Significance
Skeletal muscle is the major tissue involved in regulating glucose uptake and body metabolism. In response to exercise, glycogen is metabolized in the liver to glucose, which is taken up by muscle to provide fuel for contraction. We show that NURR1, an orphan nuclear receptor, is up-regulated in response to exercise and strongly stimulates glucose uptake. Genetic overexpression of Nurr1 in skeletal muscle of mice enhances physical performance. In obese mice, overexpression of Nurr1 or treatment with putative chemical activators of NURR1 prevents hyperglycemia and hepatic steatosis and mimics the effect of exercise. Our work provides insights into a transcriptional link between exercise and metabolism, and identifies NURR1 agonists as potential therapeutics to ameliorate obesity and other metabolic abnormalities.
Keywords: exercise, Mediator complex, metabolic syndrome, nuclear receptor, obesity
Abstract
Skeletal muscle plays a central role in the control of metabolism and exercise tolerance. Analysis of muscle enhancers activated after exercise in mice revealed the orphan nuclear receptor NURR1/NR4A2 as a prominent component of exercise-responsive enhancers. We show that exercise enhances the expression of NURR1, and transgenic overexpression of NURR1 in skeletal muscle enhances physical performance in mice. NURR1 expression in skeletal muscle is also sufficient to prevent hyperglycemia and hepatic steatosis, by enhancing muscle glucose uptake and storage as glycogen. Furthermore, treatment of obese mice with putative NURR1 agonists increases energy expenditure, improves glucose tolerance, and confers a lean phenotype, mimicking the effects of exercise. These findings identify a key role for NURR1 in governance of skeletal muscle glucose metabolism, and reveal a transcriptional link between exercise and metabolism. Our findings also identify NURR1 agonists as possible exercise mimetics with the potential to ameliorate obesity and other metabolic abnormalities.
While the beneficial effects of exercise on metabolism and overall organismal health are well-known, much remains to be learned about the mechanistic basis of the benefits of physical activity and the systemic interactions among tissues and organs. Skeletal muscle accounts for ∼40% of body mass in healthy individuals, and represents the major site of glucose uptake and metabolism in the body (1, 2). During exercise, glycogen is metabolized in the liver to yield glucose, which is taken up by skeletal muscle to provide energy for contraction (3–5). Conversely, under conditions of caloric excess, glucose and fatty acids are directed to the liver, where energy is stored as triglycerides, causing hepatic steatosis, a growing health concern (6, 7).
Uptake of glucose by skeletal muscle is mediated by GLUT4, the major glucose transporter in the sarcolemma (8–10). Exercise induces expression of GLUT4 and its translocation from intracellular stores to the sarcolemma (11). A variety of exercise-responsive signal transduction pathways culminate in the nucleus to modulate the expression of GLUT4 and other metabolic genes (12). Key among the signal transducers in these pathways are AMPK and several calcium-sensitive kinases that regulate transcription by targeting class II histone deacetylases (HDACs), which relieves their repressive influence on MEF2 and other transcription factors (13–16). Integration of metabolic gene regulation also occurs through regulatory interactions between MEF2 and the nuclear coactivator PGC-1, which associates with PPAR and other nuclear receptors to enhance metabolic gene expression (17).
Recently, we showed that MED13, a component of the Mediator complex, modulates systemic metabolism in skeletal muscle by suppressing the expression of Glut4 and other genes involved in glucose uptake and glycogen storage (18, 19). Among a collection of genes up-regulated in skeletal muscle of MED13 mutant mice was the orphan nuclear receptor Nr4a2/Nurr1. NURR1 has been implicated in survival of dopaminergic neurons (20) and other processes (21–24), but its potential involvement in skeletal muscle metabolism has not been previously investigated.
Here, using in vivo genome-wide epigenetic analysis of enhancers and MEF2-activated genomic regions in muscle after exercise, we show that binding sites for NURR1/NR4A2 are among the most common motifs in exercise-responsive enhancers. NURR1 is up-regulated in skeletal muscle after exercise, and transgenic mice that overexpress NURR1 in skeletal muscle display increased physical performance in a regimen of voluntary wheel running. Our results reveal that NURR1 can potently stimulate muscle glucose uptake, thereby normalizing glucose and insulin levels and conferring resistance to hepatic steatosis in obese mice. Moreover, treatment of genetically obese ob/ob mice or mice on a high-fat diet (HFD) with putative NURR1 agonists enhances systemic energy metabolism and prevents obesity and hepatic steatosis. These findings reveal a transcriptional basis for the beneficial effects of exercise on systemic metabolism, and suggest a potential pharmacologic approach for normalizing metabolism under conditions of caloric excess via the activation of skeletal muscle NURR1.
Results
Identification of Exercise-Responsive Enhancers and MEF2 Targets.
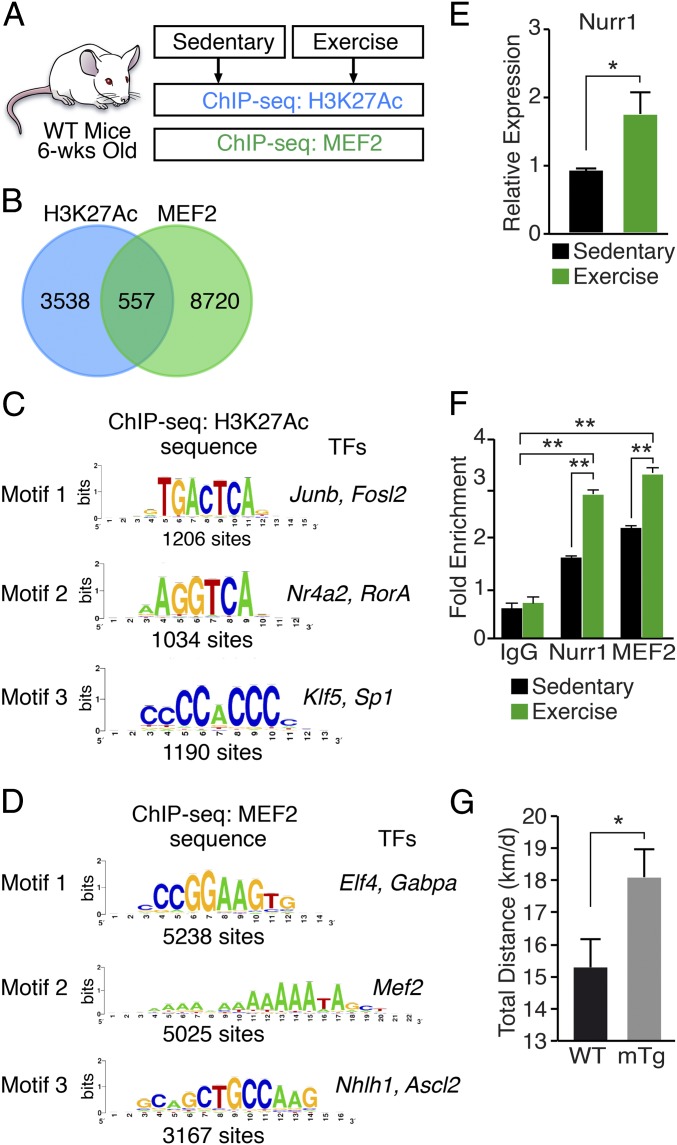
Enhancers, marked by extended genomic binding of histone H3 acetylated on lysine 27 (H3K27Ac), act as primary determinants of gene activation (25, 26). To explore the regulation of muscle enhancers in response to exercise, we performed chromatin immunoprecipitation of H3K27Ac followed by next-generation sequencing (ChIP-seq) using skeletal muscle from mice that were subjected to a regimen of voluntary wheel running for 8 wk compared with sedentary controls (Fig. 1A). The H3K27Ac mark identifies cell type-specific and active enhancers. A total of 3,955 enhancer peaks associated with 4,095 genes were identified by elevated H3K27Ac occupancy in exercised compared with sedentary muscle (Fig. 1B and SI Appendix, Fig. S1 A and B). Gene ontology analysis showed that many identified peaks of H3K27Ac DNA binding were associated with genes involved in regulation of protein phosphorylation, cell motility, glucose metabolism, and other processes (SI Appendix, Fig. S1 C and D).
Fig. 1.
Chromatin analysis in response to exercise. (A) Schematic of chromatin immunoprecipitation experiments using tibialis anterior muscle from sedentary mice or mice following 8 wk of voluntary wheel running beginning at 6 wk of age. (B) Venn diagram showing overlap of 557 exercise-responsive genes associated with ChIP-seq peaks for H3K27Ac and MEF2 in skeletal muscle. For H3K27Ac ChIP-seq, 3,955 enhancer peaks were associated with 4,095 genes. For MEF2 ChIP-seq, 10,032 peaks were associated with 9,277 genes. (C) De novo motif discovery from peaks of the H3K27Ac ChIP-seq reads. The three most abundant sequence motifs are shown. (D) De novo motif discovery from peaks of the MEF2 ChIP-seq reads. The three most abundant sequence motifs are shown. (E) Nurr1 mRNA normalized to 18S RNA in quadriceps muscle of sedentary mice, and mice following 8 wk of voluntary wheel running. n = 6. (F) ChIP-seq assays showing binding of NURR1 and MEF2 to the Glut4 promoter in quadriceps muscle of sedentary mice, and mice following 8 wk of voluntary wheel running. Antibodies against NURR1 or MEF2 were used in ChIP-seq assays for the Glut4 promoter. Graphs display mean quantification of ChIP-seq data (percentage of input) normalized to IgG control. IgG control, n = 3. Sedentary mice, n = 6. Exercised mice, n = 6. (G) WT and Nurr1-mTg mice were subjected to a voluntary wheel-running regimen for 8 wk. Nurr1-mTg mice showed enhanced physical performance, measured by total distance run in km/d. n = 6. Data are represented as mean ± SEM. *P < 0.05, **P < 0.005.
MEF2 is a key regulator of gene expression and metabolism in response to exercise (13–16). To identify direct transcriptional targets of MEF2 involved in the response of muscle to exercise, we performed MEF2 ChIP-seq analysis on muscle samples from sedentary and exercised mice. The exercised samples showed 10,032 enriched peaks associated with 9,277 genes compared with sedentary (Fig. 1B and SI Appendix, Fig. S1 E and F). Gene ontology analysis showed that many identified peaks of MEF2 DNA binding were associated with genes involved in ATPase binding, cAMP binding, and cyclic nucleotide binding (SI Appendix, Fig. S1 G and H). Comparing the genes associated with H3K27Ac ChIP-seq peaks with the peaks of MEF2 binding yielded 557 common genes associated with the peaks (Fig. 1B). Gene ontology analysis showed that these peaks were associated with genes involved in fundamental molecular functions, such as growth factor, protein kinase, carbohydrate derivative binding, and others (SI Appendix, Fig. S1I).
To identify potential transcriptional mediators of exercise, we examined the H3K27Ac and MEF2 peaks for enrichment of distinctive sequence elements by de novo motif analysis (Fig. 1 C and D). For the MEF2 subset, consensus binding sites for Elf4, MEF2, and Nhlh1 represented the top three most enriched sequences. For the H3K27Ac subset, the top three most enriched sequences corresponded to binding sites for JUNB, NR4A2/NURR1, and KLF5. Interestingly, among the peaks of MEF2 genomic binding that were induced by exercise was the Nr4a2/Nurr1 locus (SI Appendix, Fig. S1J), which encodes an orphan nuclear receptor that has been reported to be among the most up-regulated genes in human muscle after intensive exercise (27, 28).
Up-Regulation of Nurr1 by Exercise.
To further investigate NURR1 regulation during exercise, we analyzed skeletal muscle samples from exercised and sedentary mice. After 8 wk of voluntary exercise, Nurr1 expression in muscle was increased approximately twofold (Fig. 1E). NURR1 binds to the Glut4 promoter and regulates its activity together with MEF2 (18). To determine if exercise promotes recruitment of NURR1 and MEF2 to the Glut4 promoter, we performed ChIP analysis using muscle samples after exercise. ChIP-qPCR analysis showed significantly higher binding of NURR1 and MEF2 to the Glut4 promoter compared with muscle from sedentary mice (Fig. 1F).
Skeletal Muscle Expression of NURR1 Enhances Physical Performance.
Given the above findings, and our prior studies highlighting the importance of NURR1 in the regulation of muscle glucose homeostasis (18), we generated transgenic mice that overexpressed Nurr1 specifically in skeletal muscle under control of the human skeletal muscle actin promoter. We selected a transgenic mouse line (referred to as Nurr1-mTg) that expressed Nurr1 ∼20-fold above normal levels in skeletal muscle for in-depth analysis (SI Appendix, Fig. S2A). When maintained on a normal chow diet, these mice showed the same weight gain as wild-type (WT) littermates, with comparable lean and fat mass (SI Appendix, Fig. S2 B and C), and displayed no overt abnormalities. Histological analysis of skeletal muscle revealed normal muscle fiber size and structure in Nurr1-mTg mice (SI Appendix, Fig. S2D). However, muscle of Nurr1-mTg mice showed an increase in expression of slow myosin (Myh7) and a mild increase in mitochondrial copy number compared with WT mice (SI Appendix, Fig. S2 E and F).
To determine whether NURR1 overexpression in Nurr1-mTg mice might impact exercise performance, we subjected WT and Nurr1-mTg mice to a regimen of voluntary wheel running for 8 wk. As shown in Fig. 1G, Nurr1-mTg mice displayed enhanced wheel-running capacity, running ∼20% farther than WT mice.
Muscle Overexpression of Nurr1 Normalizes Glucose Handling in Response to HFD.
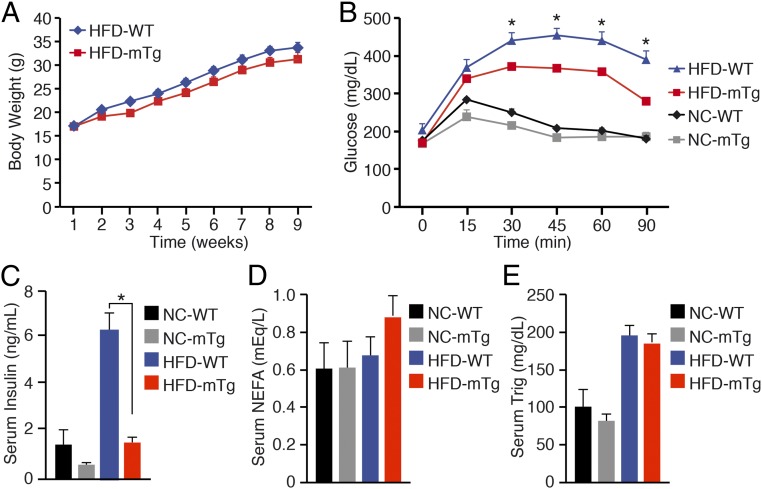
When subjected to an HFD, Nurr1-mTg mice gained weight comparable to WT littermates (Fig. 2A). Glucose tolerance tests showed modest improvement in glucose tolerance of Nurr1-mTg mice compared with WT littermates on normal chow, whereas there was a pronounced improvement in glucose tolerance of Nurr1-mTg mice on an HFD (Fig. 2B). HFD-induced obesity commonly leads to hyperinsulinemia, which was observed in WT mice on an HFD for 10 wk (Fig. 2C). Remarkably, however, serum insulin levels of Nurr1-mTG mice on an HFD were significantly lower than in WT mice (Fig. 2C), whereas serum free fatty acid and triglyceride levels were similar between Nurr1-mTg and WT mice on an HFD for 10 wk (Fig. 2 D and E). Thus, although Nurr1-mTg and WT mice showed comparable weight gain on an HFD, Nurr1-mTg mice displayed improved glucose tolerance and protection from hyperinsulinemia.
Fig. 2.
Nurr1-Tg mice display enhanced skeletal muscle glucose metabolism. (A) Weekly body weight measurement of WT and Nurr1-mTg mice on an HFD beginning at 6 wk of age. (B) Glucose tolerance tests of WT and Nurr1-mTg mice after 8 wk on a normal chow diet or HFD. (C) Serum insulin levels after 10 wk on NC or HFD in the postprandial state. (D) Serum nonessential free fatty acid (NEFA) levels after 10 wk on NC or HFD in the postprandial state. (E) Serum triglyceride (Trig) levels after 10 wk on NC or HFD in the postprandial state. Data are represented as mean ± SEM. n = 6. *P < 0.05.
Resistance of Nurr1-mTg Mice to Hepatic Steatosis.
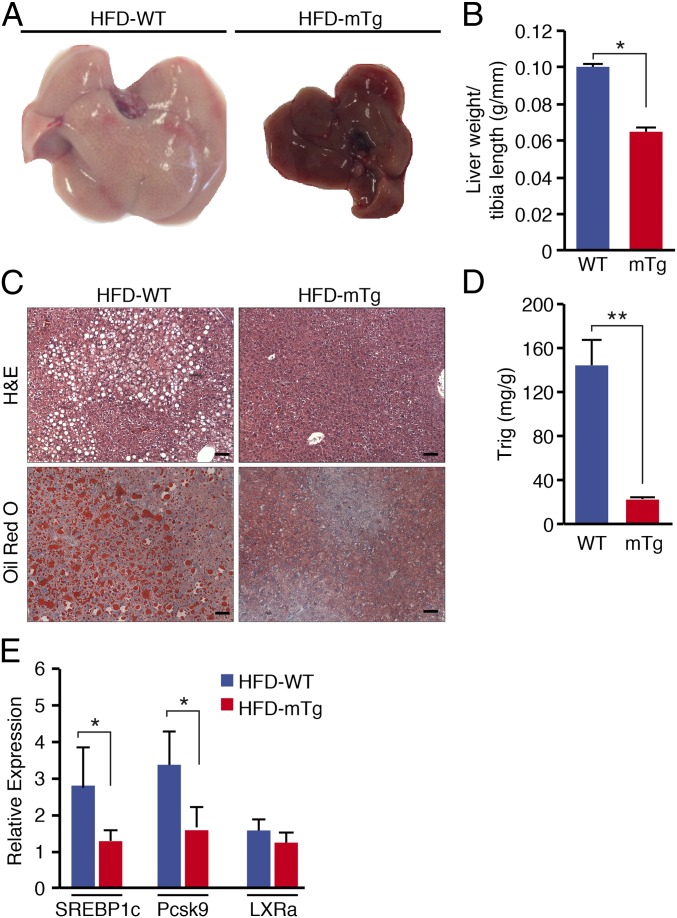
One of the consequences of HFD-induced obesity is hepatic steatosis, due to the accumulation of triglyceride in the liver. WT mice develop severe hepatic steatosis when maintained on an HFD for 10 wk (Fig. 3A). In contrast, livers from Nurr1-mTg mice on an HFD appeared normal (Fig. 3A). Nurr1-mTg mice also displayed a dramatic reduction in liver weight compared with WT mice on an HFD for 10 wk (Fig. 3B). Detection of liver triglyceride accumulation by hematoxylin and eosin (H&E) and oil red O staining confirmed the resistance of Nurr1-mTg mice to hepatic steatosis (Fig. 3C). Biochemical measurements also revealed a pronounced 75% decrease in triglyceride levels in livers from Nurr1-mTg mice compared with WT mice on an HFD for 10 wk (Fig. 3D). In contrast, Nurr1-mTg mice showed no difference in adipocyte size, fat deposition, or expression of markers of browning, thermogenesis, or adipogenesis in white or brown adipose tissues (SI Appendix, Fig. S2 G and H).
Fig. 3.
Resistance of Nurr1-mTg mice to hepatic steatosis. (A) Livers of WT and Nurr1-mTg mice after 10 wk on an HFD. (B) Liver weights after 10 wk on an HFD normalized to tibia length. n = 6 for both groups. (C) Histological sections of livers of WT and Nurr1-mTg mice after 10 wk on an HFD stained with H&E (Upper) or oil red O (Lower). (Scale bars, 50 μm.) (D) Quantification of liver Trig levels in WT and Nurr1-mTg mice after 10 wk on an HFD. (E) Expression of genes involved in liver lipid metabolism: SREBP1c, Pcsk9, and LXRa after 10 wk on an HFD normalized to the level of expression on NC, as measured by real-time qRT-PCR. Data are represented as mean ± SEM. n = 6 for each genotype. *P < 0.05, **P < 0.005.
Reduced hepatic triglyceride accumulation could result from decreased fatty acid import, diminished de novo fatty acid lipogenesis, increased fatty acid oxidation, or enhanced very low density lipoprotein-mediated triglyceride export (29–32). Enzymes involved in fatty acid and cholesterol synthesis and gluconeogenesis are regulated by several transcription factors, such as liver X receptor alpha (LXRα), sterol regulatory element-binding protein 1c (SREBP1c), and carbohydrate-responsive element-binding protein (ChREBP) (33–36). To determine whether these pathways might be altered in Nurr1-mTg mice, we analyzed the expression profile of selected metabolic genes in the liver (SI Appendix, Fig. S3A). SREBP1c and proprotein convertase subtilisin/kexin (Pcsk9) showed a significant decrease in hepatic expression in Nurr1-mTg mice compared with WT mice on an HFD (Fig. 3E).
To determine if changes in food intake or body temperature might contribute to the resistance of Nurr1-mTg mice to hepatic triglyceride accumulation, we performed metabolic cage studies. Nurr1-mTg and WT mice showed similar activity, O2 consumption, CO2 production, and respiratory exchange ratios (RERs) (SI Appendix, Fig. S3 B–D). Together, these results demonstrate that Nurr1 overexpression in skeletal muscle leads to protection from hepatic steatosis independent of body weight or adiposity changes.
Changes in Muscle Gene Expression in Nurr1-mTg Mice.
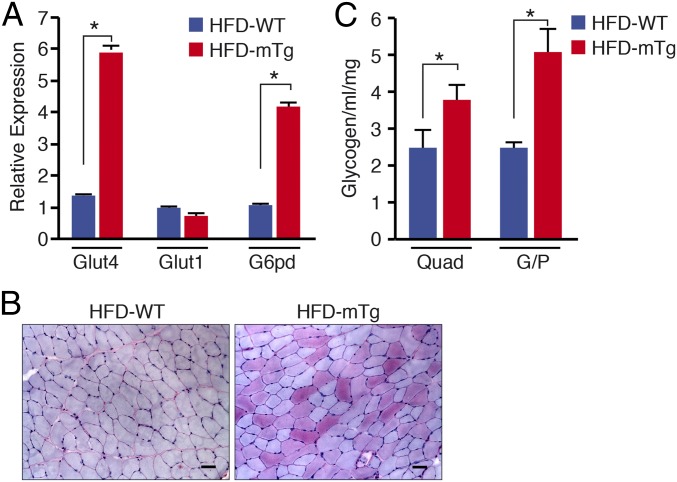
To explore the mechanistic basis of resistance to hepatic steatosis in Nurr1-mTg mice, we compared the expression profiles of glucose regulatory genes in skeletal muscle from WT and Nurr1-mTg mice on an HFD for 10 wk. GLUT4 is the main effector of insulin-stimulated glucose transport in skeletal muscle (8–11, 37–39). GLUT4 mRNA was specifically up-regulated in skeletal muscle of Nurr1-mTg mice, whereas expression of GLUT1 was unchanged (Fig. 4A). Skeletal muscle from Nurr1-mTg mice also showed increased expression of glucose-6-phosphate dehydrogenase (G6pd), pointing to enhanced glucose metabolism.
Fig. 4.
Changes in skeletal muscle glucose-handling gene expression and glycogen storage of Nurr1-mTg mice. (A) Expression of Glut4, Glut1, and G6pd mRNA in tibialis anterior muscle of WT and Nurr1-mTg mice after 10 wk on an HFD as detected by qRT-PCR. (B) Detection of glycogen in tibialis anterior muscle of WT and Nurr1-mTg mice by periodic acid–Schiff (PAS) staining after 10 wk on an HFD. (Scale bars, 50 μm.) (C) Quantification of glycogen content of quadriceps and gastrocnemius/plantaris (G/P) muscle of WT and Nurr1-mTg mice after 10 wk on an HFD. Data are represented as mean ± SEM. n = 6 for each genotype. *P < 0.05.
Histological staining of skeletal muscle for glycogen or detection of glycogen by biochemical assays confirmed that skeletal muscle of Nurr1-mTg mice had higher glycogen content (Fig. 4 B and C). These findings suggest that Nurr1 overexpression in skeletal muscle leads to enhanced glucose uptake and storage as glycogen in skeletal muscle. Gene expression profiles generated by RNA-seq revealed that cellular metabolic processes, carbohydrate metabolism, and glycogen biosynthesis gene networks were substantially changed in muscle of Nurr1-mTg mice on an HFD compared with WT mice on an HFD (SI Appendix, Fig. S4).
A Putative NURR1 Agonist Enhances Metabolism and Prevents Hepatic Steatosis.
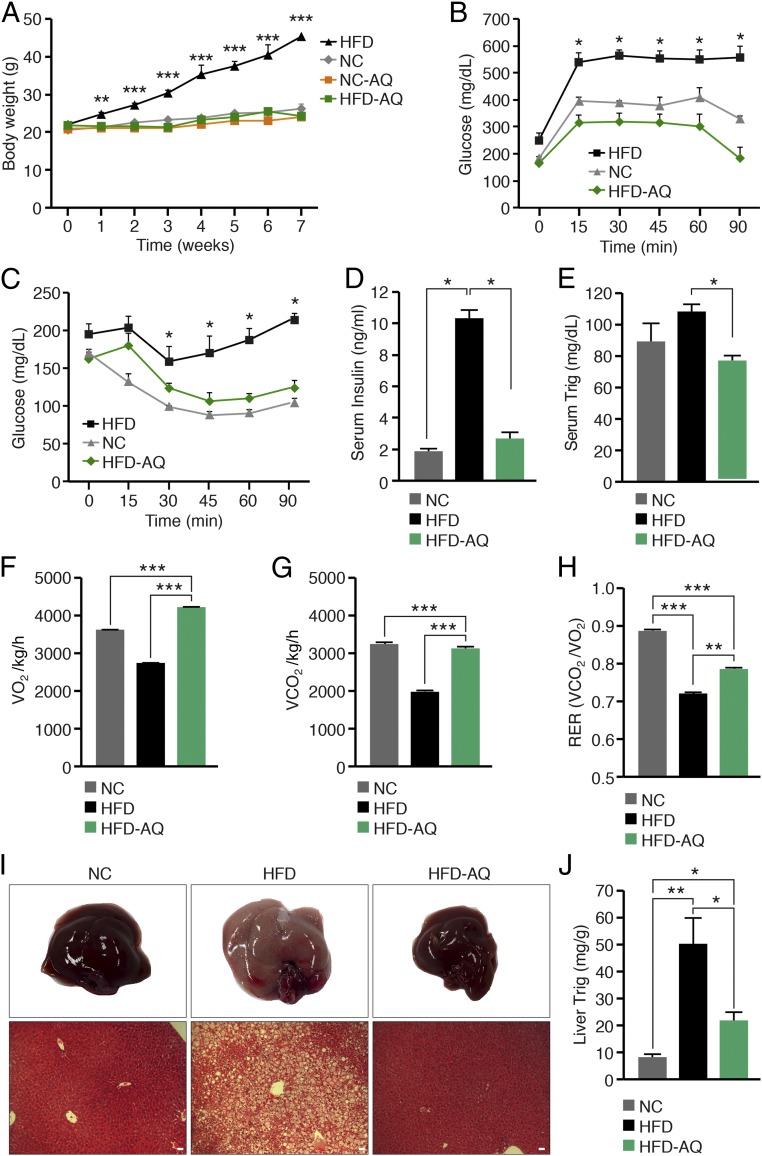
NURR1 has been reported to be specifically activated by three cyclic compounds sharing a 4-amino-7-chloroquinoline scaffold (40). Among these compounds, amodiaquine (AQ) showed the highest activity. AQ is an orally available drug that is well-tolerated in mice without adverse effects. We therefore provided AQ in the drinking water ad libitum of WT mice maintained on normal chow and an HFD for 10 wk (SI Appendix, Fig. S5A). When mice were maintained on normal chow, AQ had no effect on weight gain (Fig. 5A). However, when mice were maintained on an HFD, AQ prevented obesity. Mice provided with AQ while on an HFD showed body weights comparable to mice on normal chow. Food consumption did not decrease significantly, nor was there a decline in lipid absorption in AQ-treated mice compared with control mice on an HFD (SI Appendix, Fig. S5 B and C). However, mice on an HFD with AQ showed a significant decrease in fat mass and an increase in lean mass composition compared with control mice on an HFD (SI Appendix, Fig. S5D). The decrease in fat mass correlated with a decrease in adipocyte size in white adipose tissue from mice treated with AQ compared with control mice (SI Appendix, Fig. S5 E and F). White adipose tissue from mice treated with AQ also showed increased expression of mitochondrial genes, such as citrate synthase (CS), ATP synthase, H+ transporting mitochondrial F1 complex beta (Atp5b), and cytochrome c oxidase subunit 4 isoform 1 (Cox4i), indicating an enhanced oxidative metabolism (SI Appendix, Fig. S5G). Additionally, AQ triggered an improvement in glucose and insulin tolerance of mice on an HFD (Fig. 5 B and C). Serum insulin and triglyceride levels of AQ-treated mice were also significantly lower than in untreated mice on an HFD (Fig. 5 D and E). To determine if the decrease in fat mass and improved glucose tolerance were associated with changes in energy expenditure, we used metabolic cages to monitor the mice on normal chow and an HFD. AQ-treated mice showed a significant increase in O2 consumption, CO2 production, and RER compared with control mice (Fig. 5 F–H).
Fig. 5.
Putative NURR1 agonist AQ confers resistance to obesity and prevents HFD-induced hepatic steatosis. (A) Weekly body weight of mice on NC or HFD in the presence or absence of AQ. (B) Glucose tolerance of mice on NC or HFD in the presence or absence of AQ. Values were measured after 7 wk. (C) Insulin tolerance of mice on NC or HFD in the presence or absence of AQ. Values were measured after 8 wk. (D) Serum insulin levels in the postprandial state of mice on NC or HFD in the presence or absence of AQ. Values were measured after 10 wk. (E) Serum Trig levels in the postprandial state of mice on NC or HFD in the presence or absence of AQ. Values were measured after 10 wk. (F–H) Average oxygen consumption per h (F), carbon dioxide production during the light/dark cycle normalized to lean mass (G), and respiratory exchange ratio (H) of mice on NC or HFD in the presence or absence of AQ. Values were measured after 10 wk. (I) AQ prevents hepatic steatosis in WT mice on HFD. Livers were analyzed after 10 wk on NC or HFD in the presence or absence of AQ. (I, Lower) H&E staining. (Scale bars, 50 μm.) (J) Quantification of liver Trig levels of mice on NC or HFD in the presence or absence of AQ after 10 wk. Data are represented as mean ± SEM. n = 6 for HFD. *P < 0.05, **P < 0.005, ***P < 0.0005.
To investigate if AQ prevented hepatic steatosis, we looked at liver triglyceride accumulation in response to an HFD. As expected, after 8 wk of an HFD, histological and biochemical analysis showed that WT mice developed hepatic steatosis with triglyceride accumulation (Fig. 5I). However, the mice treated with AQ were resistant to hepatic steatosis on an HFD and showed a 50% decrease in hepatic triglyceride levels (Fig. 5J).
To further explore the mechanistic basis of the resistance of AQ-treated mice to hepatic steatosis, we analyzed the expression of selected metabolic genes in the liver. Genes involved in gluconeogenesis [such as glucose 6-phosphatase c (G6Pc) and G6Pd], cholesterol synthesis [such as HMG-CoA-synthase (HCoASynt), HMG-CoA-reductase (HCoARed), and Pcsk9], and fatty acid synthesis [such as acetyl-CoA carboxylase alpha (Acca)] showed similar expression in the livers of mice on an HFD treated with AQ and control mice (SI Appendix, Fig. S5H). The free fatty acid transporter CD36/FAT, stearoyl-CoA desaturase (Scd1), fat-specific protein 27 (Fsp27), SREBP1c, and Pcsk9 genes were up-regulated in mice on an HFD (SI Appendix, Fig. S5 I and J). In contrast, these genes were down-regulated in livers from mice on an HFD treated with AQ. These results show that AQ treatment prevents the development of HFD-induced obesity and associated metabolic disorders such as hyperglycemia, hyperinsulinemia, and hepatic steatosis.
Skeletal Muscle Gene Expression Changes in Response to AQ.
To further investigate the influence of AQ on skeletal muscle metabolism, we examined gene expression profiles in skeletal muscle from WT mice on an HFD in the presence and absence of AQ. Gene expression profiles generated by RNA-seq revealed numerous metabolic processes that were altered by AQ, including genes associated with monosaccharide metabolism, carbohydrate metabolism, and ketone metabolism (SI Appendix, Fig. S5 K and L). Comparative analysis of gene expression profiles generated by RNA-seq analysis of muscle from Nurr1-mTg and WT mice treated with AQ revealed 290 genes that were commonly regulated in muscle from these groups. Interestingly, among these 290 genes, protein phosphatase 1 regulatory (inhibitor) subunit 1a (Ppp1r1a), protein phosphatase 1, and regulatory (inhibitor) subunit 3C (Ppp1r3c) were significantly decreased in muscle from both Nurr1-mTg and AQ-treated mice. Decreased Ppp1r1a expression leads to activation of protein phosphatase 1 activity and the inhibition of glycogenolysis and increased glycogen synthesis (41). Thus, overexpression of Nurr1 or treatment with AQ regulates common targets that are involved in regulation of glycogenolysis. However, there are also many genes that are differentially regulated by AQ that likely contribute to its effects.
AQ Enhances Metabolism and Reverts Hepatic Steatosis in Obese Mice.
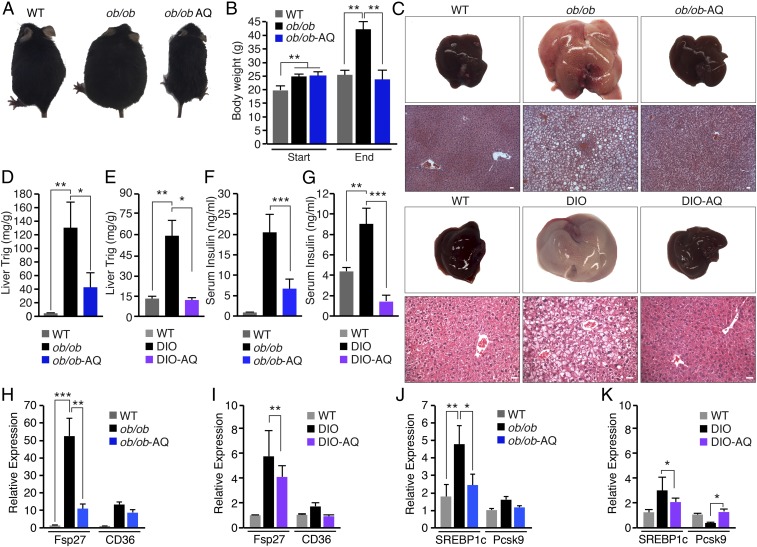
To determine if AQ could revert metabolic disease in obese mice, we tested the responsiveness to AQ of leptin-deficient (ob/ob) mice and diet-induced obese (DIO) mice after 8 wk of an HFD. Leptin is secreted by adipocytes and acts on the central nervous system to control energy consumption and food intake. We used ob/ob mice to assess whether the metabolic actions of AQ depended on the leptin pathway. Adult ob/ob and DIO mice were provided with AQ in drinking water ad libitum with normal chow and compared with WT mice on normal chow for 4 wk (Fig. 6A and SI Appendix, Fig. S6A). At the start of the study, ob/ob mice on normal chow weighed 25% more than WT mice (Fig. 6B). Similarly, the DIO mice weighed 20% more than WT mice on normal chow (SI Appendix, Fig. S6B). The ob/ob and DIO mice treated with AQ for 4 wk lost weight and were resistant to weight gain despite consuming comparable quantities of chow (Fig. 6B and SI Appendix, Fig. S6 B–D). The decrease in body weight was correlated with a decrease in adipocyte size in white adipose tissue from ob/ob and DIO mice treated with AQ compared with the respective control mice (SI Appendix, Fig. S6E).
Fig. 6.
NURR1 agonist reverts hepatic steatosis in obese mouse models. (A) WT and ob/ob mice on normal chow. ob/ob mice were administered AQ (ob/ob AQ) in drinking water from 5 to 9 wk of age. (B) Body weight of WT, ob/ob, and ob/ob mice administered AQ (ob/ob-AQ) at the start (5 wk of age) and the end (9 wk of age) of the experiment. (C) Livers showed that AQ treatment for 4 wk reverts hepatic steatosis in ob/ob and DIO mice to normal WT levels. (C, Lower) H&E staining. (Scale bars, 50 μm.) (D and E) Quantification of liver Trig levels of WT, ob/ob, and ob/ob-AQ mice after 4 wk of AQ treatment (D) and WT, DIO, and DIO-AQ mice after 4 wk of AQ treatment (E). (F and G) Serum insulin levels of WT, ob/ob, and ob/ob-AQ mice after 4 wk of treatment (F) and WT, DIO, and DIO-AQ mice after 4 wk of treatment (G). (H and I) Expression of genes involved in fatty acid synthesis fat-specific protein 27 and transport (CD36/FATP) in liver tissue of WT, ob/ob, and ob/ob-AQ after 4 wk of treatment (H) and WT, DIO, and DIO-AQ after 4 wk of treatment (I), as measured by real-time qRT-PCR. (J and K) Expression of genes involved in liver lipid metabolism, SREBP1c and Pcsk9, in liver tissue of WT and ob/ob mice in the absence or presence of AQ after 4 wk (J) and WT and DIO mice in the absence or presence of AQ after 4 wk (K), as measured by real-time qRT-PCR. Data are represented as mean ± SEM. n = 6. *P < 0.05, **P < 0.005, ***P < 0.0005.
To determine if AQ was able to reverse hepatic steatosis, we looked at liver triglyceride accumulation. Histological and biochemical analysis showed a significant decrease in size and triglyceride accumulation, respectively, with a 66% decrease in triglyceride levels in ob/ob mice treated with AQ compared with nontreated ob/ob mice, and a 79% decrease in triglyceride levels in DIO-mice treated with AQ compared with untreated mice on an HFD (Fig. 6 C–E). Obesity commonly leads to hyperinsulinemia, which was observed in WT mice on an HFD and ob/ob mice (Figs. 2C and 6 F and G). Remarkably, serum insulin levels of AQ-treated mice were significantly lower than in untreated ob/ob and DIO mice (Fig. 6 F and G). Analysis of expression of selected metabolic genes in the liver also revealed that the Fsp27 gene was up-regulated in ob/ob and DIO mice, whereas it was down-regulated in livers from ob/ob and DIO mice treated with AQ (Fig. 6 H and I). Additionally, SREBP1c expression was significantly decreased in livers from ob/ob and DIO mice treated with AQ (Fig. 6 J and K). In contrast, ob/ob and DIO mice treated with AQ showed no difference in expression of genes involved in gluconeogenesis (such as G6Pc and G6Pd), cholesterol synthesis (such as HCoARed and Pcsk9), and fatty acid synthesis (Acca2) in liver tissue (SI Appendix, Fig. S6 F and G). AQ had a similar metabolic effect on ob/ob and DIO mice, suggesting that it acts in a leptin-independent manner.
We examined whether AQ activation might act via central nervous system-dependent mechanisms by delivering it through an intracerebroventricular route to mice connected to a minipump attached via a catheter to a cannula containing the compound. We administered leptin via intracerebroventricular delivery as a positive control for the procedure, which has been previously shown to impact the body weight of mice on an HFD (SI Appendix, Fig. S6 H–M). Mice administered AQ via an intracerebroventricular route while on an HFD showed body weights comparable to mice receiving vehicle alone (SI Appendix, Fig. S6I). As a positive control, mice were treated with leptin via intracerebroventricular delivery while on an HFD and showed a decrease in body weight, as previously described (42, 43). Additionally, mice receiving vehicle, AQ, or leptin showed no difference in food consumption or glucose levels (SI Appendix, Fig. S6 J–L). AQ has been shown to activate gene expression in dopaminergic neurons (40), so we analyzed the expression of NURR1 target genes described previously to validate that the drug indeed reached neuronal targets (SI Appendix, Fig. S6M).
Taken together, our results show that a NURR1 agonist reverts obesity-associated metabolic disorders such as hyperglycemia, hyperinsulinemia, and hepatic steatosis in ob/ob and DIO mice. Moreover, the finding that intracranial delivery of AQ had no metabolic effects suggests that it acts outside the nervous system to evoke its metabolic actions.
Beneficial Metabolic Effects of a Second Putative NURR1 Agonist.
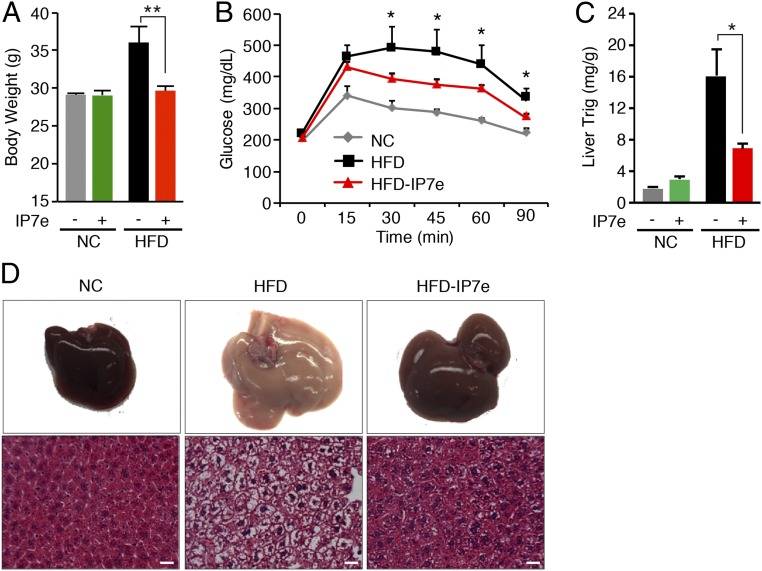
While AQ has been shown to act as a direct agonist of NURR1 and our results show that AQ mimics certain of the metabolic actions of Nurr1 overexpression, this compound may also act through additional mechanisms (44). Therefore, we compared the effect of a second NURR1 agonist, isoxazolo-pyridinone 7e (IP7e), that is structurally dissimilar to AQ (45). IP7e is a cell-permeable, isoxazolo-pyridinone compound that acts as a potent activator of Nurr1-dependent transcriptional activity. We provided IP7e through oral gavage to adult mice maintained on normal chow and an HFD for 4 wk (SI Appendix, Fig. S7A). When mice were maintained on a normal chow diet, IP7e had no impact on weight gain. However, when mice were maintained on an HFD, IP7e prevented obesity, despite no difference in food consumption compared with control mice on an HFD (Fig. 7A and SI Appendix, Fig. S7 A and B). Moreover, IP7e-treated mice on an HFD showed a significant decrease in fat mass, which correlated with a decrease in size of white adipocytes compared with untreated mice on an HFD (SI Appendix, Fig. S7C). Additionally, IP7e elicited an improvement in glucose handling of mice on an HFD (Fig. 7B).
Fig. 7.
IP7e confers resistance to obesity and prevents hepatic steatosis. (A) Body weight at the end of the study of mice on NC or HFD in the presence or absence of IP7e. (B) Glucose tolerance measurements of mice on NC or HFD in the presence or absence of IP7e after 4 wk. (C) Quantification of liver Trig levels in mice on NC or HFD in the presence or absence of IP7e after 4 wk. (D) IP7e prevents hepatic steatosis in WT mice on an HFD. Livers of mice on NC and HFD in the absence or presence of IP7e were analyzed after 4 wk. (D, Lower) H&E staining. (Scale bars, 50 μm.) Data are represented as mean ± SEM. n = 6 for each group. *P < 0.05, **P < 0.005.
We also examined the potential effect of IP7e on liver triglyceride accumulation in response to an HFD. Indeed, IP7e mice were resistant to hepatic steatosis and showed a 48% decrease in hepatic triglyceride levels on an HFD compared with untreated mice on an HFD (Fig. 7 C and D). Thus, IP7e treatment prevents the development of HFD-induced obesity and associated metabolic disorders such as hyperglycemia and hepatic steatosis, mimicking the effects of AQ.
Discussion
The results of this study reveal the components of a gene regulatory program for the control of glucose uptake and utilization by skeletal muscle and a potential modulator of the metabolic effects of exercise. Up-regulation of Nurr1 in response to MED13 deletion (18) or transgenic Nurr1 overexpression confers a beneficial metabolic phenotype in mice, normalizing glucose and insulin levels under conditions of caloric excess and preventing hepatic steatosis.
NURR1 as a Potential Mediator of Exercise.
Nurr1 has been identified as one of the most up-regulated genes in human skeletal muscle in response to exercise (27, 28). Expression of Nurr1 in skeletal muscle has also been shown to decline in mice exposed to an HFD (46). Changes in expression of Nurr1 and the related orphan nuclear receptor genes Nr4a1 (Nur77) and Nr4a3 (Nor1) have also been associated with alterations in lipid, carbohydrate, and energy homeostasis in mice (47–51). Transgenic overexpression of Nur77 in skeletal muscle has been reported to increase glycogen content, oxidative metabolism, and fatigue resistance (52, 53). Overexpression of Nor1 in skeletal muscle also enhanced oxidative metabolism, increased glycogen storage in skeletal muscle, increased physical performance, and reduced adiposity (49, 54). Muscle-specific deletion of Nur77 also results in reduced muscle mass and increased susceptibility to diet-induced obesity and insulin resistance (55, 56). However, the potential causal role of NURR1 in the control of energy homeostasis and hepatic steatosis in vivo has not been previously explored.
Our findings reveal that NURR1 is a potential transcriptional mediator of exercise and regulates the expression of genes involved in carbohydrate metabolism and glycogen biosynthesis in skeletal muscle. These findings are consistent with the results of Nurr1 overexpression in skeletal muscle, which leads to enhanced glucose uptake and storage as glycogen in skeletal muscle.
The Metabolic Effects of AQ and IP7e.
Members of the NR4A family are inducible by a range of metabolites (47), exercise (27, 28), and environmental stimuli (48) such as adrenoreceptor agonists (49), glucose (50), insulin (51), and others (57). However, unlike many other nuclear receptors, NURR1 does not contain a classical ligand-binding domain (LBD) cavity (58), which has complicated the identification of compounds that might directly activate NURR1. However, various compounds including AQ, an antimalarial and antiinflammatory drug, and glafenine, a pain-relieving drug, were identified as novel NURR1 activators (40). These drugs bind directly to the NURR1 LBD, leading to the activation of NURR1 and increased expression of its target genes. IP7e has also been identified as an activator of NURR1 (45). AQ and related compounds sharing a 4-amino-7-chloroquinoline scaffold are well-known antimalarial drugs, and have also been shown to display neuroprotective activity by protecting dopaminergic neurons from injury by environmental toxins and microglia-dependent neuroinflammation (40). Additionally, these compounds have been shown to regulate autophagy such that long-term use of these compounds would need to be carefully evaluated (59). Beneficial metabolic effects of these compounds have not, to our knowledge, been reported.
NURR1 activators have been shown to bind to the noncanonical ligand-binding domain of NURR1, enhancing transcriptional activity (40). While our findings are consistent with NURR1 agonism in skeletal muscle as a molecular basis for the beneficial metabolic effects of AQ, it is also likely that AQ exerts its systemic metabolic effects by acting on tissues in addition to skeletal muscle. Our finding that direct intracranial delivery of AQ does not evoke metabolic effects appears to rule out an activity through the central nervous system. We have also not ruled out the possibility that AQ might act through NURR1-independent mechanisms. In this regard, the similarity in metabolic actions of AQ and IP7e, which lack chemical similarity but both activate NURR1, suggests that NURR1, at least in part, may mediate their actions in vivo. However, we also observed a key difference between the actions of AQ and IP7e versus NURR1 overexpression in skeletal muscle, as the two compounds prevented weight gain on an HFD whereas NURR1 overexpression selectively prevented hepatic steatosis but not obesity. Whether this additional effect of these compounds on overall weight gain represents a specific response to NURR1 activation or alternative mechanisms cannot in nonmuscle tissues be resolved from the present studies. Despite these uncertainties, the metabolic actions of AQ and IP7e suggest the potential benefits of this class of compounds in metabolic disorders.
It is noteworthy that millions of people have been treated with chloroquine, a close chemical relative of AQ, as an antimalarial drug. Three clinical observational studies showed that patients with autoimmune inflammatory arthritis treated with hydroxychloroquine, a 4-amino-7-chloroquinoline scaffold compound, had a lower risk of developing diabetes (60). In addition, two clinical trials reported that treatment with high doses of hydroxychloroquine lowered glucose levels in individuals with type 2 diabetes (61). However, the mechanisms by which hydroxychloroquine may affect blood glucose and reduce the risk for type 2 diabetes have not been established. In our study, we show that AQ prevents the development of metabolic disorders associated with obesity and restores insulin levels to normal.
A Transcriptional Pathway for Muscle Metabolism.
Chronic exercise stimulates the activity of MEF2, which directly regulates the expression of GLUT4 and other metabolic genes (62–67). MEF2 and NURR1 physically interact and bind to the GLUT4 promoter and regulate in synergy its activity (18). In addition, MEF2 activates expression of PGC-1 and serves as a coactivator of PGC-1 to enhance metabolism (17, 64). Exercise enhances MEF2 activity through activation of calcium-dependent kinases that phosphorylate class II HDACs, promoting their export from the nucleus and derepression of MEF2 (14, 65, 66, 68). However, Nurr1 overexpression or activation did not affect the expression of PGC-1 or PPAR, suggesting an independent pathway for the control of glucose metabolism.
Skeletal muscle displays a range of fiber types with distinctive metabolic and contractile properties. Type I myofibers, which display a slow contractile phenotype, are associated with improved metabolic function. MEF2 drives the slow myofiber gene program (15, 16). However, in Nurr1 transgenic mice or mice treated with AQ, we did not observe an increase in slow myofiber number. Thus, the beneficial metabolic actions of NURR1 expression or treatment with AQ appear to reflect more direct actions on the expression of GLUT4 and possibly other genes. In this regard, overexpression of GLUT4 in skeletal muscle enhances glucose disposal and insulin action (37–40, 67).
Taken together, our findings reveal a glucose regulatory pathway that depends on NURR1 and its downstream target genes in skeletal muscle. The ability of the putative NURR1 agonists AQ and IP7e to mimic the beneficial metabolic effects of Nurr1 up-regulation raises interesting possibilities with respect to pharmacologic manipulation of metabolic syndrome, obesity-associated pathologies, and diabetes.
Materials and Methods
Animals.
Mice were housed in a pathogen-free barrier facility with a 12-h light/dark cycle and maintained on standard chow (2916; Teklad Global). Nurr1-mTg mice were derived by pronuclear injection of mouse embryos. Briefly, the Nurr1 coding sequence, obtained from the Invitrogen library, was cloned into a human skeletal muscle actin promoter-driven plasmid (69) with a polyadenylation sequence from the human growth hormone gene. The plasmid was injected into the pronucleus of mouse embryos and then implanted in a surrogate dam for gestation. F0 generation pups were selected by the presence of transgene by PCR from a tail biopsy and bred to the C57/BL6J wild type. The Nurr1-mTg mice were backcrossed with C57/BL6J mice for more than three generations. Male mice were used in all experiments. The ob/ob mice (B6.Cg-Lepob/J) were purchased from the Jackson Laboratory. For HFD (60% fat calories; D12492; Research Diet), mice were fed from the age of 5 wk to the indicated times. Normal chow diet consisted of 16% protein (Teklad regular rodent diet). Tissues were taken in the fed state except when otherwise mentioned. Animal work described in this manuscript was approved and conducted under the oversight of the University of Texas (UT) Southwestern Institutional Animal Care and Use Committee.
Drug Treatments.
AQ was purchased from Acros Organics, dissolved in water (7.5 mM), placed in water bottles, and provided to mice ad libitum during the experimental period. IP7e was purchased from Calbiochem, Millipore and dissolved in Tween 80 in a 10× stock solution. To obtain the final concentration (1×; 10 mg/kg), IP7e was dissolved in saline solution (0.9% NaCl). Gavage treatments were performed once a day. Control animals received the Tween 80-dissolved saline solution (0.9% NaCl; vehicle) once a day. For AQ treatment, we delivered the drug through an intracerebroventricular route. Mice were connected to an s.c. implanted minipump attached via a catheter to a cannula that delivered the compound as previously described (43). The minipump was filled with 7.5 mM AQ, and a volume of 0.11 μL/h was infused into the mice. The control group received vehicle via intracerebroventricular delivery.
Antibodies.
Antibodies to NURR1 (1:1,000; Ab41917; Abcam), MEF2 (1:1,000; sc-313; Santa Cruz Biotechnology), and GFP (1:1,000; A11122; Life Technology) were used.
RNA Analysis.
RNA was isolated from mouse tissues using TRIzol reagent (Invitrogen). Reverse-transcription PCR (RT-PCR) was performed to generate cDNA. Primers for ribosomal 18S RNA served as internal controls for the quality of RNA. The sequences of primers are listed in SI Appendix, Table S1. Illumina RNA-seq analysis was performed by the University of Texas Southwestern Microarray Core Facility using RNA extracted from tissues of 12-wk-old WT or Nurr1-mTg mice on an HFD or normal chow (NC) diet.
RNA-Seq Analysis.
Quality assessment of the RNA-seq data was done using the NGS QC Toolkit (70). Reads with more than 30% nucleotide with Phred quality scores less than 20 were removed from further analysis. Quality-filtered reads were then aligned to the mouse reference genome GRCm38 (mm10) using the TopHat (v 2.0.12) (https://www.encodeproject.org/software/tophat/) aligner using default settings except for –library-type = fr-firststrand. Aligned reads were counted using featureCounts (v 1.4.6) (71) per gene ID. Differential gene expression analysis was done using the R package edgeR (72) (v 3.8.6). For each comparison, genes were required to have 1 cpm in at least three samples to be considered as expressed. They were used for normalization factor calculation. Differential gene expression analysis was performed using the GLM approach following edgeR official documentation. Cutoff values of fold change greater than 1.5 and FDR less than 0.05 were then used to select for differentially expressed genes between sample group comparisons. Normalized gene cpm values were averaged within groups for heatmap generation. RNA-seq data is available on the Gene Expression Omnibus (GEO) under accession ID GSE130840 (73).
ChIP Experiments.
ChIP assays were performed as previously described (74). Briefly, quadriceps muscle powder was cross-linked with 1% formaldehyde for 15 min at room temperature. Chromatin fragmentation was performed by sonication using a Diagenode Biorupter (7 min with 30 s on/off). Proteins were immunoprecipitated by using 3 μg anti-MEF2, anti-NURR1 antibody, or IgG control. The antibody–chromatin complexes were left to rotate end-to-end overnight at 4 °C. Antibody–chromatin complexes were pulled down using Dynabeads protein G-conjugated magnetic beads (Life Technologies). Chromatin was washed, eluted, and reverse cross-linked, and then treated with protease. Chromatin fragments were then analyzed by quantitative PCR using SYBR Green fluorescence. Primers are listed in SI Appendix, Table S1. All values are expressed as mean ± SEM. Statistical analysis was performed using a two-tailed Student’s unpaired t test. Results were considered significant when P < 0.05.
ChIP-Seq Experiments.
ChIP assays were performed on powdered tibialis anterior muscle using ChIP-IT High Sensitivity (53040; Active Motif) according to the manufacturer’s instructions. ChIP-seq libraries were constructed using the Kapa Hyper Prep Kit (KK8500; Roche). Raw reads were mapped using Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) to obtain about 50 million uniquely mapped single-end reads per library. Peak calling was performed by MACS/MACS2 (liulab.dfci.harvard.edu/MACS/). The resulting output files included the peak list in BED format, ChIP signal in bedGraph format, and mapped reads in BAM format. All files were subjected to integrative in-depth analysis with published data and data generated by the Encode project using the Bioconductor software (https://www.bioconductor.org) project site. De novo motif discovery in H3K27Ac and MEF2 ChIP-seq peak regions was identified using Regulatory Sequence Analysis Tools (RSAT) (rsat.sb-roscoff.fr). Top motif 1 Pearson correlation (0.956): k-mer sig, 73.94; evalue, 1.1e-74; top motif 2 Pearson correlation (0.95): k-mer sig, 31.96; evalue, 1.1e-32; top motif 3 Pearson correlation (0.93): k-mer sig, 22.47; evalue, 3.4e-23. ChIPseq data analysis is available on GEO under accession ID GSE130840 (73).
Histology.
White adipose tissue, brown adipose tissue, and liver were isolated and fixed in 4% paraformaldehyde (PFA) and processed for H&E staining. For oil red O staining, liver tissues were fixed in 4% PFA overnight, incubated in 12% sucrose for 12 h and then in 18% sucrose overnight, and cryoembedded and sectioned by the UT Southwestern Histology Core Facility. For skeletal muscle fiber analysis, tissues were frozen in liquid nitrogen-precooled isopentane, and 8-μm sections were used for H&E and fiber-type staining.
Metabolic Chambers and Whole-Body Composition Analysis.
Metabolic phenotyping was performed using TSE metabolic chamber analysis by the Mouse Metabolic Phenotyping Core Facility at UT Southwestern. The mice on an HFD for 6 wk were placed in TSE metabolic chambers for an initial 5-d acclimation period, followed by a 4.5-d experimental period with data collection. Whole-body composition parameters were measured by magnetic resonance imaging using a Bruker Minispec mq10 system.
Lipid Absorption.
For analysis of fecal lipids, feces were collected over a 72-h period from individually housed mice on week 1 of the AQ HFD dietary study. The lipid content of feces and diet was extracted using chloroform:methanol (2:1) solvent. Lipid content of diet and stool was determined gravimetrically and used to calculate the fraction of consumed lipid that was absorbed. Total dietary lipid absorption was determined by calculating the difference between the dietary lipid ingested (defined as lipid content of diet during 72-h food intake) and fecal lipid output (defined as fecal lipid content during 72-h fecal collection).
Plasma and Tissue Chemistry.
Blood was collected using a 1-mL syringe coated with 0.5 M K2EDTA, and serum was collected by centrifugation for 20 min at 1,000 × g. Insulin and leptin levels were measured by ELISA. Serum triglyceride levels were measured using the Ortho Vitros 250 chemistry system. To measure triglyceride in the liver and skeletal muscle, tissue specimens were frozen immediately after isolation and pulverized in liquid nitrogen with a cell crusher. Serum and tissue triglyceride levels were measured by the Mouse Metabolic Phenotyping Core Facility at UT Southwestern.
Glucose Tolerance and Insulin Tolerance.
Glucose tolerance and insulin tolerance tests were performed as previously described (18). For glucose tolerance tests, mice were fasted for 6 h and injected intraperitoneally with a glucose solution (0.15 g/mL; 158968; Sigma-Aldrich) at 1.5 g/kg body weight. Blood glucose concentrations were measured before and 15, 30, 60, and 90 min after glucose injection. For insulin tolerance tests, mice were fasted for 6 h and injected intraperitoneally with insulin (human insulin I9278; Sigma-Aldrich) at 1.0 U/kg body weight. Blood glucose concentrations were measured before and 15, 30, 60, and 90 min after insulin injection.
Glycogen Measurements.
A glycogen colorimetric/fluorometric assay kit (Abcam; 65620) was used according to the manufacturer’s protocol to measure the quadriceps glycogen content in WT and Nurr1-mTg mice on an HFD and NC diet.
Voluntary Wheel Running.
Six-week-old Nurr1-mTg and corresponding WT littermates were randomly assigned to housing in individual cages with or without a running wheel for a total of 8 wk. Completed wheel revolutions and time spent running were continuously monitored and recorded. Run distance over 24-h periods was determined at the end of the experiment.
Statistical Analysis.
All values are given as mean ± SEM. Differences between two groups were assessed using unpaired two-tailed Student’s t tests. P < 0.05 was regarded as significant. Statistical analysis was performed in Excel (Microsoft).
Supplementary Material
Acknowledgments
We thank John Shelton for his help with histology. We thank Joyce Repa for her assistance with lipid absorption experiments. We thank Beibei Chen and Wenduo Ye for their expertise and assistance with bioinformatics analysis. We also thank Spencer Barnes (Department of Bioinformatics) for help with bioinformatics. We thank Jose Cabrera, Cheryl Nolen, John McAnally, Dylan Tennison, Evelyn Tennison, and Jennifer Brown for their assistance. We appreciate the services of the UT Southwestern Mouse Metabolic Phenotyping Core Facility. This work was supported by grants from the NIH (AR-067294, HL-130253, and DK-099653), American Diabetes Association (1-16-PDF-006 to L.A.), American Heart Association (14SDG17950008 to T.F.), and Robert A. Welch Foundation (1-0025 to E.N.O.).
Footnotes
Conflict of interest statement: L.A., R.B.-D., and E.N.O. have filed a patent application (PCT/US2017/029575) that has been published (https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2017189686). E.N.O. and L.A.L. are coauthors on a 2018 commentary paper.
Data deposition: RNA-seq and ChIP-seq assays are available on the Gene Expression Omnibus (accession ID GSE130840).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902490116/-/DCSupplemental.
References
- 1.DeFronzo RA, et al. (1981) The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes 30:1000–1007. [DOI] [PubMed] [Google Scholar]
- 2.Zurlo F, Larson K, Bogardus C, Ravussin E (1990) Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest 86:1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koopman R, et al. (2005) A single session of resistance exercise enhances insulin sensitivity for at least 24 h in healthy men. Eur J Appl Physiol 94:180–187. [DOI] [PubMed] [Google Scholar]
- 4.Lee AD, Hansen PA, Holloszy JO (1995) Wortmannin inhibits insulin-stimulated but not contraction-stimulated glucose transport activity in skeletal muscle. FEBS Lett 361:51–54. [DOI] [PubMed] [Google Scholar]
- 5.Mikines KJ, Sonne B, Farrell PA, Tronier B, Galbo H (1988) Effect of physical exercise on sensitivity and responsiveness to insulin in humans. Am J Physiol 254:E248–E259. [DOI] [PubMed] [Google Scholar]
- 6.Ranalletta M, et al. (2005) Altered hepatic and muscle substrate utilization provoked by GLUT4 ablation. Diabetes 54:935–943. [DOI] [PubMed] [Google Scholar]
- 7.Turner N, et al. (2013) Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia 56:1638–1648. [DOI] [PubMed] [Google Scholar]
- 8.Birnbaum MJ. (1989) Identification of a novel gene encoding an insulin-responsive glucose transporter protein. Cell 57:305–315. [DOI] [PubMed] [Google Scholar]
- 9.Charron MJ, Brosius FC III, Alper SL, Lodish HF (1989) A glucose transport protein expressed predominately in insulin-responsive tissues. Proc Natl Acad Sci USA 86:2535–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James DE, Strube M, Mueckler M (1989) Molecular cloning and characterization of an insulin-regulatable glucose transporter. Nature 338:83–87. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy JW, et al. (1999) Acute exercise induces GLUT4 translocation in skeletal muscle of normal human subjects and subjects with type 2 diabetes. Diabetes 48:1192–1197. [DOI] [PubMed] [Google Scholar]
- 12.Egan B, Zierath JR (2013) Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 17:162–184. [DOI] [PubMed] [Google Scholar]
- 13.Kim MS, et al. (2008) Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol Cell Biol 28:3600–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGee SL, Sparling D, Olson AL, Hargreaves M (2006) Exercise increases MEF2- and GEF DNA-binding activity in human skeletal muscle. FASEB J 20:348–349. [DOI] [PubMed] [Google Scholar]
- 15.Potthoff MJ, Olson EN (2007) MEF2: A central regulator of diverse developmental programs. Development 134:4131–4140. [DOI] [PubMed] [Google Scholar]
- 16.Wu H, et al. (2000) MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J 19:1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, et al. (2002) Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418:797–801. [DOI] [PubMed] [Google Scholar]
- 18.Amoasii L, et al. (2016) A MED13-dependent skeletal muscle gene program controls systemic glucose homeostasis and hepatic metabolism. Genes Dev 30:434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amoasii L, Olson EN, Bassel-Duby R (2018) Control of muscle metabolism by the Mediator complex. Cold Spring Harb Perspect Med 8:a029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saucedo-Cardenas O, et al. (1998) Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA 95:4013–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadkhodaei B, et al. (2009) Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci 29:15923–15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volakakis N, et al. (2015) Nurr1 and retinoid X receptor ligands stimulate ret signaling in dopamine neurons and can alleviate α-synuclein disrupted gene expression. J Neurosci 35:14370–14385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassett MH, Suzuki T, Sasano H, White PC, Rainey WE (2004) The orphan nuclear receptors NURR1 and NGFIB regulate adrenal aldosterone production. Mol Endocrinol 18:279–290. [DOI] [PubMed] [Google Scholar]
- 24.Kim K-S, et al. (2003) Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J Neurochem 85:622–634. [DOI] [PubMed] [Google Scholar]
- 25.Creyghton MP, et al. (2010) Histone H3K27Ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zentner GE, Tesar PJ, Scacheri PC (2011) Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catoire M, et al. (2012) Pronounced effects of acute endurance exercise on gene expression in resting and exercising human skeletal muscle. PLoS One 7:e51066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA (2005) Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J 19:1498–1500. [DOI] [PubMed] [Google Scholar]
- 29.Brown MS, Goldstein JL (2008) Selective versus total insulin resistance: A pathogenic paradox. Cell Metab 7:95–96. [DOI] [PubMed] [Google Scholar]
- 30.Denechaud P-D, et al. (2008) ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J Clin Invest 118:956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi SH, Ginsberg HN (2011) Increased very low density lipoprotein (VLDL) secretion, hepatic steatosis, and insulin resistance. Trends Endocrinol Metab 22:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon Y-A, et al. (2012) The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab 15:240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benhamed F, et al. (2012) The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J Clin Invest 122:2176–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C (2013) Novel insights into ChREBP regulation and function. Trends Endocrinol Metab 24:257–268. [DOI] [PubMed] [Google Scholar]
- 35.Lund EG, et al. (2006) Different roles of liver X receptor α and β in lipid metabolism: Effects of an α-selective and a dual agonist in mice deficient in each subtype. Biochem Pharmacol 71:453–463. [DOI] [PubMed] [Google Scholar]
- 36.Cohen JC, Horton JD, Hobbs HH (2011) Human fatty liver disease: Old questions and new insights. Science 332:1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treadway JL, et al. (1994) Enhanced peripheral glucose utilization in transgenic mice expressing the human GLUT4 gene. J Biol Chem 269:29956–29961. [PubMed] [Google Scholar]
- 38.Hansen PA, Wang W, Marshall BA, Holloszy JO, Mueckler M (1998) Dissociation of GLUT4 translocation and insulin-stimulated glucose transport in transgenic mice overexpressing GLUT1 in skeletal muscle. J Biol Chem 273:18173–18179. [DOI] [PubMed] [Google Scholar]
- 39.Tsao TS, Burcelin R, Katz EB, Huang L, Charron MJ (1996) Enhanced insulin action due to targeted GLUT4 overexpression exclusively in muscle. Diabetes 45:28–36. [DOI] [PubMed] [Google Scholar]
- 40.Kim C-H, et al. (2015) Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc Natl Acad Sci USA 112:8756–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fong NM, et al. (2000) Identification of binding sites on protein targeting to glycogen for enzymes of glycogen metabolism. J Biol Chem 275:35034–35039. [DOI] [PubMed] [Google Scholar]
- 42.Halaas JL, et al. (1997) Physiological response to long-term peripheral and central leptin infusion in lean and obese mice. Proc Natl Acad Sci USA 94:8878–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujikawa T, Chuang JC, Sakata I, Ramadori G, Coppari R (2010) Leptin therapy improves insulin-deficient type 1 diabetes by CNS-dependent mechanisms in mice. Proc Natl Acad Sci USA 107:17391–17396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solomon VR, Lee H (2009) Chloroquine and its analogs: A new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol 625:220–233. [DOI] [PubMed] [Google Scholar]
- 45.Montarolo F, et al. (2014) Effects of isoxazolo-pyridinone 7e, a potent activator of the Nurr1 signaling pathway, on experimental autoimmune encephalomyelitis in mice. PLoS One 9:e108791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Y, Luo L, Luo N, Zhu X, Garvey WT (2007) NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: Potential role in insulin resistance. J Biol Chem 282:31525–31533. [DOI] [PubMed] [Google Scholar]
- 47.Roche E, et al. (1999) Palmitate and oleate induce the immediate-early response genes c-fos and nur-77 in the pancreatic beta-cell line INS-1. Diabetes 48:2007–2014. [DOI] [PubMed] [Google Scholar]
- 48.Kanzleiter T, et al. (2009) Regulation of the nuclear hormone receptor nur77 in muscle: Influence of exercise-activated pathways in vitro and obesity in vivo. Biochim Biophys Acta 1792:777–782. [DOI] [PubMed] [Google Scholar]
- 49.Pearen MA, et al. (2008) The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology 149:2853–2865. [DOI] [PubMed] [Google Scholar]
- 50.Susini S, Roche E, Prentki M, Schlegel W (1998) Glucose and glucoincretin peptides synergize to induce c-fos, c-jun, junB, zif-268, and nur-77 gene expression in pancreatic beta(INS-1) cells. FASEB J 12:1173–1182. [PubMed] [Google Scholar]
- 51.Wu X, et al. (2007) The effect of insulin on expression of genes and biochemical pathways in human skeletal muscle. Endocrine 31:5–17. [DOI] [PubMed] [Google Scholar]
- 52.Chao LC, et al. (2012) Skeletal muscle Nur77 expression enhances oxidative metabolism and substrate utilization. J Lipid Res 53:2610–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pearen MA, Muscat GEO (2012) Orphan nuclear receptors and the regulation of nutrient metabolism: Understanding obesity. Physiology (Bethesda) 27:156–166. [DOI] [PubMed] [Google Scholar]
- 54.Pearen MA, et al. (2013) Transgenic muscle-specific Nor-1 expression regulates multiple pathways that effect adiposity, metabolism, and endurance. Mol Endocrinol 27:1897–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chao LC, et al. (2009) Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes 58:2788–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tontonoz P, et al. (2015) The orphan nuclear receptor Nur77 is a determinant of myofiber size and muscle mass in mice. Mol Cell Biol 35:1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearen MA, Muscat GEO (2010) Minireview: Nuclear hormone receptor 4A signaling: Implications for metabolic disease. Mol Endocrinol 24:1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, et al. (2003) Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423:555–560. [DOI] [PubMed] [Google Scholar]
- 59.Liang X, Tang J, Liang Y, Jin R, Cai X (2014) Suppression of autophagy by chloroquine sensitizes 5-fluorouracil-mediated cell death in gallbladder carcinoma cells. Cell Biosci 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wasko MCM, et al. (2015) Antidiabetogenic effects of hydroxychloroquine on insulin sensitivity and beta cell function: A randomised trial. Diabetologia 58:2336–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quatraro A, et al. (1990) Hydroxychloroquine in decompensated, treatment-refractory noninsulin-dependent diabetes mellitus. A new job for an old drug? Ann Intern Med 112:678–681. [DOI] [PubMed] [Google Scholar]
- 62.Thai MV, Guruswamy S, Cao KT, Pessin JE, Olson AL (1998) Myocyte enhancer factor 2 (MEF2)-binding site is required for GLUT4 gene expression in transgenic mice. Regulation of MEF2 DNA binding activity in insulin-deficient diabetes. J Biol Chem 273:14285–14292. [DOI] [PubMed] [Google Scholar]
- 63.Mora S, Pessin JE (2000) The MEF2A isoform is required for striated muscle-specific expression of the insulin-responsive GLUT4 glucose transporter. J Biol Chem 275:16323–16328. [DOI] [PubMed] [Google Scholar]
- 64.Michael LF, et al. (2001) Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci USA 98:3820–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ojuka EO, et al. (2002) Intermittent increases in cytosolic Ca2+ stimulate mitochondrial biogenesis in muscle cells. Am J Physiol Endocrinol Metab 283:E1040–E1045. [DOI] [PubMed] [Google Scholar]
- 66.Ojuka EO, Goyaram V, Smith JAH (2012) The role of CaMKII in regulating GLUT4 expression in skeletal muscle. Am J Physiol Endocrinol Metab 303:E322–E331. [DOI] [PubMed] [Google Scholar]
- 67.Ren JM, et al. (1995) Overexpression of Glut4 protein in muscle increases basal and insulin-stimulated whole body glucose disposal in conscious mice. J Clin Invest 95:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKinsey TA, Zhang CL, Olson EN (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci USA 97:14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miniou P, et al. (1999) Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res 27:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel RK, Jain M (2012) NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y, Smyth GK, Shi W (2014) featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amoasii L, et al. (2019) NURR1 activation in skeletal muscle controls systemic energy homeostasis (RNA-seq data and ChIP-seq data). Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130840. Deposited May 7, 2019. [DOI] [PMC free article] [PubMed]
- 74.Tuteja G, Jensen ST, White P, Kaestner KH (2008) Cis-regulatory modules in the mammalian liver: Composition depends on strength of Foxa2 consensus site. Nucleic Acids Res 36:4149–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.