Significance
HIV-1 arose following the cross-species transmission of a simian immunodeficiency virus of chimpanzees (SIVcpz) to humans. In this study, we looked at genetic diversity in chimpanzee CD4, which encodes the main receptor for both HIV and SIVcpz. We identified multiple CD4 alleles circulating in chimpanzee populations, some of which impede cellular entry of HIV-1 and SIVcpz. We demonstrate that some chimpanzees would be significantly protected from virus infection because their CD4 protein is modified by the addition of glycans to the virus binding surface. These glycans create a shield that protects the receptor from being engaged by viruses, demonstrating a powerful form of host resistance against deadly primate lentiviruses.
Keywords: arms race, virus entry, population genetics, selection
Abstract
Pandemic HIV-1 (group M) emerged following the cross-species transmission of a simian immunodeficiency virus from chimpanzees (SIVcpz) to humans. Primate lentiviruses (HIV/SIV) require the T cell receptor CD4 to enter into target cells. By surveying the sequence and function of CD4 in 50 chimpanzee individuals, we find that all chimpanzee CD4 alleles encode a fixed, chimpanzee-specific substitution (34T) that creates a glycosylation site on the virus binding surface of the CD4 receptor. Additionally, a single nucleotide polymorphism (SNP) has arisen in chimpanzee CD4 (68T) that creates a second glycosylation site on the same virus-binding interface. This substitution is not yet fixed, but instead alleles containing this SNP are still circulating within chimpanzee populations. Thus, all allelic versions of chimpanzee CD4 are singly glycosylated at the virus binding surface, and some allelic versions are doubly glycosylated. Doubly glycosylated forms of chimpanzee CD4 reduce HIV-1 and SIVcpz infection by as much as two orders of magnitude. Full restoration of virus infection in cells bearing chimpanzee CD4 requires reversion of both threonines at sites 34 and 68, destroying both of the glycosylation sites, suggesting that the effects of the glycans are additive. Differentially glycosylated CD4 receptors were biochemically purified and used in neutralization assays and microscale thermophoresis to show that the glycans on chimpanzee CD4 reduce binding affinity with the lentiviral surface glycoprotein, Env. These glycans create a shield that protects CD4 from being engaged by viruses, demonstrating a powerful form of host resistance against deadly primate lentiviruses.
The origins of HIV-1 can be traced back to an ancient reservoir of simian immunodeficiency viruses (SIVs) found in African primates (1). SIVs have transmitted to humans twice through infection by chimpanzees (giving rise to HIV-1 groups M and N) (2–4). The pandemic form of HIV-1 (group M) subsequently established sustained transmission chains in humans, resulting in ∼70 million infected worldwide. SIVs constitute a viral reservoir with ongoing zoonotic potential.
Current estimates suggest that primates have been infected with lentiviruses for more than 10 My (5, 6). Viruses and their hosts exist in a constant state of genetic conflict, where what is advantageous for one party is often disadvantageous for the other. This can drive tit-for-tat rounds of evolution in both viral and host genomes, which play out in gene regions corresponding to interaction interfaces between host and viral proteins (reviewed in refs. 7–9). For example, host genes encoding lentiviral restriction factors, including TRIM5α, APOBEC3G, and Tetherin, all possess signatures of rapid evolution in regions of the proteins that make contact with lentiviral proteins (10–12). Presumably, these genes have experienced selection in favor of new allelic forms that had greater ability to restrict viral replication. The CD4 receptor of T cells plays an essential role in SIV/HIV attachment and entry into host cells, and the CD4 gene has also evolved under positive selection during primate speciation (13, 14). Given the essential role of CD4 during infection by primate lentiviruses, it is tempting to speculate that SIVs may have been responsible for driving selection for alleles encoding new forms of the CD4 protein with heightened resistance to SIV entry.
A potent antibody-evasion strategy employed by viruses is the so-called “glycan shield” present on many viral glycoproteins, which masks neutralizing antibody epitopes. These protective glycan shields have been demonstrated for diverse viruses, such as HIV-1 (15, 16), Ebola virus (17), human coronavirus HCoV-NL63 (18), Hendra virus (19), and Lassa virus (20). Based on the effectiveness of this strategy, one might think that hosts would experience selection in favor of mutations that add glycans to cell surface receptors. Glycans are bulky and could provide physical distance between virus attachment proteins and host cellular receptors, impeding virus binding. However, there are relatively few examples of this type of host resistance mechanism. In one example, a substitution acquired before the evolutionary split of dogs and coyotes introduced a single N-linked glycan to the surface of transferrin receptor (TfR), and this glycan powerfully impedes entry by carnivore parvoviruses (21–23). Dogs and coyotes were presumably protected from parvoviruses for millions of years by this glycan on TfR (23). Then, in the mid-1970s, a variant of parvovirus arose with a small number of capsid substitutions that allowed the virus to reestablish use of this receptor, despite its glycan, and this new canine parvovirus quickly swept the globe as a new pandemic in dogs that persists to this day (21–24). Similarly, the receptors for New World arenaviruses, mammalian coronaviruses, and various retroviruses all have acquired a single glycan on their virus binding surface, which impedes viral entry (25–28). In all of these examples, the addition of a receptor-associated glycan creates a highly effective shield that significantly compromises viral infection. The economy of this defense is notable, employing only a single glycan, and is in stark contrast to the heavy levels of glycosylation usually seen on virus surface proteins.
Here, we show that the CD4 receptors encoded by chimpanzees bear either one or, in some cases, two glycans on the virus binding surface. In a survey of the CD4 alleles of 50 individuals, we find that all chimpanzee CD4 receptors are modified with a unique, N-linked glycan on the SIV binding surface of CD4 that is not present in human or gorilla CD4. In addition, we identify a single-nucleotide polymorphism (SNP) in chimpanzees that results in a second glycan being added to this surface. We show that the effects of these two CD4 glycans are additive, and result in poor binding of primate lentiviruses to glycosylated CD4. Given that the primate lentivirus reservoir is ancient (5, 29), and because some chimpanzee subspecies are themselves endemically infected with SIV (30, 31), it is likely that the chimpanzee genome may have been under intense pressure in favor of SIV resistance mutations such as these. We demonstrate that the addition of chimpanzee-specific glycans on the virus binding surface of CD4 creates a protective shield against primate lentiviruses, highlighting a powerful form of host resistance against deadly primate lentiviruses.
Results
Polymorphisms in Chimpanzee CD4 Residues That Engage Lentiviruses.
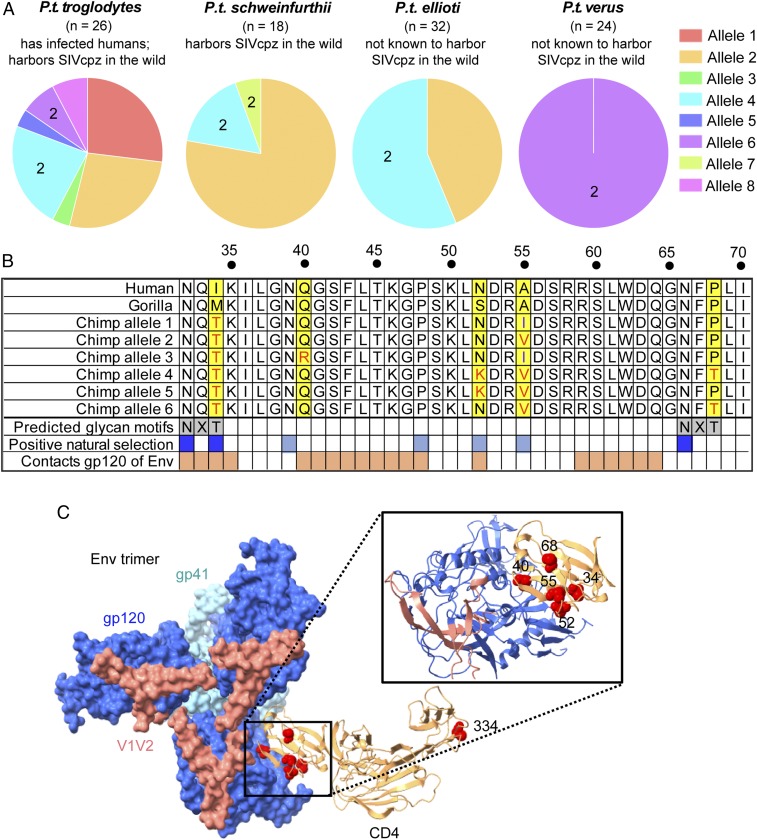
Chimpanzees (Pan troglodytes) are divided into four subspecies: P.t. troglodytes, P.t. schweinfurthii, P.t. ellioti, and P.t. verus. SIVs have transmitted to humans twice through infection by P.t. troglodytes, giving rise to HIV-1 groups M and N (2–4). To characterize genetic variation in the chimpanzee CD4 gene, we obtained CD4 sequence data from several sources. First, we processed raw whole-genome shotgun sequencing reads (Methods) obtained from the Great Apes Genome Project (32) and then performed SNP calling and genotyping for each individual. From this processed whole-genome sequencing data, we then extracted CD4 gene sequences. This resulted in 24 CD4 sequences, never before reported, covering all four chimpanzee subspecies. In addition, we obtained CD4 genotype data from a previous study on chimpanzee CD4 (33), and also sequenced CD4 from four additional individuals ourselves [P.t. troglodytes (n = 1), P.t. schweinfurthii (n = 1), and P.t. verus (n = 2)]. This final dataset consisted of CD4 sequence from 50 chimpanzee individuals (n = 100 haplotypes) (SI Appendix, Table S1). Allele structures were inferred using phase2.1 in DnaSP (34–36) (SI Appendix, Table S2). For the purpose of this study, we then combined alleles differing by only synonymous substitutions, because these alleles will encode the same CD4 protein. Using this approach, we identified eight unique alleles circulating within the four P. troglodytes subspecies, each of which encodes a unique CD4 protein (Fig. 1A and SI Appendix, Table S2).
Fig. 1.
CD4 is polymorphic in chimpanzees. (A) Alleles encoding eight unique CD4 proteins were identified in four chimpanzee subspecies. The number (n) of haplotypes investigated is shown above each pie chart. A number “2” within the pie chart indicates alleles encoding two glycosylation sites in the D1 domain. (B) An amino acid alignment of part of the CD4 D1 domain from human, gorilla, and chimpanzee alleles 1–6, identifying unique chimpanzee residues (yellow boxes), chimpanzee specific predicted glycan motifs (gray boxes), sites that have evolved under positive selection (blue boxes) (from refs. 13 and 14), and sites that contact Env (dark orange) (from ref. 39). Sites in dark blue are part of two chimpanzee-specific glycosylation motifs that have also been the target of positive selection. CD4 alleles 7 and 8 do not differ from the represented alleles in the region shown, and are therefore not illustrated. (C) Cryo-EM structure of an HIV-1 Env trimer in complex with CD4 (PDB ID code 5U1F) (39) was visualized in ChimeraX (76). Individual gp120 and gp41 subunits are colored in dark blue and light blue, respectively. V1V2 regions of Env are highlighted in pink and CD4 D1–D4 is depicted as the tan ribbon structure. Residues specific to chimpanzees (yellow highlight in B) were shown on the human sequence as red spheres, which is valid because the chimpanzee and human CD4 structures are predicted to be highly homologous (Methods). Residue 334 is an additional nonsynonymous polymorphism in chimpanzee CD4, specific to allele 8.
The allele frequencies in each chimpanzee subspecies are shown (Fig. 1A). Several alleles are shared between subspecies, with alleles 2 and 4 being found in three of the four chimpanzee subspecies. P.t. troglodytes, the source of all known human infections (1), harbors the greatest number of alleles (seven of the eight total), although the reasons for this aren’t immediately obvious. These alleles are differentiated by 10 SNPs, 6 of which are nonsynonymous. Four of these nonsynonymous SNPs are within the N-terminal D1 domain of CD4, which mediates interaction with HIV-1 Envelope (Env) (Fig. 1 B and C) (37–39). In the D1 domain alignment shown in Fig. 1B, only six alleles are shown because alleles 7 and 8 do not differ from the represented alleles in the region shown. These alleles contain nonsynonymous SNPs in the signal peptide (allele 7) and D4 domain (allele 8), substitutions that are not expected to play a role in virus entry. Besides the D1 domain SNPs, another position of interest is residue 34, where all chimpanzee alleles encode a unique threonine that chimpanzees’ close relatives, human and gorilla, do not encode (Fig. 1B). All of the five sites of chimpanzee-specific variation (yellow highlight in Fig. 1B) fall at or near residues that have been targeted by positive natural selection (blue boxes in Fig. 1B) (13, 14), suggesting that substitutions at these sites could have functional significance.
CD4 Is Differentially Glycosylated in Humans, Chimpanzees, and Gorillas.
Computational modeling suggested that the chimpanzee-specific threonines at positions 34 and 68 create N-linked glycosylation motifs (“NXT” motifs) (Fig. 1B). All chimpanzee CD4 alleles are predicted to encode the glycosylation motif at positions 32–34, while only alleles 4, 6, and 7 are predicted to have the second glycosylation motif at 66–68. We noted that the two subspecies of chimpanzees that are not infected with SIVcpz in the wild, P.t. ellioti and verus (40–42), have the highest prevalence of CD4 alleles predicted to encode both of these glycosylation motifs (Fig. 1A). This gave rise to our hypothesis that these glycans may convey some level of protection against infection. Human and gorilla encode neither of the glycosylation motifs that are predicted in the chimpanzee CD4 D1 domain. Gorilla CD4 was predicted to have a unique glycosylation site in the D1 domain at position 15 (NCT motif), while human CD4 was predicted to have no glycosylation sites in the D1 domain (a full alignment of the D1 domain is shown; see SI Appendix, Fig. S1). There are two additional glycosylation motifs predicted in CD4, but both fall outside of the D1 domain (one in D3, the other in D4) and are shared between human, chimpanzee (all alleles), and gorilla.
To determine whether these CD4 proteins are actually differentially glycosylated, we purified the D1–D2 domains of CD4 from human, gorilla, and chimpanzee, after expression in human (293T) cells. These CD4s are referred to as soluble CD4 (sCD4) because the D1 and D2 domain does not encompass the transmembrane region of the CD4 receptor, and this protein is secreted from the cell. The sCD4 proteins encoded by human, gorilla, and chimpanzee (allele 6) are predicted to have 0, 1, or 2 glycosylation sites in the D1 domain, respectively, as discussed above. We analyzed migration patterns of these purified proteins by SDS/PAGE (Fig. 2A). Consistent with the glycan predictions (Fig. 2A, bottom of gel), human sCD4 ran at ∼20 kDa, chimpanzee sCD4 at a larger size (∼23 kDa), and gorilla sCD4 ran predominantly at a size in between. This is consistent with a previous study that showed that chimpanzee CD4 migrates at a higher molecular weight in an acrylamide gel compared with human CD4 (43). The purified sCD4s were then treated with peptide-N-glycosidase (PNGase) F, which is highly specific for removing N-linked oligosaccharides from glycoproteins. PNGase F treatment resulted in all three sCD4 proteins running at approximately the same size, suggesting that these sCD4s are indeed differentially glycosylated (Fig. 2B). Western blotting and PNGase F treatment confirmed that these CD4s are glycosylated similarly when expressed in CD4+ T cells (Hut 78 cells) (SI Appendix, Fig. S2).
Fig. 2.
Human, chimpanzee, and gorilla CD4 are differentially glycosylated in their D1 domains. sCD4 (comprised of domains D1–D2 of CD4) was purified from 293T cells transfected with sCD4 expression constructs. (A) Protein stain of concentrated imidazole eluted fractions of human, chimpanzee (allele 6), and gorilla sCD4. The number of predicted Asn-specific glycosylation sites, indicated below the gels, was determined computationally (67). (B) PNGase F treatment of samples purified in A.
Chimpanzee CD4 Variants Differentially Impede Entry of Primate Lentiviruses.
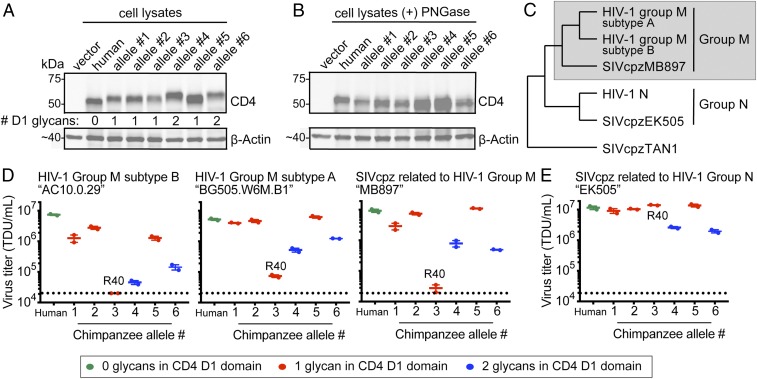
To look for functional differences between chimpanzee-specific forms of CD4, we next used retroviral transduction to stably integrate chimpanzee CD4 alleles 1–6 into Cf2Th (canine) cells that were also stably expressing human CCR5. Western blotting of cell lysates confirmed similar CD4 expression between cell lines (Fig. 3A; CD4 and CCR5 expression profiles by flow cytometry are shown in SI Appendix, Fig. S3). The differential migration of the proteins produced by these alleles matches the predicted number of glycosylation motifs in each allelic sequence (Fig. 3A). Cell lysates were then treated with PNGase F, resulting in similar CD4 migration between all samples (Fig. 3B). While glycosylation patterns can sometimes be tissue-specific (44), the CD4 glycosylation patterns observed were consistent between CD4 receptors expressed in canine cells (Fig. 3 A and B), purified from 293T cells (Fig. 2), or expressed in CD4+ T cells (SI Appendix, Fig. S2).
Fig. 3.
Chimpanzee CD4 alleles differentially support entry of HIV-1 and SIVcpz. (A) Lysate from Cf2Th cells stably expressing human or chimpanzee CD4 alleles 1–6 were probed for CD4 expression by Western blotting. The number of predicted Asn-specific glycosylation sites within the D1 domain of CD4 was determined computationally. (B) PNGase F treatment of cell lysates from A, followed by Western blotting. (C) Cladogram shows the relationship between viruses used in this study, which is consistent with a previous report (ref. 45). SIVcpzTAN1 isolated from P.t. schweinfurthii served as the outgroup. The gray box denotes the SIVcpz clade that gave rise to group M HIV-1. (D and E) HIV-1 ΔEnv-GFP viruses were pseudotyped with Envs (top of graphs) from (D) group M-related HIV-1 and SIVcpz variants, or (E) group N-related SIVcpz. Cf2Th cells stably expressing human CCR5 and various CD4s (x axis) were infected with various volumes of these pseudoviruses and then analyzed by flow cytometry 48 h postinfection. GFP+ cells were enumerated and virus titers (TDU/mL) were determined for those samples falling within the linear infection range (n = 2 titration points). The mean virus titers obtained from each of two independent experiments were plotted (dots), with error bars representing the SEM. Dotted lines represent the limit of detection for this assay. The data are presented for CD4 proteins with zero glycosylation sites in the D1 (green), one glycan in the D1 domain (red), or two glycans in the D1 domain (blue). The CD4 proteins encoded by alleles 3 and 1 are identical, except for the R encoded at position 40 by allele 3. Thus, we have denoted “R40” near the allele three data points because this mutation is rendering CD4 defective for the entry of group M-related viruses (D), while remaining functional for entry of SIVcpz related to group N (E).
Next, we sought to infect these cells with primate lentiviruses that are relevant to chimpanzees. Experiments with SIVcpz have, to date, been performed with live virus, making it hard to assess just viral entry. For this reason, we developed a pseudotyping system for SIVcpz Env that is similar to what is used for HIV-1 Env. Briefly, the genomic region of SIVcpz encompassing both the rev and env genes was amplified by PCR from different SIVcpz infectious molecular clones, and cloned into a mammalian expression plasmid. This plasmid was then cotransfected along with a plasmid encoding full-length HIV-1ΔEnv-GFP to generate pseudotyped viruses bearing SIVcpz Env on their surface. Using this approach, we produced pseudoviruses representing SIVcpz isolates “MB897” and “EK505.” SIVcpz MB897 is closely related to HIV-1 group M, while SIVcpz EK505 is closely related to HIV-1 group N (45) (Fig. 3C). We also produced similar pseudoviruses with the Envs of two different subtypes of HIV-1 group M, which arose when chimpanzees infected humans with an SIVcpz of the MB897/HIV-1 group M clade (gray box in Fig. 3C).
The cell lines stably expressing human CCR5 and various CD4 proteins were then infected with each of these pseudoviruses. The percent of GFP+ (i.e., infected) cells was measured by flow cytometry, and converted to transducing units per milliliter (TDU/mL). Because there were some differences in receptor expression from one stable cell line to the next (Fig. 3A and SI Appendix, Fig. S3), samples were first gated for live cells, then gated for a fixed range of CD4/CCR5 receptor expression between all samples, and finally GFP+ cells were scored in this population. Error bars represent the mean + SEM in virus titers from two independent experiments (Fig. 3 D and E). All group M-related viruses tested (SIVcpz MB897 and the two HIV-1 group M isolates) showed a very similar receptor use profile (Fig. 3D). Furthermore, both SIVcpz Envs tested were fully capable of utilizing human CD4 (Fig. 3 D and E). This is consistent with previous reports that SIVcpz can utilize human CD4 (45–48), collectively supporting the idea that human CD4 was not a barrier in either of the zoonotic transmissions leading to HIV-1 group M and group N.
Nonetheless, while human CD4 supported entry of all viruses tested, some versions of chimpanzee CD4 dramatically limited virus infection. For group M-related viruses, virus entry differed by one to two orders of magnitude depending on the specific chimpanzee CD4 allele (Fig. 3D), even though these alleles all encode CD4 proteins that differ by only 1–3 amino acids from each other. Chimpanzee CD4 proteins with two glycosylation sites in D1 (Fig. 3D, blue data points), or the Q40R substitution specific to allele 3, significantly impeded infection of all three group M-related viruses. These differences are more pronounced for the group M-related viruses (Fig. 3D) than they are for the one group N-related virus that we tested (Fig. 3E). Because the Q40R SNP was only identified in one individual (SI Appendix, Table S2), we did not pursue the characterization of this SNP further. However, this polymorphism was also identified by another recent study (49), confirming that alleles containing this protective SNP are indeed circulating in chimpanzee populations. It is interesting to note that entry mediated by SIVcpz EK505 Env (related to HIV-1 group N) (45) was at least partially impeded by CD4 alleles encoding two glycosylation sites, yet unaffected by the Q40R substitution (allele 3) (Fig. 3E). This Q40R substitution in chimpanzee CD4 warrants further exploration, because it appears to interact differently with group M- versus group N-related lentiviruses. Unlike other residue sites explored in this study, position 40 in CD4 is not under positive selection, in fact it is highly constrained as the ancestral glutamine (Q) in the CD4 of all primate species for which CD4 sequences are available.
A Glycan Shield on Chimpanzee CD4 Impedes Virus Binding and Infection.
Chimpanzee CD4 allele 6, which encodes two glycosylation sites in D1 (Figs. 1–3), was modified by site-directed mutagenesis to encode the human residues at positions 34 and 68, ablating one or both of the predicted glycosylation motifs. These receptors were stably expressed in Cf2Th cells expressing human CCR5, and then infected with an HIV-1ΔEnv-GFP pseudotyped with Env from two different HIV-1 group M subtypes. Ablating both glycosylation sites in chimpanzee CD4 resulted in a restoration of HIV-1 infection, while single mutations had intermediate effects (black bars in Fig. 4). In addition, we made the corresponding reciprocal mutations in human CD4 (I34T and P68T together, and each single mutant), producing versions of human CD4 that impede HIV-1 entry (gray bars in Fig. 4). These data imply that the amino acids encoded at positions 34 and 68 of CD4, establishing or destroying glycosylation motifs, each influence lentiviral entry in an additive fashion.
Fig. 4.
The effects of CD4 D1-domain glycans are additive. Cf2Th cells stably expressing human CCR5 and the indicated CD4s (x axis) were infected with HIV-1 ΔEnv-GFP pseudotyped with a subtype B or subtype A HIV-1 group M Env (top of graphs) and then analyzed by flow cytometry 48 h later. Samples were first gated for live cells, then gated for a fixed range of CD4/CCR5 receptor expression between all samples, and finally GFP+ cells were scored in this population. The percent cells infected (GFP+) was normalized to the percent cells infected in the control cell line expressing human CD4. Error bars represent the SEM from two independent experiments, each conducted in triplicate.
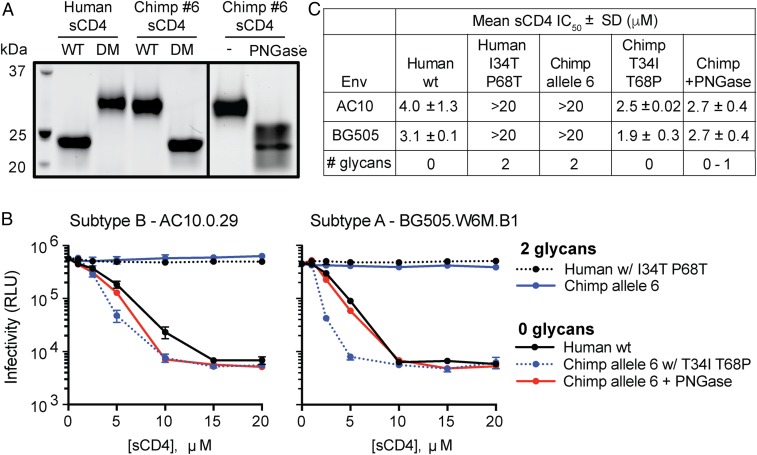
We next confirmed that the mutations at sites 34 and 68 change the glycosylation status of CD4. We purified versions of sCD4 representing human, chimpanzee (allele 6), and also the human-to-chimpanzee (I34T, P68T) and chimpanzee-to-human (T34I, T68P) double mutants (“DM” in Fig. 5A). When mutations were introduced into chimpanzee or human sCD4, these proteins migrated at similar molecular weights as the wild-type version of the other species, confirming that the two mutated sites destroy or create the two glycosylation sites in CD4 (Fig. 5A).
Fig. 5.
CD4 glycosylation inhibits HIV-1 entry by reducing Env-CD4 binding affinity. (A) Protein stain of purified soluble CD4 (sCD4) representing human, chimpanzee, and the DM described in Fig. 4. WT, wildtype. (B) Infectivity of two HIV-1 Env (top of graph) pseudotyped viruses that were preincubated with increasing concentrations of various human or chimp sCD4s (legend) as a competitive inhibitor. In one of these experiments (red lines), chimpanzee sCD4 was first treated with an Asn-specific glycosidase (PNGase F), thus effectively removing glycans (see right side in A). Infectivity was determined by measurement of luciferase activity in TZM-bl cells. Error bars represent the SD from n = 4 technical replicates, and the results shown are representative of two independent experiments. (C) Table of IC50 values from sCD4 neutralization assays in B, where the mean and SD values reflect the variation of the IC50 calculations from two independent experiments.
We then assessed the binding of variably glycosylated sCD4 to HIV-1 Env. We preincubated two different HIV-1 pseudoviruses with increasing concentrations of each purified sCD4, and then infected TZM-bl cells. TZM-bl cells express human CD4 and CCR5, as well as an integrated Tat-inducible luciferase reporter. Neutralization of infection occurs when the sCD4 binds to Env of the pseudovirus, thus preventing virus interaction with CD4 expressed on the surface of TZM-bl cells. This results in a reduction in Tat-inducible luciferase signal (relative luminescence units, RLU). When HIV-1 was preincubated with human sCD4, we observed a dose-dependent reduction in RLUs as expected (black line in Fig. 5B). Chimpanzee sCD4 with two glycans (from allele 6), however, did not neutralize HIV-1 infection even at the highest sCD4 concentrations, indicating that HIV-1 binds poorly to the doubly glycosylated CD4 receptor (blue line in Fig. 5B). Engineering chimpanzee sCD4 to lack glycosylation sites restored binding to HIV-1, while introducing the chimpanzee glycosylation sites into human sCD4 destroyed binding (Fig. 5B). Based on this, if the human CD4 had these two glycans, it appears that HIV-1 would have much more difficulty entering human cells.
To definitively show that glycans are impeding virus binding to chimpanzee CD4, rather than the underlying protein sequence changes inherent to mutagenesis studies, we treated our purified chimpanzee sCD4 (harboring two glycans) with PNGase F and then repurified the enzyme-treated protein. Chimpanzee sCD4 treated with PNGase F migrated similarly to human sCD4, but as a smear, indicating a mixed preparation of fully and partially deglycosylated sCD4 proteins (right box in Fig. 5A). Preincubation of PNGase F-treated chimpanzee sCD4 with HIV-1 pseudovirus restored binding and virus neutralization in the TZM-bl assay (red line in Fig. 5B). The IC50 values for human, chimpanzee double mutant (T34I, T68P), and PNGase F-treated chimpanzee sCD4s were nearly equivalent, suggesting that the ability of HIV-1 to interact with CD4 is dependent on the absence of high molecular weight glycans on CD4 (Fig. 5C).
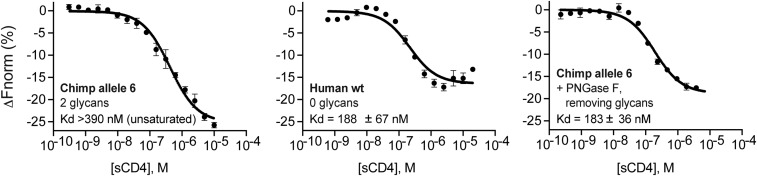
As another metric for quantifying Env interaction with our purified sCD4 proteins, we measured sCD4 binding affinity with purified HIV-1 gp120 monomer using microscale thermophoresis. The binding constant (Kd) for human sCD4 (0 glycans) was lower than that of chimpanzee sCD4 (allele 6, two glycans), which reflects an overall higher affinity of HIV-1 Env for human CD4 (Fig. 6). Furthermore, it should be noted that for chimpanzee CD4, saturation was not reached even at the highest sCD4 concentration tested (Fig. 6). Thus, the Kd value reported (>390 nM) underestimates the true Kd value. The importance of glycosylation in reducing Env-CD4 binding was further confirmed with PNGase F-treated chimpanzee sCD4, which had a similar affinity for gp120 as human sCD4 (Fig. 6). Collectively, this suggests that glycans on the surface of CD4 inhibit virus infection by significantly reducing Env-CD4 binding.
Fig. 6.
Kd values for the interactions between HIV Env and CD4 proteins with different levels of glycosylation. Purified soluble human or chimp CD4 (sCD4) proteins (bold type within graphs) were serially diluted and incubated with a fixed concentration (25 nM) of purified gp120 monomer (from HIV-1 isolate AC10.0.29) (50) that we labeled with Alexa488. Following incubation, the samples were loaded into glass capillaries and the thermophoretic mobility was measured using the Monolith NT.115 (NanoTemper). The derived ΔFnorm (%) = Fhot/Fcold curves from at least two independent experiments were combined to create one dataset, and the averaged data points, with SEM shown, were fitted using the Kd model in the MO.Affinity Analysis Software (NanoTemper). The Kd values and confidence intervals are shown in each plot.
Here, we identify two glycosylated residues in chimpanzee CD4 that provide a protective barrier against infection by at least some primate lentiviruses. By surveying the sequence and function of CD4 alleles from 50 chimpanzee individuals representing four subspecies, we find that all chimpanzee CD4 alleles encode a fixed, chimpanzee-specific substitution in CD4 (34T) that creates a glycosylation site on the virus binding surface of CD4. Additionally, we identified a SNP that has arisen in CD4 (68T) that is not yet fixed, but instead alleles containing this SNP are circulating at variable frequencies within the four chimpanzee populations studied. This substitution creates a second glycosylation site on the virus binding surface of chimpanzee CD4. As a result, all allelic versions of chimpanzee CD4 are at least singly glycosylated at the virus binding surface, and some allelic versions are doubly glycosylated. Using complementary biochemical and mutagenic approaches, we have shown that the glycans on chimpanzee CD4 reduce binding affinity with lentiviral Env, impeding cellular entry of at least some primate lentiviruses. In addition, full restoration of virus infection in cells bearing chimpanzee CD4 requires reversion of both threonines at sites 34 and 68, which destroys both of the glycosylation sites. This suggests that the effects of the glycans are additive. Based on signatures of positive selection that align with these substitutions in chimpanzee CD4 (Fig. 1B) (13, 14), and the clear advantage that comes with these glycan-introducing substitutions, we conclude that chimpanzees have experienced selection in favor of the creation of a potent host deterrent to virus infection, in this case in the form of a glycan shield on chimpanzee CD4. Based on the allele frequencies shown in Fig. 1A, it appears that anywhere from 20 to 100% of chimpanzees, depending on the subspecies, would have one or two copies of a CD4 allele bearing the two glycans that diminish infection.
Population-level genetic variation in host cellular receptors has previously been shown to limit or prevent virus infection in some but not all individuals within a species. In the case of HIV-1, some humans of European descent encode a deletion in CCR5 (CCR5Δ32) that prevents HIV-1 infection (51, 52). Additionally, human SNPs in cellular receptors have been shown to convey protection against filoviruses and hepatitis B virus (53, 54). Our results allow us to speculate that certain chimpanzee individuals, based on their CD4 genotype, may be capable of sustaining higher SIVcpz titers than others. Higher viral titers would likely impact virus transmissibility, suggesting the intriguing possibility that certain chimpanzee individuals with permissive CD4 genotypes may have been the source for the zoonotic emergence of HIV-1. We can’t say for sure that the SIVcpz variants that existed in chimpanzees ∼100 y ago, when the transmission to humans occurred (55), engaged with the CD4 receptor in the same way as the modern day SIVcpz isolates used in this study. However, because both modern-day group M-related SIVcpz and HIV-1 show very similar patterns of receptor engagement (Fig. 3D), it is reasonable to conclude that the virus that is the common ancestor of both had the same pattern of interaction. Identifying genetic variation that alters inter- and intraspecies host susceptibility, like those experiments with CD4 described herein, will be crucial to expanding our understanding of disease susceptibility and also viral zoonosis.
While glycosylation of chimpanzee CD4 protects against infection by HIV/SIVcpz, these glycans may also protect against additional primate viruses that utilize the CD4 receptor for entry. For example, human herpesvirus 7 (HHV-7) utilizes the CD4 receptor for entry into T cells, and this interaction can be blocked by treatment with soluble forms of HIV-1 gp120 (suggesting that both viruses utilize a similar binding interface) (56). Importantly, roseoloviruses closely related to HHV-7 have been identified in chimpanzees (57). Further investigation is warranted to determine whether chimpanzee-specific CD4 glycans influence entry of viruses other than HIV/SIVcpz. Our study adds to a growing body of evidence that cellular receptor-associated glycans can defend host cells against viral infection (21–23, 25–28). Furthermore, our results add to previous studies showing that glycans have important biological effects in the three-way relationship between HIV-1 Env, CD4, and host antibodies (58–60).
While this paper was under evaluation, Bibollet-Ruche et al. (49) published a study that similarly analyzed the genetic diversity of CD4 in chimpanzee populations. Both studies are in agreement with regard to the alleles identified, the approximate frequencies at which these alleles circulate within the four chimpanzee subspecies, and share the findings that allelic variants encoding a second predicted glycosylation motif (T68) impede SIVcpz entry. In this study, we use biochemical approaches to explore the significance of the glycosylation motifs in CD4, which complement and are in concordance with the infection experiments in both studies. Through CD4 protein purification, virus neutralization assays, and biochemical analysis of binding affinities, we directly demonstrate that CD4 glycosylation significantly reduces Env–CD4 binding affinity, explaining the reduced infectivity of group M-related HIV-1 and SIVcpz with glycosylated forms of the CD4 receptor.
Collectively, our data and that of a related study (49) show that glycans on chimpanzee CD4 diminish susceptibility to infection by at least some primate lentiviruses. It appears that natural selection has favored a growing glycan shield on chimpanzee CD4. Given the economy and effectiveness of this defense strategy on the part of the host, it is interesting to consider why more heavily glycosylated receptors are not more ubiquitously observed as a defense strategy employed by mammals. There may be a cost to coating these important host receptors with glycans, while viruses can employ a full glycan shield and still perform the functions that they need to perform, like attaching to receptors and entering cells. The CD4 receptor plays an essential role in immunity and, while it is unlikely that the 68T SNP (introducing a second glycan) would have been maintained if the normal functions of CD4+ T cells were disrupted, follow-up investigation is required to confirm this.
Methods
Genotype and Allele Determination of CD4 from Chimpanzees.
Chimpanzee CD4 sequencing data were obtained from several sources. First, short-read data available through the National Center for Biotechnology Information’s (NCBI) Short Read Archive (BioProject PRJNA189439) were mapped onto the P. troglodytes genome (panTro4) using BWA-MEM (61). We applied GATK base quality score recalibration, indel realignment, duplicate removal, and SNP discovery and genotyping in each individual separately (62). Joint genotyping and variant recalibration was performed in a species-specific manner and in accordance to the GATK best practices recommendations (63, 64). Variant recalibration was performed using SNPs called by the neighbor quality score method of ssahaSNP on capillary sequencing runs from NCBI’s Trace Read Archive (65), dbSNP (if available), and high-quality SNPs called on the hg18 genome lifted over to the assembly used for mapping (32). Postprocessing was performed using custom scripts written in Python. These sequences are summarized in SI Appendix, Table S2.
Second, sequence data from a population-level survey of chimpanzee CD4 diversity was obtained from GenBank (EF437437–EF437442, EF437451, and EF437460–EF437475) (33). Four additional chimpanzees were evaluated using archived RNA samples originating from the Chimpanzee Biomedical Research Resource (NIH8U42OD011197) supported through a cooperative agreement with the NIH. These archived RNA samples, which had previously been utilized for other genetic analyses in 2013 (66), were derived from blood collected as part of each animal’s annual veterinary medical examination under the approval of the MD Anderson Cancer Center’s Institutional Animal Care and Use Committee. Extracted RNA was used as a template for oligo(dT) primed reverse transcription (SuperScript III RT; Thermo Fisher). PCR amplification of CD4 was performed using Phusion High-Fidelity PCR Master Mix (New England Biolabs; #M0531S) containing ∼50 ng of cDNA and 0.2-μM final concentration of each PCR primer CD4-5′ UTR forward 5′-GGGCAAGAAAGACGCAAGC-3′ and CD4-3′ UTR reverse 5′-ATCTGCCTGGCCTCGTG-3′ in a final volume of 50 μL. PCR clean-up was performed using Exonuclease-I and recombinant Shrimp Alkaline Phosphatase treatment (Affymetrix) for 15 min at 37 °C, and then the cleaned-up products were Sanger sequenced using the following sequencing primers: CD4-forward 5′-GGGCAAGAAAGACGCAAGC-3′, 5′-TTCTGCGGGCTCAGGTCC-3′, 5′-TAGTGTTCGGATTGACTGC-3′, 5′-TACCCAGGACCCTAAGC-3′, and CD4-reverse 5′-ATCTGCCTGGCCTCGTG-3′, 5′-TTTACCCCTTGGACTCC-3′, 5′-GGTTCACTTCCTGATGC-3′. These sequences were deposited as GenBank IDs MK184950–MK184953. CD4 sequences from all three sources were parsed by subspecies and combined. These sequences were then truncated to the first 1.311 nucleotides (of 1,377 total), partially omitting the C terminus of CD4, which was poorly resolved by next generation sequencing and could not be accurately phased (32). The resulting genotype information was used to phase SNPs using Phase 2.1 in DNAsp v5 (34–36). Glycosylation sites were predicted with NetNGlyc (67).
Receptor Expression Constructs and Site-Directed Mutagenesis.
Chimpanzee CD4 (allele #6; NM_001009043.1) was synthesized (IDT GeneBlock) and cloned into the pCR8/GW/TOPO TA plasmid (Thermo Fisher). Each chimpanzee CD4 allele was then reconstructed by sequential site-directed mutagenesis reactions using overlapping PCR primers containing the mutation of interest, following standard methods. Alleles were analyzed by Sanger sequencing to verify the desired nucleotide changes. CD4 inserts were then shuttled into a Gateway-converted pLPCX retroviral packaging vector (Clontech).
Env Clones.
HIV-1 Env AC10.0.29 was obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH (#11024) from David Montefiori and Feng Gao, Duke University School of Medicine, Durham, NC (68). HIV-1 Env BG505.W6M.B1 (69) was a gift from Julie Overbaugh, Fred Hutchinson Cancer Research Center, Seattle, WA. SIVcpz MB897 and EK505 molecular clones were a gift from Brandon Keele, Frederick National Laboratory for Cancer Research, Frederick, MD. The RevEnv cassettes were amplified by PCR using the following primer pairs: MB897 RevEnv (EF535994.1) forward 5′-tcgccaccATGGCAGGAAGAAGCGGAAGC-3′, reverse 5′- TTATAGCAAAGCTCTTTCTAAACCTTG-3′ and EK505 RevEnv (DQ373065.1) forward 5′- tcgccaccATGGCAGGAAGAAGCGGAGT-3′ and reverse 5′- TTATAAAAGTGCTCTTTCTAGACCTTGT-3′. PCR products were cloned into the pCR8/GW/TOPO TA plasmid (Thermo Fisher) and then shuttled into a Gateway-converted pCDNA3.1 mammalian expression vector (Invitrogen).
Cell Culture and Generation of Stable Cell Lines.
HEK293T (ATCC CRL-11268) were cultured in DMEM (Invitrogen) with 10% FBS, 2 mM l-glutamine, and 1% Pen Strep (complete medium) at 37 °C and 5% CO2. Hut78 cells (70) were cultured in RPMI-1640 complete medium (ATCC). Cf2Th cells stably expressing human CCR5 were cultured in DMEM complete supplemented with 250 μg/mL hygromycin. To produce retroviruses for transduction, HEK293T cells plated in antibiotic free media (1 × 106 cells per well in a six-well plate) were transfected with 2 μg of pLPCX transfer vector, 1 μg of pCS2-mGP (MLV gag/pol), and 0.2 μg of pC-VSV-G (VSV-G envelope) using a 3:1 ratio of TransIT 293 (Mirus) transfection reagent to DNA according to the manufacturer’s directions. Forty-eight hours posttransfection, retrovirus was collected, filtered through 0.22-μm cellulose acetate filters, and stored at −80 °C in single-use aliquots. Cf2Th cells were plated at 2 × 104 cells per well of a 12-well dish (∼15% confluent) and 24-h later, transduced with 500 μL of retroviral supernatant by spinoculation at 1,200 × g for 75 min in the presence of 5 μg/mL polybrene. The following day, the cells were placed in complete medium containing antibiotic (3 μg/mL puromycin, 250 μg/mL hygromycin) and cultured until stable outgrowth was noted (>1 wk). Stable cell lines were maintained indefinitely in selection media.
Single-Cycle HIV-1 Pseudovirus Infections.
To produce HIV-1ΔEnv-GFP reporter viruses, 13 × 106 HEK293T cells were seeded into a 15-cm dish in antibiotic free media and 24 h later transfected with 5.3 μg of Q23ΔEnv-GFP (group M backbone) (71) and 2.7 μg of Env plasmid. Forty-eight hours posttransfection, the cell supernatant was harvested, concentrated (∼100-fold) using Amicon Ultracel 100K filters (Millipore), and stored at −80 °C in single-use aliquots. Virus titers were determined by GFP FACS analysis, using a titration of different virus volumes on Cf2Th cells stably expressing CD4/CCR5. From the virus titration, the linear range of the infectivity curve was determined, and two points within the linear range were selected to calculate the mean virus titers in TDU/mL. Where viral infections were normalized to human CD4 (Fig. 4), viral titers were first determined on human CD4/CCR5-expressing cells, and then all cell lines were infected with an equivalent multiplicity of infection. Briefly, Cf2Th cells stably expressing CD4 and CCR5 were plated at 3 × 104 cells/well of a 48-well plate (∼60% confluent) 24 h before infection. The cells were then infected with HIV-1 pseudoviruses at a multiplicity of infection of ∼0.6 in the presence of 5 μg/mL of polybrene by spinoculation at 1,200 × g for 75 min. Forty-eight hours postinfection, the cells were harvested from the plate and fixed in 2% paraformaldehyde. The fixed cells were washed three times with PBS and resuspended in 50 μL FACS buffer (1× PBX buffer containing 2% FBS and 1 mM EDTA) and stained for 30 min at 4 °C with the following antibody mixture: 1:200 APC mouse α-human CD4 clone L200 (BD #551980) and 1:200 PeCy7 mouse α-human CD195 2D7 (CCR5) (BD #557752). Stained cells were analyzed on a CyAn ADP (Beckman Coulter) flow cytometer. Following live cell gating, CD4- and CCR5-expressing gates were drawn to capture cells with equivalent CD4 and CCR5 expression between all samples, and then the percent GFP+ cells was enumerated within the double-positive population. The data from ∼1 × 104 live cells was analyzed using FlowJo v10.
sCD4 Purification and Virus Neutralization Assays.
sCD4s were purified following a strategy from a previous study (72). Human, chimpanzee allele 6, and gorilla CD4 cloned into the pCR8 plasmid served as a template for sCD4 PCR. Briefly, the D1–D2 domain (nucleotide positions 75–603) was amplified by PCR and cloned into pHL-sec (73) (Addgene # 99845), which has been optimized for protein production in mammalian cells. Next, 13 × 106 293T cells (15-cm dishes) were transfected with 30 μg of sCD4 expression plasmids using TransIT-293 (Mirus), and cell supernatant containing secreted sCD4 was harvested at days 3 and 6 posttransfection. Cell supernatant was spun down at 1,200 × g for 5 min to remove cell debris and then filtered using a 0.45-μm cellulose acetate membrane. Cleared cell supernatant was then incubated with 1-mL bed volume of Ni-NTA agarose beads (Qiagen, #30210) equilibrated in wash buffer (25 mM Na3PO4 pH 7.4, 500 mM NaCl, 20 mM imidazole) for 2 h at 4° C. The mixture was then added to a gravity-flow chromatography column and washed with 50 mL of wash buffer. Bound protein was eluted in wash buffer containing 250 mM imidazole. Fractions containing sCD4 were pooled, washed 3× with PBS, and concentrated to 500 μL using an Amicon Ultra 15-mL centrifugal filter with a 10-kDa molecular mass cutoff (EMD Millipore, #UFC901008). The sample was purified to homogeneity on a Superdex 75 Increase 10/300 GL (GE Healthcare, #29-1487-21) in PBS. Purified samples were concentrated to 1 mg/mL using an Amicon Ultra 0.5-mL centrifugal filter with a 10-kDa molecular mass cutoff, aliquoted for single-use, and flash-frozen in liquid nitrogen.
PNGase F (New England Biolabs, #P0704S) treatment of sCD4 was carried out under denaturing (Fig. 2B) or nondenaturing (Fig. 5A) reaction conditions as described in the manufacturer’s protocol. For reactions carried out under denaturing conditions, 1 μg of sCD4 eluted from Ni-NTA beads was used as input and the entire reaction was analyzed using the TGX stain-free system (Bio-Rad). For reactions carried out under nondenaturing conditions, 1 mg of chimpanzee sCD4 eluted from gel filtration was incubated with 5,000 units of PNGase F at 30 °C for 16 h. This reaction was then subjected to the purification protocol described above to remove PNGase F and ensure that fully folded chimpanzee sCD4 was retained.
For the neutralization assays, each virus (normalized by equivalent TDU/mL) was incubated with serial twofold dilutions of sCD4 (range 20 μM to 1 μM final concentration) at 37 °C for 1 h. Following incubation, the sCD4 treated and untreated viruses were spinoculated (1,200 × g for 75 min) onto 1 × 104 TZM-bl cells plated in 96-well plates, in the presence of 5 μg/mL of polybrene. Following spinoculation, the cells were washed 3× with PBS and given fresh media. At 48 h postinfection, infectivity was measured by firefly luciferase assays following the manufacturers protocol (Promega). Luminescence was determined using the BMG Clariostar plate reader.
Glycosylation State of Full-Length CD4 by Western Blotting.
Human, chimpanzee, and gorilla CD4 cloned into the pCR8 plasmid served as a template for cloning into a pLPCX plasmid encoding a C-terminal HA-tag using Gibson assembly. The glycosylation state of HA-tagged CD4 proteins was then determined in human immortalized CD4+ T cells as follows: 1 μg of each HA-tagged CD4 and turbo GFP (nucleofection control; Evrogen #FP521) expression plasmids were nucleofected into Hut78 cells following the manufacturer’s directions (Amaxa cell line nucleofector Kit R, Program V-001). At 48 h postnucleofection, cells were lysed in Nonidet P-40 buffer [150 mM NaCl, 50 mM Tris⋅HCl pH 7.4, 1% Nonidet P-40, 1 mM DTT, 1 μL/mL Benzonase (Sigma-Aldrich #E1014), and protease inhibitor mixture (Sigma-Aldrich, #11873580001)] by resuspending the cell pellet and incubating on ice for 30 min. Cell lysate was cleared by centrifugation at maximum speed for 15 min. Whole-cell extracts were quantified using the Bradford assay and 10 μg was subjected to PNGase F treatment according to the manufacturer’s protocol (New England Biolabs, #P0704S). Whole-cell extracts and PNGase F treated whole cell extracts (5 μg per lane) were resolved on a 10% TGX Stain-free polyacrylamide gel (Bio-Rad, #1610182) by applying 180V until protein loading dye ran off the gel. Protein was transferred to a nitrocellulose membrane (GE Healthcare Biosciences, #10600002) using a wet transfer apparatus set at 100V for 45 min. The membrane was blocked with 3% milk in TBST (0.1%) for 30 min at room temperature. Primary antibodies were diluted in TBST (0.1% Tween-20) and incubated with the membrane for 30 min at room temperature (1:5,000 anti-HA-HRP Sigma-Aldrich #1201381900, 1:5,000 anti-TurboGFP Thermo Fisher Scientific #PA5-22688). After primary incubation, cells were washed 1 × 1 × 2 × 3 min in TBST (0.1% Tween-20). Secondary antibodies were diluted in 3% milk in TBST (0.1% Tween-20) and incubated with the membrane for 20 min at room temperature (1:10,000 anti-mouse-HRP Promega #W402B). After secondary incubation, membranes were washed 1 × 1 × 2 × 4 min in TBST (0.1% Tween-20), developed using ECL reagent (Sigma-Aldrich, #GERPN2232), and imaged on a Bio-Rad ChemiDoc Imaging System.
Expression of full-length CD4 was monitored in Cf2Th cells by Western blotting. Briefly, Cf2Th cells were pelleted, lysed in Nonidet P-40 buffer, and treated with PNGase F, as described above. Western blotting was carried out as described above using a 1:1,000 dilution of anti-CD4 antibody raised in rabbit (Abcam, #133616) along with a 1:10,000 dilution of a secondary anti-rabbit HRP (Promega, #W4011), and a 1:5,000 dilution of anti-Actin raised in mouse (Cell Signaling Technology, #3700S).
Gp120 Labeling and Microscale Thermophoresis.
HIV-1 AC10.29 gp120 Avi His recombinant protein was obtained from NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (cat #13055) from Xueling Wu, Aaron Diamond AIDS Research Center, New York, NY (50) and was labeled as follows: soluble Env in PBS was mixed with 1 M sodium bicarbonate to increase the pH above 8. Alexa Fluor 488 NHS Ester (ThermoFisher, #A20000) was resuspended in anhydrous DMSO and added at an Env:dye molar ratio of 1:10. Samples were wrapped in foil and incubated at room temperature for 1 h. Reactions were brought up to 500 μL with PBS, moved to an Amicon Ultra-0.5 mL centrifugal filter with a 50-kDa molecular mass cutoff (EMD Millipore, #UFC503008), and washed 4× in PBS to remove any unconjugated dye. Using the same centrifugal filter, labeled gp120 was then concentrated to 500 μg/mL
Microscale thermophoresis was performed using Monolith NT.115 (NanoTemper Technologies). The MST power and LED excitation power were both set at 60%. Measurements were carried out in buffer (PBS, 0.05% tween 20, and 0.5 mg/mL BSA) and standard capillaries. The concentration of Alexa488 labeled gp120 was kept constant at 25 nM and sCD4 ligands were serial-diluted twofold (15–16 points per curve). Binding curves were obtained by plotting the normalized fluorescence [Fnorm (%) = Fhot/Fcold; interval time = 20 s] versus the logarithm of sCD4 concentrations. The curve fit and binding affinity was calculated using the Kd model in the MO. Affinity Analysis software v2.3. All experiments were performed in at least duplicate and data plotted using Prism 8 for macOS.
Structural Comparison of Human and Chimpanzee CD4.
Human CD4 D1-D4 (PDB: 5U1F) was used to generate a structural prediction of chimpanzee CD4 allele 6 using SWISS-MODEL (74, 75). Human and the predicted chimpanzee CD4 allele six structures were compared in Mac PyMOL (The PyMOL Molecular Graphics System, v1.7.4.5 Schrödinger) revealing that these protein orthologs are highly homologous in structure (root mean square deviation = 0.14 Å).
Supplementary Material
Acknowledgments
We thank Colin Parrish for discussions about this manuscript. This work was supported by NIH Grants DP1 DA046108 and R01 AI137011 (to S.L.S.), F32 GM125442 and T32 AI007447-25 (to C.J.W.), F30 AI112277 (to A.C.S.), and Instrumentation Grant S10OD21603; and by the Burroughs Wellcome Fund (N.R.M. is a Postdoctoral Enrichment Program fellow; S.L.S. is an Investigator in the Pathogenesis of Infectious Disease).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MK184950–MK184953).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813909116/-/DCSupplemental.
References
- 1.Sharp P. M., Hahn B. H., Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1, a006841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao F., et al. , Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Keele B. F., et al. , Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon F., et al. , Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4, 1032–1037 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Compton A. A., Malik H. S., Emerman M., Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gifford R. J., et al. , A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 20362–20367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyerson N. R., Sawyer S. L., Two-stepping through time: Mammals and viruses. Trends Microbiol. 19, 286–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren C. J., Sawyer S. L., How host genetics dictates successful viral zoonosis. PLoS Biol. 17, e3000217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sironi M., Cagliani R., Forni D., Clerici M., Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 16, 224–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawyer S. L., Wu L. I., Emerman M., Malik H. S., Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A. 102, 2832–2837 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawyer S. L., Emerman M., Malik H. S., Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, E275 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNatt M. W., et al. , Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5, e1000300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z. D., Weinstock G., Gerstein M., Rapid evolution by positive Darwinian selection in T-cell antigen CD4 in primates. J. Mol. Evol. 66, 446–456 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Meyerson N. R., et al. , Positive selection of primate genes that promote HIV-1 replication. Virology 454–455, 291–298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei X., et al. , Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Stewart-Jones G. B. E., et al. , Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165, 813–826 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennemann N. J., et al. , Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. MBio 5, e00862-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walls A. C., et al. , Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 23, 899–905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradel-Tretheway B. G., Liu Q., Stone J. A., McInally S., Aguilar H. C., Novel functions of Hendra virus G N-glycans and comparisons to Nipah virus. J. Virol. 89, 7235–7247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sommerstein R., et al. , Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog. 11, e1005276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palermo L. M., Hueffer K., Parrish C. R., Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J. Virol. 77, 8915–8923 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palermo L. M., Hafenstein S. L., Parrish C. R., Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 80, 8482–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaelber J. T., et al. , Evolutionary reconstructions of the transferrin receptor of Caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog. 8, e1002666 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hueffer K., et al. , The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77, 1718–1726 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radoshitzky S. R., et al. , Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc. Natl. Acad. Sci. U.S.A. 105, 2664–2669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wentworth D. E., Holmes K. V., Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): Influence of N-linked glycosylation. J. Virol. 75, 9741–9752 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiden M. V., Farrell K., Wilson C. A., Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J. Virol. 68, 626–631 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marin M., Lavillette D., Kelly S. M., Kabat D., N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor functions. J. Virol. 77, 2936–2945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katzourakis A., Tristem M., Pybus O. G., Gifford R. J., Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U.S.A. 104, 6261–6265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp P. M., Shaw G. M., Hahn B. H., Simian immunodeficiency virus infection of chimpanzees. J. Virol. 79, 3891–3902 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santiago M. L., et al. , SIVcpz in wild chimpanzees. Science 295, 465 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Prado-Martinez J., et al. , Great ape genetic diversity and population history. Nature 499, 471–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hvilsom C., et al. , Genetic subspecies diversity of the chimpanzee CD4 virus-receptor gene. Genomics 92, 322–328 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Stephens M., Smith N. J., Donnelly P., A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Librado P., Rozas J., DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Stephens M., Donnelly P., A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arthos J., et al. , Identification of the residues in human CD4 critical for the binding of HIV. Cell 57, 469–481 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Ryu S. E., et al. , Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature 348, 419–426 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q., et al. , Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 24, 370–378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prince A. M., et al. , Lack of evidence for HIV type 1-related SIVcpz infection in captive and wild chimpanzees (Pan troglodytes verus) in West Africa. AIDS Res. Hum. Retroviruses 18, 657–660 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Van Heuverswyn F., et al. , Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368, 155–171 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Leendertz S. A. J., et al. , No evidence for transmission of SIVwrc from Western red colobus monkeys (Piliocolobus badius badius) to wild West African chimpanzees (Pan troglodytes verus) despite high exposure through hunting. BMC Microbiol. 11, 24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camerini D., Seed B., A CD4 domain important for HIV-mediated syncytium formation lies outside the virus binding site. Cell 60, 747–754 (1990). [DOI] [PubMed] [Google Scholar]
- 44.Parekh R. B., Tse A. G., Dwek R. A., Williams A. F., Rademacher T. W., Tissue-specific N-glycosylation, site-specific oligosaccharide patterns and lentil lectin recognition of rat Thy-1. EMBO J. 6, 1233–1244 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bibollet-Ruche F., et al. , Efficient SIVcpz replication in human lymphoid tissue requires viral matrix protein adaptation. J. Clin. Invest. 122, 1644–1652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takehisa J., et al. , Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J. Virol. 81, 7463–7475 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan Z., et al. , Recapitulating cross-species transmission of simian immunodeficiency virus SIVcpz to humans by using humanized BLT mice. J Virol 90:7728–7739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato K., et al. , Experimental adaptive evolution of simian immunodeficiency virus SIVcpz to pandemic human immunodeficiency virus type 1 by using a humanized mouse model. J. Virol. 92, e01905–e01917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bibollet-Ruche F., et al. , CD4 receptor diversity in chimpanzees protects against SIV infection. Proc. Natl. Acad. Sci. U.S.A. 116, 3229–3238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia M., Lu H., Markowitz M., Cheng-Mayer C., Wu X., Development of broadly neutralizing antibodies and their mapping by monomeric gp120 in human immunodeficiency virus type 1-infected humans and simian-human immunodeficiency virus SHIVSF162P3N-infected macaques. J. Virol. 90, 4017–4031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marmor M., et al. ; HIV Network for Prevention Trials Vaccine Preparedness Protocol Team , Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J. Acquir. Immune Defic. Syndr. 27, 472–481 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Huang Y., et al. , The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2, 1240–1243 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Kondoh T., et al. , Single-nucleotide polymorphisms in human NPC1 influence filovirus entry into cells. J. Infect. Dis. 218(Suppl 5):S397–S402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan H., et al. , Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J. Virol. 88, 3273–3284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worobey M., et al. , Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455, 661–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lusso P., et al. , CD4 is a critical component of the receptor for human herpesvirus 7: Interference with human immunodeficiency virus. Proc. Natl. Acad. Sci. U.S.A. 91, 3872–3876 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lavergne A., et al. , African great apes are naturally infected with roseoloviruses closely related to human herpesvirus 7. J. Virol. 88, 13212–13220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song R., Oren D. A., Franco D., Seaman M. S., Ho D. D., Strategic addition of an N-linked glycan to a monoclonal antibody improves its HIV-1-neutralizing activity. Nat. Biotechnol. 31, 1047–1052 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Julien J.-P., et al. , Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9, e1003342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malenbaum S. E., et al. , The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74, 11008–11016 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 (16 March 2013).
- 62.McKenna A., et al. , The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DePristo M. A., et al. , A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van der Auwera G. A., et al. , From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinf. 43:11.10.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ning Z., Cox A. J., Mullikin J. C., SSAHA: A fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lou D. I., et al. , Rapid evolution of BRCA1 and BRCA2 in humans and other primates. BMC Evol. Biol. 14, 155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gupta R., Jung E., Brunak S.,Prediction of N-glycosylation Sites in Human Proteins. Preparation (2004). http://www.cbs.dtu.dk/services/NetNGlyc/. Accessed 5 November 2018.
- 68.Li M., et al. , Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X., et al. , Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80, 835–844 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu L., Martin T. D., Vazeux R., Unutmaz D., KewalRamani V. N., Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J. Virol. 76, 5905–5914 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Humes D., Overbaugh J., Adaptation of subtype a human immunodeficiency virus type 1 envelope to pig-tailed macaque cells. J. Virol. 85, 4409–4420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morgan R. A., et al. , Retroviral vectors expressing soluble CD4: A potential gene therapy for AIDS. AIDS Res. Hum. Retroviruses 6, 183–191 (1990). [DOI] [PubMed] [Google Scholar]
- 73.Aricescu A. R., Lu W., Jones E. Y., A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bienert S., et al. , The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 45, D313–D319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.