Abstract
Germline genes that are aberrantly expressed in nongermline cancer cells have the potential to be ideal targets for diagnosis and therapy due to their restricted physiological expression, their broad reactivation in various cancer types, and their immunogenic properties. Among such cancer/testis genes, components of the PIWI-interacting small RNA (piRNA) pathway are of particular interest, as they control mobile genetic elements (transposons) in germ cells and thus hold great potential to counteract genome instability in cancer. Here, we systematically investigate the potential reactivation of functional piRNA-silencing mechanisms in the aberrant context. While we observe expression of individual piRNA-pathway genes in cancer, we fail to detect the formation of functional piRNA-silencing complexes. Accordingly, the expression of a PIWI protein alone remains inconsequential to the cancer cell transcriptome. Our data provide a framework for the investigation of complex aberrant gene-expression signatures and establish that reactivation of piRNA silencing, if at all, is not a prevalent phenomenon in cancer cells.
Keywords: RNA, piRNA, germline, cancer, transposon
Mobile genetic elements (transposons) threaten genomic integrity and must be restrained (1). Transposon restriction is imperative, especially in germ cells, where transposition mutagenesis has the potential to change the identity of a species. Thus, germ cells employ a specialized small RNA-silencing pathway—PIWI-interacting small RNAs (piRNAs) and their PIWI protein partners—to establish lasting epigenetic restriction of transposons (2, 3). Recently, aberrant expression of select piRNA-pathway genes has been observed in nongermline cancers, in line with the well-established phenomenon of cancer/germline genes, which describes the aberrant expression of germline-specific genes in nongermline cancers (4–6). Such cancer/germline genes have the potential to be ideal targets for diagnosis and therapy because of their restricted physiological expression and their broad reactivation in various cancer types. However, their function in the aberrant context remains largely elusive. Three consequences of cancer/germline gene expression could be envisioned: (i) cancer cells could reactivate germline programs such as piRNA-mediated transposon silencing; (ii) individual germline genes could assume novel function in the aberrant context; and (iii) the aberrant expression of an individual germline gene in the absence of cofactors could be nonfunctional and without biological consequence. Here, we systematically investigate the potential aberrant reactivation of functional piRNA-silencing mechanisms in cancer considering all three hypotheses.
Results and Discussion
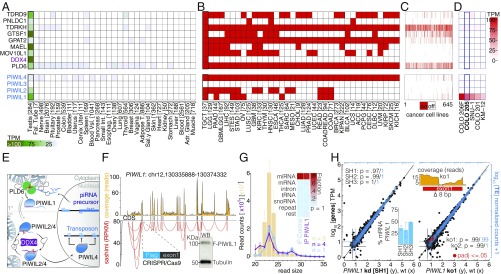
Biogenesis of piRNAs and formation of functional PIWI–piRNA-silencing complexes (piRISCs) requires the coordinated action of biogenesis and effector factors (3, 7), most of which are only expressed in the germline of healthy individuals (Fig. 1A) (8). To evaluate the potential formation of functional piRISCs in cancer, we first characterized the expression of conserved piRNA-pathway genes in tumor samples (Fig. 1B) and in cancer cell lines (Fig. 1 C and D) using available gene-expression data (9, 10). The human genome encodes four PIWI proteins—PIWI-like (PIWIL)1, -2, -3, and -4 (11, 12)—that are either fueled by PLD6/ZUCCHINI(ZUC)-dependent piRNA biogenesis or “ping-pong” processing, which requires the slicer activity of PIWIL2 (HILI) and the RNA helicase DDX4/VASA (13) (Fig. 1E). We did not observe aberrant expression of DDX4/VASA in any nongermline cancer (Fig. 1 B–D), eliminating the possibility of reactivated ping-pong processing, and thus focused our analysis on potential PLD6/ZUC-dependent piRNA production. Because of high sequence diversity and lack of sequence conservation, piRNAs are best defined by their interaction with PIWI proteins, according to their initial definition as PIWI-interacting RNAs (11, 14). Echoing the phenomenon of cancer/germline gene-expression in primary tumor samples [The Cancer Genome Atlas (TCGA)], some cancer cell lines exhibit aberrant expression of piRNA-pathway components. These cell lines represent a trackable model to directly probe for the presence of bona fide piRNAs. For the molecular and functional characterization of aberrant PIWIL1 complexes, we chose the colon cancer cell line COLO205, which is the top available cell line that robustly expresses PIWIL1 but no other PIWIL gene (Fig. 1D). To unambiguously detect the expression of PIWIL1 protein in COLO205, we inserted a FLAG (F-) tag in-frame with PIWI’s start codon into the endogenous genomic locus using CRISPR-Cas9 genome editing. Western blotting analysis showed expression of endogenously tagged F-PIWIL1 with an expected molecular mass of ∼100 kDa, in agreement with the presence of full-length, wild-type messenger RNA (Fig. 1F). To identify if aberrantly expressed PIWIL1 interacts with small RNAs, we purified F-PIWIL1 and characterized associated RNAs by next-generation sequencing. RNA fragments copurifying with F-PIWIL1 were indistinguishable from background pull-down both by nucleotide size distribution and by genomic annotation, and mainly contained background from the abundant class of microRNAs (Fig. 1G). None of our experiments detected potentially functional piRISCs in COLO205 cells. Accordingly, neither knockdown (Fig. 1H, Left) nor knockout (Fig. 1H, Right) of PIWIL1 had any effect on gene expression or abundance of transposon transcripts in the aberrant context.
Fig. 1.
Individual piRNA-pathway genes are aberrantly expressed in cancer without evidence for the reactivation of functional piRNA silencing. (A and B) Gene expression of core piRNA-pathway genes in human tissues (Genotype-Tissue Expression) (A) and tumor samples (TCGA) (B); red indicates “on”: 25th percentile RSEM >5. Abbreviations of tumor types are according to TCGA, with the number of individual samples indicated in brackets. (C) Expression of select piRNA-pathway genes in 645 human cancer cell lines (on: RPKM >5). (D) The top five cell lines ranked by aberrant PIWIL1 expression. (E) A simplified model of mammalian piRNA pathways highlighting conserved piRNA biogenesis factors. (F) Expression of full-length PIWIL1 in COLO205 by RNA-seq (read coverage track), and Western blotting (WB) (Inset) of an endogenously FLAG-tagged allele. (G) Length distribution (nucleotides) of total small RNAs [yellow bars; read counts normalized to total microRNAs (miRNAs) (×106)] and small RNAs that coimmunoprecipitate (IP) with F-PIWIL1 (purple, n = 4) or a negative control (blue, n = 1) [read counts normalized to total reads (×106)], and their genomic annotation (Inset) (Pearson: P = 1). (H) Differential gene expression [log10 (TPM + 1)] and transposon transcript abundance [log 10 (normalized counts + 1)] upon PIWIL1 knockdown (kd) (y axis, Left) and knockout (ko) (y axis, Right) compared with wild-type (wt) COLO205 (x axis). One representative example and Pearson correlation coefficients for three kd (SH1, -2, -3) (Left) and two ko (ko1, ko2) (Right) are shown [red: adjusted P (padj) ≤ 0.05)]. PIWIL1 kd efficacy (Inset Left) and ko1 mutation (Inset Right). RPKM, reads per kilobase million; RSEM, RNA-seq by expectation maximization; SH, short hairpin; snoRNA, small nucleolar RNA; TE, transposable element; TPM, transcripts per million; tRNA, transfer RNA.
Despite our effort, we did not find any evidence for the presence of bona fide piRNAs or a piRNA-independent function of PIWIL1 in cancer cells. Our careful experimental analyses support a recent report that identified abundant RNA fragments as ubiquitous background of small RNA-sequencing (RNA-seq) data (15). Such fragments have sometimes been misinterpreted as piRNAs and contaminate databases that attempt to catalog individual piRNA sequences from different experimental sources. While we do not wish to generalize from our limited set of experiments in COLO205 cells, we aim to raise awareness for the experimental requirements to define piRNAs as “PIWI-interacting RNAs,” and caution against hasty interpretation of RNA fragments. The careful study of aberrant cancer/germline gene expression holds great promise for identifying modulators of cancer biology (16) and for discovering novel diagnostic markers and therapeutic targets (5). However, not every aberrant expression results in functional products with biological impact.
Materials and Methods
Cancer Cell Lines: Knockdown, Knockin, and Knockout.
COLO205 cells (American Type Culture Collection CLL-222); knockdown (TransOMIC #TRHSU2000); Fendo-PIWIL1 [PIWIL1-exon1 was targeted using CRISPR-Cas9 (guide: 5′-CTGACCGCGGGCCCTTCCTC-3′) and repaired using a donor construct containing a FLAG tag in-frame with PIWIL1’s ATG (based on Addgene #68375)]. CRISPR-Cas9–induced missense mutations generated knockout alleles.
Illumina Sequencing and Data Analysis.
RNA extraction (Direct-zol RNA MiniPrep, Zymo Research); RNA-seq samples (NEBNext Ultra II DNA Library Prep). Small RNA preparations were previously described (17). Illumina data were aligned to human genome (hg38) and quantified (Hisat v2.2.1.0 and Stringtie v1.3.4d for gene expression; STAR v2.5.2b and Tetoolkit v2.0.3 for transposons). Statistical analyses and visualizations (R, v3.5.1). Data are available at https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128526).
Acknowledgments
We thank Drs. Zissimos Mourelatos, Alexei Aravin, and John V. Moran and the members of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Board of Scientific Counselors for encouraging us to share these data. We also thank the NIDDK and National Heart, Lung, and Blood Institute genomics cores and the NIH High-Performance Computing group. This work was supported by the intramural research program of the NIDDK.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA sequencing data and small RNAsequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128526).
References
- 1.Kazazian H. H. Jr, Moran J. V., Mobile DNA in health and disease. N. Engl. J. Med. 377, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siomi M. C., Sato K., Pezic D., Aravin A. A., PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Czech B., et al. , piRNA-guided genome defense: From biogenesis to silencing. Annu. Rev. Genet. 52, 131–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson A. J. G., Caballero O. L., Jungbluth A., Chen Y.-T., Old L. J., Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 5, 615–625 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Rousseaux S., et al. , Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 5, 186ra66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross R. J., Weiner M. M., Lin H., PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozata D. M., Gainetdinov I., Zoch A., O’Carroll D., Zamore P. D., PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 20, 89–108 (2019). [DOI] [PubMed] [Google Scholar]
- 8.GTEx Consortium , The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstein J. N., et al. ; Cancer Genome Atlas Research Network , The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klijn C., et al. , A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33, 306–312 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Aravin A., et al. , A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Williams Z., et al. , Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 13, 854–863 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Xiol J., et al. , RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157, 1698–1711 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Girard A., Sachidanandam R., Hannon G. J., Carmell M. A., A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Tosar J. P., Rovira C., Cayota A., Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol. 1, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla S. A., et al. , Cancer-germline antigen expression discriminates clinical outcome to CTLA-4 blockade. Cell 173, 624–633.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein CB, et al. , Decoding the 5′ nucleotide bias of PIWI-interacting RNAs. Nat. Commun 10, 1, 828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]