Significance
The present crystal structure of phenylalanine hydroxylase (PAH) provides the 3D structure of the full-length human PAH, both unbound and complexed with the tetrahydrobiopterin (BH4) cofactor. The BH4-bound state is physiologically relevant, keeping PAH stable and in a precatalytic state at low L-Phe concentration. Furthermore, a synthetic form of BH4 (Kuvan) is the only drug-based therapy for a subset of phenylketonuria patients. We found two tetramer conformations in the same crystal, depending on the active site occupation by BH4, which aid to understand the stabilization by BH4 and the allosteric mechanisms in PAH. The structure also reveals the increased mobility of human, compared with rat, PAH, in line with an increased predisposition to disease-associated mutations in human.
Keywords: human phenylalanine hydroxylase, phenylketonuria, X-ray crystallography, cryo-EM, allosteric regulation
Abstract
Phenylalanine hydroxylase (PAH) is a key enzyme in the catabolism of phenylalanine, and mutations in this enzyme cause phenylketonuria (PKU), a genetic disorder that leads to brain damage and mental retardation if untreated. Some patients benefit from supplementation with a synthetic formulation of the cofactor tetrahydrobiopterin (BH4) that partly acts as a pharmacological chaperone. Here we present structures of full-length human PAH (hPAH) both unbound and complexed with BH4 in the precatalytic state. Crystal structures, solved at 3.18-Å resolution, show the interactions between the cofactor and PAH, explaining the negative regulation exerted by BH4. BH4 forms several H-bonds with the N-terminal autoregulatory tail but is far from the catalytic FeII. Upon BH4 binding a polar and salt-bridge interaction network links the three PAH domains, explaining the stability conferred by BH4. Importantly, BH4 binding modulates the interaction between subunits, providing information about PAH allostery. Moreover, we also show that the cryo-EM structure of hPAH in absence of BH4 reveals a highly dynamic conformation for the tetramers. Structural analyses of the hPAH:BH4 subunits revealed that the substrate-induced movement of Tyr138 into the active site could be coupled to the displacement of BH4 from the precatalytic toward the active conformation, a molecular mechanism that was supported by site-directed mutagenesis and targeted molecular dynamics simulations. Finally, comparison of the rat and human PAH structures show that hPAH is more dynamic, which is related to amino acid substitutions that enhance the flexibility of hPAH and may increase the susceptibility to PKU-associated mutations.
Phenylalanine hydroxylase (PAH) catalyzes the tetrahydrobiopterin (BH4)-dependent hydroxylation of l-phenylalanine (l-Phe) to l-tyrosine (l-Tyr), which is the rate-limiting step in the degradation of l-Phe and l-Tyr. In humans this reaction is vital for preventing the accumulation of neurotoxic amounts of l-Phe, which is the hallmark of the disease phenylketonuria (PKU) (1–3). Patients must follow a life-long protein-controlled diet to avoid severe neurological symptoms, but in a subset of the patients the diet can be alleviated by sapropterin dihydrochloride (Kuvan), a synthetic form of the cofactor BH4 (1) that shows multifactorial effects on PAH mutants, including their stabilization (4). As most of the mutations in PAH cause protein instability, leading to misfolding and loss-of-function (3), it is of medical interest to discover and optimize other compounds with pharmacological chaperone potential that can specifically prevent PAH misfolding and extend the range of mutations that can be rescued (5, 6). Structural information is essential in this endeavor.
Mammalian PAH is tetrameric, and its 52 kDa subunits are composed of a regulatory domain (RD, residues 1–110) with an unstructured N-terminal tail (N term; residues 1–29), a catalytic domain (CD, residues 111–410), and an oligomerization domain (OD, residues 411–452) responsible for dimerization and subsequent tetramerization (SI Appendix, Fig. S1). This oligomeric and multidomain arrangement allows the complex regulation of PAH activity, including (i) activation by the substrate l-Phe, which elicits a positive cooperative response on PAH, ensuring that neurotoxic accumulation of l-Phe is avoided, and (ii) decreasing PAH activity at low substrate concentration to preserve a lower threshold value of l-Phe for protein synthesis (for reviews, see refs. 2, 7, and 8). At low, physiological concentration of l-Phe, PAH is complexed with the cofactor BH4 (present in hepatocytes at equal concentrations as PAH subunits) that acts as a negative regulator of the activation by l-Phe (8, 9). BH4 also stabilizes PAH and increases the steady state levels of the enzyme in vivo (10, 11). While the activation by l-Phe induces a large conformational change (8), recently proposed to involve dimerization of RDs (12–14), BH4 binding elicits a limited conformational change proposed to be mostly constrained to the N-term tail (15, 16). The importance of these N-terminal 29 residues in both the inhibitory regulation by BH4 and the activation by l-Phe is evident, as PAH lacking this tail is not regulated by either BH4 or l-Phe and is constitutively active (15).
Until now, the available mammalian PAH structures are truncated, lacking one or two of the domains (17–19), or full-length without ligands, the latter only of rat PAH (rPAH) (12, 14). Understanding of the structural defects associated to PKU mutations and development of specific stabilizing therapies would highly benefit from the availability of the structure of human PAH (hPAH), both with and without ligands. Thus, we aimed to solve the long-awaited full-length structures of hPAH both in the absence of ligands and with the cofactor BH4 and have obtained two structures at 3.18-Å resolution from the same crystal, which present different occupancy by BH4. The full-length structure in absence of BH4 has been also obtained at medium resolution by cryo-EM. We here provide the unknown structures of complete hPAH with all domains and the unknown structures of full-length mammalian PAH with ligand.
Results and Discussion
Crystallization of hPAH with Two Tetramers in a Single Crystal.
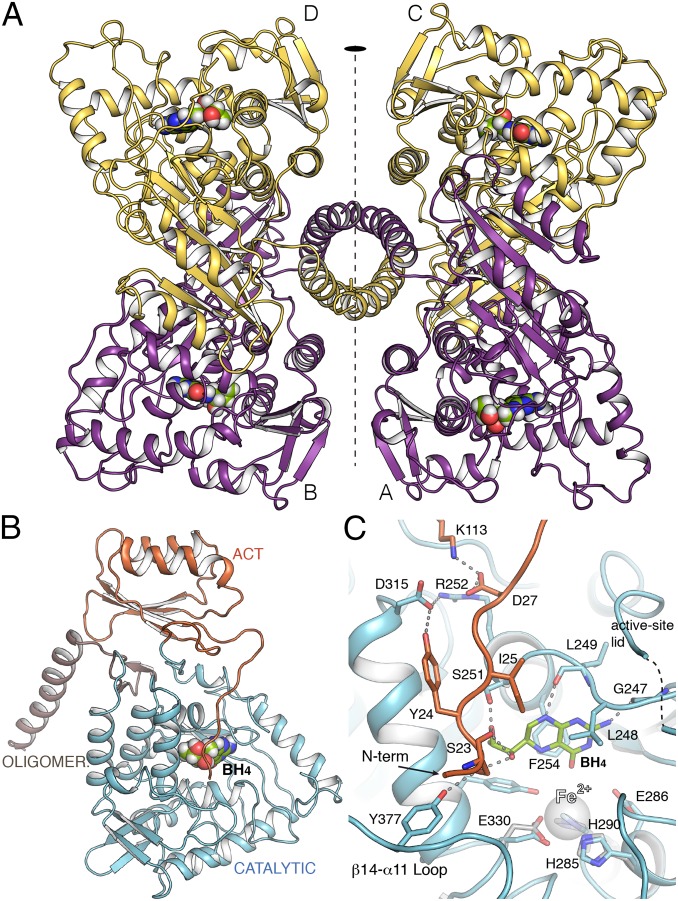
Both full-length hPAH and ∆N13-hPAH, the latter produced by cutting MBP-hPAH with factor Xa protease (20, 21), were cocrystallized with BH4 and produced crystals with similar appearance at the same conditions (see Materials and Methods). However, datasets collected from hundreds of similar looking crystals showed both low resolution and severe anisotropy, suggesting that there was still heterogeneity in the conformation of tetrameric hPAH. Nevertheless, with ∆N13-hPAH and DTT (22) we eventually obtained a crystal with a degree of anisotropy that allowed us to obtain the structural determination of hPAH at 3.18-Å resolution (Fig. 1 and SI Appendix, Fig. S2 and Table S1). Comparison with the structure of the dimeric catalytic domain (hPAH-CD)—that we also solved at 1.67-Å resolution (SI Appendix, Table S1) with previously published conditions (17) (SI Appendix, Fig. S2B)—shows that the absence of Fe in our structure does not alter the arrangement of the iron-binding 2-His-1-carboxylate facial triad (Fig. 1C). Four independent monomers of PAH were found in the asymmetric unit belonging to two different tetramers, Tet1 and Tet2, each created as a dimer of dimers by a twofold crystallographic axis. While Tet1 presents the BH4 bound in two active sites (chains C, D) and the other two (chains A, B) without the cofactor, Tet2 shows BH4 bound to all four active sites (Fig. 1). All four independent monomers in both tetramers present a similar overall structure for the backbone (RMSD ∼ 0.6 Å for Cα superimposition), with hPAH arranged as a tetramer through the C-terminal oligomerization helix, following an approximate 222 symmetry between monomers, only the twofold symmetry between dimers being strict (Fig. 1A). Each monomer presented electron density for all domains (residues 22–450) (Fig. 1A and SI Appendix, Fig. S2). The tetramers can be roughly encased in a rectangle with a short edge along the dimer (∼70 Å) and a long edge (∼110 Å) between the two dimers, while differences in the long edges are observed between Tet1 and Tet2 (SI Appendix, Fig. S3). The RD is essential for this specific oligomeric arrangement in hPAH as deduced by comparison with the human truncated form lacking RD (PDB 2PAH) (18) (SI Appendix, Fig. S4).
Fig. 1.
Crystal structure of human PAH. (A) Ribbon representation of hPAH in complex with BH4 at the four active sites (Tet2). The tetramer is formed as a dimer of dimers by a twofold symmetry (depicted). BH4 cofactor is drawn as spheres. (B) The hPAH monomer with each domain colored differently. (C) Detailed view of the active site of hPAH in complex with BH4 (green sticks). Relevant residues are represented as capped sticks and labeled. The FeII cation, not present in the structure, is represented as a transparent sphere for comparison purposes. Its location is obtained by direct superposition with the CD of hPAH (obtained in this work). Residues from this structure involved in the interaction with FeII cation are shown in gray sticks.
The presence of unbound and BH4-bound states—from here on referred to as apo and holo states, respectively—in the same crystal allowed a direct comparison between both conformations. Until now, investigations of how cofactor binding affects the conformation of the RD have come from modeling through superimposition of truncated structures (apo rPAH containing RD and CD, PDB 1PHZ) onto hPAH-CD with ligands (i.e., with BH4, PDB 1J8U, and with both BH4 and with substrate analog thienylalanine, THA, PDB 1MMK). These modeling analyses have predicted interactions of BH4 with S23 at the flexible and unstructured N term (15, 16, 23), a segment that has been termed an intrasteric autoregulatory sequence (IARS) (19). Comparing our apo and holo structures with each other and with the previous ligand-bound truncated forms reveals main events implicated in the negative regulation of PAH by BH4 and derived enzyme stabilization, as well as in the conversion to the active BH4-bound structure in the presence of substrate. As detailed below, the conformational events involve so far unidentified networks of residues, far beyond the IARS.
Structural Effects of BH4 Binding Associated to Negative Regulation of Activation by l-Phe and Enzyme Stabilization.
The BH4 binding-site is flanked by the N term (residues 21–32), the active-site lid (130–150), the FeII-coordinating residues, the β6-α7 loop (residues 245–251), and F254 (Fig. 1C). BH4 is sandwiched between hydrophobic residues, F254 from one side and L248 and I25 from the other (Fig. 1C). Besides, some polar interactions are observed between BH4 and residues from the IARS and the β6-α7 loop (Fig. 1C). The direct interaction of S23 in the N term with BH4, via H-bonds with both O1′ and O2′ in the dihydroxypropyl side-chain, is observed in our BH4-bound structure and confirms previously predicted interactions with these hydroxyls in the natural (6R)-BH4 configuration (15, 16, 23). These interactions appear related to the inhibitory effect of BH4 on l-Phe activation of PAH and explain why the cofactor analog 6-methyl-tetrahydropterin (6-MPH4) is not inhibitory, and (6S)-BH4 is less inhibitory (24).
The activation of PAH by l-Phe requires a slow (seconds-to-minutes timescale with 1 mM l-Phe, depending on the temperature) global conformational change, and the activation rate is slower for the BH4-preincubated than for the unbound enzyme (8–10, 23), associated to the negative regulation of PAH activation exerted by BH4. Recent structural analyses have shown that full activation involves the shift and dimerization of the RDs, with concomitant movement of the N-term IARS that releases from the CD (12, 14). In apo hPAH, S23 H-bonds with Y377 in the CD, whereas in the holo structure S23 is also engaged in a larger H-bond network involving BH4 and extends to residues in the CD and the RD (Fig. 1C). Structural comparison between apo- and holo-hPAH monomers reveals other BH4-induced changes, beyond the N-term IARS (Fig. 2A). Surprisingly, the conformation of the N term is similar for apo and holo hPAH (Fig. 2A) and also for apo rPAH (12), showing that binding of BH4 does not induce large conformational changes on the N term. Comparative analyses of apo- and holo-hPAH monomers by calculating differences in interresidue distances between the apo and all holo monomers (see Materials and Methods) revealed many distances that facilitate or abolish interactions upon BH4 binding, most involving residues in the CD (SI Appendix, Fig. S5 and Table S2). Out of the 35 largest distance differences, about half engage intra-CD residue pairs (19 pairs), most concerning residues around the BH4 binding site (SI Appendix, Table S2). These are followed by intra-RD residues (six pairs), between RD and CD (five pairs), between RD and OD (three pairs), and intra-OD and between CD and OD (one pair each). Thus, upon BH4 binding, changes are observed that link the CD with the OD, both directly and via the RD. Actually, the largest change (∼6 Å) is the increased distance between K73 in the RD and L430 at the start of the tetramerization helix in the OD, indicating conformational changes affecting oligomerization and allostery (see below). In the active site, the most apparent change involves residues L248 and F254 in the BH4 binding-site (Fig. 2 A, 1 and SI Appendix, Table S2). In the apo subunit, these two residues show a direct van der Waals interaction (∼3 Å) and partly occupy the BH4 binding-site, whereas in the BH4-bound active site they move to sandwich the cofactor (Fig. 2A). Interestingly, binding of BH4 changes the interaction network between the N term and the CD (Fig. 2 A, 1). In the apo state, residue D27 makes H-bond interactions with α7 residues (notably, S251), while in the holo state, it makes a salt-bridge interaction with R252 and K113 instead. The salt-bridge interaction between E26 and K113 observed in apo state is lost in holo state (Fig. 2 A, 1 and SI Appendix, Fig. S5). Interactions of BH4 with the N term and the CD, as well as the network of polar and salt-bridge interactions linking the RD (Y24, D27, K113) with the CD (D315, R252) (Fig. 1C), appear determinant for the regulatory inhibitory effect and the stabilization of BH4. Differential scanning calorimetry (DSC) experiments at 200 °C/h with the BH4 analog 7,8-dihydrobiopterin (BH2) (25) (SI Appendix, Fig. S6A) support the stabilization effect of BH4 beyond the CD. Hence, while apo hPAH presents the expected unfolding transition temperatures at ∼49 and 58 °C, previously associated to the RD and the CD, respectively (26), both transition temperatures increased ∼2 °C in holo hPAH (SI Appendix, Fig. S6A).
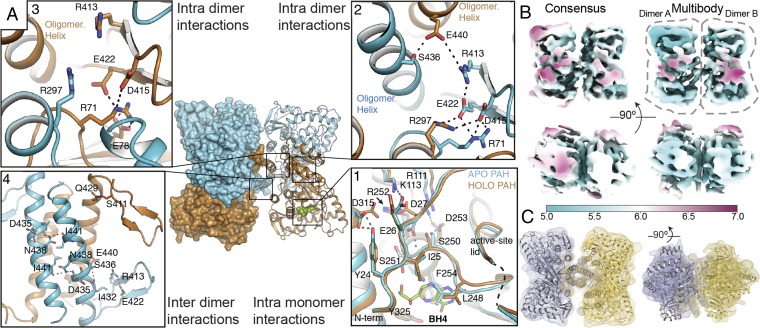
Fig. 2.
Long-range effects in hPAH upon BH4 binding and cryo-EM of apo hPAH. (A) Tet1 with monomers colored differently (apo in cyan and holo in ochre). (A1) Structural comparison of the active site in apo hPAH vs. BH4-bound hPAH. (A2 and A3) Detailed view of the apo–holo dimer interface showing network of salt-bridge interactions. (A4) Intermolecular polar interactions in the OD of Tet1. (B) Top and side view of the cryo-EM reconstruction of apo-hPAH tetramer (Left) and multibody reconstruction of two independent dimers (Right). Map color represents the map local-resolution estimation (see color guide). Note that the multibody refined map is a composite of two dimers refined independently. (C) Rigid body structural fitting of Tet2 dimer onto the cryo-EM multibody reconstruction of two independent dimers.
Long-Range Structural Effects of BH4 Binding.
A different disposition of the monomers in Tet1 and Tet2 is observed depending on occupancy by BH4 that changes the overall dimensions of the tetramer, the distances between monomers, and the relative disposition (∼5-Å gliding) of one dimer versus the other through the oligomerization helices (SI Appendix, Fig. S3). Considering that all monomers presented a nearly identical backbone we reasoned that the observed differences should rely on different structural arrangements inside the dimers that would promote changes in the tetramer. Superposition of the holo monomers in apo–holo and holo–holo dimers shows a rotation of ∼3° and displacements up to 3 Å between monomers. Thus, the changes observed inside the active site of a monomer upon BH4 binding (Fig. 2 A, 1) must also propagate to the dimeric partner (intradimer interactions) and to the tetramer (interdimer interactions). Structural analysis of Tet1 and Tet2 provides clues about such long-range effects of BH4 binding (Fig. 2A). Changes are observed at the dimer interface between the apo–holo or holo–holo dimers (SI Appendix, Fig. S7). It is worthwhile to mention residue R297 that participates in an intradimer interaction network with residues E422, D415 (OD), and R71 (RD) only when BH4 is bound (Fig. 2 A, 2 and 3 and SI Appendix, Fig. S7). Another relevant residue in this network is R413 that, in the apo monomer, can establish a salt-bridge interaction with the oligomerization helix (residue E440) of the dimeric partner (Fig. 2 A, 2). This interaction is not seen between holo monomers (SI Appendix, Fig. S7). As previously mentioned, the quaternary structure of hPAH is also affected by BH4 binding; the most significative is a gliding of ∼5 Å between dimers along the OD helices (Movies 1 and 2) changing the network of polar interactions between the OD helices in the tetramer (Fig. 2 A, 4 and SI Appendix, Fig. S8). Contrary to tyrosine hydroxylases or tryptophan hydroxylases the OD helices in mammalian PAH lack a proper Leu zipper and present abundance of polar residues inside the coiled-coil structure, as has already been noticed in the structure of rPAH (12). The lack of a classic leucine zipper explains why PAH can dissociate to dimers as well as the conformational variability in the C-terminal helices.
To further investigate the dynamics of apo-hPAH tetramer, i.e., with all four subunits in the apo state, we performed single-particle cryoelectron microscopy (cryo-EM) studies on the same full-length construct but in the absence of cofactor (see Materials and Methods and SI Appendix, Table S3). A consensus refinement of the hPAH tetramer yielded a cryo-EM map of resolution ranging from 5 to 7 Å due to the high mobility of the monomers within the tetramer (Fig. 2B). Three-dimensional classification allowed for the separation of different conformations in the dataset (SI Appendix, Fig. S9), but these still harbored the same flexibility-related problems as the consensus refinement. To address these flexibility issues, we performed multibody refinement by masking individual dimers (Fig. 2B) (27, 28). This procedure improved resolution to ∼5 Å for most of the protein except for the RD (Fig. 2B), resulting in an excellent fit of the crystal structure onto the resulting maps (Fig. 2C). Moreover, variance analysis using principal component analysis showed that most of the orientational differences between dimers can be described by three main components (SI Appendix, Fig. S10). Interestingly, the amplitude distribution along these components is monomodal, indicating that these are continuous movements in which dimers rotate around each other mainly as rigid bodies (SI Appendix, Fig. S10C and Movie 3). Thus, in absence of BH4, hPAH dimers present a large range of motion inside the tetramer. While there are no noticeable changes in the internal structure of the CD, the RD seems to present extra flexibility.
In agreement with crystallographic and cryo-EM data, molecular dynamics (MD) simulations (eight 100-ns-long runs) of hPAH performed with and without BH4 (see Materials and Methods and SI Appendix, Fig. S11) show a highly dynamic tetrameric structure in both apo and holo states, with large-scale interdimer motions, but relatively small intradimer fluctuations. We calculated the fluctuation profile (root-mean-square fluctuation; RMSF) for each monomer in each of the simulations. Comparative analysis shows that BH4 stabilizes significantly (P < 0.05) several regions in the CD, but increases flexibility in loop α14–β11 (residues 374–384) that contains Y377 residue (Fig. 1C and SI Appendix, Fig. S12). This loop, important for embracing the substrate, moves ∼5 Å closer to the active site upon substrate binding (29). The Y138 loop, termed the active site lid, also moves considerably (>10 Å) toward the active site upon l-Phe binding (29), but, contrary to loop α14–β11, it loses mobility upon BH4 binding (SI Appendix, Fig. S12).
Significance of the PAH:BH4 Precatalytic Complex and the Role of Y138.
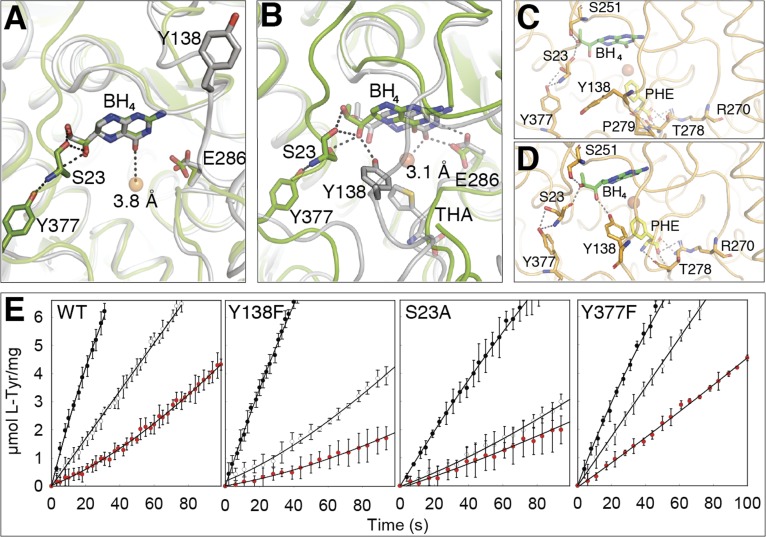
In the holo-hPAH subunits, BH4 is bound, placing its O4 atom, the site of hydroxylation of the pteridine ring during catalysis, at 3.8 Å from the catalytic iron (Fig. 3A), 0.7 Å longer than the required distance to form the FeII–OO–BH4 adduct and generation of the FeIV = O hydroxylating intermediate (29, 30, 31). The cofactor bound hPAH structure presented in this work thus corresponds to a precatalytic conformation, which is of high significant physiological relevance, as PAH and BH4 coexist in hepatocytes in stoichiometric amounts, forming a stable complex at normal low concentrations of l-Phe in vivo (≤54 µM) (10). BH4-bound hPAH presents lower initial catalytic activity and is activated by l-Phe at a slower rate compared with the ligand-free enzyme (8–10, 23) (SI Appendix, Fig. S6B and see below). The physiological significance of the negative regulation of PAH activation by BH4 is related to the maintenance of a low hydroxylating activity at low substrate concentrations to preserve a basal level of l-Phe required for protein synthesis (9, 10). Furthermore, the stabilization of PAH by BH4 binding (SI Appendix, Fig. S6A) contributes to maintaining the in vivo half-life of the enzyme, as supported by the decrease of steady state levels of PAH caused by BH4 loss in cells and in liver (11, 32, 33). No atomic structural information is available on the large conformational changes associated to complete l-Phe activation, including the dimerization of the ACT domains (12, 14). Nevertheless, our structures provide insights into the substrate-induced conformational changes around the active site related to the transition from the precatalytic to the catalytic state upon entrance of the substrate at the active site. Superposition of holo hPAH with the binary hPAH-CD:BH4 and ternary hPAH-CD:BH4:THA complexes reveals a possible role of Y138 in this transition by interacting with the O1′ hydroxyl and contributing to the displacement of the cofactor 0.7 and 2.6 Å toward the iron and Glu286, respectively (Fig. 3 A and B). To probe plausible transition pathways occurring upon substrate binding, we performed a 2-ns-long targeted MD (TMD) simulation of Tet2 in complex with l-Phe and BH4 (Movie 4). With this simulation technique we gradually steer one catalytic domain of Tet2 to a target structure defined by the ternary hPAH-CD:BH4:THA complex (PDB 1MMK). Of note, the binding site of l-Phe was solvent-accessible, and there was no clash of the bound substrate with residues in hPAH. At ∼0.8 ns of the simulation, Y138 has moved ∼10 Å from its original position toward the active site and forms a transient H-bond with the carbonyl O of P279. After ∼1.4 ns (Fig. 3C), an H-bond is formed between the side-chain of T278 and the amino group of l-Phe, and the translation of Y138 toward the active site is accelerated; and at the end of the simulation, an H-bond is formed between the phenol O of Y138 and the O1′ of BH4 in its precatalytic position (Fig. 3D). At that point the cofactor only maintains a single H-bond interaction with S23, which gets further weakened at the end of the simulation when S23 interacts with S251. To further investigate the role of the hydroxyl group of Y138 in initiating the movement of BH4 toward the catalytic binding mode, as well as the relevance of S23 and its interacting CD-residue Y377 (Figs. 1C and 3 A–D), we prepared the hPAH mutants Y138F, S23A, and Y377F, which were expressed and purified as tetramers, but at lower yield than wild-type (WT). Other residues that interact with BH4 in the precatalytic conformation (region 245–255, 286, 322, and 325) also interact with BH4 in the catalytic conformation, and, in addition, these sites are actually associated with severe destabilization of PAH (www.biopku.org), and thus not suitable for mutagenesis investigations. The initial reaction rates at 37 °C, comparative to the hPAH-WT, were measured for the four enzymes in three initial states, i.e., l-Phe-activated (preincubated with 1 mM l-Phe), nonactivated (not preincubated with either substrate or cofactor), and precatalytic BH4-bound (preincubated with 75 µM BH4) (Fig. 3E and SI Appendix, Fig. S6B). The concentrations of substrate and cofactor were otherwise identical at the start of the reactions. Activated Y138F shows reduced activity, to 70% of WT, but more remarkable is the large reduction in activity for the nonactivated and precatalytic states for this mutant (to 35–45% of WT). This reduction is not associated to different steady state kinetic parameters, as the Y138F mutant presents similar Km(BH4), S0.5(l-Phe), and hill coefficient (h) as hPAH-WT (SI Appendix, Fig. S6C), supporting a role of the Y138 hydroxyl group in disengaging BH4 from the precatalytic conformation in the full-length enzyme. The other two mutants, S23A and Y377F, were prepared to validate the effect of the H-bonding interactions centered at S23 to stabilize the enzyme–cofactor complex in the precatalytic state. Both mutants, and, notably, S23A, show reduced activity for the l-Phe–activated state and increased S0.5(l-Phe) (SI Appendix, Fig. S6C), indicating the participation of these residues in substrate activation, through the side-chain O in the case of S23. Moreover, the mutant S23A is expected to maintain the negative regulatory interactions with O1′ and O2′ of BH4 through its backbone carbonyl and amide groups, explaining the low basal activity of the BH4-bound state. On the other hand, as the function of Y377 appears related to its interaction with S23 for proper localization of this residue and the IARS through H-binding interactions (Fig. 3 A–C), the rupture of the H-bond with S23 in the Y377F mutant results in an increased activity for the unactivated states (Fig. 3E and SI Appendix, Fig. S6B).
Fig. 3.
Transition to active cofactor binding. (A) Superposition of hPAH:BH4 (Tet2A; green) with hPAH-CD:BH4 (PDB 1J8U; gray). In both, BH4 is in precatalytic position, with the O4 atom 3.8 Å from the FeII and Y138 (not visible in Tet2) ∼20 Å away. H-bonds shown as dashed lines. (B) Superposition of hPAH:BH4 (Tet2A; green) with hPAH-CD:BH4:THA (PDB 1MMK; gray). BH4 is at the catalytic position, interacting with E286 and with the O4 3.1 Å from FeII and generates the FeII–O2–BH4 adduct. The movement of Y138, induced by substrate analog (THA) binding, positions the hydroxyl group of Y138 2.9 Å away from O1′ in the dihydroxypropyl side-chain of BH4. (C and D) Snapshots from the TMD of l-Phe binding to full-length hPAH (Tet2A). At ∼1.41 ns, l-Phe H-bonds to T278, and the translation of Y138 is accelerated (C). At the end of the 2-ns simulation the phenol O of Y138 H-bonds with the O1′ of BH4 (D). (E) Initial reaction rates for hPAH-WT and mutants, starting the reaction as l-Phe–activated (black circles), nonactivated (white circles), and precatalytic BH4-bound (red circles).
Human and Rat PAH Present Significant Structural Differences despite High Sequence Homology.
PAH from human and rat share a high sequence similarity (96%). Among the 452 residues of hPAH there are only 16 nonconservative and 14 conservative mutations compared with rPAH (SI Appendix, Fig. S13). Conservative mutations are distributed among the three domains, and nonconservative mutations are located in RD and CD. Considering differences in size of RD and CD, mutations are strongly concentrated in the RD (8% of residues of RD are mutated between hPAH and rPAH vs. 2% of the CD), supporting differences in the regulatory properties of both enzymes (34).
Overall structures of the human and rat PAH monomers are very similar (RMSD values for the backbone ranging from 0.8 to 1.1 Å) (SI Appendix, Fig. S14A). Major differences are seen at the interface between RD and CD [loop β1–α1 (44–46)], in α2 (88–100) of the RD, residues 115 in the hinge region 110–118 (loop β4–β5), 143, 338 in the CD, and at the beginning of the OD helix (L427-K438) (SI Appendix, Figs. S14 and S15) that provoke differences in tetramer arrangement (SI Appendix, Fig. S15). Regions with largest conformational changes in the RD are located around nonconservative mutations, i.e., C29 (S29 in rPAH), L88 (K88 in rPAH), K114 (E114 in rPAH), and D116 (N116 in rPAH) (SI Appendix, Fig. S14B). In the RD/CD interface it is worthwhile to mention the salt-bridge in rPAH between E114 and K88 (K114 and L88 in hPAH), linking α2 with loop 110–118, that obviously is absent in hPAH, where a different interaction pattern is observed in this region. In the apo state of hPAH, salt-bridges are observed between D122 and both K114 and K85 and D116 and R99. Consequently, both sides of the α2 helix in the RD, i.e., the C-terminal (K99) side in hPAH and the N-terminal (K85/K88) side in rPAH, are involved in interactions with the important loop 110–118, located at the end of the RD, at the boundary between the RD and the CD. Differences are also observed in the conformation of the OD helices, i.e., in rPAH the C-terminal helices of the four monomers adopt two distinct orientations with respect to the CD, differing by a tilt of ∼10°, and with the monomers situated across the diagonal of the tetramer containing similarly positioned helices (12). Structural analysis reveals that the different conformations relate with a different pattern in polar interactions at the beginning of the OD helices depending on whether Q428 is making an H-bond with N73 or is inserted in the interface between the CD and the RD (SI Appendix, Fig. S15). In hPAH there are no such distinct orientations for the C-terminal OD helices that are placed in between the two conformations observed in rPAH (SI Appendix, Fig. S14). In hPAH the interaction network in this region is affected by the presence of the BH4 cofactor in the active site. In the apo, Q428 makes H-bond interactions with both the backbone atoms of the region connecting the OD helix with the protein core and with residue E78 (SI Appendix, Fig. S14C), while in the holo, Q428 interacts with S70 and, interestingly, K73 (N73 in rat) is salt-bridging with D425, thus contributing to the locked, inhibited structure in the holo state of hPAH.
The effect of mutations between human and rat PAH was studied by parallel MDs with our hPAH structures and existing rPAH structure(s) (see Materials and Methods and SI Appendix, Fig. S16). In agreement with our structural results, fluctuation analyses showed that, overall, hPAH is more dynamic, notably, in the regions with significant structural differences (SI Appendix, Fig. S17), such as RD, showing nonconservative mutations (SI Appendix, Fig. S13B), and the area around the OD helix.
Relevance for HPA and BH4-Responsive PKU.
Increased understanding of the high vulnerability of human enzymes—notably, those highly regulated, associated with central metabolic pathways—to disease-associated mutations points to evolutionary divergence from consensus amino acids that confer stabilization (35). This evolutionary mechanism also appears to describe the adaptation of hPAH toward a less stable and highly mobile enzyme (SI Appendix, Fig. S17), most probably associated with the acquisition of the flexibility required for prompter response to changes in substrate and cofactor concentration along the evolution of the neuronal system (2). Flexibility is, however, often traded off with an increased susceptibility to mutations (35). Indeed, for the more than 900 hPAH mutations causing HPA/PKU (www.biopku.org), several studies have shown the relationship between hPAH protein stability and both remaining activity and allelic phenotype (3, 36). The enzyme destabilization caused by each mutation is actually highly predictive of patient phenotype and therapeutic response to BH4 supplementation (3). As here reported, the stabilization by BH4 is initiated at its binding-site in the CD but also affects the whole structure through networks that propagate to the RD and OD both inside the monomer (Fig. 2A and SI Appendix, Fig. S5) and intradimer (Fig. 2). HPA/PKU-associated mutations are located throughout the hPAH structure, but seem to be concentrated in hotspot regions with highly destabilizing mutations (235–281, corresponding to exon 7, and 282–330 to exons 8 and 9 in the PAH gene) (36). These regions include the flexible loop β6–α7 (244–250), whose conformation is modified (Fig. 2 A, 1 and SI Appendix, Fig. S5) and rigidified (SI Appendix, Fig. S12) upon BH4 binding and presents a high mutation frequency, with several mutations associated with moderate-to-severe (residues P244, V245, A246, G247, and L249) or mild (L248) HPA/PKU phenotype (www.biopku.org; ref. 36). The hotspot regions also include mutations at R252, F254, and D315 (involved in changes from apo to holo conformation) (www.biopku.org; ref. 36). Together with S251, R252 appears essential for the stabilizing networks established with RD residues D27 in the IARS upon BH4 binding (Fig. 2 A, 1 and SI Appendix, Fig. S5). Likewise, the propagation of BH4 binding from the CD to the OD through the RD toward the other subunits in the dimer also involves regions with high mutation frequency, such as the β2–β3 loop, which interacts with R297, D415, and E422 (Fig. 2A).
In conclusion, the identification of the networks that transmit the BH4-bound state from the binding site at the CD to the other regions in the monomer, dimer, and tetramer of hPAH increments our understanding of the negative regulation of PAH by its cofactor and the stabilization caused by BH4-binding, observed both for WT and mutant hPAH (4, 11). Thus, the structures of hPAH tetramers totally and partially bound with BH4 presented here provide a rationale for BH4-responsive PKU by sapropterin dihydrochloride and pave the way to new stabilizing/chaperoning therapeutic approaches to address PKU.
Materials and Methods
Descriptions of the protein production; crystallization, crystallographic, and cryo-EM structural determinations; of the computational methods; of activity measurements and differential scanning calorimetry; and of the reagent preparations are in SI Appendix.
Accession Numbers.
The atomic coordinates for hPAH have been deposited in the PDB with codes 6HYC and 6HPO, for the full-length hPAH and catalytic domain, respectively. The EM maps have been deposited in the EMDB with accession number EMD-4605 (consensus refinement).
Supplementary Material
Acknowledgments
The work was supported by grants from the MICINN Spanish Ministry of Research, Innovation & Universities BFU2017-90030-P (to J.A.H.) and BFU2017-87316 (to R.F.-L.); programs Forny (248889/O30) and FRIMEDBIO (261826) from the Research Council of Norway (to A.M.); the Western Norway Regional Health Authorities (Helse Vest Projects 911959 to M.I.F. and 912246 to A.M.); and the K.G. Jebsen foundation (to M.I.F. and A.M.). We thank Kay Diederichs and Pavel Afonine for discussions and help in data processing and refinement, the staff from ALBA synchrotron facility (Barcelona) and the European Synchrotron Radiation Facility (ESRF, Grenoble) for support during data collection, the Midlands Regional Cryo-EM Facility at Leicester Institute of Structural and Chemical Biology, and Christos Savva for assistance during data acquisition. We thank the EM units from CNIO and Centro Nacional de Biotecnología-Centro de Investigaciones Biológicas (CSIC) for support with the EM facilities, and Peter Gimeson from Malvern Panalytical for help with DSC experiments and discussions. We are very grateful to Prof. Torgeir Flatmark for critical discussions on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.B. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6HYC and 6HPO). The EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) (http://www.ebi.ac.uk/pdbe/emdb) with accession code EMD-4605.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902639116/-/DCSupplemental.
References
- 1.Blau N., van Spronsen F. J., Levy H. L., Phenylketonuria. Lancet 376, 1417–1427 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Flydal M. I., Martinez A., Phenylalanine hydroxylase: Function, structure, and regulation. IUBMB Life 65, 341–349 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Wettstein S., et al. , Linking genotypes database with locus-specific database and genotype-phenotype correlation in phenylketonuria. Eur. J. Hum. Genet. 23, 302–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erlandsen H., et al. , Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations. Proc. Natl. Acad. Sci. U.S.A. 101, 16903–16908 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pey A. L., et al. , Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. J. Clin. Invest. 118, 2858–2867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos-Sierra S., et al. , Novel pharmacological chaperones that correct phenylketonuria in mice. Hum. Mol. Genet. 21, 1877–1887 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Fitzpatrick P. F., Allosteric regulation of phenylalanine hydroxylase. Arch. Biochem. Biophys. 519, 194–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufman S., The phenylalanine hydroxylating system. Adv. Enzymol. Relat. Areas Mol. Biol. 67, 77–264 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Xia T., Gray D. W., Shiman R., Regulation of rat liver phenylalanine hydroxylase. III. Control of catalysis by (6R)-tetrahydrobiopterin and phenylalanine. J. Biol. Chem. 269, 24657–24665 (1994). [PubMed] [Google Scholar]
- 10.Mitnaul L. J., Shiman R., Coordinate regulation of tetrahydrobiopterin turnover and phenylalanine hydroxylase activity in rat liver cells. Proc. Natl. Acad. Sci. U.S.A. 92, 885–889 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thöny B., Ding Z., Martínez A., Tetrahydrobiopterin protects phenylalanine hydroxylase activity in vivo: Implications for tetrahydrobiopterin-responsive hyperphenylalaninemia. FEBS Lett. 577, 507–511 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Arturo E. C., et al. , First structure of full-length mammalian phenylalanine hydroxylase reveals the architecture of an autoinhibited tetramer. Proc. Natl. Acad. Sci. U.S.A. 113, 2394–2399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffe E. K., New protein structures provide an updated understanding of phenylketonuria. Mol. Genet. Metab. 121, 289–296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meisburger S. P., et al. , Domain movements upon activation of phenylalanine hydroxylase characterized by crystallography and chromatography-coupled small-angle X-ray scattering. J. Am. Chem. Soc. 138, 6506–6516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings I. G., Teh T., Kobe B., Essential role of the N-terminal autoregulatory sequence in the regulation of phenylalanine hydroxylase. FEBS Lett. 488, 196–200 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Pey A. L., Thórólfsson M., Teigen K., Ugarte M., Martínez A., Thermodynamic characterization of the binding of tetrahydropterins to phenylalanine hydroxylase. J. Am. Chem. Soc. 126, 13670–13678 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Erlandsen H., et al. , Crystal structure of the catalytic domain of human phenylalanine hydroxylase reveals the structural basis for phenylketonuria. Nat. Struct. Biol. 4, 995–1000 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Fusetti F., Erlandsen H., Flatmark T., Stevens R. C., Structure of tetrameric human phenylalanine hydroxylase and its implications for phenylketonuria. J. Biol. Chem. 273, 16962–16967 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Kobe B., et al. , Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 6, 442–448 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Martinez A., et al. , Expression of recombinant human phenylalanine hydroxylase as fusion protein in Escherichia coli circumvents proteolytic degradation by host cell proteases. Isolation and characterization of the wild-type enzyme. Biochem. J. 306, 589–597 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Døskeland A. P., Martinez A., Knappskog P. M., Flatmark T., Phosphorylation of recombinant human phenylalanine hydroxylase: Effect on catalytic activity, substrate activation and protection against non-specific cleavage of the fusion protein by restriction protease. Biochem. J. 313, 409–414 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez A., Olafsdottir S., Haavik J., Flatmark T., Inactivation of purified phenylalanine hydroxylase by dithiothreitol. Biochem. Biophys. Res. Commun. 182, 92–98 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Solstad T., Stokka A. J., Andersen O. A., Flatmark T., Studies on the regulatory properties of the pterin cofactor and dopamine bound at the active site of human phenylalanine hydroxylase. Eur. J. Biochem. 270, 981–990 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Haavik J., Døskeland A. P., Flatmark T., Stereoselective effects in the interactions of pterin cofactors with rat-liver phenylalanine 4-monooxygenase. Eur. J. Biochem. 160, 1–8 (1986). [DOI] [PubMed] [Google Scholar]
- 25.Erlandsen H., Bjørgo E., Flatmark T., Stevens R. C., Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase. Biochemistry 39, 2208–2217 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Thórólfsson M., et al. , L-phenylalanine binding and domain organization in human phenylalanine hydroxylase: A differential scanning calorimetry study. Biochemistry 41, 7573–7585 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakane T., Kimanius D., Lindahl E., Scheres S. H., Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen O. A., Flatmark T., Hough E., Crystal structure of the ternary complex of the catalytic domain of human phenylalanine hydroxylase with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine, and its implications for the mechanism of catalysis and substrate activation. J. Mol. Biol. 320, 1095–1108 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Andersen O. A., Flatmark T., Hough E., High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin. J. Mol. Biol. 314, 279–291 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Panay A. J., Lee M., Krebs C., Bollinger J. M., Fitzpatrick P. F., Evidence for a high-spin Fe(IV) species in the catalytic cycle of a bacterial phenylalanine hydroxylase. Biochemistry 50, 1928–1933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguado C., Pérez B., Ugarte M., Desviat L. R., Analysis of the effect of tetrahydrobiopterin on PAH gene expression in hepatoma cells. FEBS Lett. 580, 1697–1701 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Gunasekera R. S., Hyland K., In vivo regulation of phenylalanine hydroxylase in the genetic mutant hph-1 mouse model. Mol. Genet. Metab. 98, 264–272 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kowlessur D., Citron B. A., Kaufman S., Recombinant human phenylalanine hydroxylase: Novel regulatory and structural properties. Arch. Biochem. Biophys. 333, 85–95 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Medina-Carmona E., et al. , Enhanced vulnerability of human proteins towards disease-associated inactivation through divergent evolution. Hum. Mol. Genet. 26, 3531–3544 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Pey A. L., Stricher F., Serrano L., Martinez A., Predicted effects of missense mutations on native-state stability account for phenotypic outcome in phenylketonuria, a paradigm of misfolding diseases. Am. J. Hum. Genet. 81, 1006–1024 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.