analysis of the indicated mRNAs in colons from the indicated BM chimeric mice on day 10 after initiation of DSS treatment. Each symbol (A, B, and D) represents an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (one-way ANOVA with post hoc Tukey multiple comparisons test). Data are representative of four independent experiments (mean ± SD in A, B, and D).
Discussion
We have shown that mice harboring recessive loss-of-function mutations in Lrba display elevated susceptibility to DSS-induced colitis. This finding contrasts with a previous report that mice carrying a distinct Lrba null allele responded similarly to wild-type mice in a Salmonella Typhimurium infection-induced model of colitis in which Lrba−/− and wild-type mice exhibited similar amounts of weight loss and bacterial load in the cecum content 2 d after infection (7). We speculate that enhanced colitis susceptibility in these Lrba−/− mice may not have been detectable due to their strain background [C57BL/6N (7)], which carries a homozygous nonfunctional Nramp1 allele that renders them highly susceptible to S. Typhimurium infection (30–32). Although spontaneous colitis was not observed in Lrba mutant mice [this paper and (7)], the DSS model represents a useful tool for the study of intestinal inflammation caused by LRBA deficiency. Nonsense mutations similar to the oscar and oscar2 mutations described here have been documented in human LRBA-deficient patients with IBD.
Treg cell deficiency, decreased canonical Treg cell markers and impaired Treg cell suppressive function have been proposed to contribute to the development of autoimmune disease and IBD in LRBA-deficient patients (1, 5, 7). However, we found that BM chimeras with a Lrba−/−Rag2−/− hematopoietic compartment developed increased intestinal inflammation compared with those with a Rag2−/− hematopoietic compartment. Moreover, FoxP3+ Treg cells in the lamina propria of Lrba−/− mice expressed normal levels of CTLA4. Although our data do not rule out a contribution of Treg cells to DSS-induced colitis in Lrba−/− mice, they demonstrate that Lrba−/− T cells are not absolutely necessary.
We attributed the colitis susceptibility of Lrba−/− mice in large part to DCs, which produced abundant type I IFN in IRF3/7- and PI3K/mTOR1-dependent manners in response to endosomal TLR stimulation. Type I IFN has shown both protective and damaging effects during DSS-induced colitis (33–36). We showed that Lrba−/− mice with a loss-of-function mutation in the IFNAR1 receptor still developed DSS-induced colitis with similar weight loss and disease activity indices as Lrba−/− mice sufficient for IFNAR1, indicating that excessive type I IFN is not causative for the enhanced susceptibility of Lrba−/− mice to DSS-induced colitis. Rather, we propose that the increased expression of IRF3/7-dependent genes, in particular, those encoding chemokines IL-8, CXCl10, RANTES, and CCL3 (37–41), predisposes Lrba−/− mice to excessive intestinal inflammation in response to DSS. In addition, the moderately elevated production by Lrba−/− BMDCs of IL-23, a cytokine implicated in IBD pathogenesis through effects on T cell proliferation and Th17 differentiation (42, 43), may contribute to intestinal inflammation in these animals. Finally, a major question is how LRBA functions in the endolysosomal pathway to limit TLR signaling. Cellular and molecular analyses probing the integrity of the endolysosomal pathway in Lrba−/− cells should yield insights into this issue.
In conclusion, our data shed light on the mechanism for hyperactivated endosomal TLR signaling in Lrba−/− DCs. More broadly, they point to dysregulation of the innate immune system as a possible source of pathology in the LRBA-deficient intestine that warrants further investigation.
Materials and Methods
Mice.
C57BL/6J mice 8- to 10-wk of age were purchased from The Jackson Laboratory. ENU mutagenesis was performed as previously described (10). Briefly, mutagenized G0 males were bred with C57BL/6J females, and the resulting G1 males were crossed with C57BL/6J females to produce G2 mice. G2 females were backcrossed with their G1 sires to yield G3 mice, which were screened for phenotypes. For the DSS-induced colitis screen, mice received 1.3% DSS (wt/vol; MP Biomedical) in the drinking water for 7 d followed by an additional 3 d off DSS. Weight was recorded daily and reported as a percentage relative to the pretreatment weight. DAI was scored cumulatively as follows: weight loss: 0 (no loss), 1 (1–10% loss of body weight), 2 (10–15% loss of body weight), 3 (15–20% loss of body weight), and 4 (>20% loss of body weight); stool consistency: 0 (normal), 2 (loose stool), and 4 (diarrhea); and bleeding: 0 (no blood), 1 (hemoccult positive), 2 (hemoccult positive and visual pellet bleeding), and 4 (gross bleeding and/or blood around the anus). Rag2−/− and CD11c-DTR+ mice were from The Jackson Laboratory. Ifnar1macro−1/macro−1 mice have been previously described (15). All mice were housed in the University of Texas Southwestern Vivarium. All procedures were approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee, and performed in accordance with institutionally approved protocols.
Sequencing and Determination of Candidate Genes.
Whole-exome sequencing and mapping were performed as described (10). Briefly, exome-enriched DNA from all G1 mice were sequenced using the Illumina HiSeq 2500 platform. All G3 mice were genotyped across coding mutations according to their pedigree using Ion Torrent AmpliSeq custom primer panels as previously described (10). To correlate phenotype with the genotyping results, the percentage of original weight (before initiation of DSS treatment) was used as a continuous variable in linkage analysis.
Generation of Lrba−/− and Unc93b1−/− Mouse Strains Using the CRISPR/Cas9 System.
To generate the Lrba−/− and Unc93b1−/− mouse strains, female C57BL/6J mice were superovulated by injection of 6.5 U pregnant mare serum gonadotropin (Millipore), followed by injection of 6.5 U human CG (Sigma-Aldrich) 48 h later. The superovulated mice were subsequently mated overnight with C57BL/6J male mice. The following day, fertilized eggs were collected from the oviducts and in vitro-transcribed Cas9 mRNA (50 ng/μL) and Lrba small base-pairing guide RNA (50 ng/μL; 5′- AATGCGAAACATGGTGGATC-3′) or Unc93b1 small base-pairing guide RNA (50 ng/μL; 5′- AGGAAGTCCCAACCAGCTGC-3′) were injected into the cytoplasm or pronucleus of the embryos. The injected embryos were cultured in M16 medium (Sigma-Aldrich) at 37 °C in 5% CO2. For the production of mutant mice, two-cell stage embryos were transferred into the ampulla of the oviduct (10–20 embryos per oviduct) of pseudopregnant Hsd:ICR (CD-1) female mice (Harlan Laboratories).
BM-Derived DCs and Macrophage Cultures.
BMDCs, BMpDCs, and BMDMs were in vitro differentiated by standard protocols (44, 45). Briefly, BM was isolated from femurs and tibias of mice, and BMDCs and BMDMs were differentiated in recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) (40 ng/mL; PeproTech) or M-CSF (30 ng/mL; PeproTech), respectively, for 7 d. Media were changed on day 3, and cells were harvested on day 7. BMDCs were further purified using the Pan DC Isolation Kit (Miltenyi Biotec). BMpDCs were differentiated in human FMS-like tyrosine kinase 3 ligand (hFLT3L) (200 ng/mL; PeproTech) for 9 to 10 d without disturbance and purified using the Plasmacytoid DC Isolation Kit (Miltenyi Biotec).
Cells were stimulated for 16 h with the following TLR ligands: poly (I:C) (200 μg/mL; Invivogen), R848 (20 ng/mL; Enzo), LPS (10 ng/mL; Enzo), CpG-A (1 μg/mL for BMpDCs, 100 μg/mL or the concentration as indicated in Fig. S5 for BMDC/BMDM/peritoneal macrophages; Invivogen), or CpG-B (200 ng/mL; Invivogen). The inhibitors Ly294002 (12.5 nM; Cell Signaling Technologies), rapamycin (10 nM; Sigma-Aldrich), or Torin1 (25 nM; Cell Signaling Technologies) were applied to BMDCs or BMpDCs in the culture medium for 3 h; cells were given fresh media for TLR stimulation. Culture supernatants were harvested for measurement of TNF, IL-6, IL-23 (p19/p40), IFN-α, and IFN-β by ELISA using cytokine-specific kits according to the manufacturer’s instructions [TNF and IL-6, eBioscience; IFN-α, InvivoGen; IL-23 (p19/p40) and IFN-β, Biolegend].
For measurement of protein expression by BMDCs or BMpDCs, cells were harvested and lysed in 1× LDS sample buffer (Thermo Fisher) with 2-mercaptoethanol (Sigma-Aldrich) and a protease and phosphatase inhibitor mixture (Thermo Fisher), and lysates were analyzed by Western blotting using standard methods. For analysis of STAT1 signaling, BMDCs, or BMpDCs were stimulated with IFN-α (100 U/mL; PBL Assay Science), IFN-β (2.5 ng/mL; R&D Systems), or IFN-γ (10 nM; R&D Systems) for the indicated times before lysis.
For adoptive transfer of BMDCs and BMpDCs, C57BL/6J mice receiving DSS in the drinking water were injected i.v. with 6 × 106 cells/mouse cultured BMDCs or BMpDCs on day 0 and day 3 with respect to initiation of DSS treatment. Body weights and DAI parameters were measured daily.
Protein Array Analysis.
BMpDCs were stimulated for 16 h with CpG-A, and culture supernatants were harvested to measure chemokines using a Proteome Profiler Mouse Chemokine Array Kit (R&D Systems) according to the manufacturer’s instructions.
CD4+ T Cell Transfer Colitis Model.
Adoptive transfer of T cells was performed as previously published (46). Briefly, naive CD4+ T cells were isolated from spleens by negative selection with the Naive CD4+ T Cell Isolation Kit (Miltenyi Biotec) or MagCellect Mouse Naive CD4+ T Cell Isolation Kit (R&D Systems). Cells were then stained with anti-CD4 (RM4-5), anti-CD45RB (C363-16A), anti-CD25 (PC61), anti-CD44 (IM7), and CD4+CD25−CD44lowCD45RBhi cells were sorted with a FACSAria II cell sorter (BD Biosciences). Some 1 × 105 purified cells were transferred into Rag2−/− mice through tail vein injection. Body weights and DAI parameters were monitored daily.
Lamina Propria Lymphocyte (LPLs) Isolation and Analysis.
LPLs were isolated as previously described (47). Briefly, intestines were dissected from mice, and Peyer‘s patches were removed. Intestines were cut into small pieces and thoroughly washed with ice-cold PBS solution. Epithelial cells were removed by incubating intestine tissues in Hank's Balanced Salt Solution supplemented with 1 mM EDTA and 1 mM DTT at 37 °C for 30 min with gentle shaking, followed by extensive washing with PBS. Residual tissues were then digested with collagenase IV (Sigma-Aldrich), DNase I (Sigma-Aldrich), and dispase (Sigma-Aldrich) at 37 °C for 60 min with gentle shaking. Cells were filtered through 100 μm cell strainers and applied onto a 40%:80% Percoll PLUS gradient (GE Healthcare) in which LPLs were found at the interface between the 40% and the 80% Percoll fractions.
For detection of Treg cells by transcription factor expression, freshly isolated LPLs were fixed and permeabilized with BD Mouse FoxP3 fixation and permeabilization buffers (BD Biosciences) per the manufacturer’s instructions and stained with anti-CD3 (17A2), anti-CD4 (RM4-5), anti-FoxP3 (MF23), and anti-CD152 (CTLA4; clone UC10-4B9). To examine mononuclear phagocyte subsets, freshly isolated LPLs cells were first blocked with anti-CD16/32 (2.4G2) and then stained with antibodies against cell surface markers CD45 (30-F11), CD11c (N418), CD11b (M1/70), and CD103 (M290).
Antibodies.
Primary antibodies used for Western blot: p-IKKα/β (Ser176/180) (16A6), IKKβ (D30C6), p-NF-κB p65 (Ser536) (93H1), NF-κB p65 (D14E12), p-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E), p44/42 MAPK (Erk1/2) (137F5), p-mTOR (Ser2448) (D9C2), mTOR (7C10), p-AKT (Thr308) (D25E6), AKT (C67E7), p-S6 (Ser240/244) (D68F8), S6 (54D2), p-4E-BP1 (Thr37/46) (236B4), 4E-BP1 (53H11), IRF3 (D83B9), p-IRF3 (Ser396) (4D4G), p-IRF7 (Ser437/438) (#14767), IκBα (L35A5), p-STAT1 (Tyr701) (D4A7), STAT1 (D1K9Y), FoxO1 (C29H4), FoxO3 (75D8), GAPDH (D16H11), Lamin A/C (4C11), and β-actin (13E5) were from Cell Signaling Technologies. IRF7 (ab109255) was from Abcam.
Primary antibodies used for flow cytometry: CD3ε (clone 145–2C11), CD4 (clone RM4-5), CD8a (clone 53–6.7), B220 (CD45R, clone RA3-6B2), NK-1.1 (clone PK136), CD25 (clone PC61.5), CD44 (clone IM7), CD11b (clone M1/70), F4/80 (clone BM8), CD111c (clone N418), FoxP3 (clone MF23), CD103 (M290) were from Biolegend or BD Biosciences. CD152 (clone UC10-4B9), MHC class II (clone M5/114.15.2), and IFN-γ (clone XMG1.2) were from eBioscience.
Histology and Microscopy.
Freshly isolated distal colons were fixed in formalin and embedded in paraffin. Hematoxylin-eosin staining was conducted using a standard protocol by the UT Southwestern Histo Pathology Core.
Quantitative RT-PCR (RT-qPCR).
Total RNA from whole colons or cultured BMpDCs was isolated using a TRIzol Plus RNA Purification Kit (Thermo Fisher) according to the manufacturer’s instructions. DNase treatment and RNA cleanup were performed with the PureLink DNase set (Thermo Fisher) to remove any excess DSS, which can interfere with the reverse transcriptase reaction. One microgram of RNA was reverse transcribed to cDNA with SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies). Transcript levels were analyzed using iTaq Universal SYBR Green Supermix (Bio-Rad) on a StepOnePlus Real-Time PCR System (Life Technologies) with the primers listed in SI Appendix, Table S1. Relative expression was calculated using the ΔΔCt standardization method using Gadph.
Antibody Responses to Immunization.
On day 0, each mouse was i.p. immunized with 2 × 106 infectious units of the recombinant Semliki Forest virus vector encoding β-galactosidase (β-gal). On day 7, each mouse was further i.p. immunized with 40 μg of 4-hydroxy-3-nitrophenylacetic aminoethylcarboxymethyl-Ficoll (Biosearch Technologies). On day 13, blood from the submandibular vein of mice was collected for ELISA analysis of β-gal-specific IgG and NP-specific IgM responses as previously described (48).
Isolation and Stimulation of Peritoneal Macrophages.
Thioglycollate-elicited macrophages were recovered 4 d after i.p. injection of 2 mL BBL thioglycollate medium, brewer modified (4% wt/vol; BD Biosciences) by peritoneal lavage with 5 mL PBS. The peritoneal macrophages were cultured in a DMEM cell culture medium [DMEM containing 10% vol/vol FBS (Gemini Bio Products), 1% vol/vol penicillin and streptomycin (Life Technologies)] at 37 °C and 95% air/5% CO2. Peritoneal macrophages were stimulated for 16 h with the following TLR ligands: poly (I:C) (200 μg/mL; Invivogen), R848 (20 ng/mL; Enzo Life Sciences), LPS (10 ng/mL; Enzo Life Sciences), CpG-A (100 μg/mL; Invivogen), or CpG-B (200 ng/mL; Invivogen). Culture supernatants were harvested for measurement of TNF and IFN-β by ELISA using cytokine-specific kits according to the manufacturer’s instructions (TNF, eBioscience; IFN-β, Biolegend).
Hematopoietic Chimeras.
Recipient mice were irradiated with two doses of 5.5 Gy γ-radiation and injected i.v. with donor BM cells (4 × 106 cells per mouse). Recipient mice were maintained for 2 wk on water containing trimethroprim-sulfamethoxazole antibiotics. Eight weeks after BM transplantation, mice were challenged with DSS. Body weights and DAI parameters were monitored daily.
To deplete DCs in CD11c-DTR+ mice, after BM reconstitution, mice were injected i.p. with DT (Sigma-Aldrich; 12 ng/g body weight) on the days indicated in Fig. 2 for a total of three doses.
Flow Cytometry.
Blood cells, spleen cells, or LPLs were isolated and stained with antibodies as previously described (49). Immune cell populations were defined by antibody staining as follows: B cells (B220+CD3−), T cells (B220−CD3+), CD4+ T cells (CD3+CD4+CD8−), CD8+ T cells (CD3+CD4−CD8+), macrophages (CD11b+F4/80+), neutrophils (CD11b+F4/80−), NK cells (CD3−NK1.1+), NK T cells (CD3+NK1.1+), CD11c+ DC (CD3−CD11c+), CD8a+ DC (CD3−CD11c+CD8+), pDCs (CD3−CD11c+B220+), and Treg (CD3+CD4+FoxP3+). Data were acquired on a LSRFortessa cell analyzer (BD Biosciences) and analyzed with FlowJo software (TreeStar). Cell sorting was performed on a FACSAria II cell sorter (BD Biosciences).
Statistical Analysis.
The statistical significance of differences between groups was analyzed by Student’s t test or one-way ANOVA with Tukey’s multiple comparisons test using GraphPad Prism software. All differences with P values < 0.05 were considered significant. P values are denoted by *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant with P > 0.05.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants AI125581 (to B.B.) and DK107886 and AI095542 (to E.E.T.), Immunology T32 Training Grant AI005184 (to W.M.), and by the Lyda Hill Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901407116/-/DCSupplemental.
References
- 1.Lo B, et al. (2015) AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science 349:436–440. [DOI] [PubMed] [Google Scholar]
- 2.Alkhairy OK, et al. (2016) Spectrum of phenotypes associated with mutations in LRBA. J Clin Immunol 36:33–45. [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Herrera G, et al. (2012) Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am J Hum Genet 90:986–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gámez-Díaz L, et al. (2016) The extended phenotype of LPS-responsive beige-like anchor protein (LRBA) deficiency. J Allergy Clin Immunol 137:223–230. [DOI] [PubMed] [Google Scholar]
- 5.Charbonnier LM, et al. (2015) Regulatory T-cell deficiency and immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like disorder caused by loss-of-function mutations in LRBA. J Allergy Clin Immunol 135:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JW, Howson J, Haller E, Kerr WG (2001) Identification of a novel lipopolysaccharide-inducible gene with key features of both A kinase anchor proteins and chs1/beige proteins. J Immunol 166:4586–4595. [DOI] [PubMed] [Google Scholar]
- 7.Gámez-Díaz L, et al. (2017) Immunological phenotype of the murine Lrba knockout. Immunol Cell Biol 95:789–802. [DOI] [PubMed] [Google Scholar]
- 8.Schneider H, et al. (1999) Cytolytic T lymphocyte-associated antigen-4 and the TCR zeta/CD3 complex, but not CD28, interact with clathrin adaptor complexes AP-1 and AP-2. J Immunol 163:1868–1879. [PubMed] [Google Scholar]
- 9.Turer E, et al. (2017) Creatine maintains intestinal homeostasis and protects against colitis. Proc Natl Acad Sci USA 114:E1273–E1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, et al. (2015) Real-time resolution of point mutations that cause phenovariance in mice. Proc Natl Acad Sci USA 112:E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berndt BE, Zhang M, Chen GH, Huffnagle GB, Kao JY (2007) The role of dendritic cells in the development of acute dextran sulfate sodium colitis. J Immunol 179:6255–6262. [DOI] [PubMed] [Google Scholar]
- 12.Arimura K, et al. (2017) Crucial role of plasmacytoid dendritic cells in the development of acute colitis through the regulation of intestinal inflammation. Mucosal Immunol 10:957–970. [DOI] [PubMed] [Google Scholar]
- 13.Abe K, et al. (2007) Conventional dendritic cells regulate the outcome of colonic inflammation independently of T cells. Proc Natl Acad Sci USA 104:17022–17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung S, et al. (2002) In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity 17:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Won S, et al. (2012) Increased susceptibility to DNA virus infection in mice with a GCN2 mutation. J Virol 86:1802–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colina R, et al. (2008) Translational control of the innate immune response through IRF-7. Nature 452:323–328. [DOI] [PubMed] [Google Scholar]
- 17.Honda K, et al. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777. [DOI] [PubMed] [Google Scholar]
- 18.Sato M, et al. (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548. [DOI] [PubMed] [Google Scholar]
- 19.Ma C, Spies NP, Gong T, Jones CX, Chu WM (2015) Involvement of DNA-PKcs in the type I IFN response to CpG-ODNs in conventional dendritic cells in TLR9-dependent or -independent manners. PLoS One 10:e0121371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiducci C, et al. (2008) PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J Exp Med 205:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaur S, et al. (2008) Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci USA 105:4808–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao W, et al. (2008) Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol 9:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei CQ, et al. (2013) FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J Biol Chem 288:12596–12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litvak V, et al. (2012) A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490:421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luron L, Saliba D, Blazek K, Lanfrancotti A, Udalova IA (2012) FOXO3 as a new IKK-ε-controlled check-point of regulation of IFN-β expression. Eur J Immunol 42:1030–1037. [DOI] [PubMed] [Google Scholar]
- 26.Brinkmann MM, et al. (2007) The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol 177:265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL (2008) UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452:234–238. [DOI] [PubMed] [Google Scholar]
- 28.Tabeta K, et al. (2006) The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol 7:156–164. [DOI] [PubMed] [Google Scholar]
- 29.Casrouge A, et al. (2006) Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314:308–312. [DOI] [PubMed] [Google Scholar]
- 30.Govoni G, et al. (1996) The Bcg/Ity/Lsh locus: Genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1 Gly169 allele. Infect Immun 64:2923–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown DE, et al. (2013) Salmonella enterica causes more severe inflammatory disease in C57/BL6 Nramp1G169 mice than Sv129S6 mice. Vet Pathol 50:867–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon MM, et al. (2013) A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol 14:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kole A, et al. (2013) Type I IFNs regulate effector and regulatory T cell accumulation and anti-inflammatory cytokine production during T cell-mediated colitis. J Immunol 191:2771–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tschurtschenthaler M, et al. (2014) Type I interferon signalling in the intestinal epithelium affects Paneth cells, microbial ecology and epithelial regeneration. Gut 63:1921–1931. [DOI] [PubMed] [Google Scholar]
- 35.McFarland AP, et al. (2011) Localized delivery of interferon-β by Lactobacillus exacerbates experimental colitis. PLoS One 6:e16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauch I, et al. (2014) Type I interferons have opposing effects during the emergence and recovery phases of colitis. Eur J Immunol 44:2749–2760. [DOI] [PubMed] [Google Scholar]
- 37.Daig R, et al. (1996) Increased interleukin 8 expression in the colon mucosa of patients with inflammatory bowel disease. Gut 38:216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ajuebor MN, Hogaboam CM, Kunkel SL, Proudfoot AE, Wallace JL (2001) The chemokine RANTES is a crucial mediator of the progression from acute to chronic colitis in the rat. J Immunol 166:552–558. [DOI] [PubMed] [Google Scholar]
- 39.Ansari N, et al. (2006) Comparison of RANTES expression in Crohn’s disease and ulcerative colitis: An aid in the differential diagnosis? J Clin Pathol 59:1066–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh UP, et al. (2016) Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine 77:44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCormack G, et al. (2001) Tissue cytokine and chemokine expression in inflammatory bowel disease. Inflamm Res 50:491–495. [DOI] [PubMed] [Google Scholar]
- 42.Ahern PP, et al. (2010) Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33:279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen D, et al. (2006) IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest 116:1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blasius AL, et al. (2010) Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci USA 107:19973–19978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diacovo TG, Blasius AL, Mak TW, Cella M, Colonna M (2005) Adhesive mechanisms governing interferon-producing cell recruitment into lymph nodes. J Exp Med 202:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ostanin DV, et al. (2009) T cell transfer model of chronic colitis: Concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol 296:G135–G146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov II, et al. (2006) The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126:1121–1133. [DOI] [PubMed] [Google Scholar]
- 48.Arnold CN, et al. (2012) A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc Natl Acad Sci USA 109:12286–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi JH, et al. (2017) IgD class switching is initiated by microbiota and limited to mucosa-associated lymphoid tissue in mice. Proc Natl Acad Sci USA 114:E1196–E1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.