Significance
Bacterial endospores are found as normal inhabitants throughout the environment, and some are vectors leading to food spoilage, food poisoning, and infectious diseases. Promoting efficient germination of spores could sensitize spores for practical inactivating measures. Specific nutrient germinants that trigger spore germination are recognized by cognate germinant receptors (GRs). In this work, we report a structure-based mechanism for GR-mediated germinant binding and recruitment. Crystal structure, molecular docking, and biophysical and genetic analyses indicate that the N-terminal domain of GR A proteins likely possesses a binding pocket that accommodates specific germinant molecules at the interface between its two subdomains. These results can be explored for the development of germinant analogues as either potentiators or inhibitors of spore germination.
Keywords: Bacillus, spores, spore germination, spore germinant receptor
Abstract
Germination of Bacillus spores is induced by the interaction of specific nutrient molecules with germinant receptors (GRs) localized in the spore’s inner membrane. GRs typically consist of three subunits referred to as A, B, and C, although functions of individual subunits are not known. Here we present the crystal structure of the N-terminal domain (NTD) of the A subunit of the Bacillus megaterium GerK3 GR, revealing two distinct globular subdomains bisected by a cleft, a fold with strong homology to substrate-binding proteins in bacterial ABC transporters. Molecular docking, chemical shift perturbation measurement, and mutagenesis coupled with spore germination analyses support a proposed model that the interface between the two subdomains in the NTD of GR A subunits serves as the germinant binding site and plays a critical role in spore germination. Our findings provide a conceptual framework for understanding the germinant recruitment mechanism by which GRs trigger spore germination.
Formation of metabolically dormant endospores is common to many species in the Firmicutes phylum, in particular Bacillales and Clostridiales (1). These spores can survive prolonged nutrient starvation and are resistant to a variety of antimicrobial treatments due to their special outer layers and unique features of the spore core. However, spores are capable of monitoring the nutritional state of their surroundings and returning to a metabolically active state through a series of events termed germination, when environmental conditions once again favor vegetative growth (2, 3). Notably, spores lose many of their resistance properties during germination and can then be easily eliminated by routine decontamination methods. Many members of Bacillales and Clostridiales genera are found in the environment and some can be human commensals; importantly, spores of some of these species can be crucial in causing food spoilage as well as some infectious diseases and intoxications of public health concern (1). Discovery and development of novel agents that promote highly efficient germination of spores in populations could facilitate efforts to render spores more susceptible to benign disinfection measures, and thus reduce transmission of spores that can cause some serious human diseases (4, 5).
In nature, Bacillus spore germination is irreversibly triggered when specific small-molecule nutrients called germinants are recognized by germinant receptors (GRs) located in the inner spore membrane, resulting in the release of dipicolinic acid (DPA) from the endospore core and degradation of the peptidoglycan cortex (3, 6, 7). All known spores of Bacillales and most Clostridiales species have GerA-type GRs whose coding genes are typically arranged in tricistronic operons that encode three protein components termed A, B, and C, and in some cases perhaps a D subunit (7, 8). Multiple GR isoforms with distinct or partially overlapping germinant specificities have been described (2, 9); SI Appendix, Table S1 lists the number of the A subunits of GRs identified in Bacillales and Clostridiales species. In Bacillus subtilis, the model organism for spore research, three functional GRs, GerA, GerB, and GerK, have been extensively studied. The GerA GR triggers spore germination in response to l-alanine or l-valine, while the GerB and GerK GRs act cooperatively to trigger germination with a cogerminant mixture of l-asparagine, d-glucose, d-fructose, and K+ ions (AGFK). Interestingly, all three B. subtilis GRs are colocalized primarily in a single cluster termed a “germinosome” in the inner membrane (IM) of spores along with the auxiliary germination protein GerD (10). Loss of any subunit of a particular GR generally destabilizes the other two subunits and abolishes the activity of the entire GR but has minimal effects on the function of other GRs (11, 12). Despite a clear understanding of their roles in spore germination, the mechanism whereby GRs recognize specific germinants and initiate the germination cascade remains largely unknown.
The A, B, and C subunits of GRs exhibit minimal sequence homology with proteins of known function, although both within and across species individual GR subunits exhibit significant sequence and predicted topology homologies with the corresponding subunits of other GRs (9) (current study). Bioinformatic and membrane topological analyses show that the A and B subunits of the GRs are integral membrane proteins, whereas the C proteins are peripheral membrane proteins with an N-terminal diacylglycerol anchor (13, 14). While the B subunits are predicted to have transmembrane (TM) helices throughout the entire protein, the A subunits consist of a core TM domain flanked by a large hydrophilic N-terminal domain (NTD) and a short C-terminal tail. The crystal structure of the C subunit of the B. subtilis GerB GR (GerBC) revealed a previously uncharacterized type of protein fold consisting of three distinct α/β mixed domains (15). A number of site-directed mutagenesis studies of A and B subunits of GRs (11, 12, 16, 17), aided by the limited sequence homology between the B subunits of GRs with the superfamily of single-component amino acid transporters (18), have led to the speculation that GRs may possess a transporter-related function. However, the precise function of each GR subunit and how they interact with germinants have remained elusive.
To further probe the role of the individual GR subunits in spore germination, we have now determined the crystal structure of the conserved N-terminal soluble domain of the A subunit of the Bacillus megaterium GerK3 GR (GerK3ANTD; lacking the extreme N-terminal 25 residues), the first structure of any GR A subunit. GerK3ANTD consists of two distinct three-layer αβα sandwich subdomains separated by a deep cleft. Interestingly, this protein shares significant structural similarity to substrate-binding proteins that serve as receptors for membrane-associated small-molecule transporters and signal transducers (19–21). Our biochemical and biophysical data strongly suggest that amino acid residues lining the cleft between the two subdomains of the NTDs of the A subunits are involved directly in the recognition and binding of germinants. This study represents a significant advance in our understanding of the structure and function of the NTD of the GR A subunit, permitting insight into the mechanism underlying the recruitment of germinants to Bacillus GRs and suggesting further avenues for study of the functions of GRs as a whole.
Results and Discussion
Crystal Structure of GerK3ANTD.
To explore the structure–function relationship of GR A subunits, we first sought to identify and produce protein construct variants exhibiting higher crystallization propensity. To this end, we cloned a total of 56 individual NTD constructs of 15 major GR A subunits from four species, B. subtilis, B. megaterium, Bacillus cereus, and Clostridium perfringens, and examined their protein expression level as well as the solubility and quality of the purified proteins. We found that the NTDs of three GR A proteins, B. cereus GerIA, and B. megaterium GerUA and GerK3A, were expressed at the highest levels and well-behaved during purification; the NTDs of B. subtilis GerAA and GerBA were largely insoluble under similar experimental conditions. The GerK3ANTD construct was selected for the structural study because we were unable to obtain diffraction-quality crystals of the NTD constructs from GerIA and GerUA.
GerK3, one of the three gerK operons identified and named in B. megaterium after the canonical B. subtilis GerK receptors, is a chromosomally encoded B. megaterium GR bearing a frameshift mutation in the B gene that results in a severely truncated B-subunit variant (22). GerK3 GR is therefore expected to be nonfunctional. However, GerK3A shares 77.4% and 60.5% sequence identity with the B. megaterium plasmid-borne GerUA and B. subtilis GerKA, respectively, with both of these latter A proteins a part of a functional GR (SI Appendix, Fig. S1). In the current study, we crystallized the 277-residue GerK3ANTD protein that lacks the N-terminal flexible region (NDR; residues 1–25) unique to GerK3A, the predicted TM domain (303–469), and the C-terminal hydrophilic tail (CDR; 470–540) (Fig. 1A). The structure of GerK3ANTD has been solved at 2.79-Å resolution by the method of single-wavelength anomalous dispersion (SAD) using a selenomethionine (SeMet) derivative. The final model was refined to an R factor of 20.5% and a free R value of 22.7% (SI Appendix, Table S2).
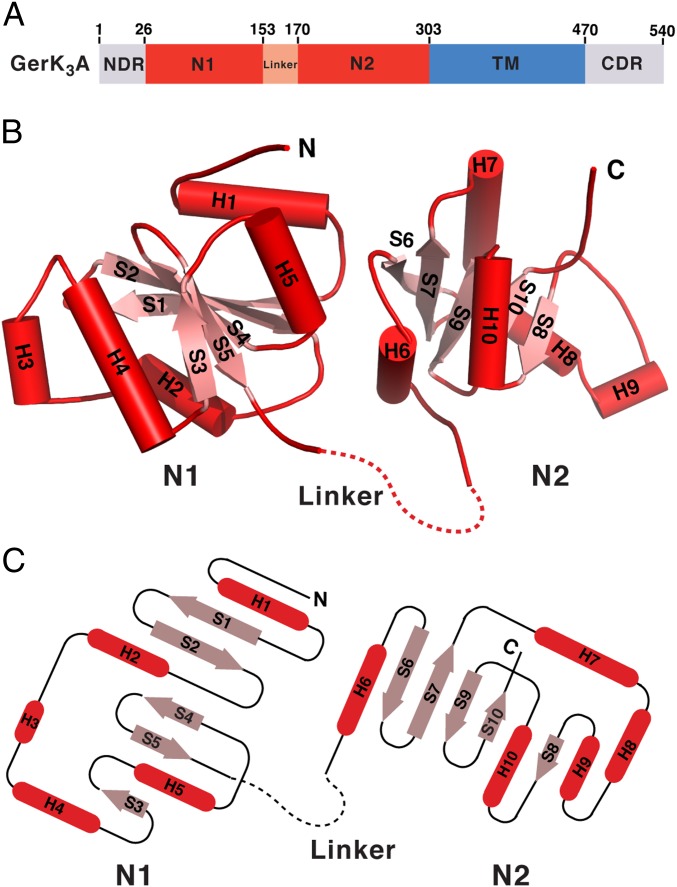
Fig. 1.
Crystal structure of GerK3ANTD. (A) Schematic representation of the domain organization of GerK3A. GerK3ANTD (residues 26–302) comprises N1, linker, and N2 domains. The boundaries of the N1 and N2 domains are defined according to results presented here. (B) Ribbon diagram of GerK3ANTD, with the secondary structure elements labeled. α-helices and β-strands are shown in red and pink, respectively. The disordered linker region is marked by a dashed line. (C) Topology diagram of GerK3ANTD. Cylinders and arrows represent α-helices and β-strands, respectively.
GerK3ANTD forms a butterfly-shaped architecture consisting of two distinct globular subdomains (N1 and N2) connected by a flexible linker (Fig. 1B). Each of these two subdomains contains a central five-stranded antiparallel β-sheet flanked by α-helices on each side (Fig. 1C). The N1 domain (residues 26–152) is formed by the central β-sheet arranged with a topology of S1S2S4S5S3, with helices H1 and H5 flanking on one side and H2, H3, and H4 on the other side. In the N2 domain (residues 170–296), the central β-sheet has the same topology (S6S7S9S10S8) surrounded by helices H6 and H10 as well as H7, H8, and H9. The 16-residue linker region (residues 153–169) between the N1 and N2 domains is not visible in the electron density map and therefore must be disordered.
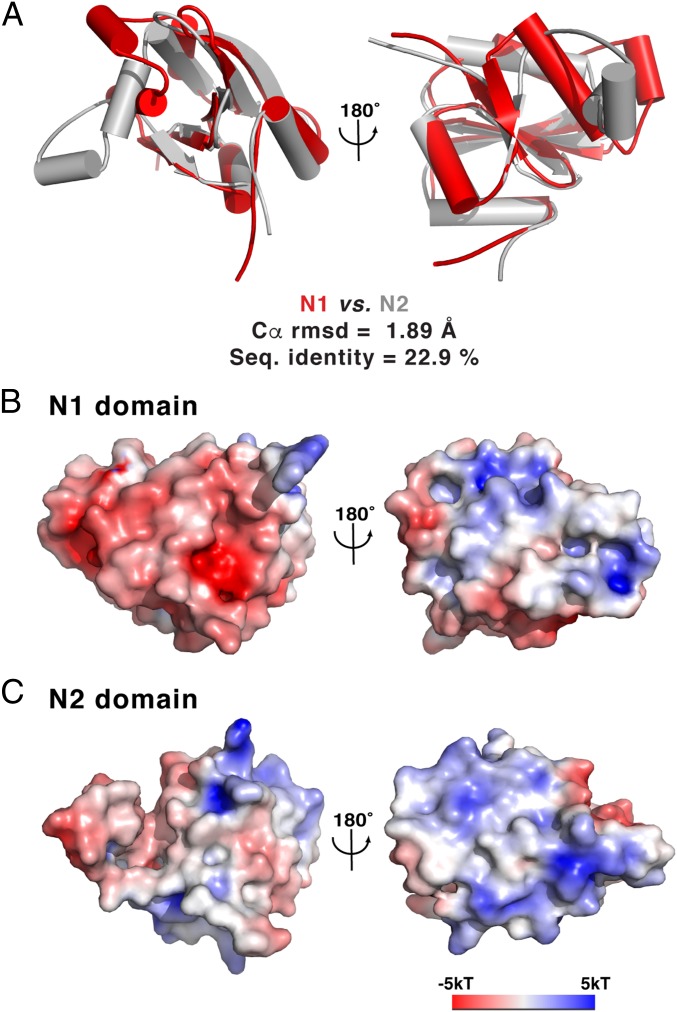
Despite their low sequence identity (19.2%), the N1 and N2 domains share essentially identical secondary-structure topology and connectivity and superimpose very well with an rmsd of ∼1.89 Å over 96 aligned Ca positions (Fig. 2A and SI Appendix, Fig. S2). The 24 invariant residues shared between the N1 and N2 domains are located throughout the entire subdomains (SI Appendix, Fig. S2). The most notable structural difference in two subdomains is in the two outer helices (H3 and H4 in the N1 domain vs. H8 and H9 in the N2 domain) in which H8 and H9 exhibit a more relaxed conformation (Fig. 1B). However, these two domains contain a similar mix of positive and negative patches on their surface-exposed regions, with a largely electronegative surface along both the H4-S3-H5 side in N1 and the H9-S8-H10 side in N2, as well as electropositive patches on the other side of the two domains (Fig. 2 B and C). Consequently, it seems likely that GerK3ANTD has evolved by an internal duplication in the ancestral GR A gene.
Fig. 2.
The N1 and N2 domains of GerK3ANTD are structurally similar to each other. (A) Superimposition of the N1 (red) and N2 (gray) domains. (B and C) Molecular surface representation of the N1 (B) and N2 (C) domains shown in similar orientations as in A and colored according to the local electrostatic potential calculated with the program ABPS (65).
A BLAST search using the B. subtilis GerAA subunit as the query sequence identified 253 GerAA homologs, with 1 to 32 coding genes in each species, in 52 completed spore-forming Bacillales and Clostridiales genomes (SI Appendix, Table S1). Pairwise sequence alignment of these A subunit homologs shows that they share a sequence identity of 21.8 to 77.4% with GerK3A (SI Appendix, Fig. S1). Consistent with the results from a previous report (22), GerK3A clusters most closely with B. megaterium GerUA and B. subtilis GerKA (discussed above) while sharing sequence identities of 37.2% and 38.3% with B. subtilis GerAA and GerBA, respectively. Secondary-structure predictions suggest that the NTDs of B. subtilis GerAA and GerBA have essentially the same secondary-structure topology as GerK3ANTD, whose predicted secondary structure elements are matched very well with the aforementioned secondary structure conformations (SI Appendix, Fig. S1). Moreover, a similar predicted secondary structure packing pattern is shared by both GerK3ANTD and the NTD of the well-characterized B. cereus GerIA GR (GerIANTD) (SI Appendix, Fig. S1). This similarity seems especially striking since our homology modeling experiments show that these two NTDs also have similar 3D structures (discussed below) and when we note that GerIA contains an additional 200-residue N-terminal extension (23). We thus conclude that the overall structure of the GerK3ANTD is conserved among the GR A subunit homologs.
Structural Relatives of GerK3ANTD.
Although primary amino acid sequence analysis failed to identify any known protein motif in GerK3ANTD, a structure homology search of the Protein Data Bank (PDB) using the National Center for Biotechnology Information’s (NCBI) Vector Alignment Search Tool (VAST+) (24) revealed that both the N1 and N2 domains of GerK3ANTD share a common fold with bacterial periplasmic-binding proteins (PeBPs) (Pfam database identification numbers CL0177 and CL0144). PeBPs serve as initial receptors for a wide variety of small ligands, such as carbohydrates, amino acids, oligopeptides, transition metal ions, and vitamins. These proteins are often associated with downstream membrane protein components to mediate chemotactic responses and selective solute uptake (19, 21, 25). The PeBP superfamily contains functionally diverse members from both gram-negative and gram-positive bacteria, many of which are the substrate-binding proteins of prokaryotic ATP-binding cassette (ABC) transporters. Despite the lack of a uniform size distribution and low sequence identity, PeBPs are believed to have evolved from a common ancestor due to their highly conserved core structural folds, ligand binding mechanisms, and the operon arrangement of their genes (26). The characteristic PeBP fold consists of two mixed αβα domains separated by a deep cleft wherein the ligand binds and is engulfed by a hinge-bending motion between the two domains. This overall fold is very similar to that of GerK3ANTD.
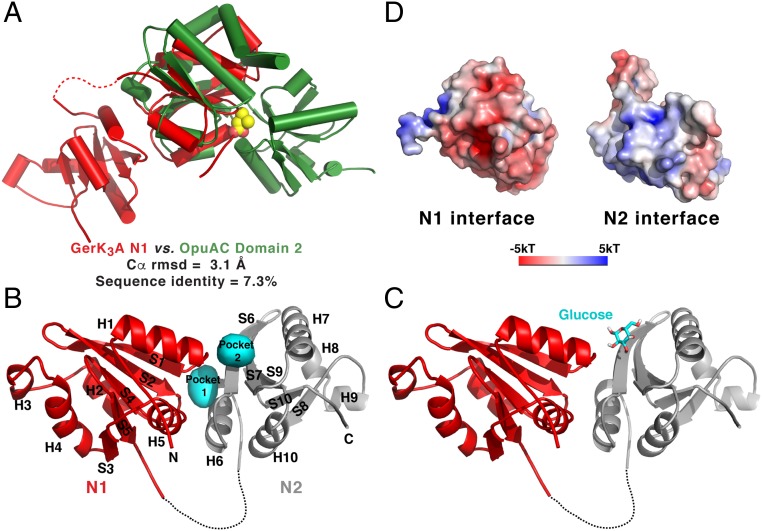
Based on structural similarity, the GerK3ANTD protein most closely resembles OpuAC, the extracellular substrate-binding protein of the B. subtilis OpuA system that belongs to the ABC transporter superfamily and mediates the uptake of the compatible solutes glycine betaine and proline betaine (27). The N1 domain of GerK3ANTD can be superimposed onto domain 2 of OpuAC bound to glycine betaine with an rmsd of 3.1 Å for 55 aligned Ca atoms, while these two proteins share only 7.3% sequence identity (Fig. 3A). Similar structural conservation also exists between the N2 domain of GerK3ANTD and domain 2 of OpuAC (SI Appendix, Fig. S3A). While the majority of their secondary-structure elements are well aligned in the superposition of the individual domains, the sequential orders and connectivities of these structural segments are completely different (compare Fig. 1C and SI Appendix, Fig. S3B). For instance, the N1 and N2 domains in GerK3ANTD fold in a continuous segment, whereas domains 1 and 2 in OpuAC, as in many other PeBPs, contain two interdomain cross-overs. Our analyses suggest that GerK3ANTD structurally resembles the overall fold of PeBPs, although this GR A protein has a different topological arrangement of its two domains than do PeBPs.
Fig. 3.
The structure of GerK3ANTD is similar to those of PeBP proteins. (A) Superimposition of the N1 domain of GerK3ANTD (red) and the domain 2 of OpuAC (green; PDB ID code 2B4L). The bound ligand for OpuAC, glycine betaine, is shown in space-filling representation. (B) Molecular surface representation of the two interfacial pockets (cyan) between the N1 (red) and N2 (gray) domains of GerK3ANTD defined by HOLLOW (66). (C) One of the top-ranked poses of the glucose molecule determined by AutoDock Vina is shown as licorice sticks in the presumed binding site in GerK3ANTD. (D) Molecular surface representation of the interface between the N1 and N2 domains of GerK3ANTD colored according to the local electrostatic potential (−5 kT to +5 kT), calculated with the program ABPS (65).
Molecular Docking of Small-Molecule Germinants onto the GerK3ANTD.
A key feature of the PeBP superfamily is that the ligand binding site in PeBP is situated between the two domains. Consequently, upon ligand binding, a domain reorientation around the hinge region produces a movement that brings the two domains together from an open conformation to enclose the ligand in a closed conformation (25). It was thus tempting to speculate that GerK3ANTD acts as the germinant-recruiting subunit in the spore germination process. Therefore, we sought to investigate whether small-molecule ligands can bind at the interface between the N1 and N2 domains of GerK3ANTD. Approximately 518 Å2 of solvent-accessible surface area of this interface is buried, suggesting that these two subdomains can potentially interact with each other. To test this notion, we expressed and purified the glutathione S-transferase (GST)-tagged N1 domain with and without the linker and performed affinity pulldown assays with the purified untagged N2 domain (SI Appendix, Fig. S4); all of the subdomain constructs of GerK3A were largely soluble after cell lysis and eluted as a single, symmetric peak on a gel filtration column. However, GST-GerK3AN1 or GST-GerK3AN1-Linker did not pull down untagged GerK3AN2 or GerK3ALinker-N2 (SI Appendix, Fig. S4, lane 8). Thus, the two subdomains of GerK3ANTD alone interact only weakly if at all with each other in vitro, consistent with the possibility of the two subdomains adopting of an open conformation in the absence of the ligand.
As noted above, the GerK3 GR is likely nonfunctional and has no known germinants that might trigger its function. However, GerK3A shares a high sequence identity with GerUA, a subunit of the B. megaterium GerU GR that can trigger spore germination in response to a number of compounds, including glucose, leucine, and proline (28). We hypothesized that GerK3A could also interact and bind to these compounds. Indeed, two well-defined pockets at the interface of the N1 and N2 domains (each ∼500 Å3) are located close by helix H6 and strand S6, respectively (Fig. 3B). As our attempts to crystallize the GerK3ANTD-ligand complex have failed, we used molecular docking to locate potential binding sites on GerK3ANTD that can accommodate glucose, leucine, and proline; these molecules have a volume of 194, 183, and 142 Å3, respectively. The small-molecule models were docked as a rigid body to the GerK3ANTD structure using AutoDock Vina (29), and the entire structural surface was searched for possible binding sites without bias. Notably, with each small molecule tested, for at least one of the top-three docked conformations (ranked by the order of the binding energy), this molecule was predicted to bind to GerK3ANTD in one of the interface pockets (Fig. 3C and SI Appendix, Fig. S5). As a potential control, we performed a similar search for a glycine betaine-binding site on the OpuAC structure alone, but none of the top-ranked glycine betaine docking sites was close to the known binding site revealed in the structure of the OpuAC–glycine betaine complex. This negative result strongly suggests that upon ligand binding OpuAC adopts a closed conformation that is inaccessible to exogenous ligands; unfortunately, the presumably ligand-free OpuAC is not available for testing. Interestingly, the electrostatics of the interfaces of the N1 and N2 domains of GerK3ANTD are highly complementary to each other, with the electronegative surface of the N1 domain and the electropositive surface of the N2 domain, suggesting that these two subdomains are capable of forming a closed conformation (Fig. 3D). Taken together, our data suggest that GerK3ANTD adopts an open conformation and its subdomain interface is capable of serving as the binding site for small-molecule germinants.
Effects of Subdomain Interface Mutations of the NTD of GR A Subunits on Spore Germination.
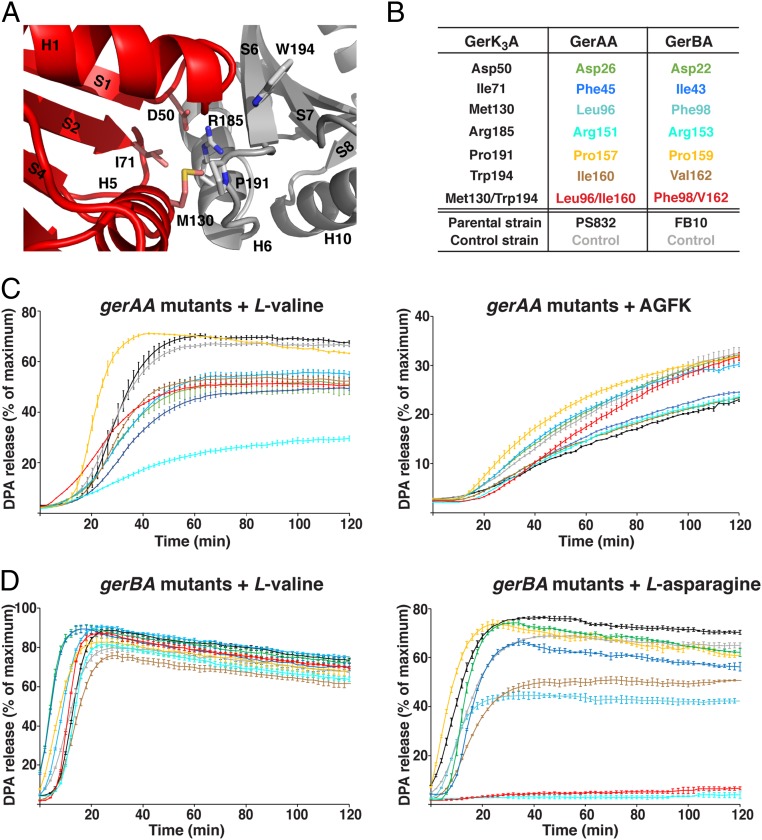
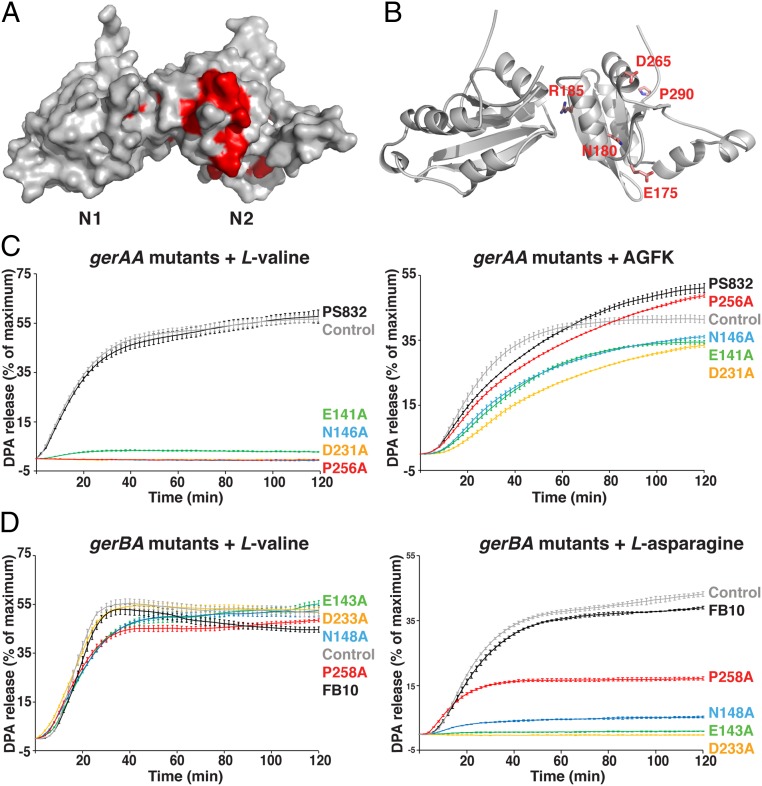
To ascertain the functional role of the NTD of the GR A subunits in spore germination, we decided to investigate the effects of alanine substitutions at the subdomain interface of GR A proteins on spore germination, with residue selection guided by the GerK3ANTD structure. We selected three residues each from the N1 (Asp50, Ile71, and Met130) and N2 (Arg185, Pro191, and Trp194) domains, all of which are located on the GerK3ANTD subdomain interface (Fig. 4A); Asp50 and Arg185 are also highly conserved among the A proteins of all GRs (discussed below). Because it is likely that GerK3 is a nonfunctional GR and its ligands are unknown, we created B. subtilis strains carrying alanine substitutions for the wild-type (WT) residues in the GerAA and GerBA proteins at predicted positions equivalent to those selected in GerK3ANTD (Fig. 4B and SI Appendix, Fig. S1) and assessed the germination activity of the mutant spores by monitoring DPA release upon addition of the germinants l-valine or AGFK (Fig. 4 C and D). Although the structures of those three A proteins are likely similar, it is worth noting that many of these interfacial residues are not conserved among GerK3A, GerAA, and GerBA (Fig. 4B and SI Appendix, Fig. S1). Since AGFK germination of spores with the parental PS832 background requires both the GerB and GerK GRs, the gerBA mutants were introduced into a B. subtilis PS832 variant strain (FB10) containing a single gerBB mutation in the gerB operon of PS832 that allows the GerB GR to function alone in response to l-asparagine (30). The WT B. subtilis gerAA and gerBA genes were also transformed to PS832 or FB10, respectively, and the resulting strains were taken as the respective control strains to assess any effects of the cloning and transformation (these strains are shown as control strains in Fig. 4 B–D). All control and mutant strains showed sporulation efficiency comparable to those of the parental strains.
Fig. 4.
Effects of B. subtilis gerAA and gerBA putative interface mutations on spore germination. (A) Close-up view of the N1–N2 interface in GerK3ANTD with selective interfacial residues of the N1 (pink) and N2 (gray) domains shown as licorice sticks. Numbers of secondary structure elements are labeled as in Fig. 1B. (B) The list of the putative N1–N2 interfacial WT residues in B. subtilis GerAA and GerBA proteins that were subjected to alanine substitution, and their equivalent residues in GerK3ANTD. Note that the WT B. subtilis gerAA and gerBA genes were cloned and transformed into PS832 or FB10, respectively, and the resulting strains were considered and labeled as control strains in this experiment to assess any effects of the cloning alone. (C) DPA release from spores with the predicted gerAA interface mutations in the PS832 background in the presence of 10 mM l-valine (Left) or 10 mM AGFK (Right). (D) DPA release from spores with the predicted gerBA interface mutations in the FB10 background in the presence of 10 mM l-valine (Left) or 10 mM l-asparagine (Right). The colors of the germination curves correspond to the mutations listed in B. For each strain, the percentage of DPA release was normalized against the RFU readings obtained from the same spores boiled in water. Data represent means ± SD for at least three independent measurements.
As expected, the spores of most gerAA mutant strains exhibited varying levels of defects in l-valine triggered germination while having similar levels of germination in response to AGFK (Fig. 4C and SI Appendix, Table S3). Five single mutants (D26A, F45A, L96A, R151A, and I160A) and one double mutant (L96A/I160A) showed an extended lag phase following l-valine addition, released only about 50% of the total DPA, and had two- to fivefold lower initial rates of germination than the parental and control spores. Interestingly, while the spores of most gerBA mutant strains exhibited levels of germination and initial germination rates comparable to the control spores, the R153A and F98A/V162A mutants showed essentially complete loss of germination with l-asparagine (Fig. 4D and SI Appendix, Table S3). Notably, the levels of the control and various GerAA proteins in spore lysates were essentially identical, indicating that these mutations do not affect protein stability and assembly; the levels of the GerAC proteins, the C protein of the B. subtilis GerA GR, were also very similar to that of the WT strain in the gerAA mutant strains (SI Appendix, Fig. S6A). We do not have antibody specific for GerBA, but the levels of the GerBC proteins in the gerBA mutant strains were comparable to that in control spores (SI Appendix, Fig. S6B), suggesting that the corresponding mutant GerBA proteins are likely stable enough to assemble the whole receptor; several studies have shown that loss of the GerAA or GerAB subunits can result in the loss of the downstream C protein (11, 12). However, we cannot rule out the possibility that the mutations we chose to make could have a direct impact on the folding of the encoded proteins and lead to a loss of function (discussed below). Together, these germination data demonstrate that the subdomain interface in the NTDs of the GR A proteins plays an important role in mediating germinant signals to induce spore germination. If this subdomain interface is indeed involved in germinant binding, the observed distinct effects of the equivalent mutations in GerAA and GerBA on germination suggest that the interface region may be important in influencing germinant binding specificity.
Interaction of Inosine with the B. cereus GerIANTD.
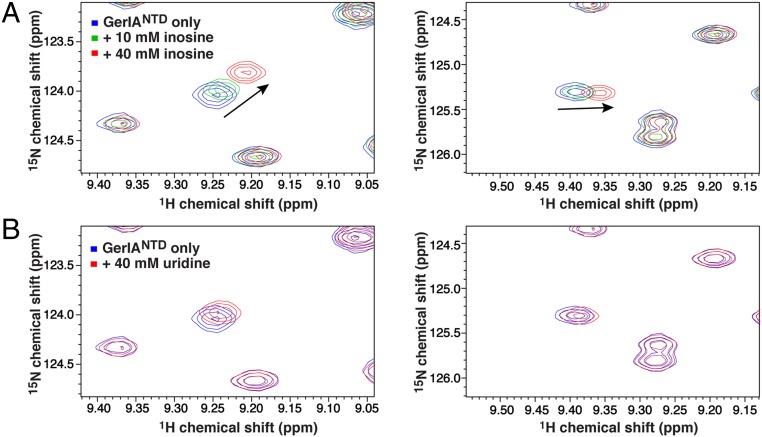
We next sought to identify a direct physical interaction between the NTD of a GR A protein and a specific germinant by NMR spectroscopy that allows the study of weak and transient interactions (31). GerIANTD was chosen as a candidate because GerI has been shown to be essential for normal inosine-mediated germination of B. cereus spores (23, 32) and because GerIANTD is soluble, stable, and well-behaved during purification. A 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of GerIANTD displays excellent peak dispersion indicative of a well-folded structural unit that is suitable for probing protein–ligand interactions by NMR despite the relatively large size of the GerIANTD protein (28.8 kDa). A total of 217 independent peaks (∼91% of the expected peaks) were identified in the HSQC spectrum (SI Appendix, Fig. S7 A and B). We next titrated 15N-labeled GerIANTD with unlabeled inosine and recorded the HSQC spectra under the exact same sample and experimental conditions. As expected, overlay of the 1H-15N HSQC spectra displays gradual shifts in amide resonance positions for a number of peaks upon titration of increasing amounts of inosine (10 mM vs. 40 mM), suggestive of a specific interaction between GerIANTD and inosine and a fast exchange between free and bound states on the NMR timescale (Fig. 5A and SI Appendix, Fig. S7A). (Unfortunately, the limited solubility of inosine precluded the measurement of the exact dissociation constant of inosine to GerIANTD.) In contrast, upon addition of 40 mM unlabeled uridine, a pyrimidine nucleoside that is not involved in GerI-induced B. cereus germination (23), very few amide peaks were shifted in the HSQC spectrum of the protein, and the magnitudes of changes in the shifted peaks were small compared with those affected by inosine, indicating that uridine does not interact with GerIANTD (Fig. 5B and SI Appendix, Fig. S7B). Indeed, the average per-peak chemical shift perturbations, ∆δ, calculated as the distance between the free and 40 mM inosine-bound GerIANTD peaks, is more than threefold larger than the average shifts between the free and 40 mM uridine-titrated peaks (SI Appendix, Fig. S7C). Collectively, our NMR data strongly suggest that GerIANTD binds specifically, albeit weakly, to the germinant inosine.
Fig. 5.
Chemical shift perturbation analyses of GerIANTD in the presence of inosine or uridine. (A) Close-up views of the two selected regions of the superimposed 1H-15N HSQC spectra of the 15N-labeled GerIANTD before (blue) and after addition of 10 mM (green) or 40 mM (red) unlabeled inosine, with the arrows marking peaks exhibiting large chemical shift perturbations. (B) The same regions of the 1H-15N HSQC spectra of the 15N-labeled GerIANTD before (blue) and after addition of 40 mM unlabeled uridine (red).
As we were unable to obtain the crystal structure of GerIANTD, we employed homology modeling and molecular docking to explore the potential binding pocket of inosine in the GerIANTD protein. Using the program MODELER (33) with the final refined GerK3ANTD structure as the template, we modeled a 3D structure of GerIANTD, with 92% query coverage based on 32.5% sequence identity between the target and the template. The model structure with the lowest DOPE score (−24,103.78320) was selected and validated using a Ramachandran plot generated by MolProbity (34), wherein 96.2% of residues were in the favored region, with additional 2.4% of the residues in the allowed region. As would be expected, superimposition of the model with the template GerK3ANTD shows marginal structural deviations with an rmsd of 0.93 Å (SI Appendix, Fig. S8A). Interestingly, the subsequent molecular docking of inosine shows that most of the top-ranked docked conformations for inosine are projected to bind to GerIANTD at the interface between the two subdomains of the GerIANTD model, although the exact locations and orientations of these inosine molecules differ (SI Appendix, Fig. S8 B–D). Combined, these results support our structural and mutagenesis studies that the GR A proteins play essential roles in germinant recognition and binding.
To probe the importance of the subdomain interface of the NTDs of GR A proteins in recruiting germinants, we generated seven GerIANTD mutants that carry alanine substitutions at various interfacial positions predicted based on the homology model of GerIANTD and the sequence alignment of GerIANTD with GerK3ANTD (SI Appendix, Figs. S1 and S9A). We have been able to express and purify two 15N-labeled mutant proteins (R388A and L394A) but not the other five mutants (D262A, Y281A, L332A, E397A, and L332A/E397A) due to low protein yield and aggregation. The 1H-15N HSQC spectra of the purified R388A and L394A proteins display similar peak dispersion as that of the WT protein but chemical shift perturbations for a number of amide resonance peaks, suggesting that the mutant proteins retain an overall fold similar to that of the WT protein (the overlaid 1H-15N HSQC spectra of the WT and R388A proteins are shown in SI Appendix, Fig. S9B). Nevertheless, addition of 40 mM inosine to the mutant proteins still caused a number of peak shifts similar to those observed in the titration of inosine to the WT protein (SI Appendix, Fig. S9 C and D). It is possible that these two residues chosen for mutation in GerIANTD, Arg388 and Leu394, are not directly involved in inosine binding in GerIA. Notably, the molecular docking results described above indicate that inosine can potentially bind to GerIA at different regions of the N1–N2 interface (SI Appendix, Fig. S8 B–D). It is also worth noting that the mutation of a single residue might not be adequate to completely disrupt germinant binding, which is in agreement with the various levels of germination defects observed in the gerAA and gerBA mutant strains (Fig. 4 C and D). Alternatively, these mutations might have effects on transduction of the signal induced by germinant binding to other GR subunits. Further studies to determine the structures of the NTDs of GR A proteins bound to their specific germinants may be required to fully elucidate the molecular basis of germinant recruitment by the GRs.
Impact of Mutations in Highly Conserved Amino Acids of GR A Proteins on Germination.
Our BLAST search has identified 253 GR A proteins in spore-forming Bacillales and Clostridiales genomes (discussed above). With the structure of GerK3ANTD in hand, we mapped the most conserved 27 residues (>80% identity among all orthologs) in the NTD of the A proteins onto the surface of the GerK3ANTD structure (Fig. 6A and SI Appendix, Fig. S1). Among them, the majority of the conserved residues are in the N2 domain, whereas only two residues are located in the N1 domain (Asp50 and Gly132) and three residues (Arg155, Glu163, and Gly168) are in the disordered linker region, suggesting that the N2 domain may play a more important role in germination even though N1 and N2 share a similar conformation. The most conserved region in the N2 domain is located on one side of the central β-sheet covering the space between helices H6 and H10 and strands S8 and S9 (the large red patch in Fig. 6A); 15 highly conserved residues are located in this region (SI Appendix, Fig. S1). To determine to what extent these conserved amino acids are critical for GR A protein function, we selected four of the most conserved residues (Glu175, Asn180, Asp265, and Pro290) in the N2 domain of GerK3ANTD for our functional analysis; these four residues share 97 to 99% identity among all orthologs (Fig. 6B). Using the same strategy described above, we created B. subtilis strains carrying single alanine substitutions of the equivalent amino acids in either GerAA (E141A, N146A, D231A, and P256A) or GerBA (E143A, N148A, D233A, and P258A). For GerAA mutant spores, l-valine–mediated germination was essentially abolished while AGFK-triggered germination remained similar to that of the control spores (Fig. 6C and SI Appendix, Table S4). Likewise, all of the GerBA mutant spores exhibited essentially same germination with l-valine as the control spores but behaved very differently when responding to l-asparagine alone: the N148A and P258A mutations led to 20% and 40% reduction in the initial germination rate and with only 5% and 15% of total DPA released, respectively, while the E143A and D233A mutations completely eliminated spore germination with l-asparagine (Fig. 6D and SI Appendix, Table S4). Interestingly, in contrast to Pro256 in GerAA and Pro258 in GerBA, alanine substitutions of the highly conserved Arg185 (>97% identity among all GR A subunit orthologs and equivalent to Arg151 in GerAA and Arg153 in GerBA), located in the interface of the N1 and N2 domains of GerK3ANTD, had a greater effect on l-asparagine–mediated germination than on l-valine germination (discussed above; Fig. 4 C and D). Importantly, the levels of the control and variant GerAA/GerAC/GerBC proteins in the spores were comparable, indicating that these mutations do not affect protein stability and GR assembly (SI Appendix, Fig. S10 A and B). These results suggest that these proline and arginine residues may be directly involved in the determination of germinant specificity. Collectively, these data clearly establish that the most highly conserved amino acids in the N2 domain are crucial for GR A protein function.
Fig. 6.
Effects of mutations of highly conserved residues in B. subtilis gerAA and gerBA on spore germination. (A) Molecular surface of GerK3ANTD with the regions colored in red representing residues at least 80% identical among all 253 GerK3ANTD orthologs (see also SI Appendix, Fig. S1). (B) The five highly conserved residues in the N2 domain selected for mutagenesis are shown in licorice stick representation. (C) DPA release from spores with the gerAA conserved mutations in the PS832 background in the presence of 10 mM l-valine (Left) or 10 mM AGFK (Right). (D) DPA release from spores with the gerBA conserved mutations in the FB10 background in the presence of 10 mM l-valine (Left) or 10 mM l-asparagine (Right). Note that the spores of PS832 and FB10 transformed with the WT gerAA or gerBA genes, respectively, are considered and labeled as the control spores in these experiments to assess any effects of the cloning alone. For each strain in C and D, the percentage of DPA release was normalized against the RFU readings obtained from the same spores boiled in water. Data represent means ± SD for at least three independent measurements.
Implications for GR–Germinant Recognition and GR-Mediated Germination.
The molecular basis for germinant recognition by GRs to trigger the signal transduction cascade in spore germination has been the subject of intensive research for the past four decades. There is considerable evidence indicating that small-molecule germinants can pass through the outermost spore layer and interact directly with specific GRs in the spore IM to activate downstream events, yet the exact mechanism of GR’s action is still unknown. The results presented in this study provide structural and biochemical evidence that the N-terminal soluble domain of GR A proteins shares structural similarity with the PeBP superfamily and is directly involved in germinant recruitment through an intradomain interface.
Our crystallographic analysis reveals that GerK3ANTD shows clear structural homology to PeBP superfamily proteins; both the sequence alignments and secondary-structure prediction suggest that the two-domain PeBP fold of GerK3ANTD is likely conserved among all GR A proteins. Molecular docking and subsequent structure-guided mutagenesis coupled with spore germination analyses provide support for a model that the interface between the two subdomains in the NTD of the GR A proteins can serve as the germinant binding site and play a critical role in germination. Consistent with this model, the observed distinct effects of mutations at the equivalent residues in the subdomain interface of various GR A proteins on germination activities may reflect effects on GRs’ germinant specificity. Furthermore, our 15N-HSQC spectra show that titration of inosine, but not uridine, to GerIANTD can induce specific chemical shift perturbations in the millimolar range, indicating a direct interaction between inosine and GerIANTD. Nevertheless, GerIANTD exhibits low affinity for inosine, perhaps for two reasons. First, the germinant binding site of GerIANTD might be occupied with inosine and/or amino acid cogerminants (23) that were copurified with the protein expressed in Escherichia coli cells. Indeed, the recombinant PnrA protein, the bacterial purine nucleoside receptor of an ABC transporter of Treponema pallidum, was found to bind to inosine when expressed in and purified from E. coli grown in Luria-Bertani broth with glucose as the primary carbon source (35). In fact, in the GerK3ANTD structure, there was a well-defined peanut-shaped peak of electron density (i.e., 2 Fo − Fc and Fo − Fc) in the interface between the N1 and N2 domains, which could not be adequately accounted for by one or two water molecules (SI Appendix, Fig. S11). This electron density peak can be modeled as a molecule of glucose, proline, or leucine, the cogerminants of B. megaterium GerUA, but we have not been able to identify an endogenous ligand bound to bacterially expressed GerK3ANTD by mass spectrometry, likely due to its noncovalent linkage with the protein. Second, the A, B, and C proteins of GRs are all essential for GR function (2, 36). Indeed, mutations altering B proteins have been found to affect both the affinity and specificity of B. subtilis and B. megaterium GRs for various germinants (17, 30, 37). It is thus possible that the B and C subunits of GRs, which are missing in our experiments, are also required for high-affinity germinant binding. In addition, the moderate affinity of small-molecule germinants for GRs may be beneficial for spores to minimize the likelihood of rushing back to life in an environment that is not nutritionally amenable to vegetative growth.
The most puzzling questions about GR’s function have always been about the precise function of individual GRs, and how the A, B, and C proteins of GRs work together to achieve this function. We have now shown that the structure of an NTD of the GR A proteins closely resembles that of a PeBP, many of which are a part of the large prokaryotic ABC transporter family mediating chemotaxis and solute uptake. It is thus tempting to speculate that the organization and function of the GRs may at least in part mirror those of the ABC transporters. All bacterial ABC transporters couple ATP hydrolysis with substrate translocation across cellular or organelle membranes (38–40). The transporters that mediate uptake generally consist of three subunits or individual proteins: the solute-binding protein that is either tethered to the cell surface through a lipid anchor or fused to the translocator itself, the membrane-spanning substrate translocator, and the cytoplasmic membrane-associated ATPase. Upon sensing the binding of the substrate to the translocator delivered by the solute-binding protein, the action of ATPase initiates ATP hydrolysis to drive conformational switches in the translocator, thereby providing alternating access from the outside and inside of the cell for unidirectional transport of the substance across the membrane. In GRs, the A protein clearly acts like the solute binding protein and is anchored to the membrane through its own TM CTD. As a putative single-component transporter, the B protein may participate in germinant binding and/or translocation (18, 30, 37). However, even though the C protein is a lipoprotein, its tertiary fold structure is very different from that of a classic ATPase (15). Therefore, GRs could not possibly act as a new type of ABC transporter. Consistent with this notion, neither transport nor metabolism of germinants, nor even consumption of energy, is required for at least the early stages of spore germination (41). However, it is still possible that GRs are evolutionarily linked to the ABC transporters.
Given the results presented in current study, a tentative model for how GR–germinant interactions might trigger the downstream events in spore germination would begin with germinants being captured and bound to the N1–N2 subdomain interface of GR A proteins that are most likely located on the outside of spore’s IM (14). This binding would drive subdomain reorientation in the GR A proteins from a germinant-free open form to a germinant-bound closed form, resulting in subsequent conformational changes in IM-bound GR B and C proteins. Such structural changes in GR molecules could then alter the local lipid arrangement and permeability of the IM whose lipids have been shown to be largely immobile in dormant spores and become free and diffusible in the early stage of spore germination (42). The resulting mobile spore IM would allow the activation or reorganization of other IM-bound proteins, most importantly the SpoVA channel proteins through which DPA and associated Ca2+ ions from the spore core are released during spore germination (3, 43). DPA release then triggers degradation of the spore’s peptidoglycan cortex by cortex-lytic enzymes to complete core rehydration and resumption of metabolism (3). While the proposed signal transduction model is quite speculative, it does provide a basis for further structural and functional studies on GRs in vitro and in spores to attain a comprehensive understanding of how early signal transduction in spore germination takes place.
Finally, it is possible that a more definitive understanding of GR signaling in spore germination might allow development of universal agents or compounds that can mimic the effects of germinants on all GRs. While again this is a very speculative possibility, if such a compound could be generated it would allow triggering of germination of all spores on surfaces. Since germinated spores are much easier to kill than dormant spores, use of this novel compound would make decontamination of spores of Clostridium difficile and Bacillus anthracis on surfaces much simpler and more effective.
Materials and Methods
Protein Expression and Purification.
The gerK3A (Biocyc ID BC4731) and gerIA (Biocyc ID BMQ_2245) genes were amplified by PCR from chromosomal DNA of B. megaterium strain QM B1551 and B. cereus strain ATCC 14579, respectively, and cloned into a modified pET15b vector containing a removable tobacco etch virus (TEV) protease recognition site. The interface mutations of GerIA were introduced into gerIA gene using an overlap PCR method (44). GerK3ANTD (residues 26–302), GerK3AN2 (residues 171–302), GerK3ALinker-N2 (residues 153–302), and GerIANTD (residues 238–484; WT and mutants) were expressed in E. coli and purified by Ni2+-nitriloacetic acid (GE Healthcare) affinity chromatography. After removal of the His6 tag by overnight cleavage with TEV protease, the samples were further purified by anion exchange (Source Q; GE Healthcare) and gel filtration (SD200; GE Healthcare) chromatography. For GST pull-down assays, the GerK3AN1 (residues 26–152) and GerK3AN1-Linker (residues 26–170) constructs were cloned into a modified pGEX vector and purified by glutathione affinity chromatography (G4B; GE Healthcare) followed by anion exchange and gel filtration chromatography; the GST tag was used to ensure that the sizes of the GST-GerK3AN1 and GerK3AN2 proteins are distinguishable on SDS/PAGE. For crystallization, GerK3ANTD was concentrated to 15 mg/mL by ultrafiltration in 15 mM Tris⋅HCl (pH 7.6) and 150 mM NaCl supplemented with 5 mM DTT. SeMet-substituted GerK3ANTD was produced following established procedures (45) and purified as described above. For NMR experiments, the WT and mutant GerIANTD proteins were expressed in M9 minimal media supplemented with 15NH4Cl as the sole nitrogen source and purified as described above. The proteins were concentrated to 0.48 mM in 20 mM Na-K phosphate (pH 7.2), 150 mM NaCl, 2 mM β-mercaptoethanol, 0.2 mM EDTA, 8% D2O, and 0.02% NaN3.
BLAST Search and Sequence Conservation Analysis.
The B. subtilis GerAA amino acid sequence was used as query sequence for the initial homology sequence search of spore-forming members of the Bacillales and Clostridiales orders on the NCBI genomic BLAST server (https://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). The hits were selected if the E-value was < e−45 and the query coverage >90%. The best hit for each species was then used as the query sequence to further search its own genome to find all possible homologs in various genomes. The number of homologs found in each species is summarized in SI Appendix, Table S1. A ClustalW alignment of GerAA homologs was performed using DNASTAR Lasergene suite 8 (DNASTAR Inc.).
Crystallization and Structure Determination.
The native and SeMet-substituted GerK3ANTD proteins were crystallized from a solution consisting of 10% PEG 6000, 0.15 M lithium sulfate, 5% glycerol, and 0.1 M Tris⋅HCl (pH 8.5) at 4 °C by using the hanging-drop vapor diffusion method. Crystals were flash-frozen in crystallization solution supplemented with 30% glycerol. Diffraction data of the native and SeMet GerK3ANTD crystals were collected at the National Synchrotron Light Source (NSLS) beamline X29A and NSLS-II beamline 17-ID-1. Data were processed using the HKL2000 suite (46) and the XDS package (47). The crystals contain two GerK3ANTD molecules in the asymmetric unit. The GerK3ANTD structure was determined by SAD by using the data collected at the selenium peak wavelength. Approximately 13 SeMet sites were identified by SOLVE (48) as implemented in PHENIX (49), and initial phases calculated from these sites were improved by density modification using RESOLVE/PHENIX. The resulting electron density map was readily interpretable and used to build two-thirds of the molecule with the program Coot (50). Iterative cycles of refinement in REFMAC (51) and autoBUSTER (52) followed by manual rebuilding in Coot were carried out until no further improvement of the Rfree factor was observed. X-ray data collection and phasing and refinement statistics are summarized in SI Appendix, Table S2. Ramachandran statistics were calculated using Molprobity (34). Molecular graphics were rendered using PyMOL (Delano Scientific LLC). Interaction surface areas were calculated by using PISA (53).
GST Affinity Pull-Down Assays.
Indicated proteins (28 μM each) were incubated at room temperature for 10 min in 25 μL of binding buffer consisting of 50 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, and 2 mM DTT before addition of 30 μL G4B resin. After 10 min of incubation, the resin was spun down, and the ∼25 μL supernatant, marked as the unbound (U) fraction in the figures, was removed. The resin was then washed three times with 0.6 mL of binding buffer and the G4B-bound proteins, marked as the bound (B) fractions in the figures, were eluted with 35 μL buffer consisting of 50 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, and 50 mM glutathione. One-third of each of the supernatant and eluted fractions was analyzed by SDS/PAGE and Coomassie staining.
Molecular Docking and Homology Modeling.
Three-dimensional coordinates of glucose, leucine, proline, and inosine were obtained from the Coot monomer library (50). Docking of glucose, leucine, and proline into the GerK3ANTD structure was performed using the iterated Local Search Global Optimization algorithm provided by AutoDock Vina (29). The PDBQT format files (required as input) of both small-molecule ligands and GerK3ANTD were generated using the AutoDock Tools package provided by AutoDock 4 (54). Each ligand was docked as a rigid body and the entire surface of the GerK3ANTD structure was searched for possible binding sites without bias. A cubic box was built around the protein with 86, 92, and 76 points as x, y, and z sizes. A spacing of 1.0 Å between the grid points was used, making the center of the protein to be the center of the cube, that is, x, y, and z centers at 36.006, 58.995, and 54.13, respectively. All other parameters were set as default as defined by AutoDock Vina.
The homology model of GerIANTD was generated using MODELER v9.20 (33) made available through NMRbox (55), and the final structure of GerK3ANTD was used as the template. The tertiary structure models were calculated by satisfaction of spatial restraints using the automodel function of MODELER with default parameters for target optimization, refinement and energy minimization protocols. The models were ranked based on the DOPE and GA341 scores and subject to validation by the Ramachandran plot criteria. Docking of inosine to the best homology model of GerIANTD was performed as described above.
Generation of B. subtilis Strains with gerAA and gerBA Mutant Genes.
Two isogenic B. subtilis strains were used as the parental strains in this aspect of the work: (i) PS832, a laboratory derivative of strain 168, and (ii) FB10, PS832 with the F269I substitution in the gerBB gene that allows the spores of this strain to germinate with l-asparagine alone (30). Mutations were introduced into gerAA (1,449 bp) and gerBA (1,452 bp) genes using an overlap PCR method (44). The PCR products were then cloned into a modified pBluescript II KS(–) vector containing a chloramphenicol-resistance (Cmr) cassette separated by two inserts from the B. subtilis gerAA or gerBA region. The first insert consisted of 500 bp within the upstream region of gerAA or gerBA (nucleotides −600 to −101 relative to the +1 gerAA or gerBA translation start site). Downstream from this insert is the Cmr cassette, followed by B. subtilis gerAA (nucleotides −100 to +1,449) or gerBA (nucleotides −100 to +1,452) variants. Because all of the mutation sites occur in the NTD region of the gerAA or gerBA gene, the C-terminal region of the gene would provide homology for a double cross-over with B. subtilis chromosomal DNA. The mutagenized plasmids were used to transform PS832 or FB10 competent cells, with selection for Cmr. Transformants in which the mutagenized gerAA or gerBA gene had integrated into the chromosome with replacement of the WT gene were identified by PCR, and the PCR-amplified regions were sequenced to confirm the presence of the mutation(s). The WT B. subtilis gerAA and gerBA genes were also cloned and transformed into PS832 and FB10 the same way and the resulting strains were used as the control strains in this set of experiments and labeled as such in Figs. 4 B–D and 6 C and D.
Spore Preparation, Purification, and Germination.
Luria broth medium was used for vegetative growth of B. subtilis strains at 37 °C (56). B. subtilis spores were prepared at 37 °C on 2× Schaeffer’s-glucose sporulation medium plates without antibiotics, and spores were harvested and purified as described previously (56). The purified spores were >98% free from growing or sporulating cells, germinated spores, and cell debris as determined by phase-contrast microscopy and stored in the dark at 4 °C.
Before germination, spores at an optical density at OD600 of 2.0 were heat-activated at 75 °C for 30 min and cooled on ice. Germination reactions were initiated by addition of heat-activated spores (final OD600 of 0.5) to a mixture containing 25 mM potassium Hepes (pH 7.4), 50 μM terbium chloride, and 10 mM germinants (l-valine, l-asparagine, or the AGFK mixture with equal concentrations of all four components) or as indicated at 37 °C for 2 h in 96-well plates in a total volume of 200 μL per well. The progress of germination was monitored by real-time measurement of DPA release based on fluorescence emission of the Tb3+–DPA complex at 490 nm (excited at 276 nm) using a Gemini EM microplate fluorescence reader (Molecular Devices) as described previously (9, 43, 57–59). Each reaction mixture was tested in quadruplicate, and the reading at zero time was used as the background. For all spores examined, the total DPA content of spores in the reaction mixture was determined from the maximum relative fluorescence units (RFU) of the same amount of spore suspensions boiled 30 min in water, and the percentage of DPA released at each time point was calculated against the maximum RFU measured at the same time. The percentages of spores that had germinated by the end of reaction incubations were also routinely checked by phase-contrast microscopy to make sure these analyses largely agreed with the corresponding RFU values. All curves generated by plotting time versus percentage of RFU were fitted using nonlinear regression to the exponential equation of the OriginPro 8 software program (OriginLab Corporation) to determine initial rates of DPA release (percentage of DPA release per min) (SI Appendix, Tables S3 and S4).
Western Blot Analyses of the Spore Total Lysates.
Total lysates of spores of various B. subtilis strains were prepared as described previously (58). Equal amounts of the total lysates were run on SDS/PAGE, and the levels of the GR subunits were detected using specific rabbit polyclonal antisera (58).
Chemical Shift Mapping by NMR.
Two-dimensional 1H-15N HSQC spectra of the 15N-labeled GerIANTD were collected at 25 °C on an Agilent VNMRS 800-MHz spectrometer equipped with an HCN cold probe. For titration experiments, the 15N-labeled GerIANTD WT and mutant proteins (final concentration of 0.25 mM) was mixed with excess inosine (final concentrations of 10 and 40 mM) or uridine (final concentration of 40 mM) prepared in the protein buffer; the pH of each mixture was adjusted to match that of the labeled apo protein. The chemical shifts of the complexes were measured by 1H-15N HSQC using the same instrument setup for the apo protein. Spectra were collected with 512 (t2) × 256 (t1) complex points, spectra widths of 7,999.36 Hz (t2) and 2,146 Hz (t1), with 16 or 64 transients. All spectra were processed with NMRPipe (60), and peak heights and locations were analyzed in CcpNmr Analysis (61) made available through NMRbox (55). The sum of chemical shift perturbations for each nonoverlapped backbone amide group peak was calculated by using the following equation (62):
where and are the 1H and 15N chemical shift differences between the bound and free states, respectively, and α denotes the chemical shift scaling factor for 15N. The scaling factor α, which is set to 0.154 (63) in this paper, was determined from the ratio of the average variances of the 1H and 15N chemical shifts observed for the 20 common amino acid residues in proteins as deposited with the BioMagResBank (64) (http://www.bmrb.wisc.edu/).
Supplementary Material
Acknowledgments
We thank W. Shi at the X29A beamline of NSLS and J. Jakoncic and A. Soares at the 17-ID-1 beamline of NSLS-II for help with data collection and V. Gorbatyuk, I. Bezsonova, and A. Rizzo for discussions on NMR data processing. This work was supported by a Department of Defense Multi-University Research Initiative award to P.S. and B.H. through the US Amy Research Laboratory and the US Army Research Office under contract number W911NF-09-1-0286 and by NIH Grant GM099948 to B.H. This study made use of NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource, which is supported by NIH Grant P41GM111135.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.wwpdb.org/ (PDB ID code 6O59).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903675116/-/DCSupplemental.
References
- 1.Setlow P., Johnson E. A., “Spores and their significance” in Food Microbiology: Fundamentals and Frontiers, Doyle M. P., Buchanan R. L., Eds. (ASM Press, Washington, D.C., ed. 4, 2012), pp. 45–79. [Google Scholar]
- 2.Moir A., Cooper G., Spore germination. Microbiol. Spectr. 3, 10.1128/microbiolspec (2015). [DOI] [PubMed] [Google Scholar]
- 3.Setlow P., Wang S., Li Y. Q., Germination of spores of the orders Bacillales and Clostridiales. Annu. Rev. Microbiol. 71, 459–477 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Nerandzic M. M., Donskey C. J., Sensitizing Clostridium difficile spores with germinants on skin and environmental surfaces represents a new strategy for reducing spores via ambient mechanisms. Pathog. Immun. 2, 404–421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohler L. J., Quirk A. V., Welkos S. L., Cote C. K., Incorporating germination-induction into decontamination strategies for bacterial spores. J. Appl. Microbiol. 124, 2–14 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Paidhungat M., Setlow P., Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182, 2513–2519 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paredes-Sabja D., Setlow P., Sarker M. R., Germination of spores of Bacillales and Clostridiales species: Mechanisms and proteins involved. Trends Microbiol. 19, 85–94 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Ramirez-Peralta A., et al. , Identification of new proteins that modulate the germination of spores of bacillus species. J. Bacteriol. 195, 3009–3021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., et al. , Structure-based functional studies of the effects of amino acid substitutions in GerBC, the C subunit of the Bacillus subtilis GerB spore germinant receptor. J. Bacteriol. 193, 4143–4152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths K. K., Zhang J., Cowan A. E., Yu J., Setlow P., Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81, 1061–1077 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper G. R., Moir A., Amino acid residues in the GerAB protein important in the function and assembly of the alanine spore germination receptor of Bacillus subtilis 168. J. Bacteriol. 193, 2261–2267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mongkolthanaruk W., Cooper G. R., Mawer J. S., Allan R. N., Moir A., Effect of amino acid substitutions in the GerAA protein on the function of the alanine-responsive germinant receptor of Bacillus subtilis spores. J. Bacteriol. 193, 2268–2275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson M. J., Carlson P. E., Janes B. K., Hanna P. C., Membrane topology of the Bacillus anthracis GerH germinant receptor proteins. J. Bacteriol. 194, 1369–1377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korza G., Setlow P., Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J. Bacteriol. 195, 1484–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Setlow B., Setlow P., Hao B., Crystal structure of the GerBC component of a Bacillus subtilis spore germinant receptor. J. Mol. Biol. 402, 8–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie G., Lazarevska M., Lowe C. R., Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J. Bacteriol. 190, 2014–2022 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christie G., Lowe C. R., Amino acid substitutions in transmembrane domains 9 and 10 of GerVB that affect the germination properties of Bacillus megaterium spores. J. Bacteriol. 190, 8009–8017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jack D. L., Paulsen I. T., Saier M. H., The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146, 1797–1814 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Berntsson R. P., Smits S. H., Schmitt L., Slotboom D. J., Poolman B., A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Tam R., Saier M. H. Jr, Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57, 320–346 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheepers G. H., Lycklama A Nijeholt J. A., Poolman B., An updated structural classification of substrate-binding proteins. FEBS Lett. 590, 4393–4401 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Gupta S., et al. , Investigating the functional hierarchy of Bacillus megaterium PV361 spore germinant receptors. J. Bacteriol. 195, 3045–3053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clements M. O., Moir A., Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J. Bacteriol. 180, 6729–6735 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madej T., et al. , MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 42, D297–D303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quiocho F. A., Ledvina P. S., Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol. Microbiol. 20, 17–25 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Fukami-Kobayashi K., Tateno Y., Nishikawa K., Domain dislocation: A change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Horn C., et al. , Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J. Mol. Biol. 357, 592–606 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Christie G., Lowe C. R., Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J. Bacteriol. 189, 4375–4383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trott O., Olson A. J., AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paidhungat M., Setlow P., Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant D-alanine. J. Bacteriol. 181, 3341–3350 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connell M. R., Gamsjaeger R., Mackay J. P., The structural analysis of protein-protein interactions by NMR spectroscopy. Proteomics 9, 5224–5232 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Hornstra L. M., de Vries Y. P., Wells-Bennik M. H., de Vos W. M., Abee T., Characterization of germination receptors of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 72, 44–53 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eswar N., et al. , Comparative protein structure modeling using modeller. Curr. Protoc. Bioinf. Chapter 5, Unit-5 6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deka R. K., et al. , The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum. J. Biol. Chem. 281, 8072–8081 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Setlow P. Germination of spores of Bacillus species: What we know and do not know. J. Bacteriol. 196, 1297–1305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christie G., Götzke H., Lowe C. R., Identification of a receptor subunit and putative ligand-binding residues involved in the Bacillus megaterium QM B1551 spore germination response to glucose. J. Bacteriol. 192, 4317–4326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidson A. L., Chen J., ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73, 241–268 (2004). [DOI] [PubMed] [Google Scholar]
- 39.van der Heide T., Poolman B., ABC transporters: One, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 3, 938–943 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkens S., Structure and mechanism of ABC transporters. F1000Prime Rep. 7, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott I. R., Ellar D. J., Metabolism and the triggering of germination of Bacillus megaterium. Use of L-[3H]alanine and tritiated water to detect metabolism. Biochem. J. 174, 635–640 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowan A. E., et al. , Lipids in the inner membrane of dormant spores of Bacillus species are largely immobile. Proc. Natl. Acad. Sci. U.S.A. 101, 7733–7738 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., et al. , Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 194, 1875–1884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heckman K. L., Pease L. R., Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J., Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993). [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terwilliger T. C., Berendzen J., Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams P. D., et al. , PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Smart O. S., et al. , Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68, 368–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Yanamala N., Tirupula K. C., Klein-Seetharaman J., Preferential binding of allosteric modulators to active and inactive conformational states of metabotropic glutamate receptors. BMC Bioinf. 9 (suppl. 1), S16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maciejewski M. W., et al. , NMRbox: A Resource for biomolecular NMR computation. Biophys. J. 112, 1529–1534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paidhungat M., Setlow B., Driks A., Setlow P., Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182, 5505–5512 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y., et al. , Activity and regulation of various forms of CwlJ, SleB, and YpeB proteins in degrading cortex peptidoglycan of spores of Bacillus species in vitro and during spore germination. J. Bacteriol. 195, 2530–2540 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y., et al. , Structural and functional analysis of the GerD spore germination protein of Bacillus species. J. Mol. Biol. 426, 1995–2008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., Jin K., Setlow B., Setlow P., Hao B., Crystal structure of the catalytic domain of the Bacillus cereus SleB protein, important in cortex peptidoglycan degradation during spore germination. J. Bacteriol. 194, 4537–4545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Vranken W. F., et al. , The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Schumann F. H., et al. , Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 39, 275–289 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Mulder F. A., Schipper D., Bott R., Boelens R., Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292, 111–123 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Ulrich E. L., et al. , BioMagResBank. Nucleic Acids Res. 36, D402–D408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho B. K., Gruswitz F., HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.