Significance
Mitochondria contain cardiolipin, a special phospholipid that strongly interacts with proteins. Cardiolipin has a unique fatty acid composition, the disturbance of which causes the human disease Barth syndrome. However, neither the mechanism by which cardiolipin acquires specific fatty acids nor their actual function has been established. In this paper, we demonstrate that the expression of the complexes of oxidative phosphorylation is responsible for inducing the specific fatty acid composition in cardiolipin. We hypothesize that the purpose of this process is the stabilization of lipid–protein interactions in the extremely crowded environment of mitochondrial membranes.
Keywords: cardiolipin, lipids, membrane, mitochondria, respiration
Abstract
Cardiolipin (CL) is a mitochondrial phospholipid with a very specific and functionally important fatty acid composition, generated by tafazzin. However, in vitro tafazzin catalyzes a promiscuous acyl exchange that acquires specificity only in response to perturbations of the physical state of lipids. To identify the process that imposes acyl specificity onto CL remodeling in vivo, we analyzed a series of deletions and knockdowns in Saccharomyces cerevisiae and Drosophila melanogaster, including carriers, membrane homeostasis proteins, fission-fusion proteins, cristae-shape controlling and MICOS proteins, and the complexes I–V. Among those, only the complexes of oxidative phosphorylation (OXPHOS) affected the CL composition. Rather than any specific complex, it was the global impairment of the OXPHOS system that altered CL and at the same time shortened its half-life. The knockdown of OXPHOS expression had the same effect on CL as the knockdown of tafazzin in Drosophila flight muscles, including a change in CL composition and the accumulation of monolyso-CL. Thus, the assembly of OXPHOS complexes induces CL remodeling, which, in turn, leads to CL stabilization. We hypothesize that protein crowding in the OXPHOS system imposes packing stress on the lipid bilayer, which is relieved by CL remodeling to form tightly packed lipid–protein complexes.
The biogenesis of mitochondrial membranes requires the assembly of proteins and lipids. While it is known that proteins form well-defined complexes (1, 2), supercomplexes (3–5), and mesoscopic structures (6, 7), the assembly of lipids, their self-organization and their interactions, are not as clearly understood. Among the mitochondrial lipids, cardiolipin (CL) is particularly important because it binds tightly to proteins (8–10) and increases substantially their propensity to cluster and to form supercomplexes (11–13). CL is therefore critical for the coassembly of lipids and proteins in mitochondrial membranes.
The biosynthesis of CL is followed by an exchange of its four fatty acids (14–16), called CL remodeling, which is accomplished by the combined action of a phospholipase (17) and tafazzin, a phospholipid-lysophospholipid acyltransferase (18). CL remodeling seems to play a role in membrane assembly because mutations in tafazzin have profound effects on membrane homeostasis, including a reduction in the CL concentration (19–21), a lower abundance of supercomplexes (22–27), reduced efficiency of oxidative phosphorylation (28, 29), and the clinical phenotype of Barth syndrome (30). We recently found that CL remodeling is a precondition for the exceptionally long half-life of CL (26).
Two unresolved issues have emerged with regard to the mechanism of CL remodeling. First, the connection between CL remodeling and membrane assembly and indeed the very function of CL remodeling have not been established and second, the observed substrate specificity of CL remodeling remains to be explained. This is because tafazzin is a promiscuous transacylase that reacts with all phospholipids and does not have any intrinsic acyl specificity (31). It is therefore not clear what is driving the incorporation of certain fatty acids specifically into CL. We have shown that the specificity of the tafazzin reaction results from the physical properties of lipids. For instance, one can force tafazzin to produce certain lipid species in vitro by altering the lipid phase state provided this alteration induces sufficient packing stress (31, 32). However, it is unknown what triggers such packing stress in vivo.
To identify the process that induces CL remodeling in vivo, we altered the expression of various mitochondrial proteins. Specifically, we searched for proteins that, like tafazzin, produce an altered CL species composition and a partial replacement of CL by monolyso-cardiolipin (MLCL). We focused on inner membrane proteins that are expected to disturb the packing order of mitochondrial lipids. These included the MICOS complex and the ATP synthase because they may induce strong membrane curvature (33–36), the fission-fusion machinery because fission and fusion create nonbilayer membrane intermediates (37), and respiratory complexes because they may entrap lipids in a crowded protein environment (38, 39). We reasoned that identifying the source of acyl specificity is likely to reveal the biological function of tafazzin.
Results
Cld1, taz1, and Mitochondrially Encoded Proteins Are Necessary for CL Remodeling in Yeast.
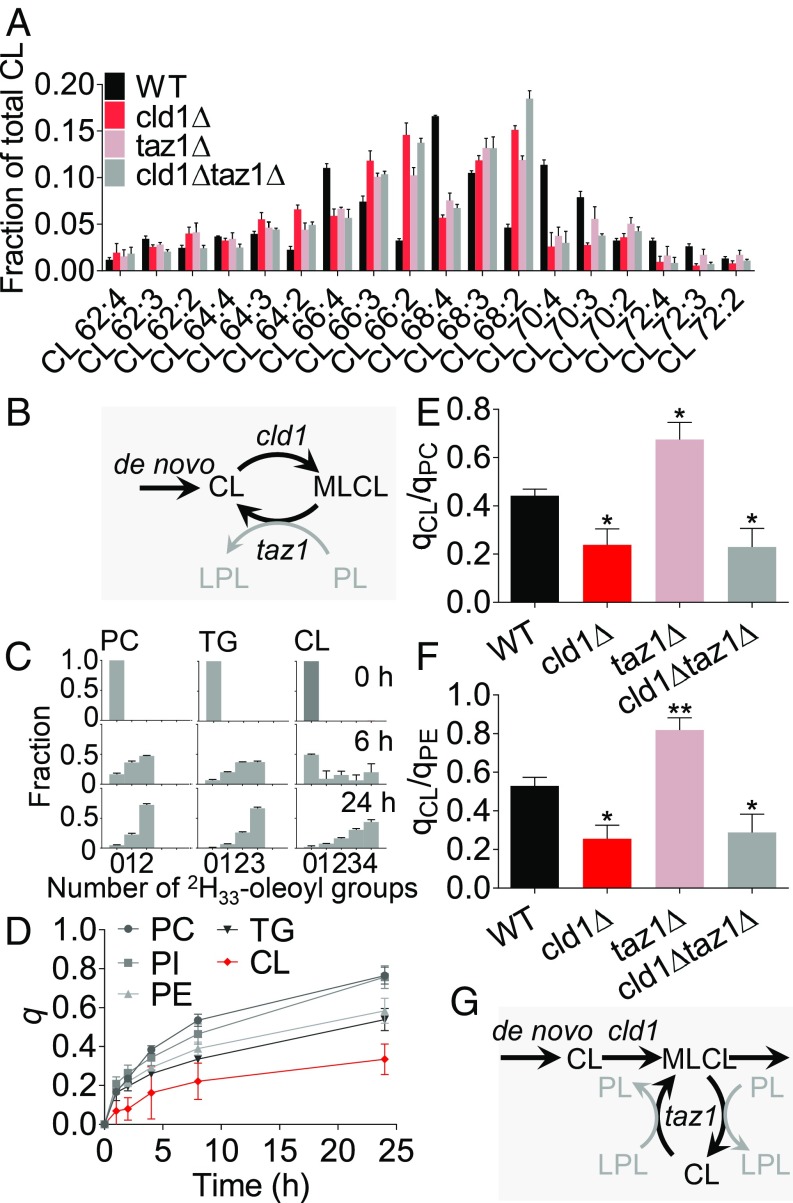
CL remodeling in yeast requires the phospholipase cld1 and the tafazzin homolog taz1 (17, 21, 28, 40–42). Consistent with that notion, we found that the deletion of either enzyme reduced the proportion of tetra-unsaturated CL in favor of diunsaturated and triunsaturated species (Fig. 1A). Since cld1 removes acyl groups from CL and taz1 is capable of reattaching them, they have been thought to work in tandem, exchanging fatty acids by repeated cycles of deacylation and reacylation (Fig. 1B). Therefore, both enzymes are expected to accelerate the turnover of CL-bound fatty acids. To test this prediction, we incubated yeast strains with 2H33-oleic acid and determined isotopomer abundance patterns of oleoyl-containing lipid species (Fig. 1C), which can be used to calculate their fractional syntheses (26). As shown previously in flies and cell cultures (26), the turnover of CL was considerably slower than the turnover of other lipids (Fig. 1D). However, deletion of cld1 further decreased the turnover of CL, whereas deletion of taz1 had the opposite effect (Fig. 1 E and F). This is not consistent with the deacylation-reacylation mechanism shown in Fig. 1B but suggests that cld1 and taz1 act separately on CL. We propose a revised mechanism, in which cld1 converts CL to MLCL and then taz1 redistributes fatty acids between MLCL, CL, and other lipids, which prolongs the half-life of CL (Fig. 1G).
Fig. 1.
Taz1 and Cld1 have opposite effects on the turnover of CL in yeast. (A) Yeast strains were grown in YPGE to the midlogarithmic phase. Lipids were analyzed by MS. Data are mean values ± SEM (n = 3). (B) In the conventional remodeling mechanism, cld1 and taz1 create a deacylation-reacylation cycle. (C–F) Yeast cells were grown in YPGE supplemented with 2H33-oleic acid. Cells were harvested at the indicated time points, and lipids were analyzed by MS to determine the isotopomer pattern of dioleoyl-phosphatidylcholine (PC), dioleoyl-phosphatidylethanolamine (PE), dioleoyl-phosphatidylinositol (PI), trioleoyl-glyceride (TG), and tetraoleoyl-CL. The fractional synthesis (q) was calculated from the isotopomer patterns. After 24 h, the ratios of fractional syntheses of CL to PC (qCL/qPC) and of CL to PE (qCL/qPE) were determined. Data are mean values ± SEM (n = 4). Comparisons between mutants and wild type were made by t test (*P < 0.05; **P < 0.01). (G) In the revised remodeling mechanism, cld1 and taz1 act consecutively. First, cld1 forms MLCL, then taz1 redistributes acyl groups between CL, MLCL, and other phospholipids (PL) and lysophospholipids (LPL). Remodeling by taz1 protects CL from degradation.
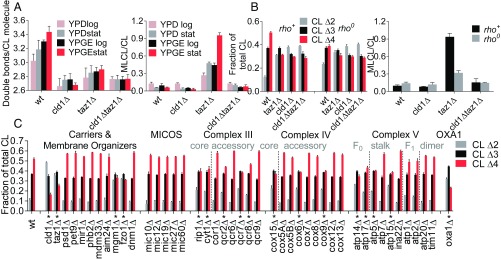
As expected, Cld1 removed saturated acyl groups from CL in vivo (SI Appendix, Fig. S1), consistent with its enzymatic specificity in vitro (42). In contrast, taz1 acquires acyl specificity through the phase state of lipids in vitro (31, 32), which raises the question of what affects the acyl specificity in vivo. To identify factors that affect the acyl specificity of CL remodeling, we measured the CL composition at different culture conditions. Conditions that promoted mitochondrial biogenesis, such as the stationary instead of the logarithmic growth phase or a nonfermentable (yeast extract-peptone-glycerol-ethanol, YPGE) instead of a fermentable (yeast extract-peptone-dextrose, YPD) carbon source, increased the abundance of CL 68:4, CL 70:3, and CL 70:4 at the expense of CL 64:2, CL 66:2, CL 66:3, and CL 68:2. Importantly, this effect was no longer present in the deletion mutants cld1∆, taz1∆, and cld1∆taz1∆ (Fig. 2A and SI Appendix, Fig. S2). Since conditions that favor mitochondrial biogenesis also promote the expression of proteins of oxidative phosphorylation (OXPHOS), we asked whether the OXPHOS system plays a role in CL remodeling. Indeed, when cells lacked mitochondrial DNA (rho0), preventing the assembly of any OXPHOS complex, the CL composition became abnormal. Deletion of mitochondrial DNA from the wild type was as effective in altering CL as deletion of TAZ1 or CLD1, but deletion of mitochondrial DNA from cld1∆, taz1∆, or cld1∆taz1∆ did not cause any further changes (Fig. 2B). These data demonstrate that besides Cld1 and Taz1, other proteins play a role in the remodeling of CL.
Fig. 2.
The expression of OXPHOS complexes controls CL remodeling in yeast. (A) Yeast strains were grown in YPD or YPGE and harvested in the midlogarithmic (log) or in the stationary (stat) phase. The average number of double bonds per CL molecule and the MLCL/CL ratio were determined by MS. Data are mean values ± SEM (n = 3). (B) Yeast cells and their corresponding rho0 strains were grown in YPD and harvested in stationary phase. Lipids were analyzed by MS to determine the proportion of diunsaturated (CL∆2), triunsaturated (CL∆3), and tetraunsaturated (CL∆4) CL species as well as the MLCL/CL ratio. Data are mean values ± SEM (n = 4). (C) Yeast strains with the indicated deletions (SI Appendix, Table S1) were grown in YPD to stationary phase, and CL was analyzed by MS. Data are mean values ± SEM (n = 3). Asterisks indicate a statistically significant difference to the wild type (P < 0.05, χ2 test).
Specific Subunits of the OXPHOS System Affect CL Remodeling in Yeast and in Drosophila.
To identify specific proteins involved in CL remodeling, we analyzed the CL composition in yeast strains that carried deletions in the OXPHOS system, the MICOS complex, solute carriers, and proteins affecting the membrane organization (listed in SI Appendix, Table S1). Among those, only the deletion of MGM1, FZO1, and some OXPHOS subunits had an effect on the CL composition but not the deletion of MICOS proteins, carriers, PHB2, MDM33, AIM24, or DNM1 (Fig. 2C). The effects of mgm1∆ and fzo1∆ were expected given that these mutants are unable to maintain mitochondrial DNA and, therefore, behave similar to rho0 cells (43–45). Accordingly, they also had a very low CL content (SI Appendix, Table S2). However, the findings in the OXPHOS system were intriguing because in all three complexes, some deletions had an effect on CL, but others did not. For instance in complex IV, the deletion of COX6 altered the CL composition but the deletion of COX12 did not, although both deletions inhibited complex IV activity as shown by the growth defect on nonfermentable carbon sources (SI Appendix, Fig. S3). Likewise, some subunits of complexes III and V were essential for CL remodeling, but others were not (Fig. 2C). Importantly, deletion of ATP20 and TIM11 did not affect CL, suggesting that the dimerization of complex V was not required to maintain a normal CL composition. To further test the involvement of the OXPHOS system, we deleted OXA1, the membrane insertion machinery that is required to initiate the assembly of OXPHOS complexes (46). OXA1 deletion altered the CL composition drastically (Fig. 2C). Thus, particular deletions in the OXPHOS system had a similar effect on the CL composition as the deletion of TAZ1, although they did not induce the formation of MLCL (SI Appendix, Fig. S4). Together, our data strongly support the idea that the OXPHOS complexes are involved in the remodeling of yeast CL.
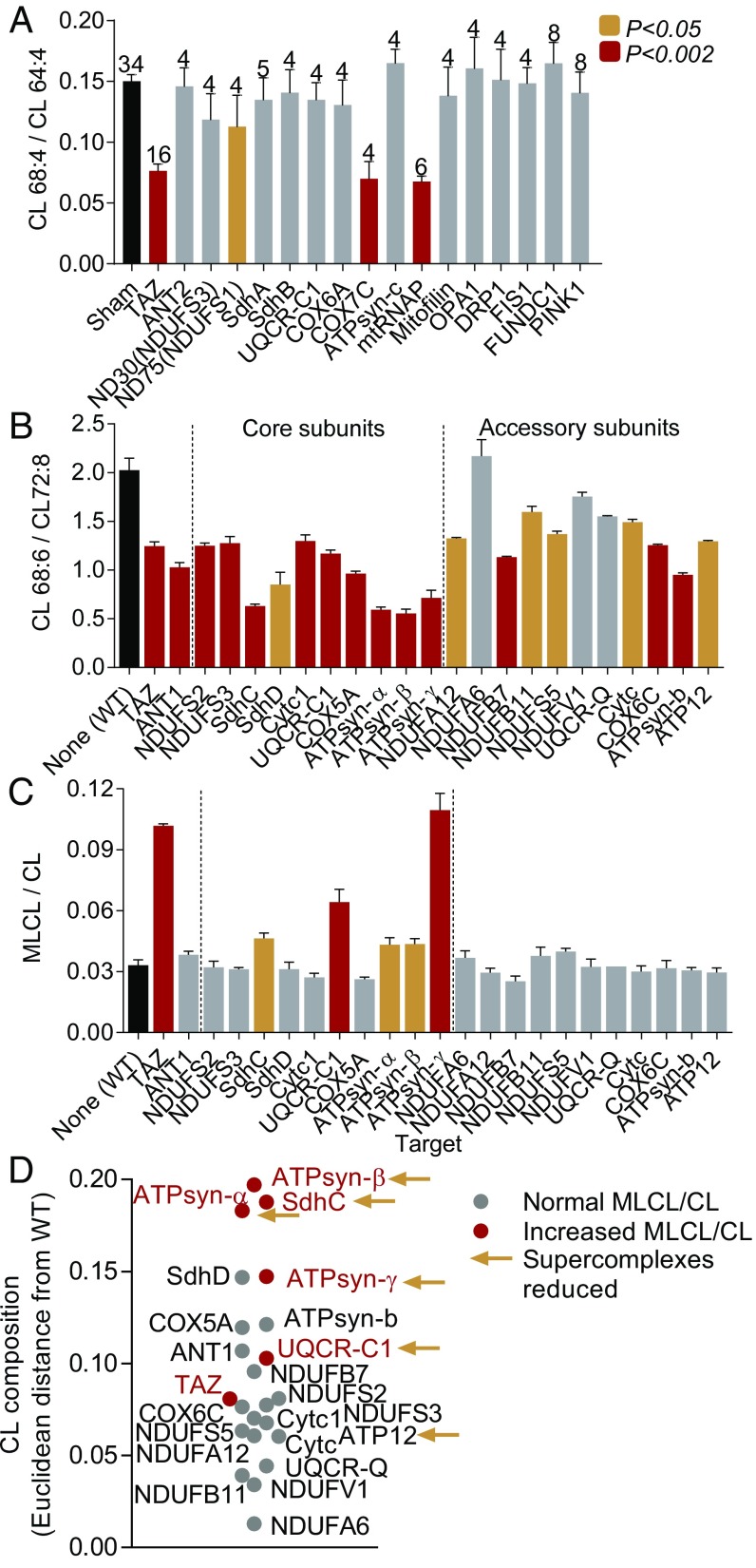
To explore the extent to which this phenomenon is conserved, we tested this result in a different organism. Specifically, we knocked down mitochondrial proteins in Drosophila S2 cells using double-stranded RNA-mediated interference (RNAi) (47). RNAi efficiency was determined by quantitative RT-PCR (SI Appendix, Table S3), and the knockdown effect on the CL species composition was determined by mass spectrometry (MS). As expected, the knockdown of tafazzin altered the composition of CL by shifting the proportion of the three main species (CL 64:4, CL 66:4, CL 68:4) from longer to shorter chains. In contrast, the knockdown of many other mitochondrial proteins, including proteins involved in mitophagy (PINK1, FUNDC1), the fission-fusion cycle, cristae biogenesis (OPA1, DRP1, FIS1), the adenine nucleotide translocase (ANT2), and the mic60 homolog mitofilin, did not cause any change in CL. However, knockdown of the mitochondrial RNA polymerase (mtRNAP), which prevents the assembly of all OXPHOS complexes, altered the CL composition, as did the knockdowns of ND75 and COX7C. The knockdowns of mtRNAP and COX7C were as effective in changing CL as the knockdown of tafazzin (Fig. 3A). Importantly, the abundance of tafazzin remained normal (SI Appendix, Fig. S5). These data confirm the importance of OXPHOS proteins for CL remodeling and document that their effect was not caused by a suppression of the tafazzin protein level.
Fig. 3.
Assembly of the OXPHOS system affects CL remodeling in Drosophila. (A) Knockdown of the indicated transcripts was performed with dsRNA in S2 Drosophila cells and the CL composition was measured by MS. The bars show mean values ± SEM with the number of experiments on Top. Data were compared by t test. (B and C) Muscle-specific knockdown of the indicated transcripts (SI Appendix, Table S1) was achieved in adult Drosophila by RNAi expression using Mhc-Gal4. Fly thoraces were dissected and processed for lipid analysis by MS. Bar graphs are mean values ± SEM (n = 4). Data were compared by t test. (D) Euclidean distances were calculated between the CL composition of the knockdown strains and the CL composition of the wild type (58).
Only Global Inhibition of the OXPHOS Expression Destabilizes CL.
To determine why some OXPHOS proteins affected CL but others did not, we performed tissue-specific knockdowns in Drosophila flight muscle, an organ with very high OXPHOS capacity (48), and measured the total amount of CL (SI Appendix, Fig. S6), the composition of CL species (SI Appendix, Table S4), and the abundance of OXPHOS complexes (SI Appendix, Fig. S7). The knockdowns of core subunits were generally more effective in altering the CL composition than the knockdowns of accessory subunits (Fig. 3B). Importantly, the knockdown of several core subunits also increased the MLCL/CL ratio, i.e., they faithfully mimicked the effect of tafazzin on CL (Fig. 3C). Those knockdowns (SdhC, UQCR-C1, ATPsyn-α, ATPsyn-β, ATPsyn-γ) also caused larger changes in the CL composition than most other knockdowns (Fig. 3D). At the same time, those knockdowns reduced the abundance of multiple OXPHOS complexes, including supercomplexes, not only the abundance of a single complex. In particular, the knockdown of UQCR-C1 and ATPsyn-γ suppressed nearly all OXPHOS complexes (SI Appendix, Fig. S7). In contrast, knockdowns that reduced only a single complex were less effective in changing CL. Thus, impaired assembly of the OXPHOS complexes led to changes in the CL composition, the severity of which were greater the more complexes were affected. To further test global involvement of OXPHOS complexes, we overexpressed ImpL2, a secreted homolog of insulin growth factor binding protein 7, which counteracts insulin signaling in flies (49, 50). Indeed, overexpression of ImpL2 caused a reduction in the abundance of all OXPHOS complexes and a near complete loss of supercomplexes in flight muscle mitochondria. The total amount of CL was also reduced, and the composition of CL was altered (SI Appendix, Fig. S8). These data suggest that it is the assembly of the entire OXPHOS system, rather than any of its specific components, that drives CL remodeling.
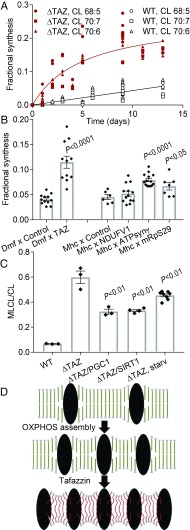
We have previously shown that the remodeling of fatty acids protects CL from degradation and have suggested that the protection may result from increased protein association (26). To determine whether it is the assembly of the OXPHOS complexes that stabilizes CL, we again used the muscle-specific Drosophila knockdown system. Adult flies were cultured in the presence of 13C6-glucose and the isotopomer pattern of CL was determined in the flight muscles (SI Appendix, Fig. S9). The fractional synthesis of CL, i.e., the proportion of newly synthesized molecules, was calculated from the isotopomer patterns of individual molecular species (26). As expected, the fractional synthesis rose very slowly in muscle CL of wild-type flies, whereas it rose quickly in flies with tafazzin deletion (Fig. 4A), and therefore, the half-life of CL was much shorter in the absence of tafazzin (SI Appendix, Table S5). Next, we analyzed flies with muscle-specific knockdowns. Not only the knockdown of tafazzin but also the knockdown of ATPsyn-γ and mitochondrial ribosomal protein S29, both of which inhibit global OXPHOS assembly, increased the turnover of CL in flight muscle. In contrast, knockdown of NDUFV1, which affects complex I assembly only, did not increase CL turnover (Fig. 4B). We confirmed these results by another labeling technique, in which flies were incubated with 2H2O. Knockdown of either tafazzin or the ATPsyn-γ accelerated the incorporation of 2H isotopes, indicating a faster turnover of CL (SI Appendix, Fig. S10). Finally, we tested whether the stimulation of OXPHOS expression, either by activating the SIRT/PGC pathway or by starvation (51, 52), can counteract the effect of tafazzin deletion. Indeed, either starvation or the overexpression of SIRT1 or PCG1, partially reversed the effect of tafazzin deletion on the MLCL/CL ratio (Fig. 4C). Since the MLCL/CL ratio is an indicator of CL degradation (26), the data further support the stabilizing effect of OXPHOS complexes on CL. Collectively, our data demonstrate that both tafazzin and the assembly of the OXPHOS system are required for the remodeling of CL, which, in turn, is a precondition for CL stability.
Fig. 4.
Both tafazzin and the OXPHOS system are required for the high stability of CL in Drosophila flight muscle. (A) Flies were cultured in the presence of 13C6-glucose to determine the fractional synthesis of CL in flight muscle. (B) Flies with the indicated knockdowns were cultured for 3 d in the presence of 13C6-glucose. The fractional syntheses of CL species were determined in flight muscle. Data are mean values ± SEM. Comparisons between knockdowns and controls were made by t test. (C) The MLCL/CL ratio was measured in transgenic flies with tafazzin deletion (∆TAZ) plus overexpression of SIRT1 or PGC1 or starvation for 10 d (starv). Bars show means ± SEM. Data were compared by t test. (D) The cartoon shows a hypothetical model describing the connection between OXPHOS assembly and CL. Assembly of the OXPHOS system causes protein crowding, which imposes curvature elastic stress on the molecular packing of lipids (54). CL remodeling relieves this stress by forming molecular species with intrinsic negative curvature. As a result, stable lipid–protein complexes emerge.
Discussion
Tafazzin-catalyzed remodeling of the acyl groups of CL has been recognized as an important step in mitochondrial biogenesis because mutations in tafazzin cause Barth syndrome, a mitochondrial disease associated with CL degradation (26), dissociation of respiratory supercomplexes (23), and inefficient oxidative phosphorylation (29). It has remained unclear, however, whether remodeled CL is superior to nonremodeled CL and, thus, what the actual function of tafazzin is (28, 41). Since in vitro, the activity and also the specificity of CL remodeling critically depend on the physical state of lipids (31), we hypothesized that in vivo CL remodeling is controlled by a mitochondrial process that affects the physical state of membrane lipids. Our data demonstrate that CL remodeling is dependent on the expression of OXPHOS complexes. In yeast, the expression of mitochondrial DNA and several nuclear-encoded OXPHOS proteins was essential for CL remodeling, and in Drosophila, knockdown of OXPHOS complexes inhibited CL remodeling. Importantly, there was a close connection between CL remodeling and CL stability. Knockdown of either tafazzin or OXPHOS complexes increased the turnover of CL, suggesting that both are necessary to prevent CL degradation.
The OXPHOS complexes are located in the cristae, which are among the protein-richest membranes of the cell. Their frequently cited lipid-to-protein mass ratio of 22:78 (53) implies that any complex, depending on its size, is surrounded by only 40–400 lipid molecules (SI Appendix, Table S6). It has been shown that such high density of proteins imposes curvature elastic stress on the molecular packing of lipids (54). We hypothesize that the function of tafazzin is to mitigate this packing stress. Because tafazzin can form any molecular species within the confines of the total fatty acid composition, it will naturally produce molecular species with the lowest free energy, i.e., the ones that minimize stress by bringing the monolayer spontaneous curvature as close as possible to the actual curvature (Fig. 4D). As a result, tafazzin lowers the energy cost to build tightly packed cristae membranes. Consistent with that, deletion of tafazzin reduced the density of complex V molecules in flight muscle mitochondria (7).
In summary, we have shown that the expression of OXPHOS complexes is a precondition for CL remodeling and that CL remodeling in turn is necessary for CL stability. This conclusion is supported by recent reports that show increased CL remodeling in response to muscle overload (increased OXPHOS expression) (55) and decreased CL remodeling in response to rapid cell proliferation (decreased OXPHOS expression) (56). Furthermore, the conclusion is consistent with the clinical presentation of Barth syndrome that afflicts organs, such as skeletal muscle and heart (30), which have high aerobic energy metabolism and, therefore, a high concentration of OXPHOS proteins. The idea that impaired lipid–protein interactions in the OXPHOS complexes are the principal underlying cause of the disease explains the established lability of supercomplexes in Barth patients and in tafazzin-deficient models (22–27).
Materials and Methods
Specimens.
Saccharomyces cerevisiae strains were previously described (41). Strains with knockouts of various mitochondrial proteins, including the wild-type parent strain BY4743, were obtained from the Yeast Knockout Collection of Dharmacon (https://dharmacon.horizondiscovery.com/cdnas-and-orfs/non-mammalian-cdnas-and-orfs/yeast/yeast-knockout-collection). Yeast cells were grown at 30 °C in YPD medium containing 10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose, or in YPGE medium containing 10 g/L yeast extract, 20 g/L peptone, 3% glycerol, and 3% ethanol. Drosophila melanogaster Schneider 2 (S2) cells were cultured at 23 °C in Schneider’s Drosophila medium. For the knockdown experiments, S2 cells were treated with double-stranded RNA (dsRNA). D. melanogaster strains are specified in SI Appendix. Flies were raised on standard fly media. To isolate flight muscles, thoraces of adult animals were dissected under the microscope.
Lipid Analysis.
Lipid extracts, supplemented by internal standards, were analyzed by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (57). Molecular species were identified by LC-MS/MS. The calculation of the Euclidean distance between two CL compositions was described in a previous paper (58). The turnover of lipids was measured by isotopomer spectral analysis, which requires the incorporation of isotope-labeled metabolites and the subsequent analysis of the samples by LC-MS/MS. Fractional syntheses were estimated by isotopomer spectral analysis (59) as described in detail in a previous paper (26).
Protein Analysis.
Blue-native polyacrylamide gel electrophoresis (BN-PAGE) was performed using NativePAGE gels from Life Technologies. Silver staining of native gels was performed with the SilverXpress staining kit from Life Technologies. For Western blot analysis, proteins were separated by SDS/PAGE, transferred to a polyvinylidene fluoride (PVDF) membrane, incubated with primary polyclonal tafazzin antibody, and visualized with fluorescent LiCor GAM-IRDye680 secondary antibodies.
Detailed methods and biological reagents are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 GM115593 (to M.S.), GM 124717 (to E.O.-A.), and R01 HL117880 (to M.L.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900890116/-/DCSupplemental.
References
- 1.Guerrero-Castillo S., et al. , The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 25, 128–139 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Garcia C. J., Khajeh J., Coulanges E., Chen E. I., Owusu-Ansah E., Regulation of mitochondrial complex I biogenesis in Drosophila flight muscles. Cell Rep. 20, 264–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cogliati S., et al. , Mechanism of super-assembly of respiratory complexes III and IV. Nature 539, 579–582 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Gu J., et al. , The architecture of the mammalian respirasome. Nature 537, 639–643 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Letts J. A., Fiedorczuk K., Sazanov L. A., The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Strauss M., Hofhaus G., Schröder R. R., Kühlbrandt W., Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27, 1154–1160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acehan D., et al. , Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 100, 2184–2192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyer K., Klingenberg M., ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry 24, 3821–3826 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Lange C., Nett J. H., Trumpower B. L., Hunte C., Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20, 6591–6600 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinzawa-Itoh K., et al. , Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M., Mileykovskaya E., Dowhan W., Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Pfeiffer K., et al. , Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Bazán S., et al. , Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J. Biol. Chem. 288, 401–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlame M., Rüstow B., Lysocardiolipin formation and reacylation in isolated rat liver mitochondria. Biochem. J. 272, 589–595 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma B. J., Taylor W. A., Dolinsky V. W., Hatch G. M., Acylation of monolysocardiolipin in rat heart. J. Lipid Res. 40, 1837–1845 (1999). [PubMed] [Google Scholar]
- 16.Xu Y., Kelley R. I., Blanck T. J. J., Schlame M., Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 278, 51380–51385 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Beranek A., et al. , Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J. Biol. Chem. 284, 11572–11578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y., Malhotra A., Ren M., Schlame M., The enzymatic function of tafazzin. J. Biol. Chem. 281, 39217–39224 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Vreken P., et al. , Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem. Biophys. Res. Commun. 279, 378–382 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Schlame M., et al. , Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann. Neurol. 51, 634–637 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Gu Z., et al. , Aberrant cardiolipin metabolism in the yeast taz1 mutant: A model for Barth syndrome. Mol. Microbiol. 51, 149–158 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Brandner K., et al. , Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: Implications for Barth syndrome. Mol. Biol. Cell 16, 5202–5214 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenzie M., Lazarou M., Thorburn D. R., Ryan M. T., Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J. Mol. Biol. 361, 462–469 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Gonzalvez F., et al. , Barth syndrome: Cellular compensation of mitochondrial dysfunction and apoptosis inhibition due to changes in cardiolipin remodeling linked to tafazzin (TAZ) gene mutation. Biochim. Biophys. Acta 1832, 1194–1206 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Huang Y., et al. , Cardiac metabolic pathways affected in the mouse model of barth syndrome. PLoS One 10, e0128561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., et al. , Loss of protein association causes cardiolipin degradation in Barth syndrome. Nat. Chem. Biol. 12, 641–647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole L. K., et al. , Impaired cardiolipin biosynthesis prevents hepatic steatosis and diet-induced obesity. Diabetes 65, 3289–3300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baile M. G., et al. , Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J. Biol. Chem. 289, 1768–1778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G., et al. , Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke S. L., et al. , Barth syndrome. Orphanet J. Rare Dis. 8, 23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlame M., et al. , The physical state of lipid substrates provides transacylation specificity for tafazzin. Nat. Chem. Biol. 8, 862–869 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlame M., Xu Y., Ren M., The basis for acyl specificity in the tafazzin reaction. J. Biol. Chem. 292, 5499–5506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbot M., et al. , Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 21, 756–763 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Bohnert M., et al. , Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 21, 747–755 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Paumard P., et al. , The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21, 221–230 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiko C., et al. , Bovine F1Fo ATP synthase monomers bend the lipid bilayer in 2D membrane crystals. eLife 4, e06119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chernomordik L. V., Kozlov M. M., Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72, 175–207 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Althoff T., Mills D. J., Popot J. L., Kühlbrandt W., Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 30, 4652–4664 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M., Gu J., Guo R., Huang Y., Yang M., Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167, 1598–1609.e10 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Baile M. G., Whited K., Claypool S. M., Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Mol. Biol. Cell 24, 2008–2020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye C., et al. , Deletion of the cardiolipin-specific phospholipase Cld1 rescues growth and life span defects in the tafazzin mutant: Implications for Barth syndrome. J. Biol. Chem. 289, 3114–3125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyurina Y. Y., et al. , Lipidomics characterization of biosynthetic and remodeling pathways of cardiolipins in genetically and nutritionally manipulated yeast cells. ACS Chem. Biol. 12, 265–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones B. A., Fangman W. L., Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 6, 380–389 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Rapaport D., Brunner M., Neupert W., Westermann B., Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273, 20150–20155 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Hermann G. J., et al. , Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hell K., Neupert W., Stuart R. A., Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20, 1281–1288 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogers S. L., Rogers G. C., Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc. 3, 606–611 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Sacktor B., Biochemical adaptations for flight in the insect. Biochem. Soc. Symp. 41, 111–131 (1976). [PubMed] [Google Scholar]
- 49.Honegger B., et al. , Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 7, 10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Owusu-Ansah E., Song W., Perrimon N., Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell 155, 699–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood J. G., et al. , Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686–689 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Lagouge M., et al. , Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127, 1109–1122 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Lotan R., Nicolson G. L., “Plasma membranes of eukaryotes” in Advanced Cell Biology, Schwartz L. M., Azar M. M., Eds. (Van Nostrand-Reinhold, Princeton, NJ, 1981), pp. 129–154. [Google Scholar]
- 54.Brown M. F., Soft matter in lipid-protein interactions. Annu. Rev. Biophys. 46, 379–410 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Fajardo V. A., Mikhaeil J. S., Leveille C. F., Saint C., LeBlanc P. J., Cardiolipin content, linoleic acid composition, and tafazzin expression in response to skeletal muscle overload and unload stimuli. Sci. Rep. 7, 2060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mürke E., Stoll S., Lendeckel U., Reinhold D., Schild L., The mitochondrial phospholipid cardiolipin is involved in the regulation of T-cell proliferation. Biochim. Biophys. Acta 1861, 748–754 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Sun G., et al. , Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80, 7576–7585 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlame M., et al. , Comparison of cardiolipins from Drosophila strains with mutations in putative remodeling enzymes. Chem. Phys. Lipids 165, 512–519 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Kelleher J. K., Masterson T. M., Model equations for condensation biosynthesis using stable isotopes and radioisotopes. Am. J. Physiol. 262, E118–E125 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.