Significance
The genomic landscape of metastatic castration-resistant prostate cancer (mCRPC) has been well-defined, but the association of genomic findings with patient clinical outcomes and with other characteristics including histology and transcriptional pathway activity remains poorly understood. Here, we describe comprehensive integrative analysis of genomic and transcriptomic profiles, histology, and clinical outcomes for 429 patients with mCRPC. Of all the molecular factors we examined, alterations in RB1 had the strongest association with poor outcome. Our study identifies molecularly defined groups of patients who may benefit from a more aggressive treatment approach, with the genomic and outcome data made available to the research community for further interrogation.
Keywords: castration-resistant prostate cancer, integrative genomics, clinical outcomes, biomarkers
Abstract
Heterogeneity in the genomic landscape of metastatic prostate cancer has become apparent through several comprehensive profiling efforts, but little is known about the impact of this heterogeneity on clinical outcome. Here, we report comprehensive genomic and transcriptomic analysis of 429 patients with metastatic castration-resistant prostate cancer (mCRPC) linked with longitudinal clinical outcomes, integrating findings from whole-exome, transcriptome, and histologic analysis. For 128 patients treated with a first-line next-generation androgen receptor signaling inhibitor (ARSI; abiraterone or enzalutamide), we examined the association of 18 recurrent DNA- and RNA-based genomic alterations, including androgen receptor (AR) variant expression, AR transcriptional output, and neuroendocrine expression signatures, with clinical outcomes. Of these, only RB1 alteration was significantly associated with poor survival, whereas alterations in RB1, AR, and TP53 were associated with shorter time on treatment with an ARSI. This large analysis integrating mCRPC genomics with histology and clinical outcomes identifies RB1 genomic alteration as a potent predictor of poor outcome, and is a community resource for further interrogation of clinical and molecular associations.
Several studies have described the genomic landscape of primary and metastatic castration-resistant prostate cancer (mCRPC), revealing distinct genomic subtypes in primary localized disease, including ETS fusion-positive and SPOP-mutated prostate cancer (1–5), and subsets of patients with advanced disease who harbor potentially clinically actionable alterations in their tumor or in the germline (4–6). Based on these findings, prospective trials are currently enrolling patients with defined genomic alterations, including PARP inhibitor studies for patients with alterations in BRCA2/1, ATM, and other DNA repair genes (NCT02952534, NCT02987543, NCT02854436), and AKT inhibitor studies for men with PI3K pathway alterations (NCT02525068, NCT03310541).
In addition, various genomic and histologic features of prostate cancer have been described as conferring a worse prognosis. Among these are the presence of neuroendocrine or small-cell characteristics in tumors, sometimes referred to as aggressive variant prostate cancer or neuroendocrine prostate cancer (7–9), the detection of androgen receptor (AR) splice variant 7 in circulating tumor cells (10, 11), and the presence of genomic alterations in TP53, RB1, DNA repair genes, AR, and PI3K pathway genes in circulating tumor DNA (12, 13). However, studies that comprehensively examine all of these characteristics—histology, genomics, and transcriptomics—and their association with outcomes in mCRPC are lacking.
Here, we expand a foundational genomic resource of mCRPC tumors (5) from 150 to 429 patients (444 tumors), and integrate the analysis of whole-exome sequencing, gene expression, and histopathology with clinical outcomes, including survival and time on treatment with the next-generation androgen signaling inhibitors (ARSIs) enzalutamide and abiraterone acetate to identify the most important prognostic markers in mCRPC within a single large multiinstitutional genomic dataset, with tumor- and patient-level data made available for additional correlative analyses.
Results
Clinical and Histopathologic Parameters.
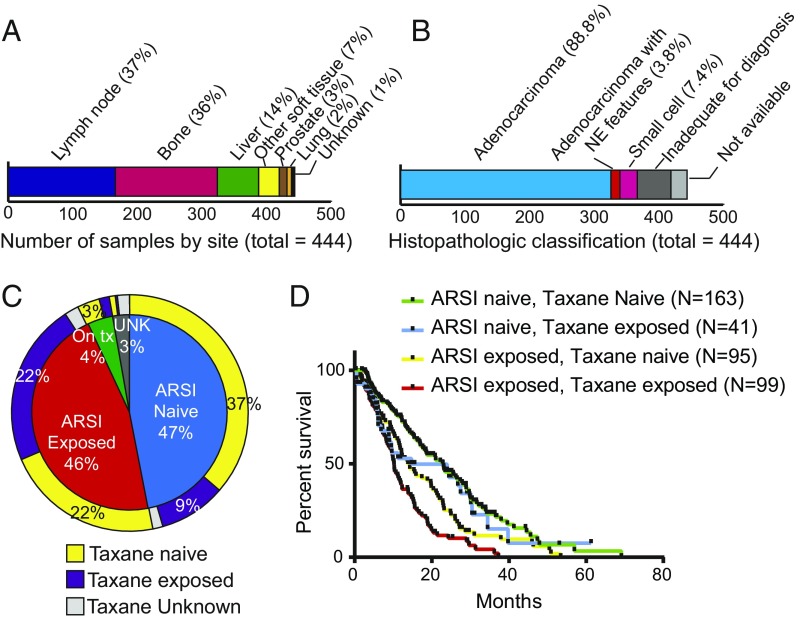
A total of 429 patients were enrolled at one of seven international consortium centers, all of whom underwent biopsy for the collection of mCRPC tissue as well as collection of blood for matched normal DNA extraction. Whole-exome sequencing was successfully performed on 444 tumors (some patients underwent multiple biopsies), and RNA sequencing (RNA-seq) was successfully performed on a subset of these (332 tumors from 323 patients). Of the 444 biopsies, 37% were lymph node, 36% were bone, and 14% were liver (Fig. 1A). Samples underwent central histopathologic review (Fig. 1B), revealing neuroendocrine (NE) or small-cell features in 11.2% (41 of 366) of evaluable cases, including from patients who were enrolled on a trial of the Aurora kinase A inhibitor alisertib (14). Median age at diagnosis with prostate cancer was 61 y, median age at biopsy of the profiled sample was 67 y, and median overall survival from the time of biopsy was 16 mo (SI Appendix, Table S1). Samples were balanced for exposure status to ARSIs (47% ARSI naive, 46% previously exposed) (Fig. 1C). Sixty-three percent of samples were acquired before exposure to a taxane. Patients who were naive to both ARSI and taxane at the time of biopsy had the longest median overall survival from the date of biopsy, whereas patients previously treated with both an ARSI and a taxane had the shortest (Fig. 1D), consistent with their more advanced disease state at the time of tissue acquisition. Of note, time on treatment with a first-line ARSI was highly associated with overall survival from the start of first-line therapy (Kendall’s τ = 0.65).
Fig. 1.
Overview of sample and patient characteristics for 444 tumors from 429 patients with mCRPC. (A) Site of mCRPC tumors profiled. (B) Histopathologic classification of profiled tumors. Tumors were classified by central review as adenocarcinoma, pure small-cell/neuroendocrine cancer, adenocarcinoma with neuroendocrine features (also included mixed acinar/neuroendocrine carcinoma), or could not be classified due to scant material or no tumor visible on the slides that were available for review despite successful sequencing. (C) Patient exposure status to next-generation AR signaling inhibitors (abiraterone acetate, enzalutamide, or ARN509) and to taxanes at the time of biopsy for the 444 profiled tumors. (D) Overall survival (OS) from the date of biopsy of the profiled tumor. OS was longer for tumors from ARSI- and taxane-naive patients compared with patients who had received an ARSI before the biopsy (P < 0.01, log-rank test). Survival was shortest when the patient had received both an ARSI and taxane chemotherapy at the time of biopsy.
Landscape of Genomic Alterations.
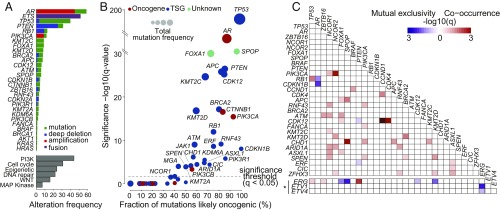
The frequency of genomic alterations was similar to that reported in prior cohorts (4, 5, 15), with AR, ETS family transcription factors including ERG and ETV1, TP53, and PTEN and RB1 emerging as the most commonly altered genes (Fig. 2A). Likewise, alterations in biological pathways (SI Appendix, Table S2) were also consistent with prior reports, with a significant (>20%) subset of patients harboring at least one alteration in a PI3K, cell-cycle, epigenetic, or DNA repair pathway gene. Single-nucleotide variants (SNVs) in the most frequently altered genes were found to be likely oncogenic (16) in the majority of cases (Fig. 2B), with a high fraction of oncogenic mutations in AR, TP53, PIK3CA, BRCA2, PTEN, APC, and CDK12. Mutations in ATM were predicted to be likely oncogenic in nearly 60% of cases, with the rest being missense mutations of unknown significance.
Fig. 2.
Landscape of genomic alterations in 444 tumors from 429 patients with mCRPC. (A) Genomic alterations of potential biologic relevance by frequency. (A, Top) Frequency of alteration by gene. Frequency of ETS gene alterations applies to the subset of 323 patients who underwent tumor RNA sequencing, where fusion status could be determined. (A, Bottom) Frequency of alteration by pathway (SI Appendix, Table S2). (B) Fraction of SNVs considered to be likely oncogenic for genes harboring mutations. Genes colored in red are putative oncogenes, genes colored in blue are putative tumor suppressor genes (TSGs), and genes colored green are currently unknown. (C) Co-occurrence or mutual exclusivity between the most commonly altered genes. The significance of the relationship is represented by gradient. Relationships with P value < 0.05 with multiple hypothesis correction are shown. Associations involving ETS genes (*) apply only to cases where RNA-sequencing data are available.
The large size of the dataset allowed for a comprehensive search for genomic alterations that are co-occurring or mutually exclusive (Fig. 2C). As expected, we found mutual exclusivity between alterations in genes of the ETS family (e.g., ERG and ETV1), and between alterations in ERG and SPOP or FOXA1, which represent distinct genomic subsets of prostate cancer (1, 3). Alterations in ERG and PTEN were co-occurring, in line with their synergistic role in promoting oncogenesis in mouse models of prostate cancer (17). We also confirmed co-occurrence between alterations in TP53 and RB1, known to occur at high frequency in neuroendocrine cancers (7, 9), and to confer aggressive behavior in prostate cancer models (18, 19). Interestingly, RB1 alteration had a tendency toward mutual exclusivity with alterations in AR. CHD1 alterations also tended to co-occur with SPOP mutations (3). We found strong co-occurrence of loss-of-function alterations in CDK12, a gene implicated in the control of genomic stability (20) whose inactivation in prostate cancer is associated with focal tandem duplications (21–23), with amplification of the cell-cycle genes CCND1 and CDK4, raising the possibility of vulnerability to CDK4/6 inhibitors for CDK12-mutated tumors. Conversely, while genomic alterations in RB1 and BRCA2, both located on chromosome 13q, 16 Mb apart, had a tendency toward co-occurrence, this association did not reach statistical significance.
Association of Genomic Alterations with Clinical Outcomes.
A key novelty of this dataset is the opportunity to correlate contemporaneously obtained comprehensive genomic profiles with clinical outcome. We focused our analysis on 18 of the most commonly altered genes and pathways. For clinical outcome, we restricted the analysis to those patients who were taxane-naive and initiating therapy with a first-line ARSI for mCRPC (n = 128).
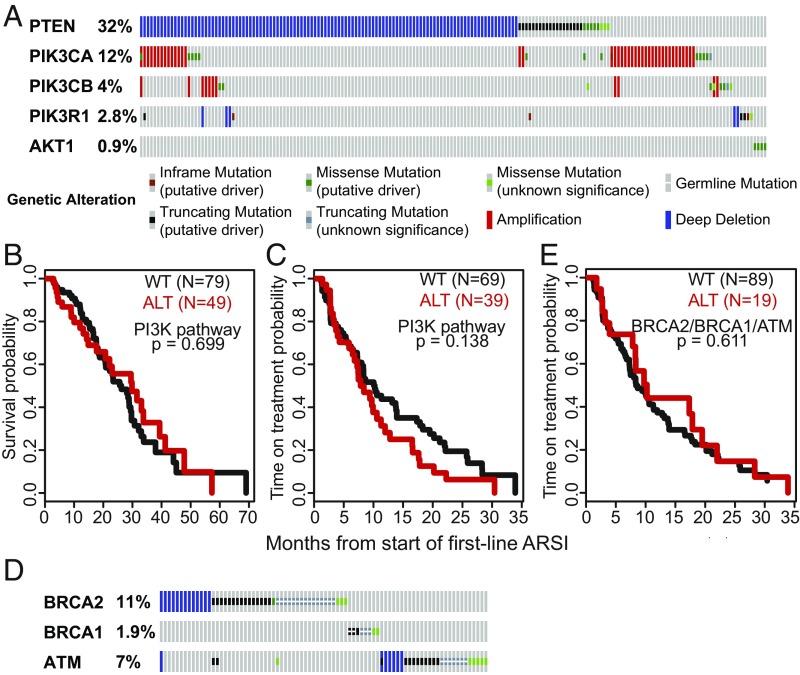
We examined the association of genomic alterations with overall survival from the start of a first-line ARSI (n = 128) and time on treatment with a first-line ARSI (subset of n = 108 patients who received the ARSI without another concurrent therapy) in univariate analysis (Table 1). In this analysis, genomic alterations in the PI3K pathway and its component genes (Fig. 3A) were not significantly associated with either time on therapy with ARSIs or with overall survival (Fig. 3 B and C), unlike prior cell-free DNA (cfDNA)-based analysis (13). Furthermore, we explored the association of genomic alterations in the DNA repair genes BRCA2, BRCA1, and ATM (Fig. 3D) with clinical outcomes, given prior conflicting reports of prognostic significance of these alterations (13, 24–26). We again found no association between alterations in these genes and time on treatment or overall survival (Fig. 3E and Table 1). Notably, we found an association between SPOP mutation and longer time on treatment with a first-line ARSI (SI Appendix, Fig. S1), consistent with prior data showing enrichment of SPOP mutations in earlier disease relative to mCRPC (1, 4) and favorable prognosis of SPOP-mutated tumors (27, 28), though this did not translate into a survival benefit. Alterations in AR and TP53 were associated with a shorter time on an ARSI (Figs. 4D and 5C), though there was no association with overall survival (Table 1). Overall, genomic alterations in RB1 showed the strongest discrimination for a shorter time on an ARSI and survival, with concordance probability estimates (CPEs) of 0.82 and 0.77, respectively (Fig. 5 A and B and Table 1). Aneuploid chromosomal status was associated with worse overall survival and time on treatment compared with diploid status (SI Appendix, Fig. S2).
Table 1.
Association of common genomic alteration with overall survival and time on treatment with first-line ARSI
| Gene/pathway alteration | Univariate P value for survival from first-line ARSI (CPE, n = 128 or as indicated) | Univariate P value for time on treatment with first-line ARSI (CPE, n = 108 or as indicated) |
| RB1 | 0.002 (CPE 0.768) | <0.001 (CPE 0.818) |
| TP53 | 0.072 (CPE 0.605) | 0.046 (CPE 0.609) |
| WNT pathway | 0.115 | 0.153 |
| ETS fusion | 0.159 | 0.206 |
| APC | 0.255 | 0.167 |
| CTNNB1 | 0.274 | 0.448 |
| ATM | 0.331 | 0.850 |
| BRCA2 | 0.327 | 0.418 |
| BRCA2/BRCA1/ATM | 0.495 | 0.611 |
| AKT1 | 0.558 | 0.053 |
| RNF43 | 0.614 | 0.844 |
| AR | 0.658 | 0.005 (CPE 0.651) |
| PTEN | 0.676 | 0.412 |
| PI3K pathway | 0.699 | 0.138 |
| PIK3CA | 0.716 | 0.165 |
| PIK3R1 | 0.752 | 0.892 |
| PIK3CB | 0.799 | 0.277 |
| BRCA1 | 0.809 | 0.998 |
| NEPC score | 0.218 (n = 99) | 0.930 (n = 80) |
| AR signaling score | 0.847 (n = 99) | 0.847 (n = 80) |
| RB1 loss score | <0.001 (n = 99) | 0.014 (n = 80) |
| CCP score | 0.002 (n = 99) | 0.045 (n = 80) |
| AR-V7 SRPM | 0.524 (n = 75) | 0.329 (n = 56) |
| AR-V7/ARpromoter1-2 | 0.475 (n = 75) | 0.378 (n = 56) |
| AR-V3 SRPM | 0.444 (n = 75) | 0.077 (n = 56) |
Univariate log-rank analysis for association of common genomic alterations with survival from the start of a first-line ARSI for mCRPC (n = 128), and with time on treatment with a first-line ARSI for mCRPC (n = 108 patients who received a first-line ARSI as monotherapy). Where indicated, analysis was limited to a subset of patients who had RNA-sequencing data either from polyA libraries or both polyA and capture libraries. P < 0.05 are highlighted in bold.
Fig. 3.
Alteration in PI3K and homologous recombination repair genes and association with clinical outcomes. (A) Oncoprint of genomic alterations in PI3K pathway genes. (B and C) Kaplan–Meier analysis showing overall survival (B) and time on treatment with a first-line ARSI (C) in PI3K pathway altered (red) versus unaltered (black) tumors. (D) Oncoprint of genomic alterations in BRCA2, BRCA1, and ATM. (E) Kaplan–Meier analysis showing time on treatment with a first-line ARSI in BRCA2/1/ATM–altered (homozygous deletion or somatic or pathogenic germline mutation) (red) versus unaltered (black) tumors.
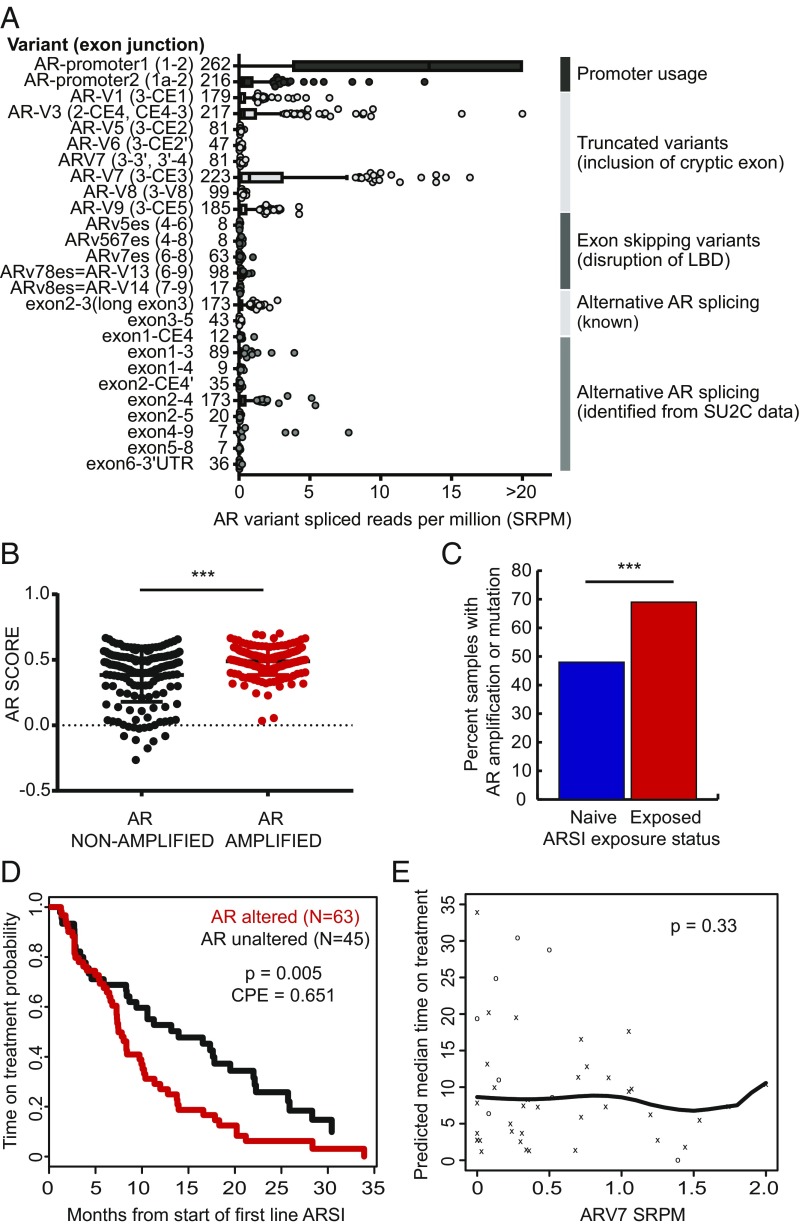
Fig. 4.
Androgen receptor alterations and outcome. (A) AR splice variant landscape. LBD, ligand binding domain. (B) AR pathway expression score in AR-amplified (n = 168) versus nonamplified (n = 159) tumors. ***P < 0.001. (C) AR amplification frequency in ARSI-naive versus exposed tumors. (D) Kaplan–Meier analysis showing time on treatment with a first-line ARSI in AR-amplified versus nonamplified tumors. (E) Association between ARV7 expression and time on treatment with a first-line ARSI. o, censored event; x, off-treatment event.
Fig. 5.
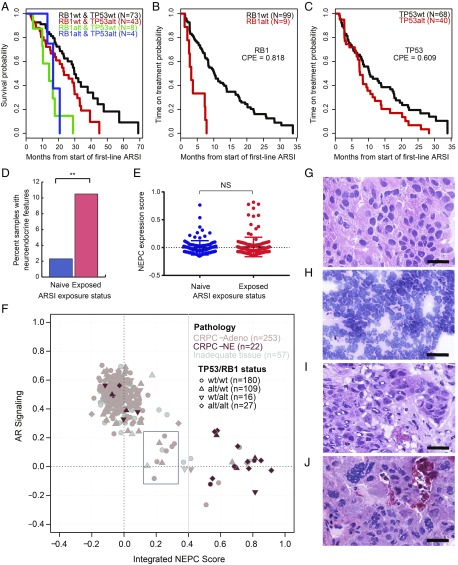
Integrative analysis incorporating histopathology, transcript-based assessment of AR signaling and NEPC score, TP53 and RB1 genomic status, and clinical outcomes. (A) Kaplan–Meier analysis showing overall survival from the start of a first-line ARSI versus genomic status for TP53 and RB1 in n = 128 patients who received a first-line ARSI and underwent tissue profiling at baseline (before or within 90 d of therapy start). (B and C) Kaplan–Meier analysis showing time on treatment with a first-line ARSI by genomic status for RB1 and TP53. P values were generated from the log-rank statistic. (D) Frequency of histopathologic neuroendocrine features in pre- versus post-ARSI samples, among patients who received an ARSI at some point during their treatment history. Patients who were not reported to have received an ARSI at any point were excluded. **P < 0.01. (E) NEPC expression score in pre- (n = 118) versus post- (n = 152) ARSI samples, as in D. NS, not significant. (F) AR and NEPC expression scores, histopathology (CRPC-Adeno, no NE features; CRPC-NE, histopathologic NE features) and TP53/RB1 genomic status (circle, wild type for both; diamond, both altered) for the 332 tumors with RNA-sequencing data. Ten cases (3%, blue box) had low AR and low NEPC expression scores. (G–J) Representative cases of CRPC-Adeno (G), CRPC-NE, small-cell type (H), CRPC-Adeno showing intermediate transcriptomic scores (I), and CRPC-Adeno showing a high NEPC score/low AR signaling score (J). Tumors represented in I and J were noted to have distinct nuclear features, including various degrees of nuclear pleomorphism, irregular nuclear membrane contours, and/or high mitotic activity. (Scale bars, 25 μm.)
Androgen Receptor Alterations.
We confirmed a high frequency of genomic alterations in AR, namely amplifications and mutations, consistent with prior reports (5, 12) (Fig. 2A). Using RNA-seq data, we also identified splice variants in AR, most commonly AR splice variant 7 (AR-V7) and variant 3 (AR-V3), both a product of splicing with cryptic exons, similar to our prior report (5) (Fig. 4A). Genomic alterations (amplification and mutation) were associated with an increased AR expression score, consistent with increased AR output (Fig. 4B). Furthermore, genomic alterations in AR were detected at higher frequency postexposure to the ARSIs compared with ARSI-naive tumors (Fig. 4C), suggesting an association with resistance to these next-generation AR-targeting agents. Consistent with this, we found an association of AR genomic alterations with a shorter time on treatment with a first-line ARSI (Fig. 4D) but not with overall survival from the start of a first-line ARSI (Table 1). Given prior data showing a strong association between AR-V7 expression in circulating tumor cells (CTCs) and clinical outcomes (10, 11), we examined the association of AR-V7 expression and outcomes in our tumor dataset. AR-V7 levels were increased in tumors exposed to taxanes and to ARSI therapy (SI Appendix, Fig. S3). However, there was no association between AR-V7 expression in tumors with either time on a first-line ARSI or overall survival (Fig. 4E and Table 1). This was true even when deriving an AR-V7 expression cutpoint that produced the maximum log-rank test (AR-V7 cutpoint for survival 1.92, P = 0.62). Similar results were observed for time on treatment, AR-V7/AR promoter 1:2 ratio, and AR-V3 (Table 1).
Integrative Analysis of Histopathology, Genomics, and Expression.
There is growing recognition that a subset of CRPC patients have a more fulminant clinical course—typically characterized by rapidly progressive visceral metastasis (versus bone), relatively low serum prostate-specific antigen (PSA) levels, and variable expression of neuroendocrine markers such as synaptophysin or chromogranin (29). However, there is a lack of consensus on how to precisely define this clinical state, particularly since the frequency of histologically defined neuroendocrine prostate cancer varies widely in different cohorts. Furthermore, there is increasing recognition that patients can develop AR-negative (PSA-low) disease that is histologically negative for neuroendocrine marker expression (30). RNA-based expression signatures have been proposed as a potential alternative diagnostic strategy to define this distinct clinical state (8, 30, 31). The availability of matched histology, whole-exome, and RNA-seq data from this cohort provides an opportunity to explore this question through an unbiased integrative approach.
As previously noted, 11.2% of tumors in our dataset had evidence of NE features on histopathologic review (Fig. 1D). Among patients who received treatment with an ARSI during their disease course, tumors with histopathologic NE features were enriched postexposure to an ARSI (10.5%) compared with ARSI-naive tumors (2.3%) (Fig. 5D). Transcript-based NE score was not significantly different between the two groups, but a subset of tumors in the post-ARSI setting displayed a higher NE expression score (Fig. 5E). Of note, unsupervised gene expression clustering identified a distinct cluster of tumors with higher NE expression score, in line with prior reports and independent of site of metastasis (SI Appendix, Fig. S4) (31).
We performed an integrative analysis incorporating histology, expression-based AR signaling and NE scores, and RB1/TP53 genomic status (7, 32) in all tumors where RNA-sequencing data were available (Fig. 5F). As expected, there was an inverse correlation between cases with a high AR signaling score and cases with a high NE score. We identified three groups based on the expression signatures. The first and largest group, characterized by high AR signaling and low NE score, consisted predominantly (86%) of adenocarcinomas without NE histologic features (Fig. 5G). The second group, demonstrating intermediate NE and AR scores (n = 17), included cases classified histologically as adenocarcinoma (59%) and cases that were “inadequate for diagnosis” (41%). The third group, demonstrating high NE score and low AR signaling, consisted predominantly (74%) of tumors harboring histologic NE features (Fig. 5H).
Although there was an association between NE expression score, histologic NE features, and RB1/TP53 loss (SI Appendix, Fig. S5), concordance between these characteristics was imperfect (Fig. 5F). We posited that some cases with discrepancy between pathology and transcriptomic classification may demonstrate a distinct morphology. To address this, a second consensus review of those cases was performed by three study pathologists. We confirmed that all discrepant cases from group 3 (high NE score but showing adenocarcinoma histology) and all cases from group 2 (intermediate AR and NE scores) displayed adenocarcinoma features. However, we noted distinct nuclear features in about half of these cases, including various degrees of nuclear pleomorphism, irregular nuclear membrane contours, and/or high mitotic activity (Fig. 5 I and J). Of note, within group 2, 10 cases (3% of total; Fig. 5F, box) had low AR and low NEPC expression scores. Of these, all histologically evaluable cases showed adenocarcinoma histology, and the majority harbored atypical nuclear features.
Given incomplete concordance between these histopathologic, genomic, and expression characteristics, we asked which of these features was most associated with clinical outcome. While there were insufficient histopathologic NE cases to make this determination, NE transcriptional score was not significantly associated with time on a first-line ARSI or overall survival (Table 1 and SI Appendix, Fig. S6). Of all molecular characteristics examined in a multivariate analysis, RB1 alteration emerged as the only variable strongly associated with survival (relative risk 3.31) and with time on treatment with a first-line ARSI (relative risk 6.56) (Table 2). Consistent with this, expression scores for RB1 loss and cell-cycle progression (CCP) were both associated with worse survival and time on treatment with a first-line ARSI (Table 1 and SI Appendix, Fig. S7).
Table 2.
Multivariate analysis evaluating the association of common genomic alterations with overall survival and time on treatment with first-line ARSI
| Clinical outcome | Gene alteration(s) | Multivariate relative risk (95% CI for RR) |
| Overall survival from start of first-line ARSI | RB1− | 1 |
| RB1+ | 3.31 (1.64, 6.67) | |
| Time on treatment with first-line ARSI | RB1− and AR− | 1 |
| RB1− and AR+ | 1.86 (1.18, 2.95) | |
| RB1+ | 6.56 (2.94, 14.62) |
Common genomic alterations listed in Table 1 were included in this analysis. Only significant associations are shown. CI, confidence interval; RR, relative risk.
Discussion
The landscape of genomic alterations in mCRPC has been established, with a subset of patients harboring potentially actionable alterations that are currently being explored in targeted prospective clinical trials. However, the majority of genomic alterations in prostate cancer do not yet have clear clinical applicability. Some studies have associated specific genomic or molecular features with clinical outcomes, though this has generally been performed in cell-free DNA or circulating tumor cells. Here, we present an integrative analysis of genomic alterations with expression and histologic assessment in tumors from patients with mCRPC, representing the clinical spectrum of advanced disease, with tissue collected pre- and posttreatment with ARSIs and taxanes.
Importantly, we find that RB1 loss is the only genomic factor that is significantly associated with both survival and time on ARSI therapy in mCRPC. It is worth noting that the association was strong despite likely underestimation of RB1 loss by examining genomic homozygous loss alone, as RB1 loss has also been shown to occur epigenetically, through structural genomic events like tandem duplication of partial exons, and focally, by immunohistochemistry (33). TP53 and AR alteration were also associated with shorter duration of ARSI therapy, though the association was not as strong as for RB1 loss and did not extend to survival. The AR findings suggest that AR targeting with the ARSIs abiraterone acetate and enzalutamide may be incomplete, and that further targeting of the protein may be clinically beneficial in patients who develop resistance to these agents.
We found no association between alterations in PI3K pathway genes or alterations in the DNA damage repair genes BRCA2, BRCA1, and ATM with overall survival and time on treatment with an ARSI. This is in contrast to a prior study that found an association between alterations in BRCA2, BRCA1, and ATM detected in cfDNA with both response to ARSIs and with survival (13). The differing conclusions may be related to differences between tumor and cfDNA profiling, potential difficulty in detecting homozygous loss in cfDNA relative to a tumor, and a possible bias introduced by requiring detectable cfDNA, though the authors accounted for cfDNA detection in a multivariate analysis. Furthermore, data for the prognostic role for BRCA2, BRCA1, and ATM alterations were previously conflicting, with another study showing better prognosis for tumors with alterations in these genes (25) and a third study showing no impact for the presence of germline DNA repair gene alterations on outcomes in mCRPC (26). Notably, we also found no association between AR-V7 in tumors and clinical outcomes, in contrast to prior CTC-based studies (10, 11). This finding requires further exploration of the concordance between AR-V7 expression in tissue versus CTCs, and the biological significance of AR-V7 detection in these contexts. Our findings suggest that AR-V7 RNA detection in tumors may have limited clinical utility. Overall, while profiling of cfDNA and CTCs offers advantages over tumor profiling, including ease of access and the ability to capture genomic heterogeneity (34, 35), blood-based profiling may be limited by lower sensitivity of detection and may reflect the fact that CTCs and cfDNA are generally detectable in patients with more advanced or aggressive disease. We recognize that association with clinical outcomes was performed in a subset of 128 patients who were initiating first-line therapy for mCRPC with an ARSI, and that a larger cohort could reveal additional associations. Nonetheless, the size of our cohort meets or exceeds previously reported datasets, and it is unclear if smaller differences in outcomes would be considered clinically meaningful.
We found that neuroendocrine histology, generally viewed as conferring more aggressive clinical behavior, is more frequent postexposure to ARSIs (10.5%), though at a lower frequency than recently reported in another study (17%) (8), despite inclusion in our cohort of patients with neuroendocrine features from a clinical trial of alisertib (14). Prostate cancers with histologic neuroendocrine differentiation (36) typically have alterations in TP53 and/or RB1, high neuroendocrine expression score, and low AR signaling score, though concordance is not complete. This is not surprising, given the complexity of defining histologic neuroendocrine differentiation, which relies on the identification of a variety of characteristic features (29). Some tumors with discordant histologic and molecular classification may be in transition from adenocarcinoma to NE differentiation, and displayed distinct nuclear features that may suggest such a transition. In such cases, paraffin-embedded tissue may aid in the classification by allowing for further examination of histomorphologic features and immunohistochemical staining of markers.
The size of our whole-exome sequencing dataset allows for genomic association analysis that was not previously possible. Through this analysis, we identified co-occurrence of alterations in CDK12, recently shown to confer immunogenic potential, and alterations in cell-cycle genes CDK4 and CCND1 (23), pointing toward a possible role for combination immune checkpoint blockade and CDK4/6 inhibition in clinical trials. Further laboratory studies will be needed to explore this and other potential biological interactions identified through genomic analysis.
In summary, we present an integrative analysis of genomic alterations, gene expression, histopathology, and clinical outcomes in the largest single mCRPC dataset to date, with the data made available to the research community for interrogation of genomic features in relation to outcomes. We find that RB1 loss is the molecular factor most strongly associated with poor clinical outcomes in a contemporary cohort, highlighting the need for further investigation into mechanisms of resistance to AR therapies induced by loss of RB, and potential therapeutic strategies targeting this mechanism.
Methods
Patients and Samples.
Subjects with mCRPC who were receiving standard-of-care therapy or treatment in a clinical trial [including trials combining AR therapies with other agents, a trial of the PARP inhibitor olaparib (37), and a trial of the Aurora kinase A inhibitor alisertib in patients with neuroendocrine features (14)] and who had disease amenable to biopsy under radiographic guidance were considered for inclusion at one of seven SU2C-PCF (Stand Up to Cancer/Prostate Cancer Foundation) International Prostate Cancer Dream Team consortium sites (Dana-Farber Cancer Institute, Karmanos Cancer Institute, Memorial Sloan Kettering Cancer Center, Royal Marsden, University of Michigan, University of Washington, and Weill Cornell Medicine) (5). All subjects included in this study provided written consent for research use of tumor tissue with institutional review board approvals or appropriate waivers (Office of Human Research Studies at the Dana-Farber Cancer Institute, Wayne State University Institutional Review Board, Memorial Sloan Kettering Cancer Center Institutional Review Board/Privacy Board, Royal Marsden Ethics Committee, University of Michigan Medical School Institutional Review Board, University of Washington Institutional Review Board, and Weill Cornell Medicine Institutional Review Board). Clinical data, including treatment history, duration of therapy, and survival, were collected using a web-based electronic data capture. All samples and clinical data were deidentified.
Histopathology.
Pathology for all available cases was reviewed centrally by three board-certified pathologists with expertise in prostate cancer pathology, who were blinded to clinical and genomic data. Review was conducted on H&E-stained frozen sections, allowing for review of the exact material that was used for nucleic acid extraction. Each slide was assessed for the ability to make a diagnosis based on the quality of the sample and presence of tumor cells; cases in which a specific pathology diagnosis could not be called were classified as inadequate for diagnosis. The remaining cases (n = 366) were classified according to a previously published system (36), by consensus in the event that all three pathologists did not agree. There was no additional material for paraffin embedding, immunohistochemistry, or other confirmatory studies.
Sequencing and Analysis.
Flash-frozen needle biopsies and matched normal samples underwent nucleic acid extraction as previously described (5). Extracted DNA underwent whole-exome library construction and somatic mutation analysis as previously described. BAM files were aligned to the hg19 human genome build. Copy-number aberrations were quantified and reported for each gene as previously described (38, 39). Amplifications and homozygous deletions for a set of 20 genes previously implicated in prostate cancer (SI Appendix, Table S3) underwent further confirmatory review of segmentation files. Annotation of known or likely oncogenic SNVs was performed using the OncoKB platform (16).
Transcriptome libraries were prepared as previously described (5), using polyA+ RNA isolation, or captured using Agilent SureSelect Human All Exon V4 reagents, or in some cases using both polyA and capture methods. Library quality assessment and sequencing were performed as previously described. Paired-end transcriptome-sequencing reads were aligned to the human reference genome (GRCh38) using STAR (40). Gene expression as fragments per kilobase of exon per million fragments mapped (FPKMs) was determined using featureCounts against protein-coding genes from the Gencode v26 reference. Fusions in ETS genes (ERG, ETV1, ETV4, ETV5, FLI1) and RAF1/BRAF were detected using CODAC (41) and assessed manually in all cases where RNA-sequencing data were available. In addition, the presence of AR splice variants was quantified as the number of reads across specific splice junctions in splice reads per million (SRPMs) and as the ratio of reads across a specific splice junction to the sum of AR promoter 1 and promoter 2 reads (a surrogate of total AR expression), separately for polyA and capture libraries.
NEPC and AR signaling scores were computed by the Pearson’s correlation coefficient between the log2-transformed FPKM values of each score’s gene list and a reference gene expression vector, as previously described (7, 32). CCP and RB loss scores were computed by the average (i.e., mean) Z score-transformed expression levels across each score’s gene list, as previously described (42, 43). A high correlation (R ≥ 0.95, P < 0.001, Pearson’s correlation test) was noted between scores derived from polyA versus capture RNA-sequencing libraries (SI Appendix, Fig. S8), allowing for joint analysis of samples sequenced with either library construction method.
All data from SNV, copy-number, and expression analysis as well as clinical characteristics and outcomes measures (Dataset S1) have been made available in cBioPortal (44) (www.cbioportal.org), and have been deposited in GitHub, https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019.
Statistics and Genomic Association with Outcomes.
Fisher’s exact tests and unpaired t tests were performed in R (3.5.0) and GraphPad Prism software as indicated. For the analysis shown in Fig. 2B, enrichment analysis using a binomial distribution test was performed as previously described (45) to identify genes that had a significant fraction of known or likely oncogenic alterations (as defined by OncoKB) among all identified SNVs. Multiple hypothesis test correction was applied using the Benjamini–Hochberg method, with q values of <0.05 considered significant for enrichment of oncogenic mutations among all SNVs identified for a gene. Kaplan–Meier analysis was performed from time of biopsy to death for all samples. Overall survival analysis was performed for the n = 128 subjects who received an ARSI (abiraterone, enzalutamide, or apalutamide) in the first-line setting before a taxane, either alone or in combination with another agent in a clinical trial, and where the profiled tissue was obtained before the start of therapy or within 90 d after starting first-line therapy. Time on treatment analysis was evaluated for a subset of n = 108 patients (of the 128 above), who received an ARSI in the first-line setting without another agent, so as not to confound the interpretation of response to the ARSI. P values for individual (univariate) association tests between genomic status and survival/time on treatment were generated from the log-rank statistic. In cases where a data-driven threshold value was used to determine the genomic status, the P value was computed from the maximum log-rank statistic. When a genomic class contained a small number of events, the P value was produced using a permutation log-rank test. A concordance probability estimate provided a metric to assess the level of separation between the Kaplan–Meier curves and is reported in relevant cases. Multivariate analyses were performed for the association of common genomic characteristics shown in Table 1 with overall survival and time on a first-line ARSI, with relative risk reported based on the Cox proportional hazards model. Kendall’s tau, derived from the Clayton copula, was used to evaluate the level of association between the time on therapy end point and overall survival.
Supplementary Material
Acknowledgments
We thank the affected individuals who participated in this study. We thank the following for their support in the conduct of the study: Jyoti Athanikar, Karen Giles, Ritika Kundra, Brigit McLaughlin, and Liangzuo (Tony) Ren. This work was supported by the Prostate Cancer Foundation and by a Stand Up to Cancer Prostate Cancer Dream Team research grant. Stand Up to Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research Award SU2C-AACR-DT0712. Additional support was provided by Prostate Cancer Foundation Young Investigator Awards (to W.A., J.M., and N.S.); Department of Defense Prostate Cancer Research Program Awards W81XWH-17-1-0124 (to W.A.), W81XWH-09-1-0147 (PCCTC), PC170510 and PC170503P2 (to C.C.P.), and W81XWH-17-1-0380 (to N.D.S.); NCI Prostate Cancer SPORE Awards P50CA186786 (University of Michigan), P50CA092629 (MSKCC), P50CA090381 (DFCI), P50CA211024 (WCMC), and P50CA097186 (University of Washington); NCI Cancer Center Award P30 CA008748 (MSKCC); NIH Award R01CA125612 (to M.A.R.); European Research Council Consolidator Grant 648670 (to F.D.); and the Nuovo-Soldati Foundation (J.C.). The RM and ICR team is supported by the Movember Foundation and Prostate Cancer UK, PCF, the ECMC network from Cancer Research UK, the Department of Health in the UK, and BRC grant funding.
Footnotes
Conflict of interest statement: W.A. has consulted for Clovis Oncology, Janssen, ORIC Pharmaceuticals, and MORE Health; has received honorarium from Caret Healthcare; has received travel funds from Clovis Oncology, ORIC Pharmaceuticals, and GlaxoSmithKline; and has received research funding from AstraZeneca, Clovis Oncology, Zenith Epigenetics, and GlaxoSmithKline. G.H., received research funding from Janssen. J.A. is currently employed at AstraZeneca. E.M.V.A. has consulted for Tango Therapeutics, Genome Medical, Invitae, Illumina, Foresite Capital, and Dynamo; has received research support from Novartis and BMS; has equity in Tango Therapeutics, Genome Medical, Syapse, and Microsoft; has received travel reimbursement from Roche/Genentech; and is a coinventor of institutional patents filed on ERCC2 mutations and chemotherapy response, chromatin mutations and immunotherapy response, and methods for clinical interpretation. H.B. has received research funding from Millennium Pharmaceuticals, AbbVie/Stemcentrx, Astellas, Janssen, and Eli Lilly; and is a consultant/advisor for Sanofi Genzyme and Janssen. S.T. is a coinventor for University of Michigan patents on ETS fusion genes, and diagnostic field of use licensed to Hologic/Gen-Probe Inc., which has sublicensed rights to Roche/Ventana Medical Systems; is a consultant for and has received honoraria from Janssen, AbbVie, Sanofi, Almac Diagnostics, and Astellas/Medivation; has performed sponsored research from Astellas/Medivation and GenomeDx; and is an equity holder in and employee of Strata Oncology. J.M. has consulted for AstraZeneca and Janssen; and has received travel support or speaker fees from Astellas, AstraZeneca, Sanofi, and IPSEN. D.B. has received honoraria and travel support from Janssen. H.I.S. has consulted for Astellas, Ferring Pharmaceuticals, Janssen Biotech, Janssen Research and Development, Sanofi Aventis, Clovis Oncology, Merck, and WCG Oncology; is a member of the board of directors for Asterias Biotherapeutics; and has received research support from Illumina Inc., Innocrin, and Janssen. P.W.K. is a board member for Context Therapeutics LLC, has consulted for BIND Biosciences, BN Immunotherapeutics, DRGT, GE Healthcare, Janssen Pharmaceutica, Metamark, New England Research Institutes, OncoCellMDx, Progenity, Sanofi-Aventis, Seer Biosciences Inc., Tarveda Therapeutics, and Thermo Fisher Scientific; serves on data safety monitoring boards for Genentech/Roche and Merck & Co.; and has investment interest in Context Therapeutics, DRGT, Seer Biosciences, Placon, and Tarveda Therapeutics. M.-E.T. has consuled for Janssen. J.S.d. is an advisory board member for AstraZeneca, Astellas, Bayer, Boehringer Ingelheim, Genentech/Roche, Genmab, GSK, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini/Silicon Biosystems, Orion, Pfizer, Sanofi Aventis, and Taiho; and his institution has received funding or other support from AstraZeneca, Astellas, Bayer, Genentech, GSK, Janssen, Merck Serono, MSD, Menarini/Silicon Biosystems, Orion, Sanofi Aventis, and Taiho. P.S.N. is a compensated advisor to Janssen, Astellas, and Roche. M.A.R. is a coinventor on a patent for Gene Fusion Prostate Cancer (Harvard/Michigan) and EZH2 (Michigan) in the area of diagnostics and therapeutics; is a coinventor on a patent filed by Cornell University on AURKA and SPOP mutations in the field of prostate cancer diagnostics; receives royalties on licensing agreements for these inventions; and receives research support from Janssen, Eli Lilly, Millenium, and Sanofi-Aventis. C.L.S. serves on the board of directors of Novartis; is a cofounder of ORIC Pharmaceuticals and coinventor of enzalutamide and apalutamide; is a science advisor to Agios, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ, Petra, and PMV; and is a cofounder of Seragon, purchased by Genentech/Roche in 2014.
Data deposition: The data reported in this paper are available in Dataset S1 and at www.cbiportal.org, and have been deposited in GitHub, https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019.
See Commentary on page 11090.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902651116/-/DCSupplemental.
References
- 1.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abida W, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol. 2017 doi: 10.1200/PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pritchard CC, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltran H, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal R, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J Clin Oncol. 2018;36:2492–2503. doi: 10.1200/JCO.2017.77.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aparicio AM, et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res. 2016;22:1520–1530. doi: 10.1158/1078-0432.CCR-15-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scher HI, et al. Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur Urol. 2017;71:874–882. doi: 10.1016/j.eururo.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conteduca V, et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: A multi-institution correlative biomarker study. Ann Oncol. 2017;28:1508–1516. doi: 10.1093/annonc/mdx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Annala M, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–457. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 14.Beltran H, et al. A phase II trial of the Aurora kinase A inhibitor alisertib for patients with castration-resistant and neuroendocrine prostate cancer: Efficacy and biomarkers. Clin Cancer Res. 2019;25:43–51. doi: 10.1158/1078-0432.CCR-18-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armenia J, et al. PCF/SU2C International Prostate Cancer Dream Team The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50:645–651. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakravarty D, et al. OncoKB: A precision oncology knowledge base. JCO Precis Oncol. 2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ku SY, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mu P, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quigley DA, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174:758–769.e9, and erratum (2018) 175:889. doi: 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viswanathan SR, et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell. 2018;174:433–447.e19. doi: 10.1016/j.cell.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YM, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell. 2018;173:1770–1782.e14. doi: 10.1016/j.cell.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abida W, Sawyers CL. Targeting DNA repair in prostate cancer. J Clin Oncol. 2018;36:1017–1019. doi: 10.1200/JCO.2017.76.6592. [DOI] [PubMed] [Google Scholar]
- 25.Hussain M, et al. Targeting androgen receptor and DNA repair in metastatic castration-resistant prostate cancer: Results from NCI 9012. J Clin Oncol. 2018;36:991–999. doi: 10.1200/JCO.2017.75.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mateo J, et al. Clinical outcome of prostate cancer patients with germline DNA repair mutations: Retrospective analysis from an international study. Eur Urol. 2018;73:687–693. doi: 10.1016/j.eururo.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boysen G, et al. SPOP-mutated/CHD1-deleted lethal prostate cancer and abiraterone sensitivity. Clin Cancer Res. 2018;24:5585–5593. doi: 10.1158/1078-0432.CCR-18-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blattner M, et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell. 2017;31:436–451. doi: 10.1016/j.ccell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beltran H, et al. Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res. 2014;20:2846–2850. doi: 10.1158/1078-0432.CCR-13-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bluemn EG, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–489.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltran H, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hieronymus H, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Nava Rodrigues D, et al. RB1 heterogeneity in advanced metastatic castration resistant prostate cancer. Clin Cancer Res. 2018;25:687–697. doi: 10.1158/1078-0432.CCR-18-2068. [DOI] [PubMed] [Google Scholar]
- 34.Wyatt AW, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst. 2017;110:78–86. doi: 10.1093/jnci/djx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romanel A, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re10. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epstein JI, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol. 2014;38:756–767. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mateo J, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brastianos PK, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen R, Seshan VE. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuzick J, et al. Transatlantic Prostate Group Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106:1095–1099. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ertel A, et al. RB-pathway disruption in breast cancer: Differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–4163. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerami E, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.